Abstract
Background & Aims
Fibroblast activation protein (FAP) is expressed on activated fibroblast. Its role in fibrosis and desmoplasia is controversial, and data on pharmacological FAP inhibition are lacking. We aimed to better define the role of FAP in liver fibrosis in vivo and in vitro.
Methods
FAP expression was analyzed in mice and patients with fibrotic liver diseases of various etiologies. Fibrotic mice received a specific FAP inhibitor (FAPi) at 2 doses orally for 2 weeks during parenchymal fibrosis progression (6 weeks of carbon tetrachloride) and regression (2 weeks off carbon tetrachloride), and with biliary fibrosis (Mdr2−/−). Recombinant FAP was added to (co-)cultures of hepatic stellate cells (HSC), fibroblasts, and macrophages. Fibrosis- and inflammation-related parameters were determined biochemically, by quantitative immunohistochemistry, polymerase chain reaction, and transcriptomics.
Results
FAP+ fibroblasts/HSCs were α-smooth muscle actin (α-SMA)-negative and located at interfaces of fibrotic septa next to macrophages in murine and human livers. In parenchymal fibrosis, FAPi reduced collagen area, liver collagen content, α-SMA+ myofibroblasts, M2-type macrophages, serum alanine transaminase and aspartate aminotransferase, key fibrogenesis-related transcripts, and increased hepatocyte proliferation 10-fold. During regression, FAP was suppressed, and FAPi was ineffective. FAPi less potently inhibited biliary fibrosis. In vitro, FAP small interfering RNA reduced HSC α-SMA expression and collagen production, and FAPi suppressed their activation and proliferation. Compared with untreated macrophages, FAPi regulated macrophage profibrogenic activation and transcriptome, and their conditioned medium attenuated HSC activation, which was increased with addition of recombinant FAP.
Conclusions
Pharmacological FAP inhibition attenuates inflammation-predominant liver fibrosis. FAP is expressed on subsets of activated fibroblasts/HSC and promotes both macrophage and HSC profibrogenic activity in liver fibrosis.
Keywords: Antifibrotic Therapy, Fibroblast Activation Protein (FAP), Hepatic Stellate Cell, Liver Fibrosis, Macrophage
Abbreviations used in this paper: α-SMA, α-smooth muscle actin; ALP, alkaline phosphatase; ALT, alanine transaminase; AST, aspartate aminotransferase; BMDM, bone marrow derived macrophage(s); bw, body weight; CCl4, carbon tetrachloride; CM, conditioned medium; DPP, dipeptidyl peptidase; ECM, extracellular matrix; FAP, fibroblast activation protein; FAPi, FAP inhibitor; FBS, fetal bovine serum; H&E, hematoxylin and eosin; HSC, hepatic stellate cell; Mdr2, multidrug resistance protein 2/canalicular phospholipids export pump; TGFβ1, transforming growth factor beta 1; PBC, primary biliary cholangitis; PCR, polymerase chain reaction; PDGFRβ, platelet-derived growth factor receptor beta; PMA, Phorbol 12-myristate 13-acetate; PSC, primary sclerosing cholangitis; rhFAP, recombinant human fibroblast activation protein; rmFAP, recombinant mouse fibroblast activation protein; RNA-seq, RNA sequencing; WT, wild type
Graphical abstract

Summary.
Fibroblast activation protein is highly expressed on a subset of activated fibroblasts and stellate cells at mesenchymal-epithelial interfaces and is associated with macrophage infiltration. Specific fibroblast activation protein inhibition modulates macrophage function, attenuates parenchymal liver fibrosis, and facilitates hepatocyte proliferation, with reduced efficacy in biliary fibrosis.
Liver fibrosis results from a protracted wound-healing response in chronic liver diseases, such as viral infections, nonalcoholic steatohepatitis, and cholestatic hepatitis. It may ultimately lead to cirrhosis, which is the 11th most common cause of death globally.1 The excessive accumulation of extracellular matrix (ECM) in advanced fibrosis/cirrhosis is associated with distortion of the hepatic architecture and severe complications, including liver failure and death.2,3 Fibrosis results from a disbalance of the dynamic processes of ECM resolution (fibrolysis) in favor of ECM accumulation (fibrogenesis).4 Experimental and clinical data indicate that even cirrhosis can regress with effective treatment of the underlying liver disease.5, 6, 7 However, there are no approved antifibrotic therapies that prevent progression or speed up regression in the majority of patients for whom causal treatment is impossible or ineffective. Meanwhile, our understanding of the complex immunological, cellular, and molecular network that determines fibrogenesis and fibrolysis has increased, suggesting therapeutic approaches targeting the ECM, or the major effector cells of hepatic ECM production, such as hepatic stellate cells (HSCs) and (myo)fibroblasts, or effector cells of ECM dissolution and removal, such as macrophages.8,9
Fibroblast activation protein (FAP) is a type II integral membrane glycoprotein and serine protease of the dipeptidyl peptidase (DPP) family, with high homology to DPPIV. Although DPPIV is expressed on hepatocytes and T cells, FAP is exclusively expressed on a subset of activated fibroblasts.10 FAP possesses both terminal dipeptidyl peptidase and endopeptidase activity. Moreover, it displays a unique substrate specificity for biologically relevant mediators like neuropeptide Y, B-type natriuretic peptide, substance P, and peptide YY.11 Using global degradomics technology, further substrates of FAP were identified, including proteins relevant in ECM remodeling, inflammation, and fibrinolysis, such as the ECM crosslinking enzyme lysyl oxidase-like-1, the chemokine CXCL-5, the cytokine CSF-1, and complement C1q tumor necrosis factor-related protein 6 (C1qT6).12
FAP is almost undetectable in healthy adult liver, but upregulated in active stroma of fibrotic livers, such as the portal-parenchymal interface of expanding portal tracts and fibrotic septa, and in the desmoplastic stroma of liver tumors.13,14 Importantly, increased FAP+ fibroblasts correlate with the histological severity of fibrosis, suggesting that FAP+ fibroblasts may drive fibrogenesis. However, direct functional evidence of such a role is missing.15 Moreover, there is little overlap of FAP+ fibroblasts with fibroblasts expressing α-smooth muscle actin (α-SMA), generally considered a marker of HSC activation and fibrogenesis.16,17 The gene expression profile of FAP+ fibroblasts is enriched for inflammatory genes,18, 19, 20, 21 suggesting that FAP-expressing fibroblasts drive inflammation during chronic wound healing. Still, the function of FAP and FAP-expressing fibroblasts and HSCs in chronic (liver) injury and fibrosis remains elusive.
Therefore, we examined FAP expression in murine and human liver fibrosis and employed recombinant FAP protein in macrophage and HSC/fibroblast cultures in vitro, to gain insight into the role of FAP and FAP+ cells in liver inflammation and fibrosis. Finally, we employed a FAP-selective inhibitor22 to assess the potential of specific FAP inhibition as antifibrotic therapy.
Results
Specific FAP Inhibition Attenuates Carbon Tetrachloride-induced Hepatic Injury
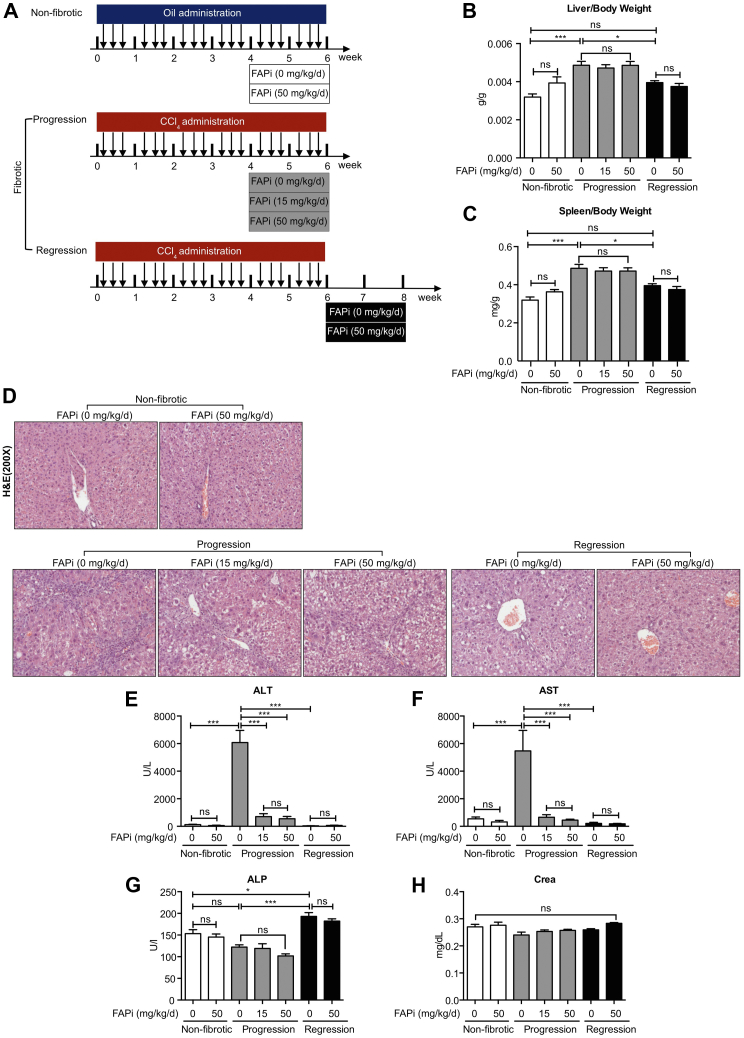
We used a specific oral FAP inhibitor (FAPi)22 in an optimized model of parenchymal liver fibrosis,23 both during fibrosis progression and during recovery (partial regression) after toxin discontinuation.6,7,23 The liver and spleen weights increased at the peak of fibrosis and returned to nearly normal after 2 weeks of recovery, regardless of FAPi treatment (Figure 1, A–C). Histopathological analysis showed pronounced mononuclear cell infiltration in fibrotic livers, which was significantly attenuated by treatment with FAPi at doses of 15 and 50 mg/kg/d. Two weeks off carbon tetrachloride (CCl4) (recovery) livers showed minimal inflammatory cell infiltration with or without FAPi treatment (Figure 1, D). Serum levels of alanine transaminase (ALT) and aspartate aminotransferase (AST) were high in the fibrosis progression group and reduced to nonfibrotic control levels by FAPi. After recovery from CCl4, ALT and AST levels were low and remained unchanged by FAPi (Figure 1, E–F). Alkaline phosphatase (ALP) levels were low in mice with fibrosis progression and increased with recovery (Figure 1, G), suggesting enhanced hepatocyte progenitor cell proliferation.24 Both doses of FAPi did not affect serum levels of AST, ALT, and creatinine in the non-fibrotic controls (Figure 1, E, F, and H).
Figure 1.
Treatment of CCl4-fibrotic mice with FAP inhibitor. (A), Scheme of experimental design. Female C56BL/6 mice received escalating doses of oral CCl4 in mineral oil (fibrotic group, n = 10) or mineral oil alone (non-fibrotic controls, n = 5) for 6 weeks. Fibrotic mice were analyzed at peak progression or after 2 weeks of regression. Other groups of fibrotic mice continued with their chow were set on chow containing FAPi at 15 or 50 mg/kg/d (n = 10 per group). (B–C), Liver/bw ratio and spleen/bw ratios. (D), Representative images of H&E-stained liver section of each group. (E–H), Serum ALT, AST, ALP, and creatinine levels. Data in (B–C, E–H) are presented as means ± standard error of the mean (SEM). Statistical analysis was performed by 1-way analysis of variance followed by Tukey’s post hoc test (∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ns, not significant).
Pharmacological FAP Inhibition Suppresses CCl4-induced Liver Fibrosis Progression, but does not Accelerate Regression
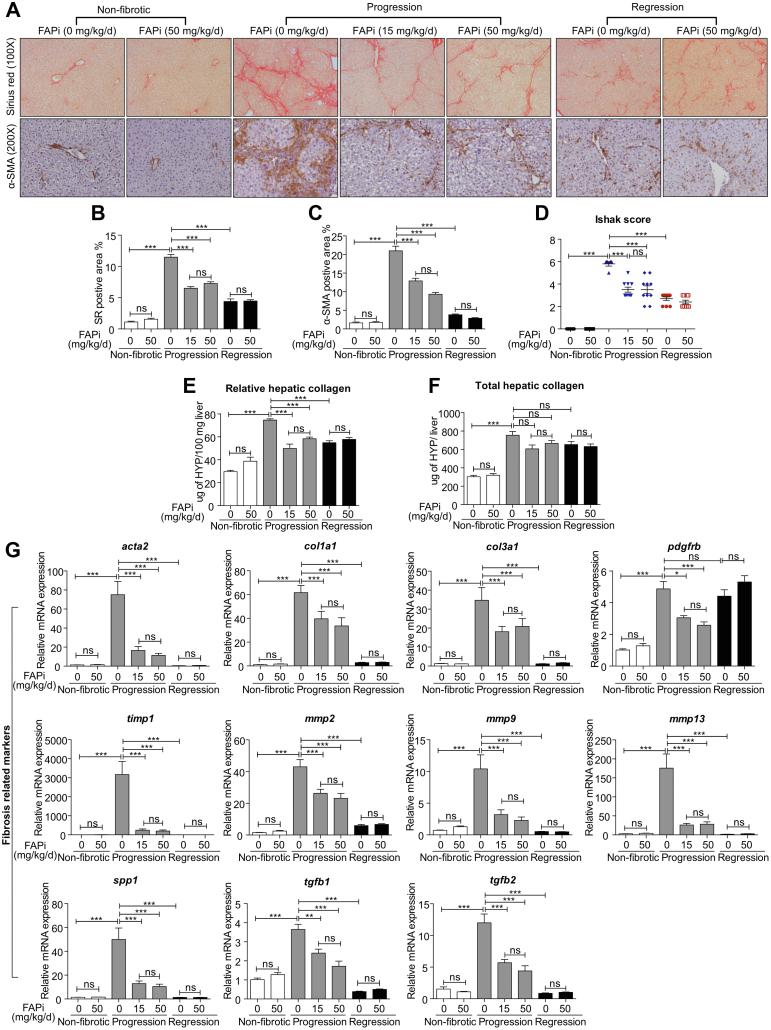
Morphometry for liver collagen and α-SMA+ activated HSC (myofibroblasts) was significantly reduced with both doses of FAPi (Figure 2, A–C), in line with an improved Ishak histological fibrosis score (Figure 2, D) and a significant decrease of relative and total liver collagen content as determined by hydroxyproline (Figure 2, E–F). Hepatic levels of key fibrosis related transcripts (acta2, col1a1, col3a1, pdgfrb, timp1, mmp2, mmp9, mmp13, spp1-ostepontin, tgfb1, tgfb2) were significantly upregulated in the untreated fibrotic mice and were reduced with both doses of FAPi, suggesting a ceiling effect at 15 mg/kg/d (Figure 2, G). FAPi did not affect any of these parameters in non-fibrotic controls (Figure 2, A–G). Consistent with our prior studies in the CCl4 model,23 characteristic histological features of reversal were observed after 2 weeks of recovery, including splitting of densely packed fibrotic septa, and a significant reduction of collagen and α-SMA densities (Figure 2, B–C), whereas fibrosis-related transcripts had normalized. However, pdgfrb expression remained upregulated, indicating a role of platelet-derived growth factor receptor beta (PDGFRβ) in fibrosis reversal.
