Abstract
Brain arteriovenous malformations (AVMs) are a disorder wherein abnormal, enlarged blood vessels connect arteries directly to veins, without an intervening capillary bed. AVMs are one of the leading causes of hemorrhagic stroke in children and young adults. Most human sporadic brain AVMs are associated with genetic activating mutations in the KRAS gene. Our goal was to develop an in vitro model that would allow for simultaneous morphological and functional phenotypic data capture in real time during AVM disease progression. By generating human endothelial cells harboring a clinically relevant mutation found in most human patients (activating mutations within the small GTPase KRAS) and seeding them in a dynamic microfluidic cell culture system that enables vessel formation and perfusion, we demonstrate that vessels formed by KRAS4AG12V mutant endothelial cells (ECs) were significantly wider and more leaky than vascular beds formed by wild-type ECs, recapitulating key structural and functional hallmarks of human AVM pathogenesis. Immunofluorescence staining revealed a breakdown of adherens junctions in mutant KRAS vessels, leading to increased vascular permeability, a hallmark of hemorrhagic stroke. Finally, pharmacological blockade of MEK kinase activity, but not PI3K inhibition, improved endothelial barrier function (decreased permeability) without affecting vessel diameter. Collectively, our studies describe the creation of human KRAS-dependent AVM-like vessels in vitro in a self-assembling microvessel platform that is amenable to phenotypic observation and drug delivery.
1. Introduction
A brain arteriovenous malformation (AVM) is an abnormal tangle of blood vessels in the brain, wherein feeding arteries are directly connected to draining veins, without an intervening capillary bed, characterized by high blood flow, and dilated, tortuous vessels [1]. Instead of a normal hierarchical vascular network, this abnormal vascular conduit, or shunt, lacks the integrity and stability characteristic of normal capillary networks. Individuals with AVMs are at increased risk of hemorrhage as patients age due to the direct shunting of high-pressure flow from the artery [2,3]. AVMs are responsible for 4% of all intracerebral haemorrhages and account for almost one third of brain bleeds in young adults [4]. Currently, there are no approved pharmacological treatments for brain AVMs, and the only available treatment options for patients are surgical resection and embolization of small AVMs or stereotactic radiosurgery for inoperable vessels to reduce vessel size. Both procedures are highly invasive with a recurrent hemorrhage risk as high as 18% in the first year. Approximately one third of all AVM cases are untreatable with currently available options [5].
Various mutations within genes encoding proteins in the mitogen-activated protein kinase (MAPK/ERK, e.g. the RAS-RAF-MEK-ERK) pathway have been implicated in AVM pathology. Recently, somatic activating KRAS mutations in endothelial cells (ECs) have been reported in as many as 60% of clinical samples [6,7]. Nikolaev et al. [7] showed that somatic activating KRAS mutations in ECs derived from patient AVMs, or expression of mutant KRAS in human umbilical vein endothelial cells (HUVECs), leads to increased ERK1/2 phosphorylation and disruption of vascular endothelial (VE)-cadherin localization in adherens junctions. This is thought to increase the permeability of the vessel wall, and to leave vessels prone to hemorrhage [7,8]. Although these genetic mutations are sufficient for the development of AVMs in zebrafish and mouse models [9], the initiation, development, and remodelling of these dynamic vessel malformations are influenced by factors in the microenvironment. Specifically, vascular endothelial growth factor (VEGF) and other pro-angiogenic factors are highly expressed in AVMs, leading to increased vessel permeability and susceptibility to rupture [10,11]. Other pathways, such as Notch and transforming growth factor (TGF)-β/bone morphogenetic protein (BMP) have also been implicated in AVM regulation [12], particularly with discontinuity of cell-cell junctions and cell proliferation, resulting in vascular dysplasia and instability [11,13,14]. Moreover, growing evidence suggests that the AVM microenvironment may promote AVM growth. Inflammation driven by local macrophages and reduction of pericyte coverage have been described as contributing factors in AVM pathogenesis, as they promote excessive angiogenic responses [15,16]. Additionally, a high flow rate is a defining feature in AVMs that leads to a positive feedback loop to increase local vessel enlargement in high-velocity vessels [17]. However, there is limited knowledge as to how environmental cues from surrounding cells, hemodynamics, and the KRAS mutation contribute to the pathogenesis of AVMs.
Animal models of the disease have achieved variable degrees of success in generating vessel lesions with characteristics that match human pathology. Several transgenic mouse models of hereditary hemorrhagic telangiectasia (HHT), an inherited disease known to cause AVMs, involve genetic inhibition or deletion of either Eng or Alk1 genes in combination with angiogenic induction to generate AVMs [18-20]. While early postnatal, endothelial specific deletion of either Alk or Eng in mice produces brain AVMs, deletion in adults fails to produce brain AVMs in the absence of angiogenic stimuli [21,22]. Critically, successful murine models have recapitulated key features of AVMs, such as vessel dilation, arteriovenous shunts, and hemorrhaging [23]. The incidence of observable AVMs can be low, and there is often high and early mortality associated with these models [23,24]. Recently, mouse and zebrafish models with the KRASG12V or KRASG12D mutation displayed enlarged, tortuous vessels that may be prone to hemorrhage [9,25]. These animal models have contributed to defining a mechanism that may underlay the majority of sporadic brain AVMs, as these activating mutations are detected in more than 50% of sporadic bAVMs. However, these models are costly, in terms of both time and resources, and have yet to be validated in a human-relevant model other than monolayer cell culture studies [7,9,26].
In recent years, organ-on-a-chip models have emerged as candidates to address the limitations of 2D models and species-specific factors in disease modeling and drug development. In comparison to cell monolayers, organ-on-a-chip models have increased architectural complexity, mimicking key aspects of in vivo tissue physiology [27]. Using a combination of adjoining channels and interfaces, they enable precise spatiotemporal control of mechanical and chemical stimuli to probe tissue responses [28,29]. Several studies have successfully produced vessel-like networks using ECs from different origins, in combination with supporting cells such as fibroblasts [30,31], pericytes [32,33], or mesenchymal stem cells [34]. Self-assembled microvessels on-chip models are formed in the presence of media flow, allowing vessel perfusion and mimicking in vivo flow dynamics and feedback [35]. Rather than having predefined geometries, the self-assembling nature of microvasculature-on-a-chip forms an organic vasculature with vessels of varying shapes and sizes. This aspect is particularly relevant when studying diseases that lead to alterations in vessel diameter, such as in AVMs [29].
Our goal was to recapitulate the key hallmarks of AVMs in vitro using human ECs harbouring a clinically relevant KRAS mutation described in patients with sporadic AVMs in a microfluidic platform that allows 3D vessel assembly and perfusion. We also aimed to test the impact of MEK inhibitors on AVM structural and functional parameters. We found that a mosaic culture of KRASG12V mutant and wildtype ECs led to irregularly shaped and enlarged microvessels, with increased vessel wall permeability. Treatment with a MEK inhibitor restored barrier function but did not reverse vessel dilation.
2. Materials and methods
2.1. Cell culture
Two separate immortalized human umbilical vein endothelial cell (Im-HUVEC) lines were used in the study. The first Im-HUVEC line constitutively expresses green fluorescent proteins (GFP). Im-HUVEC-GFPs (a gift from Dr. Roger Kamm at MIT [36]) were grown in VascuLife VEGF Endothelial Medium (Lifeline Cell Technology). The second Im-HUVEC line (a gift from Dr. Arnold Hayer at McGill University [37]) was used to generate a stable cell line with doxycycline-inducible expression of either mScarlet-tagged KRAS4AWT or KRAS4AG12V proteins (no expression of GFP). Primary HUVECs and Im-HUVECs transfected with KRAS4AWT and KRAS4AG12V were grown in Endothelial Cell Medium (ScienCell). Normal human lung fibroblasts (NHLF; Lonza) were grown in DMEM (ThermoFisher) supplemented with 20% fetal bovine serum (FBS). NHLF of passage 5–7 were used in experiments. All cells were cultured in cell culture flasks coated with 1% gelatin until they reached 85–90% confluency.
For assays performed in monolayers, Im-HUVECs-GFP or KRAS4AG12V HUVECs were seeded at 2,000 cells/mm2 in a 96 well plate. KRAS4AG12V HUVECs were induced prior to seeding and grown for 24 h at confluence. For drug treatment groups, KRAS4AG12V HUVECs were treated with 1 μM MEK inhibitor (MEKi, SL327, Sigma), 10 μM PI3K inhibitor (PI3Ki, LY294002, Sigma), or 0.1% DMSO control for 24 h. Cells were fixed and stained for analysis.
2.2. Generation of stable, Tet-On KRAS4AG12V HUVECs
Telomerase-immortalized human umbilical vein endothelial cells (Im-HUVECs, or hTERT-HUVECs) were kindly provided by Dr. Arnold Hayer (McGill University) [37]. A piggyBac ITR flanked plasmid, pB-TA-ERN - a gift from Knut Woltjen (Addgene plasmid # 80474) [38] was combined with either pDONR221-V5-mScarlet-I-hsKRAS4AWT (addgene #156405) or pDONR221-V5-mScarlet-I-hsKRAS4AG12V (addgene #156406) [9] and LR recombinase to generate pB-TetOn-mScarlet-KRAS4AWT/G12V by Gateway recombination. To generate the stable Im-HUVEC tunable KRASWT/G12V line, 1.5 μg of pB-TetON -mScarlet-KRAS4A-WT (JDW 890) or pB-TetON-mScarlet-KRAS4A -G12V (JDW 891) and 1 μg of a plasmid encoding a hyperactive piggyBac transposase (hyPBase) [39] were co-electroporated into Im-HUVEC using the P5 Primary Cell Kit (Lonza) and a 4D-nucleofector (Lonza). The program used was CA167. After electroporation, 100 μL of FBS was immediately added to the cuvette and incubated at 37 °C for 5 min. Cells were transferred to a 12-well plate after incubation and grown for 2 days in antibiotic-free media. For antibiotic selection, 0.3 mg/mL G418 was added for 5 days with media change every other day. After selection, cells (KRAS4AWT and KRAS4AG12V HUVECs) were expanded and passaged.
2.3. Device fabrication
AutoCAD was used for creating the device design (Supplemental Fig. 1A), which was written onto a chrome photomask (Heidelberg upg501 Mask Writer). The standard lithography process supplied by the photoresist manufacturer was used to generate the silicone molds. Briefly, SU-8 2050 photoresist (MicroChem) was spun onto a silicon mold to achieve a 100 pm thick layer. The photomask was used to polymerize (240 mJ/cm2 exposure energy) the pattern onto the wafer. An optical profiler was used to confirm surface and edge finishes of each silicon wafer mold. Silicone mold heights ranged from 90 to 120 μm.
