Abstract
PD-L1 (CD274) amplification occurs in a small subset of malignancies and may predict anti-PD-1/PD-L1 immunotherapy responsiveness. We hypothesized that both copy number (CN) and focality of cancer-related PD-L1 amplifications impact protein expression, and, thus, analyzed solid tumors that underwent comprehensive genomic profiling between March 2016 and February 2022 at Foundation Medicine. PD-L1 CN alterations were detected using a comparative genomic hybridization-like method. PD-L1 CN changes were correlated with PD-L1 protein expression (DAKO 22C3 antibody) by immunohistochemistry (IHC). Overall, 60,793 samples were analyzed (most frequent histologies: lung adenocarcinoma (20%), colon adenocarcinoma (12%), lung squamous carcinoma (8%)). Using a definition of CD274 CN ≥ specimen ploidy +4 (6 copies), 1.21% of tumors (738/60,793) were PD-L1 amplified. Focality category distribution was as follows: <0.1 mB (n=18 (2.4%)), ≥0.1 to <4 mB (n=230 (31.1%)), ≥4 to <20 mB (n=310 (42%)), ≥20mB (n=180 (24.4%)). Lower levels of PD-L1 amplification (below specimen ploidy +4) were more frequently non-focal amplifications compared to higher levels. In addition, more focal amplification (<0.1 mB) correlated with higher PD-L1 IHC expression. Median tumor proportion score (TPS) for samples with PD-L1 amplification (ploidy ≥+4) according to focality were 87.5% (<0.1 mB), 80% (≥0.1 to <4 mB), 40% (≥4 to <20 mB), 1% (≥20mB). In specimens with PD-L1 ploidy less than +4, but highly focal (<0.1 mB), the 75th percentile of PD-L1 expression by TPS was 80%. Conversely, non-focal (≥20 mB) PD-L1 amplification (ploidy ≥+4) can present high PD-L1 expression (TPS≥50%), albeit infrequently (0.09% of our cohort). In conclusion, PD-L1 expression measured by IHC is influenced by PD-L1 amplification level and focality. Further correlation between amplification, focality, protein expression and therapeutic outcome for PD-L1 and other targetable genes warrants exploration.
Keywords: Biomarkers, Tumor; Genetic Markers; Immunohistochemistry
Background
Programmed cell death protein pathway (PD-1/PD-L1) inhibitors are frequently employed for cancer treatment. A plethora of biomarkers of response and resistance to these agents are subject of intense research and new strategies are needed.1–3 PD-L1 protein overexpression can predict responses to PD-1/PD-L1 inhibitors, although several biological and technical challenges are still unsolved.4 PD-L1 (CD274) amplification can be present in approximately 1% of solid tumors, and may also predict high response rates to immune checkpoint inhibitors (ICIs), independent of other immune biomarkers.5–7 In fact, Hodgkin’s disease is exquisitely responsive to ICIs, and a hallmark of the disease is amplification of PD-L1.8
The potential effects of a cancer gene amplification on its RNA and protein expression are not completely understood. Several technical considerations as well as biological phenomena can influence the detection of gene amplification using next generation sequencing (NGS),9 as well as the subsequent transcription and translation of genes to RNA and protein. One possible mechanism is epigenetic modulation of genes, though other mechanisms may also be operative. For instance, 13% of putative driver somatic single nucleotide variants were unexpectedly not expressed as RNA in cancer specimens.10 Moreover, a recent paper demonstrated that 6% of cancer-related gene amplifications (≥6 copies) were silenced (ie, not expressed at RNA level compared with adjacent normal tissue).11 Counterintuitively, focal amplifications (defined as <0.1 megabases (mB) in that report) were more frequently silenced than non-focal amplifications (9% vs 5%). The relationship between copy number (CN) changes, focality and protein expression may vary on a gene-by-gene basis and depending on the definition of ‘focality’. Because protein expression may have therapeutic implications, in the current study, we analyzed PD-L1 protein (by immunohistochemistry (IHC)) vs CD274 (PD-L1) gene amplification CN and focality (thresholds of 0.1, 4 and 20 mB) using an NGS platform with 60,793 solid tumor specimens.