Figure 2.
FAP inhibition attenuates parenchymal liver fibrosis. (A–C), Livers of mice (n = 5–10/group) treated with FAPi during fibrosis progression or regression vs fibrotic untreated controls were analyzed by quantitative Sirius red and α-SMA immunohistochemistry, performed in 10 random high-power fields per mouse using ImageJ software. (D), Histological fibrosis score of liver sections. (E–F), Hepatic hydroxyproline concentration. (G), Hepatic transcript of fibrosis related transcripts. Data in (B–G) are means ± standard error of the means (SEMs). Statistical analysis was performed as for Figure 1.
FAP Inhibition Attenuates Macrophage Infiltration and Activation in Liver Fibrosis
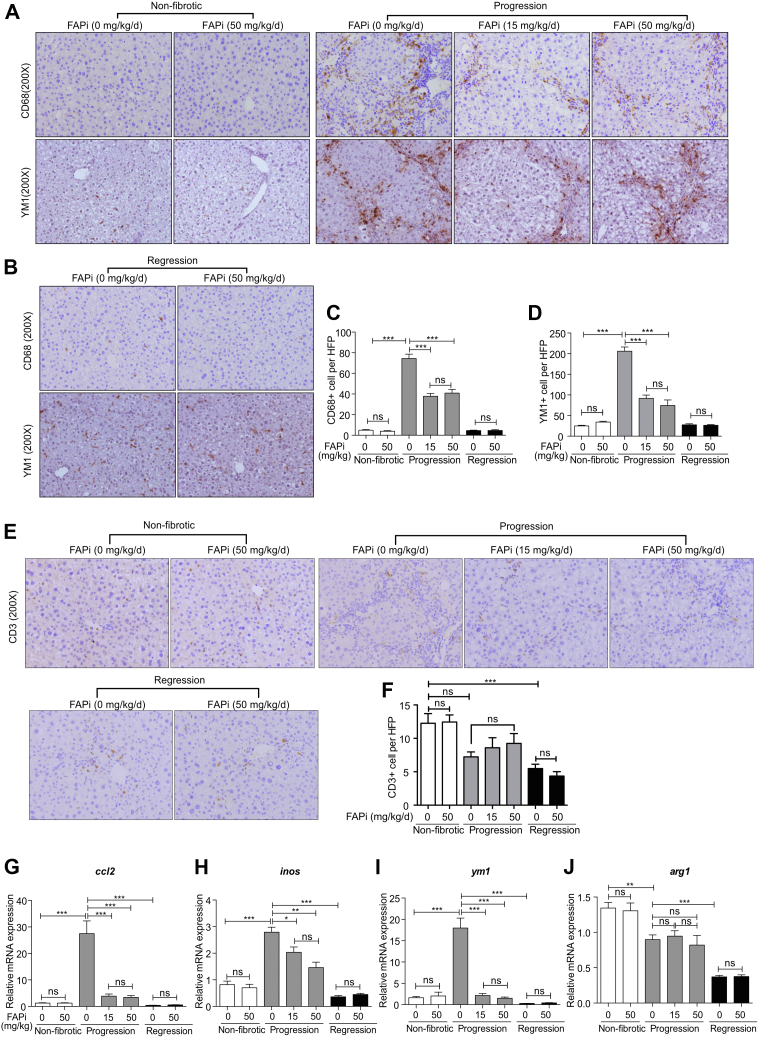
In non-fibrotic livers, CD68+ and M2-type YM1+ macrophages were sparse and randomly distributed, but significantly increased within septa with active fibrogenesis. FAPi reduced CD68+ and YM1+ macrophages by about 50% compared with untreated fibrotic controls (Figure 3, A–D), whereas intrahepatic CD3+ lymphocyte counts remained unchanged (Figure 3, E–F). In line with reduced macrophage numbers, hepatic transcript levels of ccl2 (encoding the macrophage chemokine MCP-1), inos (pro-inflammatory macrophages/M1-type), and ym1 (anti-inflammatory/M2-type) were suppressed by FAPi during fibrosis progression, whereas the M2-marker arg1 remained unchanged (Figure 3, G–J). During fibrosis regression, macrophage numbers and respective transcript levels were low and remained unchanged by FAPi treatment (Figure 3, A–J). Therefore, FAPi reduced total monocyte/macrophage recruitment and activation of scar-associated macrophages in progressive CCl4-induced liver fibrosis but did not affect macrophages during regression.
Figure 3.
Effect of FAP inhibition on hepatic inflammation in CCl4-induced fibrosis. (A–D), Livers from non-fibrotic mice and mice with fibrosis progression or regression (n = 5–10/group) treated with FAPi and controls were analyzed for macrophage markers CD68 and YM1 using immunohistochemistry in 10 random high-power fields per liver. (E–F), Representative images and quantification of CD3+ T cells in 10 random fields (×200) from the central right lobe of each liver. (G–J), Hepatic transcript levels of inflammation related genes. Data in (C–D, F, G–J) are means ± standard error of the means (SEMs). Statistical analysis was performed as for Figure 1.
FAP Inhibition Promotes Liver Regeneration in Parenchymal Liver Fibrosis Progression
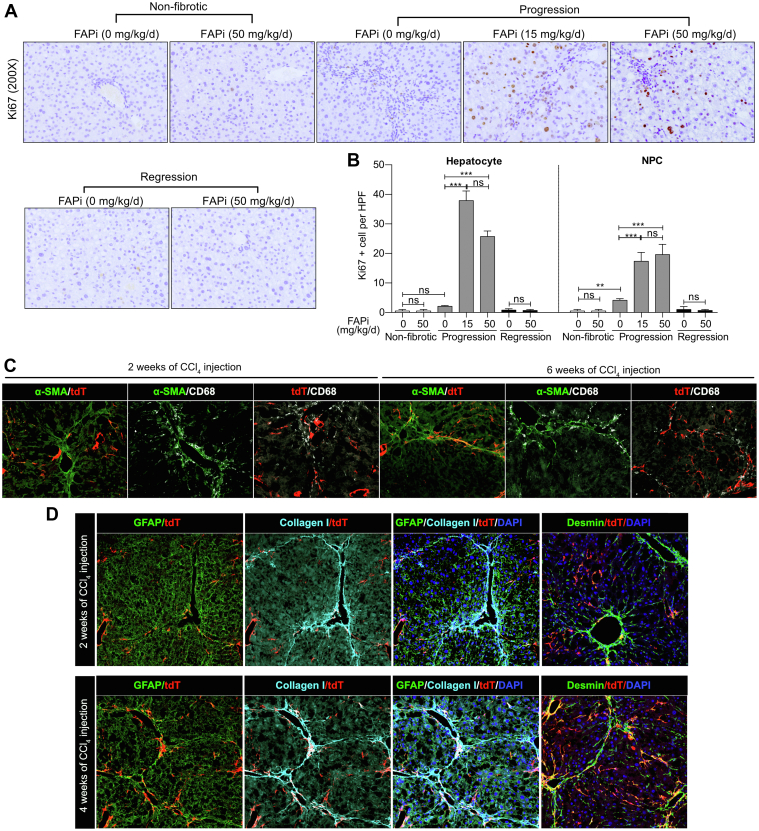
Ki67, a cell proliferation marker, was almost absent in cells of non-fibrotic and CCl4-fibrotic mice. In active fibrosis, FAPi increased the number of Ki67+ hepatocytes more than 10-fold, and of Ki67+ nonparenchymal cells more than 3-fold, suggesting that FAP prominently regulates hepatocyte proliferation and possibly liver regeneration (Figure 4, A–B).
Figure 4.
FAP inhibition increases hepatocyte and nonparenchymal liver cell proliferation in CCl4-induced fibrosis. (A–B), Representative images of Ki67 expressing cells and morphometrical quantification in 10 random high-power fields per liver. (C), Co-localization of FAP+ fibroblasts, α-SMA+ (myo-)fibroblasts, and CD68+ macrophage in livers of mice with CCl4-induced fibrosis. (D), Co-localization of FAP+ fibroblasts with type I collagen, GFAP-, and desmin-positive HSCs and (myo-)fibroblasts in livers of mice with CCl4-induced fibrosis. Data are presented as means± standard error of the means (SEMs) (n = 5 mice per non-fibrotic, and n = 10 mice per fibrotic subgroups). Statistical analysis was performed as for Figure 1.
FAP+ Fibroblasts do not Colocalize With α-SMA-expressing (Myo)fibroblasts
We used mice expressing tomato red under the control of the FAP-promoter to demonstrate that FAP+ fibroblasts formed a distinct population not overlapping with α-SMA+ (myo)fibroblasts. FAP+ cells were found at the periphery of portal fields and in (expanding) septa, whereas α-SMA+ cells were abundant in the more established ECM. Both FAP+ and α-SMA+ fibroblasts and (myo)fibroblasts were adjacent to CD68+ macrophages. (Figure 4, C). Because α-SMA only defines a subpopulation of activated HSC/myofibroblasts, we used the markers GFAP, desmin, and type I collagen to better define their phenotype. Interestingly, in early-stage fibrosis (2 weeks of CCl4 induction), only a minor population of FAP+ fibroblasts was colocalized with GFAP+ cells, and there was no colocalization with desmin or collagen fibers. Notably, in advanced fibrosis (4 weeks of CCl4), GFAP colocalization did not change significantly, whereas desmin-positive HSCs/fibroblasts now largely colocalized with FAP+ cells, prominently in collagen-rich fibrous septa and portal areas (Figure 4, D). These results indicate that FAP+ cells are a subset of fibroblasts that do not express α-SMA but mainly desmin, and that they are linked to collagen production in advanced rather than in early-stage fibrosis.
FAP Inhibition Accelerates ECM Remodeling and Modestly Attenuates ECM Deposition in Biliary Fibrosis
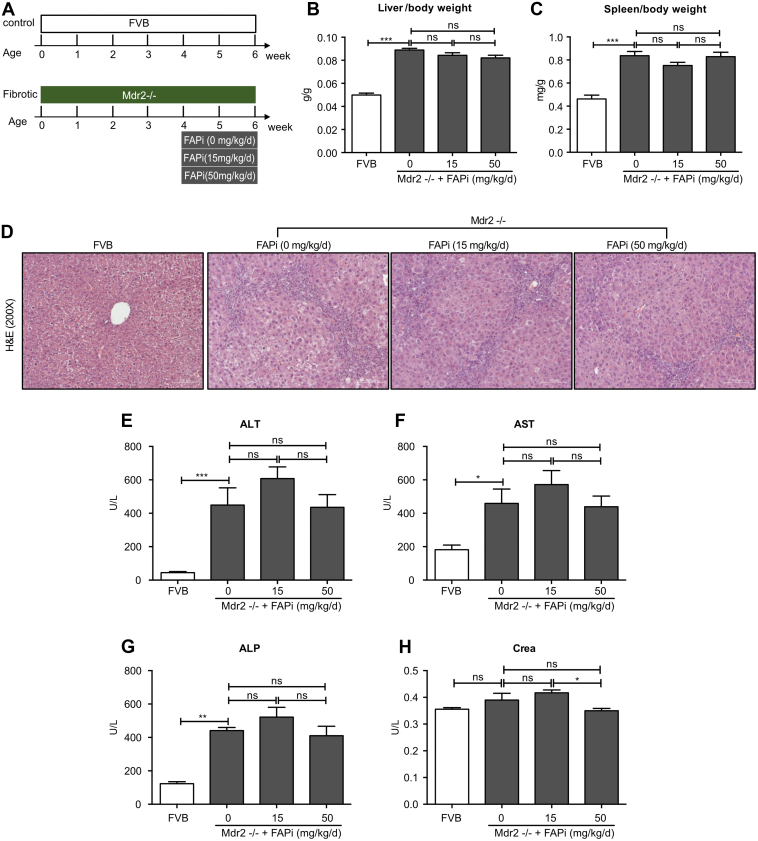
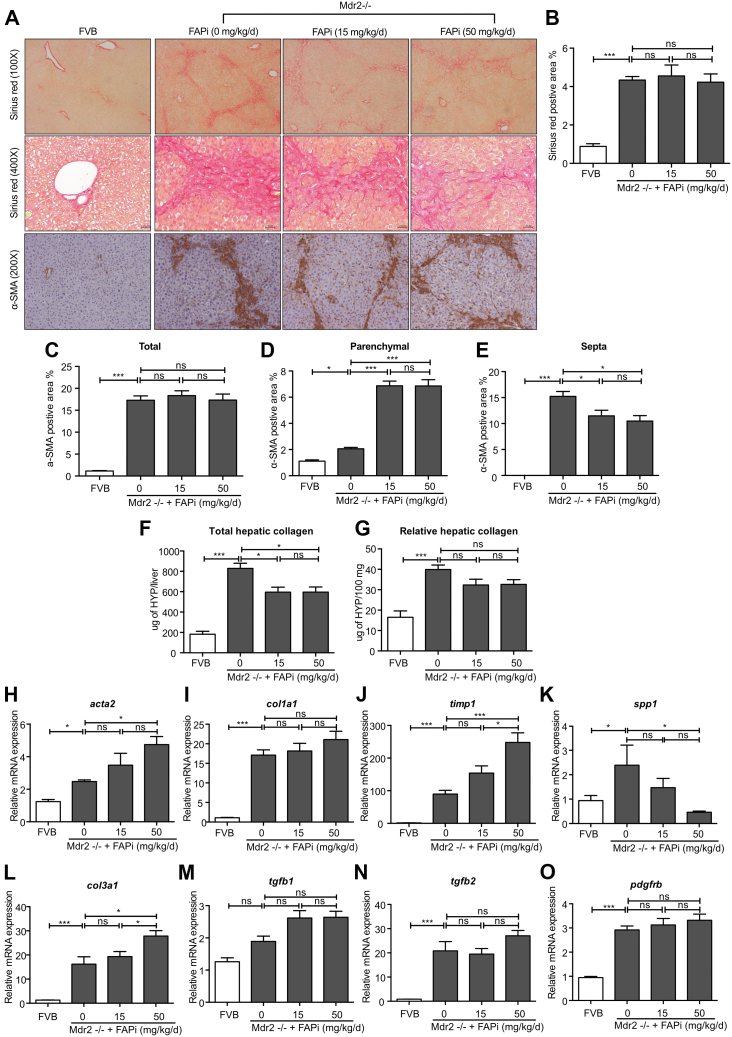
When multidrug resistance protein 2/canalicular phospholipids export pump (Mdr2)−/− mice, a model of human primary sclerosing cholangitis, and their nonfibrotic wild-type (WT) controls were treated with FAPi, effects were less pronounced. Thus, liver and spleen weight, hematoxylin and eosin (H&E) histology and scoring, and elevated liver enzymes were unchanged by FAPi treatment (Figure 5, A–H). Total and relative liver collagen content was less pronouncedly (Figure 6, F–G), but significantly reduced with FAPi compared with the CCl4 model. Collagen morphometry was unchanged (Figure 6, A–B), indicating a decrease of collagen that was proportionate to the decrease of liver mass with FAPi treatment (Figure 5, B). This was accompanied by a significant reduction of α-SMA+ myofibroblasts in the portal/septal but not in the parenchymal areas of the fibrotic livers (Figure 6, A and C–E). Moreover, the fibrotic septa of the FAPi-treated Mdr−/− mice appeared wider and collagen fibrils more loosely organized compared with their untreated Mdr2−/− mice, similar to progressive parenchymal fibrosis (Figure 6, A). Transcripts of fibrosis related genes were highly upregulated in untreated Mdr2−/− vs WT mice. FAPi did not affect col1a1 tgfb1, tgfb2, and pdgfbr expression, slightly increased acta2, col3a1, and timp1, and decreased spp1 expression that encode α-SMA, type III procollagen, TIMP1, and osteopontin, respectively (Figure 5, H−O). Collectively, treatment with FAPi had a limited to modest effect on biliary fibrosis but promoted ECM remodeling and affected fibrosis related gene expression partly in contrasting ways.
