Abstract
Previous studies demonstrated that the naturally occurring electrophile and PPARγ ligand, nitrolinoleic acid (NO2-LA), exists as a mixture of four regioisomers [Alexander, R. L., et al. (2006) Biochemistry 45, 7889–7896]. We hypothesized that these alternative isomers have distinct bioactivities; therefore, to determine if the regioisomers are quantitatively or qualitatively different with respect to PPARγ activation, NO2-LA was separated into three fractions which were identified by NMR (13-NO2-LA, 12-NO2-LA, and a mixture of 9- and 10-NO2-LA) and characterized for PPARγ interactions. A competition radioligand binding assay showed that all three NO2-LA fractions had similar binding affinities for PPARγ (IC50 ) 0.41–0.60 µM) that were comparable to that of the pharmaceutical ligand, rosiglitazone (IC50 ) 0.25 µM). However, when PPARγ-dependent transcription activation was examined, there were significant differences observed among the NO2-LA fractions. Each isomer behaved as a partial agonist in this reporter gene assay; however, the 12-NO2 derivative was the most potent with respect to maximum activation of PPARγ and an EC50 of 0.045 µM (compare with the rosiglitazone EC50 of 0.067 µM), while the 13-NO2 and 9- and 10-NO2 derivatives were considerably less effective with EC50 values of 0.41–0.62 µM. We conclude that the regioisomers of NO2-LA are not functionally equivalent. The 12-NO2 derivative appears to be the most potent in PPARγ-dependent transcription activation, whereas the weaker PPARγ agonists, 13-NO2 and 9- and 10-NO2, may be relatively more important in signaling via other, PPARγ-independent pathways in which this family of nitrolipid electrophiles is implicated.
Unsaturated nitrated fatty acids are naturally occurring electrophiles that are implicated in the modulation of several signaling pathways (1). The mechanisms by which some of the diverse activities of these nitroalkenes are mediated are incompletely understood, activities that include vasodilation, inhibition of inflammatory cells and their activation, and induction of heme oxygenase-1 expression (1–5). In addition, as a consequence of their redox and electrophilic reactivities, nitroalkenes can inhibit NFkB transactivation via alkylating its p65 subunit (6) and can activate ARE-dependent gene transcription via influencing the Nrf2/Keap1 pathway (7).
One of the more striking observations is that the nitroalkenes, nitrolinoleic (NO2-LA)1 and nitrooleic acid (NO2-OA), are potent endogenous ligands of PPARγ which can induce significant PPARγ-dependent transcription at nanomolar concentrations (4, 8). The potential physiological importance of these nitroalkenes is underscored by reports suggesting that their aggregate concentration can exceed 1 µM in human plasma and erythrocytes (9). Previously, we demonstrated that NO2-LA, one of the abundant naturally occurring nitroalkenes, exists as a mixture of four regioisomers (10). While it is expected that some of the activities associated with nitroalkenes that depend upon their electrophilic properties, alkylation and perturbation of redox status, may be relatively nonspecific and, hence, permissive for most or all nitroalkene species, it is likely that activities that depend upon precise molecular interactions with protein active sites, such as the ligand binding domain of PPARγ, may be relatively more specific, or selective, for particular nitroalkene species and isomers. To examine the potential isomer selectivity of PPARγ, we separated the regioisomers of NO2-LA into three fractions (a mixture of 9- and 10-NO2-LA, 12-NO2-LA, and 13-NO2-LA) and characterized their interactions with PPARγ. Our results indicate that the isomers are not equivalent with respect to PPARγ activation: the 12-NO2-LA isomer was significantly more potent in terms of maximum agonist activity and EC50. The results have implications for the structure-activity determinants of PPARγ activation and for quantitatively, and perhaps qualitatively, differential roles of the alternative nitroalkene species and isomers in PPARγ-dependent signaling.
EXPERIMENTAL PROCEDURES
Separation and NMR Characterization of NO2-LA Isomers.
