Abstract
Background
Neurofilament light (NfL) is a widely used biomarker for neurodegeneration. NfL is prone to oligomerisation, but available assays do not reveal the exact molecular nature of the protein variant measured. The objective of this study was to develop a homogeneous ELISA capable of quantifying oligomeric NfL (oNfL) in cerebrospinal fluid (CSF).
Methods
A homogeneous ELISA, based on the same capture and detection antibody (NfL21), was developed and used to quantify oNfL in samples from patients with behavioural variant frontotemporal dementia (bvFTD, n=28), non-fluent variant primary progressive aphasia (nfvPPA, n=23), semantic variant PPA (svPPA, n=10), Alzheimer’s disease (AD, n=20) and healthy controls (n=20). The nature of NfL in CSF, and the recombinant protein calibrator, was also characterised by size exclusion chromatography (SEC).
Results
CSF concentration of oNfL was significantly higher in nfvPPA (p<0.0001) and svPPA patients (p<0.05) compared with controls. CSF oNfL concentration was also significantly higher in nfvPPA compared with bvFTD (p<0.001) and AD (p<0.01) patients. SEC data showed a peak fraction compatible with a full-length dimer (~135 kDa) in the in-house calibrator. For CSF, the peak was found in a fraction of lower molecular weight (~53 kDa), suggesting dimerisation of NfL fragments.
Conclusions
The homogeneous ELISA and SEC data suggest that most of the NfL in both the calibrator and human CSF is present as a dimer. In CSF, the dimer appears to be truncated. Further studies are needed to determine its precise molecular composition.
Keywords: FRONTOTEMPORAL DEMENTIA, CSF, SEMANTIC DEMENTIA, NEUROCHEMISTRY
Introduction
Neurofilament light (NfL) is a 68 kDa type IV intermediate filament with structural functions in the neuronal cytoskeleton, mainly found in axons.1 Under physiological conditions, there is a basal release of NfL into the cerebrospinal fluid (CSF), which increases with age.2 However, after axonal damage in neurodegenerative conditions and other acute neurological conditions such as traumatic brain injury or stroke, its release greatly increases.3 Previous studies indicate NfL as a general marker of neurodegeneration, with its concentration in CSF and blood being increased in several neurological diseases including Alzheimer’s disease (AD),4 multiple sclerosis (MS),5 amyotrophic lateral sclerosis,6 Huntington’s disease7 and frontotemporal dementia (FTD).8 NfL is a well-established biomarker for neurodegeneration and several assays, either immunoassay or mass spectrometry (MS) based, have previously been developed intended to quantify it in body fluids. In a recent study, Budelier et al showed that, in CSF, no full-length NfL was found. Instead, it was detected by immunoprecipitation-MS analysis as various truncated species,9 shedding some light on the actual molecular state of this protein in CSF.
The hypothesis in this study is that NfL could be present in CSF as an oligomer. Our goal was to develop a homogeneous ELISA, using the same capture and detection antibody, to test this hypothesis and to quantify these oligomers in CSF. We also set out to examine the apparent size of NfL by size exclusion chromatography (SEC) in CSF and the calibrator used in the ELISA.
Methods
Participants
This study included FTD patients (n=61) from different variants, including behavioural variant FTD (bvFTD, n=28), non-fluent variant primary progressive aphasia (nfvPPA, n=23) and semantic variant primary progressive aphasia (svPPA, n=10). Among the FTD patients, 17 were mutation carriers affecting progranulin (GRN), chromosome 9 open reading frame 72 (C9orf72) or microtubule-associated protein tau (MAPT) and 44 were sporadic cases from the University College London Genetic FTD Initiative (GENFI) and Longitudinal Investigation of FTD (LIFTD) studies. Assessment was performed based on standardised history and examination and the patients were classified as symptomatic if they met consensus diagnostic criteria.10 11 AD patient samples (n=20) were obtained from the H70 Clinical Study at Sahlgrenska University Hospital, and healthy controls (n=20) from the H70 Birth Cohort Study at the University of Gothenburg.12 Demographics and oligomeric NfL (oNFL) concentrations are shown in table 1.
