ABSTRACT
Background
Catheter-related bloodstream infections (CRBIs) remain a major cause of mortality in haemodialysis (HD) patients with central venous catheters (CVCs), especially because of the non-specific symptomatology and the delay in microbiological diagnosis with possible use of non-optimal empiric antibiotics. Moreover, empiric broad-spectrum antibiotics increase antibiotic resistance development. This study aims to evaluate the diagnostic performance of real-time polymerase chain reaction (rt-PCR) in suspected HD CRBIs compared with blood cultures.
Methods
A blood sample for rt-PCR was collected simultaneously with each pair of blood cultures for suspected HD CRBI. The rt-PCR was performed on the whole blood, without any enrichment stage and with specific DNA primers: 16S (universal bacterial), Staphylococcus spp., Staphylococcus aureus and mecA. Each successive patient with a suspected HD CRBI in the HD centre of Bordeaux University Hospital was included. Performance tests were used to compare the result obtained in each rt-PCR assay with its corresponding routine blood culture.
Results
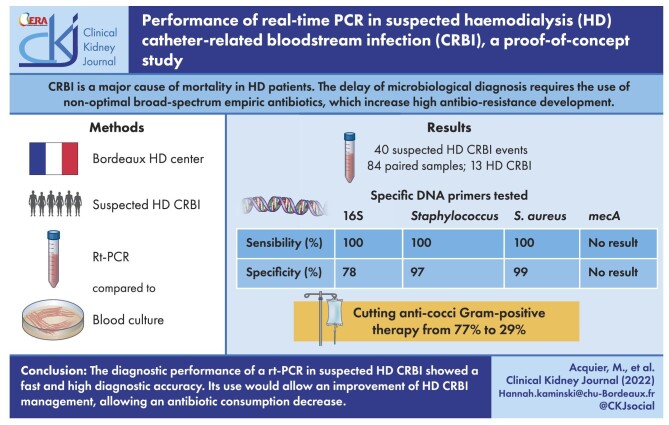
Eighty-four paired samples were collected and compared for 40 suspected HD CRBI events in 37 patients. Among these, 13 (32.5%) were diagnosed as HD CRBI. All rt-PCRs except mecA (insufficient number of positive samples) showed high diagnostic performances within 3.5 h: 16S (sensitivity 100%, specificity 78%), Staphylococcus spp. (sensitivity 100%, specificity 97%), S. aureus (sensitivity 100%, specificity 99%). Based on the rt-PCR results, antibiotics could be more appropriately targeted, thus cutting anti-cocci Gram-positive therapy from 77% to 29%.
Conclusions
The performance of rt-PCR in suspected HD CRBI events showed fast and high diagnostic accuracy. Its use would improve HD CRBI management with an antibiotic consumption decrease.
Keywords: bloodstream infection, haemodialysis, real-time PCR
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Patients treated by haemodialysis (HD) with a central venous catheter (CVC) as vascular access are at risk of infections [1]. Catheter-related bloodstream infections (CRBIs) in HD remains a major cause of morbidity and mortality [2]. It can result in life-threatening complications, including septic shock, endocarditis, septic arthritis, osteomyelitis and epidural abscess [3]. Identifying and treating CRBIs is particularly challenging due to the complex clinical features of such infections, which have a non-specific symptomatology [4]. CRBIs can be caused by Gram-positive and Gram-negative bacteria. The majority of HD CRBIs are caused by Staphylococcus spp., and Staphylococcus aureus is associated with increased morbidity and mortality [5, 6]. Moreover, patients treated with HD are at high risk of methicillin-resistant Staphylococcus (MRS) [7]. The implementation of empiric antibiotic therapy is based on Infectious Diseases Society of America guidelines published in 2010 proposing the combination of molecule targeting of MRS and a beta-lactam active agent on Pseudomonas aeruginosa [8]. The diagnostic difficulty leads to extensive use of broad-spectrum antibiotic therapy. Patients treated with HD are exposed to multiple antibiotic courses, thus increasing the risk of infections with multidrug-resistant organisms. Moreover, in patients with non-severe symptoms, a fast negative result for bacteraemia would avoid unnecessary hospitalization and antibiotic use.
