ABSTRACT
Background
Idiopathic calcium nephrolithiasis (ICN) is a common condition with a complex phenotype influenced by both environmental and genetic factors. In our study we investigated the association of allelic variants with the history of nephrolithiasis.
Methods
We genotyped and selected 10 candidate genes potentially related to ICN from 3046 subjects participating in the INCIPE survey cohort (Initiative on Nephropathy, of relevance to public health, which is Chronic, possibly in its Initial stages, and carries a Potential risk of major clinical End-points), a study enrolling subjects from the general population in the Veneto region in Italy.
Results
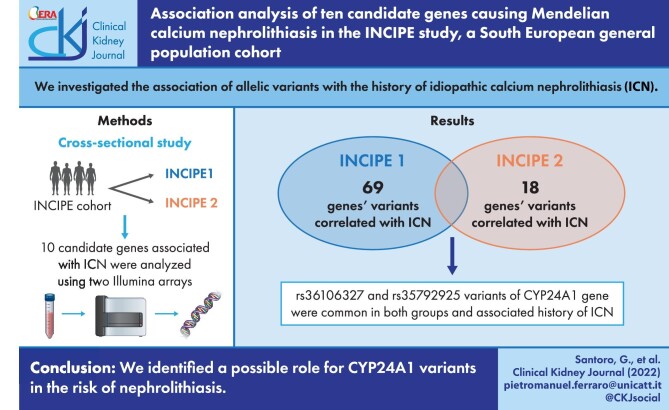
Overall, 66 224 variants mapping on the 10 candidate genes were studied. A total of 69 and 18 variants in INCIPE-1 and INCIPE-2, respectively, were significantly associated with stone history (SH). Only two variants, rs36106327 (chr20:54 171 755, intron variant) and rs35792925 (chr20:54 173 157, intron variant) of the CYP24A1 gene were observed to be consistently associated with ICN. Neither variant has been previously reported in association with renal stones or other conditions. Carriers of CYP24A1 variants showed a significant increase in the ratio of 1,25 (OH)2 vitamin D to 25 (OH) vitamin D compared with controls (P = .043). Although not associated with ICN in this study, the rs4811494 CYP24A1 variant that was reported to be causative of nephrolithiasis was very prevalent in heterozygosity (20%).
Conclusion
Our data suggest a possible role for CYP24A1 variants in the risk of nephrolithiasis. Genetic validation studies in larger sample sets will be necessary to confirm our findings.
Keywords: cross-sectional study, CYP24A1 gene, genetic variants, nephrolithiasis
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Idiopathic calcium nephrolithiasis (ICN) is a frequent condition occurring in 7–12% of individuals in the general population [1, 2]. It has been generally considered a complex phenotype influenced by both environmental and genetic factors [3]. Several observations support the role of the latter. A study has shown that the risk of calcium stones is significantly higher in consanguineous relatives than in spouses, with a decreasing risk moving from parents to 5th degree relatives [4]. A further study has shown in twins a higher disease status concordance of nephrolithiasis in monozygotic than in dizygotic twins [3].
It has been proposed that the ICN genetic risk is polygenic. Actually, genome-wide association studies (GWAS) have recognized the association of a number of genes with history of renal stones [5]. None of these studies was performed in southern Europe: those done in Europe included British and Icelandic cohorts [6, 7]. Japanese cohorts have also been investigated [6, 8, 9].
This study has been performed with the goal of investigating the association of several known candidate genes with nephrolithiasis in the INCIPE cohort (Initiative on Nephropathy, of relevance to public health, which is Chronic, possibly in its Initial stages, and carries a Potential risk of major clinical End-points). INCIPE is a general, Caucasian-only population cohort that was enrolled in 2006 to determine the prevalence of chronic kidney disease (CKD) in North-Eastern Italy [10]. This cohort is very different from the Caucasians (non-Hispanic whites) of NHANES to which it was previously compared, in terms of diabetes, obesity, metabolic syndrome, hypertension and CKD prevalence. Thus, we may argue that the nutritional habits are different and this might make for different GWAS results due to the existence of gene–environment interactions and their different genetic background.
A number of clinical variables, including data on medical history, were collected in the INCIPE study. In the present study, 10 candidate genes have been addressed. They were chosen among those causing some of the most frequent inherited forms of calcium nephrolithiasis (Table 1). We limited our analysis to genes potentially related to calcium handling, since calcium stones are the most prevalent in the general population [11].