Table 1.
Demographic characteristics and CSF oligomeric NFL concentrations of the study subgroups
| bvFTD | nfvPPA | svPPA | AD | Controls | P value | |
| No | 28 | 23 | 10 | 20 | 20 | |
| M/F | 20/8 | 13/10 | 7/3 | 8/12 | 7/13 | |
| Age, years | 62.6 (59.4–68.2) | 66.3 (63.2–72.1) | 65.2 (57.4–68.7) | 66.9 (63.6–70.1) | 70 (70–70) | 0.0023 |
| Disease duration, years | 6.1 (9.4–3.5) | 4.0 (5.4–2.7) | 5.8 (7.7–1.8) | - * | NA | 0.054 |
| CSF oNfL, pg/mL | 12 554 (18 169–6023) | 22 762 (34 442–12452) | 22 890 (34 447–14525) | 8899 (15 491–5525) | 7150 (10 889–5745) | <0.0001† |
Data are shown as median (IQR).
One-way ANOVA was applied to all study subgroups.
*Samples used for AD diagnostic (early-stage AD).
†bvFTD versus nfvPPA p<0.001, nfvPPA vs AD p<0.01, vs controls p<0.0001, svPPA vs controls p<0.05.
AD, Alzheimer’s disease; ANOVA, analysis of variance; bvFTD, behavioural variant frontotemporal dementia; CSF, cerebrospinal fluid; NA, not applicable; NfL, neurofilament light; nfvPPA, non-fluent variant primary progressive aphasia; oNfL, oligomeric NfL; svPPA, semantic variant primary progessive aphasia.
Homogeneous NFL ELISA
The homogeneous ELISA for oNfL is based on a previously developed sandwich assay13 in which in-house anti-NfL antibodies, NfL21 and NfL23 (raised against the rod domain of NfL) were used to quantify CSF NfL. Here, we verified that either antibody could be used in a homogeneous set-up to detect an NfL signal in CSF, indistinguishable from that obtained using the regular ELISA. For the oNfL assay, we chose NfL21+NfL21 (but NfL23+NfL23 worked as well, online supplemental figure 1). In brief, 96-well microtiter plates were coated with capture NfL21 antibody at a final concentration of 0.5 µg/mL (100 µL/well) in bicarbonate buffer (50 mM NaHCO3, pH 9.6), overnight at +4°C. Washes were performed between every step with 4×350 µL PBS with 0.05% Tween20, and all incubations at room temperature. Unspecific binding was blocked with 1% bovine serum albumin (BSA) in PBS (0.01 M phosphate buffer, 0.14 M NaCl, pH 7.4; 250 µL/well) for 1 hour. The assay buffer used for dilutions was optimised to PBS-Tween20 0.01% with 1% BSA. After blocking, eight-step serial twofold dilutions of calibrator, prepared from bovine brain tissue (range 1562–2 00 000 pg/mL), blank, quality controls (QCs) and samples were added to the plate (100 µL/well). Samples and QCs were diluted 1:1 in assay buffer. A 3-hour incubation period preceded the addition of biotinylated NfL21 detection antibody (1 µg/mL in assay buffer) with a 1-hour incubation. After that, enhanced streptavidin horseradish peroxidase (Enhanced Streptavidin-HRP, 4740 n, Kem En Tech) diluted 1:20 000 in assay buffer was added (100 µL/well) and incubated for 30 min. 3,3’,5,5’-Tetramethylbenzidine (One Solution, 4380A, Kem En Tech; 100 µL/well) was used as substrate. Finally, after 30 min incubation in the dark, the reaction was stopped with 0.2 M H2SO4 (100 µL/well), and the absorbance measured at 450 nm (with 650 nm reference) using an ELISA plate reader (Sunrise, Tecan Trading AG).
