Abstract
An α-galactosidase gene from Thermus brockianus ITI360 was cloned, sequenced, and expressed in Escherichia coli, and the recombinant protein was purified. The gene, designated agaT, codes for a 476-residue polypeptide with a calculated molecular mass of 53,810 Da. The native structure of the recombinant enzyme (AgaT) was estimated to be a tetramer. AgaT displays amino acid sequence similarity to the α-galactosidases of Thermotoga neapolitana and Thermotoga maritima and a low-level sequence similarity to α-galactosidases of family 36 in the classification of glycosyl hydrolases. The enzyme is thermostable, with a temperature optimum of activity at 93°C with para-nitrophenyl-α-galactopyranoside as a substrate. Half-lives of inactivation at 92 and 80°C are 100 min and 17 h, respectively. The pH optimum is between 5.5 and 6.5. The enzyme displayed high affinity for oligomeric substrates. The Kms for melibiose and raffinose at 80°C were determined as 4.1 and 11.0 mM, respectively. The α-galactosidase gene in T. brockianus ITI360 was inactivated by integrational mutagenesis. Consequently, no α-galactosidase activity was detectable in crude extracts of the mutant strain, and it was unable to use melibiose or raffinose as a single carbohydrate source.
α-Galactosidases catalyze the hydrolysis of α-1,6-linked α-galactose residues from oligosaccharides such as melibiose (galactose-α-1,6-glucose), raffinose (galactose-α-1,6-sucrose), and stachyose (galactose α-1,6-raffinose) and from polymeric galactomannans (37, 38, 48, 49, 58). Some α-galactosidases are also known to catalyze transgalactosylation, especially at a high concentration of substrate (21, 22). α-Galactosidases have been isolated from a variety of eucaryotes and bacteria. The known eucaryotic enzymes display a significant degree of amino acid sequence homology and have been placed in family 27 in the classification of glycosyl hydrolases (23, 24). The exceptions are the fungal α-galactosidases AGLII from Trichoderma reesei (38) and AglB from Aspergillus niger (14a), which resemble bacterial α-galactosidases of family 36. α-Galactosidases from the hyperthermophilic bacteria Thermotoga neapolitana and Thermotoga maritima were recently cloned and expressed in Escherichia coli (31, 35). These enzymes display a low level of amino acid sequence similarity with α-galactosidases of family 36, and they have a lower molecular mass, with a subunit size of ∼62 kDa versus ∼80 kDa for the family 36 representatives. The eucaryotic α-galactosidases of family 27 are considerably smaller than the bacterial counterparts of family 36, with an average 50-kDa subunit molecular mass. Only a limited degree of amino acid sequence similarity occurs between the α-galactosidases of these two families. The only shared consensus pattern, [LIVMFY]-x(2)-[LIVMFY]-x-[LIVM]-D-[DS]-x-[WY], is near the amino-terminal end of eucaryotic enzymes (family 27) and within the central region of the bacterial enzymes (family 36). Its presence indicates a similar reaction mechanism or a substrate binding site of the enzymes of both families. The E. coli melibiase (gene melA) (36) represents the third family of α-galactosidases (family 4) (23). The E. coli melibiase requires NAD+ and manganese ions as a cofactor (9) and is structurally related to neither family 27 nor family 36, with the consensus pattern described above being missing.
Many α-galactosidases of eubacterial and eucaryotic sources have been studied extensively regarding their biochemical and physical properties, and a number of sequences are available in sequence databases. Furthermore, work dealing with the crystallization and X-ray analysis of eucaryotic enzymes has been reported (19, 41). Still, the structure and a detailed catalytic mechanism remain to be solved.
α-Galactosidases have found a practical value in biotechnology. In the pulp and paper industry, the use of hemicellulases, including α-galactosidase, has gained interest (51, 58). Also, the potential application of α-galactosidases in the processing of soy molasses and soybean milk has been demonstrated (31, 52). Furthermore, they have been used for the elimination of d-raffinose from sugar beet molasses in the sugar industry in order to facilitate the crystallization and consequently improve the yield of sucrose (17, 40, 48). Due to the elevated temperatures used during these processes, thermostability is an important and desirable quality.
Thermophilic bacteria are known to be a source of thermostable hydrolytic enzymes, including α-galactosidases. Several α-galactosidases from thermophilic bacilli have already been studied regarding their potential as hydrolyzing enzymes for different substrates (13, 16, 17, 30, 51). Furthermore, α-galactosidases from the hyperthermophilic bacteria Thermotoga neapolitana and Thermotoga maritima were recently cloned and characterized (31, 35). Bacteria of the genus Thermus are known to exhibit α-galactosidase activity (5) and an α-galactosidase gene has already been cloned along with a β-galactosidase gene from the Thermus strain T2 (33). Still, no paper describing enzyme properties or a primary structure of Thermus α-galactosidase has been published.
The thermophilic bacteria of Thermus strain ITI360, isolated in Iceland, exhibited an α-galactosidase activity and were able to utilize melibiose or raffinose as a single carbohydrate source. The strain was identified as a T. brockianus species by multilocus enzyme electrophoresis and 16S rRNA analysis (26). Due to our interest in thermostable α-galactosidases with reference to the required properties for industrial application, we decided to study further the enzyme(s) accounting for the α-galactoside hydrolyzing activity in strain ITI360. Here, we report the cloning and sequencing of an α-galactosidase gene from this thermophilic bacterium and the purification and characterization of the recombinant enzyme. Furthermore, the insertional inactivation of the α-galactosidase gene in strain ITI360 is described.
MATERIALS AND METHODS
Bacterial strains and plasmids.
T. brockianus ITI360 was obtained from a collection of thermophilic bacteria, Technological Institute of Iceland (IceTec, Keldnaholt, Reykjavik, Iceland). The E. coli strains TAP90 (supE44 supF58 hsdR pro leuB thi-1 rpsL lacY1 tanA1 rec D1903::minitet) (43) and HB101 F′lac (::Tn1739tnpR) supE44 hsdS20 (r−B, m−B) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 F′ (::Tn1739 Cmr lac) (2) were used as hosts for λ-RES phage and RES plasmid respectively. E. coli JM109 [supE44Δ(lac-proAB) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] (F′ traD36 proAB lacIqZΔM15) (55) and RM448 (supE supF rpsL gyrA hsdR thi ΔlacX74), isogenic with LA108 (44), were used as hosts for sequencing and expression plasmids. λ-RESIII and the plasmids pUC18/19 and pBTac1 are described elsewhere (see references 2, 57, and 8, respectively).
Media, culture conditions, and transformation procedure.
T. brockianus ITI360 was grown at 65°C under strong aeration in mineral medium 162 (12) with 0.25% tryptone and 0.25% yeast extract at pH 7.5. Growth on single carbon sources was tested on agar plates with minimal medium 162 containing 0.05% NH4Cl, biotin (50 μg liter−1) and thiamine (1 mg liter−1). The method of Koyama et al. (32) was used for the Thermus transformation with a slight modification. The cells were grown at 65°C in the medium described above with MgCl2 and CaCl2 concentrations increased to 2 mM. Plasmid DNA (1 μg) was added to 0.5 ml of early-log-phase cells and incubated for 2 to 3 h at 60°C under strong aeration. The cells were then washed with 0.9% NaCl and plated on agar plates (2% agar) containing the nutrient agar medium 162 with 15 μg of kanamycin ml−1. The plates were incubated for 48 h at 60°C. The E. coli strains were grown in Luria-Bertani medium at a relevant temperature. When necessary, selective antibiotic was added (100 μg ml−1 for ampicillin, 25 μg ml−1 for kanamycin). E. coli transformation was performed according to the transformation and storage solution method (10).
Enzyme assays.
