Abstract
By combining docking and molecular dynamics simulations, we explored a library of 65 mostly axially chiral naphthylisoquinoline alkaloids and their analogues, with most different molecular architectures and structural analogues, for their activity against SARS-CoV-2. Although natural biaryls are often regarded without consideration of their axial chirality, they can bind to protein targets in an atroposelective manner. By combining docking results with steered molecular dynamics simulations, we identified one alkaloid, korupensamine A, that atropisomer-specifically inhibited the main protease (Mpro) activity of SARS-CoV-2 significantly in comparison to the reference covalent inhibitor GC376 (IC50 = 2.52 ± 0.14 and 0.88 ± 0.15 μM, respectively) and reduced viral growth by five orders of magnitude in vitro (EC50 = 4.23 ± 1.31 μM). To investigate the binding pathway and mode of interaction of korupensamine A within the active site of the protease, we utilized Gaussian accelerated molecular dynamics simulations, which reproduced the docking pose of korupensamine A inside the active site of the enzyme. The study presents naphthylisoquinoline alkaloids as a new class of potential anti-COVID-19 agents.
Keywords: Alkaloids, Natural products, Naphthylisoquinolines, SARS-CoV-2, Mpro, Steered molecular dynamics
Graphical abstract
1. Introduction
Since the first identified cases of the greatly contagious coronavirus disease 2019 (COVID-19) in December 2019, the world has been facing a critical fight against this newly discovered disease, with more than 622 million diagnosed cases and more than 6.5 million deaths up till November 2022 [1,2]. The COVID-19 pandemic is caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [3], triggering an acute disease that can eventually result in respiratory failure and death [4]. The SARS-CoV-2 infection has a high transmission rate but a relatively low death rate (1.0–3.5%), except for older people with other comorbidities. Severe pneumonia is observed in 15–20% of the infected people, and 5–10% of the patients require critical-care services [5,6]. SARS-CoV-2 is characterized by a spherical morphology with spike projections on its surface. The spike glycoprotein of the virus controls its entry into the human host cell [7]. SARS-CoV-2 infects lung type II alveolar cells, which may explain the severe alveolar damage during the infection [8]. Although a number of vaccines against SARS-CoV-2 have been successfully developed and administered [9,10], the world is still lacking an effective antiviral treatment against the disease [11].
Medical drugs currently in use are, so far, remdesivir, an antiviral agent that demonstrated benefits in decreasing the rate and the duration of hospitalization [12,13], and dexamethasone, a broad-spectrum anti-inflammatory drug offering assistance to patients with respiratory failure requiring respiratory support through the inhibition of a cytokine storm [14,15]. Also, nirmatrelvir, an orally active 3C-like protease inhibitor in combination with ritonavir, also known as paxlovid, is an antiviral drug against COVID-19 [16].
SARS-CoV-2 contains several polypeptides that promote proteolytic breakdown to produce 20 additional proteins during their lifecycle; one of them is the main protease (Mpro), which is essential for virus replication [17]. It consists of 306 amino acid residues and resembles the Mpro of SARS-CoV in structure and sequence [11,18,19]. This essential protease is a dimer, and mutations that prevent its conserved dimerization greatly reduce its catalytic activity. There are three domains in each monomer (domains I-III), of which domains I and II form the catalytic binding pocket, and domain III is responsible for enzyme dimerization [20,21]. The functional Mpro must have a conserved HIS-41-CYS-145 catalytic dyad in its active site. As a result, the catalytic activity of the enzyme will degenerate to the point where it is no longer functional if these catalytic residues are altered [22]. Several non-competitive inhibitors have been reported to form covalent bonds with CYS-145 like e.g., nirmatrelvir, ensiltrelvir, narlaprevir, and boceprevir, but there has been much less research into competitive non-covalent inhibitors [23,24]. Therefore, efforts have focused on investigating the main protease of that virus, to discover new specific non-covalent inhibitors against COVID-19 [7,25]. For more details on the discovery of inhibitors of various SARS-CoV-2 proteins, including Mpro, see Refs. [[26], [27], [28]].
A particularly interesting – and stereochemically intriguing – group of natural products are the naphthylisoquinoline (NIQ) alkaloids like korupensamine A (1a, see Fig. 1 ) [29]. They are composed of a naphthalene and an isoquinoline moiety. Following the principle of phenol-oxidative coupling, the two molecular portions are joined together in different ways, always ortho or para to oxygen functions [30,31]. These coupling types determine the molecular shapes of the alkaloids; Fig. 1 shows a selection of differently coupled NIQs, with representatives linked via the 5,8′-positions (like in compounds 1a/1b-4), via 5,1' (NIQs 5, 6, and ent-6), 7,8' (compound 7), or 7,1' (alkaloids 8 and 9). NIQ alkaloids are chiral compounds, characterized by the presence of stereogenic centers and a usually likewise chiral C,C- or N,C-axis between the two molecular moieties, the isoquinoline and the naphthalene portions. Due to the existence of more or less bulky ortho-substituents, most of these alkaloids show the phenomenon of hindered rotation about that central biaryl axis, which – in most cases - leads to rotationally stable atropisomers [[30], [31], [32], [33]]. Atropisomerism as an important structural feature of these alkaloids constitutes a synthetic challenge for their directed, stereoselective production that has been addressed in numerous elegant total syntheses [34,35]. One motivation for these synthetic efforts is the fact that, depending on their individual structures, naphthylisoquinoline alkaloids exhibit pronounced anti-malarial [32,[36], [37], [38], [39]], anti-trypanosomal [30,32,40], anti-leishmanial [31,32,[40], [41], [42], [43]], anti-bacterial [32,44], anti-fungal [32], larvicidal [45], molluscicidal [46,47], insecticidal [48,49], antibabesial [50], or cytotoxic activities against leukemia [51] and various cancer cell lines [[52], [53], [54], [55]]. Moreover, some dimeric naphthylisoquinolines, like michellamine B [56] (for the structure, see Table S1), show potent antiviral activities; they display anti-HIV effects by inhibition of certain viral enzymes like, for instance, reverse transcriptases (RTs) of both HIV type 1 and 2, as well as human alpha and beta DNA polymerases. Additionally, they can interfere with the late stages of the virus replication cycle by inhibiting cellular fusion and syncytium formation [[56], [57], [58], [59]].
Fig. 1.
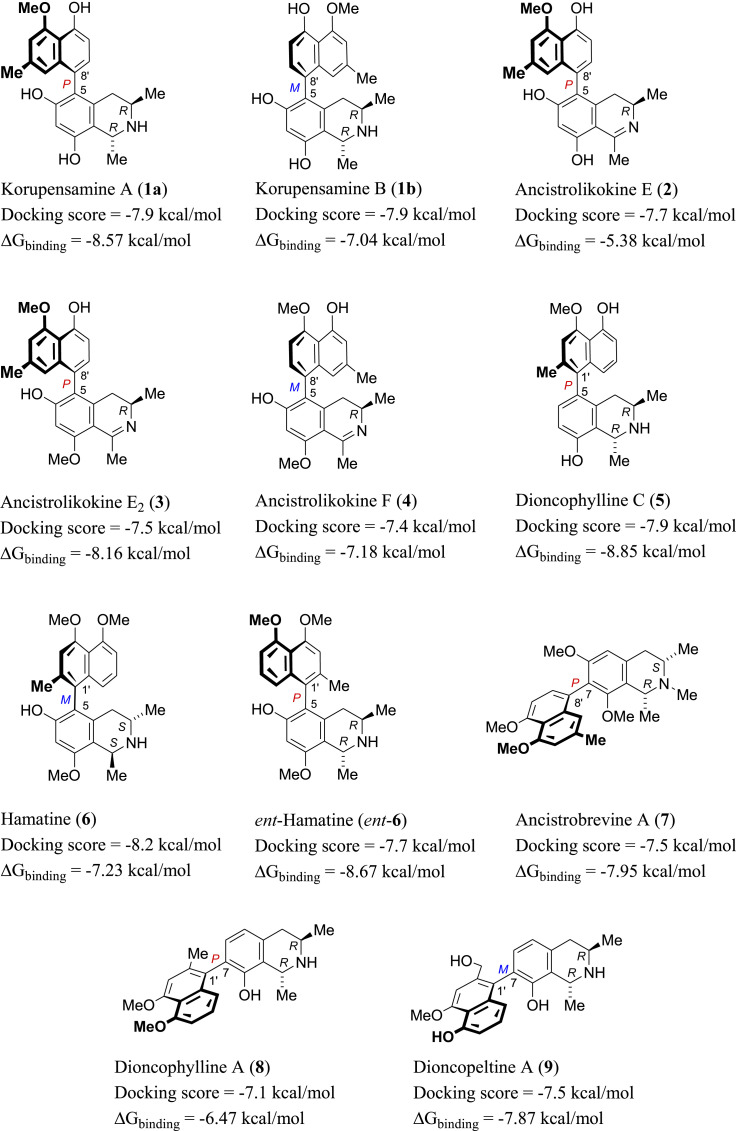
Structures of the eleven out of 65 selected top-scoring compounds with different molecular shapes retrieved from the docking step, among them 5,8′-coupled alkaloids (compounds 1a/1b-4) as well as representatives with 5,1’- (5, 6, and ent-6), 7,8’- (7), or 7,1′-axes (8, 9).