bmjno-2022-000395supp001.pdf (133.4KB, pdf)
Size exclusion chromatography
SEC was performed using an Ettan LC system (Amersham Biosciences) equipped with a Superdex 200 10/300 GL column (GE Healthcare) connected to a fraction collector (Frac-950, Amersham Biosciences). The column was equilibrated with a 10 mM ammonium bicarbonate buffer, which was also used in the isocratic separation. Injected sample volume was 500 µL and the flow rate 0.5 mL/min, while the fractions collected were 1 mL. Calibration of the column was performed using gel filtration markers (Sigma, MWGF70).
Statistical analysis
One-way analysis of variance with Tukey’s test correction was used to determine significant differences between groups, with p<0.05 considered statistically significant. The analysis was performed using GraphPad Prism software, V.9.4.1.
Results
CSF oNfL group differences
Two homogeneous setups with the in-house monoclonal antibodies were tested, namely NfL21+NfL21 and NfL23+NfL23. Both assays correlated strongly (r=0.9913, p<0.0001, online supplemental figure 1) with the only difference being the slightly higher concentration obtained with the NfL21 setup, which was the assay selected for further testing.
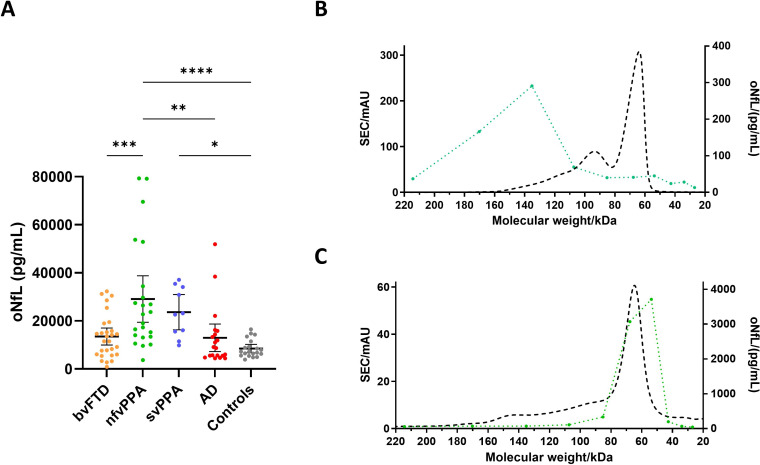
To assess any differences in oNfL concentrations, samples representing several neurodegenerative diseases were included in a pilot study. All samples were randomised and analysed blinded, under the same conditions. Results showed significant increases in oNfL concentrations in nfvPPA (2.90e+4 ± 4.66e+3 pg/mL, p<0.0001) and svPPA (2.36e+4 ± 3.24e+3 pg/mL, p<0.05) compared with controls (8.50e+3 ± 8.22e+2 pg/mL). nfvPPA also displayed increased oNfL compared with bvFTD (1.35e+4 ± 1.72e+3 pg/mL, p<0.001) and AD (1.30e+4 ± 2.74e+3 pg/mL, p<0.01). However, neither bvFTD nor AD had any significant differences compared with controls (figure 1A).
Figure 1.
Oligomeric NfL (oNfL) concentration (pg/mL) measured with the homogeneous assay in different neurodegenerative diseases and healthy controls (A) and after SEC on in-house calibrator (B) or CSF (C). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, by one-way ANOVA test; bars represent 95% CIs; green dotted line, measured concentrations in collected fractions and dashed black line, absorbance at 280 nm. AD, Alzheimer’s disease; ANOVA, analysis of variance; bvFTD, behavioural variant frontotemporal dementia; CSF, cerebrospinal fluid; nfvPPA, non-fluent variant primary progressive aphasia; SEC, size exclusion chromatography; svPPA, semantic variant primary progressive aphasia.
