Abstract
Environmental DNA libraries from three different soil samples were constructed. The average insert size was 5 to 8 kb and the percentage of plasmids with inserts was approximately 80%. The recombinant Escherichia coli strains (approximately 930,000) were screened for 4-hydroxybutyrate utilization. Thirty-six positive E. coli clones were obtained during the initial screen, and five of them contained a recombinant plasmid (pAH1 to pAH5) which conferred a stable 4-hydroxybutyrate-positive phenotype. These E. coli clones were studied further. All five were able to grow with 4-hydroxybutyrate as sole carbon and energy source and exhibited 4-hydroxybutyrate dehydrogenase activity in crude extracts. Sequencing of pAH5 revealed a gene homologous to the gbd gene of Ralstonia eutropha, which encodes a 4-hydroxybutyrate dehydrogenase. Two other genes (orf1 and orf6) conferring utilization of 4-hydroxybutyrate were identified during subcloning and sequencing of the inserts of pAH1 and pAH3. The deduced orf1 gene product showed similarities to members of the DedA family of proteins. The sequence of the deduced orf6 gene product harbors the fingerprint pattern of enoyl-coenzyme A hydratases/isomerases. The other sequenced inserts of the plasmids recovered from the positive clones revealed no significant similarity to any other gene or gene product whose sequence is available in the National Center for Biotechnology Information databases.
Naturally occurring assemblages of microorganisms often encompass a bewildering array of physiological, metabolic, and genetic diversity. In fact, it has been estimated that >99% of microorganisms observable in nature typically cannot be cultivated by standard techniques (1). Thus, a large fraction of the diversity in an environment is still unknown due to difficulties in enriching and isolating microorganisms in pure culture. Correspondingly, the diversity of enzymes catalyzing a certain reaction is only partially known. The classical and cumbersome approach for isolating enzymes from environmental samples is to enrich, isolate, and screen a wide variety of microorganisms for the desired enzyme activity. The enzyme is then recovered from the identified organism. An alternative approach is to use the genetic diversity of the microorganisms in a certain environment as a whole to encounter previously unknown genes and gene products for various purposes. One way to exploit the genetic diversity of various environments is the construction of DNA libraries. The DNA used for the preparation of the libraries is isolated from different soil samples without the culturing of the organisms present. The DNA isolation methods for soils and sediments are based either on recovery of bacterial cells and subsequent lysis (14, 26) or on direct lysis of cells in the sample followed by DNA purification (29). Higher yields of DNA from soils and sediments are usually obtained with the direct lysis method, because nonbacterial and extracellular DNA is also extracted (24).
The main goal of this study was to investigate the potential of the prepared libraries and to test the accessibility of the genetic diversity in an environment by direct cloning of environmental DNA. In this basic study, the DNA was prepared by the direct lysis method, and environmental libraries were constructed using Escherichia coli as the host. Subsequently, the libraries were screened for the presence of genes conferring the utilization of 4-hydroxybutyrate. E. coli is unable to use 4-hydroxybutyrate as a carbon and energy source for growth (28). Nevertheless, the organism possesses two different coenzyme A (CoA)-independent succinate semialdehyde dehydrogenases, which directly catalyze the oxidation of succinate semialdehyde to succinate. These enzymes are involved in the catabolism of γ-aminobutyric acid or of p-hydroxyphenylvaleric acid (11, 12). One of the succinate semialdehyde dehydrogenases is produced if succinate semialdehyde is present (12). Therefore, the ability of E. coli to grow with 4-hydroxybutyrate depends only on the presence and expression of a gene conferring 4-hydroxybutyrate dehydrogenase activity. The only known genes encoding a 4-hydroxybutyrate dehydrogenase are from Clostridium kluyveri (4hbd) (23) and Ralstonia eutropha (gbd) (28). The enzymes participate in succinate plus ethanol fermentation and in 4-hydroxybutyrate catabolism, respectively.
MATERIALS AND METHODS
Sample sites, bacterial strains, and plasmids.
For the construction of environmental DNA libraries, soil samples from a sugar beet field near Göttingen, Germany, a meadow near Northeim, Germany, and the valley of the river Nieme, Germany, were collected.
