Abstract
Cryptosporidium is globally established as a contaminant of drinking and recreational waters. A previously described cell culture infectivity assay capable of detecting infectious oocysts was adapted to quantify viable oocysts through sporozoite invasion and clustering of foci. Eight experiments were performed by using oocysts less than 4 months of age to inoculate host HCT-8 cell monolayers. Oocysts were diluted in a standard 5- or 10-fold multiple dilution format, levels of infection and clustering were determined, and the most probable number (MPN) of infectious oocysts in the stock suspension was calculated. The MPN was compared to the initial oocyst inoculum to determine the level of correlation. For oocysts less than 30 days of age, the correlation coefficient (r) was 0.9726 (0.9306 to 0.9893; n = 20). A two-tailed P value (alpha = 0.05) indicated that P was less than 0.0001. This strong correlation suggests that the MPN can be used to effectively enumerate infectious oocysts in a cell culture system. Age affected the degree of oocyst infectivity. Oocyst infectivity was tested by the focus detection method (FDM)-MPN assay and in BALB/c mice before and after treatment with pulsed white light (PureBrite). The FDM-MPN assay and animal infectivity assays both demonstrated more than a 4 log10 inactivation. Municipal water systems and a host of other water testing organizations could utilize the FDM-MPN assay for routine survival and disinfection studies.
Cryptosporidium parvum was first associated with human illness in the 1970s (5, 6). The organism’s environmentally stable oocysts are transmitted by the fecal-oral route, often through contaminated drinking water. Outbreaks have occurred as a result of oocyst resistance to disinfectants commonly used in drinking water treatment. Within the last 12 years, 19 documented outbreaks in unrelated geographic areas have occurred in the United States, Canada, United Kingdom, and Japan, affecting more than an estimated 427,000 individuals (4, 31). Consequently, this organism has become a major concern to public health and the drinking water industry.
Cell culture technology has developed into a tool that can be used to study C. parvum in an environment most similar to the situation in vivo without using animal models (13, 29, 30, 32–34, 36). The research described here uses the previously described focus detection method (FDM) set up in a multiple dilution format (27–29). The autoinfectious nature of the Cryptosporidium life cycle was observed to begin 12 h after incubation, and by 48 h, an average ratio of 17.9 foci to each oocyst was observed (29). Presumably, nonsynchronous excystation and infection were occurring during this complex life cycle, and the parasites produced clusters of reproductive stages in the host monolayer after an extended incubation period. At first, this confounded the ability to directly count the number of infected foci in the host cells. Rather than counting the individual foci, a presence or absence observation was made after 48 h of incubation and was used to detect one infectious oocyst or fewer (based upon dilution) per well in vitro.
The most-probable-number (MPN) method of enumerating microorganisms (specifically, bacteria) was adopted as a method to quantify the number of infectious oocysts. The method entails making a dilution series, plating in replicate, incubating the samples for an adequate period, and scoring the presence of growth. In the cell culture system, cluster presence (and absence) can easily be detected with the FDM and used to determine the number of viable organisms (in this case, infectious oocysts) per milliliter in a sample by the MPN method. Tables made by Halvorson and Ziegler (15) have traditionally been consulted for the MPNs; however, computer programs have enabled researchers to obtain a more accurate number and allow nonstandard volumes and dilutions to be used (11, 21). The objective of this study was to standardize and test statistically the reproducibility of the MPN method by using the cell culture FDM for infectious oocysts.
MATERIALS AND METHODS
C. parvum oocysts less than 4 months old were obtained from Pleasant Hill Farm, Iowa. The original strain was obtained from a naturally infected cow. The oocysts were perpetuated in newborn calves, purified by an ethyl ether and Percoll-sucrose gradient procedure (according to a protocol that can be obtained from Pleasant Hill Farm), and stored at 4°C in phosphate-buffered saline (PBS) with antibiotics (penicillin and streptomycin). Upon delivery, each oocyst lot number was counted with a hemacytometer and concentrations were recorded. All lot numbers were in concentrations greater than 106 oocysts per ml. Aliquots used for cell culture infection were processed as described below. Cell culture infectivity testing was performed simultaneously with in vitro excystation and vital dye exclusion to compare the viabilities determined by the three assays (3, 25).
Eight oocyst lots (from different calves) were evaluated, with three to eight replicate experiments/lot, in a total of 34 experiments in order to determine the variability of lot numbers and the FDM-MPN method. Each experiment was set up in a multiple dilution format, and 10- or 5-fold dilutions with three to six replicates per dilution were used.
In vitro cell culture.