Synthesis of NO2-LA was accomplished according to the method of Baker et al. (8) with modifications as described by Alexander et al. (10). Sequential preparative and semipreparative reverse phase HPLC was used to separate this mixture of four regioisomers into three fractions. For preparative scale separation, a Varian ProStar system with an absorbance detector and a Varian Microsorb reverse phase C18 column (250 mm × 21.4 mm) was used. The mobile phases consisted of 50% acetonitrile and 0.025% trifluoroacetic acid (solvent A), and 100% acetonitrile and 0.025% trifluoroacetic acid (solvent B). HPLC separation began at 100% solvent A followed by a linear gradient to 44% solvent B over 40 min at 40 mL/min. NO2-LA fractions eluting at 31, 32, and 33 min were collected and further purified by semipreparative scale HPLC using a reverse phase C18 column (Beckman Ultrasphere, 10 mm × 25 cm, Beckman Coulter, Fullerton, CA); chromatography was begun with a linear gradient from 100% solvent A to 31.7% solvent B which was then held at 31.7% solvent B at 5 mL/min. NO2-LA fractions eluted at ~45, 46, or 47 min and were collected. All HPLC profiles were measured at 274 nm. Final cleanup of NO2-LA fractions was accomplished using Oasis HLB 3 cm3 cartridges (Waters, Milford, MA); samples were bound and washed in 10% methanol and 0.1% formic acid, eluted with 100% methanol, dried under N2, and stored at −20 to −80 °C.
1H NMR spectra were recorded on a Bruker DRX-500 spectrometer using a BBO or TBI probe equipped with z axis gradients and processed with XWINNMR version 3.6. All acquisitions were carried out at 25 ± 1 °C. All spectra were referenced to the residual solvent peak of d-chloroform (1H 7.26 ppm and 13C 77.00 ppm). One-dimensional (1D) 1H and 13C experiments were conducted using standard Bruker parameter sets. The gradient-selected two-dimensional (2D) COSY, HMQC, and HMBC experiments were conducted with 512 points in F1 and 2048 points in F2. The 2D data were processed to 1024 × 1024 points using standard apodization functions in both dimensions.
Plasmids and Expression Vectors.
The pRevTet-Off and pRevTRE vectors were from Clontech (Palo Alto, CA). A tetracycline/doxycyline-regulated expression vector, pRevTRE/PPARG1, encoding the human PPARγ cDNA was constructed by inserting the 1842 bp NotI fragment derived from pcDNA3-PPARγ (11) (kindly provided by V. K. K. Chatterjee) into the HpaI site of pRevTRE. Recombinant human PPARγ and PPARα were expressed in Escherichia coli using plasmids pGEX-hPPARγ and pGEX-hPPARα (12).
Cell Lines and Cell Culture.
All cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and ciprofloxacin or penicillin/streptomycin.
Tetracycline/doxycycline-repressible PPARγ expressing derivatives of MCF7 cells, MCF7/RTO/PPARG1–4ʹ, were developed as follows. All gene transfers were accomplished using PA317 packaging cells to generate retroviral particle-containing supernatants that were then used to stably transduce MCF7 cells as described previously (13, 14). Briefly, MCF7 cells were transduced with pRevTet-Off-derived retroviral particles and selected in 1.5 mg/mL G418 (Invitrogen, Carlsbad, CA). A clone expressing particularly high-level doxycycline-repressible transactivator activity was identified and used for stable transduction by pRevTRE/PPARG1-derived retroviral particles. Cells were selected in 0.4 mg/mL hygromycin B (Calbiochem, San Diego, CA) and 1.5 mg/mL G418 and maintained in 1 µg/mL doxycycline. Colonies were screened for PPARγ expression by Western blot: from whole cell lysates, 30 µg protein samples were separated by SDS-PAGE (10% acrylamide), transferred to nitrocellulose, blocked, and examined for PPARγ expression using mouse monoclonal anti-PPARγ primary IgG (SC-7273, Santa Cruz Biotechnology, Santa Cruz, CA) and goat anti-mouse secondary antibody (SC-2005). Blots were developed using enhanced chemiluminescence (PerkinElmer, Waltham, MA) according to the manufacturer’s recommendations. For the clone MCF7/RTO/PPARG1–4ʹ, robust PPARγ expression was observed in the absence of doxycycline, while PPARγ expression was repressed in the presence of 1 µg/mL doxycycline. This clone was chosen for PPARγ-dependent transactivation studies. The cells were maintained in 1.5 mg/mL G418 and 0.2 mg/mL hygromycin B with or without 1 µg/mL doxycycline until just prior to experiments when selecting drugs (G418 and hygromycin B) were removed.