PDMS was mixed at a 1:10 ratio (elastomer to base) and left to de-gas for 20 min. PDMS was poured onto the wafers inside an acrylic ring so that the final PDMS mold was easily removable. PDMS was baked for 3 h in the oven at 80 °C. After baking, the PDMS was left overnight to cool before removing the PDMS mold. This was to prevent the SU-8 features from peeling off while removing the PDMS. Access holes of 2 mm for the side channels and 1 mm for the tissue chamber were cut out using biopsy punches. PDMS coated glass slides were prepared by spreading 2 mL of PDMS onto a 75 × 50 mm glass slide and baking for 1 h. Prior to bonding, PDMS molds and PDMS-coated glass slides were covered with packing tape to prevent dust accumulation. The devices were bonded together using plasma treatment (Harrick Plasma). Media reservoirs were created by attaching cryovials (that had their bottoms cut out) using PDMS (Supplemental Fig. 1B). The entire device unit was autoclaved before use (Supplemental Fig. 1C).
2.4. Microfluidic device cell seeding
Twenty-four hours after KRAS4AG12V HUVECs were plated into cell culture flasks, doxycycline (Sigma-Aldrich, D3072) was added to the media (final concentration: 5 μg/mL) to induce KRAS4AG12V expression (Supplemental Fig. 2). For all experiments in microfluidic devices, we used Im-HUVECs expressing GFP (a gift from Dr. Roger Kamm at MIT) for visualization of microvascular network formation. Im-HUVECs and NHLFs were seeded in fibrin gels (initial concentration: 10 mg/mL bovine fibrinogen) at a 4:1 ratio (Im-HUVECs to NHLF) for a concentration of 1 million total cells/mL. To generate AVM-like microvessels, KRAS4AG12V HUVECs were combined with Im-HUVEC:NHLF cell suspension at the described ratios (i.e. 83K KRAS4AG12V HUVECs/mL was used to achieve 10% KRAS and 208K KRAS4AG12V HUVECs/mL was used to achieve 25% KRAS). The cell suspension was gently pipetted into one side of the tissue chamber until the chamber was filled. Devices were placed in the incubator for 15 min for gel polymerization, after which Im-HUVECs were seeded to the side channels (10 million total cells/mL) coated with 0.1% gelatin in PBS (BioShop, cat# GEL771.500) for 2 h in Vasculife media. During this time, devices were routinely checked, and bubbles removed. To establish perfusion, Vasculife media was added in the following volumes to the media reservoir #1: 1500 μL, #2: 1000 μL, #3: 500 μL, #4: 200 μL to achieve an approximate pressure gradient of #1: 23 mm H2O, #2: 18 mm H2O #3: 8 mm H2O, #4: 3 mm H2O, as previously reported [31]. Media reservoirs were refilled every 24 h. The media flow volume across the tissue chamber was reversed (1&3 ↔ 2&4) every 2 days to prevent ECs from migrating to side channels (Fig. 1A).
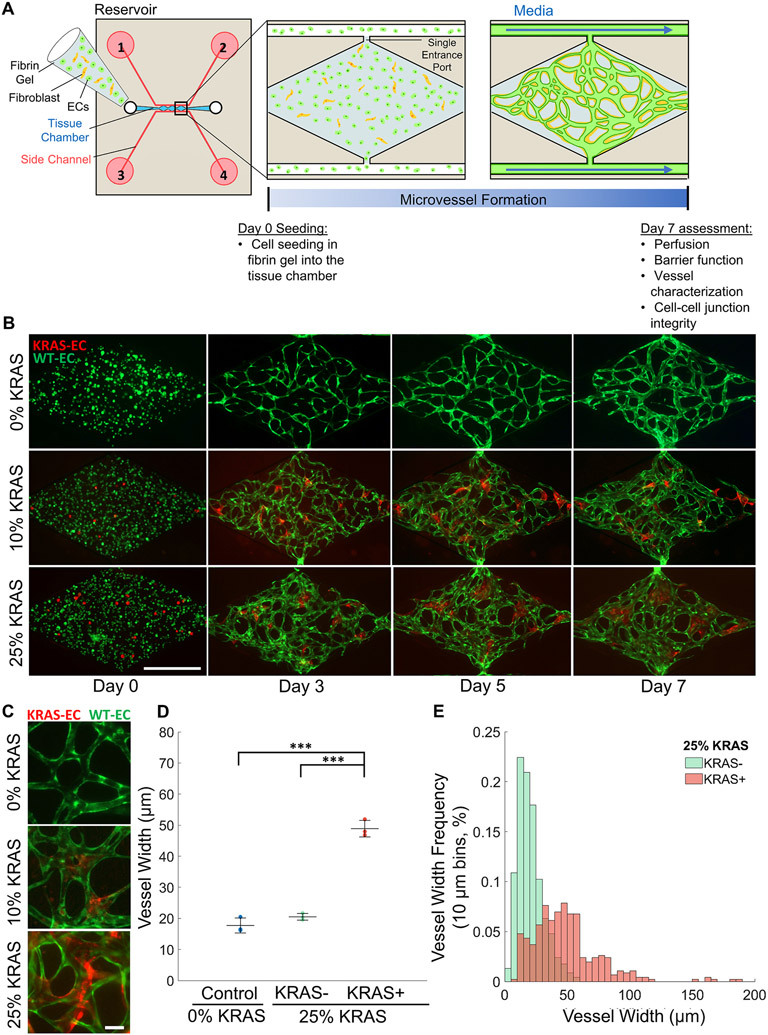
Fig. 1. Generation of a perfused human AVM-like vasculature-on-a-chip.
(A) Schematic representation of the device, experimental design and timelines. A combination of endothelial cells and fibroblasts were seeded in fibrin hydrogels for up to 7 days. (B) Representative time course of vessel network formation using different ratios of wild-type (WT) and KRAS4AG12V ECs. Images of cell suspension immediately after seeding on day 0. WT ECs (green) and KRAS4AG12V ECs (red) are round and evenly distributed within the fibrin gel. On day 3, tube-like structures begin to form as cells migrate towards each other. By day 5, microvessel connections and network shape is observable. By day 7, the microvessel network is fully interconnected, lumenized and perfusable. (Scale bar 500 μm). (C) Higher magnification fluorescent images of microvessel segments of 0% KRAS4AG12V (control), 10% KRAS4AG12V, and 25% KRAS4AG12V vasculatures (scale bar, 50 μm). (D) Quantification of the average individual microvessel width in vessel networks of 0% KRAS4AG12V ECs (blue) or 25% KRAS4AG12V vasculatures. Assessment of 25% KRAS4AG12V vasculatures was subdivided into vessel segments with (red, KRAS+) or without (green, KRAS−) KRAS4AG12V ECs. (One-way ANOVA with Tukey’s multiple comparison test; ***p < 0.001, N = 3; mean ± SD) (E) Distribution of microvessel widths in 25% KRAS4AG12V vasculatures between segments without KRAS4AG12V ECs (green) and segments with some visible KRAS4AG12V (red) (bin size: 10 μm). KRAS4AG12V positive vessel distribution is much wider and variable compared to KRAS4AG12V negative vessels.
2.5. Microvessel network perfusion and permeability
To assess microvessel perfusion and permeability, 1 mg/mL of 70 kDa dextran conjugated to rhodamine b isothiocyanate (Rhod B; Sigma) was introduced from one media reservoir. Once the dextran solution had perfused the entire vessel network (t = 1) images of each diamond within the tissue chamber were acquired at 10 min intervals, for 30 min total. Images were acquired using a Zeiss AxioObserver Widefield microscope at 5X magnification within a 37 “C temperature-controlled incubation chamber (5% CO2).
2.6. Immunofluorescence staining
Immunostaining was performed in situ, by perfusing all reagents into the device through the side channels. The devices were fixed using 4% paraformaldehyde (PFA) for 20 min at room temperature and washed with PBS overnight. Devices were then permeabilized with 0.05% Tween 20 in PBS and washed for 2–3 h with PBS. Then, a blocking solution (5% BSA) was added and incubated overnight at 4 °C. VE-cadherin primary antibody (Santa Cruz cat# sc-9989, 1:200 dilution) or Ki67 primary antibody (BD Biosciences cat# 550609, 1:200 dilution) was incubated overnight at 4 °C. Devices were washed with PBS for 4–6 h and subsequently incubated with Alexa Fluor 647-conjugated goat anti-mouse IgG (H + L) (ThermoFisher cat# A32728, 1:300 dilution) and Hoechst 33342 (Sigma-Aldrich cat# B2261, 1:10000 dilution) in blocking solution overnight at 4 °C. Devices were washed with PBS and imaged using the Zeiss AxioObserver Widefield microscope.
For immunofluorescence staining of cell monolayers, primary HUVEC and Im-HUVEC cells were washed with PBS and fixed with 4% PFA at room temperature for 20 min. Cells were then permeabilized with 0.5% Triton X-100 for 10 min, followed by blocking with 5% BSA for 1 h at room temperature. Primary antibodies were added to the cells and incubated at 4 °C overnight. Primary antibodies used were VE-cadherin (Santa Cruz cat# sc-9989; 1:200 dilution) and Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (Cell Signaling Technology cat# 9101S; 1:200 dilution). On the following day, primary antibodies were removed, and cells were washed with PBST (0.1% Triton X-100 in PBS). Secondary antibodies were incubated at room temperature for 2 h, covered in foil. Secondary antibodies used were Alexa 488 goat anti-mouse (Invitrogen cat# A11001; 1:200 dilution) and Alexa 647 goat anti-rabbit (Invitrogen A21245; 1:200 dilution). After secondary antibody incubation, cells were washed with PBST and Hoechst (ThermoFisher cat# 62249) staining was done at 1:500 dilution for 10 min at room temperature. Then, cells were washed with PB S and mounted with ProLong™ Gold Antifade Mountant (Invitrogen cat# P36930). Confocal imaging of primary HUVEC and Im-HUVEC cells grown in 2D was done with an Olympus Fluoview 1000 Confocal microscope Olympus IX81 inverted stand (Olympus, CA, USA) using the Plan Apo 40x/1.35 NA oil immersion objective (NA 1.3). Excitation wavelengths were 405 nm for Hoechst, 473 nm for Alexa Fluor 488, 559 nm for mScarlet and 635 nm for Alexa Fluor 647. Image processing was done uniformly across all conditions with the FV10-ASW 4.2 Viewer (Olympus, CA, USA) and FIJI (v2.1.0/1.53c).
2.7. Treatment of microvessel network
For drug treatment experiments, 25% KRAS4AG12V vasculatures were treated with 1 μM MEK inhibitor (MEKi, SL327, Sigma) or 10 μM PI3K inhibitor (PI3Ki, LY294002, Sigma) in Vasculife media on days 3 or 5. Treatment was maintained until endpoint (day 7). DMSO (0.1%) was used as a vehicle control.