α-Galactosidase activity was determined by two different methods, depending on the substrate. (i) The rate of hydrolysis of para-nitrophenyl-α-d-galactoside (pNP-α-galactoside [4 mg ml−1]) was measured in 0.1 M potassium buffer (pH 6.5). The reaction was stopped by addition of sodium borate (pH 9.8) to a final concentration of 0.5 M. The concentration of p-nitrophenol (molar extinction coefficient, 18,500 liters mol−1 cm−1) liberated was determined by A405. One unit of activity is defined as the amount of enzyme which liberates 1 μmol of p-nitrophenol per min under the given assay conditions. (ii) The rate of d-raffinose or melibiose hydrolysis was determined by assessing the amount of d-galactose released by high-performance liquid chromatography (HPLC). The HPLC apparatus consisted of a 2200 pump (Bischoff, Leonberg, Germany) and an ESA Coulochem II electrochemical detector (Bischoff). The sugars were separated on a Hamilton RCX-10 column (250 by 4.1 mm), with 0.1 M sodium hydroxide solution, with a flow rate of 0.75 ml min−1. The eluted sugars were detected by pulsed amperometry with an analytical cell (5040, ESA Coulochem II; Bischoff). The protein concentration of crude extracts or fraction of purification was estimated by the method of Bradford (6) with bovine serum albumin as a standard.
Cloning and sequencing of the T. brockianus α-galactosidase gene, agaT.
Recombinant DNA techniques were performed by conventional protocols (46). DNA was extracted from the thermophilic bacteria, and size-fractional fragments produced by partial digestion with XhoII were used to construct a genomic phage λ-RESIII library (2). The library was amplified in E. coli TAP90. The λ-RESIII vector allows excision of cloned fragments by site-specific recombination from the λ DNA and the conversion into autonomously replicating plasmids. This was achieved by infection of an E. coli strain harboring the transposon Tn1739tnpR on an F′ plasmid (2). The strain HB101 F′lac::Tn1739tnpR was infected with ∼2,000 recombinant phage from the Thermus genomic library. Following infection and plasmid conversion, the cells were divided into 20 portions, and each portion (each containing ∼100 recombinants) was grown in 5 ml of LB medium containing 25 μg of kanamycin ml−1. An aliquot of the cells (4 ml) was washed and resuspended in 100 μl of lysis buffer (4 mg of lysozyme ml−1, 25 mM EDTA, 0.1% Triton X-100 [pH 8]) and incubated for 30 min at 37°C. One milliliter of 0.1 mM potassium phosphate buffer (pH 6.5) containing pNP-α-galactoside (0.8 mg ml−1) was added, and the suspension was incubated for 15 h at 55°C. The enzyme reaction was stopped with 2 ml of borate buffer (0.4 M [pH 9.3]), and liberated p-nitrophenol was measured spectrophotometrically at 405 nm. An HB101 strain harboring a λ-RESIII plasmid with its original insert served as a control. The culture belonging to the part displaying the highest hydrolyzing activity was diluted, and a portion containing ∼200 CFU was divided into 20 parts. Each part was grown in 5 ml of LB medium containing 25 μg of kanamycin ml−1, and the same procedure as before was repeated. The culture exhibiting the highest α-galactosidase activity was diluted and plated. Single colonies were picked and checked for α-galactosidase activity. One clone with α-galactosidase activity was investigated further. The recombinant plasmid of this clone, designated pOF932, contained a Thermus α-galactosidase gene, designated agaT, on an ∼8.4-kb cloned XhoII fragment. The plasmid was analyzed by restriction mapping, and fragments were subcloned into pUC18/19 for activity tests and sequencing. DNA sequencing reactions with double-stranded DNA were carried out according to the dideoxy chain termination method (47), and DNA was analyzed with an automated laser fluorescent A.L.F. sequencer (Pharmacia) by using labeled primers or labeled dATP. The nucleotide sequence was analyzed on a Sun workstation with programs from the University of Wisconsin Genetics Computer Group package, version 8.01 (14). All database searches were run with the program BLASTX on a server from the National Center for Biotechnology Information, Bethesda, Md. (3).
Alignment and construction of phylogenetic tree.
Amino acid sequences of the following enzymes were used for alignment with the amino acid sequence of the T. brockianus α-galactosidase (AgaT) and construction of a phylogenetic tree: GalA of T. maritima (35), accession no. 2660642; Agl1 of T. neapolitana (31), accession no. 3237318; AgaN of Bacillus stearothermophilus NUB3621 (16), accession no. AF130985; Aga of S. mutans (1), accession no. P27756; RafA of E. coli (4), accession no. P16551; AgaR and AgaS of Pediococcus pentosaceus, accession no. L32093; and AglII of T. reesei (38), accession no. Z69254. The amino acid sequence of AglA of Aspergillus niger (13a), accession no. P28351, was added to the alignment by the construction of the phylogenetic tree. Amino acid sequences of these enzymes were retrieved from the protein databases (according to the sequence accession numbers) and aligned by using the CLUSTAL W, version 1.60 (53). A distance measure for the sequences was computed with the program PROTDIST (maximum-likelihood estimates) of the Phylip package (15). The measures were used to construct a phylogenetic tree by the neighbor-joining method (45) by using the program Neighbor (Phylip package). AglA of A. niger was used as an outgroup.
Purification of recombinant Thermus α-galactosidase, AgaT.
E. coli JM109 harboring the plasmid pOF1037 (a subclone of pOF932, agaT on a 2,375-bp BglII-HindIII fragment) was used as a source of recombinant enzyme. The recombinant strain was grown with shaking overnight at 37°C in 100 ml of LB medium containing 100 μg of ampicillin ml−1. The cells were harvested by centrifugation and resuspended in 20 ml of potassium phosphate buffer (10 mM [pH 6.5]). Crude extract was prepared by sonication (Sonicator W-385, Ultrasonics microtip; 2 × 30 s; duty cycle, 50% s−1), and debris was removed by centrifugation. The majority of the E. coli proteins were removed by thermal precipitation of the crude extract at 65°C for 15 min and centrifugation. Proteins were fractionated by fast protein liquid chromatography with an anion-exchange column, (MonoQ, HR5/5 [1 ml]; Amersham, Pharmacia Biotech) equilibrated with 10 mM phosphate buffer (pH 6.5). Proteins were eluted with an NaCl gradient (0 to 1 M) in the same buffer. One milliliter fractions were collected and tested for α-galactosidase activity. Active fractions were tested further for purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (34).
Determination of molecular mass.
The molecular mass of AgaT subunits was determined by SDS-PAGE. Before loading the gel, denaturation of E. coli extracts containing AgaT was done at 100°C for 30 min in order to denature the enzyme completely. The molecular mass of the native enzyme was determined by gel filtration chromatography with a Smart-System apparatus (Amersham Pharmacia Biotech). Purified enzyme was applied on a Superdex 75 column (Amersham Pharmacia Biotech) equilibrated with 10 mM sodium acetate buffer (pH 5.5). The column was calibrated with the following proteins: alcohol dehydrogenase from yeast (Sigma), approximate molecular mass, 150,000 Da; β-amylase from sweet potato (Sigma), approximate molecular mass, 200,000 Da; and bovine serum albumin, approximate molecular mass, 66,000 Da.
N-terminal amino acid sequence analysis of AgaT.
Purified AgaT was lyophilized and subjected to N-terminal amino acid sequencing by an Edman degradation procedure. A model 470A Sequencer (Perkin-Elmer—Applied Biosystems, Weiterstadt, Germany) was employed in accordance with the manufacturer’s procedure. The sequencing was performed at the Institut für Lebensmittel Technologie, Universität Hohenheim, Stuttgart, Germany.
Expression of AgaT in E. coli RM448.
