Abstract
T cell-derived pro-inflammatory cytokines are a major driver of rheumatoid arthritis (RA) pathogenesis. Although these cytokines have traditionally been attributed to CD4 T cells, we have found that CD8 T cells are strikingly abundant in synovium and make more interferon (IFN)-γ and nearly as much tumor necrosis factor (TNF) as their CD4 T cell counterparts. Further, using unbiased high-dimensional single-cell RNA-seq and flow cytometric data, we found that the vast majority of synovial tissue and synovial fluid CD8 T cells belong to an effector CD8 T cell population characterized by high expression of granzyme K (GzmK) and low expression of granzyme B (GzmB) and perforin. Functional experiments demonstrate that these GzmK+ GzmB+ CD8 T cells are major cytokine producers with low cytotoxic potential. Using T cell receptor repertoire data, we found that CD8 GzmK+ GzmB+ T cells are clonally expanded in synovial tissues and maintain their granzyme expression and overall cell state in blood, suggesting they are enriched in tissue, but also circulate. Using GzmK and GzmB signatures, we found that GzmK-expressing CD8 T cells were also the major CD8 T cell population in the gut, kidney, and coronavirus disease 2019 (COVID-19) bronchioalveolar lavage fluid, suggesting that they form a core population of tissue-associated T cells across diseases and human tissues. We term this population tissue-enriched expressing GzmK or TteK CD8 cells. Armed to produce cytokines in response to both antigen-dependent and -independent stimuli, CD8 TteK cells have the potential to drive inflammation.
INTRODUCTION
Rheumatoid arthritis (RA) is an autoimmune disease characterized by both local and systemic inflammation. The discovery of human leukocyte antigen (HLA)-DRB1 associations with RA have focused interest on CD4 T cells (1–3), and Th1 and Th17 CD4+ helper cell subsets have long been considered the predominant cytokine-producing T cells in RA synovium (4, 5). However, CD8 T cells are also markedly expanded in RA synovial tissue and fluid compared to osteoarthritis (OA) synovium, and they express CD69 and HLA-DR, indicating that they are activated and performing effector functions (6, 7). Previous work has shown that depletion of CD8 T cells in human synovial explants disrupted synovial architecture (8), suggesting that these cells play a vital role in the cellular interactions that drive RA. Moreover, HLA-B associations have been noted for both seropositive and seronegative RA (2, 9). Recently, we found that CD8 T cells from synovial tissue express interferon (IFN)-γ and tumor necrosis factor (TNF) transcripts at a similar or higher frequency compared to CD4 T cells from the same tissue sample, suggesting they may play an equal or even more important role than CD4 T cells in the tissue cytokine response in RA (10).
The Accelerating Medicines Partnership (AMP) RA/systemic lupus erythematosus (SLE) Network recently applied single-cell RNA-sequencing (scRNA-seq) to characterize the cellular composition of inflamed RA synovium (10). As part of this study, we found that CD8 T cells in synovial tissue have an unexpected transcriptomic profile, with high abundance of granzyme K (GzmK) and IFN-γ gene expression and low expression of granzyme B (GzmB). This finding was surprising, as GzmK is typically considered to be transiently expressed in memory CD8 T cell populations and subsequently downregulated as CD8 T cells differentiate toward the GzmB+ effector cell phenotype seen in cytotoxic T lymphocytes (CTLs) (11, 12). Unlike GzmB, GzmK does not cleave caspases to initiate target cell apoptosis, so the function of these GzmK-expressing CD8 T cells in RA synovium is unclear.
Here, we present a dedicated investigation of CD8 T cells and other granzyme-expressing lymphocytes in inflamed synovial fluid and tissue from patients with RA. We found that the most abundant tissue CD8 T cell population expressed high concentrations of GzmK with intermediate or low concentrations of GzmB and perforin. These CD8 T cells were neither senescent nor exhausted, produced high concentrations of IFN-γ and TNF, and were clonally expanded in synovial tissues. Importantly, GzmK+ GzmB+ CD8 T cells could be rapidly activated by cytokines, similar to innate-like T cells. In fact, this CD8 T cell population was transcriptionally similar to mucosal-associated invariant T (MAIT) cells and invariant natural killer T (iNKT) cells. Together, our findings point toward a previously undescribed tissue CD8 T cell population with innate-like T cell properties that suggest a key role for these cells in driving inflammation, rather than in granule-mediated killing in inflamed tissues.
RESULTS
CD8 T cells are abundant in RA synovium and produce more IFN-γ than CD4 T cells
To determine the nature of T cell populations in RA, we analyzed mass cytometry data from the AMP RA/SLE Network and found that CD8 and CD4 T cells exhibit a similar magnitude of increase among total synovial cells in inflamed RA compared to OA (Fig. 1A). Since inflamed RA synovium contains drastically higher cell numbers due to an increase of both stromal and bone marrow-derived cells, we also assessed CD8 T cell frequency among total leukocytes. CD8 T cells represent 10 to 35% of all leukocytes in inflamed synovial biopsies from patients with RA, compared to only 5% of total leukocytes in OA (Fig. 1B). To address whether migration or proliferation patterns of CD8 T cells in synovial tissue and fluid matched those of CD4 T cells, we compared the frequencies of CD8 T cells among all T cells in RA blood, synovial fluid, and synovial tissue. CD8 T cells were enriched among T cells in the joint compared to blood, with frequencies nearly twice as high in synovial tissue and even higher in synovial fluid compared to blood from patients with RA (33.8 ± 10.1% and 43.9 ± 12.1% versus 17.6 ± 6.8%, respectively (mean ± SD)) (Fig. 1C).
Fig. 1. CD8 T cells in RA synovial tissue and fluid make TNF and IFN-γ and predominantly express GzmK, whereas only a small portion express GzmB alone.
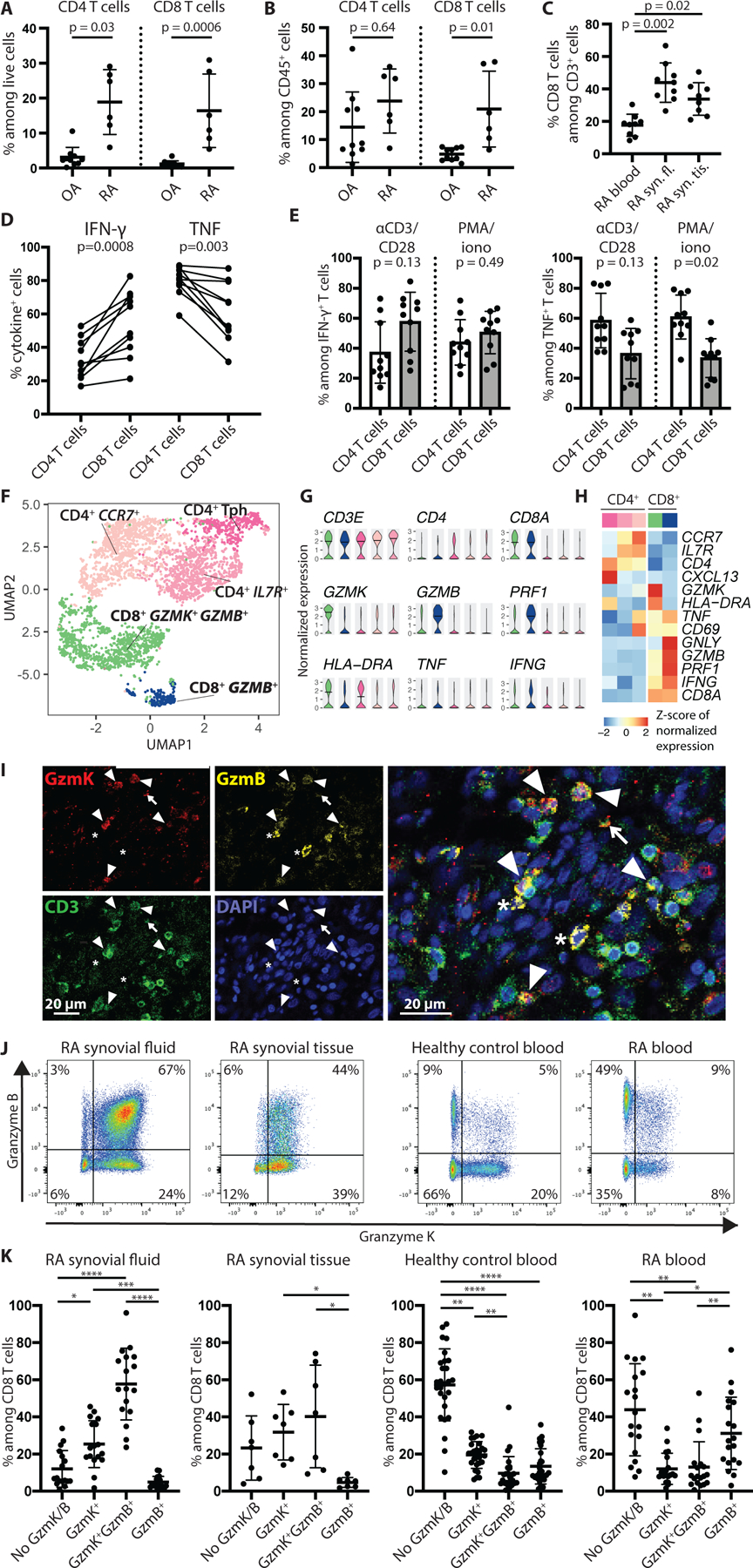
(A and B) Frequency of CD8 T cells among total live cells (A) and CD45+ cells (B) is shown in synovial tissue from patients with active RA (n=6) compared to OA (n=10) in a published synovial tissue mass cytometry dataset (10). (C) Frequency of CD8 T cells among total T cells in blood (n=9), synovial fluid (syn. fl., n=9), and synovial tissue (syn. tis., n=8) of RA patients is shown. (D) The frequency of CD4 and CD8 T cells producing IFN-γ and TNF is shown for seropositive RA synovial fluid (n=10) after stimulation with PMA and ionomycin (PMA/iono) for three hours. Paired CD4 and CD8 T cell data from the same patient are connected by a line. (E) The percentage contribution to total IFN-γ- or TNF-producing T cell pool by CD4 versus CD8 T cells from seropositive RA synovial fluid (n=10) is shown after stimulation with anti-CD3/CD28 antibody-coated beads or PMA and ionomycin for three hours. (F) A UMAP plot of Louvain clustering of 4,111 single-cell RNA-seq profiles synovial tissue T cells is shown. (G) Violin plots are shown illustrating expression of selected genes by cells in each cluster from the UMAP plot in (F). (H) A pseudobulk heatmap shows expression of selected genes by the CD4 and CD8 T cell clusters shown in (F). (I) Immunofluorescent staining of RA synovial tissue is shown. White arrowheads indicate GzmK+ GzmB+ T cells. The arrow points to a GzmK+ T cell, and the asterisks indicate GzmB+ cells, which do not stain for CD3. (J and K) Representative flow cytometry plots (J) and cumulative data (K) show GzmK and GzmB expression among CD8 T cells from synovial fluid (n=17), synovial tissue (n=7), and blood (n=21) from patients with RA and blood from healthy controls (n=27). Bars in (A to C, E, and K) represent mean ± SD. Statistics by Mann-Whitney U-test (A and B), Kruskal-Wallis test (C), two-tailed ratio paired t-test (D and E) and Friedman test with Dunn’s multiple comparisons test (K) are shown. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
To assess production of key cytokines by T cells in RA, we stimulated synovial fluid T cells with phorbol 12-myristate 13-acetate (PMA) and ionomycin. Upon stimulation, synovial fluid CD8 T cells produced large amounts of both IFN-γ and TNF, with a mean of 55% of CD8 T cells producing IFN-γ and 61% producing TNF (Fig. 1D). Importantly, synovial fluid CD8 T cells produced IFN-γ at a 1.6-fold (95% confidence interval 1.31–2.05) higher frequency than CD4 T cells from the same synovial fluid sample, and they represent more than half of all IFN-γ-producing cells on average despite being 40% of T cells (Fig. 1D and E). A smaller percentage of CD8 T cells produced TNF compared to CD4 T cells, yet the CD8 T cells still represent about 35% of TNF-producing T cells (Fig. 1D and E). These findings indicate that CD8 T cells have the potential to play a major role in T cell cytokine-driven inflammation in RA synovium, a role previously ascribed primarily to CD4 T cells.
The majority of CD8 T cells in RA synovial tissue and fluid express GzmK
To further investigate the nature of RA synovial tissue T cells, we assembled a dataset of 4,111 scRNA-seq profiles of synovial tissue T cells by integrating data from several newly collected synovial tissue cell samples with publicly available scRNA-seq data (fig. S1A, table S1). Integration and unbiased clustering of the cells yielded five clusters (Fig. 1F). One of the three CD4 T cell clusters expressed CXCL13, characteristic of T peripheral helper (Tph) cells (13), and the remaining two CD4 T cell clusters expressed CCR7 and IL7R, markers of naïve and central memory T cell populations (Fig. 1G and H). The two CD8 T cell clusters, on the other hand, both exhibited activated effector cell profiles, with low expression of CCR7 and IL7R and high expression of transcripts encoding HLA-DR, CD69, IFN-γ, and TNF (Fig. 1G and H). Importantly, the largest CD8 T cell cluster was characterized by high expression of GZMK along with intermediate expression of GZMB and PRF1 (encoding perforin). In contrast, the smaller cluster expressed high abundance of PRF1, GNLY (encoding granulysin), and GZMB, consistent with a classical CTL phenotype (Fig. 1G and H).