Scientists have embarked on developing an effective and safe treatment for COVID-19 [26], which stimulated us to conduct the present study to search for new drug leads against SARS-CoV-2 infection. In this paper, we describe the first investigation of a library of structurally most different mono- and dimeric NIQ alkaloids and selected structural analogues for their activity against SARS-CoV-2. Additionally, we investigated an integrated docking-MDS (molecular dynamics simulations) approach for speeding up and improving the selection of potentially active compounds among a group of closely related structures.
2. Results and discussion
2.1. General approach
In the frame of our continued search for potential anti-SARS-CoV-2 therapeutics [21,[60], [61], [62], [63], [64]], we investigated a small library of bioactive NIQs in the quest for inhibitors of the Mpro of SARS-CoV-2. Most of the compounds of this library (53 out of 65 structures) are atropisomerically well-defined biaryls (Table S1). Since atropisomers are stereoisomers resulting from impeded rotation about a single bond, their structures are intrinsically less flexible than those of the corresponding freely rotating conformers, and hence they are of lower entropy. Drugs with lower entropy are likely to have a higher affinity for receptors and enzymes, as the binding process will result in less loss of entropy. From this point, we started a molecular docking-based screening of the NIQ library against the structure of the Mpro of SARS-CoV-2 to find possible binders and, thus, potential inhibitors.
2.2. Docking-based screening
At the beginning of the docking step, we validated the docking protocol by AutoDock Vina [65], re-docking the co-crystallized ligands of two previously reported [66,67] SARS-CoV-2 Mpro structures (PDB ID: 7DPP and 7LTJ). The resulting top-scoring docking poses of these inhibitors aligned with the co-crystallized ones with acceptable root-mean-square deviation (RMSD) values of 1.26 Å and 0.89 Å, respectively.
The modeled structures of the compounds of the library were prepared and docked into the active site of Mpro using the AutoDock Vina software [65]. Since all of the compounds in the used library contained basic - secondary or tertiary - amino functions, we also investigated the protonated forms of the compounds for the docking step. AutoDock Vina does not take steric hindrance into consideration during the docking process, where it makes the structures of the molecules free to rotate independently of the energetics of the whole molecules. For this reason, the resulting poses for each structure in the library were filtered according to the correct and low-energy conformations. A binding score of −7 kcal/mol was another filter criterion for selection, according to which the structures and poses of lower scores were neglected.
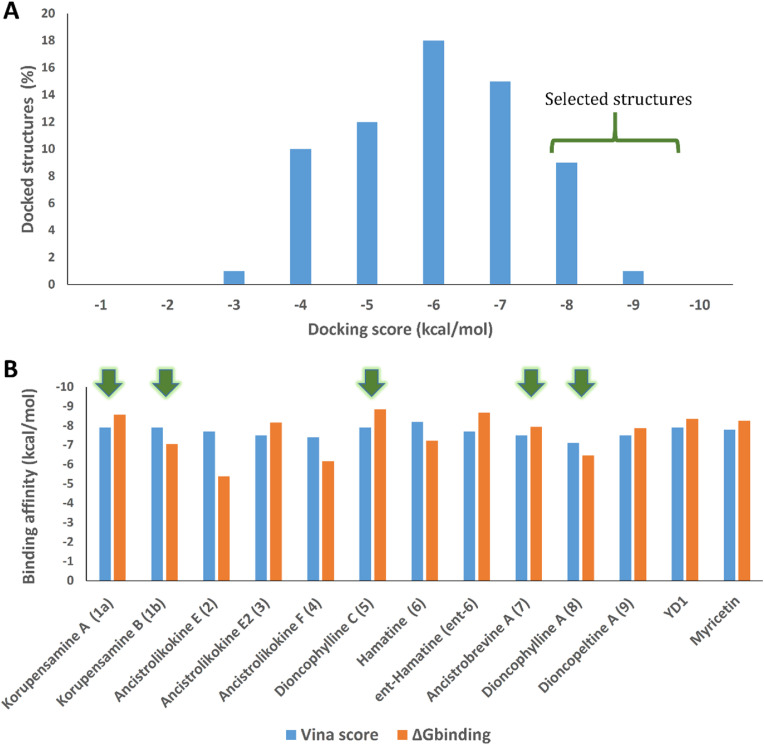
As a result, 11 out of the 65 initially considered structures were selected as top candidates for the subsequent in silico and in vitro testing (Figs. 1 and 2A ). It is worth noting that all generated poses for each of the top-scoring structures had almost the same binding mode. They aligned perfectly with each other, with a low RMSD value of 0.947 Å. This observation can be attributed to the low flexibility of the docked structures, where the number of available conformations that can generate different poses was limited. Such a uniformity of the docking poses indicated that the top-ranked pose of each docked structure was likely to be the correct one. However, further refinement, based on molecular dynamics simulation, was required, particularly the calculation of ΔG binding for each generated pose. This docking-based step managed to filter out ancistrocladine (docking score = −6.7 kcal/mol; for the structure, see Table S1) from its atropisomer, hamatine (6) (−8.2 kcal/mol), while it was not able to discriminate between the atropo-diastereomeric alkaloids korupensamine A (1a) and korupensamine B (1b), where both structures got the same score (−7.9 kcal/mol).
Fig. 2.
(A) Score distribution of the docked structures in the NIQ library against the Mpro of SARS-CoV-2. Structures with docking scores < -7 kcal/mol (17.5%) were selected for validation by MDS and ΔGbinding calculation. (B) Docking scores in comparison with the ΔGbinding values of the top-scoring compounds selected from the docking step, along with those of the co-crystallized inhibitors YD1 and myricetin. Green arrows indicate compounds that were selected for in vitro antiviral testing.
2.3. ΔGbinding-based refinement
Using the alchemical Free Energy Perturbation (FEP) method [68], we then determined the ΔG binding for the top candidates. By this procedure, ΔG binding values were extracted by running a series of MDS experiments. The 11 selected structures (Fig. 1) exhibited ΔG binding values ranging from −5.38 to −8.85 kcal/mol (Figs. 1 and 2B), indicating good affinities towards the active site of Mpro and, in turn, potential antiviral activity against SARSCoV-2. Based on the binding affinity values, it was hard to discriminate between these selected structures regarding their potential activity. Moreover, the two atropo-diastereomeric alkaloids korupensamine A (1a) and korupensamine B (1b), possessing the same gross structure, showed no significant difference in the calculated ΔG binding values (ΔΔG binding = −1.5 kcal/mol, see Fig. 1). Such a minor difference was not large enough to prefer one atropisomer over the other. This was not the case with the second pair of atropisomers investigated, ancistrocladine (for the structure, see Table S1) and hamatine (6), whose calculated ΔG binding values were significantly different (−3.88 kcal/mol and −7.23 kcal/mol, respectively; hence ΔΔG binding = −3.35 kcal/mol).
2.4. Steered molecular dynamics simulation
For this reason, we present herein the advantage of using steered molecular dynamics (SMD) in the discrimination between related congeners in terms of their affinity towards the active site of Mpro, by the determination of their relative binding affinity.
