Abstract
Isolates of the obligately biotrophic fungus Uncinula necator cluster in three distinct genetic groups (groups I, II, and III). We designed PCR primers specific for these groups in order to monitor field populations of U. necator. We used the nucleotide sequences of the gene that encodes eburicol 14α-demethylase (CYP51) and of the ribosomal DNA internal transcribed spacer 1 (ITS1), ITS2, and 5.8S regions. We identified four point mutations (three in CYP51 and one in ITS1) that distinguished groups I and II from group III based on a sample of 132 single-spore isolates originating from Europe, Tunisia, Israel, India, and Australia. We developed a nested allele-specific PCR assay in which the CYP51 point mutations were used to detect and distinguish groups I and II from group III in crude mildewed samples from vineyards. In a preliminary study performed with samples from French vineyards in which isolates belonging to genetic groups I and III were present, we found that a shift from a population composed primarily of group I isolates to a population composed primarily of group III isolates occurred during the grapevine growing season.
Powdery mildew is the most economically important fungal disease of grapes (Vitis vinifera L.) throughout the world. The causal agent is the haploid, heterothallic ascomycete Uncinula necator (Schw.) Burr. This obligately biotrophic fungus develops only on green organs of plants belonging to the family of Vitaceae (3). In Europe, India, and Australia, the only member of the family Vitaceae is the cultivated grapevine, V. vinifera L.). In temperate climates, the asexual mycelium of U. necator may overwinter on leaf primordia inside dormant buds (17, 18). The fungus begins growing again shortly after bud break, which results in the appearance of shoots covered with white, sporulating mycelium; these shoots are called “flagshoots” (3). U. necator also may overwinter as cleistothecia (the ascigerous stage of the fungus) on the bark of grapevines (11, 16). In this case, primary infections are caused by ascospores released during the spring (12). Although both flagshoots and cleistothecia are observed in Europe, cleistothecia are considered the most important way that the fungus overwinters in Europe (23).
Recent randomly amplified polymorphic DNA (RAPD) (22) studies showed that U. necator isolates collected from plants with typical flagshoot symptoms (group I isolates) had RAPD patterns very different than the RAPD patterns of isolates that presumably arose from ascospores (group III isolates) (5, 7). A similar situation seems to exist in Australia (10), but the population structure of U. necator in India seems to be more complex; in India a third genetic group (group II) has been identified (7). The genetic variation between groups is considerably greater than the genetic variation within groups, suggesting genetic isolation (7).
In European vineyards in which U. necator isolates belonging to group I and group III are both present, the fungal population may shift from predominantly group I isolates to predominantly group III isolates during the grapevine growing season (5, 7). For large-scale field studies, molecular markers that allow workers to detect isolates belonging to each genetic group are needed. As U. necator is an obligate biotroph, large-scale field studies in which RAPDs are used are extremely time- and labor-intensive. Specific PCR primers that distinguish the genetic groups of U. necator are needed for such field studies.
Our primary objective was to develop PCR primers specific for groups I and III. A secondary objective was to obtain preliminary data on changes in the populations of U. necator in vineyards in which isolates belonging to both of these genetic groups are present. We cloned and sequenced the single-copy gene encoding eburicol 14α-demethylase (CYP51), a highly conserved cytochrome P-450 enzyme essential for sterol biosynthesis (2; for a review see reference 24). CYP51 is the only U. necator gene whose sequence is known. We previously found a point mutation in CYP51 that could be used to identify U. necator isolates that are resistant to a CYP51-inhibiting fungicide (8). In other studies (1, 15, 19), the ribosomal DNA (rDNA) region encompassing internal transcribed spacers (ITS) (21) and 5.8S rDNA has been used to distinguish fungal species, subspecies, or isolates. We cloned and sequenced both CYP51 and the ITS-5.8S rDNA region from U. necator isolates belonging to all three genetic groups in order to identify mutations that distinguish the genetic groups.
MATERIALS AND METHODS
Isolates of U. necator.