The cell culture system has previously been described (27, 28). A brief description (with changes) is presented here. Human ileocecal adenocarcinoma cells (HCT-8 cells) were maintained in 75-cm2 tissue culture flasks and passaged every 2 or 3 days. LabTech II (Nalgene Nunc, Naperville, Ill.) eight-well chamber slides were seeded with 5 × 105 cells per well and grown to approximately 60 to 80% confluency in a 5% CO2 atmosphere at 37°C for 24 to 72 h. HCT-8 maintenance medium contained RPMI 1640 supplemented with 5% fetal bovine serum (Atlanta Biologicals), 2% 1 M HEPES, and 1% 200 mM l-glutamine. No antibiotics were used.
Oocyst preparation.
Once the monolayer was established, C. parvum oocysts were prepared. Oocysts were pretreated with 10% (vol/vol) bleach (5.2% sodium hypochlorite) (4°C) and washed by centrifugation after 5 to 8 min in the bleach solution. Stock microscopic oocyst counts of the oocyst suspensions were determined by a direct hemacytometer count, and 10- or 5-fold oocyst dilutions in growth medium were prepared in sterile polypropylene microcentrifuge tubes. Each dilution was pipetted onto cell monolayers in three or six replicate wells. The entire volume of each diluted oocyst suspension was inoculated. The cultures were incubated in a 5% CO2 atmosphere at 37°C for 48 h (the 90-min washing step previously described [29] was excluded because no deleterious effects to the host cells from the toxicity caused by the oocysts excysting have been noted in the diluted oocyst suspensions). Controls assayed concurrently for each experiment included uninoculated cell monolayers in two wells on each slide as negative controls. Uninoculated wells also served to determine if the chamber slides were leaking or if any transfer between wells occurred.
Antibody labeling with the FDM.
Well chamber slides were fixed with 100% methanol and labeled by an indirect antibody procedure as previously described (29). Rat antisporozoite polyclonal antibody, generously provided by Steve Upton (Kansas State University, Manhattan, Kans.) and Waterborne, Inc. (New Orleans, La.), was used as the primary antibody, while anti-rat fluorescein isothiocyanate-conjugated antibody (Sigma) was used as the secondary antibody.
Enumeration.
Fixed and labeled slides were examined by epifluorescence microscopy at a ×200 magnification, and each well was scored as either positive or negative. Positive infection was determined by the presence of visible sporozoite invasion that produced an infection focus, and clustering, a result of secondary infection, was determined when numerous life stages were present. When cells appeared positive for infection, a confirmation of Cryptosporidium life stages (meronts and microgametes, etc.) was made at ×400 magnification and/or under oil at a ×1,000 magnification. Both invasion and replication in vitro were required to score the well as positive. When both invasion and replication were not present, the well was scored as negative.
The MPN of infectious oocysts was determined by using the information collection rule (ICR) general purpose Most Probable Number Calculator, version 1.00 (20). The ICR program was modified from a previously published version specifically for the application of total cultivable viruses (MPNv) for samples processed under the ICR (19). The program enables the user to enter the number of replicates and dilutions, volumes used, and dilution factors to generate an MPN with confidence intervals. The program is easy to use and is preferred to referring to tables. The MPN data were statistically analyzed with the GraphPad Prism software (San Diego, Calif.) and Excel.
Percent infectivity was determined by the following formula: percent infectivity = (MPN per ml/microscopic oocyst count per milliliter of the stock) × 100.
In vitro viability assays.
Excystation and vital dye exclusion methods were also used to evaluate viability (3, 25). The inclusion and/or exclusion of the fluorogenic vital dyes DAPI (4′,6-diamidino-2-phenylindole) and propidium iodide (PI) within oocysts was examined microscopically with an Olympus model BH2 microscope equipped with a UV filter block (350-nm excitation and 450-nm emission) for DAPI and a green filter block (500-nm excitation and 630-nm emission) for PI. Proportions of ruptured (ghost), PI(+), DAPI(+)-PI(−), and DAPI(−)-PI(−) oocysts were quantified (3).
Excystation procedures included the use of excysting fluid (0.5% trypsin and 1.5% sodium taurocholate in tissue culture with PBS) to incubate oocysts for 90 min at 37°C (25). Sporozoites, intact oocysts, and oocyst shells were counted under ×400 magnification by Nomarski differential interference contrast microscopy. A total of 300 oocysts were counted per experiment. As an additional experimental control, the sporozoite yield viability was determined by dividing the sporozoite yield by 4 (the theoretical number of sporozoites per oocyst) and multiplying the resulting factor by 100.
Animal infectivity.