To do so, the top-ranked docking poses for each structure of the previously selected 11 compounds alongside the co-crystallized inhibitors (myricetin and YD1) were prepared and subjected to a number of SMD runs, where an external force was applied to each structure to pull it out of the Mpro active pocket, and hence, these applied forces can be traced over the course of the steering simulation.
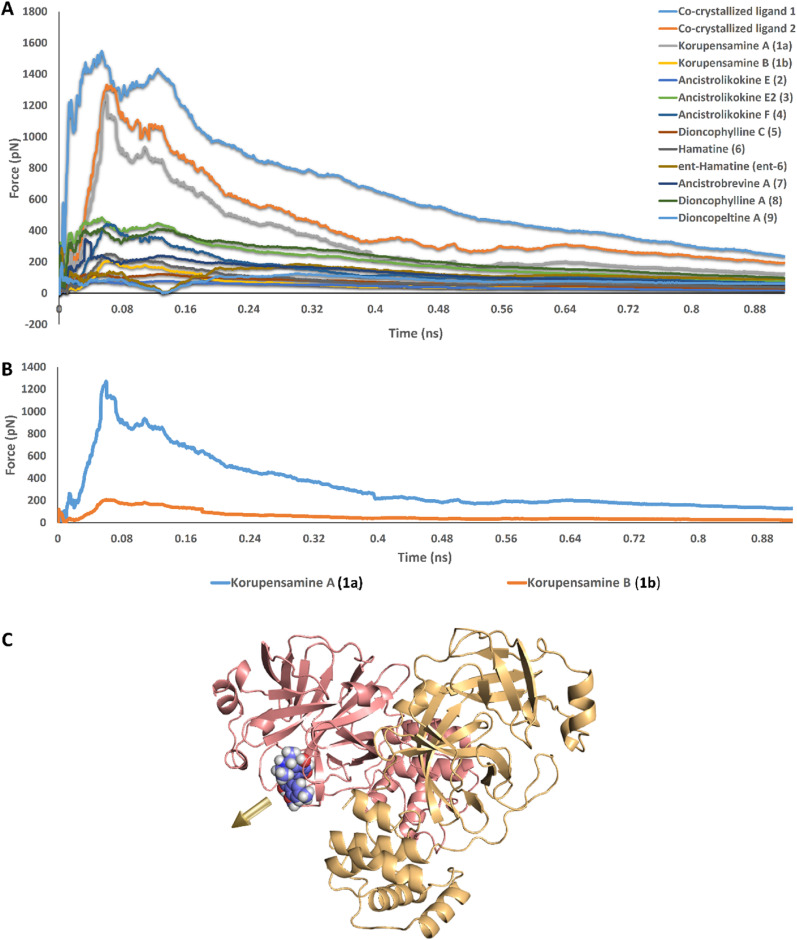
Remarkably, the force needed to unbind korupensamine A (compound 1a, 1283 pN) was comparable to those needed to unbind the previously reported co-crystallized inhibitors myricetin and YD (1522 pN and 1323 pN, respectively). In addition, their steering force profiles clustered together away from those of all of the remaining structures, including the structurally so closely related korupensamine B (1b). The unbinding forces of those other structures did not exceed 480 pN (Fig. 3 A and B).
Fig. 3.
(A) Comparison of the unbinding force profiles of the selected 11 structures shown in Fig. 1, alongside that of the co-crystallized inhibitors (myricetin and YD1; co-crystallized ligands 1 and 2, respectively). The plots show the resulting mean values from averaging the force profiles from three different SMD runs (n = 3) for each structure. (B) Unbinding force profiles of korupensamine A (1a) versus its atropisomer korupensamine B (1b), showing the significant difference in the relative affinity of the two structures toward the active site of Mpro. (C) The golden-yellow arrow indicates the pulling direction for the ligand-unbinding pathway.
2.5. Atropo-selective Mpro inhibitory activity of korupensamine A (1a) in vitro
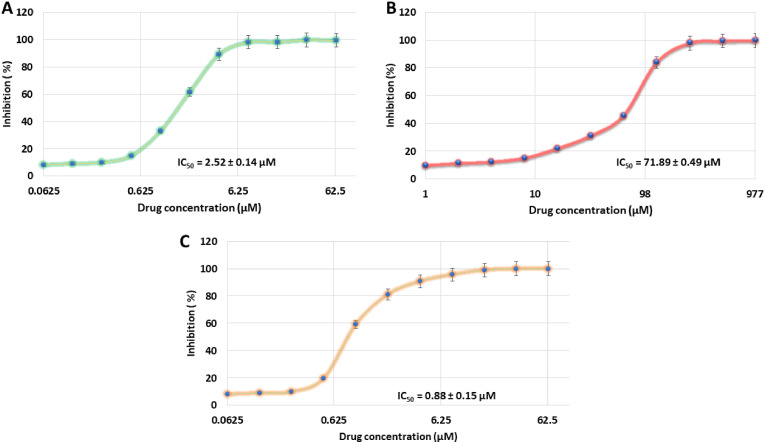
In vitro validation on the Mpro activity assay supported these findings (Fig. 4 ), where korupensamine A (1a) suppressed the catalytic activity of the enzyme in a dose-dependent manner, with an IC50 value of 2.53 ± 0.138 μM (K i = 1.42 ± 0.12 μM), while its atropo-diastereomer, korupensamine B (1b), i.e., with the same constitution as 1a, and possessing the same absolute configuration at both stereocenters, differing only by the axial configuration, was inactive up to 50 μM (IC50 = 71.89 ± 0.49 μM).
Fig. 4.
Inhibition of the activity of the SARS-CoV-2 Mpro by korupensamine A (1a) and korupensamine B (1b) at increasing concentrations of the inhibitor. The enzyme activity is reduced by 50% at a korupensamine A (1a) concentration of 2.52 ± 0.14 μM (A) in comparison to that of korupensamine B (1b) at a concentration of 71.89 ± 0.49 μM (B) and that of the reference inhibitor GC376 [70] at a concentration of 0.88 ± 0.15 μM (C).
These enzyme inhibitory results clearly showed the superior affinity of korupensamine A (1a) over its atropisomer korupensamine B (1b) – and over all of the other remaining compounds. This result emphasized, once again, the impact of axial chirality for the bioactivity of rotationally hindered biaryl agents in general, and in this particular case, the pronounced structure-specificity of the inhibitory activity of korupensamine A (1a). Additionally, the results highlighted the efficacy of the SMD approach in the differentiation between atropisomers in terms of their biomolecular interactions.
SMD had previously been reported to be an effective method of discerning between flavonoids in terms of their enzyme inhibition activity [69]. Here, once again, this fast and reliable in silico method proved its ability in pointing out the active anti-SARS-CoV-2 representative among a group of structurally most similar potential candidates.