Figure 5.
Effect of FAP inhibition on liver/spleen weight and general liver inflammation in Mdr2−/− mice. (A), Scheme of FVB control and Mdr2−/− biliary fibrotic mice (n = 5 and n = 10 mice per group, respectively) treated with FAPi (15 or 50 mg/kg bw) or vehicle for 2 weeks. (B−C), Liver/bw ratio and spleen/bw ratio. (D), Representative images of H&E staining from liver sections. (E−H), Serum ALT, AST, ALP, and creatinine levels. Data in (B−C, E−H) are means ± standard errors of the mean (SEMs). Statistical analysis was performed as for Figure 1.
Figure 6.
FAP inhibition attenuates hepatic fibrosis in Mdr2−/− mice. (A–E), Livers of nonfibrotic FBV control and biliary fibrotic Mdr2−/− mice (n = 5–10/group) were treated with FAPi (15 or 50 mg/kg/d) for 2 weeks and compared with respective untreated mice. Sirius red stained area and α-SMA-expressing cells in the parenchymal vs septal areas were assessed by quantitative morphometry in 10 high-power fields. (F–G), Total and relative hepatic hydroxyproline concentration. (H–O), Hepatic transcript levels of fibrosis related genes. Data in (B–O) are presented as means ± standard error of the means (SEMs). Statistical analysis was performed as detailed in Figure 1.
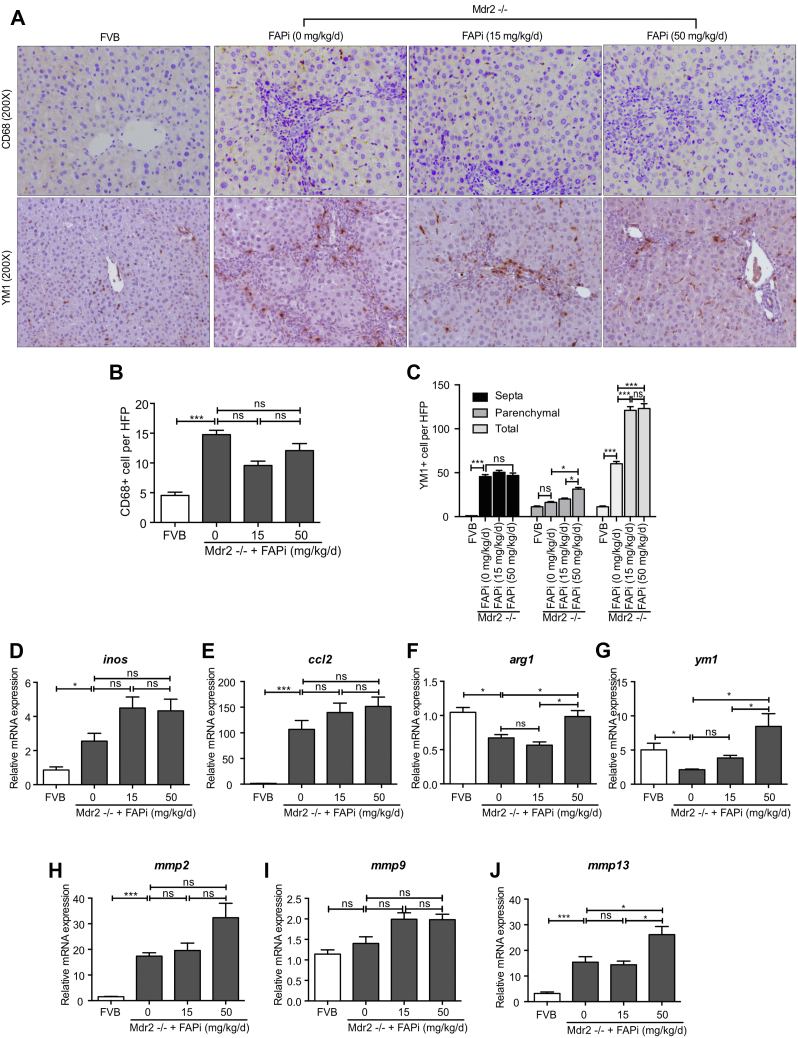
FAP Inhibition Promotes M2-type Macrophage Polarization in the Parenchyma of Mdr2−/− Mice
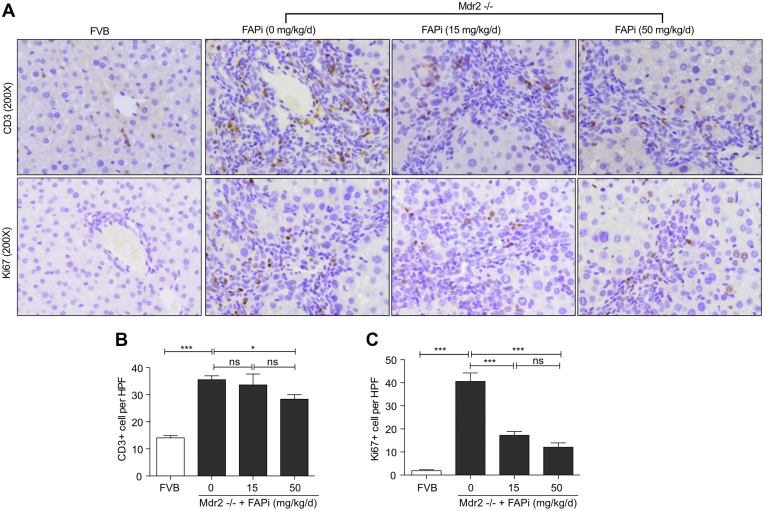
Increased infiltration of CD68+ (total) and YM1+ (M2-type) macrophages, and CD3+ lymphocytes was observed in Mdr2−/− mice, being mainly located in the portal areas and fibrotic septa (Figure 7, A−C; Figure 8, A−C). FAPi did not affect CD68+ and portal-septal YM1+ macrophage infiltration but increased parenchymal YM1+ macrophages at the highest dose (Figure 7, A−C). This increase was most pronounced when their number per unit area of liver tissue was assessed (Figure 7, A and C), suggesting that FAPi favored M2-type macrophage polarization in the otherwise less affected parenchymal regions of biliary fibrotic livers. High-dose FAPi decreased CD3+ cells and Ki67+ cells in the (active) fibrotic areas in Mdr2−/− mice (Figure 8, A−C). The vast majority of these Ki67+ cells were compatible with portal/septal (scar-associated) macrophages, with only few Ki67+ hepatocytes/biliary epithelia. With FAPi treatment, transcript levels of macrophage markers were either unchanged (inos) or dose-dependently upregulated (arg1, ym1), whereas expression of ccl2, mmp2, and mmp9 was unchanged and that of mmp13 was elevated at the highest FAPi dose (Figure 7, D−J). These data indicate that, in experimental biliary fibrosis, FAPi induced a modest beneficial ECM remodeling and M2-type macrophage polarization, coupled with an attenuation of nonparenchymal cell proliferation.
Figure 7.
Effects of FAP inhibition on hepatic inflammation in Mdr2−/− mice. (A–C), Livers of control and Mdr2−/− fibrotic mice (n = 5–10/group) treated with FAPi as in Figure 5 were assessed for numbers of CD68 and YM1 expressing macrophages in 10 random high-power fields per mouse. (D–J), Hepatic transcript levels related to macrophage activation, polarization, and MMP expression were determined by qPCR. Data in (B–J) are presented as means ± standard error of the means (SEMs). Statistical analysis was performed as detailed in Figure 1.
Figure 8.
FAP inhibition does not modify T cell infiltration and suppresses cell proliferation in Mdr2−/− mice. (A–C), Representative images and quantification of CD3- and Ki67-positive cells in 10 random fields (×200) from the central right lobe of each liver. Data in (B–C) are means ± standard error of the means (SEMs). Statistical analysis was performed as detailed in Figure 1.