2.8. Real-time cell analysis (RTCA)
Prior to the start of experiments, 16-well E-plates (ACEA Biosciences cat # 300 600 890) were coated with 0.1% (v/v) gelatin attachment factor for an hour at 37 °C. After coating, attachment factor was removed and replaced with 150 μL of culture media (ScienCell). Baseline media reading was recorded every 5 min for an hour on the xCelligence system (software version 2.0.0.1301). Following baseline measurements, Im-HUVECs cell lines (with or without 5 μg/mL doxycycline) were seeded in monolayers in duplicates into the 16-well E-plate at 15,000 cells in 150 μL of media per well. Cell recordings were taken every 10 min over the next 96 h. Cell index was normalized based on media baseline values. Cell doubling time was calculated by the software between the time range of cell settlement and cell confluency. The xCelligence system was inside a 37 °C incubator with 5% CO2 during experiments.
2.9. Western blot
Inducible Im-HUVEC KRAS4AWT and KRAS4AG12V cells (using Im-HUVECs gifted from Dr. Arnold Hayer at McGill University) were treated with 5 μg/mL doxycycline for different periods of time (6, 24, 48 and 72 h) and serum starved in 0.1% FBS culture media for 6 h prior to collection. Cells were collected from cell culture plates with 2X RIPA Lysis buffer (Millipore cat # 20–188) plus 1X protease inhibitors (Roche cat# 11836170001) and 1X phosphatase inhibitors (ThermoFisher cat# A32957). BCA assay was performed to measure protein concentration using the Pierce BCA Protein Assay Kit (ThermoFisher cat# 23227), following manufacturer’s instructions. Equal amount of protein lysate was further mixed with 2X Laemmli Sample Buffer (Bio-Rad cat# 1610737) and 2.5% of β-Mercaptoethanol (Acros Organics cat# 60-24-2). Samples were heated at 95 °C for 10 min and run on a 4–20% gradient precast gel (Bio-Rad cat # 4561094). Proteins were transferred onto a PVDF membrane (Cytiva cat# 10600023) using the Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). Blocking was done for 1 h at room temperature with 5% BSA in TBST (1XTBS-0.2% Tween 20) for phosphorylated proteins and 5% milk in TBST for total proteins. Primary antibodies were diluted in 2% BSA or milk in TBST and incubated on a shaker at 4 °C overnight. Primary antibodies used were p-p44/42 ERK1/2 (Thr202/Tyr204) (CST cat# 9101S, 1:1000 dilution), total p44/42 ERK1/2 (CST cat# 4695S, 1:1000 dilution), p-AKT (Ser473) (CST cat# 4060S, 1:1000 dilution), total AKT (CST cat# 9272S, 1:1000 dilution) and GAPDH (CST cat# 5174S, 1:1000 dilution). On the following day, membrane was washed with TB ST and incubated with Anti-rabbit IgG, HRP-linked Antibody (CST cat# 7074S, 1:3000 dilution) for 1 h at room temperature. After secondary antibody, membrane was washed and developed using the SuperSignal™ West Pico PLUS Chemiluminescent Substrate (ThermoFisher cat# 34580) for 5 min at room temperature. Membranes were imaged using the ChemiDoc Imaging System (Bio-Rad). Membrane stripping was done with Restore PLUS Western Blot Stripping Buffer (ThermoFisher cat# 46430). Densitometric quantification was performed using FIJI (v2.1.0/1.53c).
2.10. Image analysis
All images were analysed using Zen blue (ZEISS) and ImageJ 1.53q (FIJI). All graphs and calculations were done using MATLAB (MathWorks, R2020). Angiotool 0.6 40 was used to assess total branch length. Diameter, junction, and branch measurements were done manually using Zen blue. To measure fluorescent intensity in perfusion assays, a binary mask was created from the FITC channel. Two separate images were generated for the intravascular and extravascular perfusion by using the AND operation of the mask and inverse mask over the TRITC channel. The intensity values were then averaged from the intravascular and extravascular images respectively. The permeability coefficient (P) was calculated using the previously described equation [40].
Where D is the average diameter, Δt is the time interval, If,Ii,Ib is the fluorescence intensity of the final intravascular, initial intravascular, and background (extravascular) intensity respectively.
2.11. Statistical analysis
All statistical analyses were performed using Prism 9 software (GraphPad). All data presented are mean ± standard deviation (SD). One-way ANOVA with Tukey’s multiple comparisons was used to assess significance between 3 groups. The paired two-tailed t-test was used to assess diameter changes over time. The unpaired two-tailed t-test was used to assess significance between 2 groups. A p value < 0.05 was considered significant. Each diamond within the tissue chamber was considered a technical replicate (indicated by n). Separate experiments (indicated by N) were considered biological replicates. At least 3 ‘N’ were used for each experimental/seeding condition.
3. Results
3.1. Generation of perfused human AVM-like vasculatures in vitro
We used a previously described microfluidic device consisting of a central tissue chamber divided into three diamond-shaped chambers, with one port on each end connected to side channels where media is delivered [31,35] (Fig. 1A). The 50 μm ports create a single entry and exit in the 1× 2 mm diamond chamber and segments the vasculature into three technical replicates (Fig. 1A and Supplemental Fig. 1A). In this platform, perfusable microvessels were reliably generated in vitro over the course of 7 days through self-assembly of endothelial cells (i.e., Im-HUVECs), supported by fibroblasts at a 4:1 ratio. Additionally, the inclusion of endothelial cells (ECs) in the side channels, generating a monolayer, helped with vessel anastomosis and prevented leakage [35] (Fig. 1A). To prevent cells from migrating out of the tissue chamber, the media flow direction was switched between the two side channels every two days by switching inlet function between reservoirs 1&3 to 2&4 (Fig. 1A).
For cell seeding (day 0), cells were loaded into the tissue chamber in a fibrin gel. By day 3, ECs extended to neighbouring cells and clustered together to form rudimentary vessels. By day 5, the network structure was clearly established and interconnected. At this time point, microvessels were marginally perfusable (not shown), indicating that the network was lumenized. By day 7, microvessels expanded in diameter to create interconnected, well-perfused, and stable vessel networks (Fig. 1B, top panel).
To create the AVM disease model, ECs containing a stably integrated, doxycycline-inducible mScarlet (mScar)-KRAS4AG12V expression construct were induced prior to seeding into the microfluidic device [9]. Expression was confirmed by the presence of mScar after the addition of doxycycline (Supplemental Fig. 2). Induction of KRAS4AG12V elevated pERK levels in the mutant ECs, indicating functional KRAS protein activity and enhanced MAPK signaling (Supplemental Figs. 3A-C). As a control, an inducible KRAS4AWT cell line was generated in the same fashion and doxycycline induction of KRAS4AWT did not lead to changes in pERK levels (Supplemental Figs. 3A-C). Both KRAS4AWT and KRAS4AG12V lines did not show significant changes in pAKT levels (Supplemental Figs. 3D-F). Consistent with our previous studies using a different cell line [5,7], mutant KRAS expression reduced VE-cadherin localization at cell-cell junctions (Supplemental Figs. 2 and 8). For seeding into the microfluidic devices, KRAS4AG12V ECs induced with doxycycline were combined with WT ECs (GFP-tagged cell line to visually distinguish WT KRAS ECs from mutant KRAS ECs) in fibrin gels, at 10% or 25% of the total EC number. WT ECs seeded without KRAS4AG12V ECs served as controls. Over the course of 7 days, heterogeneous KRAS4AG12V and WT-EC co-cultures effectively self-assembled into perfusable microvessels (Fig. 1B, middle and bottom panels). In devices containing KRAS4AG12V ECs, mScar-positive cells were present within the vessel network, confirming that the KRAS4AG12V ECs had integrated into the vasculature (Fig. 1B and C). As expected, vasculatures with KRAS4AG12V ECs had distinct aberrant vessel structures. Vascular networks containing 25% KRAS4AG12V ECs resembled a cluster of shortened vessel structures rather than the typical microvascular networks formed using WT ECs (Fig. 1B and C).
To quantify potential changes in vessel diameter associated with the expression of mutant KRAS, the width of individual microvessels in the 25% KRAS vasculatures were measured on day 7. In line with human brain AVMs, we found that vessel segments in 25% KRAS microvessels containing KRAS4AG12V ECs (KRAS+) were significantly wider than vessel segments within the same network that lacked KRAS4AG12V ECs (KRAS−), measuring 48.9 ± 2.6 μm and 20.5 ± 1.1 μm, respectively (Fig. 1D). There was no significant difference in vessel width between KRAS− microvessels in 25% KRAS vasculatures (20.5 ±1.1 μm) and vessels in wild-type monocultures (0% KRAS; 17.7 ± 2.4 μm) (Fig. 1D). Vessel width in the 25% KRAS microvessels was larger and highly heterogenous compared to 0% KRAS, resulting in a larger vessel width distribution (Fig. 1E). This demonstrated that vessel enlargement only occurs where KRAS4AG12V ECs are present and does not affect surrounding vessels formed by WT ECs within the same network. Unfortunately, it was not possible to assess vessel width of KRAS4AG12V EC vessels in the 10% KRAS networks, as KRAS4AG12V ECs mainly concentrated in vessel junctions when seeded at low ratios (Fig. 1C). Vasculatures generated with 0%, 10%, and 25% KRAS4AG12V ECs at time of seeding will be referred to henceforth as 0%, 10% and 25% KRAS vasculatures respectively.
3.2. Changes in vessel network parameters towards AVM-like morphology
To characterize the morphological and functional changes in vessel networks formed with KRAS4AG12V ECs at day 7 post-seeding (Fig. 2A), we evaluated branch length, junction number, and vessel area coverage. As a result of vessel enlargement, the total area covered by the vessel network was greater in 10% and 25% KRAS microvessels (50.0 ± 3.2% and 53.4 ± 3.5%, respectively), and significantly higher than 0% KRAS microvessels (42.1 ± 6.9%) (Fig. 2B).