2.6. Dynamic mode of interaction
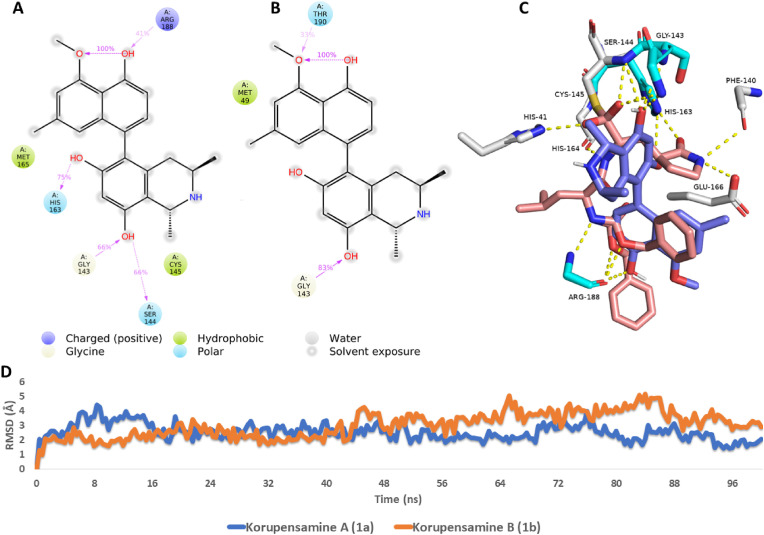
To get some insight into the dynamic binding interactions of korupensamine A (1a) inside the active site of Mpro in comparison to those of its atropo-diastereomer (i.e., korupensamine B, 1b), the selected poses of both compounds were subjected to 100-ns MDS runs. The dynamic interactions of the two structures were then extracted and analyzed (see Fig. 5 ). As shown in Fig. 5D, the binding stability of the two compounds inside the active site of the enzyme was comparable to an average RMSDs of 2.43 Å and 3.72 Å for korupensamines A (1a) and B (1b), respectively. On the one hand, both structures formed stable H-bonds with GLY-143 over the course of the simulation (Fig. 5A and B), but, on the other hand, only korupensamine A (1a) formed three additional stable H-bonds with SER-144, HIS-163, and ARG-188, while only korupensamine B (1b) established another single H-bond with THR-190. Moreover, korupensamine A (1a) had stable hydrophobic interactions with MET-165 and CYS-145, while korupensamine B (1b) revealed a single stable hydrophobic interaction with MET-49. Accordingly, korupensamine A (1a) exhibited thermodynamically more stable interactions inside the Mpro active site than its atropisomer, korupensamine B (1b). This explains why much more force was required to steer 1a out of the active site of the enzyme than the one needed in the case of 1b (Fig. 3B). Alignment of the structure of korupensamine A (1a) with that of the reference inhibitor GC376 (Fig. 5C) indicated that both structures have four common hydrophilic interactions (i.e., H-bonds) with GLY-143, SER-144, HIS-163, and ARG-188. Accordingly, H-bonding to these residues might be essential for good binding and inhibition of the Mpro of SARS CoV-2.
Fig. 5.
Ligand-protein interactions extracted from the 100-ns long MDS experiments of (A) korupensamine A (1a) and (B) korupensamine B (1b). 3D binding mode of korupensamine A (1a, in blue) in alignment with the reference inhibitor GC376 (brick-red structures) inside the active site of Mpro (C). The cyan-colored amino acid residues interact with both structures (i.e., korupensamine A and GC376), while white-colored amino acid residues interact with GC376 only. RMSDs of korupensamines A (1a) and B (1b) over the course of their MDS runs (D). Colored arrows in (A) and (B) indicate the percentage of interaction with each amino acid residue during the MDS runs.
2.7. Gaussian accelerated molecular dynamics (GaMD) simulations
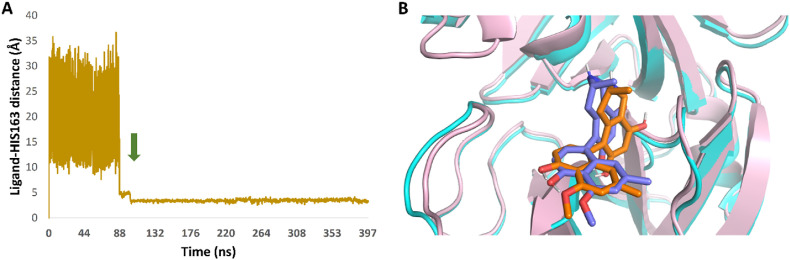
In order to explore the binding pathway of korupensamine A (1a) to the active site of Mpro, three independent Gaussian accelerated molecular dynamics simulations (GaMD) were carried out. In each 400-ns long simulation, five replicates of the structure of korupensamine A (1a) were placed in the solvent box at least 20 Å away from the modeled Mpro structure. In one of the three simulations, ligand binding was observed after 115.92 ns and remained stable inside the Mpro active site until the end of the simulation (Fig. 6 A). Interestingly, the final binding state (Fig. 6B) was almost identical to that of the top-scoring docking pose selected earlier (RMSD = 1.63 Å), indicating that this binding pose is the most likely one.
Fig. 6.
(A) Binding event of korupensamine A (1a) with the Mpro active site observed in GaMD simulation after 115.92 ns (the green arrow indicates the beginning of the binding event). (B) Structure alignment of the docking pose of korupensamine A (1a, orange structure) on its most populated structure after its binding inside the active site of Mpro during the GaMD simulation (blue structure). RMSD value between the two states = 1.63 Å. A video showing the binding event of korupensamine A (1a) inside the active site of Mpro can be found on the Zenodo website: https://zenodo.org/record/7312007#.Y35sVXbP2M9.
Additionally, the four amino acid residues that had earlier been found to interact with korupensamine A (1a) via H-bonding (GLY-143, SER-144, HIS-163, and ARG-188) were also the interacting ones upon korupensamine A binding.
2.8. Korupensamine A (1a) inhibits SARS-CoV-2 in cell culture
Due to a quantity limitation, we selected five out of the top potentially active 11 alkaloids (Figs. 1 and 2B), along with another three compounds predicted to be inactive - the dimeric alkaloids michellamine A, which is the 6′,6″-dimer of korupensamine A (1a), michellamine B, i.e. the mixed dimer of korupensamines A (1a) and B (1b), and plumbagin, the oxidized naphthalene part of the presented NIQs (for the structures, see Table S1) - for in vitro antiviral testing against SARS-CoV-2. Surprisingly, only korupensamine A (1a) was active against SARS-CoV-2, it reduced the viral replication at 30 μM by five orders of magnitude (EC50 = 4.23 ± 1.31 μM; CC50 = 35.1 μM) (Fig. S2). From this observation it can be concluded that docking and ΔG binding calculation were good in predicting active compounds, but not sufficient to discriminate between compounds of convergent scores and between atropisomers, here korupensamine A (1a) versus korupensamine B (1b), whereas SMD demonstrated its efficacy in identifying the one anti-SARS-CoV-2 agent from of a pool of structurally similar compounds.
3. Conclusion
Understanding the interactions between ligands and proteins is essential for a rational structure-based drug design. In this work, several methods combining docking and MDS (molecular docking simulations) have been utilized to investigate the mode of interaction of pharmaceutically relevant complexes in the field of axially chiral naphthylisoquinoline (NIQ) alkaloids and their analogues. To identify potential ligands for the SARS-CoV-2 Mpro, we used docking-based virtual screening in conjunction with ΔG binding calculations to examine a library of 65 structures of such alkaloids.
The top-scoring structures were subjected to binding free energy and SMD experiments to determine their relative affinity towards the active site of the enzyme and hence, distinguishing the most probably active candidate, korupensamine A (1a). Upon in vitro validation, 1a was found to be the only actual Mpro inhibitor, possessing an IC50 value of 2.52 μM, with significant antiviral activity against SARS-CoV-2, comparable to that of the reference inhibitor GC376 (IC50 = 0.88 μM). Accordingly, from a collection of structurally related, atropisomerically defined NIQ alkaloids, SMD simulation was found to be a fast and accurate method for predicting the best Mpro inhibitor.
Finally, using Gaussian accelerated molecular dynamics (GaMD) simulations over 400 ns, we explored the binding pathway and the mode of recognition and interaction of korupensamine A (1a) within the active site of Mpro.
Intriguingly, GaMD simulations managed to simulate the binding process of korupensamine A (1a) and to reproduce its docking position within the active site of the enzyme, demonstrating the validity of our approach from the outset. The study shows that using computational chemistry methods helps the drug discovery process move forward. Our strategy presented here could effectively supplement commonly used docking simulations, especially when there are only minor structural differences within a series of analogues, where docking studies may have failed to properly rank the binding affinities.
The work furthermore reveals naphthylisoquinoline alkaloids as a highly promising new class of potential anti-SARS-CoV-2 agents. The highlight among these axially chiral alkaloids is korupensamine A (1a), which does not only bind strongly to the main protease of the virus in silico, but is also active in vitro, with a high structure specificity: All of the numerous other alkaloids, whether ring-opened, ring-contracted or quinoid, as well as their non-coupled isoquinoline and naphthalene moieties (see Table S1), were inactive, also the respective dimers, even including the closely related korupensamine B (1b), which differs from 1a only by the configuration at the rotationally hindered biaryl axis. This pinpoints, once again, the importance of axially chirality, which dictates the molecular shape of a bioactive compound, and thus, determines its bioactivity.