Size exclusion chromatography
SEC analysis of in-house calibrator showed a peak fraction (~135 kDa) that was consistent with full-length NfL dimers (figure 1B). Still, in CSF, the peak was found in fractions corresponding to lower molecular weight (~53 kDa), suggesting that oligomers in CSF are formed of truncated NfL (figure 1C).
Discussion
This study shows that most NfL in CSF and the assay calibrator is a multimer (likely a dimer). None of the currently used NfL immunoassays give any information regarding the multimeric state of the protein. In the newly developed homogeneous assay, where the capture and detector antibodies are the same, the binding of the capture antibody to the target protein blocks the epitope for the detector antibody, thus nullifying the output signal if the captured protein is in monomeric form.14 However, if the captured protein is in oligomerised form, there will still be epitopes available for detector antibody-binding which consequently results in measurable absorbances.
CSF NfL concentration, measured using a standard ELISA, is increased in several neurodegenerative diseases, FTD in particular.15 CSF NfL even distinguishes FTD variants; in a recent meta-analysis, higher NfL levels were noted in semantic PPA patients when compared with bvFTD and non-fluent PPA patients.16 Here, we replicate most of these findings using an immunoassay selective for oNfL. In fact, our data suggest that most of the NfL signal in earlier studies may have been derived from oNfL rather than NfL monomers.
It is also important to emphasise that our assay may not detect all forms of oNfL, as oligomerisation might mask NfL21 epitopes in certain oligomeric species. However, from earlier studies, we are positive that this type of assay only detects oligomers of the target protein,14 and hence our results highlight this novel aspect of NfL as a biomarker. To examine the relative amount of oligomeric vs monomeric forms of NfL, as currently available assays cannot distinguish the two, an assay specific to monomeric NfL needs to be developed.
According to Budelier et al, NfL is present in the CSF as several different fragments, of which rod domain 2B fragments were identified and found to be correlating with results from the most widely used NfL ELISA (UmanDiagnostics).9 Our SEC results are in line with these findings, as in CSF, the peak fraction cannot correspond to full-length NfL, but rather oligomers of truncated NfL.9 Furthermore, we also observed that our calibrator eluted as a full-length dimer, confirming the oligomerised state of the protein. Previous studies, which also support our findings, state that the central part human NfL (detected by NfL21) has a strong propensity for self-assembly.17
One question that remains to be answered is whether the oligomerisation occurs prior to or after release of the protein from the axon, and if oligomerisation is a part of the neurodegenerative process. We hypothesise that the protein is released as a dimer in both physiological and pathological conditions, given the strong propensity for oligomerisation of the coiled coil domain of NfL.17 To address this hypothesis, further research, including cell-based experiments and/or studies on acute axonal injury, is needed.
Nevertheless, the novel homogeneous ELISA described here, along with the SEC data, provide strong evidence of the presence of oNfL in CSF. In fact, the data suggest that most of NfL in calibrators and human CSF is a dimer; an important result if we are to standardise NfL assays by reference measurement procedures for which full characterisation of the target analyte is a prerequisite.
Footnotes
HK and HZ contributed equally.
Contributors: FJM and UA contributed to the study design, acquisition and interpretation of data, and drafted the manuscript. HK and HZ contributed to the study design, supervision, interpretation of data and drafted the manuscript. KK, IjS, AS-E, JDR, AD, IS and SK provided the samples and revised the manuscript. BB and KB contributed to the study design and revised the manuscript.