The pBTac1 expression vector was used for the constitutive expression of agaT in E. coli RM448. The agaT gene was amplified with the forward primers f1 (GCG AAT TCG ATG CGG GTA AAG GTG GG) with original Thermus codons or f2 (GCG AAT TCG ATG CGT GTA AAG GTT GTT AGC CTG GAG GTG) with exchanged codons and the reverse primer S733 (GGG AAG CTT GTG GCG TTT AAA GAA GGG). The PCR amplification was performed with 0.5 U of Taq DNA polymerase (Biomaster), 10 ng of plasmid pOF1037, a 0.1 μM concentration of each synthetic primer, a 0.2 mM concentration of each deoxynucleoside triphosphate, 1.5 mM MgCl2 in the buffer recommended by the manufacturer, and 5% dimethyl sulfoxide. A total of 30 cycles were performed; each cycle consisted of denaturing at 94°C for 50 s, annealing at 50°C for 40 s, and extension at 72°C for 80 s. Subsequently, the amplified products from the f1 and f2 primers were cloned into the EcoRI and HindIII sites of pBTac1 to produce pOF3822 and pTR4, respectively.
Determination of kinetic parameters.
Michaelis-Menten kinetics of hydrolyzing reactions were verified by plotting reaction rates against substrate concentration. The Km and Vmax values were determined by nonlinear regression analysis of the plots and graphically from Lineweaver-Burk plotting of the initial cleavage rate. Enzyme assays were performed in 100 mM potassium phosphate buffer (pH 6.5) at 90°C for pNP-α-galactoside and at 80°C for melibiose and raffinose. Substrate concentrations were in the range of 0.05 to 4, 0.1 to 20, and 1 to 40 mM for pNP-α-galactoside, melibiose, and raffinose, respectively.
Temperature optimum and stability.
The temperature optimum was determined by performing pNP-α-galactosidase assays at a temperature range from 25°C to 100°C. Hydrolysis of raffinose at different temperatures was monitored at a raffinose concentration of 100 mM. Thermal stability was determined by the following procedure. After thermal precipitation of E. coli crude extract (1 mg ml−1) as described before, the enzyme was preincubated in 100 mM phosphate buffer (pH 6.5) at various temperatures (92, 86, 80, and 75°C) for different periods of time and then assayed for residual activity at 37°C.
pH optimum.
The α-galactosidase activity against pNP-α-galactoside was measured over a pH range from 2.0 to 9.0 by using 0.1 M potassium phosphate buffers (range, pH 4.6 to 9.0) and sodium citrate buffers (range, pH 2 to 4.8).
Effect of metal ions and other substances on enzyme activity.
Purified AgaT was preincubated with various metal ions and reagents at a 1 mM concentration (except with EDTA, which was 10 mM; and NAD+, which was 0.05 mM) in 100 mM potassium phosphate buffer (pH 6.5) at 37°C for 15 min. The residual enzyme activity was assayed at 37°C.
Insertional inactivation of agaT in T. brockianus ITI360.
For the inactivation of agaT, the following chromosomal integration cassette was constructed. The gene for the thermostable kanamycin resistance protein (kan) (39) from pYK134 (25), kindly supplied by T. Hoshino, was amplified by PCR. The forward primer, S765 (CCG CTC GAG GAG GAA TAA TGA ATG GACC) contained an XhoI restriction site and a Thermus Shine-Dalgarno sequence 4 bp upstream of the ATG start codon, as apparent by agaT. The reverse primer S718 (CGG GAT CCG TCA TCC GTT CAA AAT GG) contained a BamHI site. The plasmid pOF1037 has a single XhoI restriction site 18 bp downstream of the ATG start codon of agaT and a single BamHI site in the 3′ region of the gene. The XhoI-BamHI fragment of pOF1037 with a partial agaT coding sequence was substituted with the amplified kan gene. The resulting plasmid, pOF545, contained kan with an ∼800-bp flanking Thermus sequence downstream from the gene and an ∼400-bp flanking sequence upstream. To extend the 5′ flanking homologous sequence, a HindIII fragment from the plasmid pOF1031 (Fig. 1) was cloned into the HindIII site of pOF545 to produce pOF642. The correct orientation of the fragment was verified by restriction analysis. T. brockianus ITI360 was transformed with plasmid pOF642. The resulting transformants, selected on 162 nutrient kanamycin agar medium at 60°C, were analyzed for α-galactosidase activity and for growth on minimal agar medium containing melibiose or raffinose as a single carbohydrate source. Furthermore, SacI- and BamHI-digested chromosomal DNA of the wild-type strain, ITI360, and a deletion strain, OF642, was analyzed by Southern hybridization. An agaT gene fragment generated by PCR amplification with primers f1 and S733 and a kan gene fragment generated by PCR amplification with primers S765 and S718, both labeled with digoxigenin (Boehringer Mannheim), were used as probes.
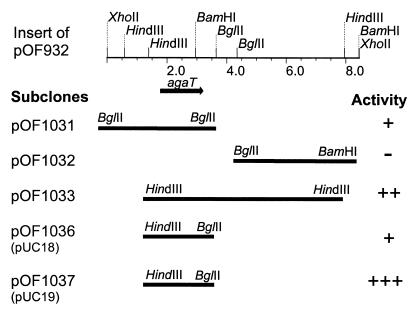
FIG. 1.
The insert of the gene library plasmid pOF932 and structure of subclones. α-Galactosidase active and inactive clones are indicated by + and −, respectively. The activity was determined by using pNP-α-galactoside as a substrate. One of the two BglII sites used for the cloning of the fragment in pOF1031 comes from the vector region (pRESIII) of pOF932.
Nucleotide sequence accession number.
The GenBank accession number for the sequence reported in this article is AF135398.
RESULTS
Cloning of T. brockianus ITI360 α-galactosidase gene, agaT.
Thermus strains, isolated in Iceland, were screened for α-galactosidase activity by using pNP-α-galactoside (data not shown). No inducers were used. Generally, the Thermus strains exhibited a low basal α-galactosidase activity: <100 mU mg−1 at temperature optimum versus 100 to 500 mU mg−1 observed for thermophilic Bacillus strains (16). On the other hand, the temperature optimum of activity with pNP-α-galactoside as a substrate was observed at >90°C with Thermus versus 65 to 80°C with the Bacillus strains.
The cloning of agaT was facilitated by the functional assay of the α-galactosidase gene product in E. coli. The activity in each portion of a divided Thermus gene library was screened against pNP-α-galactoside as a substrate. Of 20 portions, 2 showed a slight yellow color development distinguishable from the other portions, after incubation overnight at 55°C, indicating the liberation of p-nitrophenol. Overnight incubation at temperatures higher than 55°C was not applied due to the thermal lability of the substrate. The portion displaying the highest α-galactosidase activity was chosen for a new round of screening. This time, 5 portions of 20 developed a yellow color in the assay. The culture exhibiting the highest α-galactosidase activity was diluted and plated on LB agar containing kanamycin (25 μg ml−1). α-Galactosidase activity was detected in 2 out of 50 colonies which were tested. Plasmid isolation and restriction analysis revealed the identity of the positive clones. Furthermore, restriction fragments of the gene library clone pOF932, which displayed α-galactosidase activity, were subcloned into pUC18 and -19. The subclones were tested for α-galactosidase activity. Hence, the α-galactosidase gene was located on a 2,375-bp HindIII-BglII fragment, cloned into both pUC18 (pOF1036) and pUC19 (pOF1037). Higher activity in E. coli JM109/pOF1037 than E. coli JM109/pOF1036 indicated the direction of the gene downstream from the HindIII restriction site (Fig. 1).
Sequence analysis of agaT and the deduced amino acid sequence.