4. Experimental
4.1. Naphthylisoquinoline alkaloids and related compounds
The NIQs experimentally investigated in the present study and their analogues were prepared by isolation from their plant sources [56,[71], [72], [73], [74], [75], [76]] or by total synthesis [77], as described previously (for further details, see Table S1), and have been deposited in our in-house natural products library.
4.2. Molecular docking
4.2.1. Structure generation
The structures of the NIQs and their analogues used in the present study were generated using ChemDraw (12.0). Their rotational-energy profiles were calculated after energy minimization using Chem3D (12.0).
4.2.2. Ligand preparation
All torsions of the collected structures were assigned and their Gasteiger charges were added for all atoms in the compound structures using the AutoDockTools v.4.2 package [78].
4.2.3. Receptor preparation
For docking screening, the Mpro structures with the co-crystallized ligands myricetin and YD1 (PDB codes: 7DPP and 7LTJ, respectively) were used. The downloaded structures were curated on PDBfixer [79] to repair lost atoms and residues, and for removing co-crystallized water molecules. Polar hydrogens and Gasteiger charges were then added to all receptor structures using AutoDockTools v.4.2 [78].
4.2.4. Structural docking
For the docking step, AutoDock Vina software integrated in the PyRx platform was used [80,81]. The binding site for the docking search was determined according to the co-crystallized ligand of the enzyme, 6G9. The coordinates of the grid box were: x = −7.58; y = 16.39; z = 2.44. The size of the grid box was set to be 10 Å3 (for small structures) and 20 Å3 (for larger structures). Exhaustiveness was set to be 24. Docking poses were analyzed and visualized using the PyMOL software [80]. The docking protocol was validated by re-docking the co-crystallized inhibitor (6G9). The resulting top-scoring pose (score = −7.12 kcal/mol) was found to achieve a good alignment with the original co-crystallizing pose, with a low RMSD value (1.26 Å). After docking the structures of the NIQ library, a cut-off value of −7.0 kcal/mol was set to select the best ligands. This cut-off value was set according to the lowest docking score of the co-crystallized ligands (6G9 and myricetin), which got scores of −7.12 and −8.82 kcal/mol, respectively.
4.3. Molecular dynamics and ΔGbinding determination
Molecular dynamics simulations (including SMD) and binding free energy determination were carried out as described earlier [82]. GaMD simulations were performed according to the previously reported method of Miao and co-workers [83]. The detailed procedures are described in the Supplementary data.
4.4. Mpro enzyme assay (in vitro)
Top-scoring compounds were assessed for their in vitro enzyme inhibition activities using 3CL Protease, tagged (SARS-CoV-2) Assay Kit (Catalogue #: 79955-1, BPS Bioscience, Inc., Allentown, PA, USA), according to manufacturer's protocol [5]. The in vitro FRET assay was monitored at an emission wavelength of 460 nm with an excitation at 360 nm, using a Flx800 fluorescence spectrophotometer (BioTek Instruments, Winooski, VT, USA).
4.5. In vitro antiviral assay
Vero cells were incubated with the compounds for 3 days to investigate cytotoxicity, and the relative cell growth was determined by automatic cell counting. Concentrations of 30 μM of 1a and 1b did not influence cell growth. The Vero cells were then incubated with the respective compounds at increasing concentrations and subsequently infected with patient-derived SARS-CoV-2 at an MOI (Multiplicity of Infection) of approximately 0.5. DMSO was used as a solvent control [84]. After 24 h, the medium was replaced by compound-containing medium, to remove inactive viruses. After 3 days, viral replication supernatants were collected, and viral RNA was extracted. Viral replication was quantified by real-time RTqPCR. All infections were performed in triplicate [84,85].
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Deraya University and the University of Würzburg. The authors thank Dr. D. Feineis and Prof. C. Sotriffer for valuable advice, and Dr. W. Shamburger for initial help with the structures.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2023.115226.
Abbreviations
- cMD
Classical Molecular Dynamics
- DMSO
Dimethyl Sulfoxide
- FEP
Free Energy Perturbation
- FF14SB
Force Field 14 Stony Brook
- ΔGbinding
Absolute Binding Free Energy
- GaMD
Gaussian Accelerated Molecular Dynamics
- GPUs
Graphics Processing Unit
- GUI
Graphical User Interface
- IC50
Half Maximal Inhibitory Concentration
- Ki
Inhibitor Constant
- MDS
Molecular Dynamics Simulation
- MOI
Multiplicity of Infection
- Mpro
Main Protease
- NAMD
Nanoscale Molecular Dynamics
- NIQ
Naphthylisoquinoline
- NPT
Constant Pressure and Temperature
- OPLS
Optimized Potentials for Liquid Simulations
- PCR
Polymerase Chain Reaction
- PDB
Protein Data Bank
- PME
Particle Mesh Ewald
- pN
Piconewton
- RMSD
Root-Mean-Square Deviation
- RMSF
Root-Mean-Square Fluctuation
- RT
Reverse Transcriptase
- RTqPCR
Quantitative Reverse Transcription Polymerase Chain Reaction
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus-2
- SHAKE
Secure Hash Algorithm Keccak
- SMD
Steered Molecular Dynamics
- TIP3P
Transferable Intermolecular Potential with 3 Points
- VMD
Visual Molecular Dynamics
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Watson O.J., Barnsley G., Toor J., Hogan A.B., Winskill P., Ghani A.C. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infect. Dis. 2022;22:1293–1302. doi: 10.1016/S1473-3099(22)00320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maxmen A. Wuhan market was epicentre of pandemic's start, studies suggest. Nature. 2022;603:15–16. doi: 10.1038/d41586-022-00584-8. [DOI] [PubMed] [Google Scholar]
- 3.Lamers M.M., Haagmans B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022;20:270–284. doi: 10.1038/s41579-022-00713-0. [DOI] [PubMed] [Google Scholar]
- 4.Halpin D.M.G., Faner R., Sibila O., Badia J.R., Agusti A. Do chronic respiratory diseases or their treatment affect the risk of SARS-CoV-2 infection? Lancet Respir. Med. 2020;8:436–438. doi: 10.1016/S2213-2600(20)30167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu T., Siegel E., Shen D. Deep learning and medical image analysis for COVID-19 diagnosis and prediction. Annu. Rev. Biomed. Eng. 2022;24:179–201. doi: 10.1146/annurev-bioeng-110220-012203. [DOI] [PubMed] [Google Scholar]
- 6.Ballering A.V., van Zon S.K.R., Colde Hartman T., Rosmalen J.G.M. For the Lifelines Corona Research Initiative, Persistence of somatic symptoms after COVID-19 in The Netherlands: an observational cohort study. Lancet. 2022;400:452–461. doi: 10.1016/S0140-6736(22)01214-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amin S.A., Banerjee S., Gosh K., Gayen S., Jha T. Protease targeted COVID-19 drug discovery and its challenges: insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorg. Med. Chem. 2021;29 doi: 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H., Zhou Y., Zhang M., Wang H., Zhao Q., Liu J. Updated approaches against SARS-CoV-2, antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00483-20. e00483-00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeyanathan M., Afkhami S., Smaill F., Lichty M.B.D., Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machingaidze S., Wiysonge C.S. Understanding COVID-19 vaccine hesitancy. Nat. Med. 2021;27:1338–1339. doi: 10.1038/s41591-021-01459-7. [DOI] [PubMed] [Google Scholar]