Culture-based methods remain the gold standard to identify the causative microorganism in sepsis, with results available within 6–72 h (longer than an HD session) [9]. Technological developments in rapid non-culture-based infection diagnosis are now seen as crucial for a more rational use of antibiotic therapy [10]. Molecular non-culture approaches based on the detection of circulating bacterial DNA using real-time polymerase chain reaction (rt-PCR) can provide rapid detection of pathogens in blood [11]. Previous studies looking into the use of rt-PCR in bloodstream infections (BSIs) have shown encouraging positive results on mortality reduction and antimicrobial ecology [12]. In recent years, few studies have been published to evaluate the diagnostic performance of non-enriched whole blood rt-PCR in specific populations compared with the reference blood culture test [13–15]. This study aims to evaluate the feasibility, diagnostic performance and potential contributions of rt-PCR in suspected HD CRBIs compared with the standard-of-care blood culture.
MATERIALS AND METHODS
Study setting
This was a descriptive transversal prospective study performed between October 2018 and October 2019 in Bordeaux University Hospital comparing the diagnostic performance of rt-PCR and blood culture in the diagnosis of HD CRBIs (NCT04026035). All participants gave their verbal consent. A protective committee evaluated and approved the study. This committee stated that our study was a non-interventional study because of the small quantity of blood additionally sampled and allowed us to have solely verbal non-opposition consent.
Participants
The inclusion criteria were patients >18 years old with suspected HD CRBI. Exclusion criteria were age < 18 years and patients passing through the emergency department with antimicrobial therapy received for >8 h. The suspicion of HD CRBI was left to the judgment of the clinician. CVC could be tunnelled or not.
Intervention
After informing the patient, an additional blood sample [7 ml ethylenediaminetetraacetic acid (EDTA) blood collection] was taken concomitantly with those taken as part of the standard management of suspected HD CRBI. The clinical samples were collected in the ward and were sent to both the research laboratory and the routine diagnostic laboratory for processing (Laboratory of Bacteriology of Bordeaux University Hospital). Once collected, the EDTA blood collection was labelled to make it anonymous, then transported to the Aquitaine Microbiology laboratory by staff of the Nephrology Transplantation Dialysis Department of Bordeaux University Hospital. The results of the study technique were compared with the reference technique, i.e. blood culture. The samples used for research were destroyed at the end of the analysis.
Routine diagnostic approach
The standard blood culture sample (15 ml for aerobic and anaerobic bottles) procedure in the routine diagnostic laboratory for suspected HD CRBI is the following: if suspected prior to HD connection, a pair of blood cultures is collected from the venous and the arterial lines of the catheter, associated with a pair of blood cultures from the peripheral vein if possible; if suspected during the session, a pair of blood cultures are collected from the HD circuit [16].
For one suspected CRBI event, one to five pairs of blood cultures were sent to the laboratory and therefore one to five rt-PCRs were performed and compared with the concomitantly collected blood culture pair. In our centre, the microbiological diagnosis of BSI was obtained with blood culture: Gram stain and culture for species identification and antibiotic susceptibility testing.
The blood culture bottles (BacT/Alert FA Plus and BacT/Alert FN Plus) were placed in an automated system (BacT/Alert; all from bioMérieux, Marcy l'Etoile, France) that detects bacterial growth. As soon as the blood culture was positive, a Gram stain was performed and species identification was carried out with mass spectrometry using the MALDI-TOF Biotyper system (Brucker, Bremen, Germany) after a 3-h incubation of the blood culture on a chocolate agar plate. Antibiotic susceptibility was determined using the Phoenix automated system (BD Diagnostics, le Pont de Claix, France) or by agar diffusion, following the recommendations of the Antibiogram Committee of the French Society of Microbiology. The total duration of the technique to obtain the antibiogram depends on the time of positivity of the blood culture, classically between 24 and 96 h.
rt-PCR design in the research laboratory
In parallel with each pair of blood cultures according to the above protocol, a 7-ml EDTA blood tube was collected. From the sample, without a culture or enrichment phase, total DNA was extracted with the QIAamp DNA Blood Mini Kit (Qiagen, Courtaboeuf, France), then the amount of DNA was adjusted with nanodrops in order to reach a DNA concentration of 20 ng/μl (dilution in sterile milliQ water or sterile diethylpyrocarbonate water).