PPAR–NO2-LA Interactions.
Relative binding affinities of test ligands for PPAR were determined by a radioligand competition scintillation proximity assay using purified recombinant GST-hPPAR receptors (12), the radioligand [3H2]nTZD3, and varying concentratioins of unlabeled test ligands (NO2-LA, linoleic acid, and rosiglitazone) as described previously by Berger et al. (15). IC50 values were calculated from the average of duplicate data points obtained at each ligand concentration.
For reporter gene analysis of ligand-dependent PPARγ activation, MCF7/RTP/PPARG1–4ʹ cells were plated in replicate 12-well tissue culture plates at a density of 4 × 104 cells/well. Twenty-four hours later, cells were cotransfected with 0.4 µg of the PPRE-containing firefly luciferase reporter gene, pPPREx3-TK-LUC (16), and 20 pg of control CMV-Renilla luciferase reporter gene, pGL4.75 (Promega, Madison, WI), using Superfect reagent (Qiagen, Valencia, CA) according to the manufacturer’s recommendations. Twenty-four hours after the start of transfections, medium was replaced with medium containing vehicle (control) or varying concentrations of inducing agent (NO2-LA isomer, linoleic acid, or rosiglitazone). Cells were harvested after being continuously exposed to vehicle or inducing agent for 24 h. Luciferase assays were conducted using the Dual Luciferase Assay System (Promega), and values were corrected for variations in transfection efficiencies and nonspecific induction as described previously (17). For dose-response transactivation experiments, EC50 values for the various ligands were estimated as follows. Fold induction (I) [(corrected firefly luciferase activity + ligand)/(corrected firefly luciferase activity - ligand)] was plotted against added ligand concentration (L). The data were fitted to the equation I = 1 + (ImaxL)/(EC50 + L), where Imax is the theoretical maximum fold induction of PPARγ-dependent reporter gene transcription for that particular ligand. All incubations, including transfections and inductions, were conducted in DMEM supplemented with 10% FBS. For some control experiments, transgenic PPARγ expression was repressed by including 1 µg/mL doxycycline in the medium. Rosiglitazone (Cayman, Ann Arbor, MI) and NO2-LA isomers were stored as stock solutions in anhydrous dimethyl sulfoxide at −80 °C. Stock solution concentrations of NO2-LA were determined from an empirically derived extinction coefficient in methanol of 3.74 mM−1 cm−1 at 274 nm.
RESULTS
Fractionation and NMR Characterization of NO2-LA Regioisomers.
Regioisomers of NO2-LA were resolved from the synthetic mixture into three fractions by reverse phase HPLC. Integration of peak areas from a representative chromatogram indicated that the relative ratio of fractions 1, 2, and 3 was ~ 1:1:2 (Figure 1A). These fractions were purified by successive preparative and semipreparative scale HPLC followed by mixed resin chromatography as described in Experimental Procedures. From these purified fractions, structural assignments were made using 1D and 2D NMR. For fraction 1, 2D COSY spectra revealed coupling of all vinyl protons to the centrally located allylic protons, indicating the presence of either 9-NO2-LA or 13-NO2-LA. Subsequent 2D TOCSY and HMBC experiments confirmed the structure of fraction 1 as 13-NO2-LA. A similar sequence of 2D NMR studies indicated that fraction 2 consisted of only 12-NO2-LA. By this process of elimination, peak 3 must contain a mixture of 9- and 10-NO2-LA; NMR experiments show that these isomers occur in an ~ 1:1 ratio, and 2D COSY spectra provided individual assignments for most of the resonances. Summaries of 1H and 13C chemical shift data for all four NO2-LA isomers are shown in Tables 1 and 2. These analyses enabled structural assignment of the fractions as 13-NO2-LA (fx 1), 12-NO2-LA (fx 2), and a 1:1 mixture of 9- and 10-NO2-LA (fx 3) (Figure 1B). Finally, chemical shift analysis and 1D homonuclear decoupling studies showed the E stereochemistry of each nitrated olefin and the natural Z stereochemistry of each unsubstituted olefin. Details of all NMR experiments, including spectra, can be found in the Supporting Information.