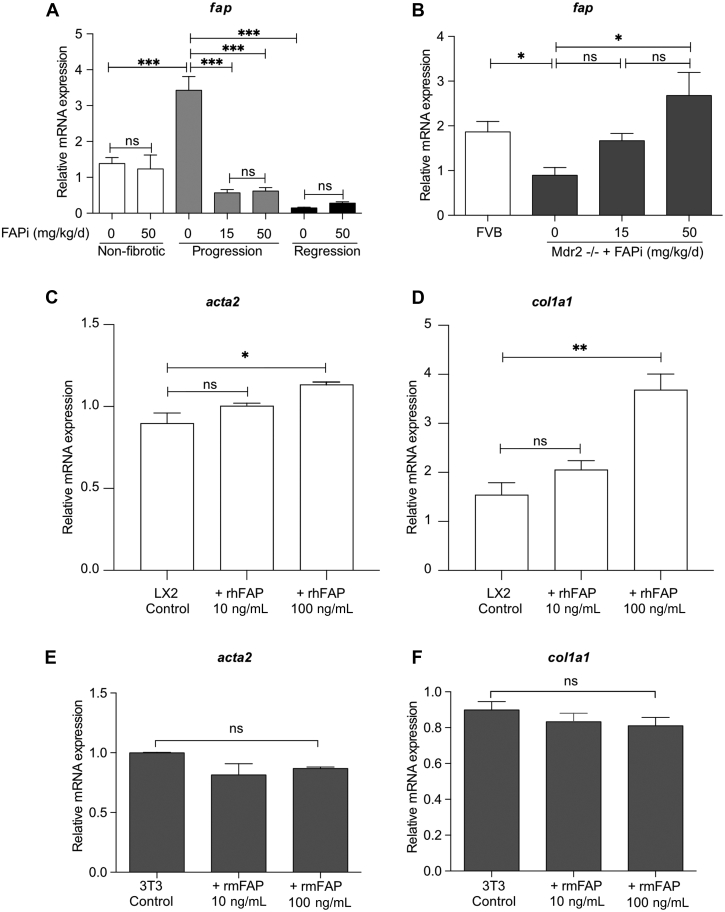
FAP Protein Regulates Macrophage Function and Polarization
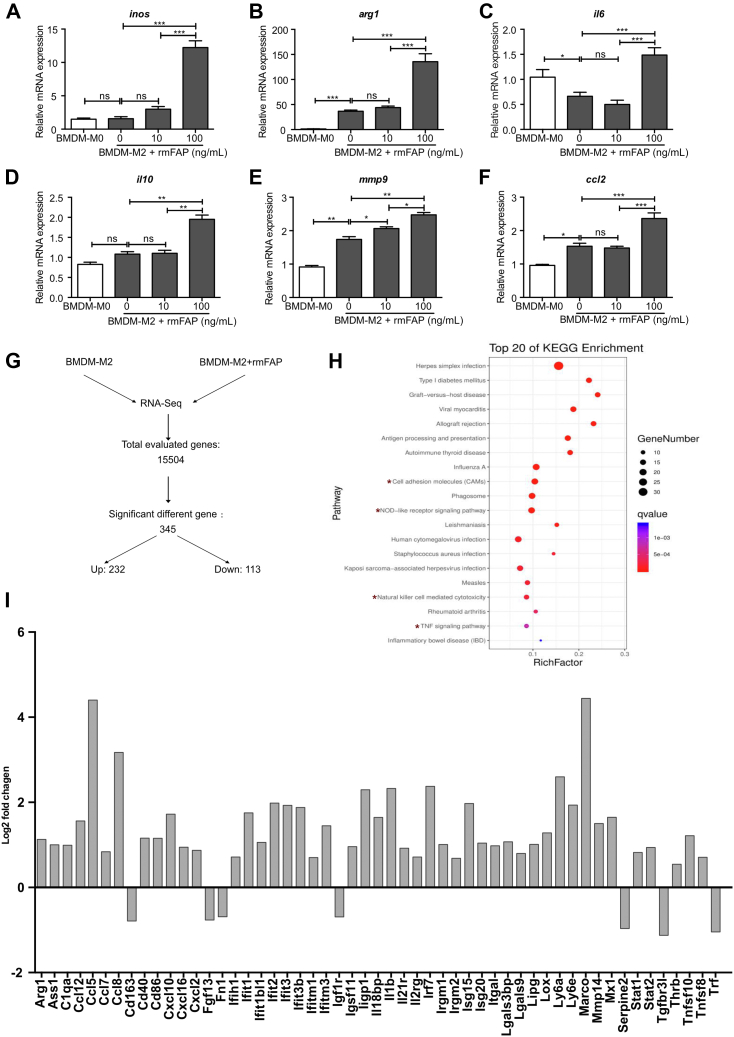
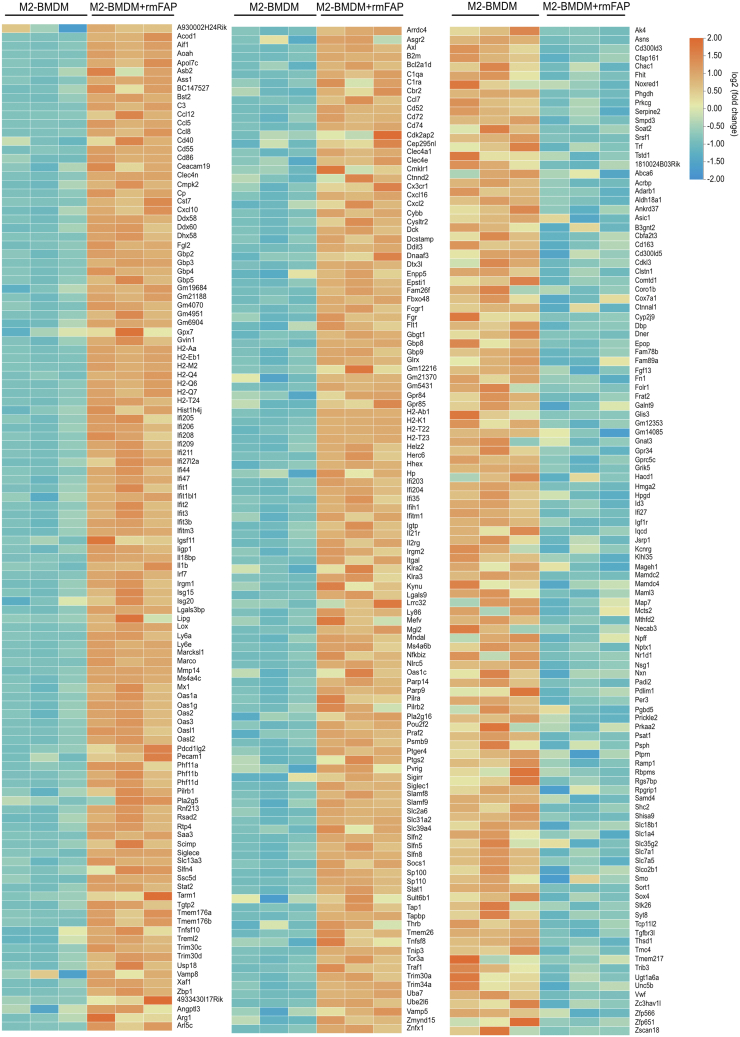
FAP expression was highly upregulated in livers of mice with parenchymal fibrogenesis, and significantly decreased after 2 weeks of FAPi treatment. In contrast, hepatic FAP expression was decreased in biliary fibrotic Mdr2−/− mice but upregulated with FAPi treatment (Figure 9, A−B). To assess the role of FAP on key cells driving both parenchymal and biliary liver fibrosis, we studied the effect of recombinant FAP on murine 3T3-fibroblasts and the human hepatic stellate line LX2 that in combination should represent major fibrogenic cells and sources of extracellular matrix proteins in the liver. Treatment of LX2 cells with recombinant human fibroblast activation protein (rhFAP) increased expression of acta2 and col1a1 transcripts (Figure 9, C−D). However, recombinant mouse fibroblast activation protein (rmFAP) had no effect on activation of or collagen production by 3T3-fibroblasts (Figure 9, E−F). Moreover, in M2-type bone marrow derived macrophages (BMDMs), rmFAP induced a unique macrophage phenotype with upregulation of both M1 genes (inos and il6) and of M2 genes (arg1 and il10), and markers of activation (ccl2 and mmp9) (Figure 10, A−F). The global gene expression profile in these M2-type BMDM following rmFAP treatment, as determined by RNA sequencing (RNA-Seq) of 15,504 transcripts, revealed significant changes in 345 transcripts (≥1.5-fold increased or decreased expression compared with untreated M2-type BMDM; q value <0.05), with 232 genes being upregulated, and 113 genes being downregulated (Figure 10, G), including 2 separate branches of up- and down-regulated genes (Figure 11). The differentially expressed genes in rmFAP-treated vs control M2-type macrophages were enriched in pathways related to cell adhesion molecules, NOD-like receptor signaling, phagosome, and natural killer cell cytotoxicity (Figure 10, H). rmFAP induced expression of macrophage markers like arg1, ass1, cd68, ly6a, ly6e, and marco, interferon-stimulated genes like iisg15, isg20, ifit1, ifit2, ifit3, and mx1), markers of ECM remodeling like mmp14 and lox, and inflammatory chemokines like il1b, ccl2, ccl5, ccl7, cxcl10, and cxcl116. Downregulated genes included cd163, fgf13, fn1, igf1r, serp2, tgfbr3, and tnfa (Figure 10, I).
Figure 9.
FAP expression in CCl4and Mdr2−/− fibrotic livers with/without FAP inhibition and effect of rhFAP/rmFAP on LX-2 HSCs and murine fibroblasts. (A–B), Transcript levels of fap. (C–D), Effect of treatment with rhFAP on LX2 HSCs activation and collagen gene expression. (E–F), Effect of treatment with rmFAP on NIH/3T3 fibroblast activation and collagen gene expression. Data in (A–F) are means ± standard error of the means (SEMs). Statistical analysis was performed as detailed in Figure 1.
Figure 10.
Effect of rmFAP on M2-type macrophage polarization and gene expression. (A–F), M0 (unpolarized)-BMDMs and M2-type BMDMs were treated with rmFAP (0–100 ng/mL, 48 hours), and macrophage activation/polarization relevant transcript levels were analyzed by qPCR. (G), RNA-Seq results in M2-BMDMs treated with 100 ng/mL rmFAP vs untreated M2-BMDMs. (H), Scatter plot for Kyoto Encyclopedia of Genes and Genomes enrichment analysis of the differentially expressed genes. Top 20 significantly enriched pathways (P < .05 by the Fisher exact test) are shown. (I), Log2-fold change of selected genes in M2-BMDMs treated with rmFAP (vs untreated M2-BMDMs). Data in (A–F) as means ± standard error of the means (SEMs). Statistical analysis was performed as detailed in Figure 1.
Figure 11.
Gene expression profiling by RNA-seq analysis of M2-polarized BMDMs with or without rmFAP treatment. Heatmaps of differentially expressed genes (differentiated as up- or downregulated by adjusted q-values <0.05 and ≥1.5 fold-change) in M2-BMDMs with vs without rmFAP treatment. Fragments per kilobase of transcript, per million mapped reads (FPKM) values were used to calculate the expression level by fold change of mRNA between the M2-BMDM and M2-BMDM+rmFAP groups, expressed as log2 (fold change) values. Differentially expressed genes (DEGs) between the M2-BMDM and M2-BMDM+rmFAP groups were identified using the Student t test. Orange color marks up-regulated and blue color down-regulated genes compared with the untreated M2-BMDM group. Side bar: X-fold change.
Conditioned Medium From FAPi-treated-HSCs Suppresses Inflammatory and Modulates MMP Expression in Macrophages
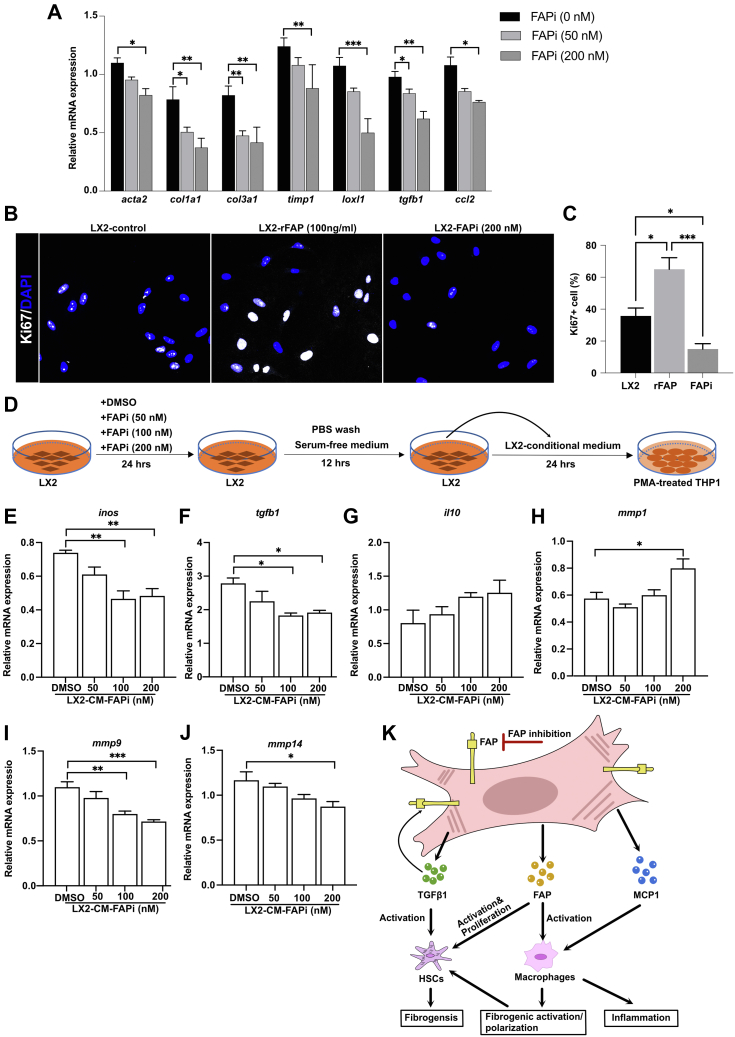
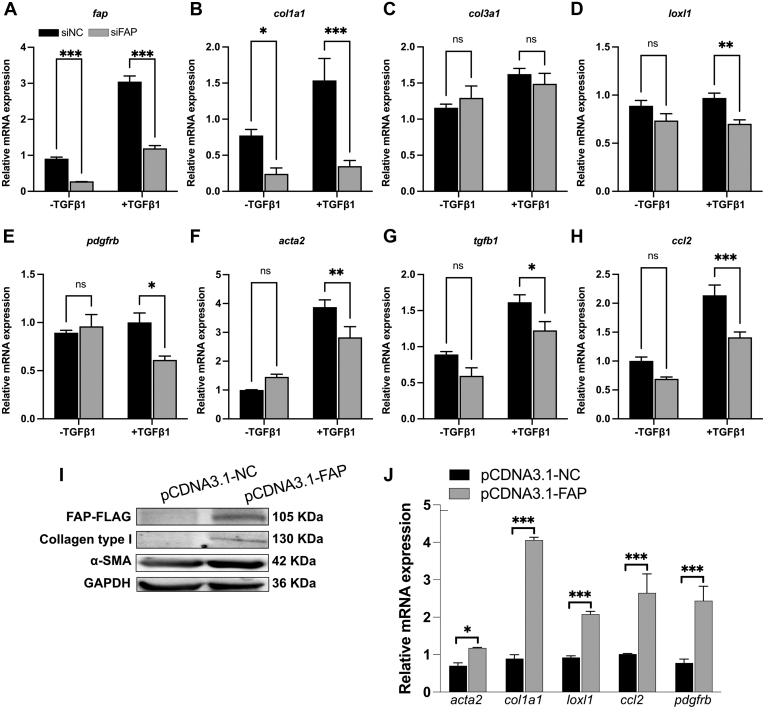
To clarify the potential mechanistic basis involved in the antifibrotic effect of FAP inhibition in murine liver fibrosis, we investigated the crosstalk between HSCs and macrophages in vitro. First, we confirmed that FAPi suppressed human LX2 HSC activation and reduced their fibrogenic gene expression in a dose-dependent manner, as reflected by downregulated transcript levels of acta2, col1a1, col3a1, timp1, loxl1, tgfb1, and ccl2 (Figure 12, A). HSC proliferation is a critical (first) step in hepatic fibrogenesis, and FAPi significantly decreased the percentage of Ki67 positive LX2 cells, while conversely Ki67 positivity increased LX2 HSC treated with rhFAP (Figure 12, B−C). For further confirmation and implication of FAP in fibrogenic gene expression by HSC, we performed small interfering RNA-mediated knockdown of FAP in LX2 cells in the presence or absence of transforming growth factor beta 1 (TGFβ1). Pre-treatment with TGFβ1 increased fibrogenic genes fap, acta2, col1a1, loxl1, tgb1, and ccl2, which was reversed by FAP knockdown. Interestingly, FAP knockdown had no effect on less activated LX2 HSC in the absence of TGFβ1, except for col1a1 (Figure 13, A−H). Conversely, overexpression of FAP upregulated α-SMA and type I collagen protein levels in LX2 cells, and increased transcript levels of acta2, col1a1, loxl1, pdgfrb, and ccl2 (Figure 13, I−J).
Figure 12.
FAPi-treated LX2 HSC modulate macrophage activation and polarization in vitro. (A), Expression of acta2, col1a1, col3a1, timp1, loxl1, tgfb1, and ccl2 in LX2 HSC treated with FAPi. (B–C), Immunofluorescent staining for Ki67 and DAPI in LX2 cells treated with rhFAP and FAPi. (D), Scheme of the experimental design of LX2 CM being added to THP1 cell-derived macrophages. (E–J), Expression of inos, tgfb1, il10, mmp1, mmp9, and mmp14 in PMA-treated (macrophage) THP1 cells after addition of culture medium from LX2 HSC pretreated with and without FAPi. (K), Working model of FAP and FAPi action during liver fibrogenesis. FAP upregulates HSC fibrogenic activation and proliferation and promotes macrophage profibrogenic activation/polarization directly as well as indirectly by production of factors that further promote HSC activation or induce monocyte recruitment (eg, tgfb1 and ccl2, respectively). Data in (A, C, E–J) are means ± standard error of the means (SEMs). Statistical analysis was performed as detailed in Figure 1.