Oocyst infectivity was evaluated by modified neonatal BALB/c mouse infectivity assay in addition to cell culture infectivity (1). Inactivation experiments were performed by using a broad-spectrum pulsed white light water treatment device (PureBrite, San Diego, Calif.) (35). Pregnant BALB/c mice were shipped overnight from Jackson Labs (Bar Harbor, Maine) to Michael Arrowood (Centers for Disease Control and Prevention, Atlanta, Ga.). Oocyst samples were packed on ice and shipped by overnight express to Michael Arrowood for inoculation into 4-day-old mice. Groups of 5 to 10 replicate mice were administered oocyst doses in a dilution series (106, 105, 104, and 103) by oral gavage and sacrificed 7 days postinoculation by CO2 inhalation. Mouse intestinal tracts were removed, and a portion of the terminal ileum from each mouse was placed in 2.5% potassium dichromate and vigorously vortexed in sterile deionized water. Oocysts were recovered and isolated by using a discontinuous sucrose gradient. The rest of the digestive tract from each dosage group was pooled and was likewise processed in an attempt to detect low numbers of oocysts that may be missed in individual mice. Each sample was incubated with a C. parvum oocyst-specific monoclonal antibody (OW50) conjugated with fluorescein isothiocyanate and analyzed by flow cytometry as previously described (1). The MPN program was used to estimate the number of viable oocysts.
RESULTS
Oocyst preparations representing eight different lots ranged from 11 to 94 days of age (Table 1). The average MPN for all lots was 5.4 × 103 (on a normalized scale) and ranged from 1.3 × 103 to 1.6 × 104 infectious oocysts per ml. The mean percent infectivity for all lot numbers and ages tested to date in the cell culture system ranged from 0.97 to 73.80%, with an average of 20.32% infectious oocysts (n = 34; standard deviation = ±21.49). Between three and seven replicate experiments were performed for each lot of oocysts. Several experiments used individually bleach-treated oocyst preparations for each dilution series, while others used a single oocyst preparation. Experiments that used the same bleach preparation for each dilution series exhibited little or no variability among the MPNs. In many cases, the same MPN was produced. For example, lot 4 oocysts (18 days of age) were bleach treated, and three separate sets of dilutions were made from the same bleached stock. The three experiments showed 1.19 × 104, 1.01 × 104, and 1.01 × 104 MPNs of infectious oocysts/ml and the proportions of infective oocysts were 47.59, 40.47, and 40.47%, respectively.
TABLE 1.
Oocyst age and infectivity
| Oocyst lot no. (no. of experiments) | Supplier lot no. (feces collection dates) | Approximate oocyst age at cell culture infection (days) | Normalized average MPN/mla (range) (103) |
|---|---|---|---|
| 1 (3) | 97-7 (3/23–27/97)b | 18–26 | 6.9 (3.9–9.6) |
| 2 (4) | 97-15 (8/9–15/97) | 11–30 | 15.9 (14.0–18.5) |
| 3 (3) | 98-1 (1/6–11/98) | 62 | 3.1 (0.8–6.3) |
| 4 (3) | 98-5 (3/1–4/98) | 18 | 10.7 (10.1–11.9) |
| 5 (8) | 98-8 (4/22–29/98) | 27–94 | 1.9 (0.3–5.8) |
| 6 (7) | 98-13 (7/21–26/98) | 12–82 | 2.3 (0.6–3.8) |
| 7 (3) | 98-319 (9/19/98) | 2–33 | 1.4 (0.2–3.4) |
| 8 (3) | 98-17 (10/11–17/98) | 34 | 1.3 (0.5–2.2) |
Data was normalized by the oocyst hemacytometer count of each undiluted sample.
Dates are presented as month/day(s)/year.
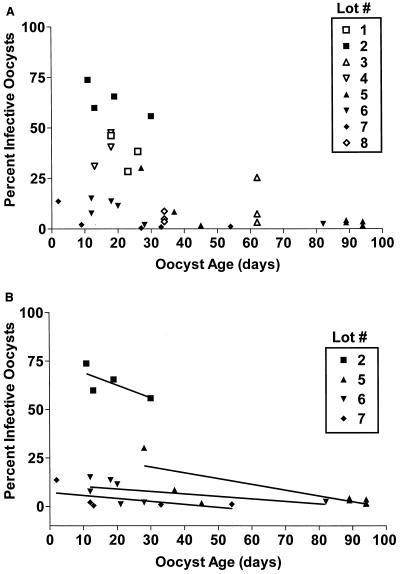
Several lots were assayed repeatedly as the oocysts aged (at 4°C in PBS). The MPNs produced revealed differences among each experiment, and subtle changes in infectivity were observed. To determine the source of variability, all experiments were plotted to show the relationship of each lot number with the percent infectious oocysts and age (Fig. 1). Figure 1A shows the percent infection plotted against age for all lots tested in the cell culture system. Figure 1B shows the regression comparison of the percent infectious oocysts with age for four oocyst lot numbers. Both lot number and age were factors associated with the variation observed with infectivity.