Table 1:
Candidate genes.
| Gene | Gene coordinates chromosome:start–end (bp)a | Genotyped and imputed variants (number) | Associated disorder |
|---|---|---|---|
| XPR1 | Chr1:180 632 022–180 890 263 | 27 356 | Distal renal tubular acidosis Tubular phosphate handling |
| ATP6V1B1 | Chr2:70 934 905–70 966 388 | 3321 | Infantile hypercalcaemia Distal renal tubular acidosis (OMIM 267 300) |
| SLC34A1 | Chr5:177 379 263–177 398 841 | 2490 | Infantile hypercalcaemia Nephrolithiasis/osteoporosis, hypophosphatemic, 1 (OMIM 616 963 and 612 286) |
| ATP6V0A4 | Chr7:138 706 308–138 799 538 | 9819 | Hereditary hypophosphatemic rickets with hypercalciuria/Fanconi syndrome Distal renal tubular acidosis (OMIM 602 722) |
| SLC20A2 | Chr8:42 416 477–42 541 922 | 12 855 | Hereditary hypophosphatemic rickets with hypercalciuria/idiopathic infantile hypercalciuria/infantile hypercalcaemia Tubular phosphate handling |
| SLC34A3 | Chr9:137 230 762–137 236 550 | 1459 | Hereditary hypophosphatemic rickets with hypercalciuria (OMIM 241 530) |
| FGF23 | Chr12:4 368 228–4 379 698 | 1381 | Hereditary hypophosphatemic rickets with hypercalciuria/hereditary hypophosphatemia and nephrocalcinosis Hypophosphatemic rickets, autosomal dominant (OMIM 193 100) |
| SLC4A1 | Chr17:44 248 394–44 268 140 | 2343 | Hereditary hypophosphatemic rickets with hypercalciuria Distal renal tubular acidosis (OMIM 179 800 and 611 590) |
| SLC9A3R1 | Chr17:74 748 634–74 769 351 | 2722 | Infantile hypercalcaemia Nephrolithiasis/osteoporosis, hypophosphatemic, 2 (OMIM 612 287) |
| CYP24A1 | Chr20:54 153 484–54 173 979 | 2478 | Infantile hypercalcaemia, 1 (OMIM 143 880) |
aGene coordinates according to GRCh38 version of human genome reference sequence.
MATERIALS AND METHODS
The INCIPE cohort consists of 3870 Caucasian subjects ≥40 years old randomly chosen from the lists of citizens of 62 randomly selected general practitioners sited in four geographical areas (Verona, two in Padova, Venice) in the Veneto region, North-East Italy. Clinical information was collected from each participant.
Subjects were classified as hypertensive when they reported having received a diagnosis of hypertension and as diabetic when they reported having received a diagnosis of diabetes or when found to have fasting plasma glucose ≥126 mg/dL. Hypercholesterolemic individuals were those with blood cholesterol levels ≥240 mg/dL. For the metabolic syndrome criteria, we used those of the American Heart Association/National Heart, Lung, and Blood Institute Scientific Statements [12]. Estimated glomerular filtration rate (eGFR) was calculated with the Chronic Kidney Disease Epidemiology Collaboration equation.
Information on kidney stone history (SH) was self-reported.
Genetics
Two independent subgroups of individuals of the INCIPE cohort were previously genotyped using either the Illumina Human Omni Express chip array (893 individuals, INCIPE-1) [13] or the Illumina Human Core Exome chip array (2153 individuals, INCIPE-2) [14]. Both INCIPE-1 and INCIPE-2 samples were preprocessed to code gene variants to the same DNA strand and filter out problematic samples and markers, and then they independently underwent the genome-wide imputation process. Imputation was performed using the TOPMed imputation server (Trans-Omics for Precision Medicine) [15]. Only variants showing a high imputation correlation value (R2 > 0.8) were retained. The genotype profiles for all variants (imputed and genotyped) belonging to the candidate genes were extracted and, when possible, annotated with the Ensembl VEP and ClinVar Databases [16, 17]. Consanguinity across samples was very low. We assessed it through the runs of homozygosity (ROHs): the inbreeding coefficient Froh resulted equal to 0.0013 ± 0.0035 (mean ± SD) and 0.0011 ± 0.0032 in INCIPE-1 and INCIPE-2, respectively, lower than the average value in the European population (0.0098 ± 0.0051) [18].