Figure 13.
FAP regulates HSC activation. (A), Knockdown of FAP inhibits TGFβ1-induced profibrotic gene expression. LX2 cells were transfected with negative control small interfering (siNC) or small interfering FAP (siFAP) following pretreatment with 5 ng/mL TGFβ1 for 24 hours. Transcript levels of fap, col1a1, col3a1, loxl1, pdgfrb, acta2, tgfb1, and ccl2 were analyzed using qPCR. (B–C), Overexpression of FAP promotes HSC activation. LX2 cells were transfected with pCDNA3.1 control plasmid or FAP plasmid for 48 hours. (B), Protein levels of FLAG, collagen type I, and α-SMA were determined by Western blot. (C), Transcript levels of col1a1, acta2, loxl1, and ccl2 were analyzed using qPCR. Data are presented as means ± standard error of the means (SEMs). Statistical analysis was performed using an unpaired 2-tailed Student t test (∗P < .05; ∗∗P < .01; ∗∗∗P < .005).
Furthermore, we evaluate how far secreted factors after FAP inhibition would impact the crosstalk between HSC and macrophages. Phorbol 12-myristate 13-acetate (PMA)-induced macrophages derived from human monocytic THP1 cells were exposed to conditioned media (CM) derived from FAPi treated LX2 HSC (Figure 12, D). Compared with CM of DMSO-treated control LX2 HSC, CM of higher dose FAPi-treated LX2 cells inhibited macrophage activation and function, as reflected by down-regulation of inos, tgfb1, mmp9, and mmp14, and upregulation of mmp1 transcripts, with a trend for il10 upregulation (Figure 12, E−J). These in vitro data support our in vivo results that FAP directly mediates proliferation and fibrogenic activation of HSC. Importantly, HSC-derived FAP also increases the numbers and induces a fibrogenic phenotype of neighboring (monocytes-)macrophages, which further enhances a profibrotic microenvironment (Figure 12, K).
FAP Expression Suggests a Comparable Role in Human Liver Fibrosis
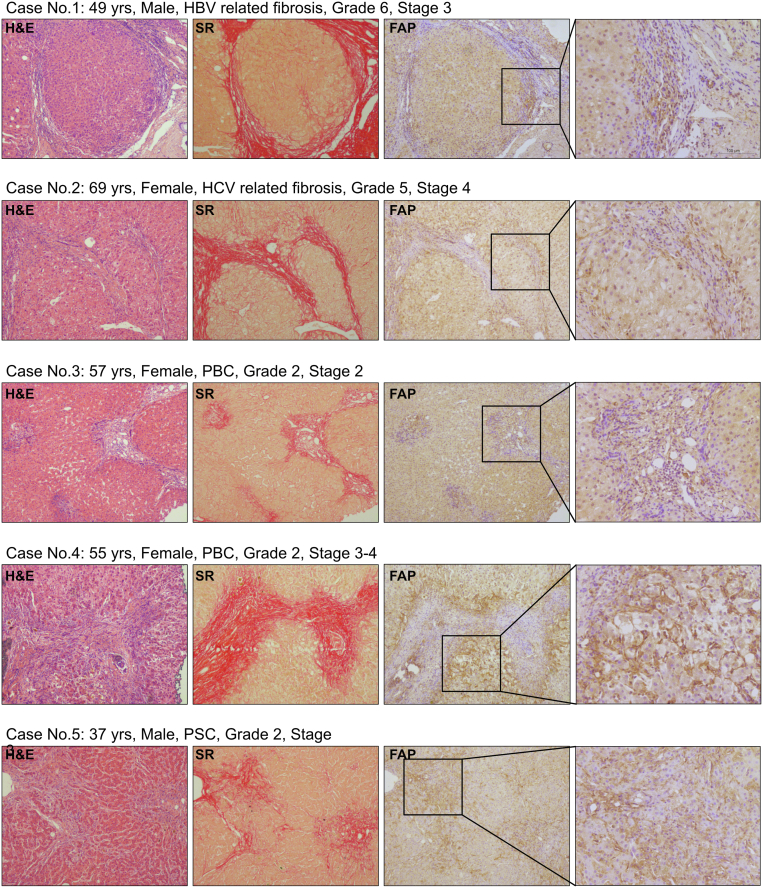
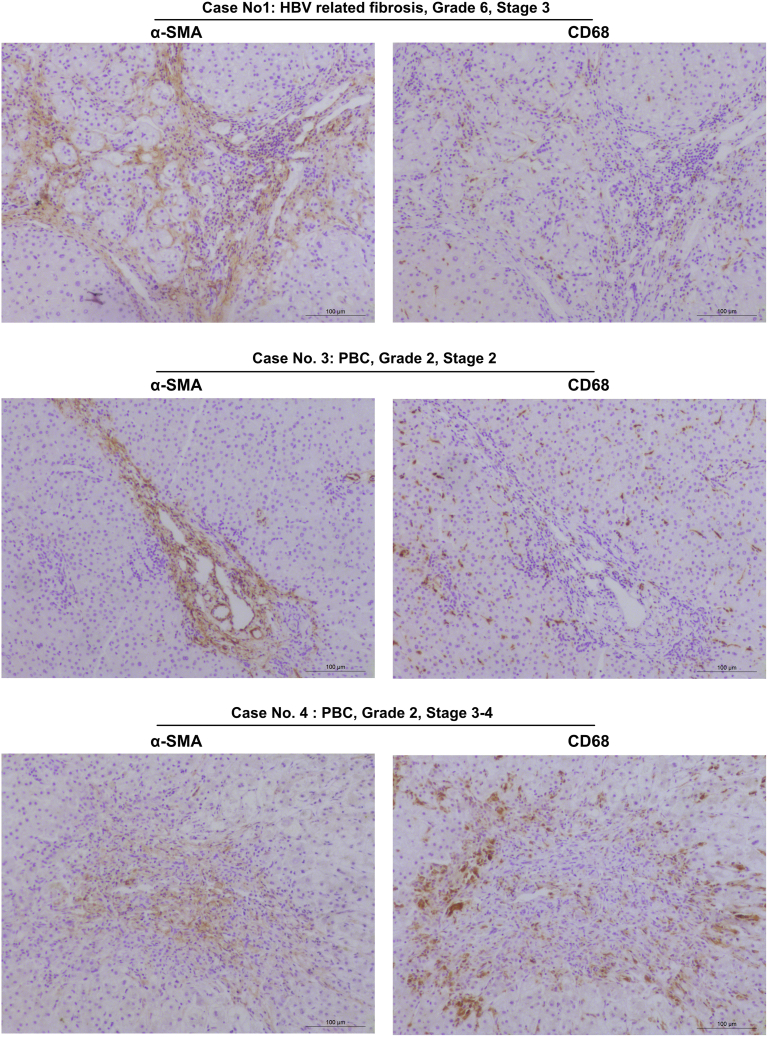
We performed FAP immunohistochemistry on livers of patients with advanced fibrosis due to hepatitis B virus/hepatitis C virus infection, primary biliary cholangitis (PBC), or primary sclerosing cholangitis (PSC), to characterize FAP expression in human vs mouse liver fibrosis. As in mice, FAP was expressed in HSC/fibroblasts at the periphery of (expanding) fibrotic septa and portal fields, whereas α-SMA was found in more central (established) fibrotic areas, and near to CD68+ macrophages (Figure 14; Figure 15). Based on our experimental data, this suggests that FAP that is expressed in areas of expanding septa plays a similar role in murine and human mesenchymal-epithelial interactions and tissue remodeling via modulation of macrophage activity and polarization.
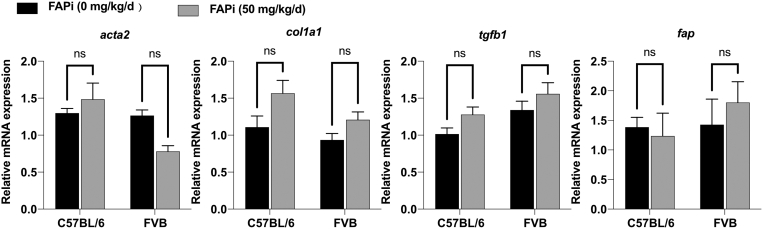
Figure 16.
In vivo FAP inhibitor administration has no effect on fibrosis related gene expression in normal C67BL/6 and FBV mouse strains. Female C56BL/6 (10 weeks old; n = 5) or FBV (4 weeks old; n = 5) background health control mice were fed chow with or without FAP inhibitor (FAPi) (50 mg/kg/d) for 2 weeks. No difference of hepatic transcript levels of acta2, col1a1, tgfb1, and fap between FAPi-treated and FAPi-treated FVB and C57BL/6 mice. Data are presented as means ± standard error of the means (SEMs). Statistical analysis was performed using an unpaired 2-tailed Student t test (ns, not significant).
Figure 14.
Expression of FAP protein in livers of patients with chronic liver diseases. Representative H&E, Sirius red, and FAP staining indicates prominent expression on HSC/fibroblasts at parenchymal-mesenchymal interfaces and adjacent to inflammatory infiltrates containing macrophages.
Figure 15.
Sequential staining CD68 and α-SMA in livers of patients with advanced chronic liver diseases. Representative images of sequential staining of liver with cirrhosis due to hepatitis B, and PBC stage 2 and stage 4. CD68+ macrophages are located peripherally of areas occupied by α-SMA+ (myo)fibroblasts, compared with a closer association of macrophages with FAP+ fibroblasts (see Figure 14).
Discussion
FAP is a membrane bound and partly shed cell membrane protein that is mainly expressed on cells of the fibroblast lineage.25 It is nearly absent in healthy tissues, but highly upregulated during tissue remodeling in embryogenesis, cancer, and fibrosis, such as liver fibrosis.24 FAP has weak terminal and stronger endopeptidase activity, can cleave certain ECM proteins, and has been shown to interact with urokinase plasminogen activator receptor that is involved in tissue remodeling.19 However, the role that FAP plays in fibrosis and cancer remains little explored. Moreover, specific pharmacological inhibition of FAP has not been tested as a potential therapy for liver fibrosis, in part due to the lack of FAP-specific inhibitors.
We showed that pharmacological FAPi, using a highly specific inhibitor,26,27 almost normalized the inflammatory response and significantly reduced HSC activation and collagen synthesis and deposition in progressive parenchymal liver fibrosis, which was accompanied by a decrease of total and M2-type macrophages. In contrast, FAPi attenuated portal but not the mild parenchymal fibrosis in biliary fibrotic Mdr2−/− mice, which was accompanied by upregulation of M2-type macrophages. These results suggest that FAP expression and functions depend on the etiology and temporospatial evolution of liver fibrosis, as well as on inflammatory activity, which is high in CCl4-induced fibrosis compared with biliary fibrosis. Previously, a constitutive FAP knockout attenuated fibrosis more pronouncedly in a high inflammatory (bleomycin) vs a low inflammatory (TGFβ1-induced) mouse model of pulmonary fibrosis,16 which is in line with the higher antifibrotic efficacy of FAPi in high FAP expressing CCl4 vs Mdr2−/− fibrotic livers.
Our finding that FAP expression and profibrotic function is highest in an inflammatory milieu is further supported by a marked FAPi-induced decrease in inflammation, together with a reduction of total and M2-type macrophages, cells implicated as drivers of fibrogenesis28, 29, 30 in CCl4-fibrotic livers. FAP expression, M2-type macrophages, inflammation, and HSC activation are rapidly downregulated once CCl4 is discontinued, coupled with a slow but steady liver remodeling and modest signs of fibrosis regression, as observed before.23 Along with downregulated FAP expression after removal of CCl4, FAPi lacked efficacy to further promote fibrosis regression, which may suggest a cooperation between FAP-expressing HSCs/fibroblasts and adjacent macrophages in generating a fibrotic response. Our prior finding that M2-type macrophages can promote both inflammation and fibrogenesis in highly inflammatory fibrosis maintained by CCl428 is in line with the findings of our present study.
Although previous findings showed that FAP is expressed in a subset of activated HSCs/fibroblasts, functional characterization suggested that FAP expressing fibroblasts likely play a distinct role in the fibro-inflammatory response to injury relative to α-SMA expressing myofibroblasts.17,21,31 Accordingly, distinct subsets of (myo)fibroblasts are responsible for mediating either inflammation or tissue damage in arthritis.21,32 Indeed, FAP expression is induced by several lymphocyte/macrophage-derived cytokines such as TNFα, MCP1, and IL6. Moreover, FAP and α-SMA are both markers of activated HSC or (myo)fibroblasts, but do not appear to colocalize.15,33 In accordance with our mouse data, we found FAP+ fibroblasts at the portal-parenchymal interface, whereas α-SMA+ (myo)fibroblasts were localized more centrally in fibrotic areas, suggesting roles in ECM remodeling vs consolidation, respectively. Therefore, targeting specific fibroblast subpopulations offers the possibility to differentially modulate inflammation and suppress fibrogenesis.
Promotion of hepatocyte regeneration and restoration of liver function are important characteristics of promising antifibrotic therapies.34 We observed low hepatocyte and mesenchymal cell proliferation in active CCl4-induced liver fibrosis, whereas FAP inhibition promoted the proliferation of hepatocytes 10-fold and of HSCs 4-fold. Mildly activated HSCs may assist liver regeneration by producing angiogenic molecules and factors that modulate endothelial cell and hepatocyte proliferation, all resulting in favorable ECM remodeling.35 Expression of PDGFRβ, an early feature of HSC activation,36 was upregulated not only during hepatic fibrogenesis, but also during spontaneous fibrosis regression. This suggests that PDGFRβ-induced (myo)fibroblast proliferation may not only result in fibrogenesis, but may also be necessary for favorable tissue regeneration, in line with prior wound healing studies.37
The Mdr2−/− model of biliary fibrosis is characterized by low level inflammation38 and modest FAP expression (compared with the inflammatory CCl4-induced parenchymal fibrosis (Figure 9, A–B). In Mdr2−/− mice, FAPi also attenuated total collagen deposition, with lesser effect on inflammation, an increase of portal-septal M2-type macrophages, with partly opposing effects on fibrosis related transcripts, and inhibition of hepatocyte proliferation. Reduction of septal α-SMA+ myofibroblasts and loosely organized collagen fibrils were accompanied by induction of mmp13, a macrophage-derived interstitial collagenase,39 in sum explaining septal fibrolytic remodeling of the FAP-expressing expanding septa by MMP-13 macrophages.