A panel of four sets of primers was used in the PCR reaction: 16S rDNA [17], Staphylococcus DNA [18], S. aureus DNA [19] and mecA DNA [20]. The primers used were ordered from Eurofins MGW Operon (Luxembourg) and are referenced. A total of 15 μl of the reagent mix was placed in each rt-PCR tube, then a 5-μl DNA sample was deposited for a final volume of 20 μl per tube. The individual tubes were deposited into the wells of the CFX 96 thermal cycler (Bio-Rad, Marnes-la-Coquette, France). Once the program was completed, the amplification and melting curves obtained were analysed using the CFX 96 software. It is therefore a SYBR Green technique with the use of melting curves to improve the specificity of the technique. The melting temperature (Tm) for the Staphylococcus spp. primers was 75°C. The Tm of the expected products with the S. aureus and mecA primers was 77–78°C. For the pair of primers targeting the bacterial 16S rDNA, the Tm was 85°C. A duplicate was systematically performed. The following sequence was used for each rt-PCR run: 98°C for 2 min once, 98°C for 5 sec, 56°C for 30 sec × 40 cycles. For the melting curve, the temperature was increased from 65° to 95°C with an increment of 0.5°C per 5 sec.
For each sample, a negative control was analysed adding water to the reaction mix. This manipulation also allowed us to check the non-contamination of the reaction mixes. A positive control of each rt-PCR was performed using a strain from an American Type Culture Collection (Manassas, VA, USA) for each patient series performed at the same time.
Positive cut-off for rt-PCR results
The rt-PCR results were expressed as an exponential curve of amplicon expansion. The cycle threshold (Ct) of positivity was set, depending on our analysis, ≤28 cycles. Considering the different curves obtained from the negative controls, where the expansion always started after 30 cycles, and the results of the rt-PCR of the samples, which were either <30 cycles or equal to 28 cycles, we fixed the threshold to 28 cycles. This setting allowed at least two cycles to differentiate the positive samples from the negative ones.
Statistical analysis
The results of both diagnostic methods were compared. A full-blind/blinded sample analysis was carried out without any knowledge of the results obtained in the routine diagnostic laboratory. In addition, the time necessary to obtain results in both approaches was assessed.
The sample size was not defined beforehand, as this was a proof-of-concept study. Qualitative variables are described in terms of numbers and percentages and quantitative variables are described in terms of number, mean, standard deviation (SD), median and quartiles. Diagnostic performances were evaluated by calculating sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) and by analysing the interrater reliability between rt-PCR and blood culture. The reference method was the blood culture. The interrater reliability between the two tests was assessed with the κ coefficient.
RESULTS
Patient demographics and study feasibility
A total of 40 suspected HD CRBI events were identified in 37 patients (3 patients had 2 different events). During the course of those 40 suspected HD CRBIs, 84 pairs of blood cultures were collected and 84 concomitant samples were analysed by rt-PCR. Among the 84 blood cultures, 27 (32%) were positive. Among the 40 suspected events, 23 (57.5%) had at least one positive blood culture. Three had only one positive blood culture for coagulase-negative Staphylococcus and were judged by the clinician to be contamination (single positive blood culture with negative controls, a growth time >48 h and a clinical improvement of patient without antibiotic treatment) and one BSI from the urinary tract was diagnosed. A total of 13 (32.5%) diagnoses of HD CRBI were made: 12 were related to a tunnelled catheter and 1 was related to a non-tunnelled catheter. The baseline characteristics of the patients are reported in Table 1. Identified microorganisms are described in the supplementary material.
Table 1:
Baseline characteristics.
| Characteristics | n |
|---|---|
| Patients | 37 |
| Suspicion of HD CRBI | 40 |
| Sample for rt-PCR | 84 |
| BSI | 14 |
| HD CRSI | 13 |
| Positive blood cultures with diagnosis of contamination | 3 |
| Empiric antimicrobial therapy | 19 |
| Empiric coverage for Gram-negative organism | 18 |
| Empiric coverage for methicillin-resistant Staphylococcus spp. | 12 |
Diagnostic performance of rt-PCR versus blood culture
The median time needed to obtain results was 3.5 h (IQR: 3 h 10 min–3 h 50 min) for rt-PCR and 12 h (IQR 9.75–21) for blood culture Gram stain.
The diagnostic performance of each rt-PCR is summarized in Tables 2–4. Supplemental Fig. 1 shows examples of three different results and their interpretation. Unfortunately, performances of the mecA amplification could not be evaluated due to the small number of positive blood cultures for MRS (n = 1).
