Abstract
Hepatitis B virus (HBV) DNA integration is an incidental event in the virus replication cycle and occurs in less than 1% of infected hepatocytes during viral infection. However, HBV DNA is present in the genome of approximately 90% of HBV-related HCCs and is the most common somatic mutation. Whole genome sequencing of liver tissues from chronic hepatitis B patients showed integration occurring at random positions in human chromosomes; however, in the genomes of HBV-related HCC patients, there are integration hotspots. Both the enrichment of the HBV-integration proportion in HCC and the emergence of integration hotspots suggested a strong positive selection of HBV-integrated hepatocytes to progress to HCC. The activation of HBV integration hotspot genes, such as telomerase (TERT) or histone methyltransferase (MLL4/KMT2B), resembles insertional mutagenesis by oncogenic animal retroviruses. These candidate oncogenic genes might shed new light on HBV-related HCC biology and become targets for new cancer therapies. Finally, the HBV integrations in individual HCC contain unique sequences at the junctions, such as virus-host chimera DNA (vh-DNA) presumably being a signature molecule for individual HCC. HBV integration may thus provide a new cell-free tumor DNA biomarker to monitor residual HCC after curative therapies or to track the development of de novo HCC.
Keywords: Hepatitis B Virus, Insertional Mutagenesis, Liver Cancer, Cell-Free Tumor DNA, Virus-Host Chimera DNA
Abbreviations used in this paper: AFP, alpha-fetoprotein; Cap-seq, capture sequencing; cfDNA, cell-free DNA; CHB, chronic HBV infection; CHIP, clonal hyperplasia with indeterminate potential; DR, direct repeat; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; NA, nucleos(t)ide analog; NGS, next-generation sequencing; PCR, polymerase chain reaction; PIVKA-II, protein induced by vitamin K absence or antagonists-II; RNA-seq, RNA sequencing; vh-DNA, virus-host chimera DNA; WGS, whole genome sequencing
Summary.
Mechanistic studies of HBV integration-induced insertional mutagenesis help develop precise HCC therapy. HBV integration provides a unique junctional DNA biomarker valuable for monitoring residual HCC after therapy or the development of de novo recurrence.
Hepatocellular carcinoma (HCC) ranks 6th among the 10 leading human cancers worldwide.1 This disease burden is especially heavy in Asia and Africa, largely because of the high prevalence of chronic hepatitis B virus (HBV) infection (CHB), ranging from 3% to 15% among the local populations. Overall, CHB is present in 50%–60% of all HCC cases worldwide.2 To control CHB, universal HBV vaccination programs have been implemented in most countries since 1986, resulting in a sharp decline in HBV carriers among children and young adults from 10% to 0.5%, as in the example of Taiwan.3,4 This is accompanied by a reduction in HCC in children and teenagers, confirming the carcinogenic effect of CHB.5 For CHB patients, active antiviral therapies such as nucleos(t)ide analog (NA) reverse transcriptase inhibitors have been widely adopted since 2000. Effective control of viral replication results in much less hepatitis activity, improving the clinical outcomes of CHB and reducing the death rate from cirrhosis, hepatic failure, and HCC to 50% of historic levels. These combined efforts have led to a significant decline in HBV-related end-stage liver diseases and death, as witnessed in the last decade.6 Despite this impressive progress in controlling liver cirrhosis and hepatic failure resulting from the use of long-term NA therapies, the absolute cases of HBV-related HCC were actually only modestly reduced, largely because of an accumulation of surviving CHB patients with advanced liver fibrosis or cirrhosis who still carry a high risk of HCC. Therefore, the major challenge for CHB patients in the NA therapy era is to decrease development of HCC.
Chronic HBV Infection in Contributing to HCC
CHB has been shown to increase HCC risk by approximately 10- to 30-fold compared with the risk in the non-HBV population.7 The lifelong risk for HCC in CHB patients is estimated to be approximately 30% in male carriers and 10% in female carriers. Both direct and indirect mechanisms are generally assumed to contribute to carcinogenesis. Chronic viral hepatitis creates a microenvironment in the liver for cycles of hepatocyte death and regeneration, favoring the growth of hepatocytes with proliferation advantages, eventually leading to cancer. The direct mechanisms include transforming viral proteins, such as the well-known HBx or deleted Pre-S,8, 9, 10 and even more important is HBV DNA integration in the genomes of HBV-related HCC cells. This review will focus on the importance of HBV DNA integration as the driving force in HCC carcinogenesis, with implications for targeted therapies and even as a novel liquid biopsy cancer biomarker.
Somatic Mutations in the Genomes of HBV-Related HCC: Host Genes and HBV DNA Integration
Similar to most human cancers, HCC is considered a disease caused by accumulated somatic mutations in the chromosomes of hepatocytes. Comprehensive whole genome sequencing (WGS) of HCC identified some well-known somatic mutations of host genes. Among them, the TERT promoter -124G>A or -146G>A point mutations were the most frequently identified, which are detected in up to 60% of non-HBV HCCs and approximately 30% of HBV-related HCCs; others are TP53, CTNNB1, AXIN, and ARID1/2.11,12 However, these common somatic mutations mostly occur in the early or advanced stages of HCC. In the micro-dissected regeneration nodules of cirrhosis, the well-accepted precursor of HCC, comprehensive genomic sequencing did not identify these common somatic mutations (not even TERT promoter point mutations). Only increased mutation at different single nucleotide positions was noted.13 Therefore, the common mutations found in early or advanced HCC are more likely the ones contributing to cancer development in concert with the initial driver mutations, presumably already occurring and existing in HCC precursors. Such driver mutation(s), which may confer capabilities for the hepatocytes to undergo clonal expansion, have not yet been identified or verified.