Because differences of genetic background between FVB and C57BL/6 mice could affect susceptibility to fibrosis and FAPi,40 we compared the profibrogenic gene expression in both nonfibrotic strains with vs without treatment with FAPi for 2 weeks. As shown in Figure 16, there was no strain difference in transcripts for col1a1, acta2, tgfb1, and fap, regardless of FAPi administration.
Figure 17.
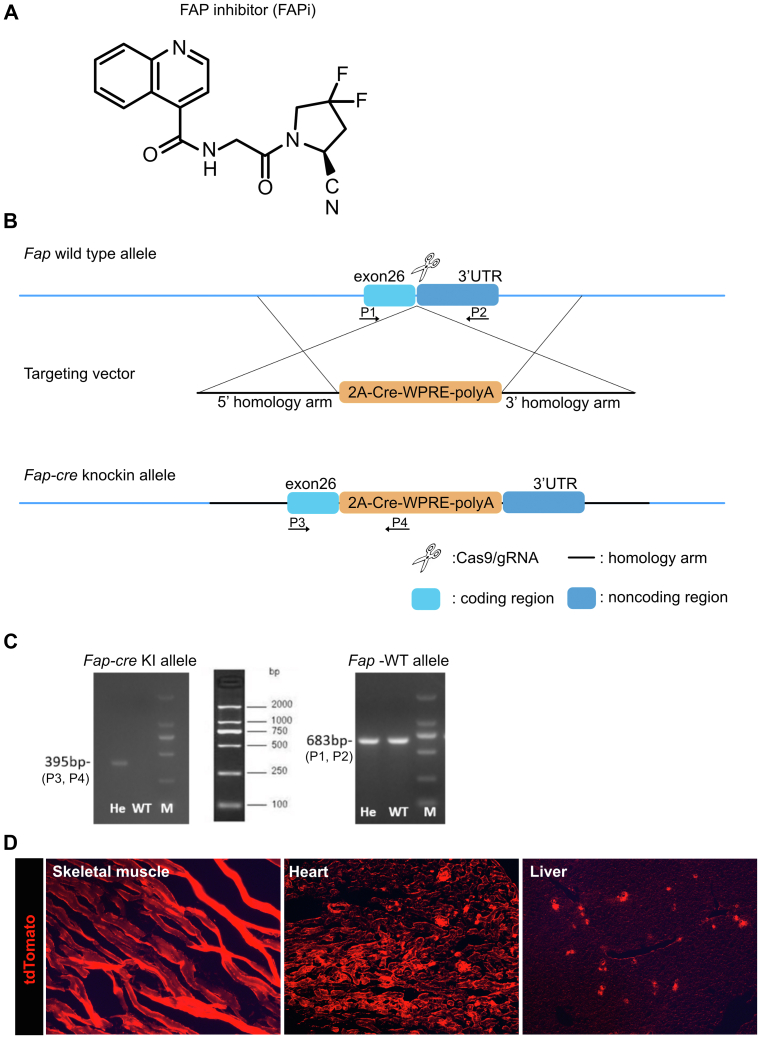
Formula and specificity of the FAP inhibitor (A) and targeting strategy to generate Fap-cre-knock in (KI) mice (B–C). (B), Diagram of the Fap-WT and Fap-Cre-KI alleles. C57BL/6 embryonic stem cells were transfected by the CRISPR/Cas9 technology using 2 ∼ 3.0 kb homologous arms on both sides of the 2A-Cre-WPRE (woodchuck posttranscriptional regulatory element)-polyA insertion sequence. Positive clones were selected with ampicillin. The Cre element in the Fap-cre-KI were inserted in exon 26, between the Fap STOP and polyadenylation signals. After embryo transfer, founder mice (F0) with successful homologous recombination were bred with C57BL/6 mice to obtain the F1 generation (Biomodel Organism Science & Technology Development, Shanghai). (C), Genotyping of WT and heterozygous (He) mice by PCR using primer pairs specific to the Fap-cre allele and Fap-WT allele confirmed correct insertion. To detect the constitutive KI allele (395-bp fragment) as well as the WT allele (683-bp fragment) by PCR. The following primers were used: (P3) 5′-CTTAATGCACCAGTTCTATC-3′, forward; (P4) 5′-GAGCATCTTCCAGGTGTG -3′, reverse; (P1) 5′-TGCCGCACTTATGCAATGAAGACAAT-3′, forward; (P2) 5′-CCCGGCAATGAACAGGTGATAAAACA-3′, reverse. 2A: ‘self-cleaving’ 2A peptide from porcine teschovirus-1. (D), tdTomato is expressed in FAP-positive cells upon crossing the reporter mice (LSL-tdTomato) with FAP-cre mice. FAP-expressing cells are detected in muscle cells of striated muscle, heart, and liver.
Fibrosis results from a dysbalance of ECM deposition vs ECM degradation. FAP has both dipeptidyl peptidase and protease activity for ECM substrates like collagens type I, III, and V, and fibrillin-2.12 Our study supports a profibrotic role in both portal and parenchymal liver fibrosis, especially in regions where inflammation dominated by macrophages prevails. We demonstrated that this profibrotic role of FAP involves the interaction of FAP expressed on fibroblasts with macrophages, because recombinant murine FAP promoted a profibrotic M2-phenotype in vitro. Thus, the role of FAP appears to go far beyond its mere activity as modestly active ECM protease, because rmFAP regulated several hundred genes in macrophages that are related to tissue remodeling and fibrosis, including spp1 encoding osteopontin, an important driver of liver fibrosis.41, 42, 43
Besides using specific pharmacological inhibition of FAP activity, depletion of FAP using genetic strategies can provide supplementary information on the role of FAP in fibrosis. To date only 2 studies have been published, where constitutive FAP knockout had a modest effect on bleomycin-induced pulmonary fibrosis progression16 or on protection from the development of atherosclerosis.44 However, constitutive FAP knockout cannot be compared with pharmacological inhibition with our highly specific FAPi.45, 46, 47 Thus, Panaro et al45 found that, although specific pharmacological FAP inhibition improved glucose tolerance in mice with diet induced obesity, FAP knockout did not protect these mice from insulin resistance, suggesting that elimination of FAP activity rather than constitutive genetic loss of FAP is essential for the beneficial effect. Importantly, pharmacological FAPi is a better proof of function than mice with a constitutive knockout that favors early adaptation to the gene deficiency, often not allowing to make conclusions about pharmacological intervention.
Although more studies need to be done, we provide molecular and functional evidence of a role of FAP in driving fibrogenesis in areas of active fibroproliferation, as characterized by high cellularity, especially in the presence of active HSC/fibroblasts and macrophages that we implicate as a fibrogenic unit that is modulated by FAP. In line with our functional and colocalization studies of (M2-type) macrophages with activated HSCs/(myo)fibroblasts in our fibrosis models, we observe a comparable colocalization of FAP-expressing immune cells/macrophages with activated HSC/(myo)fibroblasts in human liver fibrosis.
Overall, pharmacological FAP inhibition has an anti-inflammatory and anti-fibrotic effect mainly in active parenchymal compared with a modest effect in biliary murine liver fibrosis, acting in interface regions where FAP expression in HSCs/fibroblasts is high. We conclude that FAP promotes (fibrogenic) activation of adjacent macrophages involving an upregulation of several genes implicated in fibrogenesis, including spp1. Because immunohistochemistry suggests a similar role of FAP in human liver fibrosis, specific FAP inhibitors may serve as a novel class of antifibrotics in active human liver fibrogenesis.
Materials and Methods
Liver Fibrosis Models
Parenchymal fibrosis
Seven- to eight-week-old female C57BL/6 mice received escalating doses of CCl4 in mineral oil via oral gavage 3 times a week for up to 6 weeks (first dose, 0.875 mL/kg; 1.75 mL/kg from week 1–3; 2.5 mL/kg from week 4 onwards).23 Mice receiving mineral oil alone (200 μL) served as nonfibrotic controls. Spontaneous fibrosis regression was assessed 2 weeks after discontinuation of CCl4 treatment.
Biliary fibrosis
Adult female Mdr2−/− mice (FVB background, bred at the Institute of Translational Immunology) that develop spontaneous PSC-like fibrosis with maximal progression between 4 and 12 weeks of age,38 were analyzed at age 6 to 7 weeks. Age-matched WT FBV mice served as non-fibrotic controls. Animal experiments were performed in accordance with institutional and German legal guidelines for animal protection. This study was approved by the Animal Protection and Care Committee of the State of Rhineland-Palatinate (approval No. 23177-07G12-1-007).
Treatment With FAP Inhibitor In Vivo
The recently published FAP-specific inhibitor (FAPi) (S)-N-(2-cyano-4,4-difluoropyrrolidin-1-yl)-2-oxoethyl)-quinoline-4-carboxamide (compound 60)22 was reconfirmed at Boehringer-Ingelheim for its FAP vs DPP-4 inhibitory activity of 1 nM vs >85 μM, respectively (Figure 17, A). Mice received FAPi mixed in the chow (Ssniff, Soest, Germany; S8251) at a concentration of 15 mg/kg body weight (bw) (n = 10) or 50 mg/kg bw (n = 10), based on an average chow consumption of 3 g/d, or chow without inhibitor as control (n = 5) for 2 weeks (week 5 to 6 of CCl4 induction, week 4 to 6 of age for Mdr2−/− mice. In the CCl4 regression model, FAPi (50 mg/kg bw) (n = 10) or untreated control (n = 10) was administered for the 2 weeks off CCl4.
Generation of Fap-cre Transgenic Mice
Fap-cre transgenic mice were generated and confirmed using the CRISPR/Cas9 technology as detailed in Figure 17, B–D. Subsequently, the Fap-cre/+ mice were crossed with a reporter mouse containing a stop cassette flanked by loxp sites upstream of tdTomato (tdT) at the Rosa26 locus (Rosa26-loxp-stop-loxp-tdTomato) to generate Fap-cre-tdTomato reporter mice on the C57BL/6 background. To localize FAP-expressing HSC/fibroblasts, Fap-cre-tdTomato reporter mice received CCl4 for 2 or 6 weeks.
In Vitro Experiments
LX2 (human hepatic stellate cells), NIH/3T3 (murine fibroblasts), THP1 (human monocytic cell line), and mouse BMDMs were used for in vitro experiments. LX2 and NIH/3T3 were cultured at 37 °C/5% CO2 in complete or in low-serum medium (Dulbecco's Modified Eagle Medium, 10% or 1% fetal bovine serum [FBS], respectively, plus 1% penicillin/streptomycin). THP1 cells were cultured at 37 °C/5% CO2 in complete medium (RPMI 1640, 10% FBS, 1% penicillin/streptomycin, 0.05 mM 2-mercaptoethanol). BMDMs were extracted from femur and tibia of C57BL/6 mice and cultured at 37 °C/5% CO2 in Iscove’s Modified Dulbecco’s Medium supplemented with 10% FBS, 1% penicillin/streptomycin, and recombinant murine M-CSF (20 ng/mL). After 7 days in culture, BMDMs were treated with 20 ng/mL IL-4 + 20 ng/mL IL-13 for M2 polarization.
For in vitro stimulation, recombinant human FAP (BioLegend, 768902), recombinant mouse FAP (R&D, 8647-SE), FAP inhibitor were used in low-serum medium. For FAP knockdown experiments, LX2 cells were pretreated with 5 ng/ml TGFβ1 for 24 hours, and then transfected with FAP small interfering or negative control (Syngentech) using EndoFectin MAX (GeneCopoeia, EF013) according to the manufacturer’s protocol. The sequences of the siRNAs are presented in Table 1. Their final concentration was 50 nM. The transfected cells were used for experiments after 24 hours. For FAP overexpression experiments, LX2 cells at 80% confluence were transfected using EndoFectin MAX following the manufacturer’s protocol. Cells were transfected with 250 ng/cm2 of pcDNA3.1-FAP plasmids or pcDNA3.1-NC plasmids (Syngentech). Two days after transfection, cells were used for experiments. To explore the effect of conditioned medium from FAPi-treated LX2 cells on the function and polarization of macrophages, THP1 cells were differentiated into macrophages with 100 nM PMA (Sigma-Aldrich, P1585) in complete medium. After 24 hours, differentiated cells were adherent, washed one time with phosphate buffered saline and treated with conditioned medium for 12 hours. All experiments were performed in triplicate.
Table 2.
Primary Antibodies Used for IF, IHC-P, IHC-Fr, and WB
| Antibody | Species | Company | Catalog no. | Dilution |
|---|---|---|---|---|
| α-SMA (IHC-P) | Rabbit anti-mouse | Abcam | ab124964 | 1:200 |
| α-SMA (IHC-Fr) | Goat anti-mouse | Novus | NB300-978 | 1:500 |
| CD68 (IHC-P/Fr) | Rabbit anti-mouse | ThermoFisher | PA5-78996 | 1:500 |
| CD3 (IHC-P) | Rabbit anti-mouse | Abcam | ab21703 | Read to use |
| YM-1 (IHC-P) | Rabbit anti-mouse | Stem cell Technologies | 60130 | 1:50 |
| Ki67 (IHC-P) | Rabbit anti-mouse | Abcam | ab21700 | Ready to use |
| Ki67 (IF) | Rabbit anti-human | Abcam | ab21700 | Ready to use |
| FAP (IHC-P) | Sheep anti-human | R&D | AF3715 | 1:15 |
| GFAP (IHC-Fr) | Mouse anti-mouse | Sigma-Aldrich | G3893 | 1:400 |
| Desmin (IHC-Fr) | Rabbit anti-mouse | ThermoFisher | PA5-16705 | 1:500 |
| Collagen type I (IHC-Fr) | Rabbit anti-mouse | ThermoFisher | PA5-95137 | 1:500 |
| CD68 (IHC-P) | Mouse anti-human | MAIXIN. BIO | MAB-0863 | Ready to use |
| α-SMA (IHC-P) | Mouse anti-human | MAIXIN. BIO | MAB-0890 | Ready to use |
| α-SMA (WB) | Rabbit anti-human | Abcam | ab5694 | 1:1000 |
| Collagen type I (WB) | Rabbit anti-human | ThermoFisher | PA5-95137 | 1:1000 |
| FLAG (WB) | Mouse anti-human | CST | 8146 | 1:1000 |
| GAPDH (WB) | Rabbit anti-human | Abcam | ab9485 | 1:5000 |
α-SMA, α-smooth muscle actin; FAP, fibroblast activation protein; IF, immunofluorescence; IHC-Fr, immunohistochemistry -frozen sections; IHC-P, immunohistochemistry -paraffin sections; WB, Western blot.