Methods
Comprehensive genomic profiling (CGP) of 60,793 solid tumor clinical cases was performed using the FoundationOne (F1) and FoundationOne CDx (F1CDx) assays (Foundation Medicine, Cambridge, Massachusetts, USA) as described previously, in a Clinical Laboratory Improvement Amendments (CLIA)-certified and College of American Pathologists (CAP) accredited laboratory, between March 2016 and February 2022.12 All de-identified research consented samples submitted for sequencing featured >20% tumor cells and yielded >50 ng of extracted DNA. CGP was performed on hybridization-captured, adapter ligation-based libraries (median exon coverage depth >800X), to identify genomic alterations (base substitutions, small insertions/deletions, CN alterations, and rearrangements) in >300 cancer-associated genes.
CD274 (PD-L1) CN alterations were detected using a comparative genomic hybridization-like method applied to NGS data.12 13 Each specimen was analyzed alongside a process-matched normal control (an internally validated mixture of 10 heterozygous diploid samples from the HapMap project), with custom algorithms to normalize the sequence coverage distribution across captured DNA regions. Log-ratios of normalized coverage data for exonic, intronic, and single nucleotide polymorphisms (SNPs) targets accounting for stromal admixture, as well as genome-wide SNP frequencies, were used to generate the CN profiles. Circular binary segmentation and proprietary algorithms further clustered groups of targets and SNP frequencies to define upper and lower bounds of genomic segments. Empirical Bayesian algorithms used a distribution of parameters including purity and base ploidy and probability matrices were derived using different statistical sampling methodologies to fit these data. Specimen-level ploidy was estimated as described by Sun et al.13 Computational models were reviewed by expert curators for each specimen.
CD274 (PD-L1) amplification was defined as ploidy +4 (CN 6), though other cut-off’s were explored such as ploidy +2 (CN 4).6 11 Amplifications were classified as ‘non-focal’ when the length of the amplified CN segment was ≥20 Mb,14 but other thresholds (0.1 and 4 Mb) were analyzed consistent with prior publications.9 11 PD-L1 IHC was run and interpreted by experienced board-certified pathologists according to the manufacturer’s instructions in a CLIA-certified and CAP-accredited laboratory (Foundation Medicine, Morrisville, North Carolina, USA) (DAKO PD-L1 IHC 22C3 pharmDx) (tumor proportion score (TPS) method).15 TPS scores cut-offs were defined as 1% and 50% as per usage in non-small cell lung cancer (NSCLC).16 17 Kruskal-Wallis χ2 test was used to test the association between the continuous PDL1 score and CD274 focality bins.
As self-reported race was not available, genomic ancestry was determined for each patient sample. For each profiling platform (F1 and F1CDx), >40,000 SNP sites sequenced by CGP were identified. To remove biases due to linkage disequilibrium (LD), LD pruning was performed using PLINK (using the –indep flag with a window size of 50, a step size of 5, and a variance inflation factor threshold of 2). A random forest classifier was trained on 1000 genomes to identify ancestral populations (European, African, Admixed American (Hispanic), East Asian, South Asian) using genetic variation at the SNP sites. Genetic variation was defined by 10 features that captured allele-count variation as determined by principal component analysis. This classifier was applied to CGP patient samples to assign them to one of the ancestral populations.18
Results
Data from 60,793 clinical samples were analyzed in a CLIA laboratory (online supplemental table 1). Samples were derived from primary (44%) and metastatic tissue (39%) (rest unknown). Median age was 66 years; 54% were women. Most samples (78%) were from patients of European genetic ancestry. The most frequent histologies included lung adenocarcinoma (20%), colon adenocarcinoma (12%), lung squamous carcinoma, (8%) and ovarian serous carcinoma (5%).
