Abstract
Hemizygous missense variants in the X-linked BGN gene, encoding the extracellular matrix protein biglycan, cause spondyloepimetaphyseal dysplasia (SEMD, biglycan type), which is clinically characterized by short stature, brachydactyly and osteoarthritis. Little is known about the pathomechanisms underlying SEMD, biglycan type. IPSC-derived chondrocyte disease models have been shown to exhibit several key aspects of known disease mechanisms of skeletal dysplasias and are therefore considered highly suitable human disease models to study SEMD, biglycan type. Prior to creating iPSC-chondrocytes, dermal fibroblasts of two male patients with SEMD, biglycan type, carrying the p.Gly259Val variant were successfully reprogrammed into iPSCs using the CytoTuneTM-iPS 2.0 Sendai Kit (Invitrogen).
1. Introduction
1.1. Resource Table:
| Unique stem cell lines identifier | CMGANTi003-A CMGANTi004-A |
|---|---|
| Alternative name(s) of stem cell lines | SEMD1 (CMGANTi003-A)SEMD2 (CMGANTi004-A) |
| Institution | University of Antwerp and Antwerp University Hospital |
| Contact information of distributor | Josephina Meester - Josephina.Meester@uantwerpen.be |
| Type of cell lines | iPSC |
| Origin | human |
| Additional origin info required | CMGANTi003-A: 52 yrs, male, Italian CMGANTi004-A: 50 yrs, male, Italian |
| Cell Source | dermal fibroblasts |
| Clonality | clonal |
| Method of reprogramming | Sendai virus |
| Genetic Modification | yes |
| Type of Genetic Modification | hereditary |
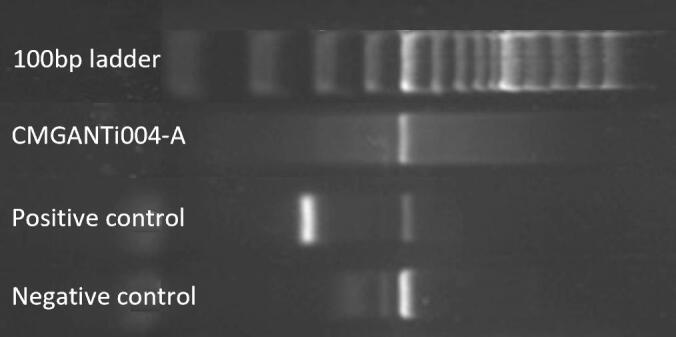
| Evidence of the reprogramming transgene loss (including genomic copy if applicable) | Absence of the Sendai virus backbone was verified with PCR and agarose gel electrophoresis. |
| Associated disease | spondyloepimetaphyseal dysplasia (SEMD), biglycan type |
| Gene/locus | BGN c.776G > T (NM_001711) |
| Date archived/stock date | December 2020 |
| Cell line repository/bank | Hpscreg https://hpscreg.eu/cell-line/CMGANTi003-A https://hpscreg.eu/cell-line/CMGANTi004-A |
| Ethical approval | Ethical committee Antwerp University Hospital, approval number: 11/8/79 2018.09.17 |
2. Resource utility
Because a cartilage biopsy is a highly invasive procedure for the patient and the regenerative capacity of cartilage tissue is limited, iPSC-derived chondrocytes provide a valuable alternative to model chondrodysplasias, including SEMD, biglycan type, and to investigate the underlying pathomechanisms.