We used 132 single-spore isolates, including 90 isolates from a previous study (7) and 42 new isolates collected from 1992 to 1998 in Australia (30 isolates), Tunisia (8 isolates), Europe (3 isolates), and Israel (1 isolate) (Table 1). Fungal material (conidia and mycelium) from all of the isolates from Europe, India, Tunisia, Israel and from the 22 Australian isolates collected in 1998 was produced and collected as previously described (5). Freeze-dried conidia of the eight remaining Australian isolates (isolates 920103, 930202, 930304b, 931201, 931302, 931502, 940301, and 940402) were obtained from B. E. Stummer (University of Adelaide, Adelaide, Australia).
TABLE 1.
Assignment of 42 single-spore U. necator isolates to genetic groups I through III on the basis of their RAPD patterns
| Isolate(s) | Geographic origin
|
Date of isolation (mo/yr) | Genetic group | |
|---|---|---|---|---|
| Country | City | |||
| GOK1.1, GOK1.2, GOK1.3 | Germany | Oppenheim | 6/1998 | III |
| IsBD1.1 | Israel | Bet-Dagan | 5/1992 | III |
| TTU1.1, TTU1.2, TTU1.7, TTU1.8 | Tunisia | Tunis | 4/1998 | I |
| TTU1.3, TTU1.4, TTU1.5, TTU1.6 | Tunisia | Tunis | 4/1998 | III |
| 920103 | Australia | Waite Campus | 11/1992 | I |
| 930202 | Australia | Nuriootpa | 1/1993 | I |
| 930304b | Australia | Nuriootpa | 1/1993 | III |
| 931201, 931502, 931302 | Australia | Summertown | 2/1993 | III |
| 940301, 940402 | Australia | Swan Valley | 1/1994 | III |
| AuLO1.1 to AuLO1.22 | Australia | Loxton | 3/1998 | III |
Powdery mildew field samples.
U. necator populations were monitored in three vineyards in France (Manosque, Nîmes, and Montirat) in which both flagshoots and cleistothecia had been observed for at least 5 years. During the 1998 grape growing season, these vineyards were not sprayed with fungicides. Mildewed tissues were collected from grapevines at different times during the grape growing season (Table 2), packed in healthy grapevine leaves and newspapers, and mailed to the laboratory. To minimize the effect of sampling, only small amounts of mildewed tissues were collected from each grapevine. Three successive samples were obtained from two grapevines in Nîmes and from two grapevines in Manosque. Four successive samples were obtained from five grapevines in Montirat. The first samples consisted exclusively of flagshoots, since they were the only early powdery mildew symptoms observed.
TABLE 2.
NAS-PCR monitoring of U. necator populations growing on nine grapevines in three French vineyards
| Vineyard | Date of collection | Total no. of samples | No. of samples for which NAS-PCR amplification was obtained with primers specific for:
|
||
|---|---|---|---|---|---|
| Groups I and II only | Groups I and II and group III | Group III only | |||
| Manosquea | 19 May 1998 | 2 | 1 | 1 | 0 |
| 10 June 1998 | 2 | 0 | 0 | 2 | |
| 30 June 1998 | 2 | 0 | 0 | 2 | |
| Nîmesa | 5 May 1998 | 2 | 2 | 0 | 0 |
| 9 June 1998 | 5 | 4 | 1 | 0 | |
| 8 July 1998 | 8 | 0 | 0 | 8 | |
| Montiratb | 5 May 1998 | 5 | 5 | 0 | 0 |
| 24 June 1998 | 12 | 0 | 7 | 5 | |
| 24 July 1998 | 9 | 0 | 0 | 9 | |
| 8 September 1998 | 9 | 0 | 0 | 9 | |
Samples were obtained from two cultivar Carignan grapevines.
Samples were obtained from five cultivar Carignan grapevines.
Leaves from grape cultivar Cinsaut grown in a greenhouse were decontaminated and placed on water agar medium (5). Mildewed samples from the field were brushed against the upper surfaces of the decontaminated grape leaves in order to contaminate them. The inoculated leaves were incubated for 14 days (5) and were transferred to fresh agar medium every 3 to 4 days in order to reduce the amount of saprophytic fungi originating from field samples that grew on the agar medium. Fourteen days after inoculation, mixtures of powdery mildew and saprophytic fungi were collected in microcentrifuge tubes from the surfaces of the leaves (6).