To confirm these findings, we examined protein expression by immunohistology and flow cytometry. First, we assessed GzmK and GzmB protein expression in tissue sections from RA synovium (Fig. 1I). We identified a number of T cells that stained for both GzmK and GzmB, in addition to scattered cells expressing only GzmB or GzmK, confirming that GzmK+ GzmB+ T cells were abundant in synovium.
Next, we used flow cytometry to quantify GzmK+ GzmB+ CD8 T cells in synovial fluid and synovial tissue from patients with seropositive RA (fig. S1B and C). Consistent with the transcriptional and imaging data, both synovial fluid and synovial tissue exhibited enrichment of GzmK+ GzmB+ CD8 T cell populations and very low frequencies of GzmB+ CD8 T cells (Fig. 1J and K). In synovial fluid, GzmK+ GzmB+ CD8 T cells represented a four-fold higher percentage of CD8 T cells compared to healthy and RA blood (57.7 ± 19.3% versus 9.6 ± 9.0% and 13.0 ± 13.6%, respectively (mean ± SD)) (Fig. 1J and K, fig. S2A). GzmK+ CD8 T cells represented the largest population in some synovial tissues, whereas GzmK+ GzmB+ CD8 T cells predominated in others; in contrast, cells expressing GzmB alone were consistently rare (31.8 ± 15.0% and 40.2 ± 27.6% versus 4.7 ± 2.7%, respectively (mean ± SD)) (Fig. 1J and K). In contrast, in healthy control blood, the majority of CD8 T cells expressed neither GzmB nor GzmK (Fig. 1J and K), consistent with the phenotype of naïve CD8 T cells. Blood from patients with seropositive RA exhibited a significant expansion of GzmB+ CD8 T cells (p=0.0075 and p=0.025 versus GzmK+ GzmB+ and GzmK+ CD8 T cells, respectively), representing about a third of all CD8 T cells, a two-fold increase compared to healthy control blood (31.1 ± 19.5% versus 13.4 ± 9.5% (mean ± SD)) (Fig. 1J and K and fig. S2A). Thus, even disregarding naïve CD8 T cells that did not expression GzmK or GzmB, the proportions of GzmK+, GzmK+ GzmB+, and GzmB+ CD8 T cells are markedly different in RA synovial fluid and tissue compared with RA blood. RA synovial tissue and fluid was enriched for GzmK+ GzmB+ CD8 T cells, whereas RA blood was enriched for GzmB+ CD8 T cells. These findings align well with a recent report that soluble GzmK concentrations are higher in synovial fluid than in serum from patients with RA (14).
We found similar profiles of GzmK and GzmB expression among CD8 T cells from synovial fluid from patients with seronegative RA and spondyloarthritis (fig. S2B), indicating that the factors driving the enrichment of GzmK+ GzmB+ CD8 T cells are shared by other inflammatory arthritides. Interestingly, the pattern of expression of GzmK and GzmB was also similar in synovial tissue CD8 T cells from patients with osteoarthritis (fig. S2C), even though these samples have low numbers of T cells (Fig. 1A and B).
GzmK and GzmB mark transcriptionally distinct T cell subsets
We hypothesized that GzmK and GzmB are markers of distinct subsets of T cells. To test this hypothesis in an unbiased way, we integrated scRNA-seq data from 11,602 cells from 19 RA synovial tissue samples, including four new RA synovial samples and 15 publicly available synovial samples (10), one RA synovial fluid sample, and three healthy control blood samples (see Methods, fig. S3A and B, table S1). Unbiased graph-based clustering of these cells yielded seven fine-grained clusters which formed three coarse-grained “metaclusters” corresponding to the differential expression of GZMK and GZMB (Fig. 2A to C). The Naïve metacluster included individual cluster 1 and was composed primarily of naïve cells that lack GZMK and GZMB but express CCR7 and SELL (CD62L) (Fig. 2C and D, fig. S3C). As expected, this metacluster was most enriched in healthy control blood (Fig. 2B). The GZMK+ metacluster was the largest metacluster in synovial tissue and fluid, comprised of individual clusters 2, 3, 4, and 5. This metacluster was characterized by high expression of GZMK with intermediate or low expression of GZMB (Fig. 2C and D). The GZMB+ metacluster, consisting of individual clusters 6 and 7, expressed GZMB largely without GZMK (Fig. 2C and D).
Fig. 2. CD8 T cells and other granzyme-expressing lymphocytes in RA synovial tissue, fluid, and blood segregate into transcriptional clusters characterized by GzmK and GzmB expression.
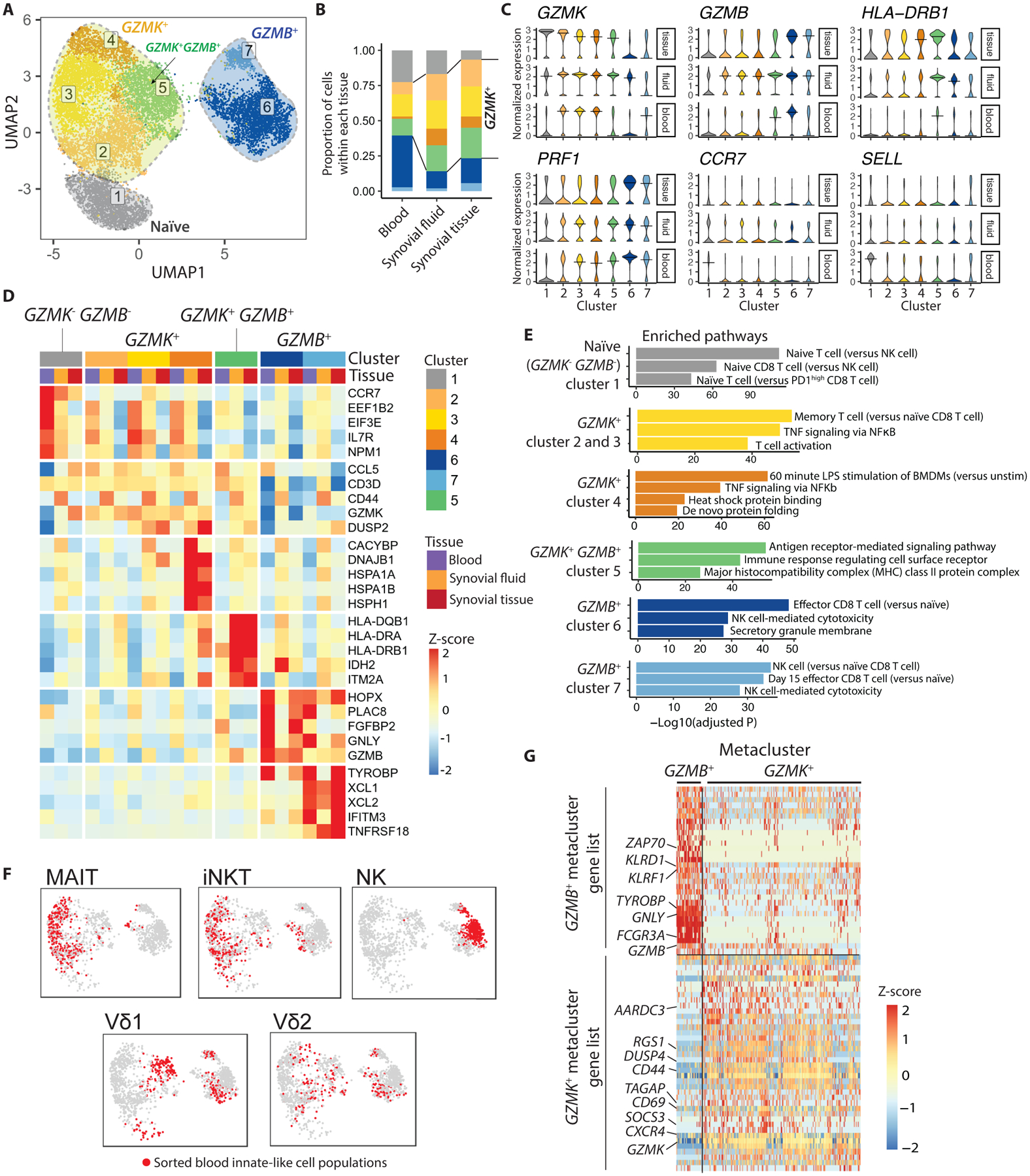
(A) Clustering of 11,602 single-cell profiles from RA synovial tissue and fluid as well as healthy control blood reveals 7 distinct clusters. Clusters of cells are projected into UMAP space. (B) Proportion of cells of each cluster is shown by tissue source. (C) Violin plots are shown depicting expression of cluster marker genes among cells from synovial tissue, synovial fluid, and blood. (D) Heatmaps show Z-scores of the average expression of selected genes, separated by tissue source. Most significantly enriched pathways per cluster are shown next to the heatmaps. (E) Most significantly enriched pathways per cluster are shown. (F) Projections of sorted innate-like T cell and NK cell populations are shown in UMAP space. (G) A heat map displaying differentially expressed genes between the GzmK and GzmB metaclusters, which make up the GzmK and GzmB gene signatures, is shown.
The GZMK+ metacluster and GZMB+ metacluster differed in expression of cytotoxic genes (Fig. 2C and D), as also seen in the synovial tissue dataset (Fig. 1F to H). Consistent with a CTL profile, the GZMB+ metacluster expressed a high abundance of transcripts encoding cytotoxic proteins granulysin (GNLY) and perforin (PRF1), and these clusters showed enrichment for gene pathways linked to CTL- and natural killer (NK) cell-mediated cytotoxicity (Fig. 2C to E, fig. S3C). On the other hand, the GZMK+ metacluster moderately expressed these genes, regardless of tissue source, and instead showed enrichment for gene sets involved in T cell activation and cytokine signaling. (Fig. 2C to E). Interestingly, individual cluster 5 expressed an intermediate abundance of GZMB (higher than clusters 2 to 4, but lower than clusters 6 and 7), and this cluster also exhibited the highest expression of genes encoding HLA-DR, suggesting that this cluster includes the most activated cells (Fig. 2C and D, fig. S3C).
The proportion of CD8 T cells in each cluster differs by tissue source (Fig. 2B), as predicted by the flow cytometry data (Fig. 1J and K). Compared to blood CD8 T cells, about twice as many synovial fluid and tissue CD8 T cells fell into the GZMK+ metacluster (Fig. 2B). Of note, some of the cells in the GZMB+ metacluster of healthy control blood and synovial tissue are likely to be NK cells, as NK cells and cytotoxic GzmB+ CD8 T cells share many similar transcriptomic gene signatures and are therefore difficult to separate in single cell RNA-seq. Synovial fluid CD8 T cells, which were sorted to gate out NK cells, fall primarily into the GZMK+ metacluster (Fig. 2B), consistent with the protein data (Fig. 1J and K).
Synovial tissue CD8 T cells share GzmK expression with innate-like T cells
NK cells and innate-like T cells such as MAIT cells and iNKT cells also express GzmK and GzmB. We hypothesized that these cell types might share transcriptomic profiles with CD8 T cell subsets that exhibit similar GzmK and GzmB expression patterns. To investigate this, we included in the scRNA-seq clustering single-cell profiles from sorted NK cells, MAIT cells, NKT cells, and Vδ1 and Vδ2 subsets of γδT cells, from a recent study of healthy blood cells by Gutierrez-Arcelus et al. (15). MAIT cells and iNKT cells localized to the GZMK+ metacluster (Fig. 2F), consistent with their expression of GzmK alone (fig. S4) (11). Importantly, none of the GZMK+ clusters were composed purely of innate-like T cells or NK cells, as the synovial fluid CD8 T cell dataset, which was sorted to exclude MAIT cells and contains very few (≤0.2%) NKT cells (16), was distributed among all GZMK+ clusters (Fig. 2B, fig. S3B). In contrast, NK cells (mainly belonging to the CD56dim subset (15)), localized to the GZMB+ metacluster, matching their expression pattern of GzmB (Fig. 2F, fig. S4) (11, 12). Vδ1 and Vδ2 cells were relatively evenly distributed among several clusters, mirroring their more diverse array of GzmK and GzmB protein expression (Fig. 2F, fig. S4). These findings indicate that there are shared transcriptomic programs among diverse lymphocyte cell types for which GzmK and GzmB act as hallmarks.
GZMK signature genes include regulators of signaling and transcription
To better characterize the differences among synovial tissue CD8 T cell subsets, we identified the top differentially expressed genes (area under the curve (AUC) > 0.6 and false discovery rate (FDR) < 0.05) that distinguish synovial tissue CD8 T cells in the GZMK+ and GZMB+ metaclusters (Fig. 2G, table S2). The GZMK+ metacluster gene list includes a number of regulators of signaling and transcription (table S2). SOCS3 is upregulated by cytokine stimulation and provides negative feedback for Janus kinase (Jak) and signal transducers and activators of transcription (STAT) signaling pathways. RGS1, RGS2, TAGAP, and AARDC3 encode proteins that regulate signaling downstream of G protein-coupled receptors. Interestingly, two genes (TAGAP and TNFAIP3) in this list are associated with increased risk of RA, and seven genes are associated with other types of autoimmune arthritis or with inflammatory bowel disease (IBD) (CREM, CXCR4, HSPA1A) or with other autoimmune diseases (CD27, CD69, RGCC, RGS1) (table S3) (17). In contrast, the GZMB+ metacluster gene list includes cytotoxic molecules such as GNLY and GZMB in addition to a number of cell surface receptors connected with activation or inhibition of cytotoxicity by CD8 T cells and NK cells, such as FCGR3A, KLRD1, and KLRF1 (table S2). The list also includes ZAP70 and TYROBP, which encode signaling molecules downstream of the TCR and receptors featuring immunoreceptor tyrosine-based activation (ITAM) motifs.