E. coli DH5α (2) was used as host, and the plasmid pBluescript SK+ (pSK+) (Stratagene, San Diego, Calif.) was employed as vector for the cloning experiments. E. coli JM109/pCK1, containing and expressing the gene encoding 4-hydroxybutyrate dehydrogenase of C. kluyveri (23), was used as the positive control for growth experiments and enzymatic analysis.
Media and growth conditions.
E. coli was routinely grown in Luria-Bertani medium at 30°C (2). Tetrazolium indicator plates (4) containing 4-hydroxybutyrate as test substrate were employed for the screening procedure. The indicator plates contained the following (grams per liter): sodium 4-hydroxybutyrate, 4.0; K2HPO4, 3.0; KH2PO4, 7.0; proteose peptone, 2.0; MgSO4, 0.1; and 2,3,5-triphenyl tetrazolium chloride, 0.025. Utilization of 4-hydroxybutyrate (4 g/liter) as a carbon source for recombinant E. coli strains was tested in M9 medium (19) supplemented with a small amount of yeast extract (0.2 g/liter), MgSO4 (2 mM), and CaCl2 (0.1 mM). All growth media for E. coli strains harboring plasmids also contained 100 μg of ampicillin/ml to maintain the presence of the plasmids.
Preparation of cell extracts.
Cells at the stationary growth phase from 500-ml cultures were harvested by centrifugation at 6,000 × g for 20 min, washed once with 100 mM potassium phosphate buffer (pH 8.0), and resuspended in 2 to 3 ml of the same buffer. The cells were disrupted by a 1-min sonication/ml of sample in a type MK2 ultrasonic disintegrator with an amplitude of 14 μm. Sonication was done in periods of 15 s each, followed by a break of 15 s. During this procedure the samples were cooled. The extract was cleared by centrifugation at 16,000 × g for 30 min at 4°C.
Enzyme assays.
4-Hydroxybutyrate dehydrogenase was assayed at 30°C in a 97 mM 2-amino-2-methyl-1,3-propanediol–HCl buffer (pH 8.5), containing 1 mM NAD+ and 20 mM 4-hydroxybutyrate (28). The reaction was started by the addition of 4-hydroxybutyrate and monitored at 334 nm (A334 = 6.3 mM−1 · cm−1). For determination of 3-hydroxybutyrate dehydrogenase activity 4-hydroxybutyrate was replaced by the same amount of racemic 3-hydroxybutyrate. Protein concentrations were measured by the method of Bradford (6) with bovine serum albumin as a standard. All enzyme activities are expressed according to the formula micromoles/(minutes × milligrams of protein).
Molecular procedures.
The DNA isolation from soil samples was based on the direct lysis method of Zhou et al. (29). Fifty grams of each environmental sample was mixed with 135 ml of DNA extraction buffer (100 mM Tris-HCl [pH 8.0], 100 mM sodium EDTA, 100 mM sodium phosphate, 1.5 M NaCl, 1% [wt/vol] cetyltrimethylammonium bromide) and 1 ml of proteinase K (10 mg/ml) in GS3 tubes by horizontal shaking at 225 rpm for 30 min at 37°C. Next, 15 ml of 20% (wt/vol) sodium dodecyl sulfate was added and the samples were incubated in a 65°C water bath for 2 h with gentle end-over-end inversion every 15 to 20 min. After centrifugation at 6,000 × g for 10 min at room temperature, the resulting supernatants were transferred into new GS3 tubes. The remaining soil pellets were extracted two more times by suspending them in 45 ml of extraction buffer and 5 ml of 20% sodium dodecyl sulfate. The subsequent incubation and centrifugation were done as described above. Supernatants from all extraction steps were combined and mixed with an equal volume of chloroform-isoamyl alcohol (24:1 [vol/vol]). The aqueous phase was recovered by centrifugation, and the DNA was precipitated with 0.6 volume of isopropanol at room temperature for 1 h. A pellet of crude nucleic acids was obtained by centrifugation at 9,000 × g for 20 min. After washing with 70% (vol/vol) ethanol, the DNA was resuspended in 2 to 4 ml of deionized water. The final purification of the DNA and the removal of coextracted humic substances were performed with the Wizard Plus Minipreps DNA Purification System (Promega, Heidelberg, Germany). The purified environmental DNA was partially digested with BamHI or Sau3AI and, in order to avoid cloning of very small DNA fragments, size fractionated by sucrose density centrifugation (10 to 40% [wt/vol]). Fractions containing DNA fragments of >2 kb were ligated into BamHI-digested pSK+ and the products were then transformed into E. coli DH5α. This strain was employed for maintenance and amplification of the environmental DNA libraries. All other manipulations of DNA, PCR, and transformation of plasmids into E. coli were done according to routine procedures (2). The Göttingen Genomics Laboratory (Göttingen, Germany) determined the DNA sequences. Sequence analysis was performed with the Genetics Computer Group software package (10).