Funding: HZ is a Wallenberg Scholar supported by grants from the Swedish Research Council (#2018-02532), the European Union’s Horizon Europe research and innovation programme under grant agreement No 101053962, Swedish State Support for Clinical Research (#ALFGBG-71320), the Alzheimer Drug Discovery Foundation (ADDF), USA (#201809-2016862), the AD Strategic Fund and the Alzheimer’s Association (#ADSF-21-831376-C, #ADSF-21-831381-C, and #ADSF-21-831377-C), the Bluefield Project, the Olav Thon Foundation, the Erling-Persson Family Foundation, Stiftelsen för Gamla Tjänarinnor, Hjärnfonden, Sweden (#FO2022-0270), the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 860197 (MIRIADE), the European Union Joint Programme – Neurodegenerative Disease Research (JPND2021-00694), and the UK Dementia Research Institute at UCL (UKDRI-1003). KB is supported by the Swedish Research Council (#2017-00915 and #2022-00732; Jan 1 2023 to Dec 31, 2026), the Alzheimer Drug Discovery Foundation (ADDF), USA (#RDAPB-201809-2016615), the Swedish Alzheimer Foundation (#AF-930351, #AF-939721 and #AF-968270), Hjärnfonden, Sweden (#FO2017-0243 and #ALZ2022-0006), the Swedish state under the agreement between the Swedish government and the County Councils, the ALF-agreement (#ALFGBG-715986 and #ALFGBG-965240), the European Union Joint Program for Neurodegenerative Disorders (JPND2019-466-236), the National Institute of Health (NIH), USA, (grant #1R01AG068398-01), the Alzheimer’s Association 2021 Zenith Award (ZEN-21-848495), and the Alzheimer’s Association 2022-2025 Grant (SG-23-1038904 QC). SK was financed by grants from the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement (ALFGBG-965923, ALFGBG-81392, ALF GBG-771071). The Alzheimerfonden (AF-842471, AF-737641, AF-929959, AF-939825). The Swedish Research Council (2019-02075), Psykiatriska Forskningsfonden, Stiftelsen Demensfonden, Stiftelsen Hjalmar Svenssons Forskningsfond, Stiftelsen Wilhelm och Martina Lundgrens vetenskapsfond.
Competing interests: HZ has served at scientific advisory boards and/or as a consultant for Abbvie, Acumen, Alector, ALZPath, Annexon, Apellis, Artery Therapeutics, AZTherapies, CogRx, Denali, Eisai, Nervgen, Novo Nordisk, Passage Bio, Pinteon Therapeutics, Red Abbey Labs, reMYND, Roche, Samumed, Siemens Healthineers, Triplet Therapeutics, and Wave, has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure, Biogen, and Roche, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work). KB has served as a consultant, at advisory boards, or at data monitoring committees for Abcam, Axon, BioArctic, Biogen, JOMDD/Shimadzu. Julius Clinical, Lilly, MagQu, Novartis, Ono Pharma, Pharmatrophix, Prothena, Roche Diagnostics, and Siemens Healthineers, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program, outside the work presented in this paper. SK has served at scientific advisory boards and/or as consultant for Geras Solutions and Biogen.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as online supplemental information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
All participants provided written informed consent at enrolment including consent to publication. The London Queen Square Research Ethics Committee approved the University College London Genetic FTD Initiative (GENFI; ID 14/0377) and Longitudinal Investigation of FTD (LIFTD; ID 15/0805). The H70 Clinical Study was approved by the Ethical Review Authority in Sweden (ID: 2019–04024) and the H70 Birth Cohort Study was approved by the Regional Ethical Review Board in Gothenburg, Sweden (ID: 869-13). The study complies with the Declaration of Helsinki.