GzmK+ GzmB+ CD8 T cells have reduced cytotoxic potential compared to GzmB+ CD8 T cells
In most CD8 T cell studies, GzmB-expressing CD8 T cells are viewed as a single cytotoxic population. Our scRNA-seq analysis instead suggests that CD8 T cells that express GzmK and GzmB together are a distinct population from CD8 T cells that express GzmB alone. One difference between these clusters is the expression of genes associated with cytotoxicity. To investigate this question at the protein level, we used flow cytometry to compare the expression of GzmB and perforin among the different CD8 T cell subsets in blood from healthy controls and patients with RA, as well as in synovial fluid from patients with RA. Indeed, the mean fluorescence intensity (MFI) of GzmB and perforin staining was significantly lower among GzmK+ GzmB+ CD8 T cells than among GzmB+ CD8 T cells from healthy control blood (p=0.004) (Fig. 3A and B). GzmK+ GzmB+ CD8 T cells in synovial fluid expressed GzmB and perforin at concentrations similar to their blood counterparts, supporting an intrinsically low cytotoxic potential of GzmK+ GzmB+ CD8 T cells compared to GzmB+ CD8 T cells. Expression of CD57 and CX3CR1, surface markers of CD8 T cells with high cytotoxic potential (12, 18–21), correlated with expression of GzmB and perforin, with a lower proportion of GzmK+ GzmB+ CD8 T cells in blood and synovial fluid expressing CD57 and CX3CR1 than GzmB+ CD8 T cells in blood (Fig. 3C, fig. S5A). These findings suggest that GzmK+ GzmB+ CD8 T cells in both blood and synovial fluid have reduced cytotoxic potential compared to GzmB+ CD8 T cells in blood.
Fig. 3. GzmK+ GzmB+ CD8 T cells have reduced cytotoxic potential in both healthy control blood and synovial fluid.
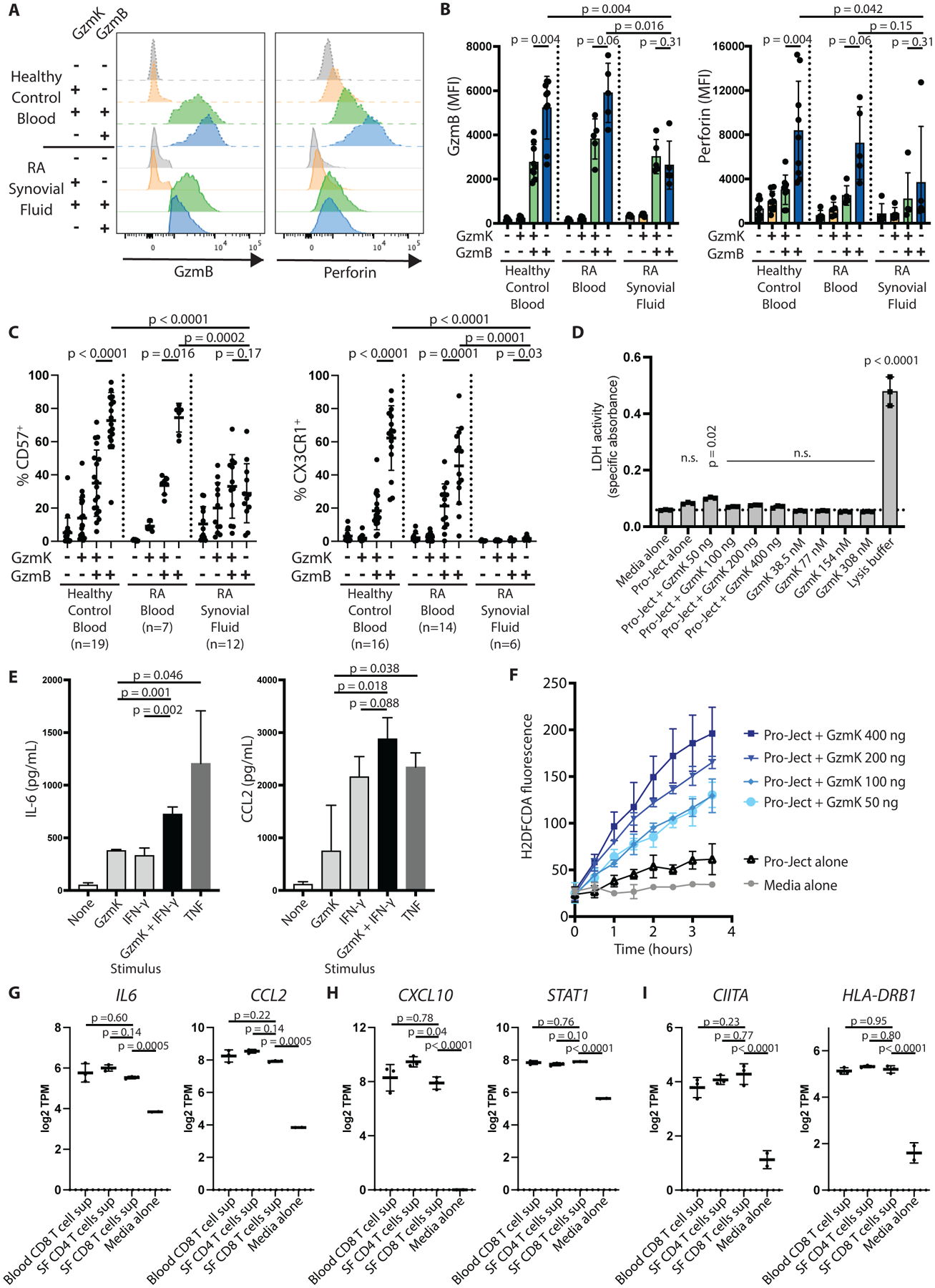
(A) Histograms show GzmB and perforin expression by the indicated CD8 T cell subset from healthy control blood (dashed lines) or RA synovial fluid (solid lines). (B) Cumulative mean fluorescence intensities (MFI) for GzmB and perforin staining by the indicated CD8 T cell subset are shown for healthy control blood (n=9), RA blood (n=5), and RA synovial fluid (n=5). Bars represent SD. (C) Expression of cytotoxic markers CD57 and CX3CR1 by the indicated CD8 T cell population are shown for healthy control blood, RA blood, or RA synovial fluid, as measured by flow cytometry. Bars represent SD. (D) RA synovial fibroblasts were incubated for 24 hours with recombinant enzymatically active GzmK in the culture supernatant or packaged into a protein transfection reagent (Pro-Ject). Cell death was measured by LDH release assay. Bars show SD of technical replicates. Representative of two independent assays. (E) RA synovial fibroblasts were incubated with recombinant GzmK, IFN-γ, or TNF for 24 hours, and IL-6 and CCL2 production was measured by enzyme-linked immunosorbent assay (ELISA). Bars show mean ± SD of three technical replicates; representative of four independent assays. (F) Recombinant GzmK was packaged into a protein transfection reagent and applied to RA synovial fibroblasts. ROS production was measured by 2’,7’-dichlorodihydrofluorescein diacetate (H2DFCDA) fluorescence over time. (G to I) Expression of selected genes involved in (G) pro-inflammatory signaling, (H) IFN-γ signatures, and (I) antigen processing and presentation are shown for low-input RNA-seq data of RA synovial fibroblasts stimulated with supernatants from the indicated T cell subset. SF, synovial fluid; TPM, transcripts per million. Bars show mean ± SD of three biological replicates. Statistical testing was done by (B and C) Wilcoxon matched-pairs signed rank test (within tissues) and Mann-Whitney test (across tissues); (D, G to I) one-way ANOVA with Dunnett’s multiple comparisons test, comparing all groups to (D) media alone or (G to I) synovial fluid CD8 T cell supernatant-stimulated cells, and (E) unpaired t-test. n.s., not significant.
In addition, the small portion of synovial fluid CD8 T cells that express GzmB alone also exhibited lower expression of GzmB, perforin, CD57, and CX3CR1 compared to GzmB+ CD8 T cells from blood (Fig. 2C and 3A to C, fig. S5A). GZMB and GNLY are also lower at the mRNA level in synovial fluid and tissue compared to blood (Fig. 2C to D), indicating that these proteins are not low due to recent release of cytotoxic granules. Thus, synovial fluid GzmB+ CD8 T cells show less cytotoxic potential that do GzmB+ CD8 T cells in blood.
GzmK activates inflammatory pathways in synovial fibroblasts
Given that GzmK+ GzmB+ CD8 T cells have reduced GzmB-driven cytotoxic potential, we next investigated the effects of GzmK itself in RA synovium. GzmK and GzmB both localize to secretory granules that are released from CD8 T cells upon activation (22). Unlike GzmB, GzmK does not cleave caspases to induce apoptotic cell death. Instead, GzmK has been reported to cleave several other target proteins on the cell surface and intracellularly to induce signaling pathways relevant to inflammatory pathogenesis (23–26). Consistent with this, we found that recombinant enzymatically active GzmK did not induce cell death among synovial fibroblasts as judged by lactate dehydrogenase (LDH) release, whether applied extracellularly or introduced into the cell cytoplasm using a protein transfection reagent (Fig. 3D). Instead, we found that GzmK can activate cultured synovial fibroblasts to produce interleukin (IL)-6 and CCL2, but not IL-8, in a dose-dependent manner, albeit at lower concentrations than TNF-stimulated fibroblasts (Fig. 3E, fig. S5B). This effect did not require perforin or any other agent to induce internalization of the GzmK, indicating that GzmK has a proteolytic target on the cell surface of these cells. When delivered intracellularly, GzmK induced a dose-dependent increase in the production of reactive oxygen species (ROS) (Fig. 3F). We did not find evidence that GzmK can induce endoplasmic reticulum (ER) stress, as has been reported using other types of fibroblasts (fig. S5C) (27). A recent study reported that murine GzmK can amplify the effect of IFN-γ on IL-6 and CCL2 production by a mouse embryonic fibroblast cell line (28). We find that human GzmK augments the ability of IFN-γ to stimulate human synovial fibroblasts to produce IL-6 and CCL2 (Fig. 3E). Together, these results demonstrate that GzmK, both alone and additively with cytokines, drives the production of cytokines, chemokines, and ROS, three molecular pathways that are aberrantly upregulated in RA (4, 29).
Next, we determined the effects of GzmK+ GzmB+ CD8 T cells on synovial fibroblasts. Because we cannot sort live cells based on intracellular granzyme expression, we compared memory CD8 T cells from synovial fluid, the majority of which are GzmK+ GzmB+ CD8 T cells, with memory CD8 T cells from blood, which are relatively enriched in GzmB+ CD8 T cells, and memory CD4 T cells from synovial fluid, which express low concentrations of granzymes. We cultured synovial fibroblasts with supernatants harvested from these different T cell subsets and then performed low-input bulk RNA-seq. The three T cell subsets had overall similar effects on synovial fibroblasts, altering fibroblast expression of hundreds of genes, including pro-inflammatory factors such as IL6, CCL2, CCL5, and IFN-signature genes such as CXCL10 and STAT1 (Fig. 3G and H, fig. S6A to C, data file S1). Synovial CD8 T cells had slightly weaker effects on most genes compared to synovial CD4 T cells, although the differences were relatively small (less than two-fold). Interestingly, genes associated with antigen processing and presentation, including CIITA, CD74, and components of the immunoproteasome (PSMB8, PSMB9, PSMB10) were as strongly upregulated by synovial fluid CD8 T cells as by synovial fluid CD4 T cells (Fig. 3I, fig. S6A and D). We also observed strong expression of genes encoding gasdermin D and caspase 1, two proteins involved in activating IL-1β, but unlike synovial fluid CD4 T cell supernatant, synovial fluid CD8 T cell supernatant did not upregulate expression of IL1B itself (fig. S6E). Supernatant from stimulated T cells also downregulated certain genes, including genes downregulated in inflamed synovium (DLX3) or enriched in subsets other than pro-inflammatory HLA+ synovial fibroblasts (AKRC12, FOS) (fig. S6F) (10).
GzmK+ GzmB+ CD8 T cells are a functionally potent tissue-resident T cell population
Our analysis of scRNA-seq, flow cytometry, and GzmK functional analysis all indicated that GzmK+ GzmB+ CD8 T cells are not classical CTLs. However, the nature of their cell state, in terms of activation, exhaustion, senescence, tissue residence, and other parameters, remained undefined. To obtain a clearer picture of GzmK+ GzmB+ CD8 T cells as defined by protein expression, we directly sorted these cells for low-input bulk RNA-seq. Since GzmK and GzmB are intracellular proteins, we developed methods for isolating RNA from fixed and permeabilized cells sorted after intracellular staining with fluorescently conjugated antibodies, based upon a recently published protocol (30). Using these methods, we sorted and isolated RNA from synovial fluid CD8 T cells expressing GzmK alone, GzmB alone, GzmK plus GzmB, or neither granzyme. Importantly, principal component analysis (PCA) separated the sorted CD8 T cell subsets based on their different granzyme expression patterns (Fig. 4A), further supporting the concept that granzyme expression defines cell populations with unique transcriptomic and thus functional properties. Review of the top differentially expressed genes confirmed their distinct nature (table S4).
Fig. 4. GzmK+ GzmB+ CD8 T cells are activated, tissue-resident CD8 T cells that do not show signs of exhaustion or senescence in blood and synovial fluid.
