Summary
Coronavirus porcine epidemic diarrhea virus (PEDV) is classified in the genus Alphacoronavirus, family Coronaviridae that encodes the only accessory protein, ORF3 protein. However, how ORF3 contributes to viral pathogenicity, adaptability, and replication is obscure. In this review, we summarize current knowledge and identify gaps in many aspects of ORF3 protein in PEDV, with emphasis on its unique biological features, including membrane topology, Golgi retention mechanism, potential intrinsic disordered property, functional motifs, protein glycosylation, and codon usage phenotypes related to genetic evolution and gene expression. In addition, we propose intriguing questions related to ORF3 protein that we hope to stimulate further studies and encourage collaboration among virologists worldwide to provide constructive knowledge about the unique characteristics and biological functions of ORF3 protein, by which their potential role in clarifying viral behavior and pathogenesis can be possible.
Subject areas: Porcine molecular biology, Protein, Virology
Graphical abstract

Porcine molecular biology; Protein; Virology
Introduction
Porcine epidemic diarrhea (PED) is an enteric disease characterized by acute diarrhea, vomiting, dehydration, and weight loss in pigs and has caused enormous economic losses in countries around the world.1 Although PED was first documented in the UK as early as 1971,2 the causing pathogen, PED virus (PEDV), was initially identified in Belgium in 1978.3 Subsequently, cases of PEDV infection were reported in several European countries, as well as Asian countries, such as China, Japan, South Korea, and Thailand (Figure 1).4 Since 2010, variants of PEDV associated with large-scale outbreaks of diarrhea have been documented in China. Suckling piglets afflicted by the severe pandemic suffered an 80–100% morbidity and a 50–90% mortality,5 which posed a serious threat to the Chinese pig industry. PEDV was first discovered in the USA in 2013.6 Since then, the virus has spread quickly across the country, killing many sick piglets and resulting in significant economic losses.7,8
Figure 1.
Geographic distribution of porcine epidemic diarrhea virus (PEDV) around the world
The white areas mean no cases, whereas the orange marked regions indicate the high prevalence of PEDV infection. The map was visualized in RStudio using the packages natural earth, sf, and ggplot2.
Coronaviruses (CoVs) are a family of enveloped, positive-sense, and single-stranded RNA viruses that belong to the Coronaviridae family of order Nidovirales and can be divided into four genera, including Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV), and Deltacoronavirus (δ-CoV).9 CoVs are major threats to humans and vertebrate species. They can infect humans, livestock, birds, bats, mice, and many other wild animals, causing respiratory, enteric, hepatic, and neurological diseases.10 Taxonomically, PEDV is classified in the genus Alphacoronavirus, family Coronaviridae. Because of the lack of effective vaccines, PEDV remains one of the biggest risks to the swine industry on a global scale.1
The detailed molecular mechanism of PEDV invading cells, especially the interactions between virus protein and cellular receptor, is largely unknown. Porcine aminopeptidase N (pAPN), a 150-kDa glycosylated type II transmembrane (TM) protein partially expressed on the brush border membrane by porcine small intestinal villous enterocytes,11,12 was initially identified as the functional cellular receptor for PEDV.13,14 However, subsequent research found that the pAPN-deficient cell line, such as the African green monkey kidney (Vero) cell, is susceptible to PEDV infection,13,15,16 challenging pAPN’s role as a functional receptor. Indeed, some recent studies disputed the conventional wisdom regarding the crucial role played by pAPN during the PEDV infection and found that though pAPN promotes PEDV infectivity via aminopeptidase activity, it still was not an acknowledged cellular receptor of PEDV.17,18 Thus, the genuine virus receptor still awaits its identification, and robust evidence must be provided to stress this fundamental virological issue in elucidating the virus pathogenesis.
The coronavirus genome is distinct from other Nidoviruses because it encodes various accessory proteins in its 3’-proximal genomic regions, which appear important for viral pathogenesis but not necessary for virus replication. In the case of PEDV, ORF3 protein, which is the only accessory protein encoded by the PEDV genome, has not been studied as thoroughly as other viral structural proteins, and much of what is known about them comes from substantial functional investigations on severe acute respiratory syndrome coronavirus (SARS-CoV),19 severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2),20 and other comparable viruses, such as Middle East respiratory syndrome coronavirus (MERS-CoV).21,22 Although ORF3 protein is not essential for PEDV replication, it plays an important role in cell cycle progression, stress responses, including apoptosis and autophagy, and innate immune responses. In addition, the evolutionary analysis indicated that the ORF3 gene tends to be mutated and truncated during cell adaptation. Some novel field PEDV strains carrying naturally truncated ORF3 gene were constantly emerging and closely related to viral pathogenesis.23,24 Therefore, extensive study of the biological function of ORF3 protein is important to further elucidate the pathogenic mechanism of PEDV.
Although a concise study about ORF3 accessory protein was reported with emphasis on the ORF3 and host interaction previously,25 there are still many unknown aspects regarding ORF3 protein’s biological characteristics, including the membrane topology, Golgi retention mechanism, potential intrinsic disordered property, functional motifs, protein post-translational modifications (PTMs), and codon usage phenotypes related to genetic evolution and gene expression. Given the important role of accessory protein in coronavirus infection and the research blind spots of PEDV accessory protein in many aspects, current knowledge of this PEDV only accessory protein, ORF3, is summarized with a focus on what is now known and which areas urgently require additional investigation.
Virion properties and structure of the PEDV genome
Porcine epidemic diarrhea virus (PEDV), belonging to the genus Alphacoronavirus in the family Coronaviridae of the order Nidovirales, causes devastating enteric disease and high mortality in neonatal piglets.1 It is an enveloped virus with an approximately 28 kb positive-sense, single-stranded RNA genome that is 5’ capped and 3’ polyadenylated.1,26 The organization of the PEDV genome is typical of CoVs, with the gene order 5’UTR-ORF1a-ORF1b-S-ORF3-E-M-N-3’UTR (Figure 2A).27 The two polyproteins (pp1a and pp1ab) are encoded by the PEDV genome’s first two-thirds, and the four structural proteins (spike protein, S; envelope protein, E; membrane protein, M; and nucleocapsid protein, N) and one accessory protein (i.e., ORF3) are encoded by the remaining one-third of the genome (Figure 2A).
Figure 2.
PEDV genome organization and viral structure
(A) PEDV genome structure shows the position of the accessory protein and viral particle structure diagram. PEDV genome is divided into two sections, with the 5′ two-thirds containing two large, overlapping open reading frames (i.e., ORF1a and ORF1b). These ORFs encode two long polyprotein precursors, pp1a and pp1ab. The remaining 3’ third of the genome encodes four structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), with an only accessory protein, ORF3. These proteins have distinct functions and each of them plays an essential role in the life cycle of the virus.
(B) Schematic diagram of a PEDV virion is shown. Structural proteins of spike (S), envelope (E), and membrane (M) proteins are embedded in the lipid bilayer envelope. The viral genomic RNA (labeled with a gray line) is assembled with the nucleocapsid (N) protein, which is enclosed by the lipid bilayer membrane. The ORF3 accessory protein is not displayed in the virion structure because it is not a structural virion component at the current cognitive level.
(C) Transmission electron microscopes (TEM) images of purified PEDV virions. The PEDV particles were negatively stained, and a virus particle is shown by an arrow. Crown-shaped spikes are visible. Scale bar: 100 nm. Magnification, ×,110,000.
PEDV particle is a membrane-enveloped virion with an average diameter of about 95–190 nm,1 which is coated in a double-layer membrane with a characteristic “crown-like” structure on its surface (Figure 2B and 2C). The S protein is a type I membrane glycoprotein composed of S1 and S2 external domains and constitutes the characteristic spikes on the outside of the virion, facilitating viral attachment to target cells and producing-neutralizing antibodies.28 The E protein upregulates interleukin-8 expression, causes endoplasmic reticulum (ER) stress in the host cell, is proposed to possess ion channel activity, and participates in virion assembly.29,30 The thick lipoprotein virion envelope is made of the M protein, which is crucial for connecting the viral nucleocapsid and envelope.31 The N protein forms the viral core, participates in the transcription of the viral genome, binds to viral RNA, and forms the main component of ribonucleoprotein (RNP).32 The only accessory protein, ORF3, promotes virus proliferation, encodes an ion channel important for viral fitness, and plays an important role in several cellular processes.33,34,35,36 Although many research groups are currently studying ORF3 protein, numerous unknown properties of ORF3 protein still need to be elucidated.
ORF3 is a multifunctional protein
The accessory proteins of various CoVs are involved in viral pathogenesis and virulence.20,37,38,39 In cell culture settings, they are not necessary for viral replication but may be crucial for controlling the host immune response, which could support viral stability and/or pathogenicity in vivo.19,40 For instance, even though the PEDV ORF3 gene is often lost during the extensive cell passaging of the virus,41 its absence was related to diminished virulence.42 However, conflicting results of PEDV field strains with a naturally occurring truncated ORF3 gene were recently found in China, and this natural deletion at the ORF3 gene was confirmed to be highly virulent with severe diarrhea and high mortality to suckling piglets,23 which indicates that ORF3 protein plays a mysterious role in PEDV pathogenicity.
The ORF3 gene has a length of 675 bp and encodes a protein of 224 amino acids. When PEDV is adapted to growth in cell culture, the protein is vulnerable to deletion or mutation.41,43 Studies showed that the deletion or variation of the ORF3 gene might be related to the virulence and cell adaptation of the virus.24,41 During the continuous passage of PEDV in vitro, a 51-nucleotide (nt) loss in the ORF3 gene is present in cell-adapted PEDV strains such as KPED-9 and P-5V, and this deletion is thought to be the primary cause of the PEDV strains’ decreased pathogenicity.44 Similarly, the attenuated DR13 and CV777 PEDV strain has also been found to have a 49-nt deletion in the ORF3 gene, which causes an early termination of translation and a reading frameshift.33 Therefore, the ORF3 gene’s ability to distinguish between cell-adapted and wild-type viruses enables it to be a valuable tool for epidemiological research of PEDV infections.41,45
Among the accessory proteins of all CoVs, PEDV ORF3 and SARS-CoV ORF3a share many similarities regarding protein structure and function. For instance, it has been demonstrated that SARS-CoV ORF3a is an ion channel protein and facilitates virus release.46 PEDV ORF3 protein is functionally analogous to SARS-CoV ORF3a and functions as a viroporin to control the generation of infectious progeny viruses.33,46,47 Besides its roles in affecting virus virulence and the cellular adaption phenotype with the truncated feature, ORF3 protein also plays multifaceted roles in vitro. Accumulating evidences suggest that ORF3 plays an essential role in numerous cellular processes. Ye et al. showed that PEDV ORF3 protein prolongs cellular S-phase and contributes to vesicle formation.36 Zou and co-workers proved that PEDV ORF3 protein triggers ER stress by upregulating GRP78 protein expression and activating the pERK-eIF2α signaling pathway,34 which further leads to autophagy in infected cells. Studies on the effect of ORF3 protein on apoptosis have also been reported. Chen et al. demonstrated that PEDV ORF3 protein could not induce apoptosis by transient transfection of ORF3 expressing-plasmid.48 However, by using artificially rescued PEDV recombinant virus strains carrying full-length ORF3 gene (rPEDV-ORF3wt, rPEDV-ORF3CV777, and rPEDV-ORF3NY) and the recombinant virus strains without ORF3 gene (rPEDV-ΔORF3), our group found that ORF3 protein can delay the cytopathic effect (CPE) formation and significantly inhibit apoptosis induced by PEDV.35 More recently, Jiang et al. reported that the plaque size and syncytia phenotypes in the ORF3-null reconstituted PEDV were larger than those of wild-type ancestor. In addition, ORF3-null virus has a relatively faster growth phenotype,49 suggesting that ORF3 is dispensable for PEDV propagation in vitro with a potential role of ORF3 in PEDV cytopathology.
Furthermore, it has been found that ORF3 protein could interact with the PEDV spike protein during the virus infection,50 which indicates the possibility that ORF3 protein may assist S protein binding to cell receptors, thus promoting virus infection of cells. Wu and colleagues proved that PEDV ORF3 protein inhibited cellular proinflammatory cytokines (IL-6 and IL-8) production through NF-κB p65 pathway blockage, which provided a fresh perspective into PEDV ORF3 protein’s involvement in the immune evasion strategy.51 Kaewborisuth et al. found that ORF3 protein could not only downregulate the IFN-β promoter activation and suppressing poly (I:C) mediated type I IFN production and induction, but also upregulate IKBKB-mediated NF-κB promoter activity.52 It should be noted that cellular apoptosis played an essential role in virus-host interaction and innate immunity, closely related to efficient virus replication and pathogenesis.53,54 It is unknown whether ORF3 protein can highjack the key participating protein in innate immune response, such as the IFN-signaling pathway, and counteract the increased innate immune response, thus reducing type I IFN production. A summary of the ORF3 functions based on current knowledge is shown in Table 1. Altogether, PEDV ORF3 protein is a multifunctional protein that participates in numerous cellular processes and may be essential in virus infection, packaging, release, virus-host interaction, and maintaining normal host immunity.