Table 2:
Evaluation of rt-PCR for 16s.
| Result | Blood culture positive | Blood culture negative | Total |
|---|---|---|---|
| rt-PCR positive | 27 | 13 | 40 |
| rt-PCR negative | 0 | 44 | 44 |
| Total | 27 | 57 | 84 |
Table 4:
Evaluation of rt-PCR for Staphylococcus aureus.
| Result | Blood culture positive | Blood culture negative | Total |
|---|---|---|---|
| rt-PCR positive | 6 | 1 | 7 |
| rt-PCR negative | 0 | 77 | 77 |
| Total | 6 | 78 | 84 |
Table 3:
Evaluation of rt-PCR for Staphylococcus spp.
| Result | Blood culture positive | Blood culture negative | Total |
|---|---|---|---|
| rt-PCR positive | 13 | 2 | 15 |
| rt-PCR negative | 0 | 69 | 69 |
| Total | 13 | 71 | 84 |
Table 5:
Comparison of the use of antimicrobial therapy with or without rt-PCR.
| Empiric therapy | Clinician judgment | Based on rt-PCR |
|---|---|---|
| No use of empiric antimicrobial therapy, n | 21 | 18 |
| In cases of no BSI, n/N (%) | 18/21 (86) | 18/18 (100) |
| In cases of BSI, n/N (%) | 3/21 (14) | 0/18 (0) |
| Empiric antimicrobial therapy, n | 19 | 22 |
| In cases of no BSI, n/N (%) | 8/19 (42) | 8/22 (36) |
| In cases of BSI, n/N (%) | 11/19 (58) | 14/22 (64) |
| Empiric coverage for Gram-negative organism, n | 18 | 15 |
| In cases of no Gram-negative BSI, n/N (%) | 12/18 (67) | 8/15 (53) |
| In cases of Gram-negative BSI, n/N (%) | 6/18 (33) | 7/15 (47) |
| Empiric coverage for Gram-positive BSI, n | 13 | 7 |
| In cases of no Gram-positive BSI, n/N (%) | 10/13 (77) | 2/7 (29) |
| In cases of Gram-positive BSI, n/N (%) | 3/13 (23) | 5/7 (71) |
The 16S rt-PCR showed excellent sensitivity (100%), with no false negatives, resulting in an excellent NPV of 1. On the other hand, the presence of 10 false positives resulted in a decreased specificity (77%) and a positive predictive value of 0.72. The κ coefficient representing the interrater reliability of the two methods was 0.72, leading to strong agreement.
The Staphylococcus spp. rt-PCR showed excellent diagnostic performance, with sensitivity and NPV at 1, as well as a very good specificity (98%) and PPV (0.92). The κ coefficient was 0.95, which corresponds to very strong agreement.
The S. aureus rt-PCR also had a sensitivity of 1 and an NPV of 1. There were no false negatives in the 64 samples with blood cultures that did not contain S. aureus. The specificity was excellent at 0.98, with a slightly poorer PPV at 0.86 in view of the few S. aureus found in total (n = 6). The κ coefficient was 0.91, corresponding to very strong agreement.
Interestingly, when analysing the agreements by events and not by samples, 34 of 40 events had 100% agreement between rt-PCR and blood culture. Among three events with two samples, the two pairs of blood cultures were discordant but the rt-PCRs were concordant with their paired blood culture. For three events with a negative blood culture, 16S rt-PCR had a false-positive result (one of two samples for the first event, one of three samples for the second event and one of five samples for the last event).
Potential added value of rt-PCR
We first focused on the potential contribution of rt-PCR regarding the time needed to obtain results, because it should, in theory, be shorter than the HD session and the sensitivity of the technique. Indeed, in three cases of HD CRBI suspected cases, the clinician did not initiate probabilistic antibiotic therapy, although the final diagnosis was HD CRBI. With the use of rt-PCR, all HD CRBIs would have been diagnosed before the end of the HD session, avoiding any delay in treatment.
Second, based on the rt-PCR results, antibiotics would be more appropriately targeted, thus cutting anti-cocci Gram-positive therapy from 77% to 29%. Indeed, with a positive result for 16S rt-PCR, combined with a negative result for Staphylococcus spp. and S. aureus rt-PCR, clinicians can safely avoid antibiotics targeting MRS.