Preparation of Conditioned Media (CM) From FAPi-treated LX2 Cells
LX2 cells were treated with different concentrations of FAPi in low-serum medium for 24 hours, washed to remove FAPi, and then received fresh serum-free medium for further incubation. After 12 hours, the medium was collected and centrifuged at 12,000 rpm for 10 minutes at 4 °C to eliminate cell debris. Each CM was stored at −80 °C for further use.
Hepatic Collagen Content
Hepatic collagen content was determined as relative hydroxyproline (μg/g liver) in 250- to 350-mg liver samples from 2 different lobes after hydrolysis in 6N HCl for 16 hours at 110 °C as described.48 Total hydroxyproline (mg/whole liver) was calculated based on individual liver weights and the corresponding relative hydroxyproline content.
Serum Biochemistry
Whole blood was collected by cardiac puncture at sacrifice, and serum stored frozen at −80 °C. ALT, AST, ALP, and creatinine were determined by the Central Clinical Laboratory of the University Medical Center Mainz with validated clinical laboratory methods.
Histology and Immunohistochemistry
Upon sacrifice, the left liver lobe was collected, fixed in formalin, paraffin-embedded, sectioned at 4 μm, and routinely processed for H&E staining. Sirius red staining and immunohistochemistry were performed as described previously.49, 50, 51 Primary antibodies used were anti-FAP, anti-α-SMA, anti-CD68, anti-YM1, anti-CD3, and anti-Ki67 (for details of primary antibodies, see Table 2) and visualized with the anti-sheep HRP-DAB Cell & Tissue Staining Kit (RD Systems, CTS019) (for FAP staining) or the ABC staining kit (Vector Labs, PK-4001) (for α-SMA, CD68, YM1, CD3, Ki67 staining). Sirius red- and α-SMA-positive areas were quantified using Image J software from 10 random, non-overlapping fields per mouse and 5 to 10 mice per group at 200× magnification. Grading and staging of fibrosis and inflammation in H&E sections was performed by an expert liver pathologist (X.Y.Z.) according to Ishak scores 0–6.52
Table 1.
Primers and Probes Used in Quantitative Real-time Polymerase Chain Reaction
| Gene | Primer sequence | TaqMan probe |
|---|---|---|
| macta2 | F: 5′-ACAGCCCTCGCACCCA-3′ | CAAGATCATTGCCCCTCCAGAACGC |
| R: 5′- GCCACCGATCCAGACAGAGT -3′ | ||
| mcol1a1 | F: 5′- TCCGGCTCCTGCTCCTCTTA -3′ | TTCTTGGCCATGCGTCAGGAGGG |
| R: 5′- GTATGCAGCTGACTTCAGGGATGT -3′ | ||
| mcol3a1 | F: 5′- AATGGTGGCTTTCAGTTCAGCT -3′ | TGGAAAGAAGTCTGAGGAAGGCCAGCTG |
| R: 5′- TGTAATGTTCTGGGAGGCCC -3′ | ||
| mpdgfrb | F: 5′- TGGTCCTTTGGAATCCTACTCTG -3′ | AAATCTTCACACTGGGTGGCACCCCTT |
| R: 5′- GGTCGTTCATGGGCAGCT -3′ | ||
| mtimp1 | F: 5′- TCCTCTTGTTGCTATCACTGATAGCTT -3′ | TTCTGCAACTCGGACCTGGTCATAAGG |
| R: 5′- CGCTGGTATAAGGTGGTCTCGTT -3′ | ||
| mmmp2 | F: 5’- CCGAGGACTATGACCGGGATAA -3′ | TCTGCCCCGAGACCGCTATGTCCA |
| R: 5′- CTTGTTGCCCAGGAAAGTGAAG -3′ | ||
| mmmp9 | F: 5′- CAGGATAAACTGTATGGCTTCTGC -3′ | CTACCCGAGTGGACGCGACCGT |
| R: 5′- GCCGAGTTGCCCCCA -3′ | ||
| mmmp12 | F: 5′- CTGCTCCCATGAATGACAGTG -3′ | |
| R: 5′- AGTTGCTTCTAGCCCAAAGAAC -3′ | ||
| mmmp13 | F: 5′- GGAAGACCCTCTTCTTCTCT -3′ | TCTGGTTAACATCATCATAACTCCACACGT |
| R: 5′- TCATAGACAGCATCTACTTTGTT-3′ | ||
| mspp1 | F: 5′- AGCAAGAAACTCTTCCAAGCAA -3′ | - |
| R: 5′- GTGAGATTCGTCAGATTCATCCG -3′ | ||
| mtgfb1 | F: 5′- AGAGGTCACCCGCGTGCTAA -3′ | ACCGCAACAACGCCATCTATGAGAAAACCA |
| R: 5′- TCCCGAATGTCTGACGTATTGA -3′ | ||
| mtgfb2 | F: 5′- GTCCAGCCGGCGGAA -3′ | CGCTTTGGATGCTGCCTACTGCTTTAGAAAT |
| R: 5′- GCGAAGGCAGCAATTATCCT -3′ | ||
| mccl2 | F: 5′- CGGAACCAAATGAGATCAGAACCTAC -3′ | - |
| F: 5′- GCTTCAGATTTACGGGTCAACTTCAC -3′ | ||
| minos | F: 5′- GGAGCAGGTGGAAGACTATTTCTT -3′ | - |
| R: 5′- CATGATAACGTTTCTGGCTCTTGA -3′ | ||
| mym1 | F: 5′- GGGCATACCTTTATCCTGAG-3′ | - |
| R: 5′- CCACTGAAGTCATCCATGTC -3′ | ||
| marg1 | F: 5′- GGTCCAGAAGAATGGAAGAGTCAG -3′ | - |
| R: 5′- CAGATATGCAGGGAGTCACC -3′ | ||
| mfap | F: 5′- CCGCGTAACACAGGATTCACTG-3′ | - |
| R: 5′- CACACTTCTTGCTCGGAGGAGA -3′ | ||
| mil6 | F: 5′- ACCAGAGGAAATTTTCAATAGGC -3′ | - |
| R: 5′- TGATGCACTTGCAGAAAACA -3′ | ||
| mil10 | F: 5′- GCTCTTACTGACTGGCATGAG -3′ | - |
| R: 5′- CGCAGCTCTAGGAGCATGTG -3′ | ||
| mgapdh | F: 5′- AGGTCGGTGTGAACGGATTTG-3′ | - |
| R: 5′- GGGGTCGTTGATGGCAACA-3′ | ||
| hfap | F: 5′- GGAAGTGCCTGTTCCAGCAATG-3′ | |
| R: 5′- TGTCTGCCAGTCTTCCCTGAAG-3′ | ||
| hacta2 | F: 5′- CTATGCCTCTGGACGCACAACT-3′ | |
| R: 5′- CAGATCCAGACGCATGATGGCA-3′ | ||
| hcol1a1 | F: 5′- GATTCCCTGGACCTAAAGGTGC-3′ | |
| R: 5′- AGCCTCTCCATCTTTGCCAGCA-3′ | ||
| hcol3a1 | F: 5′- TGGTCTGCAAGGAATGCCTGGA-3′ | |
| R: 5′- TCTTTCCCTGGGACACCATCAG-3′ | ||
| htgfb1 | F: 5′- TACCTGAACCCGTGTTGCTCTC-3′ | |
| R: 5′- GTTGCTGAGGTATCGCCAGGAA-3′ | ||
| hccl2 | F: 5′- AGGTCGGTGTGAACGGATTTG-3′ | |
| R: 5′- GGGGTCGTTGATGGCAACA-3′ | ||
| hil10 | TCTCCGAGATGCCTTCAGCAGA | |
| TCAGACAAGGCTTGGCAACCCA | ||
| hloxl1 | F: 5′- ACAGCACCTGTGACTTCGGCAA-3′ | |
| R: 5′- CGGTTATGTCGATCCACTGGCA-3′ | ||
| htimp1 | F: 5′- AGGTCGGTGTGAACGGATTTG-3′ | |
| R: 5′- GGGGTCGTTGATGGCAACA-3′ | ||
| hinos | F: 5′- AGGTCGGTGTGAACGGATTTG-3′ | |
| R: 5′- GGGGTCGTTGATGGCAACA-3′ | ||
| hmmp1 | F: 5′- ATGAAGCAGCCCAGATGTGGAG-3′ | |
| R: 5′- TGGTCCACATCTGCTCTTGGCA-3′ | ||
| hmmp9 | F: 5′- GCCACTACTGTGCCTTTGAGTC -3′ | |
| R: 5′- CCCTCAGAGAATCGCCAGTACT -3′ | ||
| hmmp14 | F: 5′- CCTTGGACTGTCAGGAATGAGG-3′ | |
| R: 5′- TTCTCCGTGTCCATCCACTGGT-3′ | ||
| hgapdh | F: 5′- GTCTCCTCTGACTTCAACAGCG-3′ | |
| R: 5′- ACCACCCTGTTGCTGTAGCCAA-3′ | ||
| siNC | 5′- UUCUCCGAACGUGUCACGUTT-3′ | |
| 5′- ACGUGACACGUUCGGAGAATT-3′ | ||
| sifap | 5′- CCUUAGCAAUGGAGAAUUUTT-3′ | |
| 5′- AAAUUCUCCAUUGCUAAGGTT-3′ |
Immunofluorescence
Immunofluorescent staining of liver tissues was performed as we reported.50 Briefly, liver tissues were fixed with 10% neutral buffered formalin and cut into sections of 5-um thickness. After the non-specific binding sites were blocked with 10% donkey serum for 1 hour at room temperature, the sections were incubated with anti-α-SMA antibody, anti-CD68 antibody, anti-desmin, anti-(GFAP), and anti-collagen type I (for details of primary antibodies, see Table 2) at 4 °C overnight, washed extensively in phosphate buffered saline, with Alexa Fluor 488/647 conjugated secondary antibodies (1:500, ThermoFisher, A-21206/A-31573) for 1 hour. Sections were then counterstained with 4, 6-diamidino-2-phenylindole (DAPI), washed, and mounted for observation under a scanning confocal microscope (Olympus, Fluoview FV1000).
Western Blot
Whole cell pellets were homogenized in ice-cold NP-40 cell lysis buffer (ThermoFisher, FNN0021) supplemented with a protease inhibitor cocktail (Roche, 4693159001). Equal amounts of cell lysate protein (20 μg) were loaded onto 12% acrylamide SDS-PAGE gels, resolved, and transferred to nitrocellulose. The membranes were incubated with primary antibodies in 5% nonfat dry milk blocking solution overnight at 4 °C and then incubated with secondary antibody. Proteins were visualized using a chemiluminescent substrate (Invitrogen, Rockford). GAPDH was used as a loading control. Primary antibodies used in Western blot are listed in in Table 2.
Gene Expression Analysis
Approximately 100 mg liver tissue from 2 different mouse liver lobes were snap frozen and homogenized using TissueLyser II (Qiagen) in Ribozol (VWR, N580). Cell pellets were obtained from 3 × 105 cells. The mRNA from whole cell pellet or liver tissue was extracted using Ribozol, followed by reverse transcription of 500 ng RNA using the qScript Reverse Transcription System (Quantabio, 95217). Relative transcript levels were quantified by quantitative polymerase chain reaction (PCR) on a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA), using the TaqMan and SYBR Green methodologies as described by us in detail.28,53 Expression levels were corrected using GAPDH as reference gene (ΔCt) and compared with the control group (ΔΔCt) using the 2C(t) method. Validated TaqMan probes and primers for qPCR are shown in Table 1.
RNA Sequencing and Data Processing
RNA-seq was performed using RNA extracted from BMDM-M2 with and without treatment with rmFAP. Each group consisted of 3 biological replicates. A cDNA library was clustered and sequenced on the Illumina NovaSeq 6000 platform. The sequencing reads were aligned to mouse genomes in ENSEMBL database. Genes were classified as differentially expressed based on a cut-off of adjusted q-value <0.05 and ≥1.5-fold change. Pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes pathway classification and functional enrichment. Clustered heatmaps were generated from all differently expressed genes using GraphPad Prism 9.3
Human Samples
Liver tissue from patients undergoing liver transplantation for cirrhosis due to hepatitis B or hepatitis C infection (each n = 1), PBC (n = 2), and PSC (n = 1) was collected at Beijing Friendship Hospital, Capital Medical University. Approved was by the Hospital’s Ethics Committee (nr. 2020-P2-118-01).
Statistical Analyses
Data are expressed as means ± standard error of the mean, and statistical analyses were performed using GraphPad Prism version 6.00 (GraphPad Software, San Diego, CA). The mean values of 2 groups were compared by an unpaired Student t test. Multiple comparisons were performed by one-way analysis of variance. Differences among selected experimental groups with P values lower than .05 were considered significant.
Acknowledgments
CRediT Authorship Contributions
Aiting Yang, PhD (Data curation: Lead; Funding acquisition: Supporting; Methodology: Equal; Writing – original draft: Lead; Writing – review & editing: Supporting)
Yong-Ook Kim, PhD (Data curation: Equal; Methodology: Equal; Writing – review & editing: Supporting)
Xu-Zhen Yan, MD (Data curation: Equal; Methodology: Equal; Writing – review & editing: Supporting)
Hiroyuki Abe, MD, PhD (Data curation: Supporting; Writing – review & editing: Supporting)
Misbah Aslam, PhD (Data curation: Supporting; Writing – review & editing: Supporting)
Kyoung-Sook Park, MSCi (Data curation: Supporting; Writing – review & editing: Supporting)
Xin-Yan Zhao, MD (Data curation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Ji-Dong Jia, MD, PhD (Supervision: Supporting; Writing – review & editing: Supporting)
Thomas Klein, PhD (Conceptualization: Equal; Investigation: Supporting; Writing – review & editing: Supporting)
Hong You, MD, PhD (Conceptualization: Supporting; Funding acquisition: Equal; Supervision: Equal; Writing – review & editing: Equal)
Detlef Schuppan, MD, PhD (Conceptualization: Lead; Funding acquisition: Lead; Investigation: Lead; Supervision: Equal; Writing – review & editing: Lead)
Footnotes
Conflicts of interest This author discloses the following: Thomas Klein is an employee of Boehringer-Ingelheim. The remaining authors disclose no conflicts.