Each mildewed sample from the vineyard was used to inoculate four leaves in petri dishes. Crude material from powdery mildew colonies and other fungi growing on the surfaces of two leaves after 14 days of incubation was collected in one 1.5-ml microcentrifuge tube, which allowed us to perform replicate molecular analyses. Microcentrifuge tubes containing dry fungal material were kept at −20°C until nucleic acids were extracted.
DNA extraction and RAPD assay.
Total genomic DNA extraction and a RAPD analysis were performed as described previously (6). The uncharacterized isolates (Table 1) were assigned to genetic groups I, II, and III by using band patterns obtained with 46 primers, as described previously (7).
CYP51 cloning and sequencing.
We considered the CYP51 sequence of a group III, fungicide-sensitive isolate (GenBank accession no. U72657) the reference sequence for U. necator CYP51. A 1,756-bp DNA fragment encompassing the entire CYP51 coding sequence was amplified by PCR by using primers C14 and C14R (8). For each isolate, both strands of three DNA inserts were sequenced by using specific primers (9).
ITS and 5.8S rDNA cloning and sequencing.
Primers ITS1 (21) and UncITS4 (5′-AATGATTCGAGGTCAACCTGTCAATCC; based on a partial sequence of U. necator rDNA kindly provided by J. Mugnier [Rhône-Poulenc Agro, Lyon, France]) were used for PCR amplification of an expected approximately 550-bp DNA fragment encompassing ITS region 1 (ITS1), 5.8S rDNA, and most of ITS2 from one isolate per genetic group. The cloning and sequencing procedures used have been described previously (9).
Allele-specific PCR assay.
Allele-specific PCR primers were designed by using the fact that a 3′ mismatch does not prime in a PCR at a specific annealing temperature (20). Primers MUT2(I-II), MUT3(I-II), and MUT4(I-II) (Table 3) were designed to specifically prime CYP51 sequences containing a T at nucleotide 110, a C at nucleotide 575, and a T at nucleotide 1587, respectively. Primers MUT2(III), MUT3(III), and MUT4(III) (Table 3) were designed to specifically prime CYP51 sequences containing a G at nucleotide 110, a T at nucleotide 575, and a C at nucleotide 1587, respectively. Primers U14DM, M1I, and M1 were based on CYP51 sequences that were identical in all of the U. necator CYP51 sequences studied. Primer MUT2(I-II) or MUT2(III) was used with primer U14DM, primer MUT3(I-II) or MUT3(III) was used with primer M1I, and primer MUT4(I-II) or MUT4(III) was used with primer M1 (Table 3); all of the primers were used at a final concentration of 0.1 μM.
TABLE 3.
Sequences of the primers used for allele-specific PCR and NAS-PCR
| Primer | Primer sequence (5′-3′) | Coordinates (bp) | Annealing temp (°C) |
|---|---|---|---|
| CYP51 allele-specific PCR | |||
| U14DM | ATGTACATTGCTGACATTTTGTCGG | 1–25 | |
| MUT2(I-II)a | CTCAATACATTTAACA | 125–110 | 52 |
| MUT2(III)b | CTCAATACATTTAACC | 125–110 | 53 |
| M1I | CGCTATCTCTCGATCAGG | 1007–990 | |
| MUT3(I-II)a | AACGGCTCTTACCAC | 561–575 | 65 |
| MUT3(III)b | AACGGCTCTTACCAT | 561–575 | 61 |
| M1 | TGATCGAGAGATAGCGC | 990–1007 | |
| MUT4(I-II)a | CTTCCCATCTATATTA | 1602–1587 | 52 |
| MUT4(III)b | CTTCCCATCTATATTG | 1602–1587 | 53 |
| rDNA allele-specific PCR | |||
| UITS | AGGATCATTACAGAGCGAGAGG | 1–22 | |
| MIT1(I-II)a | CAAAATACATATCGA | 84–70 | 53 |
| MIT1(III)b | CAAAATACATATCGG | 84–70 | 55 |
Primers specific for the allele in isolates belonging to groups I and II.
Primers specific for the allele in isolates belonging to group III.