jitc-2022-006311supp001.pdf (75.1KB, pdf)
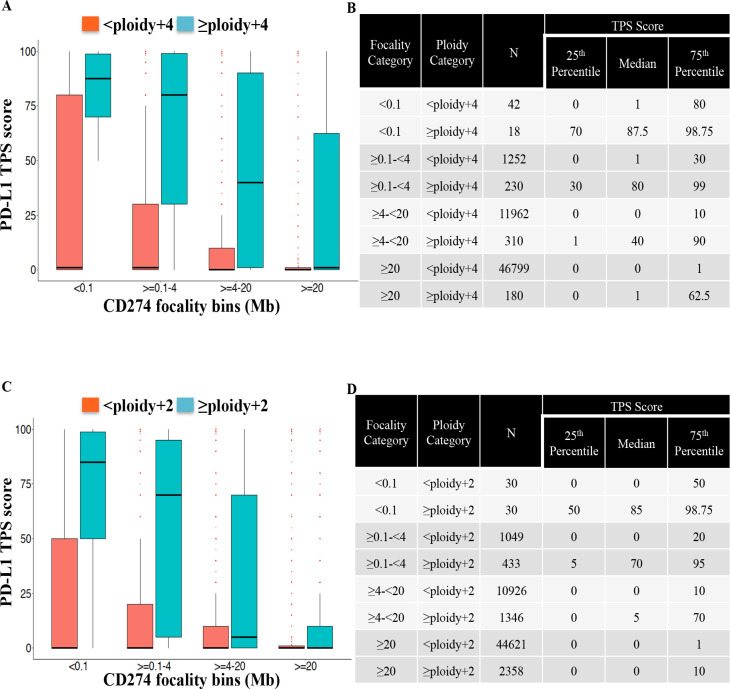
Prevalence of PD-L1 amplification varied according to ploidy category adopted. Using a definition of CD274 CN≥specimen ploidy +4 (equivalent to >6 CN), 738 of 60,793 (1.21%) were considered PD-L1 amplified (figure 1 and online supplemental table 2). These 738 samples were distributed according to focality category, from more to less focal: <0.1 mB (n=18 (2.4%)), ≥0.1 to <4 mB (n=230 (31.1%)), ≥4 to <20 mB (n=310 (42%)), ≥20 mB (n=180 (24.4%)). Using a less stringent threshold for determining CN positivity of CD274 CN≥specimen ploidy +2 (CN >4) not only increased the prevalence of PD-L1 amplification (n=4,167 (6.9% of all samples)), but also altered the distribution in focality categories: <0.1 mB (n=30 (0.7%)), ≥0.1 to <4 mB (n=430 (10.4%)), ≥4 to <20 mB (n=1346 (32.3%)), ≥20 mB (n=2,380 (56.6%)) (online supplemental table 3). From this analysis, lower levels of PD-L1 amplification (below specimen ploidy +4) are more frequently non-focal amplifications compared with higher levels.
Figure 1.
Levels of PD-L1 expression as measure by immunohistochemistry using TPS score and CD274 (PD-L1) amplification focality. Distribution of PD-L1 TPS score (numbers represent per cent) over CD274 focality bins at a CD274 ploidy gain threshold of +4 (CN 6) (A) and +2 (CN 4) (C). Numbers of patient samples according to focality and ploidy category and interquartile distribution of TPS score for ploidy gain threshold of +4 (CN 6) (B) and +2 (CN 4) (D). Association between the continuous PD-L1 score and CD274 focality bins (<0.1, ≥0.1–4, ≥4–20, ≥20) at ploidy+2 and ploidy+4 was significant at p value <2.2e−16 (Kruskal-Wallis χ2). CN, copy number; mB, megabases; TPS, tumor proportion score.