Biochemistry
Frozen plasma at −80°C was thawed and the following parameters were determined in one session to reduce biological variability: calcium and phosphate, intact parathyroid hormone (PTH) (Elecsys PTH, Roche Diagnostics, Mannheim, Germany), 25 (OH) vitamin D and 1,25 (OH)2 vitamin D (Liaison kits, DiaSorin, Saluggia, Italy). Samples from all but one INCIPE-1 subjects with the two identified variants (chr20:54 171 755:C:A rs36106327 and chr20:54 173 157:G:A rs35792925) of the CYP24A1 gene, and paired-matched controls without the two variants in a proportion 1:2 were analyzed. The matching was for age (±2 years), gender, menopausal status and CKD stage.
Statistics
Associations of SH with continuous and categorical variables were tested using the Student's t-test or χ2 test, respectively. Generalized linear models were used to test the association when including into the model the most relevant covariates (Supplementary data, Table S1). Statistical significance with P-value <.05 was used to determine the clinical variables significantly associated with the outcome.
Genetic association analysis was performed on the genetic variants having an estimated minor allele frequency >0.01.
For each individual variant, the analysis was conducted by comparing the mean dosage value of the reference allele (additive genetic model) in cases and controls. Adjustment for multiple testing (all the single nucleotide polymorphisms from all the candidate genes) was done using a shrinkage t-test [19].
Linkage disequilibrium was investigated by the Linkage Disequilibrium Calculator of Ensembl [16].
The software R was used to perform the analysis. Association tests with the genetic factors were done using the ‘st’ library of the R package (CRAN) [19].
The biological impact of the variants that resulted statistically significant (<.05) was annotated by the Ensembl Variant Effect Predictor (VEP) [16].
INCIPE-1 and INCIPE-2 were the discovery and validation cohorts, respectively. In reference to genetic variants shown to be associated with history of renal stones SH, only those confirmed in the validation cohorts were considered as significant, i.e. consistently associated.
RESULTS
Prevalence of SH in the INCIPE cohort was 12.4% (8.5% in females, 15.2% in males). Compared with subjects who did not report stone history (NSH), those with SH had higher body mass index (BMI), waist circumference, blood pressure, insulin levels and lower high-density lipoprotein (HDL) cholesterol (Table 2). eGFR did not differ between the two groups (Table 2).
Table 2:
Anthropometric, clinical and biochemical parameters of the investigated subjects according to SH.
| NSH, n = 3386 | SH, n = 478 | P-value | |
|---|---|---|---|
| BMI, kg/m2 | 26.65 ± 5.42 | 26.96 ± 4.08 | .14 |
| Waist circumference, cm, mean (SD) | 92.90 ± 14.70 | 94.65 ± 13.75 | <.01 |
| Systolic blood pressure, mmHg, mean (SD) | 136.81 ± 20.13 | 139.08 ± 18.30 | .012 |
| Diastolic blood pressure, mmHg, mean (SD) | 84.65 ± 10.72 | 86.00 ± 9.52 | <.01 |
| Triglycerides, mmol/L, mean (SD) | 1.24 ± 0.95 | 1.28 ± 0.71 | .33 |
| HDL cholesterol, mmol/L, mean (SD) | 1.48 ± 0.33 | 1.43 ± 0.31 | <.01 |
| LDL cholesterol (Friedwald) mmol/L, mean (SD) | 3.84 ± 0.90 | 3.86 ± 0.93 | .894 |
| Fasting glucose, mmol/L, mean (SD) | 5.44 ± 1.42 | 5.57 ± 1.28 | .05 |
| Insulin, mIU/mL, mean (SD) | 9.95 ± 7.09 | 11.68 ± 11.89 | .006 |
| eGFR, mL/min/1.73 m2, mean (SD) | 88.62 ± 18.66 | 88.18 ± 18.74 | .63 |
| MS, n (%) | 22.2 | 24.5 | .291 |
eGFR is calculated by the Chronic Kidney Disease Epidemiology Collaboration formula.
MS, metabolic syndrome; LDL, low-density lipoprotein.