Nucleotide sequence accession numbers.
The nucleotide sequences of the inserts of pAH1, pAH5, and pAH6 have been deposited in the GenBank database under accession nos. AF148264 to AF148266.
RESULTS AND DISCUSSION
Construction of environmental DNA libraries.
Three different soil samples (from a meadow, a sugar beet field, and the valley of the river Nieme) were used for the construction of environmental DNA libraries. The DNA was isolated from the samples by direct lysis of microorganisms present without previous enrichment or extraction of microbial cells. The extraction of DNA from the three different soils resulted in coextraction of other soil components, which caused a brownish color in the crude DNA solutions obtained. This color indicates the presence of humic acids, which are typically coextracted during direct preparation of DNA from soils (25). Further purification of the DNA with the Wizard Plus Minipreps DNA Purification System resulted in an almost complete decoloring of the DNA extracts. Approximately 15 μg of DNA per g of soil was obtained. This yield is in the same range as that described for the isolation of DNA from other soils (27, 29). The purified DNA was partially digested and size fractionated. DNA fragments of >2 kb were ligated into pSK+ and then transformed into E. coli (see Materials and Methods). Approximately 2,000 recombinant E. coli strains per μg of isolated soil DNA were obtained. The low yield was probably caused by humic substances, which remain in the purified environmental DNA and interfere with the restriction digestion or the ligation reaction (25). The quality of the three different environmental libraries produced was controlled by determination of the average insert size and the percentage of recombinant plasmids containing inserts. The three libraries revealed average insert sizes of 5 to 8 kb. The percentage of plasmids containing inserts was approximately 80%. No significant differences between the three different soils were observed during the preparation of the libraries.
Screening for genes conferring 4-hydroxybutyrate dehydrogenase activity.
To test the potential of the environmental libraries, the recombinant E. coli strains of three different libraries were screened for utilization of 4-hydroxybutyrate. Since E. coli has two CoA-independent succinate semialdehyde dehydrogenases, the use of 4-hydroxybutyrate as carbon and energy source depends only on the presence and heterologous expression of a 4-hydroxybutyrate dehydrogenase-encoding gene. The screening was performed on tetrazolium indicator plates (4) containing 4-hydroxybutyrate as test substrate. Tetrazolium in its oxidized state is soluble in water and appears colorless or faintly yellow in solution. Upon oxidation of the test substrate, it is reduced. This yields to the formation of a insoluble deep red formazan, which is precipitated in the cells. The reduction of tetrazolium is a result of electrons passing from the test substrate, through the enzymatic machinery of central metabolism, the electron transport chain, and ultimately on to tetrazolium (4). Thus, E. coli colonies capable of catabolizing 4-hydroxybutyrate reduce tetrazolium and produce a deep red formazan, whereas colonies failing to catabolize 4-hydroxybutyrate remain uncolored.