However, the most common mutation in HBV-related HCC is HBV DNA integration into chromosomes. This was first discovered in the 1980s via the classic Southern blot hybridization of restriction enzyme-digested HCC chromosomes with an HBV probe.14,15 Approximately 90% of HBV-related HCC cells contain integrated HBV DNA, usually at multiple chromosomal sites.11,16, 17, 18, 19, 20 Although HBV, like a retrovirus, replicates through reverse transcription of viral pregenomic RNA, its final product is an episomal, covalently close circular DNA21 instead of the retroviral DNA provirus that is integrated into the chromosomes of infected cells. HBV DNA integration is not a mandatory step for viral replication but is considered to arise from dead-end viral double-strand linear DNA, which probably integrates into the chromosome via a nonviral integrase-dependent process.22 Therefore, HBV DNA integration is an incidental event during HBV infection and can be found in only 0.1%–1% of infected hepatocytes but becomes the most common mutation in the majority of HBV-related HCC cases.23
Platforms for Comprehensively Identifying HBV DNA Integrations in HCC
The platforms for discovering HBV integration sites in the chromosomes of HCC cells have progressed rapidly from classic Southern blot to inverse polymerase chain reaction (PCR), which allows for a rapid estimation of copies of HBV integrations in HCC or precancer lesions but has limited value in identifying the details of HBV integration at the nucleotide sequence level. With the advent of the genomics era and the development of mass parallel sequencing technology, multiple next-generation sequencing (NGS) platforms have been used to generate exact sequence information of HBV integrations. Such platforms include WGS, RNA sequencing (RNA-seq), and capture sequencing (Cap-seq).
WGS provides unbiased genome-wide sequencing information to identify HBV integrations at single-nucleotide resolution but requires large sequencing capacity to consistently detect HBV integration regions in the entire human genome. The sequencing of detected HBV integration regions in previous WGS studies has usually required a total output of 60–100 GB or even more for a single sample, of which less than 5% of sequenced reads are HBV-related.16,24,25
On the other hand, RNA-seq provides sequencing information of the transcriptome and identifies HBV integrations that result in transcribed hybrid mRNA transcripts and may have notable effects. The detection limit of HBV integrations by RNA-seq faces a similar issue as genome-wide DNA sequencing and is dependent on overall gene expression levels at the integration sites. Furthermore, HBV integrations within introns and regulatory regions are not within the RNA-seq detection scope.
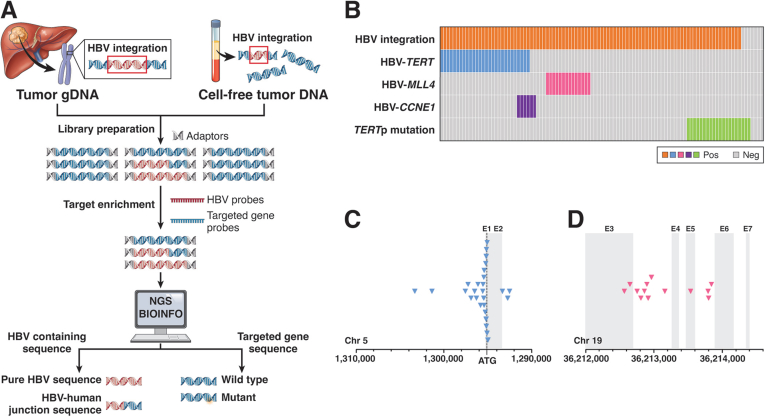
Cap-seq uses specially designed capture probes based on HBV genomic sequences to enrich HBV-containing DNA fragments through capture hybridization (Figure 1A) by removing most nonviral human genome sequences. The advantage of this Cap-seq platform is a more efficient reading of junctional DNA fragments (from the HBV integration sites) with lower sequencing output (1–5 GB to be sufficient) and more robust identification of junction sites by higher sequence readings than WGS. Recent studies have used Cap-seq to detect trace amounts of HBV integration fragments from not only HCC tissues20 but also circulating cell-free DNA (cfDNA) collected from blood samples of HCC patients.26 Although Cap-seq detection of HBV junction fragments may be biased because of the HBV sequence used for designing the capture probes, it remains the most efficient platform for identifying them from HCC tissues or blood (Figure 1A).
Figure 1.
HBV DNA integration in HBV-related HCCs. (A) Flowchart depicts the process of HBV-probe Cap-seq identification of virus-host chimera DNA from junction fragments of HBV DNA integrations in HCC, liver tissues, or circulating cell-free DNAs. Human sequences are in blue, but viral sequences are in red, and DNA fragments with blue and red sequences are junction fragments. (B) Distribution of HBV DNA integration, integration hotspots, and TERT promoter mutations in 101 HBV-related HCCs. Each column represents 1 case, and the solid color indicates its somatic mutations detected. (C) Distribution of HBV integration breakpoints (blue arrowhead) in the TERT gene promoter and exons in HBV-related HCCs. (D) Distribution of HBV integration breakpoints (pink arrowhead) in MLL4 (KMT2B) exons and introns. Modified from reference 19.
HBV integrations can be directly detected by nucleotide sequencing. Direct detection of HBV integrations, such as that described in HIVID27 or Li et al,19 is performed by mapping sequencing reads to both the host human genome and the viral HBV genome. Therefore, HBV integration sites can be exactly identified at a single-nucleotide level from overlapping, multiple reads that map to the same position of the viral genome in one site and the human genome in the other. Currently, direct detection methods are more informative with sufficient sequencing depth because they can provide definitive positional information of the integration site while the resolution differences will be minimal.