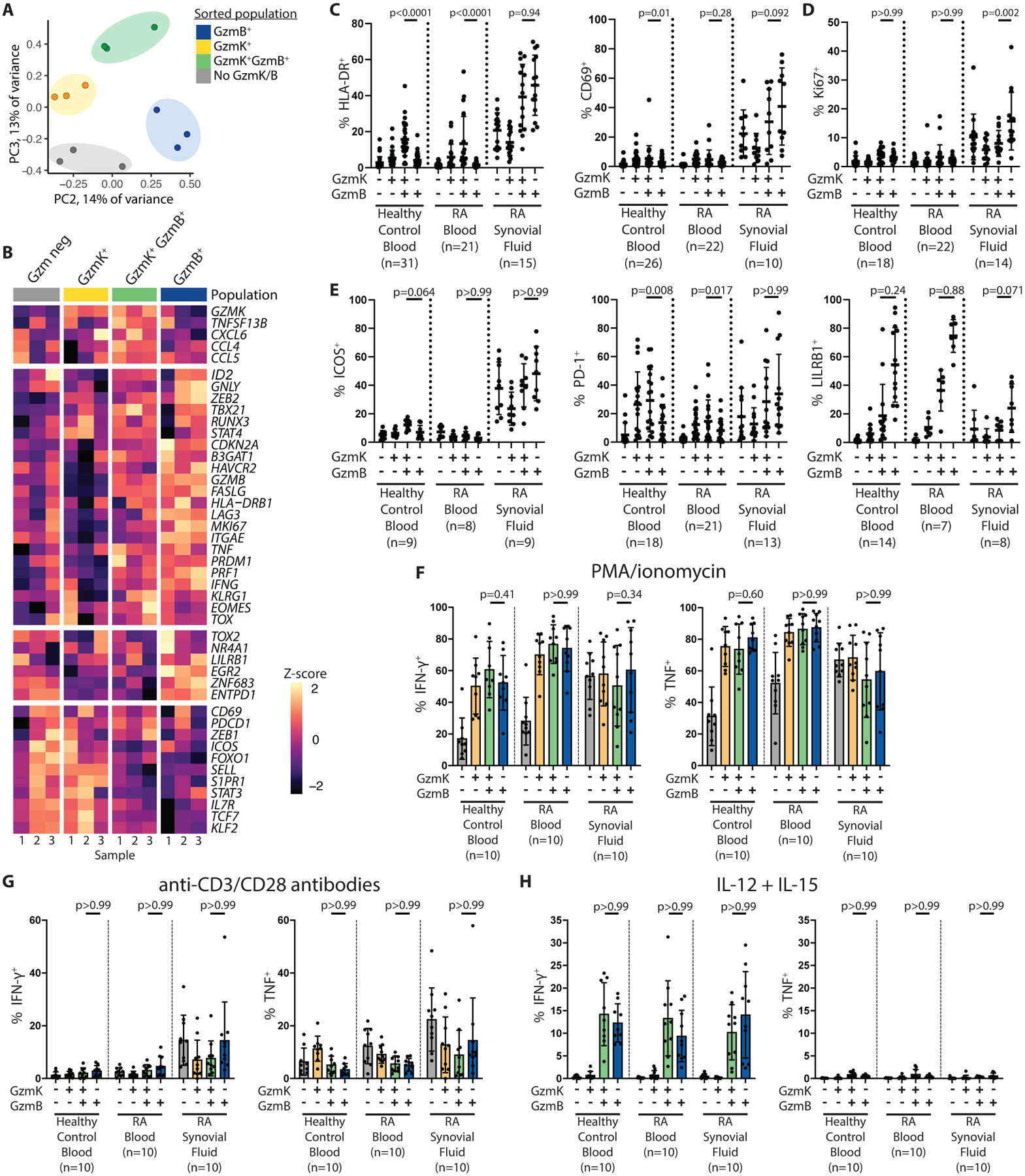
(A) A PCA plot is shown for low-input bulk RNA-seq data obtained from fixed and intracellularly stained RA synovial fluid CD8 T cells sorted for GzmK and GzmB expression. (B) Z-score heat maps of expression of selected genes in the low-input bulk RNA-seq dataset are shown. (C to E) The proportions of cells expressing markers of (C) activation (HLA-DR and CD69), (D) proliferation (Ki67), and (E) exhaustion or senescence (ICOS, PD-1, LILRB1) are shown for the indicated CD8 T cell subset in healthy control blood, seropositive RA blood, or seropositive RA synovial fluid, as measured by flow cytometry. (F to H) The proportions of cells within the indicated CD8 T cell subset producing IFN-γ and TNF are shown for healthy control blood (n=9), seropositive RA blood (n=10), or seropositive RA synovial fluid (n=10) after four hours of stimulation with (F) PMA and ionomycin or (G) anti-CD3/CD28 antibody-coated beads, or with (H) overnight stimulation with IL-12 and IL-15, as measured by intracellular cytokine staining. Statistical testing was done by Friedman test. Due to space constraints, only selected statistically significant differences are labeled. Bars represent mean ± SD.
We used this GzmK- versus GzmB-sorted low-input RNA-seq dataset along with flow cytometry to query genes and proteins associated with a number of CD8 T cell functions and states. Both GzmK+ GzmB+ CD8 T cells and GzmB+ CD8 T cells show downregulation of S1PR1 and SELL (CD62L), genes that must be turned off to allow cells to remain in tissue, compared to naïve and GzmK+ CD8 T cell subsets (Fig. 4B, fig. S7A). Both subsets also show downregulation of genes associated with memory precursor effector cells (MPECs) and central memory T cells, such as IL7R and TCF7 (31, 32) compared to CD8 T cells expressing only GzmK. Instead, GzmK+ GzmB+ CD8 T cells and GzmB+ CD8 T cell subsets both upregulated genes associated with effector T cells (32, 33), although they express different subsets of these markers. For example, GzmK+ GzmB+ and GzmB+ subsets express a similar abundance of KLRG1 and ID2, whereas only GzmB+ CD8 T cells exhibit elevated EGR2 and ZEB2 (Fig. 4B, fig. S7A). GzmK+ GzmB+ CD8 T cells instead trended toward higher expression of STAT4.
We also asked whether the GzmK+ GzmB+ CD8 T cell subset might include the narrowly defined resident memory T (Trm) cell subset, a small population of non-circulating CD8 T cells best characterized in murine models and in human skin (32, 34). Two Trm-associated genes, RUNX3 and PRDM1 (Blimp1), are expressed equally by GzmB+ and GzmK+ GzmB+ CD8 T cells, whereas ZNF683 (Hobit) and ITGAE (CD103) are mainly expressed by the GzmB+ subset (Fig. 4B, fig. S7A). However, it is difficult to draw any firm conclusions since it is not known whether these genes specifically identify Trm cells in human synovial tissue and fluid. Nonetheless, the data indicate that GzmK+ GzmB+ CD8 T cells are a tissue-associated effector CD8 T cell population and not merely transient circulating cells.
In terms of markers of activation, GzmK+ GzmB+ CD8 T cells similarly expressed HLA-DR and CD69 compared to GzmB+ CD8 T cells, but they express Ki67 (MKI67), a marker of proliferation, at a lower frequency than GzmB+ CD8 T cells (Fig. 4B to D, fig. S7B). This pattern of expression of HLA-DR differs from the single-cell RNA-seq datasets in Fig. 1F to H and 2A to D, in which genes encoding HLA-DR were expressed at the highest abundance in the GZMK+ GZMB+ cluster. Those scRNA-seq datasets used unbiased clustering to classify CD8 T cell populations – clustering driven in part by HLA class II genes – whereas the data in Fig. 4C to D categorized CD8 T cells based on protein abundance of GzmK and GzmB. The GzmK+ GzmB+ CD8 T cells identified based on protein expression and the GZMK+ GZMB+ cluster may represent overlapping but not identical cell subsets.
Given this apparent low degree of proliferation, we next investigated whether GzmK+ GzmB+ CD8 T cells might represent an exhausted or senescent subset, since such cells do not proliferate upon stimulation. Instead, the transcriptional data indicated that GzmK+ GzmB+ CD8 T cells similarly expressed genes associated with exhaustion and senescence, such as HAVCR2 (Tim3), LAG3, and CDKN2A (p16) compared to GzmB+ CD8 T cells (Fig. 4B, fig. S7A). Similarly, at the protein level, GzmK+ GzmB+ CD8 T cells from synovial fluid expressed similar concentrations of Tim3, ICOS, PD-1, LILRB1 (CD85j) and CD57 compared to GzmB+ CD8 T cells (Fig. 3C, Fig. 4E, fig. S8A and B). We also investigated CD29 and CD40L, which are associated with IFN-γ-producing cytotoxic cells and with non-cytotoxic helper cells, respectively (35–37). We did not observe any differences in expression of these markers between GzmK+ GzmB+ and GzmB+ CD8 T cells (fig. S8C and D). All granzyme-expressing populations expressed CD29, whereas CD40L was primarily observed on granzyme-negative CD8 T cells (fig. S8C and D).
We performed functional assays to directly measure the potential of these CD8 T cell subsets to produce cytokines in response to chemical and TCR-mediated stimulation. GzmK+ GzmB+ and GzmB+ CD8 T cells from synovial fluid produced IFN-γ and TNF at similar frequencies after stimulation with PMA and ionomycin or anti-CD3/CD28 antibody-coated beads, indicating that they have similar functional potential (Fig. 4F and G, fig. S9). The synovial fluid CD8 T cell subsets responded with similar strength to both types of stimulation as the corresponding blood CD8 T cells, further indicating these cells do not represent an exhausted phenotype. Taken together, these data confirm that synovial GzmK+ GzmB+ CD8 T cells are a functionally potent effector population, transcriptionally distinct from GzmB+ CD8 T cells (CTLs). We propose to call these cells tissue-enriched expressing GzmK-expressing T cells or TteK cells, reflecting their tissue enrichment, cell state, and characteristic expression of GzmK.
Given the close transcriptional relationship of GzmK+ CD8 T cells with MAIT cells and other innate-like T cells, we asked if these CD8 T cell subsets might differ in their responses to cytokine-mediated activation, a well-known mechanism of activation among CD8 T cells and other GzmK-expressing cell types, such as iNKT cells, MAIT cells, and CD56bright NK cells (38–43). After stimulation with IL-12 and IL-15, GzmK+ GzmB+ CD8 T cells responded by producing IFN-γ, but not TNF, at concentrations similar to GzmB+ CD8 T cells (Fig. 4H, fig. S9), consistent with the pattern reported by prior studies of cytokine-activated CD8 T cells (38, 43). Interestingly, GzmK+ GzmB− and Gzm-negative CD8 T cells did not respond to cytokine stimulation, indicating that this is a function only certain CD8 T cell subsets are “licensed” to perform. We observed similar patterns of high IFN-γ production and low TNF production after cytokine stimulation, but not PMA and ionomycin or TCR-mediated stimulation, among NK cells and MAIT cells, pointing toward shared downstream effects of cytokine stimulation (fig. S10).
GZMK+ GZMB+ CD8 T cell clones are transcriptionally associated with GZMK+ CD8 T cells in blood
We next investigated how GzmK+ GzmB+ CD8 T cells become enriched in synovial tissue and fluid. Given the similarities in protein and transcriptomic characteristics, we hypothesized that GzmK+ GzmB+ CD8 T cells in synovium belong to the same cell state as GzmK+ GzmB+ CD8 T cells in blood, as opposed to undergoing transcriptional changes within synovial tissue. To test this hypothesis, we analyzed a large single-cell TCR repertoire and RNA-seq dataset from the AMP RA/SLE Network, consisting of CD8 T cell profiles from 12 RA synovial tissue samples and 10 matched blood samples. The dataset consists of 6,555 synovial CD8 T cells and 4,475 blood CD8 T cells, which together form seven clusters (Fig. 5A and B, fig. S11A to C). The largest cluster in this independent data set consists of GZMK+ CD8 T cells with moderate expression of GZMB and PRF1 (Fig. 5A and B), confirming our finding regarding the GZMK+ cluster seen in Fig. 1F in an independent data set. Most cells in this cluster come from synovial tissue (fig. S11D). There are also naive cell and GZMB+ PRF1+ clusters, both of which are mainly composed of blood CD8 T cells (Fig. 5A and B, fig. S11D), consistent with our flow cytometry data from RA blood and synovial tissue (Fig. 1J and K). A small MKI67+ proliferating cluster was composed mostly of synovial tissue cells (Fig. 5A and B, fig. S11D).
Fig. 5. TCR repertoire data connects synovial GZMK+ CD8 T cells with GZMK+ CD8 T cells in blood.