FIG. 1.
Percent infectivity by age of C. parvum oocysts. (A) All lot numbers tested with the cell culture FDM-MPN assay (n = 34). (B) Comparison of the linear regression analysis from four lot numbers. Percent infectivity is determined by the following formula: [(MPN/milliliter)/(stock oocyst count/milliliter)] × 100.
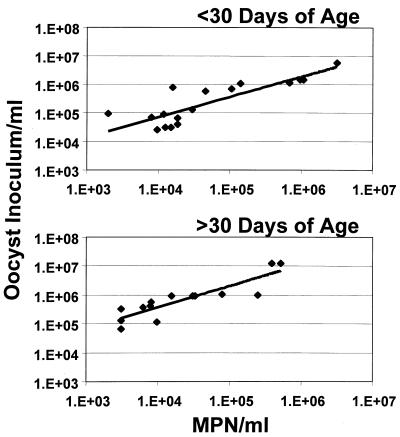
Figure 2 shows the relationship between the hemacytometer count (number of oocysts/ml) and the MPN/ml for oocysts less than or greater than 30 days of age. Correlation coefficients (r) were 0.9726 and 0.9241 for oocysts less than and greater than 30 days of age, respectively (P < 0.0001). The r value for all experiments (all ages) combined was 0.4081 (95% confidence interval [CI], 0.0811 to 0.6558 [n = 34]) (P = 0.0166). Stock oocyst counts for oocysts less than 30 days of age were statistically equivalent to the MPNs calculated; however, the relationship between the oocyst inoculum and the MPN diverged as the oocysts aged.
FIG. 2.
Linear regression comparison of the number of oocysts/milliliter inoculated and the MPN/milliliter calculated for oocysts less than or greater than 30 days of age.
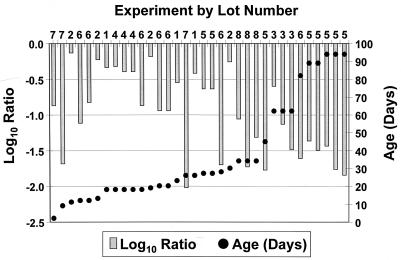
The log10 of the ratio between the stock oocyst count/milliliter and the MPN/milliliter were compared with age for each experiment (Fig. 3). As the oocyst age increased, the ratio of the inoculum/milliliter to the MPN/milliliter generally became more negative. Oocysts beyond 30 days of age had a difference in ratios >−1.0 log10 in 92% (12 in 13) and >−1.5 log10 in 46% (6 in 13) of the experiments. Oocysts 30 days of age or less had only 19% (3 in 21) of ratios that were >−1.0 log10 different.
FIG. 3.
Comparison of the log of the ratio of stock oocyst count/milliliter and MPN/milliliter with age for all experiments.
Table 2 compares the average percent infectivity and viability of various oocyst lot numbers. The average infectivity (by the FDM) and viability (by excystation, sporozoite yield, and DAPI-PI) varied considerably for each lot number. Correlation values (r) of the FDM with excystation, sporozoite yield, and DAPI-PI were 0.1340, −0.2249, and −0.8295, respectively, which demonstrates a significant difference between oocyst in vitro infectivity and surrogate viability. Correlation (r) between excystation and DAPI-PI was 0.0335 for this study.
TABLE 2.
Infectivity and viability of various oocyst lot numbers
| Oocyst lot no. | % Infectivity and/or viabilitya (SD)
|
|||
|---|---|---|---|---|
| FDM | Excystation | Sporozoite yield | DAPI-PI | |
| 1 | 37.7 (7.3) | 58.2 (3.9) | 65.1 (3.5) | 66.7 (12.1) |
| 2 | 63.5 (7.8) | 71.2 (7.6) | 58.5 (12.0) | 76.0 (9.3) |
| 3 | 8.5 (11.8) | 33.2 (7.8) | 76.5 (17.6) | 82.6 (4.5) |
| 4 | 42.7 (4.1) | ND | ND | ND |
| 5 | 4.7 (12.2) | 42.6 (16.6) | 48.0 (18.5) | 86.7 (8.8) |
| 6 | 8.7 (4.7) | 58.9 (5.7) | ND | ND |
| 7 | 3.1 (7.0) | 89.0 (2.7) | 84.0 (1.0) | 92.3 (3.1) |
| 8 | 5.4 (2.7) | ND | ND | ND |
The percent infectivity and viability is the geometric mean of the individual percents calculated for each lot number (n = 2 to 8). ND, no data recorded.