Current Illumina-based sequencing platforms generate 150 bp sequencing reads and are capable of detecting HBV integrations but are unable to retrieve the complete sequence of the integrated HBV genomic region within the host human genome. Inferring the complete integrated HBV region from detected HBV integration sites is not plausible because of the extensive sequence recombination events during the HBV integration process. However, third-generation sequencing platforms, such as PacBio and Nanopore can generate contiguous, long reads exceeding 20 kb or even 4 Mb and provide future potential to be applied for the full characterization of the integrated HBV region. Recent studies revealed frequent rearrangements of HBV DNA and translocation between chromosomes that cannot be found by the Illumina platform.20,28, 29, 30 The ability to sequence the full genomic content of the integrated HBV will allow us to better understand the role of integrated HBV toward HCC development.
HBV DNA Integration Database
Multiple online databases have been established that include detailed information on HBV integration sites. The Viral Integration Site DataBase (VISDB, https://bioinfo.uth.edu/VISDB) is a manually curated database that extracts HBV (among other viruses) integration site information from original published studies with additional data incorporated from other biological databases, including oncogene and tumor suppressor gene status, gene expression analysis, miRNA interactions, epigenetic modifications, and gene ontology.31
Different HBV genotypes or haplotypes with specific viral mutations have different integration frequencies and may have distinct effects on HCC occurrence. However, most studies focus on the human genomic site of the HBV integration event and have limited knowledge of the actual integrated HBV sequence or information of any HBV viral mutations within the integrated region. The ViMIC database (http://bmtongji.cn/ViMIC/index.php) is a database that is similar to VISDB, because it also extracts HBV integration site information from published studies.32 Although ViMIC also lacks direct nucleotide sequence information of the integrated HBV regions, it provides indirect viral mutation information by identifying and functionally annotating HBV reference genomes that were identified with HBV integration site information.
Unlike VISDB and ViMIC, which only contain HBV integration information collected from published studies, VIS Atlas (http://www.dsvisdb.tech/) is a database that not only includes reported HBV integration sites but also provides computed HBV integration sites from NGS sequencing data collected from sequencing databases and in-house-developed VIPA bioinformatic pipelines.
General Features of HBV DNA Integration Sites in Viral or Human Genomes
HBV integration is the insertion of viral double-strand linear DNA that is bound by terminal direct repeat (DR) sequences into human genome. Thus, the integrated HBV breakpoints are frequently located around the DR region (approximately 40%–50%), and the remaining integration sites are evenly distributed around viral genomes.11,16, 17, 18, 19,33 As noted in human HCCs, woodchuck HCC with WHV integrations, and adult T-cell leukemia–lymphoma with HTLV integrations, viral genomes frequently undergo complicated recombination or rearrangement, disrupting most viral genes but maintaining intact viral promoters such as enhancer regions.34, 35, 36 In addition, HBV evolves into different genotypes (A to I) and different variants (such as basal core/precore mutants) that have been shown to be relevant to HCC risk.37 Analysis of HBV integration by incorporating these viral variants may elucidate their contribution to carcinogenesis.
HBV DNA integration can be detected in the genome of both nontumor and tumor tissues with HBV-related HCC. In nontumor tissue, integration in the genome was enriched in open chromatin regions with active transcription (more around genes rather than intergenic regions).20 Relative to nontumor tissue, HBV integration is further enriched in the promoter and exon regions in HCC,33 supporting the idea that HBV DNA integration contributes to host gene activation. The detected clonal HBV integration in nontumor tissue is low because the tissue is composed of multiple, small hyperplasia clones. In contrast, a much higher clonal integration detected in HCC reflects expansion from a single lineage.20,38
Evidence for HBV DNA Integrations Important for Carcinogenesis: Positive Selection and Integration Hotspots
As an incidental event in the HBV infection cycle, HBV integration is only present in about 0.1%–1% of HBV infected hepatocytes.23 The integration takes place in the early stage of HBV infections, and hepatocytes with HBV integrations gradually expand clonally as CHB patients age.39,40 These clones are likely the HCC precursors, resulting in 90% of HBV-related HCC containing HBV integrations.11,16, 17, 18, 19, 20 The dramatic enrichment of the HBV integration proportion from 0.1% to 1% in the infection stage to 90% in the HCC stage indicates a strong advantage of HBV-integrated DNA-containing hepatocytes progressing to cancer.
The other evidence for the carcinogenesis potential of HBV integration is the observations of HBV integrations in chronic hepatitis B liver tissues occurring at random in the chromosomes, but there are HBV DNA integration hotspots in the chromosomes of HCC tissues (Figure 1B).11,16,18,19,41 Two major hotspots for HBV integrations have been identified in the genomes of HBV-related HCC: first in the TERT gene promoter (25% of patients) and second in exons 3–6 of the histone methyltransferase MLL4 (12% of patients). A few less common hotspots are cyclin E1 and cyclin A2. The emergence of these HBV integration hotspots suggested positive selection from a library of infected hepatocytes with HBV integration at random. To support this, in rare cases of HCC, another DNA virus AAV, was also found to integrate into the TERT promoter.42,43
Inserted HBV DNA can up-regulate the immediate downstream human gene through viral enhancers and promoters. HBV-TERT integration in tumor tissue tended to occur in the TERT promoter region (Figure 1C), which interrupted the tight suppression of TERT transcription in normal hepatocytes, resulting in the activation of TERT expression.16,19 The expression of telomerase is required for telomere-lengthening enzyme activity to prevent replicative senescence in response to chronic liver damage. Telomerase can also promote proliferation through cooperation with β-catenin and promote WNT/β-catenin signaling pathways.44 The concurrent HBV-TERT/TERT promoter mutation with CTNNB1 mutation in HBV-related HCC and hepatitis C virus–related HCC emphasizes the importance of the synergy between TERT and WNT/β-catenin pathways in viral hepatocarcinogenesis.45,46
Integration of HBV can also regulate the transcription of truncated human gene transcripts or virus-human fusion transcripts. In HCC, HBV integration in MLL4 distributed in exons 3–6 (Figure 1D) leads to the expression of N´-terminal truncated MLL4 and/or HBV-MLL4 fusion transcripts.47, 48, 49, 50 Although the oncogenic mechanism of HBV-MLL4 is not yet well-elucidated, it possibly occurs through epigenetic regulation because the DNA-binding domain and histone methyltransferase enzymatic domain of MLL4 remain intact after HBV integration. The menin binding motif of MLL4 can bring the menin histone methyltransferase complex to specific gene loci targeted by menin. MLL4 recruitment to specific gene loci may therefore be altered by HBV integration (with resulting loss of the MLL4 menin binding motif), leading to aberrant epigenetic regulation of a wide range of target genes. Another example is HBx-LINE1, which is reported as a virus-human functional fusion transcript in HBV-related HCC,51 which promotes the β-catenin signaling pathway by acting as a molecular sponge to prevent miR-122 from targeting WNT1 transcription.52 Local epigenetic regulation may also be changed to turn on transcription because chr.8p11.21 LINE1 region is transcriptionally silent without HBV integration.