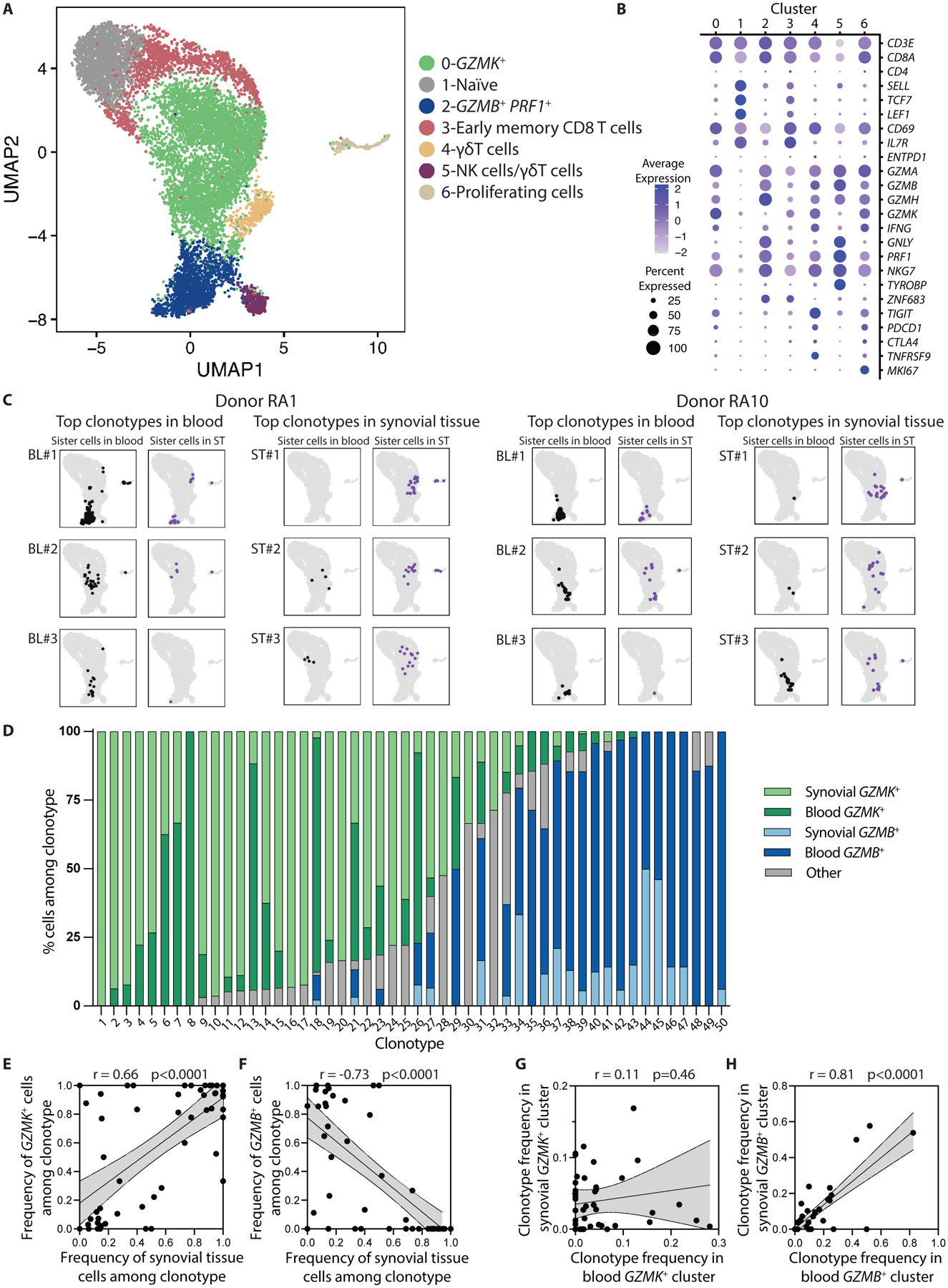
(A) A UMAP plot of Louvain clustering of 11,030 single-cell profiles from RA synovial tissue (6,555 cells) and matched blood (4,475 cells) reveals 7 distinct clusters. (B) Expression of selected markers is shown for the individual clusters shown in (A). (C) Mapping of sister cells belonging to the top 3 most frequent clonotypes in blood and synovial fluid from two representative patients shows that cells within a clonotype tend to map to the same location within the UMAP, whether they are in blood or synovial tissue (ST). (D) Transcriptional cluster and tissue origin is shown for clonal cells in the 50 analyzed clonotypes. Cells mapping to any of the five clusters other than GZMK+ or GZMB+ were categorized as “other.” (E and F) Correlations are shown for the frequency of synovial tissue cells among each of the 50 analyzed clonotypes and the frequency of clones in the GZMK+ (E) or GZMB+ (F) cluster in that clonotype. (G and H) Correlations are shown for the frequency of each of the 50 analyzed clonotypes among blood and synovial tissue CD8 T cells in the GZMK+ (G) or GZMB+ (H) cluster in that individual. Statistical testing was done by Pearson correlation.
To study patterns of CD8 T cell enrichment in this cross-sectional dataset, we used the TCR sequences as barcodes to identify clones that derive from a single naive T cell as clonotypes. We selected the top 2 to 3 blood and synovial tissue clonotypes from each donor for a total of 50 clonotypes to model CD8 T cell behavior across blood and tissue in this patient cohort. We then classified the cells belonging to each of these clonotypes based on transcriptional cluster and location (blood versus tissue) (Fig. 5A). This analysis revealed that cells from each clonotype, whether from blood or synovial tissue, primarily mapped to the same cluster, indicating that they have similar transcriptomic phenotypes (either GZMK+ or GZMB+) (Fig. 5C). Specifically, of the 50 clonotypes, 26 were >75% GZMK+, 17 were >75% GZMB+, and only 7 (15%) were mixed (<75% either GZMK+ or GZMB+) (Fig. 5D). In other words, there was homogeneity in granzyme type and cluster classification among cells within a clonotype across blood and synovial tissues.
We next determined whether these CD8 T cell clonotypes exhibited the same enrichment of GZMK+ CD8 T cells in synovium as seen in the total CD8 T cell population. Indeed, there was a strong correlation between the frequency of clonotypes in the GZMK+ cluster and the frequency of that clonotype in synovium (Fig. 5E). In contrast, clonotypes skewed toward the GZMB+ cluster were rare in synovial tissue (Fig. 5F). Thus, GZMK expression identifies CD8 T cell clones that are enriched in tissues, reinforcing our proposal to term these cells TteK cells.
We then investigated the mechanisms by which GZMK+ CD8 T cells accumulate in tissues. There are several non-mutually exclusive possibilities, including preferential proliferation, migration, and differentiation of GZMK+ CD8 T cells. We first compared proliferation by GZMK+ and GZMB+ CD8 T cells. Synovial CD8 T cells in the GZMK+ cluster exhibited greater clonal overlap with the proliferating cluster than did the synovial GZMB+ PRF1+ CD8 T cells (0.243 versus 0.073), suggesting that the majority of proliferating cells in synovium are GZMK+ (fig. S11E). We corroborated this finding with flow cytometry of Ki67 expression by synovial fluid CD8 T cells. Although the small number of GzmB+ synovial fluid cells express Ki67 at a higher frequency than other CD8 T cell subsets (Fig. 4D), the majority of proliferating CD8 T cells belong to the GzmK+ GzmB+ CD8 T cell subset (fig. S11F), suggesting that proliferative expansion may play a role in the accumulation of GzmK+ GzmB+ CD8 T cells in synovial tissue.
We also used clonal expansion as a measure of possible antigen-specific proliferation. GZMK+ CD8 T cells exhibited similar clonal expansion in blood and synovium, which at first glance suggests a lack of clonal expansion in synovial tissue (fig S11G). However, the GZMK+ clones most expanded in synovium are different than those most expanded in blood, as shown by a lack of association between the frequency of each individual clone in blood and synovium (Fig. 5G). In contrast, GZMB+ CD8 T cells are less clonal in synovial tissue than in blood (fig. S11G), and their clonal frequency in synovium is strongly correlated with the frequency in blood (Fig. 5H). This suggests that GZMB+ CD8 T cells do not undergo antigen-specific expansion in synovial tissue.
To address whether GzmK+ and GzmB+ CD8 T cells have different migratory potential, we measured their expression of selected chemokine receptors using flow cytometry. We found that GzmK+ and GzmK+ GzmB+ CD8 T cells preferentially expressed CCR2, CCR5, and CXCR3, whereas GzmB+ CD8 T cells instead expressed CX3CR1; CD8 T cells lacking GzmK and GzmB (naive CD8 T cells) expressed CCR7 (Fig. 3C, fig. S12). The findings indicate that GzmK+ and GzmK+ GzmB+ CD8 T cells have unique migratory potential distinct from GzmB+ CD8 T cells.
We did not observe any discrete transitional CD8 T cell populations in our analysis to suggest that CD8 T cell differentiation to a GZMK+ phenotype occurs in synovial tissue. However, it is possible that this differentiation in RA synovium occurs so rapidly that the transitional population is too small to discern in this dataset. Alternatively, GZMK+ CD8 T cells may differentiate outside of the synovium or at another point in time and then migrate into synovium using the chemokine receptors described above.
GZMK+ CD8 T cells outnumber GZMB+ CD8 T cells in many different tissues and disease states
Since GzmK+ GzmB+ CD8 T cells are the predominant CD8 T cell subset in RA synovium and synovial fluid, we next examined whether similar CD8 T cells are also enriched in other diseased tissues using scRNA-seq. To do this, we aggregated 26,625 scRNA-seq CD8 T cell profiles obtained from diseased and unaffected tissue from patients with inflammatory bowel disease (IBD) (44, 45), lupus nephritis (LN) (46), RA (10, 47), as well as healthy tissue from controls (Fig. 6A, table S5). We also included a dataset from bronchoalveolar lavage fluid (BALF) from healthy controls and patients with coronavirus disease 2019 (COVID-19), as an example of an acute infectious illness (48). Clusters comprising primarily CD8 T cells were included in the analysis. To identify shared cell states driven by T cell heterogeneity regardless of tissue difference, we corrected tissue and donor effects (49) and identified GzmK-associated cell states in all disease tissues (Fig. 6B and C). We found that the GZMK+ CD8 T cells were more frequent than GZMB+ CD8 T cells in many diseases and tissues. Specifically, the percentage of GZMK+ CD8 T cells was significantly higher than the percentage of GZMB+ CD8 T cells in RA synovium as well as inflamed bowel from patients with Crohn’s disease and ulcerative colitis (UC) (p=1×10−5, p=2×10−5, and p=5×10−5, respectively, Fig. 6C). Interestingly, non-inflamed bowel also had higher frequencies of GZMK+ CD8 T cells than GZMB+ CD8 T cells, with healthy kidney and osteoarthritis synovium showing a similar trend. These findings suggest that the frequency of GZMK expression is high among tissue CD8 T cells even in healthy states, although we note that sample and CD8 T cell numbers from OA synovium and healthy kidney were low, limiting generalizability. CD8 T cells from kidneys with lupus nephritis had similar frequencies of GZMK+ and GZMB+ CD8 T cells, in concordance with the individual T cell cluster frequencies reported for this dataset (46). We expected to find high frequencies of GZMB+ CD8 T cells in BALF from patients with COVID-19. However, the frequency of GZMB+ cells did not exceed that of GZMK+ cells in either mild or severe COVID-19 (Fig. 6C).
Fig. 6. GZMK is prominently expressed by CD8 T cells isolated from multiple healthy and diseased tissues.
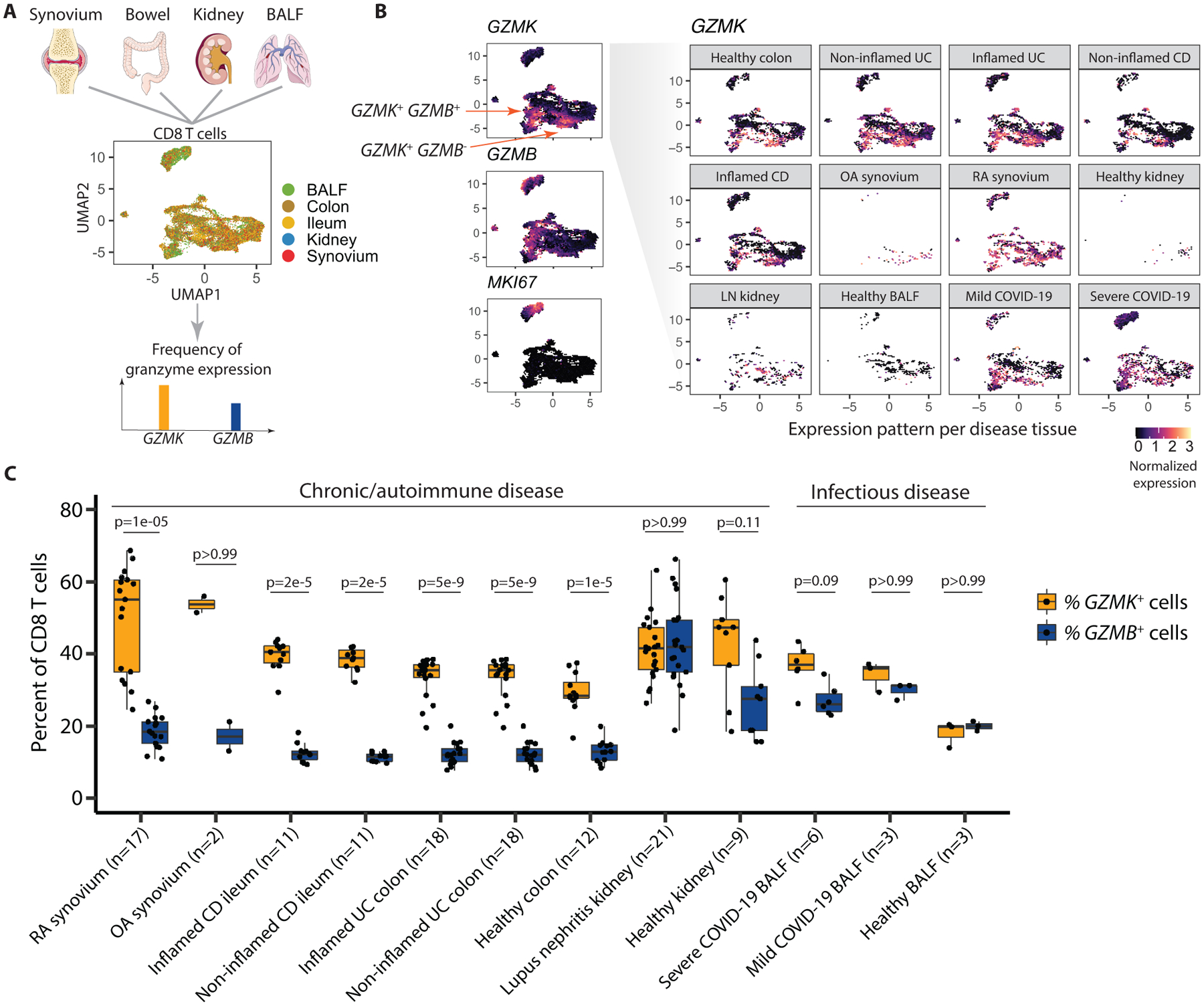
(A) The diagram illustrates the unbiased integration of 26,625 single-cell profiles from CD8 T cells in RNA-seq datasets from healthy or diseased tissues from RA synovium, Crohn’s disease (CD) ileum, ulcerative colitis colon, lupus nephritis (LN) kidney, and COVID-19 bronchoalveolar lavage fluid, where colors represent different tissue sources. The frequency of GZMK and GZMB gene expression was then assessed. (B) Expression pattern of GZMK, GZMB, and MKI67 (Ki67) is shown for the integrative dataset in UMAP space. The UMAP depicts expression of GZMK by cells from each individual tissue and disease state. (C) The percent of CD8 T cells from the indicated samples expressing GZMK or GZMB mRNA is shown. Data were analyzed by a two-tailed Wilcoxon rank-sum test. Boxplots summarize the median, interquartile range and 95% quantile range.