FIGURE 1:
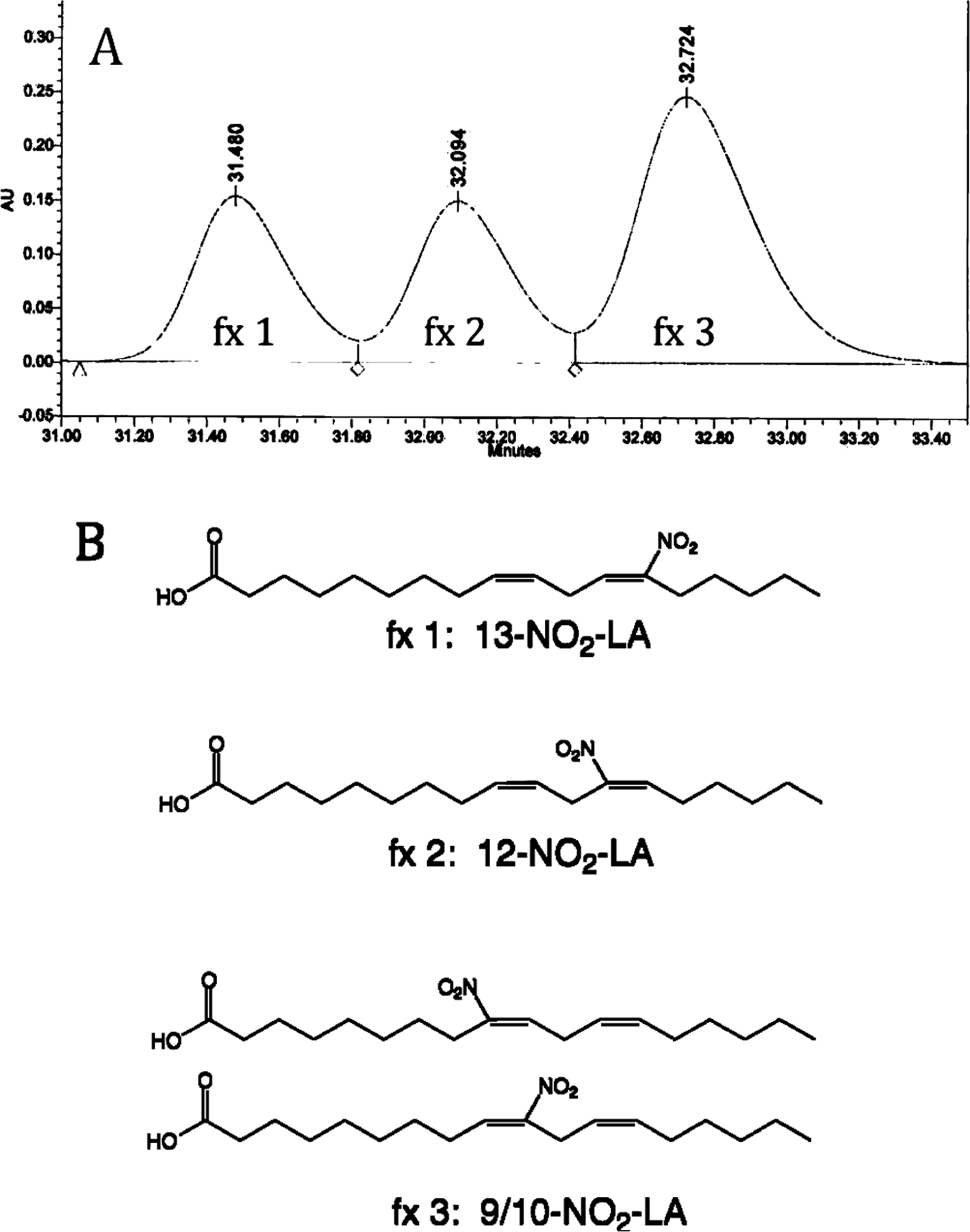
Fractionation and structural assignment of NO2-LA regioisomers. (A) Shown is a representative preparative scale HPLC elution profile of the mixture of NO2-LA regioisomers synthesized as described in Experimental Procedures. Elutions were monitored at 274 nm. Integration of the peak areas indicated that the relative ratio of fractions (fx) 1, 2, and 3 was ~1:1:2. (B) Structures assigned by NMR analysis to the three fractions (see the text and Supporting Information).