In addition to modulating local gene transcription, HBV integration is also involved in architectural changes in the genome, such as gene rearrangement, copy number variation, and interchromosomal fusion. The higher-order structural changes can lead to loss of the tumor suppressor TP53, amplification of the oncogenes TERT and MYC, and telomeric deletion.20,29 Such gross genome aberration may predispose individuals to tumorigenesis and escalate the heterogeneity of individual HCC through different mechanisms.
Production of viral protein is also noted in HBV integration events. The integration breakpoint in HBV is generally located near the DR region, so the commonly expressed viral proteins encoded by HBV integration are HBx and hepatitis B surface antigen (HBsAg) in the form of C´-terminal truncation, full-length, or HBx-human chimeric protein.41 HBx is known to maintain HBV transcription and impede DNA homologous recombination repair through the degradation of Smc5/6,53,54 and HBsAg is known to manipulate the immune system and might increase the survival of hepatocytes with HBV DNA integration. Thus, the viral proteins encoded by some integrated HBV can contribute to carcinogenesis through different mechanisms.
The progression from HBV infection to HCC takes decades, and the infected hepatocytes need to overcome many host defense barriers to be transformed.55 Initially, as hepatocytes harboring integrated HBV DNA are infected by HBV, they are under host immune surveillance and clearance. Determining how integrated HBV DNA allows host cells to survive immune clearance may be a first step in understanding the microenvironment. After HBV clearance or when chronic HBV enters the inactive carrier phase, HBV DNA replication is restricted and evades immune surveillance. However, integrated HBV can still promote carcinogenesis by its regulatory function through DNA insertional mutagenesis. From single hepatocytes to clonal hyperplasia, HBV DNA integrations may enhance cell proliferation with the aid of other accompanying somatic mutations. Subsequently, clonal hyperplasia outgrowth requires functional angiogenesis and the potential for metastasis to progression to HCC. It is imperative to develop experimental systems to investigate the above-mentioned carcinogenic mechanisms by assessing hotspot integration.
Recently, the hydrodynamic injection of target genes56 with sleeping beauty transposase constructs has been adopted to efficiently integrate the target genes into the chromosomes of mouse hepatocytes.57 Follow-up found that overexpression of the TERT gene was insufficient to lead to HCC in mice, but a combination of TERT overexpression with an activated β-catenin gene cassette might succeed. This interesting experimental model may duplicate the carcinogenic process via HBV DNA integrations and allow us to explore the detailed immune or molecular mechanisms during the transformation process. If it can be robustly duplicated, the system will become a useful model to study both immune and carcinogenic processes. It can even be further expanded for screening HCC-specific lead compounds or conducting preclinical cancer treatment evaluation.
Signature Junction Fragments From HBV Integration Sites of Individual HCC as Cancer or Clonal Hyperplasia Biomarkers
Integrated HBV DNA as a Clonality Marker for HCC
Before the genomics era, the HBV DNA integration sites, only revealed by patterns of Southern blot, were unique to an individual HCC, so they could be used as the signature of individual HCCs. Therefore, in the setting of HCC recurrence after surgical resection, the HBV DNA integration patterns of origin versus recurrence can be compared. If the Southern blot patterns of HBV integrations are identical, recurrent HCC definitely arose from residual original HCC. In contrast, a different pattern indicates that the recurrence was a de novo HCC.58 Using this and other similar approaches, approximately 40% of recurrent HCC cases were shown to be de novo cases.59
HCC Clonal Marker in Blood: Junction DNA Fragments (Virus‒Host Chimera DNA) of HBV Integration Sites of HCC in Circulating Cell-free Tumor DNA
HBV DNA integrations of one HBV-related HCC are unique in terms of the exact nucleotide sequences of the viral-host junctions (vh-DNA).60 Junctional DNA fragments can be released into the circulation as cfDNA, similar to host cells or other cancers.61 Therefore, it is interesting to detect junctional fragments in the circulation of HBV-related HCC patients as the liquid biopsy approach adopted for lung or breast cancers.62,63
To explore this possibility, we conducted a study to test whether HBV integration junction fragments can be detected in the blood at the time of HCC resection.64 From the resected HCC tissues (Figure 2A, designated therapy), the unique junctions of individual HCC were identified by the Cap-seq platform (Figure 1A). Next, primers were designed for signature junction fragment for individual HCC for a case-specific droplet digital PCR to assay their presence in blood collected at the same time (Figure 2A, the virus‒host chimera DNA in red). The results were quite encouraging, because for HCC with sizes larger than 1.5 cm, their signature junction DNA fragments could be detected in the circulation, and their abundance correlated well with HCC size.64
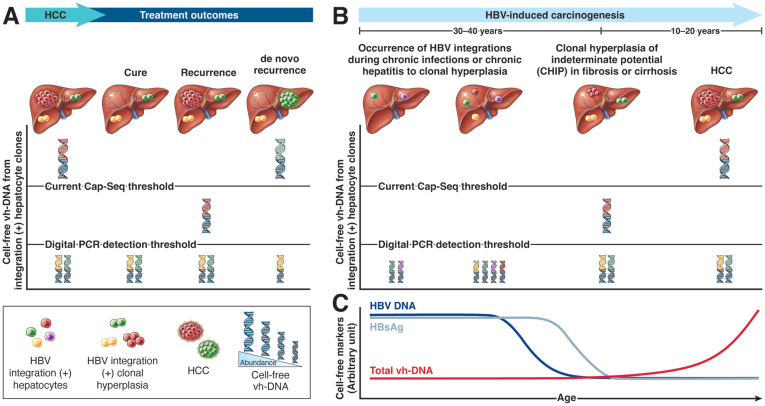
Figure 2.