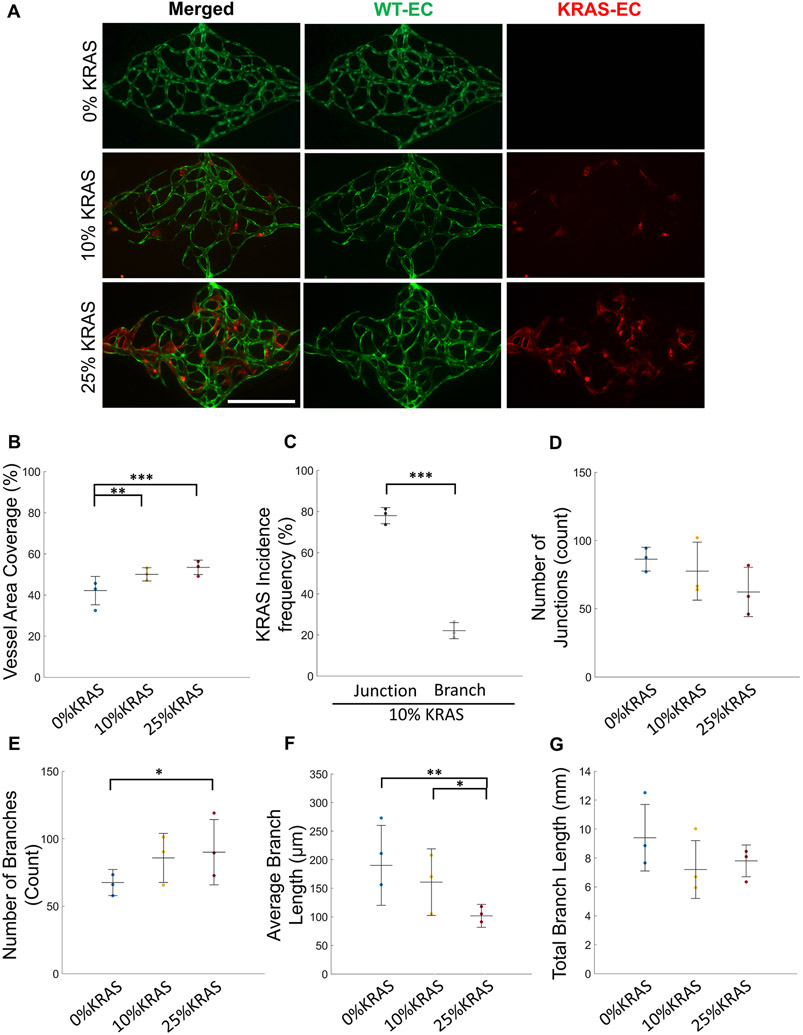
Fig. 2. Quantification of AVM-like microvessel morphological characteristics.
(A) Representative images of day 7 microvessel networks (WT ECs, green; KRAS4AG12V ECs, red) (Scale bar 500 μm). (B) Total vessel area coverage of 0% (blue), 10% (yellow), and 25% KRAS4AG12V (red) microvasculatures. (One-way ANOVA with Tukey’s multiple comparison test; **p < 0.01, ***p < 0.001, N = 3; mean ± SD) (C) Frequency of KRAS4AG12V EC location in vessel junctions (left) and branches (right) in 10% KRAS4AG12V microvasculatures shows preference for junction localization. (Paired Student’s t-test; ***p < 0.001, N = 3; mean ± SD). Characterization of the vascular networks by (D) quantification of number of junctions, (E) number of branches, (F) average branch length, and (G) total branch length of 0% (blue), 10% (yellow), and 25% KRAS4AG12V (red) microvasculatures. (One-way ANOVA with Tukey’s multiple comparison test; *p < 0.05, **p < 0.01, N = 3; mean ± SD).
We next visualized where the mutant KRAS cells localize in the network. In 10% KRAS vasculatures, KRAS4AG12V ECs preferentially localized in vessel junctions at a significantly higher rate than expected if this was a random process (Figs. 1C, 2 A and 2C), and were depleted in branches. Seventy-eight ± 9.0% of KRAS4AG12V ECs localized at junctions, compared to 22.0 ± 9.0% localizing at vessel branch segments (Fig. 2C). Despite the increased frequency of mutant KRAS ECs at the junctions, the number of junctions in each vasculature was not significantly different (Fig. 2D). Conversely, there was a significant increase in the number of branches between 0% KRAS (67.5 ± 9.7) and 25% KRAS (90.0 ± 24.2) vasculatures (Fig. 2E), indicating that there were more branches extending from a single junction.
The average branch length in the overall network decreased from 190.1 ± 70.0 μm in 0% KRAS to 160.7 ± 58.3 μm in 10% KRAS and 101.8 ± 20.1 μm in 25% KRAS vasculatures (Fig. 2F). As a result of the inverse relationship between average branch length and number, the total branch length, being the length of all the vessel segments end-to-end, was not significantly different. The total branch length was 9.4 ± 2.3, 7.2 ± 2.0, and 7.8 ± 1.1 mm in the 0%, 10%, and 25% KRAS microvessels, respectively (Fig. 2G). Taken together, we found that inclusion of KRAS4AG12V ECs in microvessel formation affected the overall vessel networks by increasing vessel width, shortening branch length, and increasing vessel density.
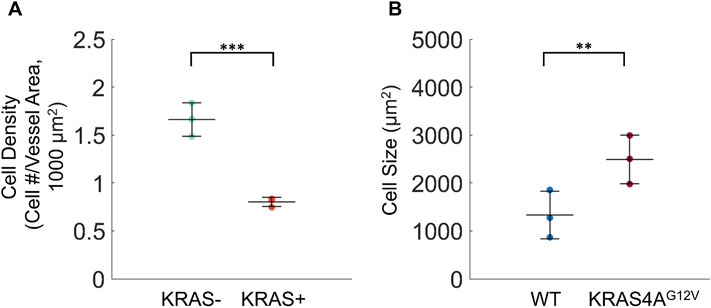
To assess if the increase in vessel width in KRAS+ microvessels was a result of an increased number of ECs in each vessel, we quantified the number of ECs in vessel segments containing only GFP or mScar cells. We determined that the average EC density in vessels made from mScar KRAS4AG12V ECs was two times lower than that of WT ECs (1.7 ± 0.5 and 0.8 ± 0.2 cells/1000 μm2, respectively) (Fig. 3A), indicating that the resulting enlargement of microvessels in 10% and 25% vasculatures is due to an increase in the overall size of KRAS4AG12V ECs and not more cells. To support this finding, we conducted 2D studies to measure size of WT ECs and KRAS4AG12V ECs. As expected, we found that KRAS4AG12V ECs were significantly larger in size compared to WT ECs (Fig. 3B; 2489.6 ± 506.8 μm2 and 1331.0 ± 496.6 μm2, respectively). We also assessed proliferation of ECs in monolayer and in the microfluidic device using Ki67 staining and confirmed there were no differences between 0% and 25% KRAS4AG12V conditions (Supplemental Figs. 4, 5, 6). From these findings we can conclude that overall vessel enlargement in the KRAS4AG12V vessels is not the result of an excessive number of ECs, but rather hypertrophy of KRAS4AG12V ECs.
Fig. 3. Vessel enlargement in KRAS4AG12V microvasculatures is due to an increase in cell size.
(A) Number of endothelial cells in WT ECs (KRAS−, green) or KRAS4AG12V ECs (KRAS+, red) vessel segments normalized by vessel area. (B) Quantification of endothelial cell size in monolayers of im-HUVECs (WT) and KRAS4AG12V ECs (Unpaired Student’s t-test; p** < 0.01, ***p < 0.001, N = 3; mean ± SD). Supplemental Fig. 8 shows enlarged cell phenotype.
3.3. Human AVM-like vessels display increased permeability
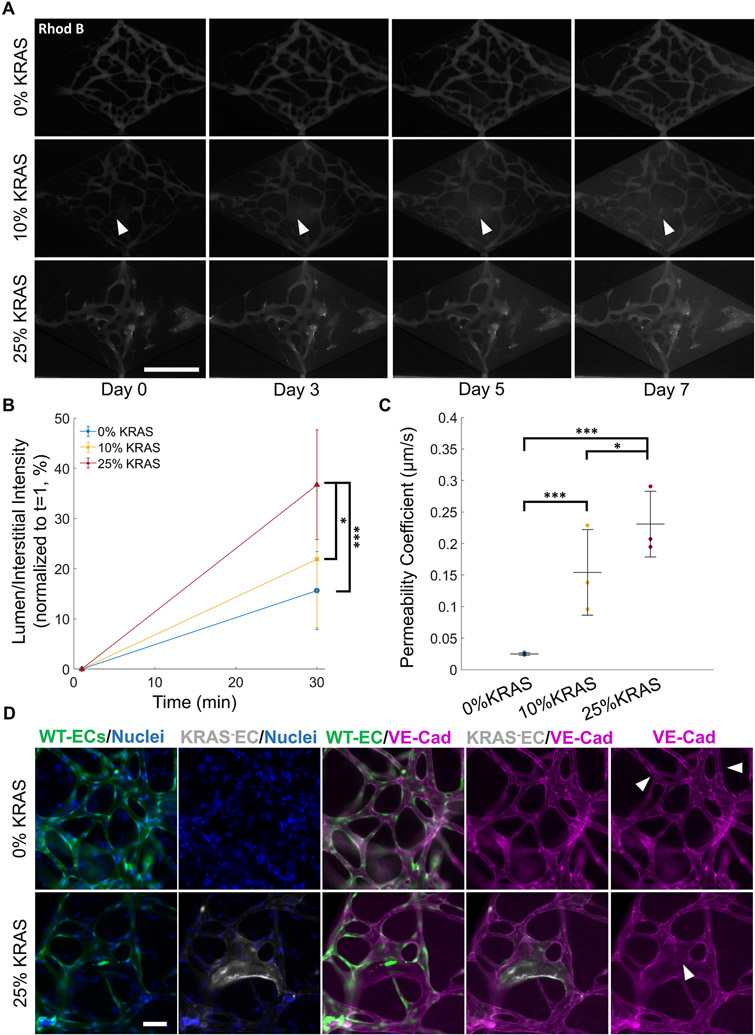
To test whether KRAS+ microvessels exhibited weakened barrier integrity, we assessed the extravasation of 70 kDa dextran conjugated to rhodamine b isothiocyanate (rhod B). On day 7, dextran-rhod B was perfused throughout the vasculature via addition to the inlet (denoted as time = 1 min). Images collected every 10 min for up to 30 min after initial dextran perfusion confirmed that the control microvessel network was intact and lumenized, showing physiological barrier properties. This also demonstrated that vessels were well connected to the inlets and outlets of the tissue chamber. No focal leakage was observed (Fig. 4A, top panel, Supplemental Fig. 7). In addition, we observed an increase in interstitial fluorescence intensity over time in both 10% and 25% KRAS vasculatures, demonstrating increased permeability (Fig. 4A, middle and bottom panel). In 10% KRAS microvasculatures, rhod B dextran was observed leaking from a singular point, originating from the vessel junction in 15% of the devices (Fig. 4A, middle panel) while the remaining 10% KRAS vasculatures showed no apparent source of focal leakage. Instead, an overall increase in interstitial fluorescence intensity was observed. In the 25% KRAS vasculatures, the overall vasculature was more permeable to rhod B dextran and there was progressive extravasation throughout the chamber due to the widespread distribution of KRAS4AG12V ECs throughout the vascular network (Fig. 4A, bottom panel). No focal leakage points were observed. Quantification of the fluorescence intensity in the vessel lumen and extravascular space normalized to the interstitial lumen intensity at 1 min demonstrated a significant increase in extravascular Dextran-rhod B in 25% KRAS microvessels compared to 0% KRAS microvessels (36.7 ± 10.9% and 15.6 ± 7.7% respectively, Fig. 4B). The permeability coefficient was also calculated and indicated that there was a significant increase in vessel permeability in both 10% and 25% KRAS vasculatures (14.3 ± 8.1 and 22.7 ± 7.6 × 10−2 μm/s) compared to 0% KRAS vasculatures (2.46 ± 1.1 × 10−2 μm/s) (Fig. 4C). The observed increased vessel permeability in 10% and 25% KRAS vasculatures is consistent with the poor vascular integrity in human AVMs.