Table 1.
Subcellular localization and functions of PEDV accessory protein
| Strain name | Features and functions | Reference |
|---|---|---|
| PEDVAVCT12 | Suppress cell-adapted PEDV replication in cultured cells; Inhibit PEDV rescue in a reverse genetics system; Inhibits the manipulation of the viral genome and PEDV rescue; Inhibits recovery of PRRSV, but not influenza virus. | (Jengarn et al.55; Wongthida et al.43) |
| icPEDV | Deletion of ORF3 causes lower diarrhea scores and partial attenuation in pigs. | (Beall et al.42) |
| CV777 | Consists of four putative transmembrane domains, functions as a potassium ion channel in both Xenopus laevis oocytes and yeast; Promotes viral replication of wild-type PEDV. | (Wang et al.33) |
| CH/YNKM-8/2013 (Field strain) | Facilitates the formation of double-membrane vesicles (DMVs); Promotes the proliferation of attenuated PEDV; Regulates the cell cycle progression by prolonging the S phase. | (Ye et al.36) |
| rPEDV | Contributes to virus proliferation and cell viability; Inhibits early cell apoptosis by suppressing Caspase-3 cleavage. | (Si et al.35) |
| Eukaryotic expressed | Causes ER stress response and promotes autophagy; Up-regulations of the GRP78 and activation of PERK-eIF2α signaling pathway. | (Zou et al.34) |
| PEDVAV12 | Interacts with S protein during PEDV replication; Contributes to PEDV replication. | (Kaewborisuth et al.50) |
| DR13, YN144 | Early termination of ORF3 could be the attenuation and cell adaptation marker of the PEDV. | (Song et al.56; Chen et al.57) |
| Eukaryotic expressed | Inhibits cellular proinflammatory cytokines IL-6 and IL-8 production; Blocks the NF-κB p65 activation and expression; Disrupts nuclear factor p65 phosphorylation and nuclear translocation. | (Wu et al.51) |
| PEDVAV12 | Interacts with the IκB kinase β (IKBKB), resulting in the up-regulation of the IKBKB-mediated NF-κB promoter activity, down-regulation of the IKBKB-mediated IFN-β promoter, and IFN-β mRNA expression. | (Kaewborisuth et al.52) |
| rPEDV | Transports through the exocytic pathway; Not a structural virion component and constitutive protein of PEDV. | (Si et al.58) |
| PEDVAV12 | Interacts with cellular VPS36 protein and associates with PEDV replication. | (Kaewborisuth et al.59) |
| Induced autophagy in IPEC-J2. | ||
| Eukaryotic expressed | Functions as the key inducers of PEDV-induced autophagy. | (Lin et al.60) |
| HN2021 | Large deletion is associated with severe diarrhea and high mortality. | (Zhang et al.23) |
| NJ strain | Used as the target for the detection of classical PEDV strains. | (Liu et al.61) |
| Eukaryotic expressed | As an IFN suppressive protein, it inhibits type I and III IFN production. | (Zhang et al.62; Zhang et al.63) |
Note: rPEDV means recombinant PEDV; ca-PEDV means cell culture adapted PEDV; icPEDV means infectious-clone-derived PEDV; Functions shown in bold is our observation.
The Golgi-resident phenotype of ORF3 accessory protein and the supposed mechanism
The ORF3 protein was initially shown in the cytoplasm,36,59 specifically in the perinuclear area50 and the Golgi apparatus.34,50,58 These results suggest that ORF3 can relate to particular cellular pathways that might regulate viral replication and pathogenesis. Subsequent plasmids transfection and PEDV infection experiments showed that the wild-type ORF3 protein aggregated in Golgi and was partially expressed on the cell membrane, it was obviously a Golgi resident protein.58 However, the truncated ORF3 protein of the cell-adaptation strain DR13 had an altered intracellular localization, which presented only in the ER and could not be expressed on the cell membrane owing to a large amino acid deletion at its C-terminus.58 We further screened the amino acid motif that determines its subcellular localization and found that the 170YLAI173 motif (amino acids 170-173) determines its Golgi/ER retention and plays a decisive role in its cell membrane translocation.58 These findings raise an important issue: the involved mechanisms responsible for the Golgi retention and the cell surface expression of ORF3 protein, which need to be elucidated in greater detail.
Up to now, there are three possible mechanisms reasonable for the proteins retained in the Golgi apparatus: (1) Hydrophobic matching hypothesis: this hypothesis proposes that the protein containing a shorter transmembrane region will reside in a thinner membrane region, whereas the protein with a longer transmembrane region will be located in a thicker region.64 In other words, the transmembrane region affects the subcellular localization of the protein. At present, this hypothesis has been confirmed: on the one hand, the Golgi retention profile of yeast proteins (such as sed5) and SARS-CoV proteins (such as ORF7b) depend on the length of their transmembrane region.65,66 On the other hand, shortening the transmembrane region of VSV-G leads to a slower export rate out of the Golgi apparatus. Thus, the long transmembrane region contributes to the efficient export of secretory proteins;67 (2) Phase separation theory: phase separation is the process by which biomolecules (e.g., proteins and/or nucleic acids) are concentrated into a separate phase forming liquid droplets.68 Signal transduction in nerve cells is processed by phase separation, and synapses are the connection points where information transfer occurs between neurons and contain a compartment with a highly aggregated protein layer, the postsynaptic density (PSD), which is responsible for the processing and transmission of brain signals.69 It has been proved that PSD-95, a protein expressed in high abundance in PSD, can undergo phase separation with SynGAP at higher concentrations in vitro, which enhances protein plasticity and allows the protein to undergo phase changes in a short period, enabling an ultra-fast release of neurotransmitters and thus facilitating neural signaling.70 Phase separation also drives cargo protein sorting in chloroplasts,71 promotes cargo proteins translocated to vesicles, and facilitates protein sorting and transporting; (3) Kin-recognition model: the “kin-recognition model” was the first model proposed for the residency of glycosyltransferases in the Golgi.72 This model suggests that different transmembrane regions form so-called homodimers or heterodimers within the cell.73 The formed oligomers are too large to enter transport vesicles that sort cargo proteins exported from the Golgi, spatially preventing the loading of these proteins into the transport vesicles and therefore causing these oligomeric proteins to retain in the Golgi.73,74 Therefore, the retention of protein in Golgi is a complex process, and multiple factors are involved in the retention, among which the amino acid composition, aggregation state, and phase separation of protein are important influencing factors.
Currently, the related research on ORF3 protein is relatively limited. When considering the possible mechanisms responsible for the Golgi retention of ORF3 protein, the question of which of the above mechanisms is involved in the Golgi retention of ORF3 protein needs to be urgently revealed. Understanding this question will benefit the functional study of ORF3 protein. To the authors’ knowledge, one or more of the above mechanisms are likely involved in the phenotype of the ORF3 protein retention in the Golgi apparatus (Figure 3), and this work is one of the main topics currently ongoing in our laboratory.
Figure 3.
Working model for the roles of ORF3 protein in PEDV replication and the proposed retention mechanisms of ORF3 protein in Golgi apparatus
After PEDV infection, the unique membrane topology of ORF3 protein affects the folding, post-translational modification and aggregation form of protein, which ultimately impacts its residency within the Golgi apparatus. On the one hand, through interprotein interaction, ORF3 protein recruits other Golgi resident proteins to form a highly complex protein interaction network; on the other hand, multi-spanning domain ORF3 proteins expressed in high abundance in cells undergo glycosylation modifications in the Golgi apparatus and promote phase separation or oligomerization through binding to membrane-anchored ion channels leading to the spontaneous formation of protein aggregates, which restrict the loading of ORF3 protein into transport vesicles and eventually lead to protein retention in the Golgi apparatus (This schematic diagram is the author’s own creation).
Membrane topology of ORF3 protein
Membrane topology affects the correct folding of proteins and impacts their function.75 Therefore, an important strategy for studying the coronavirus proteins in virus pathogenicity and infection is to clarify their subcellular location and topology. The coronavirus accessory protein has a complex membrane topology and is closely related to its cellular location and biological function. In most studies, SARS-CoV ORF3a has been used as an example to investigate the ORF3 function. The SARS-CoV ORF3a protein is predominantly located in the Golgi apparatus, with its N-terminus facing the extracellular matrix (Ntlum) and its C-terminus facing the cytoplasm (Ctcyt).76,77 It is a triple-spanning transmembrane (TM) protein that resembles the M protein in topology.78 In addition, it can form tetramers and potassium channels, regulate virus release,46,79 and is connected to a pro-apoptotic function.47 Similarly, a triple-spanning membrane topology structure was also found in SARS-CoV-2 ORF3a protein that forms cation channels in lipid nanodiscs and showed dimers and tetramers under cryoelectronic microscopy.80 Furthermore, TM1-TM2 and TM2-TM3 were connected by intracellular and extracellular short linkers, respectively.80 Between the S and E genes, the hCoV-NL63 genome encodes the ORF3 protein (Figure 4A), which is 225 aa long and has three putative transmembrane domains (TMDs) at aa positions 39-61, 70-92, and 97-116, respectively.81 Topology studies indicate that its N-terminal domain (NTD) is located in the extracellular region, whereas the C-terminal domain (CTD) is located in the intracellular region.81
Figure 4.
Characteristics of PEDV ORF3 and comparison to homologous genes in other coronaviruses
(A) Localization of ORF3 gene within the PEDV genome and comparison of nucleotide (nt) identity based on multiple sequence alignments with prototype strains of CoV groups alpha, beta, gamma. The sequence of ORF3 (protein_id = AFE85963) was analyzed using BLAST and MEGA-X software (version 10.1.8). Note that IBV ORF3a and 3b were fused to one ORF3ab.
(B) Putative analysis on ORF3 protein membrane topology and glycosylation features. The putative four transmembrane (TM) domains, which including TM1 (Gln-40 to Ser-63), TM2 (Arg-75 to Ile-97), TM3 (Tyr-116 to Tyr-139), and TM4 (Gly-150 to Ile-173), are diagrammatically presented according to the previous study. Q, Glutamine (Gln); S, Serine (Ser); R, Arginine (Arg); I, Isoleucine (Ile); Y, Tyrosine (Tyr); G, Glycine (Gly); Predicted N-linked glycosylation sites are indicated by an “N” at the respective localizations with an index number identifying the amino acid position. No O-linked glycosylation sites are predicted.
The CoVs accessory protein is mostly found in intracellular membranes between the ER and Golgi compartments,82,83 where it plays important roles in the intracellular trafficking of the viral proteins and virus adsorption, invasion, packaging, budding, and release stages through an unidentified mechanism. In the case of PEDV accessory protein, it has been demonstrated that ORF3 protein is essentially accumulated in the Golgi area of the cell, similarly in infected and transfected cells.58 According to bioinformatic predictions, the PEDV ORF3 protein is a viroporin with four transmembrane domains and a potassium ion channel protein that controls virus release (Figure 4B).33 Results from our group showed that PEDV ORF3 protein is a Golgi resident protein, which diffusely aggregates around the nucleus.58 Although ORF3 protein promotes viral replication,35 it does not incorporate into virions purified by sucrose gradient ultracentrifugation.58 The above results indicated that the membrane topology of coronavirus proteins is closely related to their functions, and clarifying the protein’s topology can help to understand their functions in depth. Therefore, the defined membrane protein topology of PEDV ORF3 protein may provide a useful framework to understand its interaction with other viral and host components and contribute to establishing the basis to tackle the pathogenesis of PEDV. Although the ORF3 protein is currently predicted to be a multi-spanning transmembrane protein and confirmed to localize in the Golgi apparatus, its specific intracellular and extracellular regions and membrane topology in eukaryotes have not been demonstrated experimentally. Therefore, it’s urgent to clearly determine the specific transmembrane domains of PEDV ORF3 protein in eukaryotic cells and, specifically, to verify which domain is involved in the function of ORF3 protein.