A total of 66 224 (genotyped and imputed) variants mapping on the 10 candidate genes were studied. Only a minority are reported in the ClinVar Archive (nearly 1500). Sixty-nine and 18 variants in INCIPE-1 and INCIPE-2, respectively, were significantly associated with SH (Supplementary data, Table S2). Among these only three variants are classified in ClinVar, one in INCIPE-1 (ATP6V0A4 rs3807154) and two in INCIPE-2 (SLC34A1 rs876661296, SLC34A3 rs28407527).
Two imputed variants [chr20:54 171 755:C:A (rs36106327) and chr20:54 173 157:G:A (rs35792925)] of the CYP24A1 gene were observed to be consistently associated with SH since they were associated in both INCIPE-1 and INCIPE-2 (Table 3). Both variants have not been previously reported in association with renal stones or other conditions and are not listed in the ClinVar database. The two variants resulted to be in absolute linkage disequilibrium (D = 1.0; R2 = 1.0).
Table 3:
Association between SH and CYP2A1 gene variants.
| Gene | Gene variant RS (reference SNP) code | Gene variant position (chrom:position:ref allele:alternative allele) | Alternative allele | Estimated alternative allele frequency (%) | Non-SH allele dosagea | SH allele dosagea | P-valueb |
|---|---|---|---|---|---|---|---|
| CYP24A1 | rs36106327 | chr20:54 171 755:C:A | A | 2.0 | 0.04 | 0.07 | .03 |
| CYP24A1 | rs35792925 | chr20:54 173 157:G:A | A | 2.0 | 0.041 | 0.08 | <.01 |
aAverage dosage of alternative allele in the sample group (range: 0–2).
bShrinked t-test P-value.
Among the 166 CYP24A1 variants that we investigated in INCIPE, the rs4811494 variant that was reported to be causative of nephrolithiasis [20] was observed in homozygosity in more than 2%, and in heterozygosity in approximately 20% (Supplementary data, Table S3). Ten other CYP24A1 variants that were investigated are reported in ClinVar as pathogenic or likely pathogenic, but the only one found in our cohorts, rs114368325 in 20 subjects, is always in heterozygosity without any association with SH (Supplementary data, Table S4).
Interestingly, the SLC34A1 variant (code rs876661296) associated with SH in INCIPE-2 has been reported in homozygousity as causing idiopathic infantile hypercalcemia [20], and also observed in stone formers [21]. No homozygous case for this variant was observed in our cohort (Supplementary data, Table S3).
In INCIPE-1, carriers of the two CYP24A1 variants shown in Table 3, all heterozygous, numbered 40 (4.5%). Although having lower 25 (OH) vitamin D, they had a ratio of 1,25 (OH)2 vitamin D to 25 (OH) vitamin D higher than in matched controls not carrying the two CYP24A1 variants (Table 4). These data do not seem to have been influenced by summer sun exposure since samples were all collected in the period November 2006 to April 2007.
Table 4:
Summary statistics of demographic and biochemical variables in individuals from INCIPE-1 carrying and non-carrying the two CYP24A1 variants shown to be associated with nephrolithiasis.
| Variables | Carriers, n = 38 | Non-carriers, n = 77 | P-value |
|---|---|---|---|
| Age, years, mean (SD) | 61.2 (14.2) | 63.4 (13.3) | .414 |
| Sex, male, n (%) | 15 (39.5) | 28 (36.4) | .905 |
| Ca, mg/dL, mean (SD) | 2.2 (0.1) | 2.2 (0.13) | .805 |
| P, mg/dL, mean (SD) | 0.9 (0.2) | 0.9 (0.2) | .775 |
| PTH, pmol/L, median (IQR) | 3.5 (2.7, 4.7) | 3.9 (2.9, 5.0) | .287 |
| eGFR, mL/min/1.73 m2, mean (SD) | 85.4 (18.4) | 84.1 (16.2) | .691 |
| 1,25 (OH)2 vitamin D, pmol/L, mean (SD) | 94.9 (25.26) | 99.3 (30.6) | .449 |
| 25 (OH) vitamin D, nmol/L, median (IQR) | 34.1 (23.4, 49.4) | 42.3 (27.8, 57.8) | .043 |
| Ratio 1,25 (OH)2 vitamin D/25 OH vitamin D, median (IQR) | 2.9 (2.0, 4.2) | 2.36 (1.6, 3.3) | .043 |
| Nephrolithiasis, n (%) | 12 (31.6) | 23 (29.9) | 1.000 |
Carriers are the subjects carrying the two CYP24A1 variants (shown in Table 3). Serum samples were not available in two cases. Non-carriers are subjects without the two variants.Ca, calcium; P, phosphate; IQR, interquartile range.