Fig. 4. AVM-like microvessels display increased permeability.
(A) Representative time course images of microvessels perfused with 70 kDa dextran conjugated to rhodamine-B (rhod B). The 1 min timepoint denotes the moment vessels are initially fully perfused with dextran. White arrows in the middle panel show focal leakage originating from the vessel (scale bar 500 μm). (B) Quantification of dextran-rhod B intensity normalized to t = 1 of 0% (blue), 10% (yellow), and 25% (red) KRAS4AG12V microvasculatures. Fluorescence intensity was measured as a ratio of interstitial to lumen intensity, then normalized to t = 1.25% KRAS4AG12V microvasculatures showed a significant increase in interstitial intensity after 30 min. (C) Permeability coefficient of 0% (blue) 10% (yellow) and 25% (red) KRAS4AG12V microvessel conditions. There was a significant increase in permeability as the percentage of KRAS4AG12V increased. (One-way ANOVA with Tukey’s multiple comparison test; N = 3, *p < 0.05, ***p < 0.001; mean ± SD) (D) Immunofluorescence images of 0% and 25% KRAS4AG12V EC microvasculatures on day 7. Cell-cell VE-cadherin (purple) is absent in areas with KRAS4AG12V EC (white arrow, bottom panel) but remains intact throughout the rest of the vessel network with normal ECs (green; white arrow, top panel), (scale bar 100 μm).
To investigate a potential cause of barrier dysfunction, we stained for the cell-cell adhesion molecule VE-cadherin, previously implicated in KRAS4AG12V EC permeability defects in monolayers [7,9]. Indeed, immunofluorescence staining showed a reduction in VE-cadherin staining at the cell junctions of KRAS4AG12V ECs (Fig. 4D, bottom panel arrow). The junctions of surrounding WT ECs remained intact and unaffected. We also assessed formation of VE-cadherin junctions in cells seeded in monolayers, where we observed a significant reduction in VE-cadherin intensity and patchy localization in cell-cell junction in KRAS4AG12V ECs compared to WT ECs (Supplemental Fig. 8). This confirms that VE-cadherin expression is disrupted at the adherens junctions of KRAS4AG12V ECs and provides a potential mechanism by which vascular permeability is increased in KRAS4AG12V networks, thus recapitulating the compromised barrier function associated with AVMs.
3.4. Improvement of vessel barrier function by treatment with a MEK inhibitor
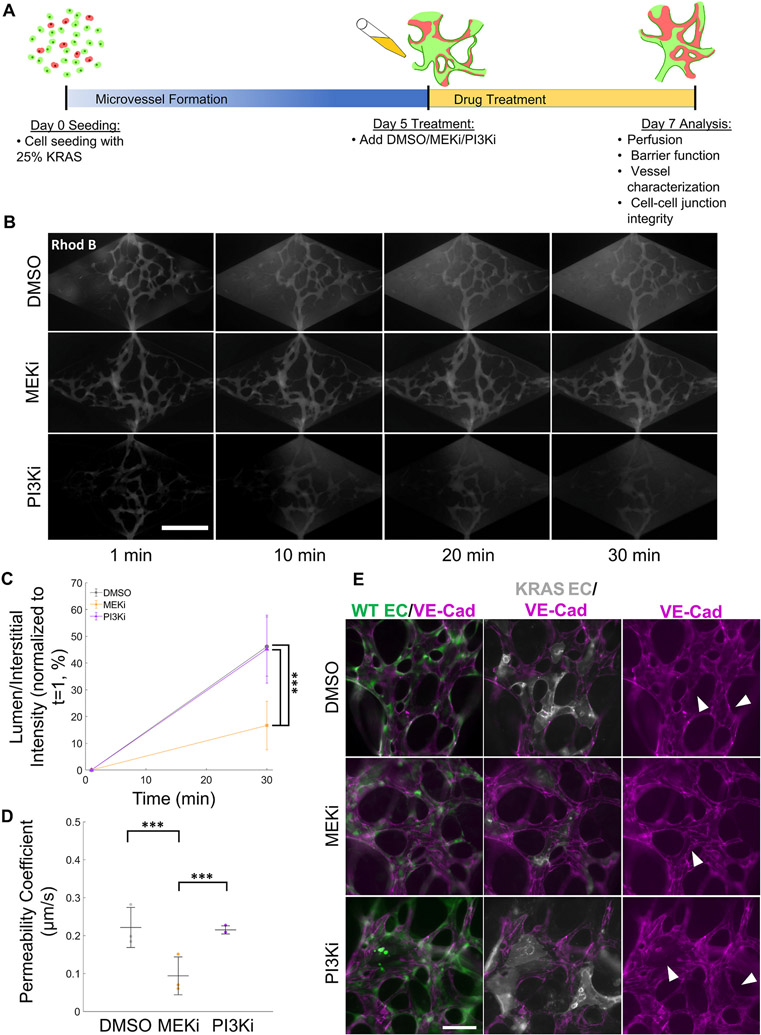
An important goal of developing an in vitro AVM model is to assess the effects of potential drugs in both structural and functional hallmarks of AVMs in a human-relevant model. Towards this goal, we treated 25% KRAS vasculatures with a MEK inhibitor (MEKi, SL327 7,9) and assessed vessel permeability. DMSO (vehicle) and a PI3K inhibitor (PI3Ki, LY294002) acted as negative controls. PI3K is a downstream effector of KRAS in other cell types, but is not activated in KRAS4AG12V ECs (Supplemental Fig. 3) [7]. To determine if MEKi would reverse established AVM defects, drug treatments started on day 5 (Fig. 5A) when vessel networks were already formed. Vessel permeability to 70 kDa MW dextran-rhod B was assessed on day 7 post-seeding.
Fig. 5. MEK inhibition restores barrier integrity of AVM-on-a-chip.
(A) Schematic of drug treatment protocol. (B) Time course images of microvessel perfusion with 70 kDa dextran-rhod B (scale bar 500 μm). (C) Quantification of microvessel 70 kDa dextran-rhod B intensity normalized to t = 1 of 25% KRAS4AG12V microvasculatures treated with different drugs starting at day 5. Fluorescent intensity was measured as a ratio of interstitial to lumen intensity normalized to t = 1. MEKi-treated AVM-like microvasculatures displayed significantly lower dextran extravasation than DMSO or PI3Ki. (One-way ANOVA with Tukey’s multiple comparison test; N = 3, ***p < 0.001; mean ± SD) (D) Permeability coefficient of 25% KRAS4AG12V microvasculatures treated with DMSO (gray), MEKi (orange) or PI3Ki (purple). The permeability of MEKi treatment microvasculatures was significantly lower than that of DMSO or PI3Ki treated AVM-like vessels. (One-way ANOVA with Tukey’s multiple comparison test; N = 3, ***p < 0.001; mean ± SD) (E) Immunofluorescence images at day 7 of day 5 treated AVM-on-a-chip. MEKi-treated AVM-like microvasculatures have an increase in VE-cadherin-rich (purple) adherens junction staining in KRAS4AG12V ECs (white, arrows indicate areas of increased VE-cadherin) compared to DMSO or PI3Ki treatment (arrows indicate areas of VE-cadherin disconnect) (scale bar, 100 μm).
Image analysis revealed that treatment with the MEKi led to a robust reduction in interstitial fluorescence intensity in the 25% KRAS vasculatures (Fig. 5B, middle panel) compared to DMSO- or PI3Ki-treated vasculatures (Fig. 5B, top and bottom panels, respectively). In the MEKi-treated microvessels, there was no apparent leakage. Quantification of the normalized fluorescent intensity showed a significantly lower permeability in MEKi-treated vasculature (Figs. 5C and 16.7 ± 9.1%) compared to the DMSO and PI3Ki treated vasculatures (Figs. 5C and 46.1 ± 11.1% and 45.2 ± 12.7%, respectively). As expected, assessment of the permeability coefficient corroborated findings from the analysis of fluorescent intensity, with the MEKi-treated networks displaying significantly lower permeability (9.0 ± 7.0 × 10−2 μm/s) than DMSO- or PI3Ki-treated networks (23.1 ± 9.5 × 10−2 and 21.6 ± 6.4 × 10−2 μm/s, respectively) (Fig. 5D). Immunofluorescent staining of MEKi-treated vasculatures showed partial restoration of cell-cell junction localization of VE-cadherin in KRAS4AG12V ECs (Fig. 5E). Additionally, assessment of VE-cadherin expression and localization in monolayers of KRAS4AG12V ECs confirmed that MEKi led to increased VE-cadherin expression and improved localization at cell-cell junctions (Supplemental Fig. 9). This suggests a role for cell-cell VE-cadherin localization in the reduction of vessel permeability. In the DMSO and PI3Ki treated microvessels, KRAS4AG12V ECs still lacked VE-cadherin, while WT ECs cell junctions remained intact (Fig. 5E).