DISCUSSION
We reported the first systematic diagnostic accuracy study of an rt-PCR assay versus blood culture in the specific setting of suspected HD CRBI. The study population was composed of 37 patients, with 40 suspected cases of HD CRBI, and we were able to analyse 84 samples. This first study demonstrated the feasibility of rt-PCR for suspicion of HD CRBI.
The accuracy of our diagnostic method is high. The Staphylococcus spp. and S. aureus rt-PCRs proved to have excellent performances and are promising for this use. Regarding the universal 16S bacterial probe, rt-PCR showed excellent sensitivity (100%) and a high NPV. However, the method seems to be too sensitive, as 10 false-positive results were observed. These characteristics are reassuring regarding its potential future application. Compared with other studies developing an rt-PCR diagnostic method directly from whole blood without any enrichment stage, we found a higher sensitivity [14, 15]. The hypotheses explaining this difference include a different DNA extraction method, without a prior enrichment step; the use of a simplex rt-PCR, which minimizes the risk of error together with increasing the sensitivity of the technique [21, 22] and the larger bacterial burden. Indeed, the sample is taken directly at the source of the infection (on the catheter or on the circuit with a high blood flow).
The diagnostic method with these four rt-PCRs on whole blood without enrichment also made it possible to extract the DNA directly from whole blood in an EDTA blood tube, resulting in a shorter amount of time needed for result delivery (median of 3.5 h). This methodology allowed us to collect important information in <4 h, which is the usual duration of an HD session. To our knowledge, this is the simplest method to acquire those results in such a short time. Thanks to this reduced time, a major challenge for the clinician confronted with a suspicion of HD CRBI can be addressed: the decision to introduce the most accurate antibiotic treatment and avoid broad-spectrum antibiotics.
It is known that exposure to antibiotic therapy is one of the main risk factors for the appearance and spread of multiresistant bacteria. Multiresistant bacteria are also associated with significant adverse events. In 2006, the National Health Safety Network reported antibiotic use in 32 ambulatory HD centres [23]. During the 12-month study period, 977 introductions were recorded, amounting to a pooled average of 3.5 introductions per 100 patient-months. Chronic HD patients with catheters had the highest rate of antibiotic therapy introduction. Vancomycin accounted for 73% of all initial antibiotic therapy prescriptions. Later, Snyder et al. [24] prospectively followed 278 HD patients over a 12-month study period. Overall, 29.8% of doses were classified as inappropriate, and the most common inappropriate antibiotic used was vancomycin. With our approach, the rt-PCR results could have resulted in significant antibiotic economy, namely a reduced use of Gram-positive anti-cocci therapy. However, rt-PCR performances are not flawless, and the severity criteria excludes the restriction of antibiotic therapy.
Our study has several limitations. First, the diagnostic method developed here does not attempt to distinguish HD CRBI from BSI related to another source or from a contamination. Indeed, the development of this diagnostic method is only appropriate in a complementary role to blood cultures: microbiological diversity makes it impossible to reach a BSI diagnosis with only rt-PCR, and the analysis of the antibiogram is crucial in the subsequent management of the disease to de-escalate treatment to a targeted antibiotic therapy.
Second, our population of patients was small. The study was carried out in a single centre, due to the constraints of delivering the samples to the laboratory performing the rt-PCR, which made it difficult to carry out a multicentre study. This small number of patients made it impossible to test the diagnostic performance of the mecA rt-PCR, preventing the provision of this crucial information to the clinician. Also, the interpretation of the results should be considered. The negativity of the sample is based on the negative control, water, which shows a curve expansion after 30 cycles in a systematic way (background noise due to SYBR Green fluorescent molecules). All curves showed either a similar curve expansion (Ct >30 cycles) or a shorter curve expansion (≤28 cycles of Ct). The reproducibility of these two results led us to set these two cut-offs. These cut-offs are therefore based on local expertise and not on the literature (non-existent in the field). The boundary we have drawn between a positive and a negative result will have to be validated on a replication cohort.