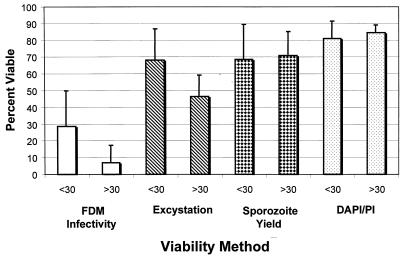
Figure 4 presents the comparison between the infectivity and viability assays for oocysts less than or greater than 30 days of age (n = 8). A statistical difference in infectivity by the FDM and viability by excystation was observed for oocysts less than 30 days of age when compared to that of oocysts greater than 30 days of age. No statistical difference in viability by DAPI-PI was observed for oocysts less than or greater than 30 days of age. It has generally been accepted that oocysts less than 90 days of age and stored in PBS at 4°C are viable as long as they exhibit ample excystation and vital dye exclusion. This study found that the percent infectious oocysts (by the FDM) decreased as the oocysts aged after 30 days and infectivity was significantly different from viability by the surrogate microscopic viability assays.
FIG. 4.
Comparison of in vitro infectivity (by the FDM-MPN method) and viability (by excystation and DAPI-PI) of oocysts less than or greater than 30 days of age. Error bars indicate standard deviations.
Table 3 shows the comparison between cell culture and BALB/c mouse infectivity. For the same lot of oocysts, the result of the FMD-MPN method was slightly higher than the mouse MPN of infectious oocysts (15 to 9.6 times greater) in untreated water samples. Cell culture and animal infectivity both showed similar reductions in the MPN/milliliter for infectious oocysts after treatment by pulsed white light (broad spectrum) (PureBrite) (16). After treatment, MPNs were 31.00 and 7.41 compared to <5.64 and 20.17/ml for cell culture and mouse infectivity, respectively. More than a 4 log10 inactivation was observed.
TABLE 3.
Cell culture FDM and BALB/c mouse infectivity by C. parvum oocysts
| Sample | MPN/ml (CI) for:
|
|
|---|---|---|
| Cell culture | BALB/c mouse | |
| Influent | 3.11 × 106 (6.89 × 105–9.03 × 106) | 1.97 × 105 (6.69 × 104–5.12 × 105) |
| 2.40 × 106 (4.80 × 105–9.70 × 106) | 2.49 × 105 (8.95 × 104–5.78 × 105) | |
| Effluenta | 31.09 (6.90–90.30) | <5.64b (0.92–19.05) |
| 7.41 (1.70–21.50) | 20.17 (3.94–42.72) | |
After treatment by pulsed white light (broad spectrum) (PureBrite).
No infection detected.
Charles N. Haas (Drexel, Pa.) performed a preliminary analysis of the cell culture data (by maximum likelihood) and computed the best-fit logistic relationship between the oocyst dose per well and the MPN infectivity response for all experiments (14). The analysis showed more scatter between data points at a given dose range than would be expected by chance alone. Based on the results of this study, the scatter is likely due to age and lot variability. However, despite the scatter, the 50% infective dose (ID50) in the cell culture system was shown to be approximately 10 oocysts.
DISCUSSION
Within the past several years, innovative cell culture technology has emerged as an alternative to various in vivo and in vitro infectivity and viability assays. In an effort to assess the inactivation of high concentrations of C. parvum oocysts, researchers are exploring the applications of infectivity assays with immunolabeling such as the FDM (27–30) and nucleic acid-based techniques such as the PCR and in situ hybridization (24, 26). The analysis presented here demonstrates the use of an in vitro assay to assess the infectivity of the protozoan parasite C. parvum, by immunolabeling infected human cells. By determining the presence or absence of infection in a multiple dilution series, the MPN of infectious oocysts can be quantified easily when used in combination with the FDM. The MPN generated by the program is provided with CIs that allow the user to analyze the differences between samples and demonstrates an application for use with survival and disinfection studies. To date, only semiquantitative in vitro infectivity assays have been used (26).
Antibody staining of infected cells provides an advantage over molecular detection methods such as PCR and in situ hybridization because the clustering of foci and specific life stages can be seen. This is a clear indication that reproduction has occurred. Sporozoites have been observed to excyst, initiate invasion of the host cells, but then not go further past that stage even after 48 h of incubation (16, 28). In this study, oocysts after pulsed broad-spectrum light treatment were observed to excyst and initiate infection (minimal) and they did form single foci. These oocysts could not proceed past the sporozoite invasion stage after 48 h of incubation, and thus, no infection clusters were observed. Molecular assays may detect the DNA or RNA produced in the invasion stage and may overestimate the total number of infectious oocysts.