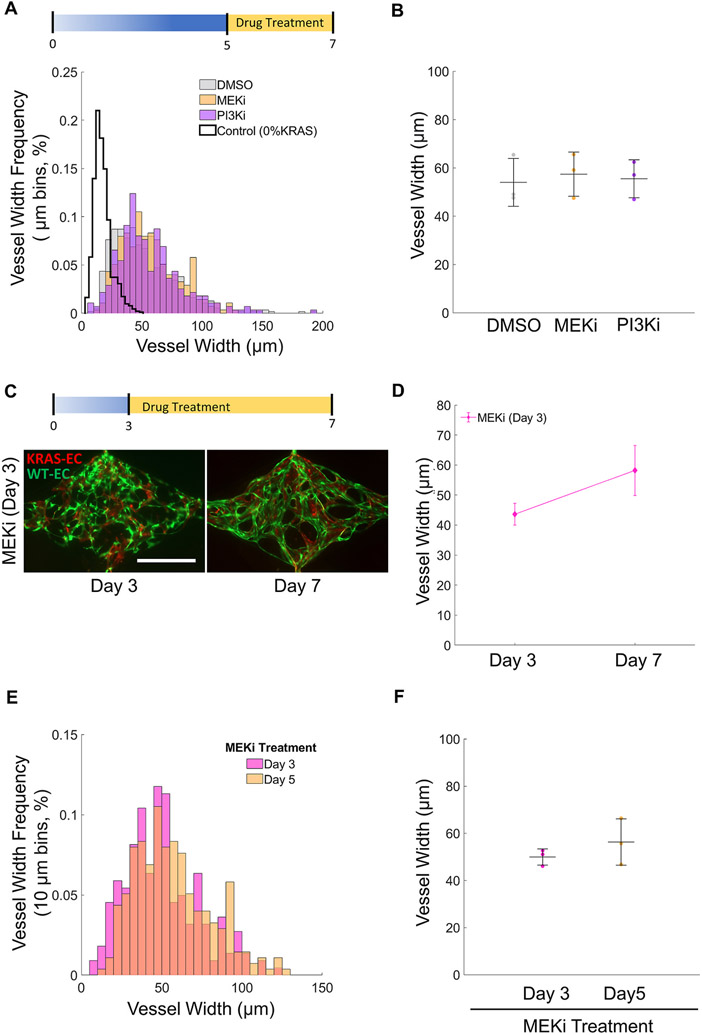
Next, we questioned if the restoration of barrier function in AVM-like vasculatures by MEKi treatment was accompanied by a concomitant decrease in vessel width. We found that vessel width was highly heterogeneous and there were no changes in width in response to drug treatment (Fig. 6A). The average vessel width was 53.9 ± 10.0, 57.4 ± 9.2, 55.5 ± 7.9 μm for the DMSO, MEKi, and PI3Ki treatments, respectively (Fig. 6B), which indicates that treatment with MEKi for 2 days is not effective in reversing vessel enlargement.
Fig. 6. MEK inhibition does not affect the width of AVM-on-a-chip microvessels.
(A) Day 7 distribution of 25% KRAS4AG12V microvessel widths treated with different drugs starting at day 5. (bin size: 10 μm). (B) Average vessel width at day 7 of 25% KRAS4AG12V microvessel treated with DMSO, MEKi, or PI3Ki starting at day 5 show no significant differences. (One-way ANOVA; N = 3; mean ± SD). (C) Day 7 images of KRAS4AG12V microvessels treated with MEKi starting at day 3 (scale bar 500 μm). (D) Average vessel width of 25% KRAS4AG12V micro vasculatures before (day 3) and after (day 7) treatment with MEKi starting day 3. (Paired Student’s t-test; N = 3; mean ± SD). (E) Day 7 vessel width distribution of 25% KRAS4AG12V microvasculatures treated with MEKi starting on day 3 or day 5. (bin size: 10 μm). (F) Average day 7 vessel width of 25% KRAS4AG12V microvasculatures treated with MEKi starting on day 3 or day 5 showing no differences (Unpaired Student’s t-test; N = 3; mean ± SD).
We then tested if treatment with MEKi at an earlier time point, when the vasculature was not yet fully formed, would prevent KRAS4AG12V EC-induced vessel enlargement. Therefore, we started drug treatment at day 3 post-seeding and assessed vessel width at day 7. Interestingly, treatment with the MEKi starting at day 3 did not prevent vessel enlargement (Fig. 6C and D) indicating that EC assembly into microvessels was not affected by MEKi treatment (Fig. 6D). In addition, a comparison of vessel widths between 25% KRAS vasculatures treated with MEKi beginning on day 3 or 5 showed that treatment with MEKi did not affect vessel width distribution (Fig. 6E) and that the average vessel width at the endpoint (day 7) was the same irrespective of when the treatment with MEKi started (Fig. 6F). Studies conducted on 2D monolayer with MEKi treated KRAS4AG12V ECs confirmed that cell size remained unchanged (Supplemental Fig. 10B). In addition, there were no changes in cell proliferation in drug-treated vasculatures (Supplemental Fig. 10C).
4. Discussion
The events that take place during sporadic AVM initiation and progression remain poorly understood. Accordingly, this lack of knowledge has stymied the development of pharmacological treatment options. In this study, we combined WT ECs with ECs harbouring a clinically-relevant KRAS4AG12V mutation and generated the first in vitro microfluidic model of AVMs. This heterogeneous mixture of mutant and normal ECs was chosen to reflect the heterogeneity found in clinical samples of AVMs [7]. Interestingly, seeding 100% KRAS4AG12V HUVECs failed to generate a perfused vasculature (data not shown). This platform focuses on the specific KRAS4AG12V contributions to the early steps in vessel formation. The resulting microvessel network recapitulates the key hallmarks of AVM: vessel dilation, tortuosity and increased permeability. Previous studies of KRAS4AG12V EC cultures reported changes in EC phenotypes, including increased cell size and irregular cell shape [7]. Moreover, we demonstrated that KRAS4AG12V-driven vessel enlargement was cell-autonomous, as mutant cell-containing vessels did not affect surrounding wildtype vessels in the same network. Thus, KRAS4AG12V mutant ECs are necessary and sufficient for eliciting the disease phenotype. The resulting enlarged vessels also increased vessel area coverage, with significantly shorter vessel branches and an increased ratio of branches to junctions. Additionally, KRAS4AG12V ECs localized preferentially to vessel junctions.
Previously, KRAS4AG12V ECs have also been noted in monolayers for their abnormally large shape [7]. Here we show that the same KRAS4AG12V-dependent alteration in EC morphology is responsible for this vessel dilation as assessment of the endothelial cell density showed that KRAS4AG12V EC-containing microvessels had in fact a significantly lower number of cells compared to microvessels containing WT ECs only. Thus, our humanized AVM model demonstrates that mutant KRAS enlarged vessel lumens are a consequence of increased EC size, rather than enhanced proliferation. Further investigation is needed to determine a potential role for shear stress in abnormal cell size in KRAS mutant cells, which could lead to abnormal cellular mechanosensory responses or create turbulent shear to disrupt vascular homeostasis [41].
One limitation to note is that when measuring the branch length in the 10% and 25% KRAS4AG12V networks, all vessels in the network were assessed, including WT-EC vessels, which is likely to have brought the average total width down. In addition, while the measured length only captures features in the XY direction, vessels were likely not parallel to the XY plane. This would affect the vessel width and length measurements. However, given the relatively short height of the chamber (100 μm) in relation to the average vessel width (20–60 μm), we assumed that the contribution of the Z component length would be very small and negligible.
Another key feature of the AVM-on-a-chip model is that it recapitulated increased permeability at the site of KRAS4AG12V positive vessel segments. Similar to vessel enlargement, the KRAS4AG12V ECs do not disrupt cell-cell junctions of surrounding WT ECs. This is in line with previously reported transwell assays that showed increased permeability due to a disruption in VE-cadherin rich adherens junctions [9]. Here, we showed that increased permeability is also a feature of AVM-like microvessels generated from KRAS4AG12V ECs despite the presence of luminal flow, which is a known cue for vessel barrier tightening [42,43]. Our findings are also consistent with barrier function results in a KRAS4AG12V zebrafish AVM model [9].
Interestingly, Nikolev et al. showed that the prevalence of cells with the KRAS mutation was highly variable, with some patients showing very high fractional abundance (nearing 100% of cells with a mutation) while other displayed very low abundance (as low as 4%) [7]. Thus, pursuing studies of the cell autonomous vs non-autonomous nature of the mutation in reductionist approaches like the current AVM-on-a-chip are important to understand brain AVM etiology. In addition, a small shunt may take several years to progress into the characteristic, tangled nidus with dilated draining veins characteristic of mature, symptomatic brain AVM. It is thus likely that the AVM-like vasculatures formed here capture only nascent AVMs that have yet to remodel into the complex nidus that is a mosaic of wild-type and KRAS mutant ECs. A limitation of this platform is that we could not grow the vessels for longer that 10 days as the microvessels would regress and deteriorate. Additionally, the fibrin gel could not be removed without destroying the vasculature, so all assessments were done in situ. An alternative explanation could be that KRAS mutations are necessary but not sufficient to induce complex bAVM, and that a “second hit” (e.g. from an environmental factor) is necessary to induce more severe presentations of the disease that are typically found in patients and occurs in other cranial AVMs and in CCM [44,45]. In the future, this platform can be used to investigate potential effects of increased flow rates/shear stress in AVM-like vessels. It is well known that disturbed flow, typically present in vessel junctions and bifurcations, results in EC dysfunction in vascular disease [46]. Considering the enlarged junctions, where KRAS4AG12V HUVECs preferentially reside, and irregularly shaped branches, these areas would be susceptible to creating disturbed flow and potentially a maladaptive mechanosensory response.
Studies have suggested that AVMs are a result of an inappropriate angiogenic response characterized by defective vessel maturation [10,14,47,48]. Indeed, brain AVMs have been previously shown to harbor increased expression of angiogenic genes and downstream VEGF signaling [49-53]. KRAS4AG12V ECs were also found to have elevated levels of VEGF and a transcriptional signature that resembles VEGF-stimulated cells [7]. VEGF (originally termed vascular permeability factor, VPF) increases vessel permeability [46], as well as angiogenesis [47]. This may explain the disruption of VE-cadherin in cell junctions described here that is typically associated with early angiogenic stages [54-56]. In addition, although no overt signs of angiogenesis were observed in the AVM-like vessels (e.g. sprouting), the increase in branch number is suggestive of increased vessel formation in KRAS4AG12V vasculatures.
MEKi has been investigated as a possible AVM treatment strategy [7,9,25,57,58]. In monocultures of KRAS4AG12V ECs, MEKi redirected VE-cadherin to cell junctions and restored normal F-actin organization and cell shape by suppressing ERK phosphorylation [7]. Transwell permeability assays using KRAS4AG12V HUVECs showed reduced leakage after 18 h of MEKi treatment. In zebrafish models, MEKi reversed vessel shunting and prevented hemorrhage [9]. Mouse models of brain AVMs resulting from delivery of brain trophic AAV-KRASG12V revealed that treatment with MEKi before AVM development led to significantly less brain AVMs and improved cognitive function [25]. We demonstrate here that treatment with MEKi improved VE-cadherin localization in KRAS4AG12V HUVECs, which resulted in improved vascular wall integrity. However, treatment with MEKi did not reverse (i.e., treatment at day 5 post-seeding when microvessels are mostly formed and there is perfusion) or prevent (i.e., treatment at day 3 post-seeding prior to the formation of an interconnected vessel network) vessel enlargement. With this platform, for the first time, we were able to uncouple vessel enlargement from vascular permeability and showed that MEKi can improve permeability without affecting vessel size, suggesting an alternative mechanism.