The insert of plasmid pOF1037 was sequenced (2,375 bp). It contained one continuous open reading frame of 1,431 bp with an ATG codon at position 396 and a TAA stop codon at position 1825. The deduced amino acid sequence of the potential α-galactosidase gene was compared with α-galactosidase amino acid sequences available from GenBank (NCIB database). There was 57.9% similarity and 29.4% identity between the T. brockianus α-galactosidase (AgaT) and GalA of T. maritima and 52.6% similarity and 28.6% identity between AgaT and Agl1 of T. neapolitana. A low-level amino acid sequence similarity was observed with α-galactosidases of family 36, mainly restricted to an amino acid sequence region (residues 380 to 460 according to AgaT numbering) containing the consensus pattern [LIVMFY]-x(2)-[LIVMFY]-x-[LIVM]-D-D-x-[WY] (Fig. 2). No similarity with the eucaryotic α-galactosidases of family 27 was observed, except for a sequence matching the α-galactosidase consensus pattern. The gene, named agaT, encodes a polypeptide (AgaT) of 476 amino acids with a calculated molecular mass of 53,810 Da. The G+C content of the open reading frame (ORF) is 62.9%. As expected for organisms with a relatively high G+C content, Arg, Pro, Ala, and Gly codons occur with a higher frequency—9.6, 8, 9, and 10%, respectively, than GC poor codons (e.g., Asn, Lys, Tyr, Phe, and Ile codons, with 1.5, 2.3, 3.3, 4.4, and 1%, frequency respectively. A putative ribosome binding site, GGAGGAGGG, was present 7 bp upstream of a ATG start codon. No potential −35/−10 consensus sequence was recognized.
FIG. 2.
Alignment of T. brockianus ITI360 AgaT (partial amino acid sequence) with the α-galactosidases of T. maritima, GalA (35), accession no. 2660642; T. neapolitana, Agl1 (31), accession no. AF011400; B. stearothermophilus NUB3621, AgaN (16), accession no. AF130985; S. mutans, Aga (in the alignment designated Aga1) (1), accession no. P27756; E. coli, RafA (4), accession no. P16551; P. pentosaceus, AgaR and AgaS, accession no. L32093; and T. reesei, AglII (38), accession no. Z69254. Hyphens indicate gaps. Alignment was achieved by using CLUSTAL W, version 1.60 (53). Identical residues are indicated by shaded boxes. The consensus pattern of eucaryotic and bacterial α-galactosidase is indicated. A conserved cysteine residue is marked with a solid triangle.
Upstream of agaT, a truncated ORF (due to the cloning procedure) was identified. The deduced amino acid sequence of this ORF displays similarity to the C-terminal amino acid sequence of β-galactosidases from Thermus strain T2 (54) (accession no. Z93773) and Thermus strain A4 (42) (accession no. g2765752). Downstream and overlapping the agaT gene, a truncated ORF was observed. The predicted amino acid sequence displays a similarity to the N-terminal part of a galactose-1-P-uridylyltransferase from T. maritima (35) (accession no. AJ001072) and T. neapolitana (31) (accession no. AF055482) (to be published elsewhere).
Purification of recombinant AgaT and its molecular mass.
The specific α-galactosidase activities in crude extracts of E. coli JM109/pOF1037 were low—about 1 and 3.4 U mg−1 without and with IPTG (isopropyl-β-d-thiogalactopyranoside) induction, respectively, at 75°C. Despite this low-level expression, the strain was used for the production of AgaT. Thermal precipitation greatly facilitated further purification, which required just one chromatographic step using a MonoQ column. SDS-PAGE of the column fraction corresponding to the peak of activity revealed a single protein band with a molecular mass of ∼54 kDa (Fig. 3), which agrees with the molecular mass of 53.8 kDa, calculated from the nucleotide sequence of agaT. The molecular mass of the native enzyme was estimated to be ∼200,000 Da on a calibrated Superdex 75 gel filtration column. These results indicate a tetrameric form of the native enzyme. AgaT, purified to homogeneity, exhibited the specific activity of 250 U mg−1 at 75°C with pNP-α-galactoside as substrate.
FIG. 3.
SDS–10% polyacrylamide gel of crude extract before and after thermal precipitation and purified recombinant T. brockianus ITI360 α-galactosidase. Lanes: 1, molecular mass markers; 2, crude extract of JM109/pOF1037 before thermal precipitation; 3, crude extract of JM109/pOF1037 after thermal precipitation; 4, column fraction of purified AgaT, corresponding to the peak of activity. The sizes of the marker proteins in kilodaltons are indicated.
N-terminal amino acid sequencing of AgaT.
The purified AgaT was subjected to peptide sequencing. A single N-terminal sequence was revealed, Met-Arg-Val-Lys-Val-Gly-Ser-Leu-Glu-Val, which corresponds to the N-terminal deduced amino acid sequence of the agaT ORF and thus confirms the suspected start of the coding region.
Expression of agaT.
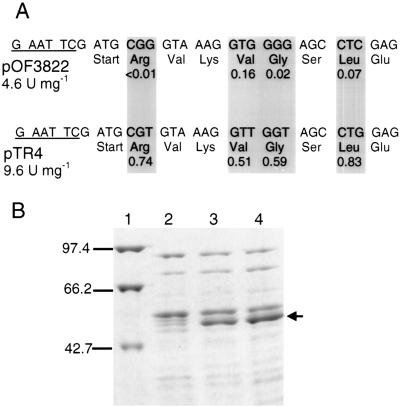
In order to improve the expression of agaT in E. coli, the AgaT coding sequence in pOF1037 was amplified by PCR with the f1 forward primer and the reverse primer as described in Materials and Methods and cloned in pBTac1. The resulting plasmid, pOF3822, was introduced into E. coli RM448 by transformation. The lac operon, including the lac repressor gene in RM448, was deleted (ΔlacX74) (44). Therefore, the expression of agaT in RM448/pOF3822 was constitutive. Although agaT was inserted downstream of the strong tac promoter in pBTac1, the specific activity was only 4.6 U mg−1 at 75°C. The sequence analysis of the agaT gene revealed a high content of rare E. coli codons (7, 27). Even among the first 21 bp after the ATG start codon, the existence of three rare codons for Arg, Gly, and Leu (27) was conspicuous. Due to the possible interference of these rare codons with translation initiation, the gene was amplified again with a forward primer, f2, with exchanged codons as described in Materials and Methods and shown in Fig. 4A. Instead of the rare Arg, Gly, and Leu codons and a Val codon, primer f2 contains corresponding codons which are more frequently used in E. coli (Fig. 4A) according to the codon usage table for E. coli genes (27) and enteric bacterial highly expressed genes (Wisconsin sequence analysis package, Genetics Computer Group, Inc.) (14). The new amplification product was cloned into pBTac1 as before, and E. coli RM448 was transformed with the resulting plasmid (pTR4). The specific activity of AgaT in crude extract of RM448/pTR4 was found to be 9.6 U mg−1 at 75°C. Higher activity in RM448/TR4 than in RM448/pOF3822 was reproducible and correlated with an increased production of AgaT in RM448/pTR4, as verified by SDS-PAGE (Fig. 4B).
FIG. 4.
Production of AgaT in E. coli RM448. (A) Exchange of rare E. coli codons within the first 21 bp of agaT after the start codon. The upper sequence shows the first 21 bp of agaT in pOF3822. The lower sequence shows the first 21 bp of agaT with exchanged codons as they occur in pTR4. The exchanged codons are marked by shaded boxes. The codon usage (fraction) according to the codon usage table for enteric bacterial highly expressed genes (Wisconsin Sequence Analysis Package, Genetics Computer Group, Inc.) (14) is indicated below the corresponding codons. The EcoRI sites, used for the ligation in pBTac1, are underlined. (B) SDS-PAGE of crude extracts of RM448 containing the plasmids pOF3822 and pTR4 after thermal precipitation. Lanes: 1, molecular mass markers; 2, RM448 without plasmid; 3, RM448 with pOF3822; 4, RM448 with pTR4. The sizes of the marker proteins (in kilodaltons) are shown. AgaT is indicated by an arrow.
Enzyme properties.