The percent viable and infectious oocysts for each lot number varied considerably, indicating that there was significant lot-to-lot variability. Several freshly isolated oocyst lot numbers were less than 10% infectious. Oocyst processing to purify the oocysts from feces may contribute to the variability (e.g., defatting feces during process using ethyl ether, etc.). Future work will evaluate different oocyst purification assays to determine the effects on the oocysts.
The infectivity of all oocyst lot numbers decreased as the oocysts aged. Belosevic et al. showed similar findings using vital dyes for oocysts isolated from different calves and stored at 4°C (2). This variation in the viability and infectivity of different oocyst lot numbers and ages suggests that the oocysts are more affected by aging than previously thought, and each lot number should be analyzed before use for survival and disinfection studies.
This is the first study to report, quantitatively, oocyst viability over time by an infectivity assay. Several previous studies on oocyst infectivity or inactivation did not account for oocyst age prior to use. This study suggests that 30 days may be the maximum storage time, prior to experimentation, for maintaining a high proportion of infectious oocysts and that age may be an important quality control issue.
In our laboratory, the FDM-MPN method has now been used in disinfection studies for the water and food industry to determine oocyst inactivation after exposure to various water activities, pulsed white light, UV light, chlorine dioxide, ozonation and ultrahigh pressure. By using the MPNs, log10 and percent reductions can be calculated (16, 28, 30). The assay has been used for environmental (secondary sewage effluent [27]) and turbid water samples, which can be a problem in other viability assays such as microscopy, PCR, and in situ hybridization due to inhibition by the particulates. Future work will test the limits of particulates in the cell culture system.
Infectious oocysts can be detected repeatedly at very low concentrations (as low as 1 oocyst per ml determined by the FDM), indicating the high-level sensitivity of this assay. For each experiment, the oocysts were diluted to less than 1 oocyst per ml. Clusters of foci were detected at these highly diluted subsamples and were obvious against the dark host monolayer. The polyclonal antibody labels all the life stages present, thus enabling detection at low concentrations. Several monoclonal antibodies were tested; however, these did not have the same specificity or ability to pick up all the life stages as the polyclonal ones (data not presented). Since the FDM-MPN assay is both cell culture and antibody based, it may be very specific to infection only by C. parvum; however, the experiments to test specificity with all isolates and Cryptosporidium species have not been performed. Cryptosporidium muris was tested, and life stages past the initial trophozoite stage and clustering were not detected. Future efforts will focus on natural isolates from humans and animals, including various genotypes (24).
Vital dye and excystation methods have been routinely used for survival and disinfection experiments (9, 17, 18, 25). Frequently, researchers use animal models to validate the surrogate in vitro results (12, 23). Oftentimes, however, only the surrogate assays are used. Campbell et al. demonstrated the usefulness of the DAPI-PI and excystation methods used in their study for small numbers of oocysts and showed a good correlation between excystation and DAPI-PI (r2 = 0.997) (3). We have recently reported a comparison of results with DAPI-PI, excystation, and FDM-MPN with animal infectivity (30). In this study and previous work, the FDM-MPN method consistently determined a lower-level viability than either vital dye staining or excystation, suggesting that these surrogates overestimate the actual oocyst infectivity, particularly for oocysts >30 days of age. The FDM-MPN method appears to be similar to animal infectivity and further comparisons are ongoing. The maximum likelihood of the cell culture data showed an ID50 of around 10 oocysts, well below any other reported ID50 in animal models, suggesting that the cell culture system is a conservative assay. Previously reported ID50s of C. parvum in mice were 79, 83, and 100 oocysts (8, 10, 22) and in humans were 132 oocysts (7). A high degree of scatter in the maximum likelihood analysis of the cell culture data was shown. The factors contributing to this are unknown; however, the variability of oocysts from different lot numbers may be a contributing factor.
In conclusion, the FDM-MPN method has been shown to be an excellent and reproducible assay for quantifying oocyst infectivity in vitro. Oocyst age is an important issue that must be addressed before proceeding with disinfection projects. It is also notable that the cell culture assay is more sensitive than surrogate in vitro viability assays. When the FDM-MPN method was compared to animal models, a similar level of infectivity was determined, suggesting that the FDM can be used as an alternative to animal assays.
ACKNOWLEDGMENTS
The research described in this article has been funded in part by the U.S. Environmental Protection Agency under assistance agreement CR.-8241580 to the University of South Florida.