DISCUSSION
The main result of this study is the association of SH with two intronic variants (rs36106327 and rs35792925) of the CYP24A1 gene. These two variants were the only variants associated with SH in both INCIPE panels. Thus, genetic findings in the INCIPE cohort put the spotlight on CYP24A1. In the INCIPE cohort we also observed more than 2% homozygous subjects, and a very high prevalence of heterozygosity (approximately 20%) for the CYP24A1 rs4811494 variant, which was reported to be causative of nephrolithiasis [20]. Homozygosity for a number of mutated CYP24A1 variants has been previously reported to be high in the general population since it was estimated to be 1960 cases per 100 000 individuals [22]. While the high prevalence of the CYP24A1 rs4811494 variant in the general population raises the possibility that the observation of this variant (reported as c.*1327G > A, benign in ClinVar) (associated with hypercalcemia) in a 5-year-old male with hypercalciuria and renal stones was a chance association, functional studies support its pathogenic role [21].
Variants of the CYP24A1 gene have been shown to associate with renal stones in previous GWAS [6, 9]. The two variants that we discovered have been neither reported to be associated with nephrolithiasis nor described as pathologic, and are likely to belong to the same haplotype. However, we cannot exclude that one of them or both may play some functional unknown role. As previously observed, inactivating mutations of this gene cause idiopathic infantile hypercalcemia, a rare autosomal recessive disease presenting in the first months of life [22]. Since CYP24A1 encodes vitamin D 24-hydroxylase, the degrading enzyme of 1,25 (OH)2 vitamin D, its defective activity due to gene mutations leads in the overt cases to increased plasma calcitriol, severe hypercalcemia, nephrocalcinosis, nephrolithiasis and renal failure. Recently, the phenotypic spectrum of this disorder has been recognized to be wider than idiopathic infantile hypercalcemia. In fact, mutations of CYP24A1 have been found in subjects with recurrent nephrolithiasis starting in early adulthood, with hypercalciuria, serum calcium only mildly or intermittently increased [23–29], and high or marginally high 1,25 (OH)2 vitamin D [25].
The CYP24A1 gene sequencing of affected individuals together with functional studies would help in clarifying whether the observed association with nephrolithiasis could be due to some effect of the gene itself or to other rare unknown gene variants in linkage disequilibrium in that chromosomal region. We have some indirect information suggesting that the first option might be true. INCIPE subjects who carried the CYP24A1 variants did not differ in terms of eGFR in comparison with non-carriers. Furthermore, they did not show any difference in plasma levels of calcium, phosphate and PTH (Table 4). However, the two variants do not seem to be neutral on the vitamin D axis regulation. In fact, although having lower absolute levels of 25 (OH) vitamin D, cases, i.e. subjects who carried the variants, had a relative higher concentration of 1,25 (OH)2 vitamin D. This suggests that akin to the known disease-causing mutations of the CYP24A1 gene, the variants associated with the history of stones in the INCIPE cohort also jeopardize the degradation of 1,25 (OH)2 vitamin D. Therefore, we speculate that in certain circumstances (sun exposure, vitamin D supplementation) these subjects might develop high levels of calcitriol. This should increase intestinal absorption of calcium leading to hypercalciuria, thus predisposing to lithogenesis. It is self-evident that these subjects should avoid sun and UV ray exposure, and vitamin D supplements.