- 11.Mei M., Tan X. Current strategies of antiviral drug discovery for COVID-19. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.671263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. For the ACTT-1 study members, remdesivir for the treatment of Covid-19 — final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monk P.D., Marsden R.J., Tear V.J., Brookes J., Batten T.N., Mankowski M., Gabbay F.J., Davies D.E., Holgate S.T., Ho L.-P., Clark T., Djukanovic R., Wilkinson T.M.A. On behalf of the Inhaled Interferon Beta COVID-19 Study Group, Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir. Med. 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The RECOVERY Collaborative Group Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2020;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson A.S., Caubel P., Rusnak J.M. For the EPIC-HR trial investigators, nirmatrelvir-ritonavir and viral load rebound in Covid-19. N. Engl. J. Med. 2022;387:1047–1049. doi: 10.1056/NEJMc2205944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 18.Yang H., Yang J. A review of the latest research on Mpro targeting SARS-COV inhibitors. RSC Med. Chem. 2021;12:1026–1036. doi: 10.1039/d1md00066g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao J., Li Y.-S., Zeng R., Liu F.-L., Luo R.H., Huang C., Wang Y.-F., Zhang J., Quan B., Shen C., Mao X., Liu X., Sun W., Yang W., Ni X., Wang K., Xu L., Duan Z.-L., Zou Q.-C., Zhang H.-L., Qu W., Long Y.H.P., Li M.-H., Yang R.-C., Liu X., You J., Zhou Y., Yao R., Li W.-P., Liu J.-M., Chen P., Liu Y., Lin G.-F., Yang X., Zou J., Li L., Hu Y., Lu G.-W., Li W.-M., Wei Y.-Q., Zheng Y.-T., Lei J., Yang S. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371:1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavlova A., Lynch D.L., Daidone I., Zanetti-Polzi L., Smith M.D., Chipot C., Kneller D.W., Kovalevsky A., Coates L., Golosov A.A. Inhibitor binding influences the protonation states of histidines in SARS-CoV-2 main protease. Chem. Sci. 2021;12:1513–1527. doi: 10.1039/d0sc04942e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alhadrami H.A., Sayed A.M., Sharif A.M., Azhar E.I., Rateb M.E. Olive-derived triterpenes suppress SARS COV-2 main protease: a promising scaffold for future therapeutics. Molecules. 2021;26:2654. doi: 10.3390/molecules26092654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang H.-P., Chou C.-Y., Chang G.-G. Reversible unfolding of the severe acute respiratory syndrome coronavirus main protease in guanidinium chloride. Biophys. J. 2007;92:1374–1383. doi: 10.1529/biophysj.106.091736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Q.A., Kato-Weinstein J., Li Y., Deng Y., Granet R., Garner L., Liu C., Polshakov D., Gessner C., Watkins S. Potential therapeutic agents and associated bioassay data for COVID-19 and related human coronavirus infections. ACS Pharmacol. Transl. Sci. 2020;3:813–834. doi: 10.1021/acsptsci.0c00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kneller D.W., Li H., Phillips G., Weiss K.L., Zhang Q., Arnould M.A., Jonsson C.B., Surendranathan S., Parvathareddy J., Blakeley M.P. Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease. Nat. Commun. 2022;13:2268. doi: 10.1038/s41467-022-29915-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg H., Wirtz Martin M.A., Altincekic N., Alshamleh I., Kaur Bains J., Blechar J., Ceylan B., de Jesus V., Dhamotharan K., Fuks C., Gande S.L., Hargittay B., Hohmann K.F., Hutchison M.T., Korn S.M., Krishnathas R., Kutz F., Linhard V., Matzel T., Meiser N., Niesteruk A., Pyper D.J., Schulte L., Trucks S., Azzaoui K., Blommers M.J.J., Gadiya Y., Karki R., Zaliani A., Gribbon P., da Silva Almeida M., Anobom C.D., Bula A.L., Bütikofer M., Caruso Í.P., Felli I.C., Da Poian A.T., Cardoso de Amorim G., Fourkiotis N.K., Gallo A., Ghosh D., Gomes-Neto F., Gorbatyuk O., Hao B., Kurauskas V., Lecoq L., Li Y., Mebus-Antunes N.C., Mompeán M., Neves-Martins T.C., Ninot-Pedrosa M., Pinheiro A.S., Pontoriero L., Pustovalova Y., Riek R., Robertson A.J., Abi Saad M.J., Treviño M.Á., Tsika A.C., Almeida F.C.L., Bax A., Henzler-Wildman K., Hoch J.C., Jaudzems K., Laurents D.V., Orts J., Pierattelli R., Spyroulias G.A., Duchardt-Ferner E., Ferner J., Fürtig B., Hengesbach M., Löhr F., Qureshi N., Richter C., Saxena K., Schlundt A., Sreeramulu S., Wacker A., Weigand J.E., Wirmer-Bartoschek J., Wöhnert J., Schwalbe H. Comprehensive fragment screening of the SARS‐CoV‐2 proteome explores novel chemical space for drug development. Angew. Chem. Int. Ed. 2022;134 doi: 10.1002/anie.202205858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan B., Joyce R., Tan H., Hu Y., Wang J. SARS-CoV-2 main protease drug design, assay development, and drug resistance studies. Acc. Chem. Res. 2022;56:157–168. doi: 10.1021/acs.accounts.2c00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh A.K., Mishevich J.L., Mesecar A., Mitsuya H. Recent drug development and medicinal chemistry approaches for the treatment of SARS‐CoV‐2 infection and COVID‐19. ChemMedChem. 2022;17 doi: 10.1002/cmdc.202200440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallock Y.F., Manfredi K.P., Blunt J.W., Cardellina J.H., II, Schäffer M., Gulden K.-P., Bringmann G., Lee A.Y., Clardy J., François G., Boyd M.R., Korupensamines A-D novel antimalarial alkaloids from Ancistrocladus korupensis. J. Org. Chem. 1994;59:6349–6355. [Google Scholar]
- 30.Lombe B.K., Feineis D., Bringmann G. Dimeric naphthylisoquinoline alkaloids: polyketide-derived axially chiral bioactive quateraryls. Nat. Prod. Rep. 2019;36:1513–1545. doi: 10.1039/c9np00024k. [DOI] [PubMed] [Google Scholar]
- 31.Tajuddeen N., Bringmann G. N,C-Coupled naphthylisoquinoline alkaloids: a versatile new class of axially chiral natural products. Nat. Prod. Rep. 2021;38:2154–2186. doi: 10.1039/d1np00020a. [DOI] [PubMed] [Google Scholar]
- 32.D. Feineis, G. Bringmann, in: A.D. Kinghorn, H. Falk, S. Gibbons, Y. Asakawa, J.-K. Liu, V.M. Dirsch (Eds.), Asian Ancistrocladus lianas as creative producers of naphthylisoquinoline alkaloids, in: Progress in the Chemistry of Organic Natural Products, vol. 119, Springer, Cham, Heidelberg, New York, Dordrecht, London, 2023, pp. 1–335. [DOI] [PubMed]
- 33.Bringmann G., Günther C., Ochse M., Schupp O., Tasler S. In: Herz W., Falk H., Kirby G.W., Moore R.E., editors. vol. 82. Springer-Verlag; Wien, New York: 2001. pp. 1–249. (Biaryls in nature: a multi-facetted class of stereochemically, biosynthetically, and pharmacologically intriguing secondary metabolites, in: Progress in the Chemistry of Organic Natural Products). [DOI] [PubMed] [Google Scholar]
- 34.Bringmann G., Gulder T., Gulder T.A.M., Breuning M. Atroposelective total synthesis of axially chiral biaryl natural products. Chem. Rev. 2011;111:563–639. doi: 10.1021/cr100155e. [DOI] [PubMed] [Google Scholar]
- 35.Tajuddeen N., Feineis D., Ihmels H., Bringmann G. The stereoselective total synthesis of axially chiral naphthylisoquinoline alkaloids. Acc. Chem. Res. 2022;55:2370–2383. doi: 10.1021/acs.accounts.2c00432. [DOI] [PubMed] [Google Scholar]