To estimate the number of tested E. coli clones and to identify positive clones, simultaneously, the recombinant E. coli strains were directly plated on tetrazolium indicator plates after transformation of the environmental libraries. Thirty-six of approximately 930,000 E. coli clones were positive during the initial screen (Table 1). Most of these clones—28—were obtained from library I, but the time necessary for the appearance of the phenotype was much longer than for positive clones of the other two libraries (Table 1). To confirm the 4-hydroxybutyrate-positive phenotype of the clones, the recombinant plasmids were isolated and retransformed into E. coli and the resulting E. coli strains were screened again on tetrazolium indicator plates containing 4-hydroxybutyrate as an additional carbon source. Only five different recombinant plasmids, designated pAH1 to pAH5, conferred a stable 4-hydroxybutyrate-positive phenotype to the resulting recombinant E. coli strains. Since all 36 initial clones harbored plasmids with inserts, the high percentage of false-positive clones was not caused by plasmid loss. Sometimes the appearance of false-positive colonies on tetrazolium indicator plates is due to crossfeeding by positive clones, which excrete catabolism intermediates in the culture medium (4). Three of the plasmids conferring a stable phenotype were obtained from library I, one was obtained from library II, and one was obtained from library III (Table 1). The insert sizes of pAH1 to pAH5 were in the range of 1,000 to 4,500 bp (Table 2). The corresponding E. coli strains (E. coli/pAH1 to E. coli/pAH5) were studied further.
TABLE 1.
Screening of three environmental libraries for genes conferring 4-hydroxybutyrate utilization
| Library | Sample site | No. of E. coli clones tested | No. of 4-hydroxybutyrate-positive E. coli clones after the initial screen | Time for appearance of phenotype (red colonies) (days) | No. of 4-hydroxybutyrate-positive E. coli clones with a stable phenotype (designation) |
|---|---|---|---|---|---|
| I | Meadow | 340,000 | 28 | 21–28 | 3 (pAH2 to pAH4) |
| II | Sugar beet field | 190,000 | 1 | 3–4 | 1 (pAH5) |
| III | Nieme River valley | 400,000 | 7 | 3–4 | 1 (pAH1) |
TABLE 2.
Characterization of pAH1 to pAH5 and the corresponding E. coli clones: insert sizes, growth rates with 4-hydroxybutyrate as sole carbon and energy source, and specific activities of 4-hydroxybutyrate and 3-hydroxybutyrate dehydrogenase in crude extracts
| Recombinant plasmid | Insert size (bp) | Result for corresponding E. coli clone
|
||
|---|---|---|---|---|
| Growth rate μ (h−1) | Specific 4-hydroxybutyrate-dehydrogenase activity (U/mg) | Specific 3-hydroxybutyrate-dehydrogenase activity (U/mg) | ||
| pAH1 | 1,002 | 0.23 | 0.35 | <0.01 |
| pAH2 | 2,499 | 0.05 | 0.04 | 0.06 |
| pAH3 | 4,495 | 0.06 | 0.04 | 0.05 |
| pAH4 | 4,347 | 0.07 | 0.04 | 0.04 |
| pAH5 | 3,211 | 0.1 | 0.20 | 0.25 |
| pCK1 | 6,600 | 0.2 | 0.12 | 0.02 |
| pSK+ | 0 | Not detectable | <0.01 | <0.01 |
Physiological and biochemical characterization of the 4-hydroxybutyrate-positive E. coli clones.
The five positive E. coli clones obtained after the screening procedure were tested for growth in M9 medium containing 4-hydroxybutyrate as the sole carbon and energy source. In contrast to the negative control, E. coli/pSK+, which harbors the plasmid used as cloning vector, all five recombinant E. coli strains showed growth under these conditions. One clone (E. coli/pAH1) revealed a similar growth rate and the other four had a slower growth rate than the positive control E. coli JM109/pCK1, which harbors the 4hbd gene from C. kluyveri (Table 2). Enzymatic analysis revealed 4-hydroxybutyrate dehydrogenase activity in crude extracts of all recombinant E. coli strains (Table 2). The recorded activities of E. coli/pAH2 to E. coli/pAH4 (0.04 U/mg) were lower than the one in crude extracts of E. coli JM109/pCK1 (0.12 U/mg). The activity of E. coli/pAH5 was slightly higher than that of the positive control. E. coli/pAH1, exhibiting a growth rate similar to that of the positive control, showed in crude extracts the highest specific 4-hydroxybutyrate dehydrogenase activity of all tested strains (0.35 U/mg). The four clones exhibiting a slow growth rate on 4-hydroxybutyrate also showed 3-hydroxybutyrate dehydrogenase activity, whereas E. coli/pAH1 revealed no significant activity (Table 2). Thus, the 4-hydroxybutyrate-positive phenotype of E. coli/pAH2 to E. coli/pAH5 might be due to a side activity of a 3-hydroxybutyrate dehydrogenase.