The inability of a MEKi to reduce vessel size in AVM-on-a-chip vessels suggests that other cell types or signaling pathways may be affected by KRAS activation in ECs [44]. KRAS is one of the most frequently mutated oncogenes in cancer and plays a central role in several growth factor receptor tyrosine kinase signaling pathways [7,8,59,60]. As a result, targeting the MAPK/ERK and KRAS protein activity for cancer treatment is of high interest. Two recent studies reported the off-label use of Trametinib, an FDA-approved MEKi, improved AVM severity, including an overall reduction in patient AVM size [61,62]. Using this platform, we can, in the future, systematically assess and isolate external factors such as perivascular cells, inflammation, increased fluid flow, and tissue-specific interactions to determine how these parameters influence AVM initiation and progression. We speculate that reduced pericyte coverage and paracrine signaling may also play a role in decreased AVM vessel stability [63]. In 3D collagen co-cultures of KRAS4AG12V HUVECs and pericytes, the KRAS4AG12V HUVECs formed abnormally wide tubes with a lack of pericyte recruitment compared to normal HUVECs. Additionally, there was a decrease of basement membrane protein deposition, suggesting a lack of pericyte-induced maturation [64]. In the future, pericytes can be included in the microfluidic devices to better mimic vessel composition [32,33,65]. In doing so, we can gain new insight into KRAS4AG12V EC-pericyte crosstalk and whether the lack of pericyte association exacerbates AVM phenotypes [16].
In conclusion, we have developed and validated an AVM-on-a-chip platform using a mosaic composition of WT and KRAS4AG12V ECs that is sufficient to drive AVM defects, namely vessel distension and hyperpermeability. These findings confirm previous results in 2D monolayer culture and animal models and provide new insight into the 3D mechanism of the KRAS4AG12V mutation [7,9]. Additionally, this platform represents an ideal method to create other vascular disease models in vitro using mutant EC cell lines. Several vascular malformations (venous, capillary, lymphatic, cavernous, telangiectasis, and arteriovenous) have been associated with genetic mutations [1,66]. A hereditary form of AVMs, HHT has been linked to mutations in BMP9/GDF2, SMAD4, ALK1, and ENG [3,7,41,42,67,68]. Additionally, there have been other mutations identified in the MAPK pathway (HRAS, BRAF, and MAP2K1) associated with AVM [6,69]. Cerebral cavernous malformations (CCMs) are caused by several different mutations in KRIT1, CCM1, 2, 3, or the gene PDCD10 [45]. It is currently unclear how mutations in these disparate genes alter EC phenotype to drive vascular malformations. This platform would be of interest to gain a better understanding of the disease phenotypes resulting from these EC mutations. In this manner, gene-specific differences in AVM formation and other vascular malformations may be revealed. Importantly, the platform described here can be used to test pharmacologic treatments for ameliorating these vascular malformations.
Supplementary Material
Acknowledgements
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) (CIHR PJT153160) and a Discovery grant from the Natural Sciences and Engineering Research Council (NSERC RGPIN 06621–2017) to S.S.N.; grants from CIHR (CIHR PJT155922), the National Institutes of Health (1R01HL159159-01A1), and the U.S. Department of Defense (W81XWH-18-1-0351) to J.D.W. and J.E.F.. S.S.N. holds the John Kitson McIvor Endowed Chair in Diabetes Research. J.E.F. is supported by a Tier 2 Canada Research Chair from CIHR. K.S. was partially supported from the Mount Sinai Hospital Graduate Scholarship in Science and Technology. M.L. was partially supported from the NSERC CREATE Training program in organ-on-a-chip engineering and entrepreneurship (TOeP) and Wildcat Graduate Scholarship. R.W. and N.K. were supported by a Canada Graduate Scholarships from CIHR. We thank Dr. R. Kamm for providing the IM-HUVECs (GFP WT ECs), Dr. A. Hayer for the hTERT-HUVECs, and O. Mourad for the support with statistical analysis.
Footnotes
Credit author statement
K.S. conducted majority of the experiments and wrote the manuscript. M.L. contributed to image analysis and experimental repeats. R.W. and N.K. developed and characterized the KRASG12V inducible IM-HUVEC line and designed the MEKi experiment. W.D.T. created and validated the inducible KRAS constructs, while J.D.W. supervised W.D.T. and designed the KRASG12V inducible constructs and also revised the manuscript. J.E.F. provided supervision to R.W. and N·K., designed experiments and revised the manuscript. S.S.N. provided supervision and guidance for this project, supervision to K.S. and M.L., manuscript writing and revisions.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biomaterials.2022.121729.
Data availability
Data will be made available on request.
References
- [1].Lawton MT, Rutledge WC, Kim H, et al. , Brain arteriovenous malformations, Nat. Rev. Dis. Prim 1 (1) (2015) 1–20, 10.1038/nrdp.2015.8. [DOI] [PubMed] [Google Scholar]
- [2].Solomon RA, Connolly ES, Arteriovenous malformations of the brain, N. Engl. J. Med 376 (19) (2017) 1859–1866, 10.1056/NEJMra1607407. [DOI] [PubMed] [Google Scholar]
- [3].Lee JM, Zhai G, Liu Q, et al. , Vascular permeability precedes spontaneous intracerebral hemorrhage in stroke-prone spontaneously hypertensive rats, Stroke 1 (2007), 10.1161/STROKEAHA.107.491621. Published online December. [DOI] [PubMed] [Google Scholar]
- [4].Rutledge WC, Ko NU, Lawton MT, Kim H, Hemorrhage rates and risk factors in the natural history course of brain arteriovenous malformations, Transl Stroke Res 5 (5) (2014) 538–542, 10.1007/s12975-014-0351-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Al-Shahi R, Warlow C, A systematic review of the frequency and prognosis of arteriovenous malformations of the brain in adults, Brain J Neurol 124 (Pt 10) (2001) 1900–1926, 10.1093/brain/124.10.1900. [DOI] [PubMed] [Google Scholar]
- [6].Al-Olabi L, Polubothu S, Dowsett K, et al. , Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy, J. Clin. Invest 128 (4) (2018) 1496–1508, 10.1172/JCI98589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nikolaev SI, Vetiska S, Bonilla X, et al. , Somatic activating KRAS mutations in arteriovenous malformations of the brain, N. Engl. J. Med 378 (3) (2018) 250–261, 10.1056/NEJMoa1709449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cheng F, Nussinov R, KRAS activating signaling triggers arteriovenous malformations, Trends Biochem. Sci 43 (7) (2018) 481–483, 10.1016/j.tibs.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fish JE, Flores-Suarez CP, Boudreau E, et al. , Somatic gain of KRAS function in the endothelium is sufficient to cause vascular malformations that require MEK but not PI3K signaling, Circ. Res 119 (2020), 316500, 10.1161/CIRCRESAHA.119.316500. Published online June 17 CIRCRESAHA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cheng P, Ma L, Shaligram S, et al. , Effect of elevation of vascular endothelial growth factor level on exacerbation of hemorrhage in mouse brain arteriovenous malformation, J Neurosurg. Published online April 26 (2019) 1–8, 10.3171/2019.1.JNS183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Walcott BP, Winkler EA, Rouleau GA, Lawton MT, Molecular, cellular, and genetic determinants of sporadic brain arteriovenous malformations, Neurosurgery 63 (Suppl 1) (2016) 37–42, 10.1227/NEU.0000000000001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fish JE, Wythe JD, The molecular regulation of arteriovenous specification and maintenance, Dev. Dyn. Off. Publ. Am. Assoc. Anat 244 (3) (2015) 391–409, 10.1002/dvdy.24252. [DOI] [PubMed] [Google Scholar]
- [13].Cunha SI, Magnusson PU, Dejana E, Lampugnani MG, Deregulated TGF-β/BMP signaling in vascular malformations, Circ. Res 121 (8) (2017) 981–999, 10.1161/CIRCRESAHA.117.309930.. [DOI] [PubMed] [Google Scholar]
- [14].Atri D, Larrivée B, Eichmann A, Simons M, Endothelial signaling and the molecular basis of arteriovenous malformation, Cell Mol. Life Sci. CMLS (2013), 10.1007/s00018-013-1475-1. Published online September 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wright R, Järvelin P, Pekonen H, Keränen S, Rauramaa T, Frösen J, Histopathology of brain AVMs part II: inflammation in arteriovenous malformation of the brain, Acta Neurochir 162 (7) (2020) 1741–1747, 10.1007/s00701-020-04328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Winkler EA, Birk H, Burkhardt JK, et al. , Reductions in brain pericytes are associated with arteriovenous malformation vascular instability, J. Neurosurg 129 (6) (2018) 1464–1474, 10.3171/2017.6.JNS17860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Murphy PA, Kim TN, Huang L, et al. , Constitutively active Notch4 receptor elicits brain arteriovenous malformations through enlargement of capillary-like vessels, Proc. Natl. Acad. Sci. USA 111 (50) (2014) 18007–18012, 10.1073/pnas.1415316111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Choi EJ, Chen W, Jun K, Arthur HM, Young WL, Su H, Novel brain arteriovenous malformation mouse models for type 1 hereditary hemorrhagic telangiectasia, PLoS One 9 (2) (2014), e88511, 10.1371/journal.pone.0088511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Milton I, Ouyang D, Allen CJ, et al. , Age-dependent lethality in novel transgenic mouse models of central nervous system Arteriovenous malformations, Stroke 43 (5) (2012) 1432–1435, 10.1161/STROKEAHA.111.647024. [DOI] [PubMed] [Google Scholar]
- [20].Tual-Chalot S, Oh P, Arthur H, Mouse models of hereditary haemorrhagic telangiectasia: recent advances and future challenges, Front. Genet 6 (2015) 25, 10.3389/fgene.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shaligram SS, Zhang R, Zhu W, et al. , Bone marrow-derived Alk1 mutant endothelial cells and clonally expanded somatic Alk1 mutant endothelial cells contribute to the development of brain arteriovenous malformations in mice, Trans. Stroke Res 21 (2021), 10.1007/s12975-021-00955-9. Published online October. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen W, Sun Z, Han Z, et al. , De novo cerebrovascular malformation in the adult mouse after endothelial Alk1 deletion and angiogenic stimulation, Stroke 45 (3) (2014) 900–902, 10.1161/STROKEAHA.113.003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nielsen CM, Huang L, Murphy PA, Lawton MT, Wang RA, Mouse models of cerebral arteriovenous malformation, Stroke 47 (1) (2016) 293–300, 10.1161/STROKEAHA.115.002869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Raj JA, Stoodley M, Experimental animal models of arteriovenous malformation: a review, Vet. Sci 2 (2) (2015) 97–110, 10.3390/vetsci2020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Park ES, Kim S, Huang S, et al. , Selective endothelial hyperactivation of oncogenic KRAS induces brain arteriovenous malformations in mice, Ann. Neurol 89 (5) (2021) 926–941, 10.1002/ana.26059. [DOI] [PubMed] [Google Scholar]