In conclusion, this study shows that rt-PCR is a promising diagnostic method for suspected HD CRBI, with high diagnostic performance. This innovative and robust rt-PCR approach should allow clinicians to better tailor the initial empirical antimicrobial therapy, crucial for both a good outcome for these vulnerable patients and for a reduction of antibiotic resistance risk. With blood culture remaining the reference method for HD, the use of rt-PCR bedside could lead to a crucial optimization of HD patient care.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to patients for consenting to participate in this study.
Contributor Information
Mathieu Acquier, Service de Néphrologie-Transplantation-Dialyse-Aphérèses, Hôpital Pellegrin, CHU de Bordeaux, Bordeaux, France.
Arnaud Zabala, UMR 5234 CNRS, Université de Bordeaux, Bordeaux, France.
Valérie de Précigout, Service de Néphrologie-Transplantation-Dialyse-Aphérèses, Hôpital Pellegrin, CHU de Bordeaux, Bordeaux, France.
Yahsou Delmas, Service de Néphrologie-Transplantation-Dialyse-Aphérèses, Hôpital Pellegrin, CHU de Bordeaux, Bordeaux, France.
Véronique Dubois, Laboratoire de Bactériologie, Hôpital Pellegrin, CHU de Bordeaux, Bordeaux, France.
Renaud de la Faille, Service de Néphrologie-Transplantation-Dialyse-Aphérèses, Hôpital Pellegrin, CHU de Bordeaux, Bordeaux, France.
Sébastien Rubin, Service de Néphrologie-Transplantation-Dialyse-Aphérèses, Hôpital Pellegrin, CHU de Bordeaux, Bordeaux, France; Unité Inserm, UMR1034, Biologie des Maladies Cardiovasculaires, Université de Bordeaux, Pessac, France.
Christian Combe, Service de Néphrologie-Transplantation-Dialyse-Aphérèses, Hôpital Pellegrin, CHU de Bordeaux, Bordeaux, France; Unité Inserm 1026 Biotis, Université de Bordeaux, Bordeaux, France.
Fatima M'Zali, UMR 5234 CNRS, Université de Bordeaux, Bordeaux, France.
Hannah Kaminski, Service de Néphrologie-Transplantation-Dialyse-Aphérèses, Hôpital Pellegrin, CHU de Bordeaux, Bordeaux, France; CNRS-UMR 5164 ImmunoConcEpT, Université de Bordeaux, Bordeaux, France.
FUNDING
This study was funded by France Rein Aquitaine.
AUTHORS’ CONTRIBUTIONS
M.A., A.Z., V.D., F.M., C.C. and H.K. were responsible for the conception and design and the analysis and interpretation of data. M.A., V.D., F.M., S.R., Y.D., V.D.P., R.D.L.F., C.C. and H.K. were responsible for drafting or revising the article.
DATA AVAILABILITY STATEMENT
The data underlying this article will be shared upon reasonable request to the corresponding author.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Nguyen DB, Shugart A, Lines Cet al. National Healthcare Safety Network (NHSN) Dialysis Event Surveillance Report for 2014. Clin J Am Soc Nephrol 2017;12:1139–46. 10.2215/CJN.11411116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sarnak MJ, Jaber BL.. Mortality caused by sepsis in patients with end-stage renal disease compared with the general population. Kidney Int 2000;58:1758–64. 10.1111/j.1523-1755.2000.00337.x [DOI] [PubMed] [Google Scholar]
- 3. Allon M. Dialysis catheter-related bacteremia: treatment and prophylaxis. Am J Kidney Dis 2004;44:779–91. 10.1016/S0272-6386(04)01078-9 [DOI] [PubMed] [Google Scholar]
- 4. Allon M. Treatment guidelines for dialysis catheter-related bacteremia: an update. Am J Kidney Dis 2009;54:13–17. 10.1053/j.ajkd.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Danese MD, Griffiths RI, Dylan Met al. Mortality differences among organisms causing septicemia in hemodialysis patients. Hemodial Int 2006;10:56–62. 10.1111/j.1542-4758.2006.01175.x [DOI] [PubMed] [Google Scholar]
- 6. Vandecasteele SJ, Boelaert JR, De Vriese AS.. Staphylococcus aureus infections in hemodialysis: what a nephrologist should know. Clin J Am Soc Nephrol 2009;4:1388–400. 10.2215/CJN.01590309 [DOI] [PubMed] [Google Scholar]