Molecular analysis.
The inserts of pAH1 to pAH5 were sequenced and compared to the sequences in the National Center for Biotechnology Information (NCBI) databases. The only genes encoding a 4-hydroxybutyrate dehydrogenase available in the databases are from C. kluyveri (4hbd) and R. eutropha (gbd) (23, 28). The 4hbd gene (1,116 bp) and the gbd gene (1,146 bp) encode polypeptides of 371 and 382 amino acids with predicted molecular masses of 41,755 and 40,495 Da, respectively. Both enzymes belong to the family of type III alcohol dehydrogenases, which are also known as iron-containing dehydrogenases. This family is very heterogeneous and distinct from the long-chain zinc-containing (type I) or short-chain zinc-lacking (type II) enzymes (for a review, see reference 21).
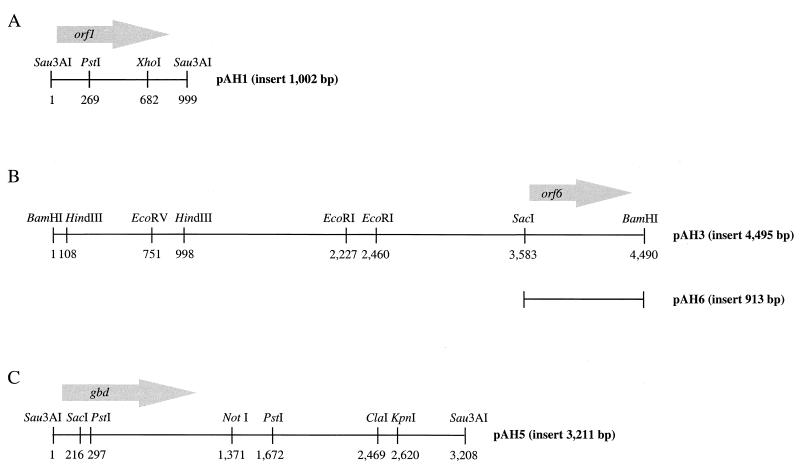
Sequencing of the 3,211-bp insert of pAH5 (Fig. 1C) revealed an open reading frame (1,137 bp) which is very similar to the gbd gene of R. eutropha. Therefore, that open reading frame was also designated gbd. The presumptive gene is preceded by a weak potential ribosome binding site, appropriately spaced from the start codon (Fig. 2). The deduced gene product (378 amino acids) with a predicted molecular mass of 39,324 Da is 69.8% identical (76.7% similar) to 4-hydroxybutyrate dehydrogenase of R. eutropha. The gbd gene product also revealed homology to the 4-hydroxybutyrate dehydrogenase of C. kluyveri (32.3% identity; 41.0% similarity) and to other type III alcohol dehydrogenases, including, e.g., Adh2 of Zymomonas mobilis (8), DhaT of Citrobacter freundii (9), and FucO of E. coli (7). No significant similarities to type I and type II alcohol dehydrogenases were found. For the detection of type III alcohol dehydrogenases, two specific fingerprint patterns are available in the PROSITE database (13). The first one is [STALIV]-[LIVF]-x-[DE]-x(6,7)-P-x(4)-[ALIV]-x-[GST]-x(2)-D-[TAIVM]-[LIVMF]-x(4)-E and the second one is [GSW]-x-[LIVTSACD]-[GH]-x(2)-[GSAE]-[GSHYQ] - x - [LIVTP] - [GAST] - [GAS] - x(3) - [LIVMT] - x - [HNS] -[GA]-x-[GTAC]. The gbd gene product showed both fingerprint patterns (amino acids 172 to 200 and 258 to 279), except that the second one was not fully retained (Fig. 2). 4-Hydroxybutyrate dehydrogenase requires NAD(H) as a cofactor, but the highly conserved NAD(H) binding fingerprint pattern G-X-G-X-X-G (15) was not present in the amino acid sequence. This is also characteristic for most type III alcohol dehydrogenases. To confirm that gbd encodes a 4-hydroxybutyrate dehydrogenase, the gene was subcloned. The resulting E. coli strain exhibited 4-hydroxybutyrate dehydrogenase activity (data not shown). Thus, gbd encodes a 4-hydroxybutyrate dehydrogenase.