Cell-free vh-DNA from HBV integration sites is a new biomarker to monitor postresection residual disease or to track clonal hyperplasia to HCC. (A) HCC-specific junction fragments in detecting postresection residual disease and recurrence. After surgical resection, HCC (red tumor in the liver) was removed (dotted circle), and its corresponding circulating vh-chimera DNA (chimera DNA with red for virus sequence and blue for human sequence) was cleared from circulation. If there is residual circulating vh-chimera DNA after resection, intrahepatic HCC is not completely removed and will likely recur (red tumor in the liver). In contrast, for those without residual vh-chimera DNA from the original HCC, recurrence HCC likely arises from a new origin (de novo HCC, green tumor in the liver). Detection of de novo HCC may require Cap-seq to analyze the circulating cell-free DNA for vh-chimeras (chimera DNA with green for the virus sequence and blue for the human sequence). (B) Progression from chronic hepatitis B and clonal hyperplasia to HCC is depicted with circulating vh-chimera DNA. As cells in HCC-prone clonal hyperplasia contain HBV integrations (cells with different colors in the liver), their specific junction fragments are released into the circulation as virus‒host chimera DNA (human sequence in blue and virus sequences from individual clones in red, green, purple, or yellow). The amount of cell-free vh-DNA is positively associated with the size of HBV integration (+) clonal hepatocytes or HCC (indicated by the height of vh-DNA). The threshold of Cap-seq and droplet digital PCR for detecting vh-chimera DNA from HCC is indicated by a straight lines. If the sensitivity and specificity of next-generation Cap-seq can be improved, it might be used to detect the low level of vh-chimera DNA from clonal hyperplasia with indeterminate potential (CHIP). This will be applied to CHIP to HCC in the future without prior knowledge of HBV integration sites. (C) Inverse course of circulating HBV DNA/HBsAg versus virus‒host chimera DNA levels in the progression from chronic hepatitis B to HCC. The level of HBV DNA or HBsAg usually declines, but that of circulating virus‒host chimera DNA increases synchronously with the development of HCC. Refer to reference 64.
Circulating Cell-free Junction Fragment (Virus‒Host Chimera DNA) to Monitor Residual Diseases and Predict HCC Recurrence After Surgical Resection
Moreover, the signature junction fragment can be used as a biomarker to study residual disease after HCC resection and as a predictor of recurrence. Post-resection recurrence of HCC is a common clinical problem, occurring at a rate of approximately 20% in 1 year and 60% in 5 years. In the previous study, blood samples were collected at 2 months after surgical resection of HCC, and the signature junction fragment of HCC was assayed by case-specific digital PCR for individual HCC (Figure 2A, HCC and chimera DNA in red color).64 As expected, the majority of patients were found not to have detectable vh-DNA in their blood, indicating no or minimal residual HCC. However, 23% of patients still had junction fragments of the original HCC in their blood samples, suggesting a notable residual HCC in the liver. Consistent with this interpretation, cases with positive droplet digital PCR results at 2 months after surgery experienced a significantly higher risk of HCC recurrence at 2 years than those negative for digital PCR (90% vs 38%). This interesting finding warrants a prospective clinical study to validate its significance in detecting minimal residual HCC after curative therapies.
New Junction Fragments in the Circulation as Biomarkers for de Novo HCC?
For those negative for droplet digital PCR for junction fragments in the postoperative blood, recurrence is likely a de novo recurrence, distinct from the original HCC (Figure 2A, the HCC and vh-DNA in green color). Therefore, in such cases, digital PCR specific to the original HCC is useless for detection because of different HBV integration sites between the 2 HCCs, even occurring in the same patient. In the absence of HCC tissues, we can try to identify new vh-DNA released from de novo HCC by applying the Cap-seq platform directly to plasma samples. Our initial experience suggested that the Cap-seq technique could discover the junctional fragments corresponding to those in some HCCs, but the success depends on the size of the HCC; assay specificity and sensitivity are good for HCCs of approximately 4–5 cm (straight line cap-seq threshold) but are not yet reliable or accurate enough for smaller HCCs. Despite this limitation of the current Cap-seq platform, it serves as proof of concept that junctional fragments from sufficient-size HCC can actually be detected in circulation.