Primers MIT1(I-II) and MIT1(III) (Table 3) were designed to specifically prime rDNA sequences containing T and C at nucleotide 70, respectively. Primer UITS was based on an rDNA sequence that was the same in all of the U. necator isolates whose rDNA were sequenced. Primer MIT1(I-II) or MIT1(III) was used with primer UITS (Table 3), all at a final concentration of 0.1 μM. Otherwise, the PCR conditions were as described previously (8).
For all of the single-spore isolates studied, amplifications were performed twice by using two independently extracted DNA samples.
NAS-PCR.
We developed a nested allele-specific PCR (NAS-PCR) assay targeting the point mutations found in CYP51. Nested PCR (4) involves two rounds of amplification by PCR. The first-round PCR was performed by using primers C14 and C14R (8). Subsequently, 1-μl aliquots of the first-round PCR mixture were subjected to a second round of PCR amplification, in which we used each of the six primer pairs designed for allele-specific PCR amplification of CYP51. In the second-round PCR the amount of Goldstar DNA polymerase (Eurogentec S. A., Seraing, Belgium) used was 0.2 U per sample, and the allele-specific PCR primers (Table 3) were used at a final concentration of 0.05 μM each. Otherwise, the PCR conditions were as described previously (8). For each combination of allele-specific second-round PCR primer pair and mildewed field sample, two independent NAS-PCR amplifications were performed by using two different DNA solutions extracted independently. Amplified DNA fragments were visualized under UV light after electrophoresis at 100 V on ethidium bromide-stained (0.4 μg/ml) 1% agarose gels electrophoresed in 0.5× Tris-borate-EDTA buffer.
DNA extracted from grapevines and from 34 filamentous fungi and yeasts collected in the field from grapevines and identified by D. Blancard and P. Lecomte (Institut National de la Recherche Agronomique, Bordeaux, France) were tested for amplification by using NAS-PCR. The yeasts and fungi tested were Acremonium sp., Alternaria alternata, Armillaria mellea, Botryotinia fuckeliana, Cephalosporium sp., Chaetomium sp., Coniella diplodiella, Coniothyrium sp., Diplodia natalensis, Elsinoe ampelina, Epicoccum sp., Eutypa lata, Fusarium roseum, Fusarium sp., Gliocladium sp., Greeneria uvicola, Guignardia bidwellii, Kloeckera apiculata, Pestalotia sp., Phellinus ignarius, Phialophora parasitica, Phomopsis viticola, Plasmopara viticola, Pseudopeziza tracheiphila, Rhizopus nigricans, Rosellinia necatrix, Saccharomyces sp., Saccharomycopsis ludwigii, Sphaeropsis sp., Stereum hirsutum, Trichoderma sp., Ulocladium sp., Verticillium dahliae, and Verticillium lecanii.
To confirm the specificity of allele-specific PCR primers for genetic groups, 13 single-spore isolates of U. necator were taken from powdery mildew colonies obtained from grapevines from each of the three vineyards and independently characterized by using allele-specific PCR and RAPD analysis.
RESULTS
RAPD amplifications.
European isolates collected in 1998 and Tunisian, Israeli, and Australian isolates were assigned either to group I or to group III on the basis of their RAPD patterns (Table 1). Australian isolates 920103 and 930202, which were assigned to PCR-restriction fragment length polymorphism group A (10), had RAPD patterns typical of group I, and the six Australian isolates assigned to PCR-restriction fragment length polymorphism group B (10) had RAPD patterns typical of group III. Thus, genetic groups A and B described previously for Australian isolates corresponded to RAPD groups I and III, respectively. The 132 isolates which we studied included 16 group I isolates, 12 group II isolates, and 104 group III isolates (7) (Table 1).
CYP51 cloning and sequencing and allele-specific PCR detection of mutations.
The CYP51 sequences of 12 group III isolates, including 7 isolates from France, 2 isolates from Portugal, 1 isolate from Switzerland, 1 isolate from Germany, and 1 isolate from Israel, were determined previously (8). In this study, we determined the CYP51 sequences of 10 additional isolates, including 3 group I isolates (isolates FCP1.1 [France] [7], GNE1.1 [Germany] [7], and 930202 [Australia] [Table 1]), 1 group II isolate (isolate IHY1.1 [India] [7]), and 6 group III isolates (isolates SLE1.1 and SNO2.1 [Switzerland] [7], IBA1.2 and IPA1.1 [India] [7], PVA2.1 [Portugal] [7], and 931502 [Australia] [Table 1]).