Table 1:
1H Chemical Shifts for 9-, 10-, 12-, and 13-Nitro Fatty Acids
| position | peak 3, 9-NO2 | peak 1, 13-NO2 | peak 3, 10-NO2 | peak 2, 12-NO2 |
|---|---|---|---|---|
| 2 | 2.35, t, 7.65 Hz | 2.35, t, 7.65 Hz | 2.35, t, 7.43 Hz | 2.37, t, 4.43 |
| 3 | 1.63, m | 1.64, m | 1.63, m | 1.64, m |
| 4 | 1.35, m | 1.33, m | 1.35, m | 1.35, m |
| 5 | 1.35, m | 1.33, m | 1.35, m | 1.35, m |
| 6 | 1.35, m | 1.33, m | 1.35, m | 1.35, m |
| 7 | 1.51, m | 1.39, m | 1.51, m | 1.40, m |
| 8 | 2.61, t, 7.88 Hz | 2.06, q, 7.20 Hz | 2.24, q, 7.65 Hz | 2.13, q, 6.98 Hz |
| 9 | a | 5.53, dtt, 10.76, 7.83, 1.71 Hz | 7.08, t, 7.88 Hz | 5.48, dtt, 11.51, 6.94, 1.58 Hz |
| 10 | 7.03, t, 8.33 Hz | 5.33, dtt, 10.76, 7.58, 1.47 Hz | a | 5.26, dtt, 11.00, 7.00, 1.58 Hz |
| 11 | 2.96, t, 7.65 Hz | 2.96, t, 7.43 Hz | 3.34, d, 7.20 Hz | 3.34, d, 6.94 Hz |
| 12 | 5.32, dtt, 10.51, 7.65, 1.58 Hz | 7.02, t, 7.88 Hz | 5.25, dtt, 11.00, 7.20, 1.58 Hz | a |
| 13 | 5.55, dtt, 10.51, 7.43, 1.58 Hz | a | 5.50, dtt, 11.00, 7.00, 1.58 Hz | 7.09, t, 7.80 Hz |
| 14 | 2.06, q, 7.43 Hz | 2.59, t, 7.65 Hz | 2.12, q, 6.98 Hz | 2.25, q, 7.40 Hz |
| 15 | 1.39, m | 1.50, p, 7.43 Hz | 1.40, m | 1.51, m |
| 16 | 1.39, m | 1.35, m | 1.40, m | 1.34, m |
| 17 | 1.39, m | 1.35, m | 1.40, m | 1.34, m |
| 18 | 0.904, t, 7.20 Hz | 0.89, t, 6.75 Hz | 0.90, t, 6.98 Hz | 0.91, t, 6.98 Hz |
Site of nitration.