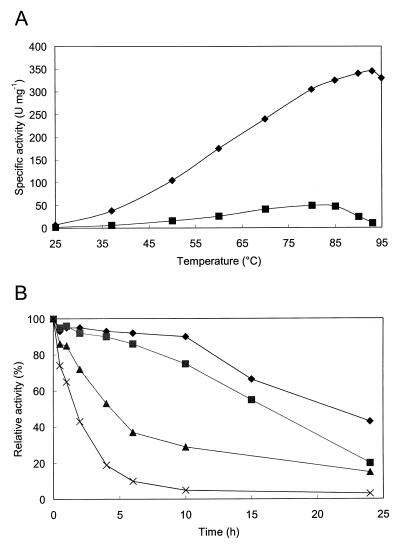
AgaT was specific for α-galactopyranosidic compounds. In contrast to pNP-α-galactopyranoside, it did not hydrolyze pNP-α-fucopyranoside, pNP-α-arabinoside, pNP-α-glucopyranoside, pNP-β-galactopyranoside, or pNP-α-mannopyranoside. The enzymatic properties of the recombinant AgaT were studied by using substrates with relevance to the sugar beet process, such as raffinose and melibiose and the artificial substrate pNP-α-galactoside. The Km and Vmax values for these substrates are listed in Table 1. The purified AgaT displayed a temperature optimum of activity for the substrate pNP-α-galactoside at 94°C, whereas the temperature optimum for raffinose hydrolysis was about 80 to 85°C (Fig. 5A). The maximum activity of AgaT was determined between pH 5.5 and 6.5. Identical enzyme activities were observed in both buffer systems used, at the overlapping pH range of 4.6 to 4.8. Thermostability of AgaT was determined by studying the kinetics of thermal inactivation. The results are represented in Fig. 5B. The half-lives of inactivation at 92, 86, 80, and 75°C were 100 min, 4.5 h, 17 h, and 22 h, respectively. The enzyme was stable at room temperature, with the activity remaining the same after 3 days. Some enzyme properties of AgaT are summarized in Table 2 along with properties of AgaN from B. stearothermophilus NUB3621 (16) and GalA from T. maritima (35) for a comparison. α-Galactosidase activity was slightly inhibited by CuCl2, CuSO4, and ZnCl2 (relative activities, 0.85, 0.74, and 0.88, respectively) and inhibited almost completely by AgNO3, HgCl2, and PCMB (relative activities, 0.12, 0.02, and 0.17, respectively). Other metal ions (Na+, K+, Li+, Mg2+, Ca2+, Mn2+, Fe2+, and Co2+, added as chloride salts; and Fe3+-citrate and NiSO4) or EDTA did not significantly affect the α-galactosidase activity, suggesting that AgaT does not have any metal cofactor requirement. Also, the α-galactosidase activity was not affected by NAD+, dithiothreitol, or mercaptoethanol.
TABLE 1.
Kinetic parameters for the hydrolysis of pNP-α-galactoside, melibiose, and raffinose
| Substrate | AgaT
|
AgaNa
|
RafAa
|
Agaa
|
||||
|---|---|---|---|---|---|---|---|---|
| Km (mM) | Vmax (U mg−1)b | Km (mM) | Vmax (U mg−1) | Km (mM) | Vmax (U mg−1) | Km (mM) | Vmax (U mg−1) | |
| pNP-α-galactose | 2.47 | 350 | 0.38 | 520 | 0.16 | 55 | 0.22 | 109 |
| Melibiose | 4.1 | 162 | 12.0 | 464 | NDc | ND | ND | ND |
| Raffinose | 11.0 | 50 | 16.4 | 230 | 17.4 | 45 | 9.0 | 299 |
Comparative data from Fridjonsson et al. (16) for AgaN of B. stearothermophilus NUB3621, RafA of E. coli, and Aga of S. mutans. Hydrolyzing reactions were performed at or near the temperature optimum of activity for each enzyme: i.e., at 90°C for pNP-α-galactoside and at 80°C for raffinose and melibiose when AgaT was examined, at 75°C for AgaN, and at 37°C for RafA and Aga.
U mg−1 = 1 μmol of substrate hydrolyzed per minute per mg of enzyme.
ND, not determined.
FIG. 5.
Effect of temperature and pH on the activity of AgaT. (A) Effect of temperature on pNP-α-galactoside hydrolysis (⧫) and raffinose hydrolysis (■). Standard assays at pH 6.5 with purified AgaT were performed. The raffinose concentration was 100 mM. (B) Thermoinactivation of recombinant AgaT. After thermal precipitation of crude extract (1 mg ml−1) as described in the text, the enzyme was preincubated in 100 mM phosphate buffer (pH 6.5) at 92°C (×), 86°C (▴), 80°C (■), and 75°C (⧫) for different periods of time and then assayed for residual activity at 37°C. All activity tests were done in triplicate. The maximum variation from the mean values (shown) was less than 5%.
TABLE 2.
Comparison of properties of AgaT with those of the thermostable α-galactosidases of B. stearothermophilus NUB3621 (AgaN) and T. maritima (GalA)a
| Protein | Temp optimum (°C) | pH optimum | Half-life of inactivation | Subunit size (kDa) | No. of subunits |
|---|---|---|---|---|---|
| AgaT | 93 | 5.5–6.5 | 100 min at 92°C, 17 h at 80°C | 53.8 | 4 |
| AgaN | 75 | 6.3–7.0 | <10 min at 80°C, 19 h at 70°C | 80.3 | 4 |
| GalA | ∼95 | 5.0–5.5 | 70 min at 90°C, 48 h at 80°C | 63.7 | 2 |
α-Galactosidase gene inactivation by integration mutagenesis.
agaT gene inactivation was carried out in order to examine whether T. brockianus ITI360 possessed α-galactosidases other than AgaT. The strain was transformed with the plasmid pOF642, which contains an integration cassette for the insertional inactivation of agaT. A low transformation frequency was observed with this plasmid (10−6 transformants per cell). The few kanamycin-resistant transformants obtained were no longer capable of growing on minimal medium containing melibiose or raffinose as the sole carbohydrate source. Also, no α-galactosidase activity was detectable in crude extracts of the mutant strains.
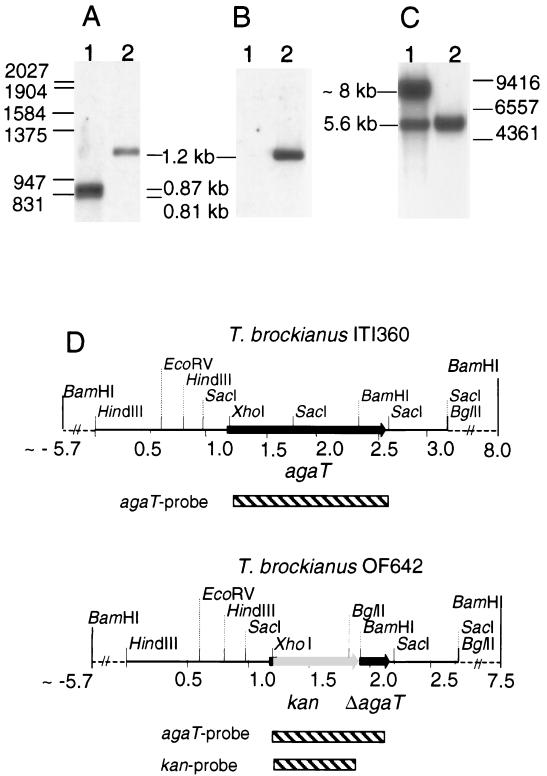
In order to verify the insertion of the kan gene into the agaT locus, SacI-digested chromosomal DNA from the wild-type and mutant strains were examined by Southern hybridization (Fig. 6A and B). According to the sequence analysis of agaT and its flanking sequences, two SacI fragments of 814 and 871 bp were expected to appear following hybridization with the agaT gene probe to the wild-type strain DNA. Due to their similar size, they appear as a single broad band of strong intensity in Fig. 6A, lane 1. On the other hand, a 1.2-kb band of lower signal intensity is detectable in Fig. 6A, lane 2, resulting from the insertion of the kan gene into the agaT locus in the mutant strain OF642 (Fig. 6A, lane 2). The labeled kan gene fragment did not hybridize to the chromosomal DNA of the wild-type strain (Fig. 6B, lane 1), but did hybridize to a fragment of 1.2 kb in strain OF642 (Fig. 6B, lane 2). In order to verify further the correct insertion, the chromosomal DNA from a wild-type strain and a mutant strain was digested with BamHI, and a Southern hybridization was performed with the agaT gene as a probe (Fig. 6C). The probe hybridized to a large fragment (∼8 kb) containing the agaT gene region upstream of a BamHI site located in agaT and to a smaller fragment (5.6 kb) containing the agaT 3′ region downstream of the BamH site in the wild-type strain (Fig. 6C, lane 1). The large fragment is not visible in OF642 due to a deletion of the agaT gene region upstream of the BamHI restriction site (Fig. 6C, lane 2). Hence, this molecular analysis revealed the insertion of the kan module into the agaT locus by a homologous recombination creating the mutant strain OF642 (ΔagaT::kan), which is both kanamycin resistant and incapable of melibiose and raffinose metabolism.