Of the 18 CYP51 sequences determined for group III isolates, 5 had a point mutation at nucleotide 462 that was associated with high levels of resistance to a fungicide that inhibits CYP51 (8). The CYP51 sequences of the 13 other group III isolates studied were identical to the sequences of fungicide-sensitive European isolates (reference sequences).
Three point mutations were found in the CYP51 sequences of the three group I isolates and one group II isolate studied (GenBank accession no. AF042067). The sequence changes were a G-T transversion at nucleotide position 110 corresponding to a Gly-37–Val substitution, a T-C transition at nucleotide position 575 resulting in a Ile-156–Thr substitution, and a C-T transversion at nucleotide position 1587 (Arg-493) that is silent.
Amplifications performed with allele-specific PCR primers U14DM and MUT2(I-II), primers M1I and MUT3(I-II), and primers M1 and MUT4(I-II) yielded fragments of the expected sizes (125, 447, and 613 bp, respectively) for all 16 group I isolates and all 12 group II isolates (Fig. 1). No amplification was obtained with DNA extracted from the 104 group III isolates, while a fragment of the expected size (1,756 bp) was obtained from these DNA samples when we used primers C14 and C14R as a control for PCR efficiency. Conversely, amplifications performed with primers U14DM and MUT2(III), primers M1I and MUT3(III), and primers M1 and MUT4(III) yielded fragments of the expected sizes for all 104 group III isolates. No amplification was obtained with DNA extracted from all 16 group I isolates and all 12 group II isolates, while a fragment of the expected size was obtained when primers C14 and C14R were used (data not shown).
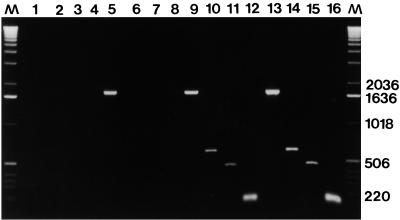
FIG. 1.
CYP51 allele-specific PCR products obtained with DNA extracted from representative U. necator isolates, including isolates FPE1.1 (group III) (lanes 5 through 8), IHY1.1 (group II) (lanes 9 through 12), and GNE1.1 (group I) (lanes 13 through 16). Lanes 1 through 4 contained an H2O negative control (no DNA). The primers used were primers C14 and C14R (lanes 1, 5, 9, and 13), primers M1 and MUT4(I-II) (lanes 2, 6, 10, and 14), primers MUT3(I-II) and M1I (lanes 3, 7, 11, and 15), and primers U14DM and MUT2(I-II) (lanes 4, 8, 12, and 16). Lanes M contained a molecular weight marker (1-kb DNA ladder [Gibco BRL]). Fragment sizes (in base pairs) are indicated on the right.
ITS and 5.8S rDNA cloning and sequencing and allele-specific PCR detection of a mutation.
A 559-bp DNA fragment encompassing ITS1, most of ITS2, and 5.8S rDNA was cloned from group I isolate FCP1.1, group II isolate ISA1.1, and group III isolate PVA2.1 (7) and sequenced. The 5.8S rDNA sequence was identical to the 5.8S rDNA sequence of Uncinula adunca, the organism that is most closely related to U. necator for which this sequence is available (14). Group I and II isolates had identical ITS and rDNA sequences (GenBank accession no. AF049332). The ITS1 sequences of these isolates differed from the ITS1 sequence of the group III isolate (GenBank accession no. AF049331) by a single C-to-T substitution at nucleotide 70.
Amplifications with primers UITS and MIT1(I-II) and primers UITS and MIT1(III) were performed with the same 132 DNA samples used for detection of mutations in CYP51. Amplification of a DNA fragment of the expected size (84 bp) was obtained with all of the group I and II isolates when we used primers UITS and MIT1(I-II) but not with any of the group III isolates. Conversely, amplification of a DNA fragment of the expected size was obtained with all group III isolates when we used primers UITS and MIT1(III) but not with any of the group I and II isolates (data not shown).
NAS-PCR analysis.