Using PD-L1 expression (DAKO 22C3 IHC assay; TPS), we demonstrated that more focal amplification (<0.1 mB) was associated with higher PD-L1 expression levels, which was observed using different ploidy categories (all p values<2.2e−16, figure 1). For samples with CD274 CN<6 (<specimen ploidy +4), but highly focal (<0.1 mB), the 75th percentile of PD-L1 expression by TPS was 80% (figure 1A and B), meaning a reasonable proportion of samples expressing PD-L1; however, median expression was 1%. Tumors with CD274 amplified (≥6 CN or ≥specimen ploidy +4) in a non-focal fashion (≥20 mB) may be currently considered as a variant of unknown significance. Even in this category, the 75th percentile of PD-L1 expression was 62.5%, suggesting a proportion of high PD-L1 expressors within this group; however, median expression level was 1%. Numerically, non-focal (≥20 Mb) CD274 amplified presenting high PD-L1 expression (TPS ≥50%) encompassed 52 samples (0.09% of our cohort; online supplemental table 2). In contrast, median PD-L1 expression level was 87.5%, 80%, and 40% for >specimen ploidy +4 with focality of <0.1, ≥0.1–4, and ≥4–20 mB.
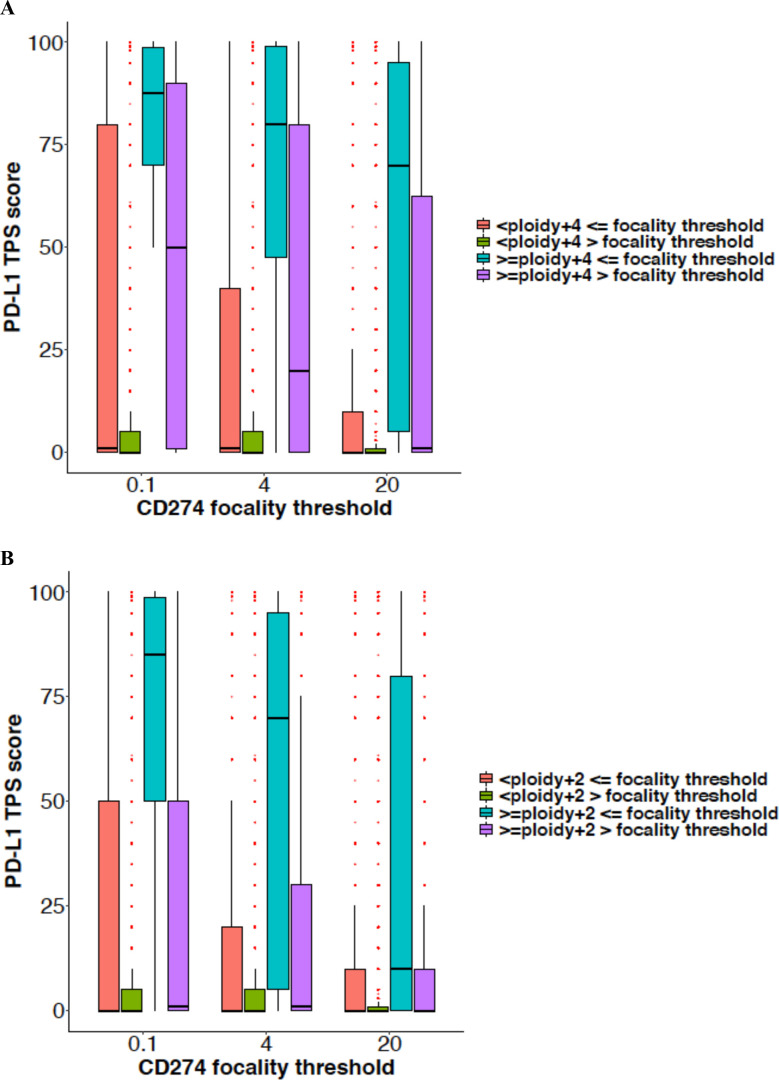
Within different CD274 focality categories, the amplification level is also driving PD-L1 expression (figure 2). The presence of PD-L1 amplification above the CN threshold is correlated to higher TPS score, and this effect is even clearer with lower levels of CD274 gene ploidy, such as +2 (figure 2B and online supplemental table 3). We analyzed the possible influence of histology on the correlation of PD-L1 gene amplification and protein expression. Separate analysis of the two most frequent histologic groups (NSCLC and colorectal cancer) and other histologies produced similar results (online supplemental table 4).