To compare the frequencies of cells with phenotypes similar to GzmK+ GzmB+ CD8 T cells compared to CTLs in a more refined way, we used the differentially expressed gene sets we obtained from synovial tissue CD8 T cell profiles in the scRNA-seq analysis presented in Fig. 2. Specifically, we calculated GZMK and GZMB gene scores based on the lists of differentially expressed genes between RA synovial tissue CD8 T cells in the GZMB metacluster and GZMK metacluster (AUC > 0.6 and FDR < 0.05) (Fig. 2G, table S2). We then measured expression of these genes among CD8 T cells in the other tissues, normalized from 0 to 1. The pattern of expression of these gene scores mirrored the frequencies of GZMK+ and GZMB+ CD8 T cells (fig. S13). Together, these findings suggest that CD8 TteK cells with a GzmK-associated phenotype are an abundant tissue CD8 T cell type in many diseases, particularly chronic autoimmune diseases, and are also present in acute infections.
DISCUSSION
GzmB-expressing CD8 T cells play a central role in killing virally infected and malignant cells through antigen-specific interactions of the TCR with major histocompatibility complex (MHC) class I-peptide complexes on target cells. In contrast, the nature of CD8 T cells in chronically inflamed autoimmune tissues is not well understood. Here, we found that the vast majority of CD8 T cells in actively inflamed synovial tissues expressed GzmK either alone or in combination with GzmB, whereas stereotypical GzmB+ CD8 CTLs were rare. GzmK+ GzmB+ CD8 T cells have poor granule-mediated cytotoxic potential, since the abundance of GzmB and perforin are low and GzmK has not been found to cleave caspases that drive apoptosis. GzmK+ GzmB+ CD8 T cells also expressed lower abundance of cytotoxic markers such as CD57 and CX3CR1 compared to GzmB+ CD8 T cells. We found that CD29, a marker of CD8 T cells that produce high amounts of IFN-γ and have cytotoxic activity (35), was expressed on all GzmK- and GzmB-expressing cells, indicating that CD29 may largely label non-naive cells. We did not see CD40L expression on either GzmK+ GzmB+ or GzmB+ CD8 T cells. CD40L has been reported to mark CD8 T cells lacking perforin and granzyme A that exhibit helper function capabilities that may play a role in synovial follicle formation in RA (8, 36, 37). Granzyme A is expressed by all GzmK- and GzmB-expressing cells (11) and accordingly, we found CD40L staining mainly among Gzm-negative CD8 T cells.
GzmK+ GzmB+ CD8 T cells appear to behave similarly to innate-like T cells, responding to TCR-independent stimulation as potently as their GzmB+ counterparts and producing cytokines to affect other cells in their environment. Indeed, in analysis by scRNA-seq, GzmK+ GzmB+ CD8 T cells cluster together with MAIT cells and iNKT cells, two innate cell types known for producing cytokines in response to TCR-independent stimulation, and not with CTLs. We call these cells CD8 TteK cells, as an effector cell type distinct from CTLs, abundantly found in several different healthy and diseased tissues, and identified by expression of GzmK. CD8 TteK cells shared transcriptional programs with MAIT cells and iNKT cells, whereas CTLs overlapped with CD56dim NK cells, reflecting differences in cytotoxicity and other functions.
We did not observe that CD8 TteK cells were enriched in RA blood, indicating that these cells were specifically enriched in tissues and not merely bystanders of systemic inflammation. The disproportionate abundance of GZMK+ CD8 T cell clonotypes in synovial tissue compared to blood suggests that CD8 TteK cells may receive antigen-specific stimuli locally in synovial tissue, driving their expansion. CD8 TteK cells may be able to migrate into and out of synovium and circulate in the periphery without changing phenotype, as GZMK+ CD8 T cells belonging to the enriched clonotypes identified in synovial tissue were typically also found in blood, albeit at lower frequencies. Additional studies are needed to test these hypotheses and identify potential synovial antigens. The drastic differences in GzmK+ versus GzmB+ subsets among CD8 T cells in RA blood and synovium serve as a warning against using RA blood as a surrogate for the populations and pathways present at the tissue sites of inflammation.
We initially detected this CD8 TteK cell population in RA synovium, but cells with a similar transcriptomic signature were also present in inflamed and healthy bowel as well as in kidneys affected by lupus nephritis. GZMK mRNA has also been detected among tumor-infiltrating T cells (50), raising the possibility that GzmK-expressing CD8 T cells may play a role in other diseases as well. Given the functional differences we have identified based on GzmK and GzmB expression, we propose that expression of these markers should be used to classify tissue CD8 T cells in other settings as well, such as tumor-infiltrating lymphocytes, as a preliminary surrogate for CD8 T cell phenotypes in these tissues. Importantly, tissue CD8 T cells that express GzmK alone or alongside GzmB should not automatically be labeled as memory cells or cytotoxic cells in single-cell RNA-seq data, as our findings indicate that such labels are not accurate descriptors of these human tissue-associated CD8 T cells.
Interestingly, CD8 T cells in lupus nephritis exhibited both strong GZMB and GZMK signatures. This finding is corroborated by the fact that clustering of these cells showed CTL-like and GZMK+ clusters of roughly the same size (46). The equal proportions of GZMK+ and GZMB+ subsets may be an organ-specific feature of inflammation in kidneys or a disease-specific feature of lupus pathogenesis. Further studies are needed to identify the factors determining GzmK versus GzmB signatures among tissue CD8 T cells.
In the context of inflamed RA synovium, GzmK itself acted like a key inflammatory factor. GzmK induced synovial fibroblasts to activate pro-inflammatory pathways, including production of IL-6, CCL2, and ROS. Stimulation of IL-6 and CCL2 production did not require internalization of GzmK, but ROS production did. We propose that the low amount of perforin expressed by GzmK+ GzmB+ CD8 T cells may allow for internalization of GzmK without producing enough pores in the plasma membrane as to lead to cell death.
The major cytokine produced by CD8 T cells is IFN-γ. IFN-γ is difficult to detect directly in biological samples, and no definitive pathogenic role for this cytokine has been identified in RA. However, the strong IFN-γ signatures in RA synovial tissue bulk and single-cell RNA-seq transcriptomes indicate that this cytokine is prominent in inflamed synovium (10, 51). IFN-γ has many possible effects in RA synovium, including upregulation of MHC class II molecules on both antigen-presenting cells and other cell types (such as synovial fibroblasts) as well as promoting regulatory T cell differentiation and inhibiting Th17 differentiation and osteoclastogenesis (52–54). IFN-γ- or IFN-γ receptor-deficient mice have more severe disease in some mouse models of RA but milder disease in others (55–58). Further, several human clinical trials have been performed using recombinant IFN-γ; small early studies showed some benefit, but a subsequent larger trial did not find any improvement compared to placebo (59, 60). Given these contradictory results, the exact role of IFN-γ in rheumatoid arthritis remains to be determined. One possible model is that the small number of CD8 TteK cells present in healthy tissues rapidly produce IFN-γ in response to cytokine stimulation early during immune responses. This IFN-γ activates beneficial antigen presentation and antiviral responses, similar to the roles of CD56bright NK cells, iNKT cells, and MAIT cells, all of which express GzmK. In inflamed RA synovium, however, a threshold may be crossed, leading to accumulation of a large number of CD8 TteK cells that produce copious amounts of IFN-γ with little or no cytotoxic effects on surrounding cells. The mechanisms that drive this process may, at a local tissue level, mimic those of familial hemophagocytic lymphohistiocytosis (fHLH), a form of primary systemic cytokine storm associated with genetic mutations in perforin and other cytotoxic granule components (61, 62). In fHLH, defective cytotoxic machinery prevents CD8 T cells and NK cells from killing macrophages and other innate immune cells to terminate the immune response. Instead, the cytokines produced by the activated macrophages and CD8 T cells form a positive feedback loop that amplifies the immune response. In RA synovium, the preponderance of CD8 TteK cells may represent a failure of CD8 T cells and NK cells to kill the innate immune cells driving the immune response, leading instead to a localized cytokine storm.
Our study has several limitations. First, although the TCR repertoire data is suggestive, we are not able to identify whether CD8 TteK cells preferentially migrate into inflamed joints or whether they differentiate or clonally expand within the synovium. We have also not been able to directly assay cytotoxic activity of these cells. CD107a degranulation assays do not differentiate between the release of cytotoxic proteins such as GzmB and perforin versus non-cytotoxic proteins, such as GzmK. Direct killing assays using LDH release or Annexin V staining to identify target cell death, are also logistically challenging since sorting the CD8 T cell subsets requires intracellular GzmK and GzmB staining, which kills the cells. Although the AMP RA/SLE Network synovial tissue samples were obtained from patients with documented elevated clinical disease activity scores (10, 63), we did not have clinical activity measures from some synovial tissues obtained from arthroplasty surgeries at Brigham and Women’s Hospital. It is possible that some of these synovial tissues lacked inflammation. Lastly, the number of samples in this study was low, limiting our ability stratify the findings by disease activity or treatment. Larger studies with more power are needed to address these issues and to further investigate CD8 T cell phenotypes in healthy tissues.
In conclusion, we found that co-expression of GzmK and GzmB defines a core tissue-associated CD8 T cell effector population. It has low cytotoxic potential but expresses high concentrations of GzmK and can produce cytokines through antigen-dependent and -independent mechanisms in RA synovium and other tissues. Further study of the differentiation and functions of CD8 TteK cells may reveal additional avenues of treatment for RA and other inflammatory diseases. Blocking GzmK may be a mechanism to reduce fibroblast activation, and blocking the cytokines that activate CD8 T cells, such as IL-12 or IL-15, may represent an effective complementary treatment approach in RA. Inducing the CD8 TteK cells to become cytotoxic could potentially shut down inflammatory pathways in RA synovium and may also yield treatment alternatives for cancer and chronic infection.
MATERIALS AND METHODS
Study design
The objectives of this project were to study CD8 T cell phenotypes in the joints (synovial tissue or fluid) of patients with rheumatoid arthritis using transcriptomic data, flow cytometric characterization of surface and intracellular proteins, and functional assays. In particular, our goal was to investigate the frequencies and phenotypes of GzmK-expressing CD8 T cells. Synovial CD8 T cells were compared to CD8 T cells from blood from patients with RA or from healthy controls. The studies used synovial tissue, synovial fluid, and blood from patients with RA or other rheumatologic diseases and blood from healthy controls. Inclusion criteria included meeting American College of Rheumatology (ACR) 2010 RA classification criteria. Exclusion criteria included active acute infection or malignancy. Perceived outliers were not excluded from analysis. There was no experimental treatment administered as part of this project, and no randomization or blinding was performed. No power analyses were performed prior to the project as each assay and outcome measure differs in robustness. The number of samples and experimental replicates are indicated in the figures or figure captions.
Human research
Research involving human subjects was performed according to the Institutional Review Board at Mass General Brigham through approved protocols with appropriate informed consent, as required. Patients with RA fulfilled the ACR 2010 Rheumatoid Arthritis classification criteria. Rheumatoid factor and anti-cyclic citrullinated peptide (CCP) antibody status and other clinical characteristics were obtained by review of electronic medical records (table S6). All RA blood and synovial fluid samples were from patients with seropositive RA unless otherwise specified. For all studies except the TCR repertoire analysis, synovial tissue samples for single-cell RNA-seq and imaging were obtained from patients undergoing arthroplasty at Brigham and Women’s Hospital or Brigham and Women’s Faulkner Hospital in Boston, MA. For the TCR repertoire analysis, synovial tissue and matched blood samples were recruited at clinical sites throughout the United States through the AMP RA/SLE Network (table S7). Synovial fluid samples were obtained as excess material from patients undergoing clinically indicated diagnostic or therapeutic arthrocentesis as directed by the treating rheumatologist. Blood samples from RA patients were obtained from patients seen at the Brigham and Women’s Hospital Arthritis Center. Blood samples from healthy controls were obtained from blood bank leukoreduction collars from anonymous platelet donors.