- 36.François G., Timperman G., Eling W., Aké Assi L., Holenz J., Bringmann G. Naphthylisoquinoline alkaloids against malaria: evaluation of the curative potentials of dioncophylline C and dioncopeltine A against Plasmodium berghei in vivo. Antimicrob. Agents Chemother. 1997;41:2533–2539. doi: 10.1128/aac.41.11.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kingston D.G.I., Cassera M.B. In: Kinghorn A.D., Falk H., Gibbons S., Asakawa Y., Liu J.-K., Dirsch V.M., editors. vol. 117. Springer Nature Switzerland AG; Cham, Switzerland: 2022. pp. 1–106. (Antimalarial natural products, in: Progress in the Chemistry of Organic Natural Products). [DOI] [PubMed] [Google Scholar]
- 38.Tajuddeen N., Van Heerden F.R. Antiplasmodial natural products: an update. Malar. J. 2019;18:404. doi: 10.1186/s12936-019-3026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moyo P., Shamburger W., van der Watt M.E., Reader J., De Sousa A.C.C., Egan T.J., Maharaj V.J., Bringmann G., Birkholtz L.-M. Naphthylisoquinoline alkaloids, validated as hit multistage antiplasmodial natural products. Int. J. Parasitol. Drugs Drug Resist. 2020;13:51–58. doi: 10.1016/j.ijpddr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lombe B.K., Bruhn T., Feineis D., Mudogo V., Brun R., Bringmann G. Antiprotozoal spirombandakamines A1 and A2, fused naphthylisoquinoline dimers from a Congolese Ancistrocladus plant. Org. Lett. 2017;19:6740–6743. doi: 10.1021/acs.orglett.7b03473. [DOI] [PubMed] [Google Scholar]
- 41.Ponte-Sucre A., Faber J.H., Gulder T., Kajahn I., Pedersen S.E.H., Schultheis M., Bringmann G., Moll H. Activities of naphthylisoquinoline alkaloids and synthetic analogs against Leishmania major. Antimicrob. Agents Chemother. 2007;51:188–194. doi: 10.1128/AAC.00936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ponte-Sucre A., Gulder T., Wegehaupt A., Albert C., Rikanović C., Schaeflein L., Frank A., Schultheis M., Unger M., Holzgrabe U., Bringmann G., Moll H. Structure–activity relationship and studies on the molecular mechanism of leishmanicidal N,C-coupled arylisoquinolinium salts. J. Med. Chem. 2009;52:626–636. doi: 10.1021/jm801084u. [DOI] [PubMed] [Google Scholar]
- 43.Ponte-Sucre A., Gulder T., Gulder T.A.M., Vollmers G., Bringmann G., Moll H. Alterations to the structure of Leishmania major induced by N-arylisoquinolines correlate with compound accumulation and disposition. J. Med. Microbiol. 2010;59:69–75. doi: 10.1099/jmm.0.014241-0. [DOI] [PubMed] [Google Scholar]
- 44.Cecil A., Rikanović C., Ohlsen K., Liang C., Bernhardt J., Oelschlaeger T.A., Gulder T., Bringmann G., Holzgrabe U., Unger M., Dandekar T. Modeling antibiotic and cytotoxic effects of the dimeric isoquinoline IQ-143 on metabolism and its regulation in Staphylococcus aureus, Staphylococcus epidermidis and human cells. Genome Biol. 2011;12:R24. doi: 10.1186/gb-2011-12-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.François G., van Looveren M., Timperman G., Chimanuka B., Aké Assi L., Holenz J., Bringmann G. Larvicidal activity of the naphthylisoquinoline alkaloid dioncophylline A against the malaria vector Anopheles stephensi. J. Ethnopharmacol. 1996;54:125–130. doi: 10.1016/s0378-8741(96)01459-6. [DOI] [PubMed] [Google Scholar]
- 46.Bringmann G., Holenz J., Aké Assi L., Zhao C., Hostettmann K. Molluscicidal activity of naphthylisoquinoline alkaloids from Triphyophyllum and Ancistrocladus species. Planta Med. 1996;62:556–557. doi: 10.1055/s-2006-957970. [DOI] [PubMed] [Google Scholar]
- 47.Bringmann G., Holenz J., Aké Assi L., Hostettmann K. Molluscicidal activity (Biomphalaria glabrata) of dioncophylline A: structure-activity investigations. Planta Med. 1998;64:485–486. doi: 10.1055/s-2006-957495. [DOI] [PubMed] [Google Scholar]
- 48.Bringmann G., Gramatzki S., Grimm C., Proksch P. Feeding deterrency and growth retarding activity of the naphthylisoquinoline alkaloid dioncophylline A against Spodoptera littoralis. Phytochemistry. 1992;31:3821–3825. [Google Scholar]
- 49.Bringmann G., Holenz J., Wiesen B., Nugroho B.W., Proksch P. Dioncophylline A as a growth-retarding agent against the herbivorous insect Spodoptera littoralis: structure–activity relationships. J. Nat. Prod. 1997;60:342–347. doi: 10.1021/np960707s. [DOI] [PubMed] [Google Scholar]
- 50.Bringmann G., Fayez S., Shamburger W., Feineis D., Winiarczyk S., Janecki R., Adaszek Ł. Naphthylisoquinoline alkaloids and their synthetic analogs as potent novel inhibitors against Babesia canis in vitro. Vet. Parasitol. 2020;283 doi: 10.1016/j.vetpar.2020.109177. [DOI] [PubMed] [Google Scholar]
- 51.Li J., Seupel R., Feineis D., Mudogo V., Kaiser M., Brun R., Brünnert D., Chatterjee M., Seo E.J., Efferth T., Bringmann G. Dioncophyllines C2, D2, and F and related naphthylisoquinoline alkaloids from the Congolese liana Ancistrocladus ileboensis with potent activities against Plasmodium falciparum and against multiple myeloma and leukemia cell lines. J. Nat. Prod. 2017;80:443–458. doi: 10.1021/acs.jnatprod.6b00967. [DOI] [PubMed] [Google Scholar]
- 52.Awale S., Dibwe D.F., Balachandran C., Fayez S., Feineis D., Lombe B.K., Bringmann G. Ancistrolikokine E3, a 5,8′-coupled naphthylisoquinoline alkaloid, eliminates the tolerance of cancer cells to nutrition starvation by inhibition of the Akt/mTOR/autophagy signaling pathway. J. Nat. Prod. 2018;81:2282–2291. doi: 10.1021/acs.jnatprod.8b00733. [DOI] [PubMed] [Google Scholar]
- 53.Fayez S., Bruhn T., Feineis D., Aké Assi L., Awale S., Bringmann G., Ancistrosecolines A-F unprecedented seco-naphthylisoquinoline alkaloids from the roots of Ancistrocladus abbreviatus, with apoptosis-inducing potential against HeLa cancer cells. J. Nat. Prod. 2020;83:1139–1151. doi: 10.1021/acs.jnatprod.9b01168. [DOI] [PubMed] [Google Scholar]
- 54.Fayez S., Cacciatore A., Sun S., Kim M., Aké Assi L., Feineis D., Awale S., Bringmann G. Ancistrobrevidines A-C and related naphthylisoquinoline alkaloids with cytotoxic activities against HeLa and pancreatic cancer cells, from the liana Ancistrocladus abbreviatus. Bioorg. Med. Chem. 2021;30 doi: 10.1016/j.bmc.2020.115950. [DOI] [PubMed] [Google Scholar]
- 55.Kushwaha P.P., Singh A.K., Prajapati K.S., Shuaib M., Fayez S., Bringmann G., Kumar S. Induction of apoptosis in breast cancer cells by naphthylisoquinoline alkaloids. Toxicol. Appl. Pharmacol. 2020;409 doi: 10.1016/j.taap.2020.115297. [DOI] [PubMed] [Google Scholar]
- 56.Boyd M.R., Hallock Y.F., Cardellina J.H., II, Manfredi K.P., Blunt J.W., McMahon J.B., Buckheit R.W., Jr., Bringmann G., Schäffer M., Cragg G.M., Thomas D.W., Jato J.G. Anti-HIV michellamines from Ancistrocladus korupensis. J. Med. Chem. 1994;37:1740–1745. doi: 10.1021/jm00038a003. [DOI] [PubMed] [Google Scholar]
- 57.Manfredi K.P., Blunt J.W., Cardellina J.H., II, McMahon J.B., Pannell L.L., Cragg G.M., Boyd M.R. Novel alkaloids from the tropical plant Ancistrocladus abbreviatus inhibit cell killing by HIV-1 and HIV-2. J. Med. Chem. 1991;34:3402–3405. doi: 10.1021/jm00116a011. [DOI] [PubMed] [Google Scholar]