Funding Aiting T. Yang was supported by a grant from Beijing Li Huanying Medical Foundation. Detlef Schuppan received project-related funding from the German Research Foundation (DFG) Collaborative Research Center (CRC) grants SFB 1066 project B3 and CRC 1292 project B10, and by EU Horizon 2020 projects under grant agreements nr. 634413 (EPoS, European Project on Steatohepatitis) and nr. 777377 (LITMUS, Liver Investigation on Marker Utility in Steatohepatitis). Hong You was supported by the National Natural Science Foundation of China (project no. 81970524 and 82130018).
References
- 1.Asrani S.K., Devarbhavi H., Eaton J., et al. Burden of liver diseases in the world. J Hepatol. 2019;70:151–171. doi: 10.1016/j.jhep.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda M., Seki E. The liver fibrosis niche: novel insights into the interplay between fibrosis-composing mesenchymal cells, immune cells, endothelial cells, and extracellular matrix. Food Chem Toxicol. 2020;143 doi: 10.1016/j.fct.2020.111556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuppan D., Afdhal N.H. Liver cirrhosis. Lancet. 2008;371:838–851. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kisseleva T., Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2021;18:151–166. doi: 10.1038/s41575-020-00372-7. [DOI] [PubMed] [Google Scholar]
- 5.Rockey D.C., Friedman S.L. Fibrosis regression after eradication of hepatitis C virus: from bench to bedside. Gastroenterology. 2021;160:1502–1520.e1. doi: 10.1053/j.gastro.2020.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kisseleva T., Cong M., Paik Y., et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci U S A. 2012;109:9448–9453. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X., Xu J., Rosenthal S., et al. Identification of lineage-specific transcription factors that prevent activation of hepatic stellate cells and promote fibrosis resolution. Gastroenterology. 2020;158:1728–1744.e14. doi: 10.1053/j.gastro.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuppan D., Kim Y.O. Evolving therapies for liver fibrosis. J Clin Invest. 2013;123:1887–1901. doi: 10.1172/JCI66028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuppan D., Ashfaq-Khan M., Yang A.T., et al. Liver fibrosis: direct antifibrotic agents and targeted therapies. Matrix Biol. 2018;68-69:435–451. doi: 10.1016/j.matbio.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Park J.E., Lenter M.C., Zimmermann R.N., et al. Fibroblast activation protein, a dual specificity serine protease expressed in reactive human tumor stromal fibroblasts. J Biol Chem. 1999;274:36505–36512. doi: 10.1074/jbc.274.51.36505. [DOI] [PubMed] [Google Scholar]
- 11.Keane F.M., Nadvi N.A., Yao T.W., et al. Neuropeptide Y, B-type natriuretic peptide, substance P and peptide YY are novel substrates of fibroblast activation protein-alpha. FEBS J. 2011;278:1316–1332. doi: 10.1111/j.1742-4658.2011.08051.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H.E., Hamson E.J., Koczorowska M.M., et al. Identification of novel natural substrates of fibroblast activation protein-alpha by differential degradomics and proteomics. Mol Cell Proteomics. 2019;18:65–85. doi: 10.1074/mcp.RA118.001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y., Kong Z., Liu P., et al. Oncogenesis, microenvironment modulation and clinical potentiality of FAP in glioblastoma: lessons learned from other solid tumors. Cells. 2021;10:1142. doi: 10.3390/cells10051142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaps L., Schuppan D. Targeting cancer associated fibroblasts in liver fibrosis and liver cancer using nanocarriers. Cells. 2020;9:2027. doi: 10.3390/cells9092027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy M.T., McCaughan G.W., Marinos G., et al. Intrahepatic expression of the hepatic stellate cell marker fibroblast activation protein correlates with the degree of fibrosis in hepatitis C virus infection. Liver. 2002;22:93–101. doi: 10.1034/j.1600-0676.2002.01503.x. [DOI] [PubMed] [Google Scholar]
- 16.Kimura T., Monslow J., Klampatsa A., et al. Loss of cells expressing fibroblast activation protein has variable effects in models of TGF-beta and chronic bleomycin-induced fibrosis. Am J Physiol Lung Cell Mol Physiol. 2019;317:L271–L282. doi: 10.1152/ajplung.00071.2019. [DOI] [PubMed] [Google Scholar]
- 17.Avery D., Govindaraju P., Jacob M., et al. Extracellular matrix directs phenotypic heterogeneity of activated fibroblasts. Matrix Biol. 2018;67:90–106. doi: 10.1016/j.matbio.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizoguchi F., Slowikowski K., Wei K., et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun. 2018;9:789. doi: 10.1038/s41467-018-02892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X., Lin Y., Shi Y., et al. FAP promotes immunosuppression by cancer-associated fibroblasts in the tumor microenvironment via STAT3-CCL2 signaling. Cancer Res. 2016;76:4124–4135. doi: 10.1158/0008-5472.CAN-15-2973. [DOI] [PubMed] [Google Scholar]
- 20.Monslow J., Todd L., Chojnowski J.E., et al. Fibroblast activation protein regulates lesion burden and the fibroinflammatory response in Apoe-deficient mice in a sexually dimorphic manner. Am J Pathol. 2020;190:1118–1136. doi: 10.1016/j.ajpath.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croft A.P., Campos J., Jansen K., et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 2019;570:246–251. doi: 10.1038/s41586-019-1263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansen K., Heirbaut L., Verkerk R., et al. Extended structure-activity relationship and pharmacokinetic investigation of (4-quinolinoyl)glycyl-2-cyanopyrrolidine inhibitors of fibroblast activation protein (FAP) J Med Chem. 2014;57:3053–3074. doi: 10.1021/jm500031w. [DOI] [PubMed] [Google Scholar]
- 23.Popov Y., Sverdlov D.Y., Sharma A.K., et al. Tissue transglutaminase does not affect fibrotic matrix stability or regression of liver fibrosis in mice. Gastroenterology. 2011;140:1642–1652. doi: 10.1053/j.gastro.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob M., Chang L., Pure E. Fibroblast activation protein in remodeling tissues. Curr Mol Med. 2012;12:1220–1243. doi: 10.2174/156652412803833607. [DOI] [PubMed] [Google Scholar]
- 25.Hamson E.J., Keane F.M., Tholen S., et al. Understanding fibroblast activation protein (FAP): substrates, activities, expression and targeting for cancer therapy. Proteomics Clin Appl. 2014;8:454–463. doi: 10.1002/prca.201300095. [DOI] [PubMed] [Google Scholar]
- 26.Koustoulidou S., Hoorens M.W.H., Dalm S.U., et al. Cancer-associated fibroblasts as players in cancer development and progression and their role in targeted radionuclide imaging and therapy. Cancers (Basel) 2021;13:1100. doi: 10.3390/cancers13051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altmann A., Haberkorn U., Siveke J. The latest developments in imaging of fibroblast activation protein. J Nucl Med. 2021;62:160–167. doi: 10.2967/jnumed.120.244806. [DOI] [PubMed] [Google Scholar]
- 28.Weng S.Y., Wang X., Vijayan S., et al. IL-4 receptor alpha signaling through macrophages differentially regulates liver fibrosis progression and reversal. EBioMedicine. 2018;29:92–103. doi: 10.1016/j.ebiom.2018.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazankov K., Jorgensen S.M.D., Thomsen K.L., et al. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat Rev Gastroenterol Hepatol. 2019;16:145–159. doi: 10.1038/s41575-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 30.Tacke F. Targeting hepatic macrophages to treat liver diseases. J Hepatol. 2017;66:1300–1312. doi: 10.1016/j.jhep.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 31.Schmidkonz C., Rauber S., Atzinger A., et al. Disentangling inflammatory from fibrotic disease activity by fibroblast activation protein imaging. Ann Rheum Dis. 2020;79:1485–1491. doi: 10.1136/annrheumdis-2020-217408. [DOI] [PubMed] [Google Scholar]
- 32.McHugh J. Functionally distinct fibroblast subsets in RA. Nat Rev Rheumatol. 2019;15:449. doi: 10.1038/s41584-019-0261-3. [DOI] [PubMed] [Google Scholar]
- 33.Levy M.T., McCaughan G.W., Abbott C.A., et al. Fibroblast activation protein: a cell surface dipeptidyl peptidase and gelatinase expressed by stellate cells at the tissue remodelling interface in human cirrhosis. Hepatology. 1999;29:1768–1778. doi: 10.1002/hep.510290631. [DOI] [PubMed] [Google Scholar]
- 34.Song G., Pacher M., Balakrishnan A., et al. Direct reprogramming of hepatic myofibroblasts into hepatocytes in vivo attenuates liver fibrosis. Cell Stem Cell. 2016;18:797–808. doi: 10.1016/j.stem.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Mabuchi A., Mullaney I., Sheard P.W., et al. Role of hepatic stellate cell/hepatocyte interaction and activation of hepatic stellate cells in the early phase of liver regeneration in the rat. J Hepatol. 2004;40:910–916. doi: 10.1016/j.jhep.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Kinnman N., Goria O., Wendum D., et al. Hepatic stellate cell proliferation is an early platelet-derived growth factor-mediated cellular event in rat cholestatic liver injury. Lab Invest. 2001;81:1709–1716. doi: 10.1038/labinvest.3780384. [DOI] [PubMed] [Google Scholar]
- 37.Lynch S.E., de Castilla G.R., Williams R.C., et al. The effects of short-term application of a combination of platelet-derived and insulin-like growth factors on periodontal wound healing. J Periodontol. 1991;62:458–467. doi: 10.1902/jop.1991.62.7.458. [DOI] [PubMed] [Google Scholar]
- 38.Popov Y., Patsenker E., Fickert P., et al. Mdr2 (Abcb4)-/- mice spontaneously develop severe biliary fibrosis via massive dysregulation of pro- and antifibrogenic genes. J Hepatol. 2005;43:1045–1054. doi: 10.1016/j.jhep.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 39.Fallowfield J.A., Mizuno M., Kendall T.J., et al. Scar-associated macrophages are a major source of hepatic matrix metalloproteinase-13 and facilitate the resolution of murine hepatic fibrosis. J Immunol. 2007;178:5288–5295. doi: 10.4049/jimmunol.178.8.5288. [DOI] [PubMed] [Google Scholar]
- 40.Walkin L., Herrick S.E., Summers A., et al. The role of mouse strain differences in the susceptibility to fibrosis: a systematic review. Fibrogenesis Tissue Repair. 2013;6:18. doi: 10.1186/1755-1536-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Mochida S., Kawashima R., et al. Increased expression of osteopontin in activated Kupffer cells and hepatic macrophages during macrophage migration in Propionibacterium acnes-treated rat liver. J Gastroenterol. 2000;35:696–701. doi: 10.1007/s005350070049. [DOI] [PubMed] [Google Scholar]
- 42.Song Z., Chen W., Athavale D., et al. Osteopontin takes center stage in chronic liver disease. Hepatology. 2021;73:1594–1608. doi: 10.1002/hep.31582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorena D., Darby I.A., Gadeau A.P., et al. Osteopontin expression in normal and fibrotic liver. altered liver healing in osteopontin-deficient mice. J Hepatol. 2006;44:383–390. doi: 10.1016/j.jhep.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Stein S., Weber J., Nusser-Stein S., et al. Deletion of fibroblast activation protein provides atheroprotection. Cardiovasc Res. 2021;117:1060–1069. doi: 10.1093/cvr/cvaa142. [DOI] [PubMed] [Google Scholar]
- 45.Panaro B.L., Coppage A.L., Beaudry J.L., et al. Fibroblast activation protein is dispensable for control of glucose homeostasis and body weight in mice. Mol Metab. 2019;19:65–74. doi: 10.1016/j.molmet.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bainbridge T.W., Dunshee D.R., Kljavin N.M., et al. Selective homogeneous assay for circulating endopeptidase fibroblast activation protein (FAP) Sci Rep. 2017;7 doi: 10.1038/s41598-017-12900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunshee D.R., Bainbridge T.W., Kljavin N.M., et al. Fibroblast activation protein cleaves and inactivates fibroblast growth factor 21. J Biol Chem. 2016;291:5986–5996. doi: 10.1074/jbc.M115.710582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S.B., Ikenaga N., Peng Z.W., et al. Lysyl oxidase activity contributes to collagen stabilization during liver fibrosis progression and limits spontaneous fibrosis reversal in mice. FASEB J. 2016;30:1599–1609. doi: 10.1096/fj.14-268425. [DOI] [PubMed] [Google Scholar]
- 49.Ikenaga N., Peng Z.W., Vaid K.A., et al. Selective targeting of lysyl oxidase-like 2 (LOXL2) suppresses hepatic fibrosis progression and accelerates its reversal. Gut. 2017;66:1697–1708. doi: 10.1136/gutjnl-2016-312473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang A., Yan X., Xu H., et al. Selective depletion of hepatic stellate cells-specific LOXL1 alleviates liver fibrosis. FASEB J. 2021;35 doi: 10.1096/fj.202100374R. [DOI] [PubMed] [Google Scholar]
- 51.Wang X., Hausding M., Weng S.Y., et al. Gliptins suppress inflammatory macrophage activation to mitigate inflammation, fibrosis, oxidative stress, and vascular dysfunction in models of nonalcoholic steatohepatitis and liver fibrosis. Antioxid Redox Signal. 2018;28:87–109. doi: 10.1089/ars.2016.6953. [DOI] [PubMed] [Google Scholar]
- 52.Farrell G., Schattenberg J.M., Leclercq I., et al. Mouse models of nonalcoholic steatohepatitis: toward optimization of their relevance to human nonalcoholic steatohepatitis. Hepatology. 2019;69:2241–2257. doi: 10.1002/hep.30333. [DOI] [PubMed] [Google Scholar]
- 53.Yang A., Yan X., Fan X., et al. Hepatic stellate cells-specific LOXL1 deficiency abrogates hepatic inflammation, fibrosis, and corrects lipid metabolic abnormalities in non-obese NASH mice. Hepatol Int. 2021;15:1122–1135. doi: 10.1007/s12072-021-10210-w. [DOI] [PubMed] [Google Scholar]