Statistical analysis
Raw, individual-level data are shown in data file S2. Details of statistical testing for each analysis are included in the corresponding figure legend. Statistical testing for flow cytometry and functional assays was performed using GraphPad Prism version 8.4 for Mac. For parametric tests, normality was assessed by Shapiro-Wilk test. We used a significance (alpha) level of 0.05. Results were considered statistically significant when p < 0.05 by Bonferroni correction.
Supplementary Material
Acknowledgments:
We thank the patients, rheumatologists, and orthopedic surgeons for providing the biological samples used in this study; Adam Chicoine and the Brigham and Women’s Hospital (BWH) Center for Cellular Profiling Flow Cytometry Core for assistance with cell sorting; Zhu Zhu and the BWH Center for Cellular Profiling Single Cell Genomics Core for assistance with single-cell transcriptomic data collection; the University of Rochester Flow Cytometry Core and Genomic Research Center for assistance with cell sorting and sequencing of the TCR repertoire project; Zach Herbert and the Molecular Biology Core Facilities at Dana Farber Cancer Institute for assistance with low-input RNA-seq of fixed cells, the National Institutes of Health (NIH) Tetramer Core Facility at Emory University for MR1 and CD1d tetramers. The MR1 tetramer technology was developed jointly by James McCluskey, Jamie Rossjohn, and David Fairlie, and the MR1 tetramers produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne. This work was supported by the Accelerating Medicines Partnership (AMP) in Rheumatoid Arthritis and Lupus Network. AMP is a public-private partnership between AbbVie Inc., the Arthritis Foundation, Bristol-Myers Squibb Company, the Foundation for the National Institutes of Health, GlaxoSmithKline, Janssen Research and Development, LLC, the Lupus Foundation of America, the Lupus Research Alliance, Merck Sharp & Dohme Corp., the National Institute of Allergy and Infectious Diseases, the National Institute of Arthritis and Musculoskeletal and Skin Diseases, Pfizer Inc., the Rheumatology Research Foundation, Sanofi, and Takeda Pharmaceuticals International, Inc. and was created to develop new ways of identifying and validating promising biological targets for diagnostics and drug development. Accelerating Medicines Partnership and AMP are registered service marks of the U.S. Department of Health and Human Services. Finally, we thank members of the Brenner and Raychaudhuri laboratories for discussions.
Funding:
Funding was provided by the Arthritis National Research Foundation, the Rheumatology Research Foundation, Joint Biology Consortium P30 AR070253, and Amgen to AHJ, the NIH grants R01 AR073290, T32 AR007530, and UH2 AR067694 to MBB; U19 AI111224-01, U01 HG009379-04, and R01 AR063759-05 to SR; and UH2-AR067676, UH2-AR067677, UH2-AR067679, UH2-AR067681, UH2-AR067685, UH2-AR067688, UH2-AR067689, UH2-AR067690, UH2-AR067691, UH2-AR067694, and UM2-AR067678 to the AMP RA/SLE Network.
Competing interests:
A.H.J. has received research support from Amgen. SR is a founder of and consultant to Mestag Therapeutics; a scientific advisor for Rheos Medicines, Janssen, and Pfizer; and a consultant to Gilead. MBB is a founder of and consultant to Mestag Therapeutics; a consultant to 4FO Ventures and Ono Pharmaceuticals; and serves on the scientific advisory board for GlaxoSmithKline. The authors declare no other competing interests.
AMP RA/SLE Network Authors
In addition to AMP RA/SLE Network members who are authors, the following AMP RA/SLE Network members are collaborators who have contributed to investigation and data curation of this manuscript:
William Apruzzese1, Joan M. Bathon11, Ami Ben-Artzi12, David L. Boyle13, S. Louis Bridges14, Vivian P. Bykerk15, Kevin Deane16, Edward DiCarlo17, Laura T. Donlin18, Andrew Filer19, Gary S. Firestein13, Lindsy Forbess12, Susan Goodman15, Ellen Gravallese1, Peter K. Gregersen20, Joel Guthridge21, V. Michael Holers16, Diane Horowitz20, Laura Hughes14, Judith A. James21, James Lederer22, Holden Maecker23, Arthur M. Mandelin II24, Mandy McGeachy25, Larry Moreland16, Harris Perlman24, Costantino Pitzalis26, Christopher Ritchlin8, William Robinson23, Paul J Utz23
Affiliations 1 to 10 can be found on the first page of the paper.
11Department of Medicine, Division of Rheumatology, Columbia University College of Physicians and Surgeons; New York, NY 10032, USA.
12Division of Rheumatology, Cedars-Sinai Medical Center; Los Angeles, CA 90048, USA.
13Division of Rheumatology, Allergy and Immunology, University of California; San Diego, La Jolla, CA 92093, USA.
14Division of Clinical Immunology and Rheumatology, Department of Medicine, University of Alabama at Birmingham; Birmingham, AL 35233, USA.
15HSS Healthcare Research Institute, Hospital for Special Surgery; New York, NY 10021, USA.
16Division of Rheumatology, University of Colorado School of Medicine; Aurora, CO 80045, USA.
17Department of Pathology and Laboratory Medicine, Hospital for Special Surgery; New York, NY 10021, USA.
18HSS Research Institute, Hospital for Special Surgery; New York, NY 10021, USA.
19Rheumatology Research Group, Institute for Inflammation and Ageing, University of Birmingham, NIHR Birmingham Biomedical Research Center and Clinical Research Facility, University of Birmingham, Queen Elizabeth Hospital; Birmingham B15 2TT, UK.
20Feinstein Institute for Medical Research, Northwell Health; Manhasset, NY 11030, USA.
21Department of Arthritis & Clinical Immunology, Oklahoma Medical Research Foundation; Oklahoma City, OK 73104, USA.
22Department of Surgery, Brigham and Women’s Hospital and Harvard Medical School; Boston, MA 02115, USA.
23Institute for Immunity, Transplantation, and Infection, Stanford University School of Medicine; Stanford, CA 94305, USA.
24Division of Rheumatology, Department of Medicine, Northwestern University Feinberg School of Medicine; Chicago, IL 60611, USA.
25Division of Rheumatology and Clinical Immunology, University of Pittsburgh School of Medicine; Pittsburgh, PA 15261, USA.
26Centre for Experimental Medicine & Rheumatology, William Harvey Research Institute, Queen Mary University of London; London EC1M 6BQ, UK.
Footnotes
Data and materials availability:
All functional data associated with this study are in the paper or supplementary materials. Single-cell and bulk sequencing data except for the immune cell repertoire dataset are available on GEO (accession numbers GSE202365, GSE202366, GSE202367, and GSE202375). The immune cell repertoire dataset from the AMP RA/SLE Network is deposited at Synapse (doi.org/10.7303/syn29837062). Source code to reproduce analyses is available on Zenodo (doi.org/10.5281/zenodo.6555398).
References and Notes
- 1.Amariuta T, Luo Y, Knevel R, Okada Y, Raychaudhuri S, Advances in genetics toward identifying pathogenic cell states of rheumatoid arthritis. Immunol. Rev 294, 188–204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, Alfredsson L, Padyukov L, Klareskog L, Worthington J, Siminovitch KA, Bae SC, Plenge RM, Gregersen PK, de Bakker PI, Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet 44, 291–296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregersen PK, Silver J, Winchester RJ, The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 30, 1205–1213 (1987). [DOI] [PubMed] [Google Scholar]
- 4.Firestein GS, McInnes IB, Immunopathogenesis of Rheumatoid Arthritis. Immunity 46, 183–196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smolen JS, Aletaha D, McInnes IB, Rheumatoid arthritis. Lancet 388, 2023–2038 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Cho BA, Sim JH, Park JA, Kim HW, Yoo WH, Lee SH, Lee DS, Kang JS, Hwang YI, Lee WJ, Kang I, Lee EB, Kim HR, Characterization of effector memory CD8+ T cells in the synovial fluid of rheumatoid arthritis. J. Clin. Immunol 32, 709–720 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Smith MD, Roberts-Thomson PJ, Lymphocyte surface marker expression in rheumatic diseases: evidence for prior activation of lymphocytes in vivo. Ann. Rheum. Dis 49, 81–87 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang YM, Zhang X, Wagner UG, Yang H, Beckenbaugh RD, Kurtin PJ, Goronzy JJ, Weyand CM, CD8 T cells are required for the formation of ectopic germinal centers in rheumatoid synovitis. J. Exp. Med 195, 1325–1336 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han B, Diogo D, Eyre S, Kallberg H, Zhernakova A, Bowes J, Padyukov L, Okada Y, Gonzalez-Gay MA, Rantapaa-Dahlqvist S, Martin J, Huizinga TW, Plenge RM, Worthington J, Gregersen PK, Klareskog L, de Bakker PI, Raychaudhuri S, Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am. J. Hum. Genet 94, 522–532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, Goodman SM, Tabechian D, Hughes LB, Salomon-Escoto K, Watts GFM, Jonsson AH, Rangel-Moreno J, Meednu N, Rozo C, Apruzzese W, Eisenhaure TM, Lieb DJ, Boyle DL, Mandelin AM 2nd, A. Accelerating Medicines Partnership Rheumatoid, C. Systemic Lupus Erythematosus, Boyce BF, DiCarlo E, Gravallese EM, Gregersen PK, Moreland L, Firestein GS, Hacohen N, Nusbaum C, Lederer JA, Perlman H, Pitzalis C, Filer A, Holers VM, Bykerk VP, Donlin LT, Anolik JH, Brenner MB, Raychaudhuri S, Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol 20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bratke K, Kuepper M, Bade B, Virchow JC Jr., Luttmann W, Differential expression of human granzymes A, B, and K in natural killer cells and during CD8+ T cell differentiation in peripheral blood. Eur. J. Immunol 35, 2608–2616 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Bengsch B, Ohtani T, Herati RS, Bovenschen N, Chang KM, Wherry EJ, Deep immune profiling by mass cytometry links human T and NK cell differentiation and cytotoxic molecule expression patterns. J. Immunol. Methods 453, 3–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao DA, Gurish MF, Marshall JL, Slowikowski K, Fonseka CY, Liu Y, Donlin LT, Henderson LA, Wei K, Mizoguchi F, Teslovich NC, Weinblatt ME, Massarotti EM, Coblyn JS, Helfgott SM, Lee YC, Todd DJ, Bykerk VP, Goodman SM, Pernis AB, Ivashkiv LB, Karlson EW, Nigrovic PA, Filer A, Buckley CD, Lederer JA, Raychaudhuri S, Brenner MB, Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan L, van den Hoogen LL, Meeldijk J, Kok HM, Jongeneel LH, Boes M, Wenink MH, Hack CE, Radstake T, van Roon JAG, Bovenschen N, Increased intra-articular granzyme M may trigger local IFN-lambda1/IL-29 response in rheumatoid arthritis. Clin. Exp. Rheumatol 38, 220–226 (2020). [PubMed] [Google Scholar]
- 15.Gutierrez-Arcelus M, Teslovich N, Mola AR, Polidoro RB, Nathan A, Kim H, Hannes S, Slowikowski K, Watts GFM, Korsunsky I, Brenner MB, Raychaudhuri S, Brennan PJ, Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions. Nat Commun 10, 687 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linsen L, Thewissen M, Baeten K, Somers V, Geusens P, Raus J, Stinissen P, Peripheral blood but not synovial fluid natural killer T cells are biased towards a Th1-like phenotype in rheumatoid arthritis. Arthritis Res. Ther 7, R493–502 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, Suveges D, Vrousgou O, Whetzel PL, Amode R, Guillen JA, Riat HS, Trevanion SJ, Hall P, Junkins H, Flicek P, Burdett T, Hindorff LA, Cunningham F, Parkinson H, The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Priol Y, Puthier D, Lecureuil C, Combadiere C, Debre P, Nguyen C, Combadiere B, High cytotoxic and specific migratory potencies of senescent CD8+ CD57+ cells in HIV-infected and uninfected individuals. J. Immunol 177, 5145–5154 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Bottcher JP, Beyer M, Meissner F, Abdullah Z, Sander J, Hochst B, Eickhoff S, Rieckmann JC, Russo C, Bauer T, Flecken T, Giesen D, Engel D, Jung S, Busch DH, Protzer U, Thimme R, Mann M, Kurts C, Schultze JL, Kastenmuller W, Knolle PA, Functional classification of memory CD8(+) T cells by CX3CR1 expression. Nat Commun 6, 8306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang SC, Theorell J, Entesarian M, Meeths M, Mastafa M, Al-Herz W, Frisk P, Gilmour KC, Ifversen M, Langenskiold C, Machaczka M, Naqvi A, Payne J, Perez-Martinez A, Sabel M, Unal E, Unal S, Winiarski J, Nordenskjold M, Ljunggren HG, Henter JI, Bryceson YT, Comparison of primary human cytotoxic T-cell and natural killer cell responses reveal similar molecular requirements for lytic granule exocytosis but differences in cytokine production. Blood 121, 1345–1356 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Nishimura M, Umehara H, Nakayama T, Yoneda O, Hieshima K, Kakizaki M, Dohmae N, Yoshie O, Imai T, Dual functions of fractalkine/CX3C ligand 1 in trafficking of perforin+/granzyme B+ cytotoxic effector lymphocytes that are defined by CX3CR1 expression. J. Immunol 168, 6173–6180 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Bade B, Boettcher HE, Lohrmann J, Hink-Schauer C, Bratke K, Jenne DE, Virchow JC Jr., Luttmann W, Differential expression of the granzymes A, K and M and perforin in human peripheral blood