Figure 2.
Distribution of PD-L1 TPS score (numbers represent per cent) over CD274 (PD-L1) focality thresholds at a CD274 ploidy gain threshold of +4 (CN 6) (A) and+2 (CN 4) (B). CN, copy number; TPS, tumor proportion score.
Discussion
PD-L1 amplification is correlated with PD-L1 expression and also responsiveness to immunotherapy. To our knowledge, this is the first data set of patients also indicating that focality of PD-L1 amplification also influences protein expression. We demonstrated that PD-L1 amplification occurred in 1.21% of patients, which is similar to the 1.1% previously reported in a large contemporary cohort.6 The focality level of a gene amplification can impact the likelihood of messenger RNA (mRNA) overexpression on a gene-by-gene basis.11 It is plausible that focality may also reflect PD-L1 protein levels. Using PD-L1 expression (DAKO 22C3 IHC assay; TPS), we demonstrated that more focal amplification (<0.1 mB) was associated with higher PD-L1 expression levels.
Tumors with PD-L1 amplified (≥6 CN or ≥specimen ploidy +4) in a non-focal fashion (≥20 mB) may be currently considered as a variant of unknown significance. Even in this category, the 75th percentile of PD-L1 expression was 62.5%, suggesting a proportion of high PD-L1 expressors within this group; however, median expression level was 1%. This finding might help for interpretation of genomic reports and can indicate that in some cases a correlation with PD-L1 protein expression is warranted. Several quantitative parameters of CN gains were suggested for NGS reports, but focality levels are currently not recommended.9 Our data suggested that focality could add additional granularity for clinical interpretation, at least for PD-L1. If focality demonstrates clinical impact on therapeutic decisions, it is feasible to incorporate this potential biomarker from NGS tests commercially available. This factor should be evaluated in prospective studies.
The directed relationship of CD274 CN gains and PD-L1 expression was described as highly correlated in multiple tumor types.6 In addition, CD274 CN ≥+4 (specimen ploidy +2) predict responses to checkpoint inhibitors in NSCLC.19 However, the role of PD-L1 gene amplification focality on clinical responses to PD-1/PD-L1 inhibitors is unknown, and merits evaluation. Assessing cancer gene amplification using NGS can offer information that is useful for therapy. For ERBB2, information about CN and gene co-amplification can be predictive of trastuzumab clinical efficacy.20 21 Mesenchymal Epithelial Transition (MET) gene amplifications can be found in multiple tumors types and, at least in lung cancer, focality of amplification is correlated with responses to MET inhibitors.22 23 Herein, we described this interplay between an additional genomic layer (focality) and protein expression of PD-L1, which could also be explored for other cancer genes. Prior data suggested that, in general, genomic biomarkers are more predictive of responses to anticancer therapies as compared with protein biomarkers,24 perhaps due to technical limitations for protein assessment, but variations are expected on a gene-by-gene basis. One limitation of our data is lack of RNA expression, and correlation between PD-L1 gene amplification and focality with PD-L1 mRNA expression and impact on proteins levels. This analysis merits further studies.