FIG. 6.
Southern blot analysis of T. brockianus ITI360 (wild type) and OF642 (ΔagaT::kan) chromosomal DNA. DNA was digested with SacI, electrophoresed on 1% agarose gel, transferred to a nylon membrane, and hybridized with an agaT gene fragment (A) and kan gene fragment (B) as probes. Furthermore, the DNA was digested with BamHI, electrophoresed and blotted as before, and hybridized with an agaT gene fragment as a probe (C). In each case, lane 1 contains ITI360 and lane 2 contains OF642. The positions of size markers are shown as horizontal lines, and their sizes are given in base pairs. (A and B) λ-EcoRI-HindIII. (C) λ-HindIII. Sizes of detected fragments are given in kilobase pairs. Restriction map of the strains based on sequence analysis and the Southern blot analysis (D). Striped bars indicates DNA fragments used as probes in the hybridization reactions. Dashed lines indicate regions which flank the sequences homologous to the Thermus sequences in pOF642 (integration cassette).
DISCUSSION
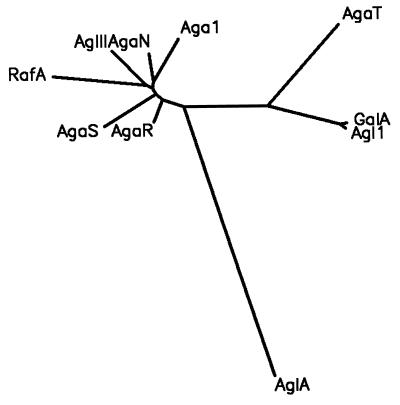
The purpose of our work was to study a thermostable α-galactosidase from Thermus bacteria, to isolate the encoding gene, and to characterize the recombinant protein. We examined α-galactosidase activity in crude extracts of different Thermus strains and chose T. brockianus ITI360 for further study. The failure of hybridization in Southern blots of T. brockianus chromosomal DNA, using an agaB gene fragment from B. stearothermophilus KVE39 (30) as a probe, indicated insufficient homology of the Thermus α-galactosidase gene to the α-galactosidase gene of the thermophilic bacillus (data not shown). Therefore, instead of using a gene probe, we screened for α-galactosidase activity in portions of a divided Thermus gene library in E. coli by using the substrate pNP-α-galactoside. By repeating this procedure for the portion displaying the highest activity, a clone with α-galactosidase was isolated. This enrichment method was successful, while other methods failed, e.g., histochemical staining with 6-bromo-2-naphthyl-α-d-galactopyranoside and Fast Blue RR (data not shown). The cloning of the α-galactosidase gene from T. brockianus ITI360 revealed a novel α-galactosidase with only limited homology to other α-galactosidases with known primary structure. The closest relatives are the α-galactosidases from the hyperthermophilic bacteria T. maritima and T. neapolitana (∼29% amino acid sequence identity). Still the Thermotoga enzymes have a different molecular mass of subunits (∼62 kDa versus the 53.8 kDa of AgaT). Furthermore, the GalA of T. maritima was estimated to be a homodimer (35), whereas in our study, AgaT was estimated to be a homotetramer. As for the Thermotoga α-galactosidases, AgaT displays only a low-level homology to α-galactosidases of family 36 in the classification of glycosyl hydrolases (20 to 25% amino acid sequence identity), mainly restricted to a central part of the enzymes containing the α-galactosidase consensus pattern described in the introduction. Also, it is smaller, with a molecular mass of the subunit of 53.8 kDa versus ∼80 kDa for the family 36 representatives. It is thus doubtful to assign AgaT to enzyme family 36. Rather, it could represent a new family of glycosyl hydrolases as reflected by Liebl et al. for GalA of T. maritima (35). Figure 7 shows a phylogenetic tree constructed according to an amino acid sequence alignment with the amino acid sequence of AglA of A. niger (an enzyme belonging to family 27) as an outgroup. The tree, constructed by the neighbor-joining method (45), shows GalA, Agl1, and AgaT branching together separate from the family 36 representatives.
FIG. 7.
Phylogenetic tree (dendrogram) showing the evolutionary relationships of α-galactosidase amino sequences according to the alignment in Fig. 2 and the amino sequence of an enzyme belonging to family 27 (AglA of Aspergillus niger), which was used as an outgroup. The tree was constructed as described in Materials and Methods.
The deduced amino acid sequence of the truncated ORFs flanking agaT displayed partial homology to the deduced amino acid sequences of genes flanking galA (lacZ and galT) and agl1 (bglA and galT) in T. maritima and T. neapolitana, respectively. This indicates a similar organization of the α- and β-galactoside utilization gene clusters in these thermophilic bacteria. Bacteria of the genera Thermotoga and Thermus both branch deeply from the tree of bacterial phylogeny (20, 56), and in nature, they both inhabit geothermal environments. Hence, the similar gene arrangements observed in these bacteria may reflect their phylogenetic and ecological relationships.
The molecular mass of the AgaT subunit corresponds to the lower molecular mass of some eucaryotic enzymes of family 27 and the melibiase of E. coli. Still, no sequence similarity was observed (except for the α-galactosidase consensus peptide pattern of family 27). Also, AgaT does not require NAD+ or Mn2+ as a cofactor as in the case of the E. coli melibiase. The cloning of α- and β-galactosidase genes from the Thermus strain T2 has been reported (33). Here, the molecular mass of the active α-galactosidase, with a yet unpublished sequence, was estimated to be 350 kDa, which corresponds better to the bacterial enzymes of family 36. No indication of additional α-galactosidases was found in T. brockianus ITI360. In fact, the agaT gene deletion led to the complete loss of α-galactosidase activity in crude extracts of the corresponding mutant strain (OF642) and the concomitant loss of the ability to use raffinose and melibiose as a single carbohydrate source.
As for many other recombinant thermostable enzymes, the purification of the α-galactosidase was easy and efficient. High-level expression in E. coli was not required to obtain sufficient amounts of purified protein due to the ease of the thermal precipitation as a purification tool. The purified AgaT was subjected to a sequential N-terminal Edman degradation in order to confirm the start of the coding region in pOF1037. This was done due to the novelty of the enzyme and the lack of homologous amino acid sequences. Having confirmed the start of the coding region, the gene was introduced into the expression vector pBTac1. This vector was used in E. coli RM448 for the expression of α-galactosidase genes from thermophilic bacilli, in which ∼10 to 30% of the soluble protein was obtained as recombinant enzyme (16, 17). However, the specific activity of AgaT in crude extract of RM448/pOF3822 was only 4.6 U mg−1 at 75°C, which is less than 2% of soluble protein. Poor expression of Thermus genes in E. coli has frequently been described (29, 50). A possible explanation for the low-level expression of agaT might be the high number of rare E. coli codons (e.g., 29 CGG arginine codons). Indeed the exchange of four rare E. coli codons among the first seven codons improved the expression of agaT.