We thank Steve Upton and Keith Woods, Kansas State University, for providing primary antiserum. We also thank Charles N. Haas, Drexel University, for analyzing the data. We are grateful for constructive criticism and guidance from Walter Jakubowski, WaltJay Consulting. We are also indebted to Michael Arrowood for animal infectivity analysis.
REFERENCES
- 1.Arrowood M J, Hurd M R, Mead J R. A new method for evaluating experimental cryptosporidial parasite loads using immunofluorescent flow cytometry. J Parasitol. 1995;81:404–409. [PubMed] [Google Scholar]
- 2.Belosevic M, Guy R A, Taghi-Kilani R, Neumann N F, Gyurek L L, Liyange L R J, Millards P J, Finch G R. Nucleic acid stains as indicators of Cryptosporidium parvum oocyst viability. Int J Parasitol. 1997;27:787–798. doi: 10.1016/s0020-7519(97)00033-7. [DOI] [PubMed] [Google Scholar]
- 3.Campbell A T, Robertson L J, Smith H V. Viability of Cryptosporidium parvum oocysts: correlation of in vitro excystation with inclusion or exclusion of fluorogenic vital dyes. Appl Environ Microbiol. 1992;58:3488–3493. doi: 10.1128/aem.58.11.3488-3493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craun G F, et al. Waterborne outbreaks of cryptosporidiosis. J Am Water Works Assoc. 1998;90:81–91. [Google Scholar]
- 5.Crawford F G, Vermund S H. Human cryptosporidiosis. Crit Rev Microbiol. 1988;16:113–159. doi: 10.3109/10408418809104469. [DOI] [PubMed] [Google Scholar]
- 6.Dupey J P, Speer C A, Fayer R. Cryptosporidiosis of man and animals. Boca Raton, Fla: CRC Press; 1990. [Google Scholar]
- 7.Dupont H L, Chappeli C L, Sterling C R, Okhuysen P C, Rose J B, Jakubowski W J. The infectivity of Cryptosporidium parvum in healthy volunteers. N Engl J Med. 1995;332:855–859. doi: 10.1056/NEJM199503303321304. [DOI] [PubMed] [Google Scholar]
- 8.Ernest J A, Blagburn B L, Lindsay D S. Infection dynamics of Cryptosporidium parvum (Apicomplexa: Cryptosporiidae) in neonatal mice (Mus musculus) J Parasitol. 1986;75:796–798. [PubMed] [Google Scholar]
- 9.Fayer R, Nerad T. Effects of low temperatures on viability of Cryptosporidium parvum oocysts. Appl Environ Microbiol. 1996;62:1431–1433. doi: 10.1128/aem.62.4.1431-1433.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finch G R, Daniels C W, Black E K, Schaefer III F W, Belosevic M. Dose response of Cryptosporidium parvum in outbred neonatal CD-1 mice. Appl Environ Microbiol. 1993;59:3661–3665. doi: 10.1128/aem.59.11.3661-3665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez J M. A general purpose program for obtaining most probable number tables. J Microbiol Methods. 1996;26:215–218. [Google Scholar]
- 12.Graczyk T K, Fayer R, Cranfield M R, Owens R. Cryptosporidium parvum oocysts recovered from water by the membrane filter dissolution method retain their infectivity. J Parasitol. 1997;83:111–114. [PubMed] [Google Scholar]
- 13.Griffiths J K, Moore R, Dooley S, Keusch G T, Tzipori S. Cryptosporidium parvum infection of Caco-2 cell monolayers induces an apical monolayer defect, selectively increases transmonolayer permeability, and causes epithelial cell death. Infect Immun. 1994;62:4506–4514. doi: 10.1128/iai.62.10.4506-4514.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas C N. Estimation of microbial densities from dilution count experiments. Appl Environ Microbiol. 1989;55:1934–1942. doi: 10.1128/aem.55.8.1934-1942.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halvorson H O, Ziegler N R. Application of statistics to problems in bacteriology. I. A means of determining bacterial population by the dilution method. J Bacteriol. 1933;25:101–121. doi: 10.1128/jb.25.2.101-121.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huffman, D. E., T. R. Slifko, M. J. Arrowood, and J. B. Rose. Inactivation of bacteria, virus and Cryptosporidium by a point-of-use device pulsed broad spectrum white light. Water Res., in press.