Conclusions
Our data indicate that PD-L1 amplification level correlates with PD-L1 expression measured by IHC. Importantly, focality of PD-L1 amplification is also important for PD-L1 expression, as more focal amplification increases PD-L1 expression in different ploidy thresholds. Nonetheless, even above higher thresholds of focality (≥20 mB), PD-L1 overexpression can be detected, especially in the presence of increased CN, although less frequently. It is therefore important to correlate genomic findings from PD-L1 amplification and focality with PD-L1 proteins levels and to further examine correlation between amplification, focality, and immunotherapy outcome. The impact of focality and amplification level on protein expression and therapeutic outcomes for other targetable genes also merits exploration.
Footnotes
Twitter: @jardim_denis, @krthkynmrgsn, @Dr_R_Kurzrock
Contributors: Conceptualization: DLJ, KM, RSPH and RK. Methodology: KM, JAE and RSPH. Investigation: DLJ, KM, JAE, RSPH and RK. Visualization: DLJ, KM, RSPH and RK. Funding acquisition: RK. Supervision: RSPH and RK. Writing—original draft: DLJ. Writing—review and editing: DLJ, KM, JAE, RSPH and RK.
Funding: This work was supported in part by National Cancer Institute grant P30 CA016672 and the Joan and Irwin Jacobs Fund philanthropic fund.
Competing interests: DLJ receives speaker fees from Roche, Janssen, Astellas, MSD, Bristol-Myers Squibb, Pfizer, AztraZeneca and Libbs, as well as consultant fees from Janssen, Bristol-Myers Squibb and Libbs. RK has received research funding from Boehringer Ingelheim, Debiopharm, Foundation Medicine, Genentech, Grifols, Guardant, Incyte, Konica Minolta, Medimmune, Merck Serono, Omniseq, Pfizer, Sequenom, Takeda, and TopAlliance; as well as consultant and/or speaker fees and/or advisory board for Actuate Therapeutics, Bicara Therapeutics, Biological Dynamics, Caris, Ilyon, NeoGenomics, Neomed, Pfizer, Roche, TD2/Volastra, Turning Point Therapeutics, X-Biotech; has an equity interest in CureMatch and IDbyDNA; serves on the Board of CureMatch and CureMetrix, and is a co-founder of CureMatch. KM, JAE, and RSPH are employees of Foundation Medicine and shareholders of Roche Holding AG.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. Not applicable.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Approval for this study was obtained from the Western Institutional Review Board (IRB protocol No. 20152817). Approval for this analysis, including a waiver of informed consent and a Health Insurance Portability and Accountability Act (HIPAA) waiver, was obtained from the Western Institutional Review Board (IRB protocol No. 20152817).
References
- 1.Jardim DL, Goodman A, de Melo Gagliato D, et al. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell 2021;39:154–73. 10.1016/j.ccell.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colen RR, Rolfo C, Ak M, et al. Radiomics analysis for predicting pembrolizumab response in patients with advanced rare cancers. J Immunother Cancer 2021;9:e001752. 10.1136/jitc-2020-001752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujii T, Naing A, Rolfo C, et al. Biomarkers of response to immune checkpoint blockade in cancer treatment. Crit Rev Oncol Hematol 2018;130:108–20. 10.1016/j.critrevonc.2018.07.010 [DOI] [PubMed] [Google Scholar]
- 4.Patel SP, Kurzrock R. Pd-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther 2015;14:847–56. 10.1158/1535-7163.MCT-14-0983 [DOI] [PubMed] [Google Scholar]
- 5.Goodman AM, Piccioni D, Kato S, et al. Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid tumors. JAMA Oncol 2018;4:1237–44. 10.1001/jamaoncol.2018.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang RSP, Murugesan K, Montesion M, et al. Pan-Cancer landscape of CD274 (PD-L1) copy number changes in 244 584 patient samples and the correlation with PD-L1 protein expression. J Immunother Cancer 2021;9:e002680. 10.1136/jitc-2021-002680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda S, Goodman AM, Cohen PR, et al. Metastatic basal cell carcinoma with amplification of PD-L1: exceptional response to anti-PD1 therapy. NPJ Genom Med 2016;1:16037. 