3. Resource details
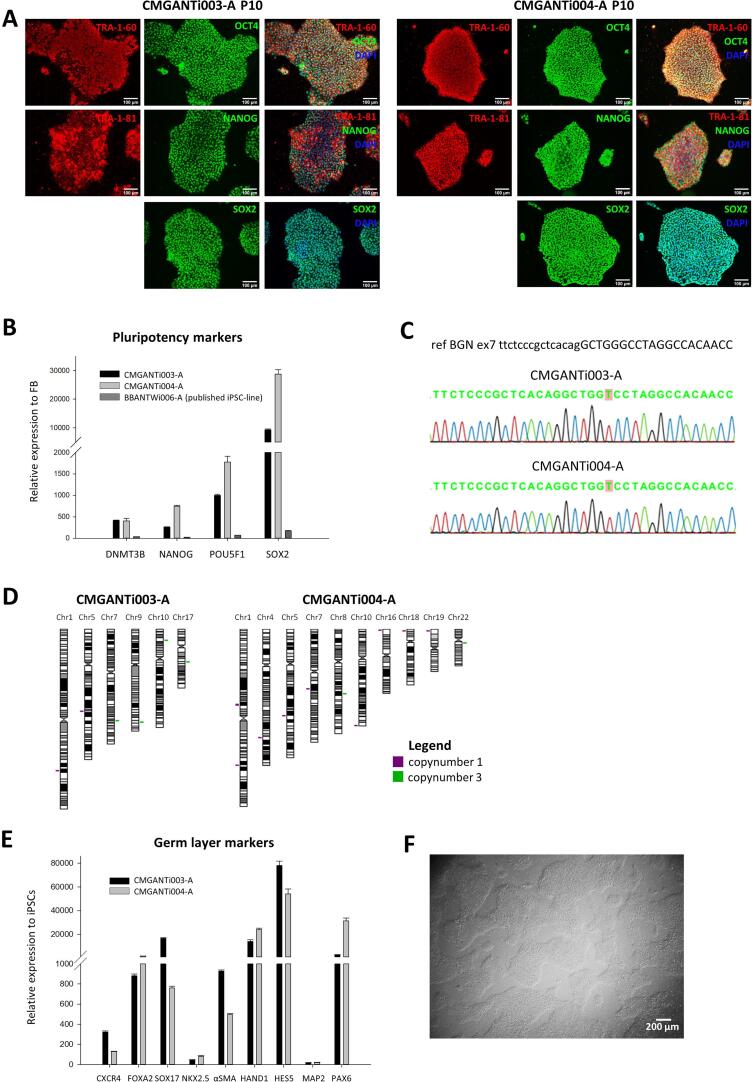
Specific hemizygous missense variants in the X-linked BGN gene, encoding the extracellular matrix protein biglycan, cause spondyloepimetaphyseal dysplasia (SEMD, biglycan type) (Cho et al., 2016), while loss-of-function pathogenic variants in this gene have been linked to an aortopathy syndrome called Meester-Loeys Syndrome (Meester et al., 2017). Clinical features of SEMD, biglycan type include short stature, brachydactyly and osteoarthritis. Some symptoms can be treated with surgery (e.g. short limbs). However, this is not without complications (e.g. risk of nerve injury), comes with high medical costs and is associated with a painful revalidation. There is thus clearly a need for curative treatments addressing the underlying pathophysiology. Little is known about the pathomechanisms underlying SEMD, biglycan type. To improve the current understanding, it is key to develop a representative human disease model. Induced pluripotent stem cell (iPSC)-derived chondrocyte disease models have been shown to exhibit several key aspects of the known disease mechanisms of skeletal dysplasias and are therefore considered highly suitable. Prior to creating iPSC-chondrocyte disease models, iPSCs need to be generated. In this article, we introduce two SEMD, biglycan iPSC-lines of two Italian brothers both carrying a missense variant in the BGN gene (p.Gly259Val). Dermal fibroblasts of the two male SEMD, biglycan type patients were reprogrammed into iPSCs using the CytoTyneTM-iPS 2.0 Sendai Kit (Invitrogen). This kit contains three Sendai viral reprogramming vectors delivering and expressing the key genetic factors OCT3/4, SOX2, KLF4 and c-MYC necessary for iPSC generation from somatic cells. Pluripotency of the resulting iPSCs was confirmed using immunocytochemistry (ICC) for the pluripotency markers OCT4, SOX2, NANOG, TRA-1–60 and TRA-1–81 (Fig. 1, A) and real-time quantitative polymerase chain reaction (RT-qPCR) for expression levels of NANOG, POU5F1, DNTM3B and SOX2 (Fig. 1, B). The iPSCs were able to differentiate into the three germ layers, i.e. ectoderm, mesoderm and endoderm, which was proven using RT-qPCR for appropriate markers of the respective germ layers (Fig. 1, E). Presence of the pathogenic variant in both iPSC lines was confirmed using Sanger sequencing (Fig. 1, C). Copy number variation (CNV) analysis using single nucleotide polymorphism (SNP) arrays verified genomic identity between the created iPSC clones and the original fibroblast cell line (Table 3). Genomic stability of the iPSC clones and the original fibroblasts was also investigated by a CNV analysis. No indels were observed in genes described in the ‘Nosology and classification of genetic skeletal disorders: 2019 revision’ of Mortier et al. (Fig. 1, D (duplications in green, deletions in purple) and Supplementary file 1) (Mortier et al., 2019). Therefore, it can be concluded that no clinically relevant CNVs were introduced during the reprogramming process. A more detailed overview of these CNVs and the involved genes can be found in Supplementary file 1. Note that this SNP array is not able to detect balanced rearrangements and low-level mosaicism. Furthermore, the iPSC clones were free of the Sendai viral backbone (Supplementary Figure 1) and mycoplasma contamination (Supplementary Figure 2, Supplementary Figure 3). In conclusion, we have successfully established two patient iPSC lines as a first step in the creation of iPSC-chondrocyte models to study and therapeutically target the disease mechanisms underlying SEMD, biglycan type.