- 58.McMahon J.B., Currens M.J., Gulakowski R.J., Buckheit R.W., Jr., Lackman-Smith C., Hallock Y.F., Boyd M.R. Michellamine B, a novel plant alkaloid, inhibits human immunodeficiency virus-induced cell killing by at least two distinct mechanisms. Antimicrob. Agents Chemother. 1995;39:484–488. doi: 10.1128/aac.39.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bringmann G., Steinert C., Feineis D., Mudogo V., Betzin J., Scheller C. HIV-inhibitory michellamine-type dimeric naphthylisoquinoline alkaloids from the Central African liana Ancistrocladus congolensis. Phytochemistry. 2016;128:71–81. doi: 10.1016/j.phytochem.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 60.Alhadrami H.A., Sayed A.M., Hassan H.M., Youssif K.A., Gaber Y., Moatasim Y., Kutkat O., Mostafa A., Ali M.A., Rateb M.E., Abdelmohsen U.R., Gamaleldin N.M. Cnicin as an anti-SARS-CoV-2: an integrated in silico and in vitro approach for the rapid identification of potential COVID-19 therapeutics. Antibiotics. 2021;10:542. doi: 10.3390/antibiotics10050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alhadrami H.A., Burgio G., Thissera B., Orfali R., Jiffri S.E., Yaseen M., Sayed A.M., Rateb M.E. Neoechinulin A as a promising SARS-CoV-2 Mpro inhibitor: in vitro and in silico study showing the ability of simulations in discerning active from inactive enzyme inhibitors. Mar. Drugs. 2022;20:163. doi: 10.3390/md20030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.ElNaggar M.H., Abdelwahab G.M., Kutkat O., GabAllah M., Ali M.A., El-Metwally M.E.A., Sayed A.M., Abdelmohsen U.R., Khalil A.T. Aurasperone A inhibits SARS CoV-2 in vitro: an integrated in vitro and in silico study. Mar. Drugs. 2022;20:179. doi: 10.3390/md20030179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orfali R., Rateb M.E., Hassan H.M., Alonazi M., Gomaa M.R., Mahrous N., GabAllah M., Kandeil A., Perveen S., Abdelmohsen U.R., Sayed A.M. Sinapic acid suppresses SARS CoV-2 replication by targeting its envelope protein. Antibiotics. 2021;10:420. doi: 10.3390/antibiotics10040420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eid R.A., Elgendy M.O., El-Gendy A.O., Elgendy S.O., Belbahri L., Sayed A.M., Rateb M.E. Efficacy of ceftazidime and cefepime in the management of COVID-19 patients: single center report from Egypt. Antibiotics. 2021;10:1278. doi: 10.3390/antibiotics10111278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandeep G., Nagasree K.P., Hanisha M., Kumar M.M.K. AUDocker LE: A GUI for virtual screening with AUTODOCK Vina. BMC Res. Notes. 2011;4:1–4. doi: 10.1186/1756-0500-4-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Su H., Yao S., Zhao W., Zhang Y., Liu J., Shao Q., Wang Q., Li M., Xie H., Shang W. Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease. Nat. Commun. 2021;12:1–12. doi: 10.1038/s41467-021-23751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clyde A., Galanie S., Kneller D.W., Ma H., Babuji Y., Blaiszik B., Brace A., Brettin T., Chard K., Chard R. High-throughput virtual screening and validation of a SARS-CoV-2 main protease noncovalent inhibitor. J. Chem. Inf. Model. 2021;62:116–128. doi: 10.1021/acs.jcim.1c00851. [DOI] [PubMed] [Google Scholar]
- 68.Ngo S.T., Tam N.M., Pham M.Q., Nguyen T.H. Benchmark of popular free energy approaches revealing the inhibitors binding to SARS-CoV-2 Mpro. J. Chem. Inf. Model. 2021;61:2302–2312. doi: 10.1021/acs.jcim.1c00159. [DOI] [PubMed] [Google Scholar]
- 69.Colizzi F., Perozzo R., Scapozza L., Recanatini M., Cavalli A. Single-molecule pulling simulations can discern active from inactive enzyme inhibitors. J. Am. Chem. Soc. 2010;132:7361–7371. doi: 10.1021/ja100259r. [DOI] [PubMed] [Google Scholar]
- 70.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fayez S., Feineis D., Mudogo V., Awale S., Bringmann G. Ancistrolikokines E–H and related 5,8′-coupled naphthylisoquinoline alkaloids from the Congolese liana Ancistrocladus likoko with antiausterity activities against PANC-1 human pancreatic cancer cells. RSC Adv. 2017;7:53740–53751. [Google Scholar]
- 72.Fayez S., Feineis D., Aké Assi L., Kaiser M., Brun R., Awale S., Bringmann G. Ancistrobrevines E-J and related naphthylisoquinoline alkaloids from the West African liana Ancistrocladus abbreviatus with inhibitory activities against Plasmodium falciparum and PANC-1 human pancreatic cancer cells. Fitoterapia. 2018;131:245–259. doi: 10.1016/j.fitote.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 73.Bringmann G., Rübenacker M., Jansen J.R., Scheutzow D., Aké Assi L. On the structure of the Dioncophyllaceae alkaloids dioncophylline A (“triphyophylline”) and “O-methyl-triphyophylline”. Tetrahedron Lett. 1990;31:639–642. [Google Scholar]
- 74.Bringmann G., Rübenacker M., Vogt P., Busse H., Aké Assi L., Peters K., von Schnering H.G. Dioncopeltine A and dioncolactone A: alkaloids from Triphyophyllum peltatum. Phytochemistry. 1991;30:1691–1696. [Google Scholar]
- 75.Bringmann G., Teltschik F., Schäffer M., Haller R., Bär S., Robertson S.A., Isahakia M.A. Ancistrobertsonine A and related naphthylisoquinoline alkaloids from Ancistrocladus robertsoniorum. Phytochemistry. 1998;47:31–35. doi: 10.1016/s0031-9422(97)00512-8. [DOI] [PubMed] [Google Scholar]
- 76.Bringmann G., Rübenacker M., Weirich R., Aké Assi L. Dioncophylline C from the roots of Triphyophyllum peltatum, the first 5,1′-coupled Dioncophyllaceae alkaloid. Phytochemistry. 1992;31:4019–4024. [Google Scholar]
- 77.Bringmann G., Ochse M., Götz R. First atropo-divergent total synthesis of the antimalarial korupensamines A and B by the “lactone method”. J. Org. Chem. 2000;65:2069–2077. doi: 10.1021/jo991634v. [DOI] [PubMed] [Google Scholar]
- 78.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eastman P., Swails J., Chodera J.D., McGibbon R.T., Zhao Y., Beauchamp K.A., Wang L.P., Simmonett A.C., Harrigan M.P., Stern C.D., Wiewiora R.P. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dallakyan S., Olson A.J. In: Chemical Biology. Methods in Molecular Biology. Hempel J., Williams C., Hong C., editors. vol. 1263. Humana Press; New York: 2015. Small-molecule library screening by docking with PyRx in; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- 82.Alhadrami H.A., Sayed A.M., Al-Khatabi H., Alhakamy N.A., Rateb M.E. Scaffold hopping of α-rubromycin enables direct access to FDA-approved cromoglicic acid as a SARS-CoV-2 MPro inhibitor. Pharmaceuticals. 2021;14:541. doi: 10.3390/ph14060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miao Y., McCammon J.A. Gaussian accelerated molecular dynamics: theory, implementation, and applications. Annu. Rep. Comput. Chem. 2017;13:231–278. doi: 10.1016/bs.arcc.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zimniak M., Kirschner L., Hilpert H., Geiger N., Danov O., Oberwinkler H., Steinke M., Sewald K., Seibel J., Bodem J. The serotonin reuptake inhibitor fluoxetine inhibits SARS-CoV-2 in human lung tissue. Sci. Rep. 2021;11:5890. doi: 10.1038/s41598-021-85049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geiger N., Kersting L., Schlegel J., Stelz L., Fähr S., Diesendorf V., Roll V., Sostmann M., König E.M., Reinhard S., Brenner D., Schneider-Schaulies S., Sauer M., Seibel J., Bodem J. The acid ceramidase is a SARS-CoV-2 host factor. Cells. 2022;11:2532. doi: 10.3390/cells11162532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.