Although this production of AgaT in E. coli was sufficient for our purposes, it had not reached the level of the Bacillus α-galactosidase production. Another reason for the low-level expression might be stable secondary structures of the transcript due to the high G+C content of agaT, which could interfere with the translation in E. coli. Such secondary structures have already been shown to suppress the translation of Thermus genes in E. coli (29). In this context, two-cistron expression systems might be helpful for the expression of agaT as shown for the leuB gene of Thermus thermophilus in E. coli (28, 50).
In our study of the enzymatic properties of AgaT, we used the artificial substrate pNP-α-galactoside and substrates with relevance to the sugar beet process, such as raffinose and melibiose. The affinity for raffinose and melibiose at 80°C is similar to that observed for α-galactosidases from thermophilic Bacillus strains at their temperature optimum of activity (16, 30). Differences in temperature optimum, stability, and pH optimum compared to those of the thermostable Bacillus enzymes were observed (16, 30). In this respect, AgaT resembles more the thermostable α-galactosidase of T. maritima and T. neapolitana. These enzymes display optimum activity at temperatures over 90°C for the hydrolysis of pNP-α-galactoside. The temperature optimum for AgaT was measured at 93°C for pNP-α-galactoside and 80 to 85°C for raffinose. The difference in temperature optimum depending on substrate may be explained by the kinetic parameters for those substrates, which are affected differently by changes in the temperature. Also, the binding of enzyme to raffinose may lead to a conformation which is less thermostable than the conformation of the enzyme bound to pNP-α-galactoside.
The half-life of inactivation at 92°C was 100 min by AgaT, and thus was longer than that by GalA of T. maritima at 90°C (70 min) (Table 2). However, during prolonged incubation periods, AgaT is inactivated faster than GalA (at 80 and 75°C). The pH optimum of AgaT (between 5.5 and 6.5) is between the optimum range of the Bacillus α-galactosidases (e.g., AgaN, 6.3 to 7.0) and GalA (5.0 to 5.5). As observed for other α-galactosidases, AgaT was inhibited by HgCl2 and PCMB, which indicates the presence of a thiol group near the catalytic site of the enzyme (11, 18). AgaT contains three cysteine residues, two of which are also found in T. maritima and T. neapolitana according to an amino acid sequence alignment (residues 161 and 336 according to AgaT numbering). Among them, the cysteine residue 336 could correspond to the conserved cysteine residue found by the family 36 representatives, as seen in the alignment in Fig. 2 (residue 483 according to E. coli RafA numbering, marked with triangle).
Concerning biotechnological aspects, AgaT may be of practical value due to its high activity and stability at temperatures over 90°C. Also, its affinity is high for melibiose and at reasonable levels for raffinose compared to the other bacterial α-galactosidases. Additional improvements in enzymatic properties, with regard to industrial applications, are being attempted by genetic engineering. In addition, we hope that further investigations of AgaT will improve our understanding of enzyme structure, function, and stability.
ACKNOWLEDGMENTS
We thank Jakob Kristjansson for strain ITI360 and Gisela Kwiatkowski for technical assistance.
This work was partly supported by the DAAD (Deutscher Akademischer Austauschdienst e. V) and by the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie.
REFERENCES
- 1.Aduse-Opoku J, Tao L, Ferretti J J, Russell R R B. Biochemical and genetic analysis of Streptococcus mutans α-galactosidase. J Gen Microbiol. 1991;137:757–764. doi: 10.1099/00221287-137-4-757. [DOI] [PubMed] [Google Scholar]
- 2.Altenbuchner J. A new λ RES vector with a built-in Tn1721-encoded excision system. Gene. 1993;123:63–68. doi: 10.1016/0378-1119(93)90540-j. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Aslanidis C, Schmid K, Schmitt R. Nucleotide sequences and operon structure of plasmid-borne genes mediating uptake and utilization of raffinose in Escherichia coli. J Bacteriol. 1989;171:6753–6763. doi: 10.1128/jb.171.12.6753-6763.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger J L, Lee L H, Lacroix C. Identification of new enzyme activities of several strains of Thermus species. Appl Microbiol Biotechnol. 1995;44:81–87. doi: 10.1007/BF00164484. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brinkman U, Mattes R E, Buckel P. High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene. 1989;85:2040–2044. doi: 10.1016/0378-1119(89)90470-8. [DOI] [PubMed] [Google Scholar]
- 8.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 9.Burstein C, Kepes A. α-Galactosidase from Escherichia coli K12. Biochim Biophys Acta. 1971;236:52–63. doi: 10.1016/0304-4165(71)90053-5. [DOI] [PubMed] [Google Scholar]
- 10.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Church F C, Meyers S P, Srinivasan V R. Isolation and characterization of α-galactosidase from Pichia guilliermondii. Dev Ind Microbiol. 1980;21:339–348. [Google Scholar]
- 12.Degryse E, Glansdorff N, Piérard A. A comparative analysis of extreme thermophilic bacteria belonging to the genus Thermus. Arch Microbiol. 1978;117:189–196. doi: 10.1007/BF00402307. [DOI] [PubMed] [Google Scholar]
- 13.Delente J, Johnson J H, Kuo M J, O’Connor R J, Weeks L E. Production of a new thermostable neutral α-galactosidase from a strain of Bacillus stearothermophilus. Biotechnol Bioeng. 1974;16:1227–1243. doi: 10.1002/bit.260160907. [DOI] [PubMed] [Google Scholar]
- 13a.den Herder I F, Rosell A M M, van Zuilen C M, Punt P J, van den Hondel C A M J J. Cloning and expression of a member of the Aspergillus niger gene family encoding α-galactosidase. Mol Gen Genet. 1992;233:404–410. doi: 10.1007/BF00265437. [DOI] [PubMed] [Google Scholar]
- 14.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.de Vries R P, van den Broeck H C, Dekkers E, Manzanares P, De Graaff L H, Visser J. Differential expression of three α-galactosidase genes and a single β-galactosidase gene from Aspergillus niger. Appl Environ Microbiol. 1999;65:2453–2460. doi: 10.1128/aem.65.6.2453-2460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP—Phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 16.Fridjonsson O, Watzlawick H, Gehweiler A, Mattes R. Thermostable α-galactosidase from Bacillus stearothermophilus NUB3621: cloning, sequencing and characterization. FEMS Microbiol Lett. 1999;176:147–153. doi: 10.1111/j.1574-6968.1999.tb13655.x. [DOI] [PubMed] [Google Scholar]
- 17.Ganter C, Böck A, Buckel P, Mattes R. Production of thermostable recombinant α-galactosidase suitable for raffinose elimination from sugar beet syrup. J Biotechnol. 1988;8:301–310. [Google Scholar]
- 18.Garro M S, de Valdez G F, Oliver G, de Giori G S. Purification of α-galactosidase from Lactobacillus fermentum. J Biotechnol. 1996;45:103–109. [Google Scholar]
- 19.Golubev A M, Neustroev K N. Crystallization of α-galactosidase from Trichoderma reesei. J Mol Biol. 1993;231:933–934. doi: 10.1006/jmbi.1993.1340. [DOI] [PubMed] [Google Scholar]
- 20.Hartmann R H, Wolters J, Kröger B, Schultze S, Specht T, Erdmann V A. Does Thermus represent another deep eubacterial branching? Syst Appl Microbiol. 1989;11:243–249. [Google Scholar]
- 21.Hashimoto H, Goto M, Katayama C, Kitahata S. Purification and some properties of α-galactosidase from Pseudomonas fluorescens H-601. Agric Biol Chem. 1991;55:2831–2838. [Google Scholar]
- 22.Hashimoto H, Katayama C, Goto M, Okinaga T, Kitahata S. Transgalactosylation catalyzed by α-galactosidase from Candida guilliermondii H-404. Biosci Biotechnol Biochem. 1995;59:619–623. doi: 10.1271/bbb.59.619. [DOI] [PubMed] [Google Scholar]
- 23.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshino T, Maseda H, Nakahara T. Plasmid marker rescue transformation in Thermus thermophilus. J Ferment Bioeng. 1993;76:276–279. [Google Scholar]
- 26.Hreggvidsson, G. Ó. (IceTec, Reykjavík, Iceland). 1998. Personal communication.