Given the observed distribution of the two CYP24A1 variants in subjects with SH and subjects with NSH (roughly 8% and 4%, respectively; see Table 3) and the prevalence of SH (∼13% in the INCIPE population) we can argue that the herein associated variants with SH are likely to be risk factors for nephrolithiasis, following a non-Mendelian model of inheritance [5]. This fits with the idea that nephrolithiasis is a complex disease. The idea is also supported by the observation that SH was observed in both carriers and non-carriers of the two CYP24A1 variants with roughly the same prevalence (31.6% and 29.9%, respectively), representing clear evidence that other genes together with several other conditions (nutritional, environmental) are involved in the common stone disease [5]. A less robust finding of our study was the association of the variant rs876661296 of the recessive SLC34A1 gene [30] with history of renal stones in INCIPE-2. Nevertheless, this result is interesting. Although in ClinVar this variant has been classified as benign, functional studies demonstrated that the codified protein, the proximal tubular sodium-dependent phosphate carrier 2A (NaPi-IIa), has a deranged function [20]; as a consequence, it was described in homozygosity as causing idiopathic infantile hypercalcemia [20]. There was no homozygous case for this variant; however, SLC34A1 single nucleotide polymorphisms have already emerged from GWAS, in Northern Europe and Asia, as associated with nephrolithiasis [6–8]. The physiopathological connection with nephrolithiasis probably flows from an altered phosphate reabsorption in the proximal tubule, and higher activation of vitamin D which stimulates intestinal absorption of calcium with the ensuing increased calciuria [31].
This study has limitations. History of stones was self-reported by participants and was not validated by confirmatory clinical records. However, we think that this self-reported diagnosis is in general terms reliable. Actually, we found in self-reported stone subjects the typical association observed in the general population between nephrolithiasis and the metabolic syndrome criteria (higher BMI, lower HDL cholesterol, higher blood pressure, higher insulin levels) [32, 33]. Furthermore, in a previous cohort study the self-reported diagnosis of stones was confirmed in 95% of cases [34]. We could not evaluate whether the history of stones was more or less severe in carriers of the CYP24A1 variants since we had no information on stone recurrence, metabolic activity, previous surgery or stone complications. INCIPE-1 and INCIPE-2 were genotyped with two different arrays. Hence the reported CYP24A1 association signals with SH are due to imputed and not really to genotyped variants. However, only the imputed variants which made it through the applied hard filtering (R2 > 80%) were selected. Due to the high level of accuracy and the statistical robustness of those variants, we expect the results reported above to be reliable.
CONCLUSIONS
Our data suggest for the first time a possible role for two new CYP24A1 variants in the risk of nephrolithiasis. It is reasonable to speculate an effect of these variants on the vitamin D axes regulation that might justify such an association. Genetic validation studies in larger sample sets and functional studies will be necessary to confirm our findings.
Supplementary Material
Contributor Information
Gloria Santoro, Division of Nephrology, Department of Medicine, University of Verona and Azienda Ospedaliera Universitaria Integrata, Verona, Italy; Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Catholic University of the Sacred Heart, Rome, Italy.
Gianmarco Lombardi, Division of Nephrology, Department of Medicine, University of Verona and Azienda Ospedaliera Universitaria Integrata, Verona, Italy.
Stefano Andreola, Division of Nephrology, Department of Medicine, University of Verona and Azienda Ospedaliera Universitaria Integrata, Verona, Italy.
Gian Luca Salvagno, University of Verona and Pederzoli Hospital, Peschiera del Garda, Verona, Italy.
Mirko Treccani, Section of Section of Biology and Genetics, Department of Neurosciences, Biomedicine and Movement Sciences, University of Verona, Verona, Italy.
Elena Locatelli, Section of Section of Biology and Genetics, Department of Neurosciences, Biomedicine and Movement Sciences, University of Verona, Verona, Italy.
Pietro Manuel Ferraro, Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Catholic University of the Sacred Heart, Rome, Italy; Division of Nephrology, Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Catholic University of the Sacred Heart, Rome, Italy.
Giuseppe Lippi, Central Laboratory, Department of Pathology and Diagnostics, University of Verona and Azienda Ospedaliera Universitaria Integrata, Verona, Italy.
Giovanni Malerba, Section of Section of Biology and Genetics, Department of Neurosciences, Biomedicine and Movement Sciences, University of Verona, Verona, Italy.
Giovanni Gambaro, Division of Nephrology, Department of Medicine, University of Verona and Azienda Ospedaliera Universitaria Integrata, Verona, Italy.
FUNDING
This study was not funded.
AUTHORS’ CONTRIBUTIONS
G.S., G.G. and G.M. contributed to the research idea and study design. G.S., S.A. and G.M. contributed to data acquisition. G.S., P.M.F., G.M.L. and G.G contributed to data analysis/interpretation. G.S., E.L. and M.T. contributed to statistical analysis. G.S., G.M.L., G.M. and G.G. drafted the paper. G.G., P.M.F., G.L. and G.L.S. contributed to supervision. Each author contributed important intellectual content during manuscript drafting for the overall work.