Prospect for Monitoring Clonal Hyperplasia in HCC via Circulating Cell-free Junctional Fragments
After decades of CHB, infected hepatocytes with HBV integrations likely gain growth advantage and become clonal hyperplasia (labeled in red, green, purple, and yellow cells in the liver, Figure 2B). To cite the framework from clonal hematopoiesis to leukemia, these clones in the liver can be labeled clonal hyperplasia with indeterminate potential (CHIP).65 Certain clones (red cells) may continue to proliferate because of accompanying somatic mutations or an appropriate microenvironment and eventually become HCC (red cells with corona). Others remain as CHIP (green or yellow cells), but rare ones may disappear (purple cells). The evolution of these clones to HCC in the liver is currently monitored by serum alpha-fetoprotein (AFP), protein induced by vitamin K absence or antagonist-II (PIVKA-II), or medical imaging,66 which are limited by the lack of information for individual clone identity. Because every CHIP likely contains unique HBV integrations in the chromosomes, such junctional fragments (short vh-DNAs) can be released into circulation. A faithful characterization of total vh-DNA may give an overall landscape of these CHIPs and provide an opportunity to follow their evolution to clinical HCC in the follow-up of CHB patients. The efficiency of coverage performance of cfDNA Cap-seq is greatly influenced by the cfDNA extraction and hybridization method as well as the subsequent sequencing platform. A recent study reported that different DNA/RNA probe types have distinct capture biases and show differences in their Cap-seq performances. The results indicated that RNA capture probes achieved higher target rates and capture efficiencies than DNA capture probes and thus have stronger binding power, whereas DNA capture probes perform better in GC-rich regions.67 This finding suggests the potential for further optimizing current Cap-seq methods by implementing a combination of DNA and RNA capture probes or other new techniques to increase the sensitivity and detection power for minimal amounts of junction fragments within cf-DNA. If the Cap-seq platform can be better refined to reliably detect the rare junction fragments coming from CHIP or early/small HCC, the task will become feasible and possible to conduct in a real-time manner.
When chronic hepatitis B patients are aging, clinically useful circulating markers for viral disease activities such as HBV DNA or HBsAg usually decline progressively (Figure 2C).68 Despite the diminishing levels of viral markers, in contrast, their HCC risk is increasing. This is because natural immunity or antiviral therapy can only remove or suppress hepatocytes or CHIP still expressing viral proteins, but the majority of CHIP, even those containing integrated HBV DNA, do not express HBV proteins and thus escape from those defense mechanisms to maintain their carcinogenesis potential. Moreover, for CHB patients who achieve functional cure (HBsAg clearance), the risk of HCC is reduced but not eliminated, especially for those with advanced fibrosis or cirrhosis or older age.69,70 Therefore, clinicians need a circulating biomarker to monitor the development of HCC among CHB patients under long-term NA therapies with declining HBsAg levels or even after HBsAg clearance.71 As the amounts of circulating junction fragments released into blood by CHIP (HCC precursors) or HCC increase in parallel with their growth, they are ideal molecules for tracking the progression of these clones and even HCC (Figure 2C). We envision circulating vh-DNA as a useful new biomarker,72 complementing AFP or PIVKA-II, to monitor the process from clonal hyperplasia evolution into HCC during or even after chronic hepatitis B cure.
Footnotes
Conflicts of interest These authors disclose the following: PJC is a consultant and received a grant from TCM Biotech. STT and YCW are employees of TCM Biotech. The remaining authors disclose no conflicts.
Funding Supported by grants from the Ministry of Science and Technology, Taiwan (MOST111-2634-F-002-017-, MOST111-2326-B-002-017-) and the “Center of Precision Medicine” from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.de Martel C., Georges D., Bray F., et al. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health. 2020;8:e180–e190. doi: 10.1016/S2214-109X(19)30488-7. [DOI] [PubMed] [Google Scholar]
- 3.Ni Y.H., Huang L.M., Chang M.H., et al. Two decades of universal hepatitis B vaccination in taiwan: impact and implication for future strategies. Gastroenterology. 2007;132:1287–1293. doi: 10.1053/j.gastro.2007.02.055. [DOI] [PubMed] [Google Scholar]
- 4.Ni Y.H., Chang M.H., Huang L.M., et al. Hepatitis B virus infection in children and adolescents in a hyperendemic area: 15 years after mass hepatitis B vaccination. Ann Intern Med. 2001;135:796–800. doi: 10.7326/0003-4819-135-9-200111060-00009. [DOI] [PubMed] [Google Scholar]
- 5.Chang M.H., Chen C.J., Lai M.S., et al. Universal hepatitis B vaccination in Taiwan and the incidence of hepatocellular carcinoma in children: Taiwan Childhood Hepatoma Study Group. N Engl J Med. 1997;336:1855–1859. doi: 10.1056/NEJM199706263362602. [DOI] [PubMed] [Google Scholar]
- 6.Choi J., Han S., Kim N., et al. Increasing burden of liver cancer despite extensive use of antiviral agents in a hepatitis B virus-endemic population. Hepatology. 2017;66:1454–1463. doi: 10.1002/hep.29321. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y.T., Jen C.L., Yang H.I., et al. Lifetime risk and sex difference of hepatocellular carcinoma among patients with chronic hepatitis B and C. J Clin Oncol. 2011;29:3643–3650. doi: 10.1200/JCO.2011.36.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi M.S., Kim D.Y., Lee D.H., et al. Clinical significance of pre-S mutations in patients with genotype C hepatitis B virus infection. J Viral Hepat. 2007;14:161–168. doi: 10.1111/j.1365-2893.2006.00784.x. [DOI] [PubMed] [Google Scholar]
- 9.Lin C.L., Liu C.H., Chen W., et al. Association of pre-S deletion mutant of hepatitis B virus with risk of hepatocellular carcinoma. J Gastroenterol Hepatol. 2007;22:1098–1103. doi: 10.1111/j.1440-1746.2006.04515.x. [DOI] [PubMed] [Google Scholar]