References
- 1.Yuan A, Rao MV, Nixon RA, et al. Neurofilaments and neurofilament proteins in health and disease. Cold Spring Harb Perspect Biol 2017;9:a018309. 10.1101/cshperspect.a018309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gafson AR, Barthélemy NR, Bomont P, et al. Neurofilaments: neurobiological foundations for biomarker applications. Brain 2020;143:1975–98. 10.1093/brain/awaa098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaetani L, Blennow K, Calabresi P, et al. Neurofilament light chain as a biomarker in neurological disorders. J Neurol Neurosurg Psychiatry 2019;90:870–81. 10.1136/jnnp-2018-320106 [DOI] [PubMed] [Google Scholar]
- 4.Zetterberg H, Skillbäck T, Mattsson N, et al. Association of cerebrospinal fluid neurofilament light concentration with Alzheimer disease progression. JAMA Neurol 2016;73:60–7. 10.1001/jamaneurol.2015.3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stilund M, Gjelstrup MC, Petersen T, et al. Biomarkers of inflammation and axonal degeneration/damage in patients with newly diagnosed multiple sclerosis: contributions of the soluble CD163 CSF/serum ratio to a biomarker panel. PLoS One 2015;10:e0119681. 10.1371/journal.pone.0119681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu C-H, Macdonald-Wallis C, Gray E, et al. Neurofilament light chain: a prognostic biomarker in amyotrophic lateral sclerosis. Neurology 2015;84:2247–57. 10.1212/WNL.0000000000001642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne LM, Rodrigues FB, Johnson EB, et al. Evaluation of mutant huntingtin and neurofilament proteins as potential markers in huntington’s disease. Sci Transl Med 2018;10:458. 10.1126/scitranslmed.aat7108 [DOI] [PubMed] [Google Scholar]
- 8.Meeter LH, Dopper EG, Jiskoot LC, et al. Neurofilament light chain: a biomarker for genetic frontotemporal dementia. Ann Clin Transl Neurol 2016;3:623–36. 10.1002/acn3.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budelier MM, He Y, Barthelemy NR, et al. A map of neurofilament light chain species in brain and cerebrospinal fluid and alterations in Alzheimer’s disease. Brain Commun 2022;4:fcac045. 10.1093/braincomms/fcac045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorno-Tempini ML, Hillis AE, Weintraub S, et al. Classification of primary progressive aphasia and its variants. Neurology 2011;76:1006–14. 10.1212/WNL.0b013e31821103e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rascovsky K, Hodges JR, Kipps CM, et al. Diagnostic criteria for the behavioral variant of frontotemporal dementia (bvFTD): current limitations and future directions. Alzheimer Dis Assoc Disord 2007;21:S14–8. 10.1097/WAD.0b013e31815c3445 [DOI] [PubMed] [Google Scholar]
- 12.Rydberg Sterner T, Ahlner F, Blennow K, et al. The Gothenburg H70 birth cohort study 2014-16: design, methods and study population. Eur J Epidemiol 2019;34:191–209. 10.1007/s10654-018-0459-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaetani L, Höglund K, Parnetti L, et al. A new enzyme-linked immunosorbent assay for neurofilament light in cerebrospinal fluid: analytical validation and clinical evaluation. Alzheimers Res Ther 2018;10:8. 10.1186/s13195-018-0339-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hölttä M, Hansson O, Andreasson U, et al. Evaluating amyloid-β oligomers in cerebrospinal fluid as a biomarker for Alzheimer’s disease. PLoS One 2013;8:e66381. 10.1371/journal.pone.0066381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridel C, van Wieringen WN, Zetterberg H, et al. Diagnostic value of cerebrospinal fluid neurofilament light protein in neurology: a systematic review and meta-analysis. JAMA Neurol 2019;76:1035–48. 10.1001/jamaneurol.2019.1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karantali E, Kazis D, Chatzikonstantinou S, et al. The role of neurofilament light chain in frontotemporal dementia: a meta-analysis. Aging Clin Exp Res 2021;33:869–81. 10.1007/s40520-020-01554-8 [DOI] [PubMed] [Google Scholar]
- 17.Carter J, Gragerov A, Konvicka K, et al. Neurofilament (NF) assembly; divergent characteristics of human and rodent NF-L subunits. J Biol Chem 1998;273:5101–8. 10.1074/jbc.273.9.5101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjno-2022-000395supp001.pdf (133.4KB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as online supplemental information.