- [26].Sun Z, Kemp SS, Lin PK, Aguera KN, Davis GE, Endothelial k-RasV12 expression induces capillary deficiency attributable to marked tube network expansion coupled to reduced pericytes and basement membranes, Arterioscler Thromb Vasc. Biol 9 (2021), 10.1161/ATVBAHA.121.316798. ATVBAHA121316798 Published online December. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Andrejecsk JW, Hughes CCW, Engineering perfused microvascular networks into microphysiological systems platforms, Curr. Opin. Biomed. Eng 5 (2018) 74–81, 10.1016/j.cobme.2018.02.002. [DOI] [Google Scholar]
- [28].Kutys ML, Chen CS, Forces and mechanotransduction in 3D vascular biology, Curr. Opin. Cell Biol 42 (2016) 73–79, 10.1016/j.ceb.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hu C, Chen Y, Tan MJA, Ren K, Wu H, Microfluidic technologies for vasculature biomimicry, Analyst 144 (15) (2019) 4461–4471, 10.1039/C9AN00421A. [DOI] [PubMed] [Google Scholar]
- [30].Whisler JA, Chen MB, Kamm RD, Control of perfusable microvascular network morphology using a multi culture microfluidic system, Tissue Eng. C Methods 20 (7) (2014) 543–552, 10.1089/ten.TEC.2013.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang X, Phan DTT, Zhao D, George SC, Hughes CCW, Lee AP, An on-chip microfluidic pressure regulator that facilitates reproducible loading of cells and hydrogels into microphysiological system platforms, Lab Chip 16 (5) (2016) 868–876, 10.1039/C5LC01563D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kosyakova N, Kao DD, Figetakis M, et al. , Differential functional roles of fibroblasts and pericytes in the formation of tissue-engineered microvascular networks in vitro, Npj Regen. Med 5 (1) (2020) 1, 10.1038/s41536-019-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kim J, Chung M, Kim S, Jo DH, Kim JH, Jeon NL, Engineering of a biomimetic pericyte-covered 3D microvascular network, PLoS One 10 (7) (2015), e0133880, 10.1371/journal.pone.0133880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jeon JS, Bersini S, Whisler JA, et al. , Generation of 3D functional microvascular networks with human mesenchymal stem cells in microfluidic systems, Integr. Biol. Quant. Biosci. Nano Macro 6 (5) (2014) 555–563, 10.1039/c3ib40267c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang X, Phan DTT, Sobrino A, George SC, Hughes CCW, Lee AP, Engineering anastomosis between living capillary networks and endothelial cell-lined microfluidic channels, Lab Chip 16 (2) (2016) 282–290, 10.1039/C5LC01050K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wan Z, Zhang S, Zhong AX, et al. , A robust vasculogenic microfluidic model using human immortalized endothelial cells and Thy1 positive fibroblasts, Biomaterials 276 (2021), 121032, 10.1016/j.biomaterials.2021.121032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hayer A, Shao L, Chung M, et al. , Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells, Nat. Cell Biol 18 (12) (2016) 1311–1323, 10.1038/ncb3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kim SI, Oceguera-Yanez F, Sakurai C, Nakagawa M, Yamanaka S, Woltjen K, Inducible transgene expression in human iPS cells using versatile all-in-one piggyBac transposons, Methods Mol. Biol. Clifton NJ 1357 (2016) 111–131, 10.1007/7651_2015_251. [DOI] [PubMed] [Google Scholar]
- [39].Yusa K, Zhou L, Li MA, Bradley A, Craig NL, A hyperactive piggyBac transposase for mammalian applications, Proc. Natl. Acad. Sci. U. S. A 108 (4) (2011) 1531–1536, 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Phan DTT, Wang X, Craver BM, et al. , A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications, Lab Chip 17 (3) (2017) 511–520, 10.1039/C6LC01422D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tzima E, Irani-Tehrani M, Kiosses WB, et al. , A mechanosensory complex that mediates the endothelial cell response to fluid shear stress, Nature 437 (7057) (2005) 426–431, 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- [42].Seebach J, Dieterich P, Luo F, et al. , Endothelial barrier function under laminar fluid shear stress, Lab. Invest 80 (12) (2000) 1819–1831, 10.1038/labinvest.3780193. [DOI] [PubMed] [Google Scholar]
- [43].Pan S, Molecular mechanisms responsible for the atheroprotective effects of laminar shear stress, Antioxidants Redox Signal. 11 (7) (2009) 1669–1682, 10.1089/ars.2009.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lapinski PE, Doosti A, Salato V, North P, Burrows PE, King PD, Somatic second hit mutation of RASA1 in vascular endothelial cells in capillary malformation-arteriovenous malformation, Eur. J. Med. Genet 61 (1) (2018) 11–16, 10.1016/j.ejmg.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Snellings DA, Hong CC, Ren AA, et al. , Cerebral cavernous malformation: from mechanism to therapy, Circ. Res 129 (1) (2021) 195–215, 10.1161/CIRCRESAHA.121.318174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chiu JJ, Chien S, Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives, Physiol. Rev 91 (1) (2011), 10.1152/physrev.00047.2009, 10.1152/physrev.00047.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Storkebaum E, Quaegebeur A, Vikkula M, Carmeliet P, Cerebrovascular disorders: molecular insights and therapeutic opportunities, Nat. Neurosci 14 (11) (2011) 1390–1397, 10.1038/nn.2947. [DOI] [PubMed] [Google Scholar]
- [48].Ryu JY, Kim YH, Lee JS, et al. , Oscillatory shear stress promotes angiogenic effects in arteriovenous malformations endothelial cells, Mol. Med 27 (1) (2021) 31, 10.1186/s10020-021-00291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Murphy PA, Lu G, Shiah S, Bollen AW, Wang RA, Endothelial Notch signaling is upregulated in human brain arteriovenous malformations and a mouse model of the disease, Lab. Invest 89 (9) (2009) 971–982, 10.1038/labinvest.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Vernimmen FJAI, Vascular endothelial growth factor blockade: a potential new therapy in the management of cerebral arteriovenous malformations, J. Med. Hypotheses Ideas 8 (2) (2014) 57–61, 10.1016/j.jmhi.2013.10.001. [DOI] [Google Scholar]
- [51].Meadows KN, Bryant P, Pumiglia K, Vascular endothelial growth factor induction of the angiogenic phenotype requires Ras activation, J. Biol. Chem 276 (52) (2001) 49289–49298, 10.1074/jbc.M108069200. [DOI] [PubMed] [Google Scholar]
- [52].Fukumura D, Gohongi T, Kadambi A, et al. , Predominant role of endothelial nitric oxide synthase in vascular endothelial growth factor-induced angiogenesis and vascular permeability, Proc. Natl. Acad. Sci. USA 98 (5) (2001) 2604–2609, 10.1073/pnas.041359198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shibuya M, Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis, Genes Cancer 2 (12) (2011) 1097–1105, 10.1177/1947601911423031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bentley K, Franco CA, Philippides A, et al. , The role of differential VE-cadherin dynamics in cell rearrangement during angiogenesis, Nat. Cell Biol 16 (4) (2014) 309–321, 10.1038/ncb2926. [DOI] [PubMed] [Google Scholar]
- [55].Altalhi W, Sun X, Sivak JM, Husain M, Nunes SS, Diabetes impairs arteriovenous specification in engineered vascular tissues in a perivascular cell recruitment-dependent manner, Biomaterials 119 (2017) 23–32, 10.1016/j.biomaterials.2016.12.003. [DOI] [PubMed] [Google Scholar]
- [56].Nunes SS, Greer KA, Stiening CM, et al. , Implanted microvessels progress through distinct neovascularization phenotypes, Microvasc. Res 79 (1) (2010) 10–20, 10.1016/j.mvr.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zhao Y, Adjei AA, The clinical development of MEK inhibitors, Nat. Rev. Clin. Oncol 11 (7) (2014) 385–400, 10.1038/nrclinonc.2014.83. [DOI] [PubMed] [Google Scholar]
- [58].Pan P, Weinsheimer S, Cooke D, et al. , Review of treatment and therapeutic targets in brain arteriovenous malformation, J. Cereb Blood Flow Metab. Off J. Int. Soc. Cereb. Blood Flow Metab 41 (12) (2021) 3141–3156, 10.1177/0271678X211026771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cox AD, Der CJ, Ras history, Small GTPases 1 (1) (2010) 2–27, 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Priemer DS, Vortmeyer AO, Zhang S, Chang HY, Curless KL, Cheng L, Activating KRAS mutations in arteriovenous malformations of the brain: frequency and clinicopathologic correlation, Hum. Pathol 89 (2019) 33–39, 10.1016/j.humpath.2019.04.004. [DOI] [PubMed] [Google Scholar]
- [61].Lekwuttikarn R, Lim YH, Admani S, Choate KA, Teng JMC, Genotype-guided successful treatment of an arteriovenous malformation in a child, JAMA Dermatol. 155 (2) (2019) 256–257, 10.1001/jamadermatol.2018.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cooke DL, Frieden IJ, Shimano KA, Angiographic evidence of response to Trametinib therapy for a spinal cord arteriovenous malformation, J Vasc Anom. 2 (3) (2021), e018, 10.1097/JOVA.0000000000000018. [DOI] [Google Scholar]
- [63].Kofler NM, Cuervo H, Uh MK, Murtomäki A, Kitajewski J, Combined deficiency of Notch1 and Notch3 causes pericyte dysfunction, models CADASIL and results in arteriovenous malformations, Sci. Rep 5 (1) (2015), 16449, 10.1038/srep16449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sun Z, Kemp SS, Lin PK, Aguera KN, Davis GE, Endothelial k-RasV12 expression induces capillary deficiency attributable to marked tube network expansion coupled to reduced pericytes and basement membranes, Arterioscler Thromb Vasc Biol. Published online December 9 (2021), 10.1161/ATVBAHA.121.316798. ATVBAHA121316798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Campisi M, Shin Y, Osaki T, Hajal C, Chiono V, Kamm RD, 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes, Biomaterials 180 (2018) 117–129, 10.1016/j.biomaterials.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Konczyk DJ, Goss JA, Smits PJ, et al. , Arteriovenous malformation associated with a HRAS mutation, Hum. Genet 138 (11–12) (2019) 1419–1421, 10.1007/s00439-019-02072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Pawlikowska L, Nelson J, Guo DE, et al. , Association of common candidate variants with vascular malformations and intracranial hemorrhage in hereditary hemorrhagic telangiectasia, Mol. Genet. Genomic Med 6 (3) (2018) 350–356, 10.1002/mgg3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Davis RB, Pahl K, Datto NC, et al. , Notch signaling pathway is a potential therapeutic target for extracranial vascular malformations, Sci. Rep 8 (1) (2018), 17987, 10.1038/s41598-018-36628-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Couto JA, Huang AY, Konczyk DJ, et al. , Somatic MAP2K1 mutations are associated with extracranial arteriovenous malformation, Am. J. Hum. Genet 100 (3) (2017) 546–554, 10.1016/j.ajhg.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.