- 27.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its proteins. J Mol Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- 28.Ishida M, Oshima T. A leader open reading frame is essential for the expression in Escherichia coli of GC-rich leuB gene of an extreme thermophile, Thermus thermophilus. FEMS Microbiol Lett. 1996;135:137–142. doi: 10.1111/j.1574-6968.1996.tb07978.x. [DOI] [PubMed] [Google Scholar]
- 29.Ishida M, Oshima T. Overexpression of genes of an extreme thermophile, Thermus thermophilus, in Escherichia coli cells. J Bacteriol. 1994;176:2767–2770. doi: 10.1128/jb.176.9.2767-2770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janz L, Ganter C, Stezowski J, Mattes R. Elucidation of functional domains in thermostable isoenzymes of α-galactosidase in Bacillus stearothermophilus. Enzymatic properties are encoded in a genetically exchangeable domain. In: Reuss M, Chmiel H, Gilles E-D, Knackmuss H-J, editors. Biochemical engineering—Stuttgart. Stuttgart, Germany: Gustav Fischer; 1991. pp. 170–173. [Google Scholar]
- 31.King M R, Yernool D A, Eveleigh D E, Chassy B M. Thermostable α-galactosidase from Thermotoga neapolitana: cloning, sequencing and expression. FEMS Microbiol Lett. 1998;163:37–42. doi: 10.1111/j.1574-6968.1998.tb13023.x. [DOI] [PubMed] [Google Scholar]
- 32.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol. 1986;166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koyama Y, Okamoto S, Furukawa K. Cloning of α- and β-galactosidase genes from an extreme thermophile, Thermus strain T2, and their expression in Thermus thermophilus HB27. Appl Environ Microbiol. 1990;56:2251–2254. doi: 10.1128/aem.56.7.2251-2254.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Liebl W, Wagner B, Schellhase J. Properties of an α-galactosidase, and structure of its gene galA, within an α- and β-galactosidase utilization gene cluster of the hyperthermophilic bacterium Thermotoga maritima. Syst Appl Microbiol. 1998;21:1–11. doi: 10.1016/s0723-2020(98)80002-7. [DOI] [PubMed] [Google Scholar]
- 36.Liljeström P, Liljeström P. Nucleotide sequence of the melA gene coding for α-galactosidase in Escherichia coli K12. Nucleic Acids Res. 1987;15:2213–2220. doi: 10.1093/nar/15.5.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luonteri E, Tenkanen M, Viikari L. Substrate specificities of Penicillium simplicissimum α-galactosidases. Enzyme Microb Technol. 1998;22:192–198. doi: 10.1016/s0141-0229(97)00170-1. [DOI] [PubMed] [Google Scholar]
- 38.Margolles-Clark E, Tenkanen M, Luonteri E, Penttilä M. Three α-galactosidase genes of Trichoderma reesei cloned by expression in yeast. Eur J Biochem. 1996;240:104–111. doi: 10.1111/j.1432-1033.1996.0104h.x. [DOI] [PubMed] [Google Scholar]
- 39.Matsumura M, Katakura Y, Imanaka T, Aiba S. Enzymatic and nucleotide sequence studies of a kanamycin-inactivation enzyme encoded by a plasmid from thermophilic bacilli in comparison with that encoded by plasmid pUB110. J Bacteriol. 1984;160:413–420. doi: 10.1128/jb.160.1.413-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattes R, Beaucamp K. DNA− Neukombination: eine praktische Anwendung in der Zuckerindustrie. Chem Zeit. 1983;2:54–58. [Google Scholar]
- 41.Murali R, Ioannou Z A, Desnick R J, Burnett R M. Crystallization and preliminary X-ray analysis of human α-galactosidase A complex. J Mol Biol. 1994;239:578–580. doi: 10.1006/jmbi.1994.1397. [DOI] [PubMed] [Google Scholar]
- 42.Ohtsu N, Motoshima H, Goto K, Tsukasaki F, Matsuzawa H. Thermostable beta-galactosidase from an extreme thermophile, Thermus sp. A4: enzyme purification and characterization, and gene cloning and sequencing. Biosci Biotechnol Biochem. 1998;8:1539–1545. doi: 10.1271/bbb.62.1539. [DOI] [PubMed] [Google Scholar]
- 43.Patterson T A, Dean M. Preparation of high titer lambda phage lysates. Nucleic Acids Res. 1987;15:6298. doi: 10.1093/nar/15.15.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pourcel C, Marchal C, Louise A, Fritsch A, Tiollais P. Bacteriophage Lambda-E. coli K12 vector-host system for gene cloning and expression under lactose promoter control. Mol Gen Genet. 1979;170:161–169. doi: 10.1007/BF00337792. [DOI] [PubMed] [Google Scholar]
- 45.Saitou N, Nei M. The neighbor-joining method, a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibuya H, Kobayashi H, Sato T, Kim W-S, Yoshida S, Kaneko S, Kasamo K, Kusakabe I. Purification, characterization and cDNA cloning of a novel α-galactosidase from Mortierella vinacea. Biosci Biotechnol Biochem. 1997;61:592–598. doi: 10.1271/bbb.61.592. [DOI] [PubMed] [Google Scholar]
- 49.Shibuya H, Kobayashi H, Park G G, Komatsu Y, Sato T, Kim W-S, Yoshida S, Kaneko R, Nagasaki H, Yoshida S, Kasamo K, Kusakabe I. Purification and some properties of α-galactosidase from Penicillium purpurogenum. Biosci Biotechnol Biochem. 1995;59:2333–2335. doi: 10.1271/bbb.59.2333. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki T, Tanaka Y, Ishida M, Ishizuka M, Yamagishi A, Oshima T. Screening of a mutant plasmid with high expression efficiency of GC-rich leuB gene of an extreme thermophile Thermus thermophilus in Escherichia coli. J Biochem. 1997;121:1031–1034. doi: 10.1093/oxfordjournals.jbchem.a021690. [DOI] [PubMed] [Google Scholar]
- 51.Talbot G, Sygusch J. Purification and characterization of thermostable β-mannanase and α-galactosidase from Bacillus stearothermophilus. Appl Environ Microbiol. 1990;56:3505–3510. doi: 10.1128/aem.56.11.3505-3510.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thananunkul D, Tanaka M, Chichester C O, Li T. Degradation of raffinose and stachyose in soybean milk by α-galactosidase from Mortierella vinacea. Entrapment of α-galactosidase within polyacrylamide gel. J Food Sci. 1976;41:173–175. [Google Scholar]
- 53.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vian A, Carrascosa A V, García J L, Cortés E. Structure of the β-galactosidase gene from Thermus sp. strain T2: expression in Escherichia coli and purification in a single step of an active fusion protein. Appl Environ Microbiol. 1998;64:2187–2191. doi: 10.1128/aem.64.6.2187-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vieira J, Messing J. The pUC plasmids and M13mp7 derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 56.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 58.Zeilinger S, Kristufek D, Arisan-Atac I, Hodits R, Kubicek C P. Conditions of formation, purification, and characterization of an α-galactosidase of Trichoderma reesei RUT C-30. Appl Environ Microbiol. 1993;59:1347–1353. doi: 10.1128/aem.59.5.1347-1353.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]