DATA AVAILABILITY STATEMENT
The data underlying this article will be shared on reasonable request to the corresponding author.
CONFLICT OF INTEREST STATEMENT
P.M.F. reports consultant/grant fees from Allena Pharmaceuticals, Alnylam, Amgen, AstraZeneca, BioHealth Italia and Vifor Fresenius. All the other authors report no disclosures.
REFERENCES
- 1. Scales CD, Smith AC, Hanley JMet al. Urologic Diseases in America Project. Prevalence of kidney stones in the United States. Eur Urol 2012;62:160–5. 10.1016/j.eururo.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Croppi E, Ferraro PM, Taddei Let al. Prevalence of renal stones in an Italian urban population: a general practice-based study. Urol Res 2012;40:517–22. 10.1007/s00240-012-0477-z [DOI] [PubMed] [Google Scholar]
- 3. Goldfarb DS, Fischer ME, Keich Yet al. A twin study of genetic and dietary influences on nephrolithiasis: a report from the Vietnam Era Twin (VET) registry. Kidney Int 2005;67:1053–61. 10.1111/j.1523-1755.2005.00170.x [DOI] [PubMed] [Google Scholar]
- 4. Edvardsson VO, Palsson R, Indridason OSet al. Familiality of kidney stone disease in Iceland. Scand J Urol Nephrol 2009;43:420–4. 10.3109/00365590903151479 [DOI] [PubMed] [Google Scholar]
- 5. Taguchi K, Yasui T, Milliner DSet al. Genetic risk factors for idiopathic urolithiasis: a systematic review of the literature and causal network analysis. Eur Urol Focus 2017;3:72–81. 10.1016/j.euf.2017.04.010 [DOI] [PubMed] [Google Scholar]
- 6. Howles SA, Wiberg A, Goldsworthy Met al. Genetic variants of calcium and vitamin d metabolism in kidney stone disease. Nat Commun 2019;10:5175. 10.1038/s41467-019-13145-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oddsson A, Sulem P, Helgason Het al. Common and rare variants associated with kidney stones and biochemical traits. Nat Commun 2015;6:7975. 10.1038/ncomms8975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Urabe Y, Tanikawa C, Takahashi Aet al. A genome-wide association study of nephrolithiasis in the Japanese population identifies novel susceptible loci at 5q35.3, 7p14.3, and 13q14.1. PLos Genet 2012;8:e1002541. 10.1371/journal.pgen.1002541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tanikawa C, Kamatani Y, Terao Cet al. Novel risk loci identified in a genome-wide association study of urolithiasis in a Japanese population. J Am Soc Nephrol 2019;30:855–64. 10.1681/ASN.2018090942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gambaro G, Yabarek T, Graziani MSet al. Prevalence of CKD in northeastern italy: results of the INCIPE study and comparison with NHANES. Clin J Am Soc Nephrol 2010;5:1946–53. 10.2215/CJN.02400310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lieske JC, Rule AD, Krambeck AEet al. Stone composition as a function of age and sex. Clin J Am Soc Nephrol 2014;9:2141–6. 10.2215/CJN.05660614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grundy SM, Cleeman JI, Daniels SRet al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute scientific statement. Circulation 2005;112:2735–52. 10.1161/CIRCULATIONAHA.105.169404 [DOI] [PubMed] [Google Scholar]
- 13. Boulygina EA, Borisov OV, Valeeva EVet al. Whole genome sequencing of elite athletes. Biol Sport 2020;37:295–304. 10.5114/biolsport.2020.96272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo Y, He J, Zhao Set al. Illumina human exome genotyping array clustering and quality control. Nat Protoc 2014;9:2643–62. 10.1038/nprot.2014.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das S, Forer L, Schönherr Set al. Next-generation genotype imputation service and methods. Nat Genet 2016;48:1284–7. 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Howe KL, Achuthan P, Allen Jet al. Ensembl 2021. Nucleic Acids Res 2021;49:D884–91. 10.1093/nar/gkaa942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Landrum MJ, Lee JM, Benson Met al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062–7. 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McQuillan R, Leutenegger A-L, Abdel-Rahman Ret al. Runs of homozygosity in European populations. Am J Hum Genet 2008;83:359–72. 10.1016/j.ajhg.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zuber V, Strimmer K. Gene ranking and biomarker discovery under correlation. Bioinformatics 2009;25:2700–7. 10.1093/bioinformatics/btp460 [DOI] [PubMed] [Google Scholar]