Potential intrinsic disordered property of ORF3 protein and the possibility of liquid-liquid phase separation (LLPS)
Intrinsically disordered proteins (IDPs), which are encoded by intrinsically disordered regions (IDRs) and earned the name “Dancing Proteins”, are a class of functional proteins lacking a fixed or organized three-dimensional structure,84,85 typically in the absence of its macromolecular interaction partners, playing crucial roles in the mechanism of virus pathogenesis, cell regulation and host signaling pathways.86,87,88,89 Overall, IDPs are characterized by their polypeptide segments, limited hydrophobic amino acids, and ability to facilitate various biological processes via mechanisms distinct from their structured counterparts.90 IDPs differ from structured proteins in many ways and tend to have distinctive functions, structure, sequence, interactions, evolution, and regulation.89,91
As obligate parasites, viruses achieve their infectious cycles through the recruitment of various host cellular components such as host proteins, nucleic acids, biological membranes, energy and metabolic machinery.92 Consequently, viral proteins frequently have multifunctional behaviors and engage in complex interactions with host ligands. During this process, IDRs could be quite helpful for viruses by ensuring binding diversity. Furthermore, IDRs endow viral proteins with considerable flexibility, whether completely disordered or partially, enabling them to quickly adapt to changing cellular conditions, survive in host immune surveillance environments, and antagonize the host’s defensive mechanism.93
It has been reported that IDPs are associated with many human diseases not limited to cancers, Down’s syndrome, Alzheimer’s disease, variants of Alzheimer’s disease, Parkinson disease, and prion disease.94,95 In addition, IDPs are also composed of various multifunctional proteins from different viral families, mainly concentrated in RNA viruses.96,97,98,99,100,101,102 For example, the SARS-CoV-2 N protein contains three dynamic disordered regions and undergoes liquid-liquid phase separation (LLPS) when mixed with RNA.103,104 IDRs are the main factor driving the formation of LLPS during this process.89,96,105 Furthermore, because they give viral proteins the ability to quickly and promiscuously bind to host proteins, the disordered regions of the SARS-CoV-2 N protein are typically linked to viral infectivity and pathogenicity.98 A recent study further proved that more than 90 disordered regions were found in SARS-CoV-2 spike protein, and the disordered regions in Omicron variants exhibit disorder-to-order transition when compared to its ancestral and the Delta variant strain, all of which indicated that the viral infectivity and pathogenicity were closely related to the IDRs of viral proteins.106 Although several therapies have been investigated, effective antiviral medications are now hard to come by. Therefore, a different approach for logical antiviral drug design could target the IDPs/IDRs in the SARS-CoV-2 genome.
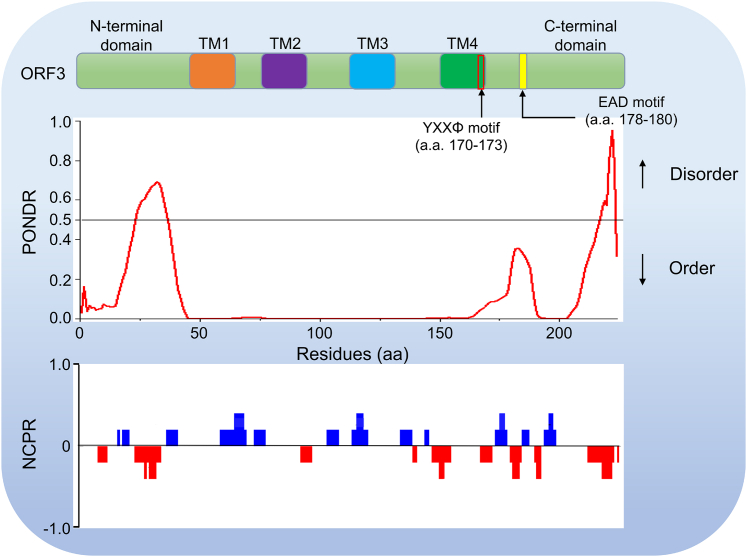
As mentioned above and in other places, the disordered regions and the resulted LLPS property in viral proteins are closely related to the infectivity and pathogenicity of the virus.88,107,108,109 In light of PEDV, ORF3 protein is an important protein affecting virus virulence. Thus it is of certain interest to know whether the IDRs exist in the ORF3 protein coding sequence. According to current knowledge, ORF3 protein has three major domains: an N-terminal domain, a central segment predicted to undergo four times transmembrane, and a C-terminal domain (Figure 5). To test the phase separation ability of the ORF3 protein, we analyzed the sequence of the ORF3 protein using the PONDR (predictor of naturally disordered regions) tool.110 The results showed that there were two IDRs, residues 24-36 and 217-223 (Table 2), in ORF3 protein-coding sequences (Figure 5), indicating that the ORF3 protein has the potential possibility to undergo LLPS. Taken together, IDPs are related to human diseases and exist in various viral proteins, including ORF3. Therefore, the IDRs in the PEDV ORF3 protein coding sequence reminds us that we should strengthen the related research about the relationship between IDRs and virus virulence or pathogenicity, which will benefit us in understanding further the new function of ORF3 protein in great detail.
Figure 5.
PEDV ORF3 protein domains aligned with results of PONDR (predictor of natural disordered regions) and NCPR (net charge per residue; 5-amino acid window) analyses
A schematic representation of the domain structure and overall topology is shown on the top. The PONDR is applied to analyze the intrinsic disorder probability, and NCPR is employed to measure the net charge per residue. For the intrinsically disordered region prediction, residues with anticipated disorder scores of more than 0.5 are regarded as inherently disordered, whereas residues with expected disorder values between 0.2 and 0.5 are considered flexible.
Table 2.
Intrinsically disordered prediction of PEDV ORF3 protein using PONDR VLXT
| Strain name/residues | Number residues disordered | Overall percent disordered | Predicted intrinsically disordered regions (IDRs) | Number of IDRs |
|---|---|---|---|---|
| DR13/224 aaa | 20 | 8.93 | [24]-[36] | 2 |
| [217]-[223] |
Note: aa means amino acid. The GenBank accession number of the DR13 protein sequence is ABS89138.
Can ORF3 protein undergo glycosylation?
Protein glycosylation is an essential and highly conserved post-translational modification (PTM) reaction responsible for various key biological processes, including protein folding, oligomerization, sorting, transport, and stability.111,112,113 During those processes, glycans can demonstrate significant interactions between glycans and proteins and incorporate them into the protein fold to stabilize the protein, for example, through interacting with surface hydrophobic residues and increasing the solubility of proteins. In addition, glycans can also promote the folding of glycoproteins via influencing chaperone contacts during production.114 Generally, N-linked glycosylation and O-linked glycosylation are the two main kinds of protein glycosylation. When an oligosaccharide (glycan) is attached to the nitrogen atom of an asparagine residue, this process is known as N-linked glycosylation. This process typically occurs on asparagine residues in Asn-X-Ser/Thr sequons (asparagine residue in the-N-X-T/S- motif), where X can be any amino acid other than proline. O-linked glycosylation is the process by which a sugar molecule is joined to a protein’s serine (Ser) or threonine (Thr) oxygen atom. O-glycosylation affects the regulation and stability of proteins in the ER or Golgi apparatus.115 Although there are two other glycosylation types, phospho-glycosylation and C-linked glycosylation, they are rarely reported to be involved in the functional regulation of viral proteins.116
As obligate parasites, viruses employ host-cell machinery to glycosylate their own proteins during viral infection and proliferation, which plays an important role in virus-host interaction and host immune response.117,118 For example, the envelope proteins of various human pathogens, including the influenza virus hemagglutinin glycoprotein (HA),119 Dengue virus (DENV) envelope glycoprotein (Env),120 human immunodeficiency virus-1 (HIV-1) Env,121 Zika envelope (E) glycoprotein,122,123,124 Japanese encephalitis virus (JEV) premembrane (prM) glycoprotein,125 West Nile virus (WNV) prM and E glycoprotein,126 Ebola virus (EBOV) glycoprotein (GP),127 and Flavivirus Env128 are extensively glycosylated. Glycosylation on viruses, however, is not exclusive to the viral genus mentioned above. Many coronavirus proteins display glycans and undergo glycosylation which is essential for their specific functions, such as the spike and envelope glycoprotein (S and E) of SARS-CoV-2,129,130,131,132 the spike glycoprotein (S) of PEDV,133 the membrane protein (M) of infectious bronchitis virus (IBV)134 and SARS-CoV,135,136 and this is also the case in multitudinous coronavirus accessory proteins, which include 3a of SARS-CoV, ORF3 of human pathogenic coronavirus NL63 (hCoV-NL63),81 7b of feline coronaviruses (FCoVs),137 and the secreted ORF8 of SARS-CoV-2.138 In addition, the glycosylation machinery in the ER-Golgi system of host cells can be hijacked by viruses to help escape the host’s immune surveillance and shield them from antibody recognition, which is crucial for multiple facets of viral pathogenesis.114,139 This is true for coronavirus, for example, it has been reported that targeting or inhibiting the protein N-glycosylation blocks SARS-CoV-2 infection.140,141,142
The active site of oligosaccharyltransferase (OST), a membrane protein complex that chooses the Asn-X-Ser/Thr consensus sequence on polypeptide chains and creates the N-glycosidic linkage between the side-chain amide of asparagine and the oligosaccharide, is only found in the lumen of the ER in eukaryotic cells.143 Therefore, neither the membrane nor the cytosol experience N-linked glycosylation.130 According to in silico analysis, although several amino acid mutations were found through the ORF3 multiple sequence alignment (Figure S1A), the ORF3 protein indeed contains three potential and conserved N-linked glycosylation motifs, asparagine-X-serine/threonine (X is any amino acid except proline), or three putative N-glycosylation sites at positions N22, N43 and N125 (Figure 4B), of which probably only the first is used because the sites at positions 43 and 125 are located inside the predicted transmembrane domains, whereas only the N22 is located in the ER lumen (Figures 4B and S1B). Up to now, there has been no prediction or study on the PEDV ORF3 protein’s glycosylation. Therefore, it is urgently needed to experimentally verify whether the ORF3 protein undergoes N-glycosylation in mammalian cells and which of the predicted N-linked glycosylation motifs is authentic for ORF3 protein. Elucidating this issue will further reveal the biological functions of ORF3 proteins.
Knowledge of functional motifs on ORF3 protein
The ER/Golgi intermediate compartment (ERGIC) or the Golgi compartment, where are the locations of CoV assembly and packaging, CoV accessory proteins were found to have similar localizations in these organelles.144 Some CoVs accessory proteins functioned as the constitutive component of virions and integrated into virus particles, such as ORF4a of human coronavirus 229E (HCoV-229E),145 3a and 7b of SARS-CoV,78,82 and ORF3 of hCoV-NL63,81 which could explain the essential role that numerous accessory proteins play in viral infection. CoVs accessory proteins, including PEDV ORF3 protein, frequently contain YXXΦ and acidic motifs generally situated at the cytoplasmic domain (C-terminus), which determine the trafficking and turnover of many membrane-spanning proteins in the cell.58,146,147,148 Based on current published data, a summary of the YXXΦ and ExD (di-acidic) motifs found in some well-known transmembrane proteins and the C-terminus of PEDV ORF3 protein are summarized in Table 3.
Table 3.
Comparison of YXXΦ and di-acidic motifs found in the well-known transmembrane proteins and the cytoplasmic domain of PEDV ORF3 protein
| Protein | NCBI identifier (No.) | YXXΦ and di-acidic motifs | Reference |
|---|---|---|---|
| ORF3 parent DR13 | EU054929 | TM-VAFVSSIDLYLAIRGRQEADLHLLRTV-COOH | (Park et al.44) |
| ORF3 Virulent DR13 | JQ023161 | TM-VAFVSSIDLYLAIRGRQEADLHLLRTV-COOH | (Park et al.149) |
| ORF3 CV777 | AF353511 | TM-VAFVSNIDLYLAIRGRQEADLHLLRTV-COOH | (Bridgen et al.150) |
| ORF3 OK10240-8 | MG334555 | TM-VAFVSSIDLYLAIRGRQEADLQLLRTV-COOH | (Zhang et al.151) |
| ORF3 USA Oh851 | KJ399978 | TM-VAFVSSIDLYLAIRGRQEADLQLLRTV-COOH | (Wang et al.152) |
| ORF3 USA PC22A | KY499262 | TM-VAFVSSIDLYLAIRGRQEADLQLLRTV-COOH | (Hou et al.153) |
| ORF3 China GD−1 | JX647847 | TM-VAFVSSINLYLAIRGRQEADLHLLRTV-COOH | (Wei et al.154) |
| ORF3 HeN−MY−2015 | KU641647 | TM-VAFVSSIDLYLAIRGRQEADLQLLRTV-COOH | (Wang et al.155) |
| VZV-GPE | P09259 | TM-NQSMYYAGLPVDDFEDSESTDTEEE-COOH | (Olson and Grose,156) |
| VSV-G | P03522 | TM-GIHLCIKLKHTKKRQIYTDIEMNRLGK-COOH | (Rose et al.157) |
| Human LAP | P11117 | TM-LLTVLFRMQAQPPGYRHVADGEDHA-COOH | (Chen et al.158) |
| Human LDLR | NM_000527 | TM-LKNINSINFDNPVYQKTTEDEVHICHN-COOH | (Ference et al.159) |
| Human ASGPR1 | P07306 | TM-MTKEYQDLQHLDNEESDHHQLRKGP-COOH | (Spiess and Lodish,160) |
| Mouse CD3 γ chain | P11942 | TM-DKQTLLQNEQLYQPLKDREYDQYSH-COOH | (Haks et al.161) |
| Mouse CD3 δ chain | P04235 | TM-QALLKNEQLYQPLRDREDTQYSRLG-COOH | (Van Den Elsen et al.162) |
| Human GLUT4 | P14672 | TM-HRTPSLLEQEVKPSTELEYLGPDEND-COOH | (Wollscheid et al.163) |
| Mouse GLUT4 | Q27994 | TM-HRTPSLLEQEVKPSTELEYLGPDEHD-COOH | (Abe et al.164) |
Note: The YXXΦ motif is underlined, and the acidic residues complying with the di-acidic [(D/E)X(E/D)] signal are boldly displayed. Sequences were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/).