- 17.Jenkins M B, Bowman D D, Ghiorse W C. Inactivation of Cryptosporidium parvum oocysts by ammonia. Appl Environ Microbiol. 1998;64:784–788. doi: 10.1128/aem.64.2.784-788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson D C, Enriquez C E, Pepper I L, Davis T L, Gerba C P, Rose J B. Survival of Giardia, Cryptosporidium, poliovirus, and Salmonella in marine waters. Water Sci Technol. 1997;35:261–268. [Google Scholar]
- 19.Klee A J. A computer program for the determination of the most probable number and its confidence limits. J Microbiol Methods. 1993;18:91–98. [Google Scholar]
- 20.Klee A J. ICR most probable number calculator, version 1.00. [Online.] Cincinnati, Ohio: Risk reduction laboratory, U.S. Environmental Protection Agency; 26 January, 1999, revision date. http://www.epa.gov/nerc/www/other.htm . 26 July 1999, last date accessed. [Google Scholar]
- 21.Koch A L. Estimation of the most probable number with a programmable pocket calculator. Appl Environ Microbiol. 1982;43:488–490. doi: 10.1128/aem.43.2.488-490.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korich D G, Marshall M M, Smith H V, Clancy J L, Fricker C R. Abstracts of the 98th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1998. Cryptosporidium oocyst infectivity: an interlaboratory comparison of the neonatal mouse model, abstr. Q-233; p. 459. [Google Scholar]
- 23.Korich D G, Mead J R, Madore M S, Sinclair N A, Sterling C R. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl Environ Microbiol. 1990;56:1423–1428. doi: 10.1128/aem.56.5.1423-1428.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan U M, Thompson R C A. PCR detection of Cryptosporidium: the way forward? Parasitol Today. 1998;14:241–245. doi: 10.1016/s0169-4758(98)01247-2. [DOI] [PubMed] [Google Scholar]
- 25.Robertson L J, Campbell A T, Smith H V. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl Environ Microbiol. 1992;58:3494–3500. doi: 10.1128/aem.58.11.3494-3500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rochelle P A, Ferguson D M, Handojo T J, DeLeon R, Stewart M H, Wolfe R L. Development of an infectivity assay for waterborne Cryptosporidium. In Proceedings of the American Water Works Associaton Water Quality Technology Conference. Denver, Colo: American Water Works Association; 1998. [Google Scholar]
- 27.Slifko T R, Friedman D E, Rose J B, Upton S J, Jakubowski W. Unique cultural methods used to detect viable Cryptosporidium parvum oocysts in environmental samples. Water Sci Technol. 1996;35:363–368. [Google Scholar]
- 28.Slifko, T. R., D. E. Friedman, J. B. Rose, J. H. Fraser, and K. M. J. Swanson. 1999. Unpublished data.
- 29.Slifko T R, Friedman D, Rose J B, Jakubowski W. An in vitro method for detecting infectious Cryptosporidium oocysts with cell culture. Appl Environ Microbiol. 1997;63:3669–3675. doi: 10.1128/aem.63.9.3669-3675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slifko T R, Friedman D E, Rose J B. Proceedings of the American Water Works Association Water Quality Technology Conference. Denver, Colo: American Water Works Association; 1998. Comparison of 4 Cryptosporidium parvum viability assays: DAPI/PI, excystation, cell culture, and animal infectivity. [Google Scholar]
- 31.Smith H V, Rose J B. Waterborne cryptosporidiosis: current status. Parasitol Today. 1998;14:14–22. doi: 10.1016/s0169-4758(97)01150-2. [DOI] [PubMed] [Google Scholar]
- 32.Upton S J, et al. A simple and reliable method of producing in vitro infections of Cryptosporidium parvum (Apicomplexa) FEMS Microbiol Lett. 1994;118:45–50. doi: 10.1111/j.1574-6968.1994.tb06801.x. [DOI] [PubMed] [Google Scholar]
- 33.Upton S J, Tilley M, Brillhart D B. Comparative development of Cryptosporidium parvum (Apicomplexa) in 11 continuous host cell lines. FEMS Microbiol Lett. 1994;118:233–236. doi: 10.1111/j.1574-6968.1994.tb06833.x. [DOI] [PubMed] [Google Scholar]
- 34.Upton S J, Tilley M, Brillhart D B. Comparative development of Cryptosporidium parvum in MDBK and HCT-8 cells under select atmospheres. Biomed Lett. 1994;49:265–271. [Google Scholar]
- 35.U.S. Environmental Protection Agency. Pesticide program guide standard and protocol for microbiological water purifiers. Fed Regist. 1986;51:19403. [Google Scholar]
- 36.Yang S, Healey M C, Du C, Zhang J. Complete development of Cryptosporidium parvum in bovine fallopian tube epithelial cells. Infect Immun. 1996;64:349–354. doi: 10.1128/iai.64.1.349-354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]