- 10.Kim C.M., Koike K., Saito I., et al. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 11.Totoki Y., Tatsuno K., Covington K.R., et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 12.Calderaro J., Couchy G., Imbeaud S., et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67:727–738. doi: 10.1016/j.jhep.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Brunner S.F., Roberts N.D., Wylie L.A., et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 2019;574:538–542. doi: 10.1038/s41586-019-1670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brechot C., Pourcel C., Louise A., et al. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286:533–535. doi: 10.1038/286533a0. [DOI] [PubMed] [Google Scholar]
- 15.Edman J.C., Gray P., Valenzuela P., et al. Integration of hepatitis B virus sequences and their expression in a human hepatoma cell. Nature. 1980;286:535–538. doi: 10.1038/286535a0. [DOI] [PubMed] [Google Scholar]
- 16.Sung W.K., Zheng H., Li S., et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto A., Totoki Y., Abe T., et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L.H., Liu X., Yan H.X., et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7 doi: 10.1038/ncomms12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C.L., Li C.Y., Lin Y.Y., et al. Androgen receptor enhances hepatic telomerase reverse transcriptase gene transcription after hepatitis B virus integration or point mutation in promoter region. Hepatology. 2019;69:498–512. doi: 10.1002/hep.30201. [DOI] [PubMed] [Google Scholar]
- 20.Peneau C., Imbeaud S., La Bella T., et al. Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut. 2022;71:616–626. doi: 10.1136/gutjnl-2020-323153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Summers J., Mason W.S. Replication of the genome of a hepatitis B–like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 22.Tu T., Zhang H., Urban S. Hepatitis B virus DNA integration: in vitro models for investigating viral pathogenesis and persistence. Viruses. 2021;13 doi: 10.3390/v13020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tu T., Budzinska M.A., Vondran F.W.R., et al. Hepatitis B virus DNA integration occurs early in the viral life cycle in an in vitro infection model via sodium taurocholate cotransporting polypeptide-dependent uptake of enveloped virus particles. J Virol. 2018;92 doi: 10.1128/JVI.02007-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Z., Jhunjhunwala S., Liu J., et al. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 2012;22:593–601. doi: 10.1101/gr.133926.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujimoto A., Furuta M., Totoki Y., et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 26.Chen W., Zhang K., Dong P., et al. Noninvasive chimeric DNA profiling identifies tumor-originated HBV integrants contributing to viral antigen expression in liver cancer. Hepatol Int. 2020;14:326–337. doi: 10.1007/s12072-020-10016-2. [DOI] [PubMed] [Google Scholar]
- 27.Li W., Zeng X., Lee N.P., et al. HIVID: an efficient method to detect HBV integration using low coverage sequencing. Genomics. 2013;102:338–344. doi: 10.1016/j.ygeno.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Ramirez R., van Buuren N., Gamelin L., et al. Targeted long-read sequencing reveals comprehensive architecture, burden, and transcriptional signatures from hepatitis B virus-associated integrations and translocations in hepatocellular carcinoma cell lines. J Virol. 2021;95 doi: 10.1128/JVI.00299-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez E.G., Demeulemeester J., Otero P., et al. Aberrant integration of hepatitis B virus DNA promotes major restructuring of human hepatocellular carcinoma genome architecture. Nat Commun. 2021;12:6910. doi: 10.1038/s41467-021-26805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Buuren N., Ramirez R., Soulette C., et al. Targeted long-read sequencing reveals clonally expanded HBV-associated chromosomal translocations in patients with chronic hepatitis B. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2022.100449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang D., Li B., Xu T., et al. VISDB: a manually curated database of viral integration sites in the human genome. Nucleic Acids Res. 2020;48:D633–D641. doi: 10.1093/nar/gkz867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Tong Y., Zhang Z., et al. ViMIC: a database of human disease-related virus mutations, integration sites and cis-effects. Nucleic Acids Res. 2022;50:D918–D927. doi: 10.1093/nar/gkab779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Zhang J., Yang Z., et al. The function of targeted host genes determines the oncogenicity of HBV integration in hepatocellular carcinoma. J Hepatol. 2014;60:975–984. doi: 10.1016/j.jhep.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 34.Ogston C.W., Jonak G.J., Rogler C.E., et al. Cloning and structural analysis of integrated woodchuck hepatitis virus sequences from hepatocellular carcinomas of woodchucks. Cell. 1982;29:385–394. doi: 10.1016/0092-8674(82)90155-6. [DOI] [PubMed] [Google Scholar]
- 35.Uren A.G., Kool J., Berns A., et al. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 36.Bushman F.D. Retroviral insertional mutagenesis in humans: evidence for four genetic mechanisms promoting expansion of cell clones. Mol Ther. 2020;28:352–356. doi: 10.1016/j.ymthe.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H.I., Yeh S.H., Chen P.J., et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:1134–1143. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mason W.S., Gill U.S., Litwin S., et al. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology. 2016;151:986–998 e4. doi: 10.1053/j.gastro.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu T., Mason W.S., Clouston A.D., et al. Clonal expansion of hepatocytes with a selective advantage occurs during all stages of chronic hepatitis B virus infection. J Viral Hepat. 2015;22:737–753. doi: 10.1111/jvh.12380. [DOI] [PubMed] [Google Scholar]
- 40.Mason W.S., Jilbert A.R., Litwin S. Hepatitis B virus DNA integration and clonal expansion of hepatocytes in the chronically infected liver. Viruses. 2021;13 doi: 10.3390/v13020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toh S.T., Jin Y., Liu L., et al. Deep sequencing of the hepatitis B virus in hepatocellular carcinoma patients reveals enriched integration events, structural alterations and sequence variations. Carcinogenesis. 2013;34:787–798. doi: 10.1093/carcin/bgs406. [DOI] [PubMed] [Google Scholar]