Those functional motifs were well studied to some extent and contributed to their intracellular transport and biological function. For example, SARS-CoV 3a protein, previously termed U274, comprised of 274 amino acids and three putative transmembrane domains, was found on the plasma membrane and perinuclear area. According to the analysis of the C-terminal domain of the 3a protein, two distinct sorting motifs, a YXXΦ (where X is any amino acid and is an amino acid with a bulky hydrophobic side chain) upstream of an ExD (di-acidic) motif, were found. The YXXΦ motif has been linked to dominating the viral protein’s intracellular localization in various host-cellular organelles,165,166,167 whereas the di-acidic motif is essential for efficient ER export.168 These two motifs’ juxtaposition seems crucial for transporting proteins to the plasma membrane.169 It was also proved that these motifs could play a significant role for SARS-CoV 3a protein to induce cellular apoptosis,170 and cell-cycle arrest,76 which was also detected in SARS-CoV-2 3a protein, was also linked to the induction of apoptosis during virus infection.148 Furthermore, previous and recent studies demonstrated that the occurrence of point mutations of the 3a proteins’ YXXΦ motif in SARS-CoV and SARS-CoV-2 showed a weakened pro-apoptotic phenotype,47,148 indicating the significant roles of these functional motifs on 3a’s pro-apoptotic function.
Similar motifs are also found in the ORF3 protein encoded by the PEDV genome.58 In our previous studies, we proved that the PEDV ORF3 protein possesses the YXXΦ and di-acidic motifs in its coding sequence. The YXXΦ motif (170YLAI173) located at the C-terminal domain was confirmed to be a key motif affecting the intracellular transport of ORF3 protein, and determining its expression on the cell membrane.58 Furthermore, ORF3 protein is closely related to PEDV-induced apoptosis.35 Therefore, it is interesting to investigate whether YXXΦ and di-acidic motifs on ORF3 protein are involved in regulating PEDV-induced apoptosis. If so, what are the similarities and differences of the regulatory mechanisms between the motifs on ORF3 and 3a protein? This work is currently ongoing in our laboratory. Though the identification and characterization of the functional motifs of PEDV ORF3 protein in regulating virus proliferation and apoptosis have not yet been revealed, ORF3 protein likely has a similar role in this regard because of its similar features (viroporin, same location between S and E, and the multi-spanning transmembrane peculiarity) to SARS-CoV and SARS-CoV-2 3a protein. Many researchers are becoming more interested in the crucial role of ORF3’s functional motifs in promoting PEDV pathogenicity and virus-host interaction because of their potential merits in antiviral targets and attenuated vaccine development. In addition, these changes were associated with virus growth and pathogenicity and whether those mutations impact PEDV field strains adaptation in cell culture is still not elucidated. Thus, those issues deserve to be explored in depth based on the current knowledge.
Codon usage of ORF3 protein and its adaptive evolution phenotype
Synonymous codons are those codons that encode the same amino acid. Synonymous codon usage is not randomly used; some codons are utilized more frequently than others.171,172 This phenomenon is known as synonymous codon usage bias (CUB) in numerous organisms, including prokaryotes, eukaryotes, and viruses.172,173,174,175,176,177,178,179 Synonymous CUB is generally influenced by factors including mutational pressure, natural selection, translational efficiency, and mammalian genome compositional restrictions.171,180 A recent report shows that some novel influencing factors, including mRNA export, transcription, and immune evasion, can also dominate the viral codon usage,173 and the virus’s CUB serves as an important influencing factor in viral adaptation to the host and determines the viral host tropism.181,182,183
It is documented that the virus’s capacity to evade host immune surveillance, its ability to survive in extreme conditions, and its genetic evolution capacity are all significantly influenced by the codon usage between the virus and the host.173,184 Therefore, understanding viral codon usage can yield valuable knowledge about gene expression and regulation based on codon adoption, which can benefit the development of a new generation of vaccines that achieve a high level of viral antigen expression to induce long-lasting immunity.185 Thus, codon usage analysis is a powerful tool to elucidate the CUB of multiple organisms. Knowledge about the CUB’s profundity and potential influencing factors is critical for elucidating the viral evolution pattern and host adaptability. Thus far, few studies have investigated how animal viruses use synonymous codons. PEDV is an animal RNA virus that exhibits a rapid evolution rate since it first appeared. Previous studies on PEDV have mostly focused on infectivity and prevalence.1,32,186 However, there are limited studies on the CUB of PEDV, which is especially true for studying PEDV ORF3 protein’s CUB.
Chen et al. identified that PEDV has a rare codon usage pattern in its genome, indicating that the frequency of synonymous codon usage is dispersed throughout the PEDV genome.187 In addition, they discovered that PEDV’s codon usage pattern is shaped by two primary factors, mutational pressure and natural selection, with the latter playing a more significant impact. Furthermore, other factors, such as geographical distribution and the dinucleotide composition, were regarded as other potential influencing factors shaping the PEDV’s codon usage pattern to some degree. Two published results proved those conclusions, indicating a conservative and invariable codon usage preference in the PEDV genome.183,188 Furthermore, our group demonstrated that PEDV had evolved a mixture of antagonistic and coincident codon usage patterns relative to Sus scrofa, which promote its host adaptation and viral replicative fitness, and the PEDV genotype II strains show the highest amount of adaptation phenotype than other divergent clades.183
Though several studies have previously mentioned the CUB of the whole PEDV genome, research on the codon usage pattern of a particular gene within the PEDV genome is still very scarce. Up to now, only three single gene’s codon usage patterns have been reported among the serious genes of the PEDV genome. Cao et al. showed a modest CUB in PEDV S protein and further demonstrated that mutational pressure, not translational selection, was the primary influence on this bias. In addition, they also discovered that the S gene’s aromaticity and hydrophobicity slightly influenced the variance in this phenomenon.189 The PEDV ORF3 genome’s codon usage patterns were then thoroughly analyzed by Xu and colleagues, who discovered that the PEDV ORF3 genome includes the CUB and that the CUB is low.190 In addition, the codon usage pattern is influenced by two main factors: mutational pressure and natural selection, with the latter having a greater impact on the CUB. Moreover, it has been discovered that other elements, such as dinucleotide composition, hydrophobicity, and aromaticity, affect the diversity in codon usage across the PEDV ORF3 genes.190 Sheikh et al. comprehensively investigated the genetic evolution among the N genes of several CoVs, including PEDV, and found the PEDV genomes have undergone rapid evolution, and the PEDV N gene’s mutation has had a significant impact on evolutionary selection.191
The above results demonstrated that although a single PEDV gene (S, N, or ORF3) and the complete PEDV genome have a similar low CUB, the codon usage pattern of a single gene cannot represent the whole PEDV genomes and vice versa. It is believed that knowledge about the codon usage patterns of the viral gene and the related influencing factors is beneficial for understanding virus evolution.192 Therefore, systematic genome analysis was needed to elucidate PEDV’s evolution mechanism and codon usage pattern. Considering the fatal harm of PEDV to pigs and the important role of the ORF3 gene in virus pathogenicity, as well as the continuous emergence of naturally occurring truncated ORF3 gene in clinically isolated PEDV strains,23,24 it is necessary to track and analyze the codon usage profile of PEDV ORF3 gene in the process of virus evolution. The pertinent findings are valuable for vaccine devolvement strategies because they will offer crucial details on virus evolution, gene transcription, regulation, and protein expression, as well as more assistance in assessing a virus’s host adaptation and evolution.191
The perspective of ORF3 biology to the field of coronaviruses in general
Given the various biological properties of ORF3 accessory protein described above, we believe that ORF3’s biology plays a potential role in the pathogenesis of PEDV, and they are ubiquitous among coronavirus accessory proteins, which are important “catalysts” for studying the pathogenesis of the coronavirus family. For example, our previous study showed that in the process of PEDV rescue by reverse genetics, deletion of the ORF3 gene and replacement with the heterologous green fluorescent protein gene (GFP) resulted in the successful rescue of PEDV and proliferation of the offspring virus on cells,35,58,193 suggesting that the accessory proteins in the coronavirus genome can be deleted or replaced with a heterologous reporter gene, which indicates that it is of great importance for the development of current vaccines for SARS-CoV-2 and other CoVs, as well as for the study of their pathogenesis. Indeed, this phenomenon has been confirmed in the current development of the live-attenuated SARS-CoV-2 vaccine and other coronavirus vaccines.43,49,193,194,195,196,197 In our recent study, we also found that the PEDV accessory protein is a Golgi resident protein, and the two functional motifs (i.e., YXXΦ and ExD) play an important role in its retension.58 Furthermore, we found that protein membrane topology, intrinsically disordered regions, functional motifs, post-translational modification reactions (especially glycosylation) and codon usage patterns that affect the biological properties of the accessory proteins are also involved in the Golgi retention mechanism of ORF3 protein. Given that the ORF3 protein shares many similarities regarding the genomic structure and is functionally analogous to those of human and other animal coronavirus accessory proteins, we speculate that some biological properties of ORF3 protein are universal to other coronavirus accessory proteins, as it has been demonstrated that the accessory protein in PEDV and SARS-CoV-2 genome can be deleted and replaced with heterologous GFP and this manipulation does not affect virus recovery and vaccine development application.49,194,196 However, many aspects have not yet been reported in studies investigating the function and properties of human and other animal coronavirus accessory proteins. From this perspective, the PEDV ORF3 biology study may open up a new avenue for decoding of other coronavirus accessory proteins’ biological function. Therefore, through this study, we call for virologists to strengthen cooperation in this regard in the future worldwide, further confirm and extend the accessory proteins’ biological characteristics to the general coronavirus field.
Concluding remarks and future perspectives
The coronavirus has been circulating in the world of veterinary medicine for a long time. Given the cross-species transmission potential of coronavirus, the continuous prevalence of coronavirus in animals poses a serious threat to human health. Therefore, to some extent, a consensus of One Health that emphasizes the relationship between humans, animals, and the environment and recognizes that human health and well-being are strongly influenced by the health of animals and their environment,198 eventually appears as an important concept and is acknowledged worldwide.
Coronavirus accessory proteins are one of the important factors affecting virus virulence, participating in multiple viral infection processes and contributing to pathogenicity. Some of them are thought to play an important role in reshaping the viral virulence and immune surveillance,199 but how they influence those viruses’ biological behaviors remains obscure. In the case of PEDV accessory protein, ORF3, although a recent mini study summarized its ability to modulate host responses, the role in viral replication, the contribution to virus pathogenicity, and host-virus interaction,25 there are still many covered spots that have not been stressed in this work, especially it failed to elaborate the biological characteristics of ORF3 protein, which is of great importance for the functional execution and viral pathogenesis. In this situation, here we focus on the research progress on unique characteristics of ORF3 accessory protein, that are important for them to play the biological role and emphasize several aspects that need to be elucidated extensively and urgently, for the purpose of reminding the virological community to pay more attention to the related research on all CoVs accessory proteins among the four genera including PEDV’s, especially the investigation on the biological characteristics of the proteins.