NAS-PCR amplifications performed with DNA extracted from grapevines and from 34 yeast and fungal species associated with grapevines in vineyards yielded no amplified fragments. All NAS-PCR amplifications performed with DNA extracted from samples containing U. necator yielded three major fragments (Fig. 2), one of which was polymorphic.
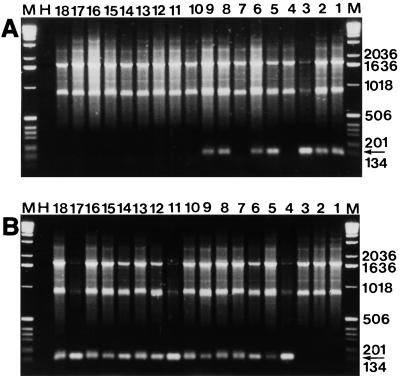
FIG. 2.
NAS-PCR products obtained from 18 DNA samples by using primers U14DM and MUT2(I-II) (A) and primers U14DM and MUT2(III) (B) in the second-round PCR. DNA was extracted from samples collected in Montirat (Table 2). The Sampling dates were 5 May (lanes 1 through 3), 24 June (lanes 4 through 9), 24 July (lanes 10 through 14), and 8 September (lanes 15 through 18). The arrows indicate the position of the 125-bp DNA fragment specifically amplified from isolates belonging to genetic groups I and II (A) or genetic group III (B). Lanes H contained an H2O control (no DNA). Lanes M contained a molecular weight marker (1-kb DNA ladder [Gibco BRL]). Fragment sizes (in base pairs) are indicated on the right.
The size of the polymorphic amplified DNA fragment was identical to the size of the CYP51 fragment which was amplified by using the allele-specific primer pairs in single-round allele-specific PCR. The identity of these fragments was verified by cloning and sequencing.
The size of the largest monomorphic PCR fragment was always identical to the size of the 1,756-bp fragment amplified with primers C14 and C14R. The size of the second monomorphic fragment amplified by NAS-PCR varied depending on the second-round PCR primers used and was always intermediate between the sizes of the fragments amplified with primers C14 and C14R and allele-specific PCR primers. When the fragment specific for genetic group III or the fragment specific for genetic groups I and II was not amplified in a NAS-PCR, the 1,756-bp fragment and the intermediate fragment provided an internal positive control for amplification of U. necator CYP51.
We used NAS-PCR to analyze 56 bulk mildewed samples collected from nine grapevines in three different vineyards. Twelve samples contained only group I and II isolates, 35 samples contained only group III isolates, and 9 samples contained both group I and II isolates and group III isolates (Table 2).
Thirteen single-spore U. necator isolates were obtained from four samples that were putatively infected by only group I and II isolates and from four mixed samples. These isolates were independently analyzed by using RAPD analysis and single-round allele-specific PCR. The RAPD analysis revealed that six isolates were group I isolates and seven isolates were group III isolates. Allele-specific PCR confirmed that the four point mutations found in CYP51 and rDNA distinguished group I from group III.
No group II isolate was found in our samples. As group II isolates have not been found in Europe, we assume that our group I and II primers detected only group I isolates in European samples.
DISCUSSION
A comparison of the CYP51 sequences of three group I U. necator isolates, one group II U. necator isolate, and 18 group III U. necator isolates identified three point mutations that distinguished groups I and II from group III. The mutation at codon 37 causes a Gly-Val substitution in the hydrophobic, N-terminal, putative transmembrane region of CYP51. The mutation at codon 156 causes an Ile-Thr substitution 15 amino acids from the end of the CR2 domain, a highly conserved region that is believed to be involved in enzyme substrate recognition (2). The mutation at codon 493 is silent. The occurrence of three point mutations in CYP51 was surprising, since this gene is highly conserved and is essential for the survival of the fungus (2, 24). Thus, mutations in CYP51 are likely to be either neutral or adaptive but probably not deleterious. The silent mutation at codon 493 is probably neutral, but the biological significance of the mutations at codons 37 and 156 in group I and II CYP51 sequences is not known.