Fig. 1.
Characterization of iPSC-line CMGANTi003-A and CMGANTi004-A.
Table 3.
Cell line identity testing.
| iPSC line | total count | correct count | errors | % identical |
|---|---|---|---|---|
| CMGANTi003-A P10 | 288,134 | 288,131 | 3 | >99.9 % |
| CMGANTi004-A P10 | 287,779 | 287,769 | 10 | >99.9 % |
4. Materials and methods
4.1. Fibroblast origin & culture and iPSC reprogramming
Dermal fibroblasts of the two SEMD, biglycan type patients were acquired from the Galliera Genetic Bank (Baldo et al., 2016). They were cultured in RPMI medium (Life Technologies) supplemented with 15 % FBS (Life Technologies), 1 % sodium pyruvate (Life Technologies), 100 U/mL Pen/Strep (Life Technologies) and 0.1 % primocin (InvivoGen Europe). The fibroblasts were reprogrammed into iPSCs using the CytoTuneTM-iPS 2.0 Sendai Kit (Life Technologies) according to the manufacturer’s protocol. In short, fibroblasts were transduced by three reprogramming vectors, which express the key genetic factors necessary for iPSC generation (i.e. OCT3/4, SOX2, KLF4 and c-MYC). Seven days after transduction, the cells were transferred to Matrigel coating (Corning) and 24 h later the medium was changed into iPSC medium. IPSC colonies were manually picked and further expanded by passaging the cells as small clumps every 4–5 days (1:5 ratio) using 0.02 % EDTA in Essential 8TM Flex medium (Life Technologies) supplemented with RevitaCell (Life Technologies) on Matrigel-coated dishes at 37 °C, 5 % CO2, 5 % O2.
4.2. Immunocytochemistry
After reprogramming, the iPSCs (passage 10) were cultured on coverslips and fixed with 100 % methanol (20′, −20 °C). Then, they were permeabilized using 0.1 % Triton X-100 solution (Sigma-Aldrich) (15′, room temperature (RT)). Non-specific binding was blocked using 5 % goat serum (Jackson ImmunoResearch) (30′, RT) and the primary antibodies were added and incubated overnight (4 °C). Hereafter, the cells were washed using 0.1 % Triton X-100 and secondary antibodies were incubated for one hour (RT). DAPI (Life Technologies) was used to visualize the cell nuclei. Coverslips were mounted on glass slides and pictures were taken using a 20x objective from Olympus BX51 fluorescence microscope.
4.3. Quantitative pluripotency marker analysis
RNA was extracted from both fibroblast and iPSC cell pellets (passage 10) using the Quick-RNATM Miniprep Kit (ZYMO Research). Subsequently, cDNA was synthesized using the SuperScriptTM III First-Strand Synthesis System (Life Technologies). Expression of the selected pluripotency markers (Table 1) was confirmed using RT-qPCR TaqMan® probes (Life Technologies) (Table 2) using a BioRad CFX384 Real-Time system (50 °C 2′, 95 °C 10′, 40x (95 °C 15′', 60 °C 1′)).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography Bright field | Normal | Fig. 1 panel F |
| Phenotype | Qualitative analysis (Immunocytochemistry) |
Staining/expression of pluripotency markers: Oct3/4, Nanog, Sox2, Tra1-60, Tra1-80. | Fig. 1 panel A |
| Quantitative analysis (RT-qPCR) | Expression of DNMT3B, NANOG, POU5F1 and SOX2 | Fig. 1 panel B | |
| Genotype | HumanCytoSNP-12 array | Resolution 72 kb, no major copy number variations | Fig. 1 panel D |
|
Identity |
HumanCytoSNP-12 array OR |
> 99.9 % identical SNPs | Table 3 |
| STR analysis | N/A | N/A | |
|
Mutation analysis (IF APPLICABLE) |
Sequencing | Hemizygous BGN c.776G > T | Fig. 1 panel C |
| Southern Blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma |
Negative | Supplementary Fig. 2, Supplementary Fig. 3 |
| Differentiation potential | Trilineage differentiation | Expression of appropriate markers of the respective germ layers, i.e. ectoderm, mesoderm and endoderm. | Fig. 1 panel E |
| List of recommended germ layer markers | Expression of these markers has to be demonstrated at mRNA (RT PCR) or protein (IF) levels, at least 2 markers need to be shown per germ layer | Endoderm: CXCR4, FOXA2, SOX17 Mesoderm: NKX2.5, αSMA (ACTA2), HAND1 Ectoderm: HES5, MAP2, PAX6 |
Fig. 1 panel E |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A |
Table 2.