In recent years, although the research on PEDV ORF3 protein has made some progress, it is still limited compared with the research progress on structural proteins, and researchers still have a poor understanding of ORF3 protein’s biological characteristics. For example, the research on its membrane topology is still in the prediction stage, so far, the predicated membrane topology of ORF3 protein is not experimentally confirmed; even some results related to PEDV replication and its genome manipulation were conflicting among the available literature35,42,55; in addition, whether the functional motifs of ORF3 protein is involved in virus proliferation, viral pathogenicity, host innate immune response and the regulation of virus-induced apoptosis is still mysterious; more importantly, though we proposed the potential retention mechanism of ORF3 protein in Golgi apparatus based on the current research progress (Figure 3), its detailed mechanism needs to be confirmed by thorough experiments; furthermore, continuously genetic evolution monitoring of ORF3 variation in novel PEDV field strains is absolutely needed, as the genetic variation of ORF3 gene is closely related to the pathogenicity and virulence of PEDV; finally yet importantly, because IDR is one of the main regulatory factors driving the occurrence of LLPS,89,105,200 it is necessary to determine whether ORF3 protein undergoes LLPS, and whether LLPS affects the PEDV’s kinetic behavior. In addition to the above-mentioned issues, it is still unknown whether ORF3 protein undergoes PTMs, such as glycosylation, phosphorylation, methylation, ubiquitylation, and palmitoylation, and how they regulate the virulence and pathogenicity of the virus during virus infection. Therefore, the gaps mentioned above should be investigated more deeply, and these posed research blind spots in the virological profiles of ORF3 will guide further studies to reveal its biological functions and benefit its counterparts in human coronavirus.
Limitations of the study
Despite the current study concerning coronavirus accessory protein’s biology is investigated in great detail and discussed in various aspects, there are still some limitations that remain. First, given that this work takes the PEDV ORF3 protein as an example to emphasize the important role of coronavirus accessory protein biology in viral behavior and pathogenesis, it is unknown whether our conclusions would be applicable to a larger number of other accessory proteins located at the 3’-proximal genomic regions; in addition, although our published data and preliminary experimental results suggest the crucial role of accessory protein biology in the coronavirus life cycle, it is still necessary to continue studying and confirming the potential mechanisms involved in this process; furthermore, in this study, we focused on the similar biological characteristics of PEDV ORF3 accessory protein and several human coronavirus accessory proteins (e.g., SARS-CoV 3a protein, SARS-CoV-2 3a protein, and HCoV-NL63 ORF3 protein), which all located between the S and E gene loci. However, because most coronavirus genomes encode several accessory proteins located at different locations of the genome, there is an urgent need to elucidate the biological properties of coronavirus accessory proteins distributed beyond the S and E gene loci. These issues are major challenges waiting to be crossed by researchers. By improving our understanding of these proteins’' biology, we can develop more effective treatments and vaccines to control the spread of various emerging and re-emerging coronavirus diseases including COVID-19.
Acknowledgments
We gratefully acknowledge the funding agencies that supported this work. This work was financially supported by the National Natural Science Foundation of China (Grant No. 32072838), Guangdong Basic and Applied Basic Foundation (Grant No. 2022A1515012239), and SAAS Program for Excellent Research Team (Grant No. 2022012). We thank the editors and reviewers, who contributed immensely to improving this publication’s quality. We are grateful to Dr. Li Jiang (Guangzhou University) for generously providing assistance with some of the figures. We also sincerely thank all the authors cited in our review for their contributions to elucidating CoV accessory proteins including PEDV’s and apologize to many investigators whose important contributions we inadvertently failed to cite because of the limited space of this manuscript.
Author contributions
F.S.: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Figures preparation, Formal analysis, Graphic design; S.S.: Funding acquisition, Writing – review and editing; R.Y.: Bibliography information collection, Review and suggestion; Z.L.: Review and suggestion; W.W.: Supervision, Writing – review and editing; C.W.: Supervision, Data interpretation, Review and editing. All authors have read and agreed to the final version of the manuscript.
Declaration of interests
The authors declare that they have no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106280.
Contributor Information
Fusheng Si, Email: mr.fusheng@163.com.
Wenqiang Wei, Email: weiwq168@163.com.
Chao Wu, Email: chaowu@wustl.edu.
Supplemental information
References
- 1.Jung K., Saif L.J., Wang Q. Porcine epidemic diarrhea virus (PEDV): an update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020;286:198045. doi: 10.1016/j.virusres.2020.198045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977;100:243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- 3.Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Gene. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., et al. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 7.Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y.W., Dickerman A.W., Piñeyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4 doi: 10.1128/mBio.00737-13. e00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M., et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam A., Ferdous J., Islam S., Sayeed M.A., Dutta Choudhury S., Saha O., Hassan M.M., Shirin T. Evolutionary dynamics and epidemiology of endemic and emerging coronaviruses in humans, domestic animals, and wildlife. Viruses. 2021;13:1908. doi: 10.3390/v13101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cong Y., Li X., Bai Y., Lv X., Herrler G., Enjuanes L., Zhou X., Qu B., Meng F., Cong C., et al. Porcine aminopeptidase N mediated polarized infection by porcine epidemic diarrhea virus in target cells. Virology. 2015;478:1–8. doi: 10.1016/j.virol.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delmas B., Gelfi J., L'Haridon R., Vogel L.K., Sjöström H., Norén O., Laude H. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B.X., Ge J.W., Li Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology. 2007;365:166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam E., Lee C. Contribution of the porcine aminopeptidase N (CD13) receptor density to porcine epidemic diarrhea virus infection. Vet. Microbiol. 2010;144:41–50. doi: 10.1016/j.vetmic.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirato K., Matsuyama S., Ujike M., Taguchi F. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J. Virol. 2011;85:7872–7880. doi: 10.1128/JVI.00464-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo D., Zhu Q., Zhang H., Sun D. Proteomic analysis of membrane proteins of vero cells: exploration of potential proteins responsible for virus entry. DNA Cell Biol. 2014;33:20–28. doi: 10.1089/dna.2013.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W., Luo R., He Q., van Kuppeveld F.J.M., Rottier P.J.M., Bosch B.-J. Aminopeptidase N is not required for porcine epidemic diarrhea virus cell entry. Virus Res. 2017;235:6–13. doi: 10.1016/j.virusres.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirato K., Maejima M., Islam M.T., Miyazaki A., Kawase M., Matsuyama S., Taguchi F. Porcine aminopeptidase N is not a cellular receptor of porcine epidemic diarrhea virus, but promotes its infectivity via aminopeptidase activity. J. Gen. Virol. 2016;97:2528–2539. doi: 10.1099/jgv.0.000563. [DOI] [PubMed] [Google Scholar]
- 19.Liu D.X., Fung T.S., Chong K.K.-L., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redondo N., Zaldívar-López S., Garrido J.J., Montoya M. SARS-CoV-2 accessory proteins in viral pathogenesis: knowns and unknowns. Front. Immunol. 2021;12:708264. doi: 10.3389/fimmu.2021.708264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comar C.E., Otter C.J., Pfannenstiel J., Doerger E., Renner D.M., Tan L.H., Perlman S., Cohen N.A., Fehr A.R., Weiss S.R. MERS-CoV endoribonuclease and accessory proteins jointly evade host innate immunity during infection of lung and nasal epithelial cells. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2123208119. e2123208119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., Zheng R., Liu S., Disoma C., Du A., Li S., Chen Z., Dong Z., Zhang Y., Li S., et al. Host E3 ligase HUWE1 attenuates the pro-apoptotic activity of the MERS-CoV accessory protein ORF3 by promoting its ubiquitin-dependent degradation. J. Biol. Chem. 2022;298:101584. doi: 10.1016/j.jbc.2022.101584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y.-H., Li H.-X., Chen X.-M., Zhang L.-H., Zhao Y.-Y., Luo A.-F., Yang Y.-R., Zheng L.-L., Chen H.-Y. Genetic characteristics and pathogenicity of a novel porcine epidemic diarrhea virus with a naturally occurring truncated ORF3 gene. Viruses. 2022;14:487. doi: 10.3390/v14030487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y., Su X., Du C., Mo L., Ke P., Wang R., Zhong L., Yang C., Chen Y., Wei Z., et al. Genetic diversity of porcine epidemic diarrhea virus with a naturally occurring truncated ORF3 gene found in guangxi, China. Front. Vet. Sci. 2020;7:435. doi: 10.3389/fvets.2020.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jantraphakorn Y., Viriyakitkosol R., Jongkaewwattana A., Kaewborisuth C. Interaction between PEDV and its hosts: acloser look at the ORF3 accessory protein. Front. Vet. Sci. 2021;8:744276. doi: 10.3389/fvets.2021.744276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S., Yang J., Zhu Z., Zheng H. Porcine epidemic diarrhea virus and the host innate immune response. Pathogens. 2020;9:367. doi: 10.3390/pathogens9050367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan Q., Liu X., Sun Y., Zeng W., Li Y., Zhao F., Wu K., Fan S., Zhao M., Chen J., Yi L. Swine enteric coronavirus: diverse pathogen–host interactions. Int. J. Mol. Sci. 2022;23:3953. doi: 10.3390/ijms23073953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W., van Kuppeveld F.J.M., He Q., Rottier P.J.M., Bosch B.-J. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016;226:117–127. doi: 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X., Zhang H., Zhang Q., Dong J., Liang Y., Huang Y., Liu H.J., Tong D. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013;10:26. doi: 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K., Xie S., Sun B. Viral proteins function as ion channels. Biochim. Biophys. Acta. 2011;1808:510–515. doi: 10.1016/j.bbamem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Haan C.A., Kuo L., Masters P.S., Vennema H., Rottier P.J. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/JVI.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol. J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K., Lu W., Chen J., Xie S., Shi H., Hsu H., Yu W., Xu K., Bian C., Fischer W.B., et al. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586:384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou D., Xu J., Duan X., Xu X., Li P., Cheng L., Zheng L., Li X., Zhang Y., Wang X., et al. Porcine epidemic diarrhea virus ORF3 protein causes endoplasmic reticulum stress to facilitate autophagy. Vet. Microbiol. 2019;235:209–219. doi: 10.1016/j.vetmic.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Si F., Hu X., Wang C., Chen B., Wang R., Dong S., Yu R., Li Z. Porcine epidemic diarrhea virus (PEDV) ORF3 enhances viral proliferation by inhibiting apoptosis of infected cells. Viruses. 2020;12:214. doi: 10.3390/v12020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye S., Li Z., Chen F., Li W., Guo X., Hu H., He Q. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Gene. 2015;51:385–392. doi: 10.1007/s11262-015-1257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortego J., Sola I., Almazán F., Ceriani J.E., Riquelme C., Balasch M., Plana J., Enjuanes L. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology. 2003;308:13–22. doi: 10.1016/s0042-6822(02)00096-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaecher S.R., Touchette E., Schriewer J., Buller R.M., Pekosz A. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J. Virol. 2007;81:11054–11068. doi: 10.1128/JVI.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majumdar P., Niyogi S. ORF3a mutation associated with higher mortality rate in SARS-CoV-2 infection. Epidemiol. Infect. 2020;148:e262. doi: 10.1017/S0950268820002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng L.-P., Gao Y.-T., Ge X.-Y., Zhang Q., Peng C., Yang X.-L., Tan B., Chen J., Chmura A.A., Daszak P., Shi Z.L. Bat severe acute respiratory syndrome-like coronavirus WIV1 encodes an extra accessory protein, ORFX, involved in modulation of the host immune response. J. Virol. 2016;90:6573–6582. doi: 10.1128/JVI.03079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song D.S., Yang J.S., Oh J.S., Han J.H., Park B.K. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3. Vaccine. 2003;21:1833–1842. doi: 10.1016/s0264-410X(03)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beall A., Yount B., Lin C.M., Hou Y., Wang Q., Saif L., Baric R. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. mBio. 2016;7:014511–e1515. doi: 10.1128/mBio.01451-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wongthida P., Liwnaree B., Wanasen N., Narkpuk J., Jongkaewwattana A. The role of ORF3 accessory protein in replication of cell-adapted porcine epidemic diarrhea virus (PEDV) Arch. Virol. 2017;162:2553–2563. doi: 10.1007/s00705-017-3390-5. [DOI] [PubMed] [Google Scholar]