FIG. 1.
(A) Restriction map of the insert of pAH1. Arrow represents length, location, and orientation of the orf1 gene. (B) Restriction map of the insert of pAH3 and localization of the insert of pAH6. Arrow represents length, location, and orientation of the orf6 gene. (C) Restriction map of the insert of pAH5. Arrow represents length, location, and orientation of the gbd gene.
FIG. 2.
Partial nucleotide sequence of the insert of pAH5. Only one strand is shown. The gene encoding 4-hydroxybutyrate dehydrogenase (gbd) has been translated by using the one-letter amino acid code; amino acid symbols are written below the first nucleotides of the corresponding codons. Putative ribosome binding site is underlined. The two conserved regions of the protein, which match the fingerprint patterns for type III alcohol dehydrogenases, are boxed. Matching amino acids of the consensus sequences for both regions according to the PROSITE database (13) are given in bold letters above the DNA sequence. The sequence of the gbd gene has been submitted to GenBank under accession no. AF148264.
E. coli/pAH1 exhibited the highest 4-hydroxybutyrate dehydrogenase activity and harbored the plasmid with the smallest insert (1,002 bp). DNA sequence analysis of the insert revealed one potential gene, designated orf1, within the sequence (Fig. 1A). A potential ribosome binding site, appropriately spaced from the start codon, preceded the presumptive gene (Fig. 3). The orf1 gene (678 bp) codes for 225 amino acids with a predicted molecular mass of 23,603 Da. The amino acid sequence deduced from orf1 was compared with deduced amino acid sequences available in the NCBI databases. This search showed similarities (44.4 and 33.0% identity and 56.1 and 40.6% similarity) to hypothetical transmembrane proteins from Synechocystis PCC6803 (SLR0305; 22,200 Da) (16) and E. coli (YdjZ; 26,200 Da) (3), respectively, which are members of the DedA family of proteins. The members of this family contain multiple predicted transmembrane regions and have not been functionally characterized so far. Hydropathy analysis (18) of the deduced orf1 protein sequence also revealed four to five hydrophobic and probably membrane-spanning regions (data not shown).
FIG. 3.
Nucleotide sequence of the insert of pAH1. Only one strand is shown. The orf1 gene has been translated by using the one-letter amino acid code; amino acid symbols are written below the first nucleotides of the corresponding codons. Putative ribosome binding site is underlined. The sequence of the orf1 gene has been submitted to GenBank under accession no. AF148265.
The sequences of the inserts of pAH2 to pAH4 revealed no significant similarity to genes encoding 4-hydroxybutyrate dehydrogenases or to any other sequence available in the databases, except for a part of the insert of pAH3 (see below). This result indicates that the constructed DNA libraries harbor genes from a wide variety of microorganisms, which mostly have not been investigated or even cultivated so far.
To identify the DNA regions on pAH2 to pAH4 which are responsible for the utilization of 4-hydroxybutyrate by the corresponding recombinant E. coli strains, the inserts were partially subcloned by restriction digestion with various enzymes or by PCR by using primers derived from sequencing and subsequent ligation into pSK+. The resulting constructs were transformed into E. coli, and the recombinant E. coli strains were screened again on tetrazolium indicator plates containing 4-hydroxybutyrate as an additional carbon source. This method was successful for the insert of pAH3. The plasmid recovered from the corresponding positive E. coli strain was designated pAH6. All attempts to subclone a DNA region conferring 4-hydroxybutyrate utilization of pAH2 and pAH4 failed. Both plasmids were not studied here any further because of the low 4-hydroxybutyrate dehydrogenase activities in crude extracts of the corresponding recombinant E. coli strains.