lymphocytes. Int. Immunol 17, 1419–1428 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Bovenschen N, Kummer JA, Orphan granzymes find a home. Immunol. Rev 235, 117–127 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Cooper DM, Pechkovsky DV, Hackett TL, Knight DA, Granville DJ, Granzyme K activates protease-activated receptor-1. PLoS One 6, e21484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma M, Merkulova Y, Raithatha S, Parkinson LG, Shen Y, Cooper D, Granville DJ, Extracellular granzyme K mediates endothelial activation through the cleavage of protease-activated receptor-1. FEBS J. 283, 1734–1747 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Zhao T, Zhang H, Guo Y, Fan Z, Granzyme K directly processes bid to release cytochrome c and endonuclease G leading to mitochondria-dependent cell death. J. Biol. Chem 282, 12104–12111 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Guo Y, Chen J, Shi L, Fan Z, Valosin-containing protein cleavage by granzyme K accelerates an endoplasmic reticulum stress leading to caspase-independent cytotoxicity of target tumor cells. J. Immunol 185, 5348–5359 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Mogilenko DA, Shpynov O, Andhey PS, Arthur L, Swain A, Esaulova E, Brioschi S, Shchukina I, Kerndl M, Bambouskova M, Yao Z, Laha A, Zaitsev K, Burdess S, Gillfilan S, Stewart SA, Colonna M, Artyomov MN, Comprehensive Profiling of an Aging Immune System Reveals Clonal GZMK(+) CD8(+) T Cells as Conserved Hallmark of Inflammaging. Immunity 54, 99–115 e112 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann MH, Griffiths HR, The dual role of Reactive Oxygen Species in autoimmune and inflammatory diseases: evidence from preclinical models. Free Radic. Biol. Med 125, 62–71 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Kunnath-Velayudhan S, Porcelli SA, Isolation of intact RNA from murine CD4(+) T cells after intracellular cytokine staining and fluorescence-activated cell sorting. J. Immunol. Methods 456, 77–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Zander R, Khatun A, Schauder DM, Cui W, Transcriptional and Epigenetic Regulation of Effector and Memory CD8 T Cell Differentiation. Front. Immunol 9, 2826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milner JJ, Goldrath AW, Transcriptional programming of tissue-resident memory CD8(+) T cells. Curr. Opin. Immunol 51, 162–169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang JT, Wherry EJ, Goldrath AW, Molecular regulation of effector and memory T cell differentiation. Nat. Immunol 15, 1104–1115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark RA, Resident memory T cells in human health and disease. Sci. Transl. Med 7, 269rv261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicolet BP, Guislain A, van Alphen FPJ, Gomez-Eerland R, Schumacher TNM, van den Biggelaar M, Wolkers MC, CD29 identifies IFN-gamma-producing human CD8(+) T cells with an increased cytotoxic potential. Proc. Natl. Acad. Sci. U. S. A 117, 6686–6696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner UG, Kurtin PJ, Wahner A, Brackertz M, Berry DJ, Goronzy JJ, Weyand CM, The role of CD8+ CD40L+ T cells in the formation of germinal centers in rheumatoid synovitis. J. Immunol 161, 6390–6397 (1998). [PubMed] [Google Scholar]
- 37.Frentsch M, Stark R, Matzmohr N, Meier S, Durlanik S, Schulz AR, Stervbo U, Jurchott K, Gebhardt F, Heine G, Reuter MA, Betts MR, Busch D, Thiel A, CD40L expression permits CD8+ T cells to execute immunologic helper functions. Blood 122, 405–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beadling C, Slifka MK, Differential regulation of virus-specific T-cell effector functions following activation by peptide or innate cytokines. Blood 105, 1179–1186 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Fauriat C, Long EO, Ljunggren HG, Bryceson YT, Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood 115, 2167–2176 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinks TSC, Zhang XW, MAIT Cell Activation and Functions. Front. Immunol 11, 1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohlgruber AC, Donado CA, LaMarche NM, Brenner MB, Brennan PJ, Activation strategies for invariant natural killer T cells. Immunogenetics 68, 649–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Tian Z, Wei H, Developmental and Functional Control of Natural Killer Cells by Cytokines. Front. Immunol 8, 930 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim TS, Shin EC, The activation of bystander CD8(+) T cells and their roles in viral infection. Exp. Mol. Med 51, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, Sud M, Andrews E, Velonias G, Haber AL, Jagadeesh K, Vickovic S, Yao J, Stevens C, Dionne D, Nguyen LT, Villani AC, Hofree M, Creasey EA, Huang H, Rozenblatt-Rosen O, Garber JJ, Khalili H, Desch AN, Daly MJ, Ananthakrishnan AN, Shalek AK, Xavier RJ, Regev A, Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730 e722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin JC, Chang C, Boschetti G, Ungaro R, Giri M, Grout JA, Gettler K, Chuang LS, Nayar S, Greenstein AJ, Dubinsky M, Walker L, Leader A, Fine JS, Whitehurst CE, Mbow ML, Kugathasan S, Denson LA, Hyams JS, Friedman JR, Desai PT, Ko HM, Laface I, Akturk G, Schadt EE, Salmon H, Gnjatic S, Rahman AH, Merad M, Cho JH, Kenigsberg E, Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 178, 1493–1508 e1420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, Chicoine A, Eisenhaure TM, Jonsson AH, Li S, Lieb DJ, Zhang F, Slowikowski K, Browne EP, Noma A, Sutherby D, Steelman S, Smilek DE, Tosta P, Apruzzese W, Massarotti E, Dall’Era M, Park M, Kamen DL, Furie RA, Payan-Schober F, Pendergraft WF 3rd, McInnis EA, Buyon JP, Petri MA, Putterman C, Kalunian KC, Woodle ES, Lederer JA, Hildeman DA, Nusbaum C, Raychaudhuri S, Kretzler M, Anolik JH, Brenner MB, Wofsy D, Hacohen N, Diamond B, S. L. E. n. Accelerating Medicines Partnership in, The immune cell landscape in kidneys of patients with lupus nephritis. Nat. Immunol 20, 902–914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephenson W, Donlin LT, Butler A, Rozo C, Bracken B, Rashidfarrokhi A, Goodman SM, Ivashkiv LB, Bykerk VP, Orange DE, Darnell RB, Swerdlow HP, Satija R, Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat Commun 9, 791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z, Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med 26, 842–844 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, Raychaudhuri S, Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, Bassez A, Decaluwe H, Pircher A, Van den Eynde K, Weynand B, Verbeken E, De Leyn P, Liston A, Vansteenkiste J, Carmeliet P, Aerts S, Thienpont B, Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med 24, 1277–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Zhang F, Mears JR, Shakib L, Beynor JI, Shanaj S, Korsunsky I, Nathan A, A. Accelerating Medicines Partnership Rheumatoid, C. Systemic Lupus Erythematosus, Donlin LT, Raychaudhuri S, IFN-gamma and TNF-alpha drive a CXCL10+ CCL2+ macrophage phenotype expanded in severe COVID-19 lungs and inflammatory diseases with tissue inflammation. Genome Med. 13, 64 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu X, Ivashkiv LB, Cross-regulation of signaling pathways by interferon-gamma: implications for immune responses and autoimmune diseases. Immunity 31, 539–550 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ivashkiv LB, IFNgamma: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol 18, 545–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schurgers E, Billiau A, Matthys P, Collagen-induced arthritis as an animal model for rheumatoid arthritis: focus on interferon-gamma. J Interferon Cytokine Res 31, 917–926 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Manoury-Schwartz B, Chiocchia G, Bessis N, Abehsira-Amar O, Batteux F, Muller S, Huang S, Boissier MC, Fournier C, High susceptibility to collagen-induced arthritis in mice lacking IFN-gamma receptors. J. Immunol 158, 5501–5506 (1997). [PubMed] [Google Scholar]
- 56.Matthys P, Vermeire K, Mitera T, Heremans H, Huang S, Schols D, De Wolf-Peeters C, Billiau A, Enhanced autoimmune arthritis in IFN-gamma receptor-deficient mice is conditioned by mycobacteria in Freund’s adjuvant and by increased expansion of Mac-1+ myeloid cells. J. Immunol 163, 3503–3510 (1999). [PubMed] [Google Scholar]
- 57.Vermeire K, Heremans H, Vandeputte M, Huang S, Billiau A, Matthys P, Accelerated collagen-induced arthritis in IFN-gamma receptor-deficient mice. J. Immunol 158, 5507–5513 (1997). [PubMed] [Google Scholar]
- 58.Finnegan A, Grusby MJ, Kaplan CD, O’Neill SK, Eibel H, Koreny T, Czipri M, Mikecz K, Zhang J, IL-4 and IL-12 regulate proteoglycan-induced arthritis through Stat-dependent mechanisms. J. Immunol 169, 3345–3352 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Veys EM, Menkes CJ, Emery P, A randomized, double-blind study comparing twenty-four-week treatment with recombinant interferon-gamma versus placebo in the treatment of rheumatoid arthritis. Arthritis Rheum 40, 62–68 (1997). [DOI] [PubMed] [Google Scholar]
- 60.Cannon GW, Pincus SH, Emkey RD, Denes A, Cohen SA, Wolfe F, Saway PA, Jaffer AM, Weaver AL, Cogen L, et al. , Double-blind trial of recombinant gamma-interferon versus placebo in the treatment of rheumatoid arthritis. Arthritis Rheum 32, 964–973 (1989). [DOI] [PubMed] [Google Scholar]
- 61.Brisse E, Wouters CH, Matthys P, Advances in the pathogenesis of primary and secondary haemophagocytic lymphohistiocytosis: differences and similarities. Br. J. Haematol 174, 203–217 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Gadoury-Levesque V, Dong L, Su R, Chen J, Zhang K, Risma KA, Marsh RA, Sun M, Frequency and spectrum of disease-causing variants in 1892 patients with suspected genetic HLH disorders. Blood Adv 4, 2578–2594 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang F, Jonsson AH, Nathan A, Wei K, Millard N, Xiao Q, Gutierrez-Arcelus M, Apruzzese W, Watts GFM, Weisenfeld D, Kang JB, Rumker L, Mears J, Slowikowski K, Weinand K, Orange DE, Rangel-Moreno J, Geraldino-Pardilla L, Deane KD, Tabechian D, Ceponis A, Firestein GS, Maybury M, Sahbudin I, Ben-Artzi A, Mandelin AM, Nerviani A, Rivellese F, Pitzalis C, Hughes LB, Horowitz D, DiCarlo E, Gravallese EM, Boyce BF, Arthritis AMPPR, Erythematosus SL, Moreland LW, Goodman SM, Perlman H, Holers VM, Liao KP, Filer A, Bykerk VP, Rao DA, Donlin L, Anolik JH, Brenner MB, Raychaudhuri S, Cellular deconstruction of inflamed synovium defines diverse inflammatory phenotypes in rheumatoid arthritis. bioRxiv, 2022.2002.2025.481990 (2022). [Google Scholar]
- 64.Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, Williamson NA, Strugnell RA, Van Sinderen D, Mak JY, Fairlie DP, Kjer-Nielsen L, Rossjohn J, McCluskey J, T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Donlin LT, Rao DA, Wei K, Slowikowski K, McGeachy MJ, Turner JD, Meednu N, Mizoguchi F, Gutierrez-Arcelus M, Lieb DJ, Keegan J, Muskat K, Hillman J, Rozo C, Ricker E, Eisenhaure TM, Li S, Browne EP, Chicoine A, Sutherby D, Noma A, R. A. S. L. E. N. Accelerating Medicines Partnership, Nusbaum C, Kelly S, Pernis AB, Ivashkiv LB, Goodman SM, Robinson WH, Utz PJ, Lederer JA, Gravallese EM, Boyce BF, Hacohen N, Pitzalis C, Gregersen PK, Firestein GS, Raychaudhuri S, Moreland LW, Holers VM, Bykerk VP, Filer A, Boyle DL, Brenner MB, Anolik JH, Methods for high-dimensonal analysis of cells dissociated from cyropreserved synovial tissue. Arthritis Res. Ther 20, 139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei K, Korsunsky I, Marshall JL, Gao A, Watts GFM, Major T, Croft AP, Watts J, Blazar PE, Lange JK, Thornhill TS, Filer A, Raza K, Donlin LT, A. Accelerating Medicines Partnership Rheumatoid, C. Systemic Lupus Erythematosus, Siebel CW, Buckley CD, Raychaudhuri S, Brenner MB, Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature 582, 259–264 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Traag VA, Waltman L, van Eck NJ, From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep 9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McInnes L, Healy J, Melville J, UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv [stat.ML] Preprint available at arsiv.org/abs/1802.03426 (2018).
- 69.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 73.van Schadewijk A, van’t Wout EF, Stolk J, Hiemstra PS, A quantitative method for detection of spliced X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic reticulum (ER) stress. Cell Stress Chaperones 17, 275–279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All functional data associated with this study are in the paper or supplementary materials. Single-cell and bulk sequencing data except for the immune cell repertoire dataset are available on GEO (accession numbers GSE202365, GSE202366, GSE202367, and GSE202375). The immune cell repertoire dataset from the AMP RA/SLE Network is deposited at Synapse (doi.org/10.7303/syn29837062). Source code to reproduce analyses is available on Zenodo (doi.org/10.5281/zenodo.6555398).