- 44.Park S.J., Moon H.J., Luo Y., Kim H.K., Kim E.M., Yang J.S., Song D.S., Kang B.K., Lee C.S., Park B.K. Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus Gene. 2008;36:95–104. doi: 10.1007/s11262-007-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su Y., Liu Y., Chen Y., Zhao B., Ji P., Xing G., Jiang D., Liu C., Song Y., Wang G., et al. Detection and phylogenetic analysis of porcine epidemic diarrhea virus in central China based on the ORF3 gene and the S1 gene. Virol. J. 2016;13:192. doi: 10.1186/s12985-016-0646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K.L., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. USA. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan C.M., Tsoi H., Chan W.M., Zhai S., Wong C.O., Yao X., Chan W.Y., Tsui S.K.W., Chan H.Y.E. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y., Zhang Z., Li J., Gao Y., Zhou L., Ge X., Han J., Guo X., Yang H. Porcine epidemic diarrhea virus S1 protein is the critical inducer of apoptosis. Virol. J. 2018;15:170. doi: 10.1186/s12985-018-1078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang G., Lee D., Lee C. Development of a next-generation vaccine platform for porcine epidemic diarrhea virus using a reverse genetics system. Viruses. 2022;14:2319. doi: 10.3390/v14112319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaewborisuth C., He Q., Jongkaewwattana A. The accessory protein ORF3 contributes to porcine epidemic diarrhea virus replication by direct binding to the spike protein. Viruses. 2018;10:399. doi: 10.3390/v10080399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Z., Cheng L., Xu J., Li P., Li X., Zou D., Zhang Y., Wang X., Wu X., Shen Y., et al. The accessory protein ORF3 of porcine epidemic diarrhea virus inhibits cellular interleukin-6 and interleukin-8 productions by blocking the nuclear factor-κB p65 activation. Vet. Microbiol. 2020;251:108892. doi: 10.1016/j.vetmic.2020.108892. [DOI] [PubMed] [Google Scholar]
- 52.Kaewborisuth C., Koonpaew S., Srisutthisamphan K., Viriyakitkosol R., Jaru-Ampornpan P., Jongkaewwattana A. PEDV ORF3 independently regulates IκB kinase β-mediated NF-κB and IFN-β promoter activities. Pathogens. 2020;9:376. doi: 10.3390/pathogens9050376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren Y., Wang A., Wu D., Wang C., Huang M., Xiong X., Jin L., Zhou W., Qiu Y., Zhou X. Dual inhibition of innate immunity and apoptosis by human cytomegalovirus protein UL37x1 enables efficient virus replication. Nat. Microbiol. 2022;7:1041–1053. doi: 10.1038/s41564-022-01136-6. [DOI] [PubMed] [Google Scholar]
- 54.Chu H., Hou Y., Yang D., Wen L., Shuai H., Yoon C., Shi J., Chai Y., Yuen T.T.-T., Hu B., et al. Coronaviruses exploit a host cysteine-aspartic protease for replication. Nature. 2022;609:785–792. doi: 10.1038/s41586-022-05148-4. [DOI] [PubMed] [Google Scholar]
- 55.Jengarn J., Wongthida P., Wanasen N., Frantz P.N., Wanitchang A., Jongkaewwattana A. Genetic manipulation of porcine epidemic diarrhoea virus recovered from a full-length infectious cDNA clone. J. Gen. Virol. 2015;96:2206–2218. doi: 10.1099/vir.0.000184. [DOI] [PubMed] [Google Scholar]
- 56.Song D.S., Oh J.S., Kang B.K., Yang J.S., Moon H.J., Yoo H.S., Jang Y.S., Park B.K. Oral efficacy of Vero cell attenuated porcine epidemic diarrhea virus DR13 strain. Res. Vet. Sci. 2007;82:134–140. doi: 10.1016/j.rvsc.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen F., Zhu Y., Wu M., Ku X., Ye S., Li Z., Guo X., He Q. Comparative genomic analysis of classical and variant virulent parental/attenuated strains of porcine epidemic diarrhea virus. Viruses. 2015;7:5525–5538. doi: 10.3390/v7102891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Si F., Chen B., Hu X., Yu R., Dong S., Wang R., Li Z. Porcine epidemic diarrhea virus ORF3 protein is transported through the exocytic pathway. J. Virol. 2020;94:e00808–e00820. doi: 10.1128/JVI.00808-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaewborisuth C., Yingchutrakul Y., Roytrakul S., Jongkaewwattana A. Porcine epidemic diarrhea virus (PEDV) ORF3 interactome reveals inhibition of virus replication by cellular VPS36 protein. Viruses. 2019;11:382. doi: 10.3390/v11040382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin H., Li B., Liu M., Zhou H., He K., Fan H. Nonstructural protein 6 of porcine epidemic diarrhea virus induces autophagy to promote viral replication via the PI3K/Akt/mTOR axis. Vet. Microbiol. 2020;244:108684. doi: 10.1016/j.vetmic.2020.108684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J., Tao D., Chen X., Shen L., Zhu L., Xu B., Liu H., Zhao S., Li X., Liu X., et al. Detection of four porcine enteric coronaviruses using CRISPR-cas12a combined with multiplex reverse transcriptase loop-mediated isothermal amplification assay. Viruses. 2022;14:833. doi: 10.3390/v14040833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 2018;92:016777-17–e1717. doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lujan P., Campelo F. Should I stay or should I go? Golgi membrane spatial organization for protein sorting and retention. Arch. Biochem. Biophys. 2021;707:108921. doi: 10.1016/j.abb.2021.108921. [DOI] [PubMed] [Google Scholar]
- 65.Banfield D.K., Lewis M.J., Rabouille C., Warren G., Pelham H.R. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J. Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schaecher S.R., Diamond M.S., Pekosz A. The transmembrane domain of the severe acute respiratory syndrome coronavirus ORF7b protein is necessary and sufficient for its retention in the Golgi complex. J. Virol. 2008;82:9477–9491. doi: 10.1128/JVI.00784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dukhovny A., Yaffe Y., Shepshelovitch J., Hirschberg K. The length of cargo-protein transmembrane segments drives secretory transport by facilitating cargo concentration in export domains. J. Cell Sci. 2009;122:1759–1767. doi: 10.1242/jcs.039339. [DOI] [PubMed] [Google Scholar]
- 68.Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu J., Shang Y., Zhang M. Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat. Rev. Neurosci. 2016;17:209–223. doi: 10.1038/nrn.2016.18. [DOI] [PubMed] [Google Scholar]
- 70.Feng Z., Zeng M., Chen X., Zhang M. Neuronal synapses: microscale signal processing machineries formed by phase separation? Biochemistry (Mosc.) 2018;57:2530–2539. doi: 10.1021/acs.biochem.8b00313. [DOI] [PubMed] [Google Scholar]
- 71.Ouyang M., Li X., Zhang J., Feng P., Pu H., Kong L., Bai Z., Rong L., Xu X., Chi W., et al. Liquid-liquid phase transition drives intra-chloroplast cargo sorting. Cell. 2020;180:1144–1159.e20. doi: 10.1016/j.cell.2020.02.045. [DOI] [PubMed] [Google Scholar]
- 72.Nilsson T., Slusarewicz P., Hoe M.H., Warren G. Kin recognition: a model for the retention of Golgi enzymes. FEBS Lett. 1993;330:1–4. doi: 10.1016/0014-5793(93)80906-b. [DOI] [PubMed] [Google Scholar]
- 73.Khoder-Agha F., Harrus D., Brysbaert G., Lensink M.F., Harduin-Lepers A., Glumoff T., Kellokumpu S. Assembly of B4GALT1/ST6GAL1 heteromers in the Golgi membranes involves lateral interactions via highly charged surface domains. J. Biol. Chem. 2019;294:14383–14393. doi: 10.1074/jbc.RA119.009539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nilsson T., Hoe M.H., Slusarewicz P., Rabouille C., Watson R., Hunte F., Watzele G., Berger E.G., Warren G. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 1994;13:562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dokholyan N.V., Li L., Ding F., Shakhnovich E.I. Topological determinants of protein folding. Proc. Natl. Acad. Sci. USA. 2002;99:8637–8641. doi: 10.1073/pnas.122076099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuan X., Yao Z., Wu J., Zhou Y., Shan Y., Dong B., Zhao Z., Hua P., Chen J., Cong Y. G1 phase cell cycle arrest induced by SARS-CoV 3a protein via the cyclin D3/pRb pathway. Am. J. Respir. Cell Mol. Biol. 2007;37:9–19. doi: 10.1165/rcmb.2005-0345RC. [DOI] [PubMed] [Google Scholar]
- 77.Tan Y.J., Teng E., Shen S., Tan T.H.P., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi N., Ye S., Alam A., Chen L., Jiang Y. Atomic structure of a Na+- and K+-conducting channel. Nature. 2006;440:570–574. doi: 10.1038/nature04508. [DOI] [PubMed] [Google Scholar]
- 80.Kern D.M., Sorum B., Mali S.S., Hoel C.M., Sridharan S., Remis J.P., Toso D.B., Kotecha A., Bautista D.M., Brohawn S.G. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021;28:573–582. doi: 10.1038/s41594-021-00619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Müller M.A., Van Der Hoek L., Voss D., Bader O., Lehmann D., Schulz A.R., Kallies S., Suliman T., Fielding B.C., Drosten C., Niedrig M. Human coronavirus NL63 open reading frame 3 encodes a virion-incorporated N-glycosylated membrane protein. Virol. J. 2010;7:6–12. doi: 10.1186/1743-422X-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y., Jin Y., Kuang L., Luo Z., Li F., Sun J., Zhu A., Zhuang Z., Wang Y., Wen L., et al. The N-terminal region of Middle East respiratory syndrome coronavirus accessory protein 8b is essential for enhanced virulence of an attenuated murine coronavirus. J. Virol. 2022;96 doi: 10.1128/JVI.01842-21. e01842–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dunker A.K., Silman I., Uversky V.N., Sussman J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 85.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 86.Mishra P.M., Verma N.C., Rao C., Uversky V.N., Nandi C.K. Intrinsically disordered proteins of viruses: involvement in the mechanism of cell regulation and pathogenesis. Prog. Mol. Biol. Transl. Sci. 2020;174:1–78. doi: 10.1016/bs.pmbts.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bondos S.E., Dunker A.K., Uversky V.N. Intrinsically disordered proteins play diverse roles in cell signaling. Cell Commun. Signal. 2022;20:20. doi: 10.1186/s12964-022-00821-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wei W., Bai L., Yan B., Meng W., Wang H., Zhai J., Si F., Zheng C. When liquid-liquid phase separation meets viral infections. Front. Immunol. 2022;13:985622. doi: 10.3389/fimmu.2022.985622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang Z., Liu H., Nix J., Xu R., Knoverek C.R., Bowman G.R., Amarasinghe G.K., Sibley L.D. The intrinsically disordered protein TgIST from Toxoplasma gondii inhibits STAT1 signaling by blocking cofactor recruitment. Nat. Commun. 2022;13:4047. doi: 10.1038/s41467-022-31720-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oldfield C.J., Uversky V.N., Dunker A.K., Kurgan L. In: Intrinsically Disordered Proteins. Salvi N., editor. Academic Press; 2019. Chapter 1 - introduction to intrinsically disordered proteins and regions; pp. 1–34. [DOI] [Google Scholar]
- 91.Van Der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T., et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Charon J., Barra A., Walter J., Millot P., Hébrard E., Moury B., Michon T. First experimental assessment of protein intrinsic disorder involvement in an RNA virus natural adaptive process. Mol. Biol. Evol. 2018;35:38–49. doi: 10.1093/molbev/msx249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Giri R., Kumar D., Sharma N., Uversky V.N. Intrinsically disordered side of the Zika virus proteome. Front. Cell. Infect. Microbiol. 2016;6:144. doi: 10.3389/fcimb.2016.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 95.Wisniewski K.E., Dalton A.J., McLachlan C., Wen G.Y., Wisniewski H.M. Alzheimer's disease in Down's syndrome: clinicopathologic studies. Neurology. 1985;35:957–961. doi: 10.1212/wnl.35.7.957. [DOI] [PubMed] [Google Scholar]