Reagents details.
|
Antibodies used for immunocytochemistry/flow-cytometry |
||||
|---|---|---|---|---|
| Antibody | Dilution | Company Cat # | RRID | |
| Pluripotency Markers | Mouse anti-TRA1-60 | 1:200 | Cell Signaling Technology Cat#4746S | AB_2119059 |
| Rabbit anti-OCT4 | 1:100 | Thermo Fisher Scientific Cat#PA596860 | AB_2808662 | |
| Rabbit anti-SOX2 | 1:500 | Merck Millipore Cat#AB5603 | AB_2286686 | |
| Mouse anti-TRA1-81 | 1:200 | Cell Signaling Technology Cat#4745S | AB_2119060 | |
| Rabbit anti-NANOG | 1:500 | ThermoFisher Scientific Cat#PA1-097 | AB_2539867 | |
| Secondary antibodies |
AF555 Goat anti-Mouse, IgM | 1:500 | Thermo Fisher Scientific Cat#A21426 | AB_2535847 |
| AF488 Goat anti-Rabbit, IgG | 1:500 | Thermo Fisher scientific Cat#A11034 | AB_2576217 | |
|
Primers |
||||
| Target | Size of band | Forward/Reverse primer (5′-3′) | ||
| Pluripotency Markers (RT-qPCR) | DNMT3B | 55 bp | Hs00171876_m1 | |
| NANOG | 99 bp | Hs04260366_g1 | ||
| POU5F1 | 77 bp | Hs04260367_gH | ||
| SOX2 | 91 bp | Hs01053049_s1 | ||
| House-Keeping Genes (RT-qPCR) | GAPDH | 93 bp | Hs02758991_g1 | |
| ACTB | 63 bp | Hs01060665_g1 | ||
| Differentiation markers (RT-qPCR) | CXCR4 | 153 bp | Hs00607978_s1 | |
| FOXA2 | 66 bp | Hs00232764_m1 | ||
| SOX17 | 149 bp | Hs00751752_s1 | ||
| NKX2.5 | 64 bp | Hs00231763_m1 | ||
| αSMA (ACTA2) | 105 bp | Hs00426835_g1 | ||
| HAND1 | 54 bp | Hs00231848_m1 | ||
| HES5 | 62 bp | Hs01387463_g1 | ||
| MAP2 | 98 bp | Hs00258900_m1 | ||
| PAX6 | 76 bp | Hs00240871_m1 | ||
| Targeted mutation sequencing | BGN c.776G > T | 319 bp | GTTTTCCCAGTCACGACAAGGGTGATGCCAGAGTCC/ CAGGAAACAGCTATGACGACTGAGGGACTGCCCG | |
| Sendai virus Plasmids (PCR) | SeV | 181 bp | GGATCACTAGGTGATATCGAGC/ACCAGACAAGAGTTTAAGAGATATGTATC | |
| KOS | 528 bp | ATGCACCGCTACGACGTGAGCGC/ ACCTTGACAATCCTGATGTGGyc | ||
| Klf4 | 410 bp | TTCCTGCATGCCAGAGGAGCCC/AATGTATCGAAGGTGCTCAA | ||
| c-Myc | 532 bp | TAACTGACTAGCAGGCTTGTCG/ TCCACATACAGTCCTGGATGATGATG | ||
4.4. SNP array (CNV analysis)
Genomic DNA was isolated from the patients’ respective fibroblast and iPSC pellets (passage 10) using the Maxwell® RSC Instrument and Maxwell® RSC Cultured Cells DNA Kit (Promega) according to manufacturer’s protocol. Subsequently, a HumanCytoSNP-12 assay (Illumina) was performed according to the Infinium HD Assay Ultra Automated Protocol using an iScan System (Illumina). The obtained data was analysed in CNV-WebStore, an in-house developed online platform to analyse and interpret microarray data, to investigate the presence of CNVs between the original cell line and the created iPSC clones (Vandeweyer et al., 2011).