The plasmid pAH6 contained a 913-bp SacI-BamHI insert (Fig. 1B). The corresponding recombinant E. coli strain (E. coli/pAH6) was able to grow in M9 medium containing 4-hydroxybutyrate as the sole carbon and energy source and showed 4-hydroxybutyrate dehydrogenase activity in crude extracts in the same range as for the original clone (E. coli/pAH3) (data not shown). The sequence of the insert of pAH6 (Fig. 4) harbored a single large open reading frame (852 bp) designated orf6 (Fig. 1B) which was preceded by a potential ribosome binding site, appropriately spaced from the start codon (Fig. 4). The deduced gene product (283 amino acids), with a predicted molecular mass of 31,750 Da, showed the signature pattern for members of the enoyl-CoA hydratase/isomerase family of proteins (amino acids 126 to 146; Fig. 4). Enoyl-CoA hydratase and enoyl-CoA isomerase are both involved in fatty acid metabolism. In E. coli (gene fadB) and Pseudomonas fragi (gene faoA), both enzymes are part of a multifunctional enzyme which contains both a 3-hydroxyacyl-CoA dehydrogenase domain and a 3-hydroxybutyryl-CoA epimerase domain (20). Database searches revealed a weak similarity (34.8% identity; 40.7% similarity) of Orf6 to the crt gene product of Clostridium acetobutylicum. CrT (261 amino acids) is a 3-hydroxybutyryl-CoA dehydratase (crotonase) involved in butyrate formation (5) and is evolutionarily related to enoyl-CoA hydratases/isomerases. In addition, the orf6 gene product revealed 34.6% identity (48.1% similarity) to the C-terminal region of the 3-hydroxyacyl-CoA dehydrogenase (HbD2) of Archaeoglobus fulgidus (17). Sequence analysis suggested the participation of CoA in the enzyme reaction, but the addition of CoA, acetyl phosphate, ATP, or combinations of these substrates to the assay mixture had no significant effect on 4-hydroxybutyrate dehydrogenase activity.
FIG. 4.
Nucleotide sequence of the insert of pAH6. Only one strand is shown. The orf6 gene has been translated by using the one-letter amino acid code; amino acid symbols are written below the first nucleotides of the corresponding codons. Putative ribosome binding site is underlined. The conserved region of the protein, which matches the fingerprint pattern for enoyl-CoA hydratases/isomerases, is boxed. Matching amino acids of the consensus sequence for this region ([LIVM]-[STA]-x-[LIVM]-[DENQRHSTA]-G-x(3)-[AG] (3)-x(4)-[LIVMST]-x-[CSTA]-[DQHP]-[LIVMFY]) (13) are given in bold letters above the DNA sequence. The sequence of the orf6 gene has been submitted to GenBank under accession no. AF148266.
Since E. coli/pAH1, E. coli/pAH3 and the subclone E. coli/pAH6 exhibited 4-hydroxybutyrate dehydrogenase activity in crude extracts, the orf1 and orf6 gene products may represent new types of 4-hydroxybutyrate dehydrogenases. Interestingly, the recorded specific 4-hydroxybutyrate dehydrogenase activities in crude extracts of the strains containing orf1 were threefold higher than those in crude extracts of the positive control, E. coli/pCK1 (Table 2). Another possibility is that the orf1 and orf6 gene products facilitate the utilization of 4-hydroxybutyrate in a way that is not yet understood. The characterization of the purified gene products will unravel the function of both enzymes.
The results presented show that it is possible to prepare DNA libraries and to clone directly functional genes from environmental soil samples. To our knowledge, no other report of such an approach has been published. The cloning of genes from environmental soil samples described by other authors involved a PCR step, i.e., cloning of genes encoding β-ketoacyl synthases (KSβ) (22) or 16S rRNA (1). The sequences of the primers used were derived from conserved regions of known genes or protein families. Thus, the identification of entirely new genes or gene products by PCR-based methods is very limited. Our direct cloning approach illustrates another way to access and to exploit the immense pool of genes from microorganisms which have not been cultivated so far. The existence of sequence information prior to cloning is not required. In addition, the existing environmental libraries can be employed for screening of various targets. This has significant implications for microbial biotechnology in the future.