- 7. Farrington CA, Allon M.. Complications of hemodialysis catheter bloodstream infections: impact of infecting organism. Am J Nephrol 2019;50:126–32. 10.1159/000501357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mermel LA, Allon M, Bouza Eet al. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 2009;49:1–45. 10.1086/599376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tabak YP, Vankeepuram L, Ye Get al. Blood culture turnaround time in U.S. acute care hospitals and implications for laboratory process optimization. J Clin Microbiol 2018;56:e00500–18. 10.1128/JCM.00500-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dark PM, Dean P, Warhurst G.. Bench-to-bedside review: the promise of rapid infection diagnosis during sepsis using polymerase chain reaction-based pathogen detection. Crit Care 2009;13:217. 10.1186/cc7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liesenfeld O, Lehman L, Hunfeld KPet al. Molecular diagnosis of sepsis: new aspects and recent developments. Eur J Microbiol Immunol 2014;4:1–25. 10.1556/EuJMI.4.2014.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Timbrook TT, Morton JB, McConeghy KWet al. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017;64:15–23. 10.1093/cid/ciw649 [DOI] [PubMed] [Google Scholar]
- 13. Fernández-Romero N, Quiles I, Jiménez Cet al. Use of multiplex PCR in diagnosis of bloodstream infections in kidney patients. Diagn Microbiol Infect Dis 2014;80:93–6. 10.1016/j.diagmicrobio.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 14. Warhurst G, Maddi S, Dunn Get al. Diagnostic accuracy of SeptiFast multi-pathogen real-time PCR in the setting of suspected healthcare-associated bloodstream infection. Intensive Care Med 2015;41:86–93. 10.1007/s00134-014-3551-x [DOI] [PubMed] [Google Scholar]
- 15. van den Brand M, van den Dungen FAM, Bos MPet al. Evaluation of a real-time PCR assay for detection and quantification of bacterial DNA directly in blood of preterm neonates with suspected late-onset sepsis. Crit Care 2018;22:105. 10.1186/s13054-018-2010-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quittnat Pelletier F, Joarder M, Poutanen SMet al. Evaluating approaches for the diagnosis of hemodialysis catheter-related bloodstream infections. Clin J Am Soc Nephrol 2016;11:847–54. 10.2215/CJN.09110815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menard A, Lehours P, Sarlangue Jet al. Development of a real-time PCR for the identification of Bordetella pertussis and Bordetella parapertussis. Clin Microbiol Infect 2007;13:419–23. 10.1111/j.1469-0691.2006.01659.x [DOI] [PubMed] [Google Scholar]
- 18. McClure JA, Conly JM, Lau Vet al. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from -resistant staphylococci. J Clin Microbiol 2006;44:1141–4. 10.1128/JCM.44.3.1141-1144.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reischl U, Linde HJ, Metz Met al. Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using real-time fluorescence PCR. J Clin Microbiol 2000;38:2429–33. 10.1128/JCM.38.6.2429-2433.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fang H, Hedin G.. Rapid screening and identification of methicillin-resistant Staphylococcus aureus from clinical samples by selective-broth and real-time PCR assay. J Clin Microbiol 2003;41:2894–9. 10.1128/JCM.41.7.2894-2899.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Santín Azcona J, Fox A, Boonham N.. Development of simplex and multiplex RT-qPCR assays for the detection of three cryptic viruses of black-grass (Alopecurus myosuroides). J Virol Methods 2022;300:114389. 10.1016/j.jviromet.2021.114389 [DOI] [PubMed] [Google Scholar]
- 22. Meritet DM, Mulrooney DM, Kent MLet al. Development of quantitative real-time PCR assays for postmortem detection of Mycobacterium spp. common in zebrafish (Danio rerio) research colonies. J Am Assoc Lab Anim Sci 2017;56:131–41. [PMC free article] [PubMed] [Google Scholar]
- 23. Klevens RM, Edwards JR, Andrus MLet al. Special report: Dialysis Surveillance Report: National Healthcare Safety Network (NHSN)–data summary for 2006. Semin Dial 2007;21:24–8. 10.1111/j.1525-139X.2007.00379.x [DOI] [PubMed] [Google Scholar]
- 24. Snyder GM, Patel PR, Kallen AJet al. Antimicrobial use in outpatient hemodialysis units. Infect Control Hosp Epidemiol 2013;34:349–57. 10.1086/669869 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared upon reasonable request to the corresponding author.