- 96.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12:1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mozzi A., Forni D., Cagliani R., Clerici M., Pozzoli U., Sironi M. Intrinsically disordered regions are abundant in simplexvirus proteomes and display signatures of positive selection. Virus Evol. 2020;6:veaa028. doi: 10.1093/ve/veaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tenchov R., Zhou Q.A. Intrinsically disordered proteins: perspective on COVID-19 infection and drug discovery. ACS Infect. Dis. 2022;8:422–432. doi: 10.1021/acsinfecdis.2c00031. [DOI] [PubMed] [Google Scholar]
- 99.Kakisaka M., Yamada K., Yamaji-Hasegawa A., Kobayashi T., Aida Y. Intrinsically disordered region of influenza A NP regulates viral genome packaging via interactions with viral RNA and host PI (4, 5) P2. Virology. 2016;496:116–126. doi: 10.1016/j.virol.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 100.Dolan P.T., Roth A.P., Xue B., Sun R., Dunker A.K., Uversky V.N., LaCount D.J. Intrinsic disorder mediates hepatitis C virus core–host cell protein interactions. Protein Sci. 2015;24:221–235. doi: 10.1002/pro.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pereira N., Cardone C., Lassoued S., Galloux M., Fix J., Assrir N., Lescop E., Bontems F., Eléouët J.F., Sizun C. New insights into structural disorder in human respiratory syncytial virus phosphoprotein and implications for binding of protein partners. J. Biol. Chem. 2017;292:2120–2131. doi: 10.1074/jbc.M116.765958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Uversky V.N., Redwan E.M., Aljadawi A.A. Protein intrinsic disorder and evolvability of MERS-CoV. Biomolecules. 2021;11:608. doi: 10.3390/biom11040608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roden C.A., Dai Y., Giannetti C.A., Seim I., Lee M., Sealfon R., McLaughlin G.A., Boerneke M.A., Iserman C., Wey S.A., et al. Double-stranded RNA drives SARS-CoV-2 nucleocapsid protein to undergo phase separation at specific temperatures. Nucleic Acids Res. 2022;50:8168–8192. doi: 10.1093/nar/gkac596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu C., Qavi A.J., Hachim A., Kavian N., Cole A.R., Moyle A.B., Wagner N.D., Sweeney-Gibbons J., Rohrs H.W., Gross M.L., et al. Characterization of SARS-CoV-2 nucleocapsid protein reveals multiple functional consequences of the C-terminal domain. iScience. 2021;24:102681. doi: 10.1016/j.isci.2021.102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bessa L.M., Guseva S., Camacho-Zarco A.R., Salvi N., Maurin D., Perez L.M., Botova M., Malki A., Nanao M., Jensen M.R., et al. The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a. Sci. Adv. 2022;8:eabm4034. doi: 10.1126/sciadv.abm4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kumar S., Thambiraja T.S., Karuppanan K., Subramaniam G. Omicron and Delta variant of SARS-CoV-2: a comparative computational study of spike protein. J. Med. Virol. 2022;94:1641–1649. doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- 107.York A. Targeting viral liquid–liquid phase separation. Nat. Rev. Microbiol. 2021;19:550. doi: 10.1038/s41579-021-00608-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu C., Holehouse A.S., Leung D.W., Amarasinghe G.K., Dutch R.E. Liquid phase Partitioning in virus replication: observations and opportunities. Annu. Rev. Virol. 2022;9:285–306. doi: 10.1146/annurev-virology-093020-013659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li H., Ernst C., Kolonko-Adamska M., Greb-Markiewicz B., Man J., Parissi V., Ng B.W.-L. Phase separation in viral infections. Trends Microbiol. 2022;30:1217–1231. doi: 10.1016/j.tim.2022.06.005. [DOI] [PubMed] [Google Scholar]
- 110.Yang P., Mathieu C., Kolaitis R.-M., Zhang P., Messing J., Yurtsever U., Yang Z., Wu J., Li Y., Pan Q., et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181:325–345.e28. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Helenius A., Aebi M., Markus Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 112.Moremen K.W., Tiemeyer M., Nairn A.V. Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Esmail S., Manolson M.F. Advances in understanding N-glycosylation structure, function, and regulation in health and disease. Eur. J. Cell Biol. 2021;100:151186. doi: 10.1016/j.ejcb.2021.151186. [DOI] [PubMed] [Google Scholar]
- 114.Watanabe Y., Bowden T.A., Wilson I.A., Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta. Gen. Subj. 2019;1863:1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reily C., Stewart T.J., Renfrow M.B., Novak J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019;15:346–366. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dang K., Jiang S., Gao Y., Qian A. The role of protein glycosylation in muscle diseases. Mol. Biol. Rep. 2022;49:8037–8049. doi: 10.1007/s11033-022-07334-z. [DOI] [PubMed] [Google Scholar]
- 117.Feng T., Zhang J., Chen Z., Pan W., Chen Z., Yan Y., Dai J. Glycosylation of viral proteins: implication in virus–host interaction and virulence. Virulence. 2022;13:670–683. doi: 10.1080/21505594.2022.2060464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nurdin J.A., Kotaki T., Kawagishi T., Sato S., Yamasaki M., Nouda R., Minami S., Kanai Y., Kobayashi T. N-glycosylation of rotavirus NSP4 protein affects viral replication and pathogenesis. J. Virol. 2023;97 doi: 10.1128/jvi.01861-22. e01861–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kobayashi Y., Suzuki Y. Evidence for N-glycan shielding of antigenic sites during evolution of human influenza A virus hemagglutinin. J. Virol. 2012;86:3446–3451. doi: 10.1128/JVI.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mondotte J.A., Lozach P.-Y., Amara A., Gamarnik A.V. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J. Virol. 2007;81:7136–7148. doi: 10.1128/JVI.00116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cao L., Pauthner M., Andrabi R., Rantalainen K., Berndsen Z., Diedrich J.K., Menis S., Sok D., Bastidas R., Park S.-K.R., et al. Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat. Commun. 2018;9:3693. doi: 10.1038/s41467-018-06121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fontes-Garfias C.R., Shan C., Luo H., Muruato A.E., Medeiros D.B.A., Mays E., Xie X., Zou J., Roundy C.M., Wakamiya M., et al. Functional analysis of glycosylation of Zika virus envelope protein. Cell Rep. 2017;21:1180–1190. doi: 10.1016/j.celrep.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Carbaugh D.L., Baric R.S., Lazear H.M. Envelope protein glycosylation mediates Zika virus pathogenesis. J. Virol. 2019;93:e00113–e00119. doi: 10.1128/JVI.00113-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guo Y., Bao L., Xu Y., Li F., Lv Q., Fan F., Qin C. The ablation of envelope protein glycosylation enhances the neurovirulence of ZIKV and cell apoptosis in newborn mice. J. Immunol. Res. 2021;2021:5317662. doi: 10.1155/2021/5317662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim J.-M., Yun S.-I., Song B.-H., Hahn Y.-S., Lee C.-H., Oh H.-W., Lee Y.-M. A single N-linked glycosylation site in the Japanese encephalitis virus prM protein is critical for cell type-specific prM protein biogenesis, virus particle release, and pathogenicity in mice. J. Virol. 2008;82:7846–7862. doi: 10.1128/JVI.00789-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hanna S.L., Pierson T.C., Sanchez M.D., Ahmed A.A., Murtadha M.M., Doms R.W. N-linked glycosylation of west nile virus envelope proteins influences particle assembly and infectivity. J. Virol. 2005;79:13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhu W., Banadyga L., Emeterio K., Wong G., Qiu X. The roles of ebola virus soluble glycoprotein in replication, pathogenesis, and countermeasure development. Viruses. 2019;11:999. doi: 10.3390/v11110999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Carbaugh D.L., Lazear H.M. Flavivirus envelope protein glycosylation: impacts on viral infection and pathogenesis. J. Virol. 2020;94:e00104–e00120. doi: 10.1128/JVI.00104-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sanda M., Morrison L., Goldman R. N-and O-glycosylation of the SARS-CoV-2 spike protein. Anal. Chem. 2021;93:2003–2009. doi: 10.1021/acs.analchem.0c03173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duart G., García-Murria M.J., Grau B., Acosta-Cáceres J.M., Martínez-Gil L., Mingarro I. SARS-CoV-2 envelope protein topology in eukaryotic membranes. Open Biol. 2020;10:200209. doi: 10.1098/rsob.200209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Reis C.A., Tauber R., Blanchard V. Glycosylation is a key in SARS-CoV-2 infection. J. Mol. Med. 2021;99:1023–1031. doi: 10.1007/s00109-021-02092-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chawla H., Fadda E., Crispin M. Principles of SARS-CoV-2 glycosylation. Curr. Opin. Struct. Biol. 2022;75:102402. doi: 10.1016/j.sbi.2022.102402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kirchdoerfer R.N., Bhandari M., Martini O., Sewall L.M., Bangaru S., Yoon K.-J., Ward A.B. Structure and immune recognition of the porcine epidemic diarrhea virus spike protein. Structure. 2021;29:385–392.e5. doi: 10.1016/j.str.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liang J.Q., Fang S., Yuan Q., Huang M., Chen R.A., Fung T.S., Liu D.X. N-Linked glycosylation of the membrane protein ectodomain regulates infectious bronchitis virus-induced ER stress response, apoptosis and pathogenesis. Virology. 2019;531:48–56. doi: 10.1016/j.virol.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Voss D., Pfefferle S., Drosten C., Stevermann L., Traggiai E., Lanzavecchia A., Becker S. Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein. Virol. J. 2009;6:79. doi: 10.1186/1743-422X-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Oostra M., de Haan C.A.M., de Groot R.J., Rottier P.J.M. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 2006;80:2326–2336. doi: 10.1128/JVI.80.5.2326-2336.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Florek D., Ehmann R., Kristen-Burmann C., Lemmermeyer T., Lochnit G., Ziebuhr J., Thiel H.J., Tekes G. Identification and characterization of a Golgi retention signal in feline coronavirus accessory protein 7b. J. Gen. Virol. 2017;98:2017–2029. doi: 10.1099/jgv.0.000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Matsuoka K., Imahashi N., Ohno M., Ode H., Nakata Y., Kubota M., Sugimoto A., Imahashi M., Yokomaku Y., Iwatani Y. SARS-CoV-2 accessory protein ORF8 is secreted extracellularly as a glycoprotein homodimer. J. Biol. Chem. 2022;298:101724. doi: 10.1016/j.jbc.2022.101724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Watanabe Y., Berndsen Z.T., Raghwani J., Seabright G.E., Allen J.D., Pybus O.G., McLellan J.S., Wilson I.A., Bowden T.A., Ward A.B., Crispin M. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020;11:2688. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Huang H.-C., Lai Y.-J., Liao C.-C., Yang W.-F., Huang K.-B., Lee I.-J., Chou W.-C., Wang S.-H., Wang L.-H., Hsu J.-M., et al. Targeting conserved N-glycosylation blocks SARS-CoV-2 variant infection in vitro. EBioMedicine. 2021;74:103712. doi: 10.1016/j.ebiom.2021.103712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Casas-Sanchez A., Romero-Ramirez A., Hargreaves E., Ellis C.C., Grajeda B.I., Estevao I.L., Patterson E.I., Hughes G.L., Almeida I.C., Zech T., Acosta-Serrano Á. Inhibition of protein N-glycosylation blocks SARS-CoV-2 infection. mBio. 2021;13 doi: 10.1128/mbio.03718-21. e0371821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huang Y.-J., Zhao H., Huang X., Deng Y.-Q., Li X.-F., Ye Q., Li R.-T., Xu Y.-P., Cao T.-S., Qin C.-F. Identification of oligosaccharyltransferase as a host target for inhibition of SARS-CoV-2 and its variants. Cell Discov. 2021;7:116. doi: 10.1038/s41421-021-00354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Braunger K., Pfeffer S., Shrimal S., Gilmore R., Berninghausen O., Mandon E.C., Becker T., Förster F., Beckmann R. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science. 2018;360:215–219. doi: 10.1126/science.aar7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994;68:6523–6534. doi: 10.1128/JVI.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang R., Wang K., Lv W., Yu W., Xie S., Xu K., Schwarz W., Xiong S., Sun B. The ORF4a protein of human coronavirus 229E functions as a viroporin that regulates viral production. Biochim. Biophys. Acta. 2014;1838:1088–1095. doi: 10.1016/j.bbamem.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Minakshi R., Padhan K. The YXXΦ motif within the severe acute respiratory syndrome coronavirus (SARS-CoV) 3a protein is crucial for its intracellular transport. Virol. J. 2014;11:75. doi: 10.1186/1743-422X-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yuan X., Li J., Shan Y., Yang Z., Zhao Z., Chen B., Yao Z., Dong B., Wang S., Chen J., Cong Y. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109:191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Park S.J., Kim H.K., Song D.S., An D.J., Park B.K. Complete genome sequences of a Korean virulent porcine epidemic diarrhea virus and its attenuated counterpart. J. Virol. 2012;86:5964. doi: 10.1128/JVI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bridgen A., Duarte M., Tobler K., Laude H., Ackermann M. Sequence determination of the nucleocapsid protein gene of the porcine epidemic diarrhoea virus confirms that this virus is a coronavirus related to human coronavirus 229E and porcine transmissible gastroenteritis virus. J. Gen. Virol. 1993;74:1795–1804. doi: 10.1099/0022-1317-74-9-1795. [DOI] [PubMed] [Google Scholar]