ACKNOWLEDGMENTS
The work was supported by the Fonds der Chemischen Industrie and by the Akademie der Wissenschaften, Göttingen, Germany.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 3.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Bochner B R, Savageau M A. Generalized indicator plate for genetic, metabolic, and taxonomic studies with microorganisms. Appl Environ Microbiol. 1977;33:434–444. doi: 10.1128/aem.33.2.434-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boynton Z L, Bennet G N, Rudolph F B. Cloning, sequencing, and expression of clustered genes encoding beta-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1996;178:3015–3024. doi: 10.1128/jb.178.11.3015-3024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Conway T, Ingram L O. Similarity of Escherichia coli propanediol oxidoreductase (fucO product) and an unusual alcohol dehydrogenase from Zymomonas mobilis and Saccharomyces cerevisiae. J Bacteriol. 1989;171:3754–3759. doi: 10.1128/jb.171.7.3754-3759.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conway T, Sewell G W, Osman Y A, Ingram L O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987;169:2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel R, Boenigk R, Gottschalk G. Purification of the 1,3-propanediol dehydrogenase from Citrobacter freundii and cloning, sequencing, and overexpression of the corresponding gene in Escherichia coli. J Bacteriol. 1995;177:2151–2156. doi: 10.1128/jb.177.8.2151-2156.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly M I, Cooper A C. Succinic semialdehyde dehydrogenases of Escherichia coli. Their role in the degradation of p-hydroxyphenylacetate and γ-aminobutyrate. Eur J Biochem. 1981;113:555–561. doi: 10.1111/j.1432-1033.1981.tb05098.x. [DOI] [PubMed] [Google Scholar]
- 12.Donnelly M I, Cooper A C. Two succinic semialdehyde dehydrogenases are induced when Escherichia coli K-12 is grown on γ-aminobutyrate. J Bacteriol. 1981;145:1425–1427. doi: 10.1128/jb.145.3.1425-1427.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann K, Bucher P, Falquet L, Bairoch A. The PROSITE database, its status in 1999. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobsen C S, Rasmussen O F. Development and application of a new method to extract bacterial DNA from soil based on separation of bacteria from soil with cation-exchange resin. Appl Environ Microbiol. 1992;58:2458–2462. doi: 10.1128/aem.58.8.2458-2462.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jörnvall H, Persson B, Jeffery J. Characteristics of alcohol/polyol dehydrogenases. The zinc-containing long-chain alcohol dehydrogenase. Eur J Biochem. 1987;167:195–201. doi: 10.1111/j.1432-1033.1987.tb13323.x. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko T, Tanaka A, Sato S, Kotani H, Sazuka T, Miyajima N, Sugiura M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 1995;2:153–166. doi: 10.1093/dnares/2.4.153. [DOI] [PubMed] [Google Scholar]
- 17.Klenk H P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 18.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 20.Nakahigashi K, Inokuchi H. Nucleotide sequence of the fadA and fadB genes from Escherichia coli. Nucleic Acids Res. 1990;18:4937. doi: 10.1093/nar/18.16.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reid M F, Fewson C A. Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol. 1994;20:13–56. doi: 10.3109/10408419409113545. [DOI] [PubMed] [Google Scholar]
- 22.Seow K-T, Meurer G, Gerlitz M, Wendt-Pienkowski E, Hutchinson C R, Davies J. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J Bacteriol. 1997;179:7360–7368. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Söhling B, Gottschalk G. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J Bacteriol. 1996;178:871–880. doi: 10.1128/jb.178.3.871-880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steffan R J, Goksoyr J, Bej A K, Atlas R M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988;54:2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torsvik V L. Isolation of bacterial DNA from soil. Soil Biol Biochem. 1980;12:15–21. [Google Scholar]
- 27.Tsai Y-L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valentin H E, Zwingmann G, Schönebaum A, Steinbüchel A. Metabolic pathway for biosynthesis of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) from 4-hydroxybutyrate by Alcaligenes eutrophus. Eur J Biochem. 1995;227:43–60. doi: 10.1111/j.1432-1033.1995.tb20358.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhou J M, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]