- 20. Schlingmann KP, Ruminska J, Kaufmann Met al. Autosomal-recessive mutations in SLC34A1 encoding sodium-phosphate cotransporter 2A cause idiopathic infantile hypercalcemia. J Am Soc Nephrol 2016;27:604–14. 10.1681/ASN.2014101025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Halbritter J, Baum M, Hynes AMet al. Fourteen monogenic genes account for 15% of nephrolithiasis/nephrocalcinosis. J Am Soc Nephrol 2015;26:543–51. 10.1681/ASN.2014040388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nesterova G, Malicdan MC, Yasuda Ket al. 1,25-(OH)2D-24 Hydroxylase (CYP24A1) deficiency as a cause of nephrolithiasis. Clin J Am Soc Nephrol 2013;8:649–57. 10.2215/CJN.05360512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schlingmann KP, Kaufmann M, Weber Set al. Mutations in CYP24A1 and idiopathic infantile hypercalcemia. N Engl J Med 2011;365:410–21. 10.1056/NEJMoa1103864 [DOI] [PubMed] [Google Scholar]
- 24. Colussi G, Ganon L, Penco Set al. Chronic hypercalcaemia from inactivating mutations of vitamin D 24-hydroxylase (CYP24A1): implications for mineral metabolism changes in chronic renal failure. Nephrol Dial Transplant 2014;29:636–43. 10.1093/ndt/gft460 [DOI] [PubMed] [Google Scholar]
- 25. Wolf P, Müller-Sacherer T, Baumgartner-Parzer Set al. A case of “late-onset” idiopathic infantile hypercalcemia secondary to mutations in the CYP24A1 gene. Endocr Pract 2014;20:e91–5. 10.4158/EP13479.CR [DOI] [PubMed] [Google Scholar]
- 26. Ferraro PM, Minucci A, Primiano Aet al. A novel CYP24A1 genotype associated to a clinical picture of hypercalcemia, nephrolithiasis and low bone mass. Urolithiasis 2017;45:291–4. 10.1007/s00240-016-0923-4 [DOI] [PubMed] [Google Scholar]
- 27. Molin A, Lemoine S, Kaufmann Met al. Overlapping phenotypes associated with CYP24A1, SLC34A1, and SLC34A3 mutations: a cohort study of patients with hypersensitivity to vitamin D. Front Endocrinol 2021;12:736240. 10.3389/fendo.2021.736240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaufmann M, Schlingmann K-P, Berezin Let al. Differential diagnosis of vitamin D-related hypercalcemia using serum vitamin D metabolite profiling. J Bone Miner Res 2021;36:1340–50. 10.1002/jbmr.4306 [DOI] [PubMed] [Google Scholar]
- 29. Molin A, Baudoin R, Kaufmann Met al. CYP24A1 mutations in a cohort of hypercalcemic patients: evidence for a recessive trait. J Clin Endocrinol Metab 2015;100:E1343–52. 10.1210/jc.2014-4387 [DOI] [PubMed] [Google Scholar]
- 30. Cogal AG, Arroyo J, Shah RJet al. Comprehensive genetic analysis reveals complexity of monogenic urinary stone disease. Kidney Int Rep 2021;6:2862–84. 10.1016/j.ekir.2021.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fearn A, Allison B, Rice SJet al. Clinical, biochemical, and pathophysiological analysis of SLC34A1 mutations. Physiol Rep 2018;6:e13715. 10.14814/phy2.13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taylor EN, Stampfer MJ, Curhan GC. Obesity, weight gain, and the risk of kidney stones. JAMA 2005;293:455–62. 10.1001/jama.293.4.455 [DOI] [PubMed] [Google Scholar]
- 33. West B, Luke A, Durazo-Arvizu RAet al. Metabolic syndrome and self-reported history of kidney stones: the National Health and Nutrition Examination Survey (NHANES III) 1988-1994. Am J Kidney Dis 2008;51:741–7. [DOI] [PubMed] [Google Scholar]
- 34. Taylor EN, Fung TT, Curhan GC. DASH-style diet associates with reduced risk for kidney stones. J Am Soc Nephrol 2009;20:2253–9. 10.1681/ASN.2009030276 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.