10.1038/npjgenmed.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roemer MGM, Advani RH, Ligon AH, et al. Pd-L1 and PD-L2 genetic alterations define classical Hodgkin lymphoma and predict outcome. J Clin Oncol 2016;34:2690–7. 10.1200/JCO.2016.66.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eijkelenboom A, Tops BBJ, van den Berg A, et al. Recommendations for the clinical interpretation and reporting of copy number gains using gene panel NGS analysis in routine diagnostics. Virchows Arch 2019;474:673–80. 10.1007/s00428-019-02555-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adashek JJ, Kato S, Parulkar R, et al. Transcriptomic silencing as a potential mechanism of treatment resistance [JCI Insight 5]. JCI Insight 2020;5:e134824. 10.1172/jci.insight.134824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boichard A, Lippman SM, Kurzrock R. Therapeutic implications of cancer gene amplifications without mrna overexpression: silence may not be golden. J Hematol Oncol 2021;14:201. 10.1186/s13045-021-01211-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–31. 10.1038/nbt.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun JX, He Y, Sanford E, et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput Biol 2018;14:e1005965. 10.1371/journal.pcbi.1005965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsui DCC, Lee JK, Frampton GM, et al. Real-world (rw) analysis of quantitative met copy number (CN) as a biomarker in NSCLC. JCO 2022;40(16_suppl):9123. 10.1200/JCO.2022.40.16_suppl.9123 [DOI] [Google Scholar]
- 15.Food and Drug Administration . PD-L1 IHC 22C3 pharmdx. rx only. SK006: 50 tests for use with autostainer link 48 food and drug administration. 2019. Available: https://www.accessdata.fda.gov/cdrh_docs/pdf15/P150013S014C.pdf
- 16.Mok TSK, Wu Y-L, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819–30. 10.1016/S0140-6736(18)32409-7 [DOI] [PubMed] [Google Scholar]
- 17.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823–33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 18.Myer PA, Lee JK, Madison RW, et al. The genomics of colorectal cancer in populations with African and European ancestry. Cancer Discov 2022;12:1282–93. 10.1158/2159-8290.CD-21-0813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murugesan K, Jin DX, Comment LA, et al. Association of cd274 (pd-l1) copy number changes with immune checkpoint inhibitor clinical benefit in non-squamous non-small cell lung cancer. Oncologist 2022;27:732–9. 10.1093/oncolo/oyac096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hino K, Nishina T, Kajiwara T, et al. Association of ErbB2 copy number and gene coalterations with trastuzumab efficacy and resistance in human epidermal growth factor receptor 2-positive esophagogastric and gastric cancer [JCO Precis Oncol 6:e2200135]. JCO Precis Oncol 2022;6:e2200135. 10.1200/PO.22.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura K, Aimono E, Oba J, et al. Estimating copy number using next-generation sequencing to determine erbb2 amplification status. Med Oncol 2021;38:36. 10.1007/s12032-021-01482-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng L-X, Jie G-L, Li A-N, et al. Met amplification identified by next-generation sequencing and its clinical relevance for met inhibitors. Exp Hematol Oncol 2021;10:52. 10.1186/s40164-021-00245-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumaki Y, Olsen S, Suenaga M, et al. Comprehensive genomic profiling of circulating cell-free DNA distinguishes focal Met amplification from aneuploidy in diverse advanced cancers. Curr Oncol 2021;28:3717–28. 10.3390/curroncol28050317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwaederle M, Zhao M, Lee JJ, et al. Association of biomarker-based treatment strategies with response rates and progression-free survival in refractory malignant neoplasms: a meta-analysis. JAMA Oncol 2016;2:1452–9. 10.1001/jamaoncol.2016.2129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-006311supp001.pdf (75.1KB, pdf)
Data Availability Statement
Data are available upon reasonable request. Not applicable.