4.5. Sanger sequencing
In the genomic DNA of patient iPSCs (passage 10) and dermal fibroblasts, exon 7 of BGN was amplified by a Touchdown PCR (94 °C 3′, 10x (94 °C 5′, 65 °C (Δ-0.5) 15′', 72 °C 15′'), 25x (94 °C 5′, 55 °C 15′', 72 °C 15′'), 72 °C 1′)) using a Verity Thermal Cycler (Applied Biosystems). Prior to Sanger sequencing, the PCR products were purified using calf intestinal alkaline phosphatase (Merck) and Exonuclease I (BioLabs). Presence of the causal mutation was verified by Sanger sequencing reactions on an ABI 3130XL Genetic Analyzer system (Applied Biosystems) according to the standard protocol.
4.6. Mycoplasma test
The absence of mycoplasma in iPSC culture medium was verified using the LookOut Mycoplasma PCR Detection Kit (Sigma-Aldrich) according to the standard protocol.
4.7. Trilineage differentiation and analysis
To proof pluripotency, iPSCs (CMGANTi003-A passage 16 and CMGANTi004-A passage 15) were differentiated into the three embryonic germ layers (mesoderm, endoderm and ectoderm) using the StemMACS Trilineage Differentiation Kit (Miltenyi Biotec) according to manufacturer’s protocol at 37 °C, 5 % CO2, 20 % O2. On day seven, cells were collected for RNA extraction and cDNA synthesis. Expression of the selected germ layer markers (Table 1) was verified using RT-qPCR as described above (Table 2).
4.8. Sendai virus detection
RNA of the iPSCs (passage 10) was extracted and cDNA was synthesized as described above. Absence of the Sendai virus backbone was verified with PCR (94 °C 5′, 94 °C 15′', 34x (60 °C 30′'), 72 °C 45′', 72 °C 10′) and agarose gel electrophoresis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge that dermal fibroblasts of the two SEMD, biglycan patients were acquired from the Galliera Genetic Bank. The research was supported by funding from the University of Antwerp (Methusalem-OEC grant “Genomed” FFB190208). PDK (1S46323N) is a predoctoral FWO fellow, JM (12X8520N) and SP (12X5422N) are postdoctoral FWO fellows. BL holds a consolidator grant from the European Research Council (Genomia – ERC-COG2017-771945) and we also acknowledge partial funding from the University of Antwerp IOF-SBO brain organoid project granted to PP.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2023.103024.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.

Loss of Sendai viral vector.
Supplementary figure 2.

Absence of mycoplasma CMGANTi003-A.
Supplementary figure 3.
Absence of mycoplasma CMGANTi004-A.
References
- Baldo C., Viotti V., Maioli E., Mogni M., Castagnetta M., Cavani S., Piombo G., Coviello D. Galliera Genetic Bank: A DNA and Cell Line Biobank from Patients Affected by Genetic Diseases. Open Journal of Bioresources. 2016;3 [Google Scholar]
- Cho S.Y., Bae J.S., Kim N.K.D., Forzano F., Girisha K.M., Baldo C., Faravelli F., Cho T.J., Kim D., Lee K.Y., Ikegawa S., Shim J.S., Ko A.R., Miyake N., Nishimura G., Superti-Furga A., Spranger J., Kim O.H., Park W.Y., Jin D.K. BGN Mutations in X-Linked Spondyloepimetaphyseal Dysplasia. Am J Hum Genet. 2016;98:1243–1248. doi: 10.1016/j.ajhg.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meester J.A., Vandeweyer G., Pintelon I., Lammens M., Van Hoorick L., De Belder S., Waitzman K., Young L., Markham L.W., Vogt J., Richer J., Beauchesne L.M., Unger S., Superti-Furga A., Prsa M., Dhillon R., Reyniers E., Dietz H.C., Wuyts W., Mortier G., Verstraeten A., Van Laer L., Loeys B.L. Loss-of-function mutations in the X-linked biglycan gene cause a severe syndromic form of thoracic aortic aneurysms and dissections. Genet Med. 2017;19:386–395. doi: 10.1038/gim.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- G.R. Mortier, D.H. Cohn, V. Cormier-Daire, C. Hall, D. Krakow, S. Mundlos, G. Nishimura, S. Robertson, L. Sangiorgi, R. Savarirayan, D. Sillence, A. Superti-Furga, S. Unger, M.L. Warman, Nosology and classification of genetic skeletal disorders: 2019 revision, Am J Med Genet A, 179 (2019) 2393-2419. [DOI] [PubMed]
- Vandeweyer G., Reyniers E., Wuyts W., Rooms L., Kooy R.F. CNV-WebStore: online CNV analysis, storage and interpretation. BMC Bioinformatics. 2011;12:4. doi: 10.1186/1471-2105-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.