Although ITS rDNA regions have been reported to be highly variable, even in closely related species (14, 15), we found only one point mutation that distinguished group I and II isolates from group III isolates. Thus, we could develop no molecular tool derived from the rDNA sequence to distinguish groups I and II.
The results of allele-specific PCR amplification experiments performed with DNA from 145 single-spore isolates of U. necator (132 isolates listed in reference 7 and Table 1 and 13 isolates obtained from our 1998 population survey) demonstrated that the four point mutations in CYP51 and ITS1 always distinguished groups I and II (34 isolates studied) from group III (111 isolates studied). Based on our sample, we are 95% certain that groups I and II are positively identified at least 91% of the time, and that isolates from group III are positively identified at least 99% of the time.
Using NAS-PCR, we found that in May, flagshoot symptoms were associated with group I isolates (Table 2). During June, group III isolates were first detected, but group I isolates were still detected in two vineyards (Nîmes and Montirat). Only group III isolates were detected in July and September, suggesting that this group represented the majority of U. necator isolates present at the end of the growing season. Our findings are consistent with the hypothesis that there are genetically isolated groups of isolates within U. necator and that these isolates occupy distinct, specialized niches (5, 7), as was shown for other phytopathogenic fungi (13).
The molecular tools which we developed can be used to reliably and efficiently distinguish groups I and II from group III. PCR primers that distinguish group I from group II might be derived from specific RAPD fragments. The NAS-PCR technique is much more suitable than RAPD analysis for field studies, however, since crude samples from the field can be directly processed. Because only 5% of the first-round PCR mixture is required for second-round, allele-specific PCR amplification, numerous second-round PCR amplifications can be performed with a single field sample. We plan to use NAS-PCR to monitor U. necator populations in vineyards in various geographic locations. More generally, NAS-PCR may be very useful for conducting large-scale population genetic studies with diverse organisms. When resistance of an organism to chemical control results from point mutations in the gene encoding the target enzyme, NAS-PCR also may be used in resistance monitoring programs.
ACKNOWLEDGMENTS
This work was supported by grant 970 307 002 from the Conseil Régional d’Aquitaine.
We thank G. Brarda (Chambre d’Agriculture de l’Aude, Carcassonne, France), H. Guillemont (Chambre d’Agriculture du Roussillon, Perpignan, France), M. Blanc (Institut Technique de la Vigne de Manosque, Manosque, France), and B. Molot (Institut Technique de la Vigne de Nîmes, Nîmes, France) for collecting and mailing grapevine mildewed samples. We thank D. Blancard and P. Lecomte (Station de Pathologie Végétale, Institut National de la Recherche Agronomique, Bordeaux, France) for providing DNA samples from yeasts and fungi associated with grapevines.
REFERENCES
- 1.Amicucci A, Zambonelli A, Giomaro G, Potenza L, Stocchi V. Identification of ectomycorrhizal fungi of the genus Tuber by species-specific ITS primers. Mol Ecol. 1998;7:273–277. [Google Scholar]
- 2.Aoyama Y, Noshiro M, Gotoh O, Imaoka S, Funae Y, Kurosawa N, Horiuchi T, Yoshida Y. Sterol 14-demethylase P450 (P45014DM) is one of the most ancient and conserved P450 species. J Biochem. 1996;119:926–933. doi: 10.1093/oxfordjournals.jbchem.a021331. [DOI] [PubMed] [Google Scholar]
- 3.Boubals D. Étude des causes de la résistance des Vitacées à l’Oïdium de la vigne (Uncinula necator (Schw.) Burr.) et de leur mode de transmission héréditaire. Ann Amelior Plant (Paris) 1961;11:401–500. [Google Scholar]
- 4.Delfau M H, Kerckaert J P, Collyn d’Hooghe M, Fenaux P, Lai J L, Jouet J P, Grandchamp B. Detection of minimal residual disease in chronic myeloid leukemia patients after bone marrow transplantation by polymerase chain reaction. Leukemia. 1990;4:1–5. [PubMed] [Google Scholar]