- 151.Zhang J., Yim-Im W., Chen Q., Zheng Y., Schumacher L., Huang H., Gauger P., Harmon K., Li G. Identification of porcine epidemic diarrhea virus variant with a large spike gene deletion from a clinical swine sample in the United States. Virus Gene. 2018;54:323–327. doi: 10.1007/s11262-018-1542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hou Y., Lin C.M., Yokoyama M., Yount B.L., Marthaler D., Douglas A.L., Ghimire S., Qin Y., Baric R.S., Saif L.J., Wang Q. Deletion of a 197-amino-acid region in the N-terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J. Virol. 2017;91:002277-17–e317. doi: 10.1128/JVI.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wei Z.Y., Lu W.H., Li Z.L., Mo J.Y., Zeng X.D., Zeng Z.L., Sun B.L., Chen F., Xie Q.M., Bee Y.Z., Ma J.Y. Complete genome sequence of novel porcine epidemic diarrhea virus strain GD-1 in China. J. Virol. 2012;86:13824–13825. doi: 10.1128/JVI.02615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang E., Guo D., Li C., Wei S., Wang Z., Liu Q., Zhang B., Kong F., Feng L., Sun D. Molecular characterization of the ORF3 and S1 genes of porcine epidemic diarrhea virus non S-INDELstrains in seven regions of China. PLoS One. 2016;11:e0160561. doi: 10.1371/journal.pone.0160561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Olson J.K., Grose C. Endocytosis and recycling of varicella-zoster virus Fc receptor glycoprotein gE: internalization mediated by a YXXL motif in the cytoplasmic tail. J. Virol. 1997;71:4042–4054. doi: 10.1128/JVI.71.5.4042-4054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rose J.K., Adams G.A., Gallione C.J. The presence of cysteine in the cytoplasmic domain of the vesicular stomatitis virus glycoprotein is required for palmitate addition. Proc. Natl. Acad. Sci. USA. 1984;81:2050–2054. doi: 10.1073/pnas.81.7.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chen R., Jiang X., Sun D., Han G., Wang F., Ye M., Wang L., Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. 2009;8:651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- 159.Ference B.A., Kastelein J.J.P., Ray K.K., Ginsberg H.N., Chapman M.J., Packard C.J., Laufs U., Oliver-Williams C., Wood A.M., Butterworth A.S., et al. Association of triglyceride-lowering LPL variants and LDL-C–lowering LDLR variants with risk of coronary heart disease. LAMA. 2019;321:364–373. doi: 10.1001/jama.2018.20045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Spiess M., Lodish H.F. An internal signal sequence: the asialoglycoprotein receptor membrane anchor. Cell. 1986;44:177–185. doi: 10.1016/0092-8674(86)90496-4. [DOI] [PubMed] [Google Scholar]
- 161.Haks M.C., Krimpenfort P., Borst J., Kruisbeek A.M. The CD3γ chain is essential for development of both the TCRαβ and TCRγδ lineages. EMBO J. 1998;17:1871–1882. doi: 10.1093/emboj/17.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Van Den Elsen P., Georgopoulos K., Shepley B.-A., Orkin S., Terhorst C. Exon/intron organization of the genes coding for the delta chains of the human and murine T-cell receptor/T3 complex. Proc. Natl. Acad. Sci. USA. 1986;83:2944–2948. doi: 10.1073/pnas.83.9.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wollscheid B., Bausch-Fluck D., Henderson C., O'brien R., Bibel M., Schiess R., Aebersold R., Watts J.D. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Abe H., Morimatsu M., Nikami H., Miyashige T., Saito M. Molecular cloning and mRNA expression of the bovine insulin-responsive glucose transporter (GLUT4) J. Anim. Sci. 1997;75:182–188. doi: 10.2527/1997.751182x. [DOI] [PubMed] [Google Scholar]
- 165.Bonifacino J.S., Traub L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 166.Marks M.S., Ohno H., Kirchnausen T., Bonracino J.S. Protein sorting by tyrosine-based signals: adapting to the Ys and wherefores. Trends Cell Biol. 1997;7:124–128. doi: 10.1016/S0962-8924(96)10057-X. [DOI] [PubMed] [Google Scholar]
- 167.Trowbridge I.S., Collawn J.F., Hopkins C.R. Signal-Dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 168.Nishimura N., Balch W.E. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 1997;277:556–558. doi: 10.1126/science.277.5325.556. [DOI] [PubMed] [Google Scholar]
- 169.Bannykh S.I., Nishimura N., Balch W.E. Getting into the Golgi. Trends Cell Biol. 1998;8:21–25. doi: 10.1016/s0962-8924(97)01184-7. [DOI] [PubMed] [Google Scholar]
- 170.Law P.T.W., Wong C.-H., Au T.C.C., Chuck C.-P., Kong S.-K., Chan P.K.S., To K.-F., Lo A.W.I., Chan J.Y.W., Suen Y.-K., et al. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- 171.Kumar N., Kaushik R., Tennakoon C., Uversky V.N., Longhi S., Zhang K.Y.J., Bhatia S. Insights into the evolutionary forces that shape the codon usage in the viral genome segments encoding intrinsically disordered protein regions. Brief. Bioinform. 2021;22:bbab145. doi: 10.1093/bib/bbab145. [DOI] [PubMed] [Google Scholar]
- 172.Arella D., Dilucca M., Giansanti A. Codon usage bias and environmental adaptation in microbial organisms. Mol. Genet. Genomics. 2021;296:751–762. doi: 10.1007/s00438-021-01771-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mordstein C., Cano L., Morales A.C., Young B., Ho A.T., Rice A.M., Liss M., Hurst L.D., Kudla G. Transcription, mRNA export and immune evasion shape the codon usage of viruses. Genome Biol. Evol. 2021;13:evab106. doi: 10.1093/gbe/evab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Qian W., Yang J.-R., Pearson N.M., Maclean C., Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bai H., Ata G., Sun Q., Rahman S.U., Tao S. Natural selection pressure exerted on “Silent” mutations during the evolution of SARS-CoV-2: evidence from codon usage and RNA structure. Virus Res. 2022;323:198966. doi: 10.1016/j.virusres.2022.198966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jiang L., Zhang Q., Xiao S., Si F. Deep decoding of codon usage strategies and host adaption preferences of soybean mosaic virus. Int. J. Biol. Macromol. 2022;222:803–817. doi: 10.1016/j.ijbiomac.2022.09.179. [DOI] [PubMed] [Google Scholar]
- 177.He Z., Qin L., Xu X., Ding S. Evolution and host adaptability of plant RNA viruses: research insights on compositional biases. Comput. Struct. Biotechnol. J. 2022;20:2600–2610. doi: 10.1016/j.csbj.2022.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Peng Q., Zhang X., Li J., He W., Fan B., Ni Y., Liu M., Li B. Comprehensive analysis of codon usage patterns of porcine deltacoronavirus and its host adaptability. Transbound. Emerg. Dis. 2022;69:e2443–e2455. doi: 10.1111/tbed.14588. [DOI] [PubMed] [Google Scholar]
- 179.Rahman S.U., Nawaz S., Ullah S., Rahman I.U., Haq M.I.U., Khan M.A., Al-Ghamdi A.A., Al-Hemaid F.M., Elshikh M.S., Aljowaie R.M., Eltayb W.A. Study of codon usage patterns and influencing factors in rice yellow mottle virus based on coding sequence data. Agronomy. 2022;12:1990. doi: 10.3390/agronomy12091990. [DOI] [Google Scholar]
- 180.Yao X., Fan Q., Yao B., Lu P., Rahman S.U., Chen D., Tao S. Codon usage bias analysis of bluetongue virus causing livestock infection. Front. Microbiol. 2020;11:655. doi: 10.3389/fmicb.2020.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Luo W., Roy A., Guo F., Irwin D.M., Shen X., Pan J., Shen Y. Host adaptation and evolutionary analysis of Zaire ebolavirus: insights from codon usage based investigations. Front. Microbiol. 2020;11:570131. doi: 10.3389/fmicb.2020.570131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Khandia R., Singhal S., Kumar U., Ansari A., Tiwari R., Dhama K., Das J., Munjal A., Singh R.K. Analysis of Nipah virus codon usage and adaptation to hosts. Front. Microbiol. 2019;10:886. doi: 10.3389/fmicb.2019.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Si F., Jiang L., Yu R., Wei W., Li Z. Study on the characteristic codon usage pattern in porcine epidemic diarrhea virus genomes and its host adaptation phenotype. Front. Microbiol. 2021;12:738082. doi: 10.3389/fmicb.2021.738082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lauring A.S., Acevedo A., Cooper S.B., Andino R. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe. 2012;12:623–632. doi: 10.1016/j.chom.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zang M., He W., Du F., Wu G., Wu B., Zhou Z. Analysis of the codon usage of the ORF2 gene of feline calicivirus. Infect. Genet. Evol. 2017;54:54–59. doi: 10.1016/j.meegid.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li Z., Ma Z., Li Y., Gao S., Xiao S. Porcine epidemic diarrhea virus: molecular mechanisms of attenuation and vaccines. Microb. Pathog. 2020;149:104553. doi: 10.1016/j.micpath.2020.104553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen Y., Shi Y., Deng H., Gu T., Xu J., Ou J., Jiang Z., Jiao Y., Zou T., Wang C. Characterization of the porcine epidemic diarrhea virus codon usage bias. Infect. Genet. Evol. 2014;28:95–100. doi: 10.1016/j.meegid.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yu X., Liu J., Li H., Liu B., Zhao B., Ning Z. Comprehensive analysis of synonymous codon usage patterns and influencing factors of porcine epidemic diarrhea virus. Arch. Virol. 2021;166:157–165. doi: 10.1007/s00705-020-04857-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cao H., Zhang H., Li D. Synonymous codon usage bias of spike genes of porcine epidemic diarrhea virus. Afr. J. Microbiol. Res. 2011;5:3784–3789. http://www.academicjournals.org/ajmr [Google Scholar]
- 190.Xu X., Li P., Zhang Y., Wang X., Xu J., Wu X., Shen Y., Guo D., Li Y., Yao L., et al. Comprehensive analysis of synonymous codon usage patterns in orf3 gene of porcine epidemic diarrhea virus in China. Res. Vet. Sci. 2019;127:42–46. doi: 10.1016/j.rvsc.2019.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sheikh A., Al-Taher A., Al-Nazawi M., Al-Mubarak A.I., Kandeel M. Analysis of preferred codon usage in the coronavirus N genes and their implications for genome evolution and vaccine design. J. Virol. Methods. 2020;277:113806. doi: 10.1016/j.jviromet.2019.113806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wong E.H.M., Smith D.K., Rabadan R., Peiris M., Poon L.L.M. Codon usage bias and the evolution of influenza A viruses. Codon usage biases of influenza virus. BMC Evol. Biol. 2010;10:253. doi: 10.1186/1471-2148-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li C., Li Z., Zou Y., Wicht O., van Kuppeveld F.J.M., Rottier P.J.M., Bosch B.J. Manipulation of the porcine epidemic diarrhea virus genome using targeted RNA recombination. PLoS One. 2013;8:e69997. doi: 10.1371/journal.pone.0069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Liu Y., Zhang X., Liu J., Xia H., Zou J., Muruato A.E., Periasamy S., Kurhade C., Plante J.A., Bopp N.E., et al. A live-attenuated SARS-CoV-2 vaccine candidate with accessory protein deletions. Nat. Commun. 2022;13:4337. doi: 10.1038/s41467-022-31930-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Thi Nhu Thao T., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A., et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020;582:561–565. doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- 196.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., Kato T., Lee R.E., Yount B.L., Mascenik T.M., et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Donaldson E.F., Yount B., Sims A.C., Burkett S., Pickles R.J., Baric R.S. Systematic assembly of a full-length infectious clone of human coronavirus NL63. J. Virol. 2008;82:11948–11957. doi: 10.1128/JVI.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Poudel U., Subedi D., Pantha S., Dhakal S. Animal coronaviruses and coronavirus disease 2019: lesson for one health approach. Open Vet. J. 2020;10:239–251. doi: 10.4314/ovj.v10i3.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Arshad N., Laurent-Rolle M., Ahmed W.S., Hsu J.C.-C., Mitchell S.M., Pawlak J., Sengupta D., Biswas K.H., Cresswell P. SARS-CoV-2 accessory proteins ORF7a and ORF3a use distinct mechanisms to down-regulate MHC-I surface expression. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2208525120. e2208525120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ainani H., Bouchmaa N., Ben Mrid R., El Fatimy R. Liquid-liquid phase separation of protein tau: an emerging process in Alzheimer's disease pathogenesis. Neurobiol. Dis. 2023;178:106011. doi: 10.1016/j.nbd.2023.106011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.