- 42.Nault J.C., Datta S., Imbeaud S., et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47:1187–1193. doi: 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- 43.La Bella T., Imbeaud S., Peneau C., et al. Adeno-associated virus in the liver: natural history and consequences in tumour development. Gut. 2020;69:737–747. doi: 10.1136/gutjnl-2019-318281. [DOI] [PubMed] [Google Scholar]
- 44.Park J.I., Venteicher A.S., Hong J.Y., et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nault J.C., Mallet M., Pilati C., et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218. doi: 10.1038/ncomms3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pezzuto F., Izzo F., Buonaguro L., et al. Tumor specific mutations in TERT promoter and CTNNB1 gene in hepatitis B and hepatitis C related hepatocellular carcinoma. Oncotarget. 2016;7:54253–54262. doi: 10.18632/oncotarget.9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saigo K., Yoshida K., Ikeda R., et al. Integration of hepatitis B virus DNA into the myeloid/lymphoid or mixed-lineage leukemia (MLL4) gene and rearrangements of MLL4 in human hepatocellular carcinoma. Hum Mutat. 2008;29:703–708. doi: 10.1002/humu.20701. [DOI] [PubMed] [Google Scholar]
- 48.Shiraishi Y., Fujimoto A., Furuta M., et al. Integrated analysis of whole genome and transcriptome sequencing reveals diverse transcriptomic aberrations driven by somatic genomic changes in liver cancers. PLoS One. 2014;9:e114263. doi: 10.1371/journal.pone.0114263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong H., Zhang L., Qian Z., et al. Identification of HBV-MLL4 integration and its molecular basis in Chinese hepatocellular carcinoma. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furuta M., Tanaka H., Shiraishi Y., et al. Characterization of HBV integration patterns and timing in liver cancer and HBV-infected livers. Oncotarget. 2018;9:25075–25088. doi: 10.18632/oncotarget.25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau C.C., Sun T., Ching A.K., et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25:335–349. doi: 10.1016/j.ccr.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 52.Liang H.W., Wang N., Wang Y., et al. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J Hepatol. 2016;64:278–291. doi: 10.1016/j.jhep.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 53.Decorsiere A., Mueller H., van Breugel P.C., et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386–389. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 54.Sekiba K., Otsuka M., Funato K., et al. HBx-induced degradation of Smc5/6 complex impairs homologous recombination-mediated repair of damaged DNA. J Hepatol. 2022;76:53–62. doi: 10.1016/j.jhep.2021.08.010. [DOI] [PubMed] [Google Scholar]
- 55.Kao J.H., Chen P.J., Chen D.S. Recent advances in the research of hepatitis B virus-related hepatocellular carcinoma: epidemiologic and molecular biological aspects. Adv Cancer Res. 2010;108:21–72. doi: 10.1016/B978-0-12-380888-2.00002-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhang G., Budker V., Wolff J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 57.Molina-Sanchez P., Ruiz de Galarreta M., Yao M.A., et al. Cooperation between distinct cancer driver genes underlies intertumor heterogeneity in hepatocellular carcinoma. Gastroenterology. 2020;159:2203–2220 e14. doi: 10.1053/j.gastro.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen P.J., Chen D.S., Lai M.Y., et al. Clonal origin of recurrent hepatocellular carcinomas. Gastroenterology. 1989;96(Pt 1):527–529. doi: 10.1016/0016-5085(89)91581-3. [DOI] [PubMed] [Google Scholar]
- 59.Chen Y.J., Yeh S.H., Chen J.T., et al. Chromosomal changes and clonality relationship between primary and recurrent hepatocellular carcinoma. Gastroenterology. 2000;119:431–440. doi: 10.1053/gast.2000.9373. [DOI] [PubMed] [Google Scholar]
- 60.Li C.L., Yeh S.H., Chen P.J. Circulating virus-host chimera DNAs in the clinical monitoring of virus-related cancers. Cancers (Basel) 2022;14 doi: 10.3390/cancers14102531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corcoran R.B., Chabner B.A. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018;379:1754–1765. doi: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 62.Newman A.M., Bratman S.V., To J., et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dawson S.J., Tsui D.W., Murtaza M., et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 64.Li C.L., Ho M.C., Lin Y.Y., et al. Cell-free virus-host chimera DNA from hepatitis B virus integration sites as a circulating biomarker of hepatocellular cancer. Hepatology. 2020;72:2063–2076. doi: 10.1002/hep.31230. [DOI] [PubMed] [Google Scholar]
- 65.Hartmann L., Metzeler K.H. Clonal hematopoiesis and preleukemia: genetics, biology, and clinical implications. Genes Chromosomes Cancer. 2019;58:828–838. doi: 10.1002/gcc.22756. [DOI] [PubMed] [Google Scholar]
- 66.European Association for the Study of the Liver Electronic address eee, European Association for the Study of the L. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 67.Zhou J., Zhang M., Li X., et al. Performance comparison of four types of target enrichment baits for exome DNA sequencing. Hereditas. 2021;158:10. doi: 10.1186/s41065-021-00171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liaw Y.F., Chu C.M. Hepatitis B virus infection. Lancet. 2009;373:582–592. doi: 10.1016/S0140-6736(09)60207-5. [DOI] [PubMed] [Google Scholar]
- 69.Kuang X.J., Jia R.R., Huo R.R., et al. Systematic review of risk factors of hepatocellular carcinoma after hepatitis B surface antigen seroclearance. J Viral Hepat. 2018;25:1026–1037. doi: 10.1111/jvh.12905. [DOI] [PubMed] [Google Scholar]
- 70.Song A., Wang X., Lu J., et al. Durability of hepatitis B surface antigen seroclearance and subsequent risk for hepatocellular carcinoma: a meta-analysis. J Viral Hepat. 2021;28:601–612. doi: 10.1111/jvh.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hsu Y.C., Suri V., Nguyen M.H., et al. Inhibition of viral replication reduces transcriptionally active distinct hepatitis B virus integrations with implications on host gene dysregulation. Gastroenterology. 2022;162:1160–1170 e1. doi: 10.1053/j.gastro.2021.12.286. [DOI] [PubMed] [Google Scholar]
- 72.Salpini R., D’Anna S., Benedetti L., et al. Hepatitis B virus DNA integration as a novel biomarker of hepatitis B virus-mediated pathogenetic properties and a barrier to the current strategies for hepatitis B virus cure. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.972687. [DOI] [PMC free article] [PubMed] [Google Scholar]