- 5.Délye C, Corio-Costet M-F. Origin of primary infections of grape by Uncinula necator: RAPD analysis discriminates two biotypes. Mycol Res. 1998;102:83–288. [Google Scholar]
- 6.Délye C, Corio-Costet M-F, Laigret F. A RAPD assay for strain typing of the biotrophic grape powdery mildew fungus Uncinula necator using DNA extracted from the mycelium. Exp Mycol. 1995;19:234–237. [Google Scholar]
- 7.Délye C, Laigret F, Corio-Costet M-F. RAPD analysis provides insight into the biology and the epidemiology of Uncinula necator. Phytopathology. 1997;87:670–677. doi: 10.1094/PHYTO.1997.87.7.670. [DOI] [PubMed] [Google Scholar]
- 8.Délye C, Laigret F, Corio-Costet M-F. A mutation in the 14α-demethylase gene of Uncinula necator that correlates with high levels of resistance to a sterol biosynthesis inhibitor. Appl Environ Microbiol. 1997;63:2966–2970. doi: 10.1128/aem.63.8.2966-2970.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Délye C, Laigret F, Corio-Costet M-F. Cloning and sequence analysis of the eburicol 14α-demethylase gene of the obligate biotrophic grape powdery mildew fungus. Gene. 1997;195:29–33. doi: 10.1016/s0378-1119(97)00141-8. [DOI] [PubMed] [Google Scholar]
- 10.Evans K J, Whisson D L, Stummer B E, Scott E S. DNA markers identify variation in Australian populations of Uncinula necator. Mycol Res. 1997;101:923–932. [Google Scholar]
- 11.Gadoury D M, Pearson R C. Initiation, development, dispersal and survival of cleistothecia of Uncinula necator in New York vineyards. Phytopathology. 1988;78:1413–1421. [Google Scholar]
- 12.Gadoury D M, Pearson R C. Germination of ascospores and infection of Vitis by Uncinula necator. Phytopathology. 1990;80:1198–1203. [Google Scholar]
- 13.Giraud T, Fortini D, Levis C, Leroux P, Brygoo Y. RFLP markers show genetic recombination in Botryotinia fuckeliana (Botrytis cinerea) and transposable elements reveal two sympatric species. Mol Biol Evol. 1997;14:1177–1185. doi: 10.1093/oxfordjournals.molbev.a025727. [DOI] [PubMed] [Google Scholar]
- 14.Hirata T, Takamatsu S. Nucleotide sequence diversity of rDNA internal transcribed spacers from conidia and cleistothecia of several powdery mildew fungi. Mycoscience. 1996;37:283–288. [Google Scholar]
- 15.Kiss L. Genetic diversity in Ampelomyces isolates, hyperparasites of powdery mildew fungi, inferred from RFLP analysis of the rDNA ITS region. Mycol Res. 1997;101:1073–1080. [Google Scholar]
- 16.Pearson R C, Gadoury D M. Cleistothecia, the source of primary inoculum for grape powdery mildew in New York. Phytopathology. 1987;77:1509–1514. [Google Scholar]
- 17.Pearson R C, Gärtel W. Occurrence of hyphae of Uncinula necator in buds of grapevine. Plant Dis. 1985;69:149–151. [Google Scholar]
- 18.Sall M A, Wrysinski J. Perennation of powdery mildew in buds of grapevines. Plant Dis. 1982;66:678–679. [Google Scholar]
- 19.Simon L, Lalond M, Bruns T D. Specific amplification of 18S fungal ribosomal genes from vesicular arbuscular endomycorrhizal fungi colonizing roots. Appl Environ Microbiol. 1992;58:291–295. doi: 10.1128/aem.58.1.291-295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sommer S S, Groszbar A R, Bottema C D K. PCR amplification of specific alleles (PASA) is a general method for rapidly detecting known single base-pair changes. BioTechniques. 1992;12:82–87. [PubMed] [Google Scholar]
- 21.White T J, Bruns T D, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal genes for phylogenetics. In: Innis M A, Gelfrand D H, Sninsky J J, White T J, editors. PCR protocols. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 22.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willocquet L. Ph. D. thesis. Paris, France: Université Paris XI; 1994. [Google Scholar]
- 24.Yoshida Y. Lanosterol 14α-demethylase (cytochrome P45014DM) In: Schenkman H, Grein K, editors. Cytochromes P 450. Berlin, Germany: Springer-Verlag; 1993. pp. 627–639. [Google Scholar]