Table 2:
13C Chemical Shifts for 9-, 10-, 12-, and 13-Nitro Fatty Acids
| position | 9-NO2 | 13-NO2 | 10-NO2 | 12-NO2 |
|---|---|---|---|---|
| 1 | 179.47 | 179.28 | 179.47 | 178.97 |
| 2 | 33.86 | 33.85 | 33.86 | 33.82 |
| 3 | 24.55 or 24.84a | 24.61 | 24.55 or 24.84a | 24.64 |
| 4 | 28.85–29.11b | 28.93–29.31c | 28.85–29.11b | 28.98–29.31c |
| 5 | 28.85–29.11b | 28.93–29.31c | 28.85–29.11b | 28.98–29.31c |
| 6 | 28.85–29.11b | 28.93–29.31c | 28.85–29.11b | 28.98–29.31c |
| 7 | 28.85–29.11b | 28.93–29.31c | 28.85–29.11b | 28.98–29.31c |
| 8 | 26.35 | 27.41 | 27.89 | 27.38 |
| 9 | 151.82 | 133.15 | 136.85 | 132.84 |
| 10 | 134.41 | 123.49 | 150.65 | 123.37 |
| 11 | 26.14 | 26.14 | 24.84 | 24.84 |
| 12 | 123.31 | 134.28 | 123.23 | 150.55 |
| 13 | 133.34 | 151.99 | 133.00 | 136.60 |
| 14 | 27.41 | 26.36 | 27.81 | 27.81 |
| 15 | 28.85–29.11b | 27.56 | 28.85–29.11b | 28.11 |
| 16 | 31.47 or 31.54a | 31.39 | 31.47 or 31.54a | 31.47 |
| 17 | 22.51 or 22.54a | 22.34 | 22.51 or 22.54a | 22.37 |
| 18 | 14.01 or 14.03a | 13.93 | 14.01 or 14.03a | 13.90 |
Assignment was ambiguous due the approximate 50:50 mixture of the 9- and 10-nitro isomers and the lack of resolution in the 2D HMBC spectra.
Due to the overlap of peaks in the 1D 13C spectra and the lack of resolution in the 2D HMBC spectra, these peaks were assigned to the range of 13C chemical shifts.
Due to the overlap of peaks in the 1D 13C spectra and the lack of resolution in the 2D HMBC spectra, these peaks were assigned to the range of 13C chemical shifts.
NO2-LA Regioisomers Are Relatively High-Affinity Ligands Selective for PPARγ.
A radioligand competition scintillation proximity assay was used to examine interactions of NO2-LA with the ligand binding domains of PPAR. As reflected in the IC50 values listed in Table 3, all three fractions of NO2-LA regioisomers interacted with PPARγ with similar binding affinities (IC50 = 0.41–0.60 µM) that were comparable to the binding affinity of the pharmaceutical ligand, rosiglitazone (IC50 = 0.25 µM). In contrast and as reported previously, the parent un-nitrated lipid, linoleic acid, exhibited a very low affinity for PPARγ. Lastly, the data indicate that NO2-LA isomers are selective ligands for PPARγ with no (9/10-NO2-LA and 13-NO2-LA) or a very low (12-NO2-LA) apparent affinity for PPARα (Table 3).
Table 3:
Scintillation Proximity Competitive Binding Assay of Ligand-PPAR Interactions
| PPAR IC50 (µM) |
||
|---|---|---|
| compound | γ | α |
| rosiglitazone | 0.25 | >15 |
| linoleic acid | 10.7 | >15 |
| 9/10-NO2-linoleic acid | 0.60 | >15 |
| 12-NO2-linoleic acid | 0.41 | 9.6 |
| 13-NO2-linoleic acid | 0.44 | >15 |
Differential Activation of PPARγ-Dependent Transcription by Regioisomers of NO2-LA.
A doxycycline-regulated PPARγ-expressing cell line, MCF7/RTO/PPARG1–4ʹ, was developed for the purpose of examining the relative potency of NO2-LA isomers in PPARγ activation. As shown in Figure 2 (inset), these cells exhibit robust expression of PPARγ when grown in the absence of doxycycline, whereas inclusion of the antibiotic completely represses expression of transgenic PPARγ. Also in Figure 2, all three isomer fractions support induction of PPRE-containing reporter gene expression. The level of induction is considerably higher in the presence of PPARγ transgene expression (without doxycycline, dark bars) than in its absence (with doxycycline, light bars), indicating that the great majority of observed NO2-LA-mediated induction is indeed PPARγ-dependent.
FIGURE 2:
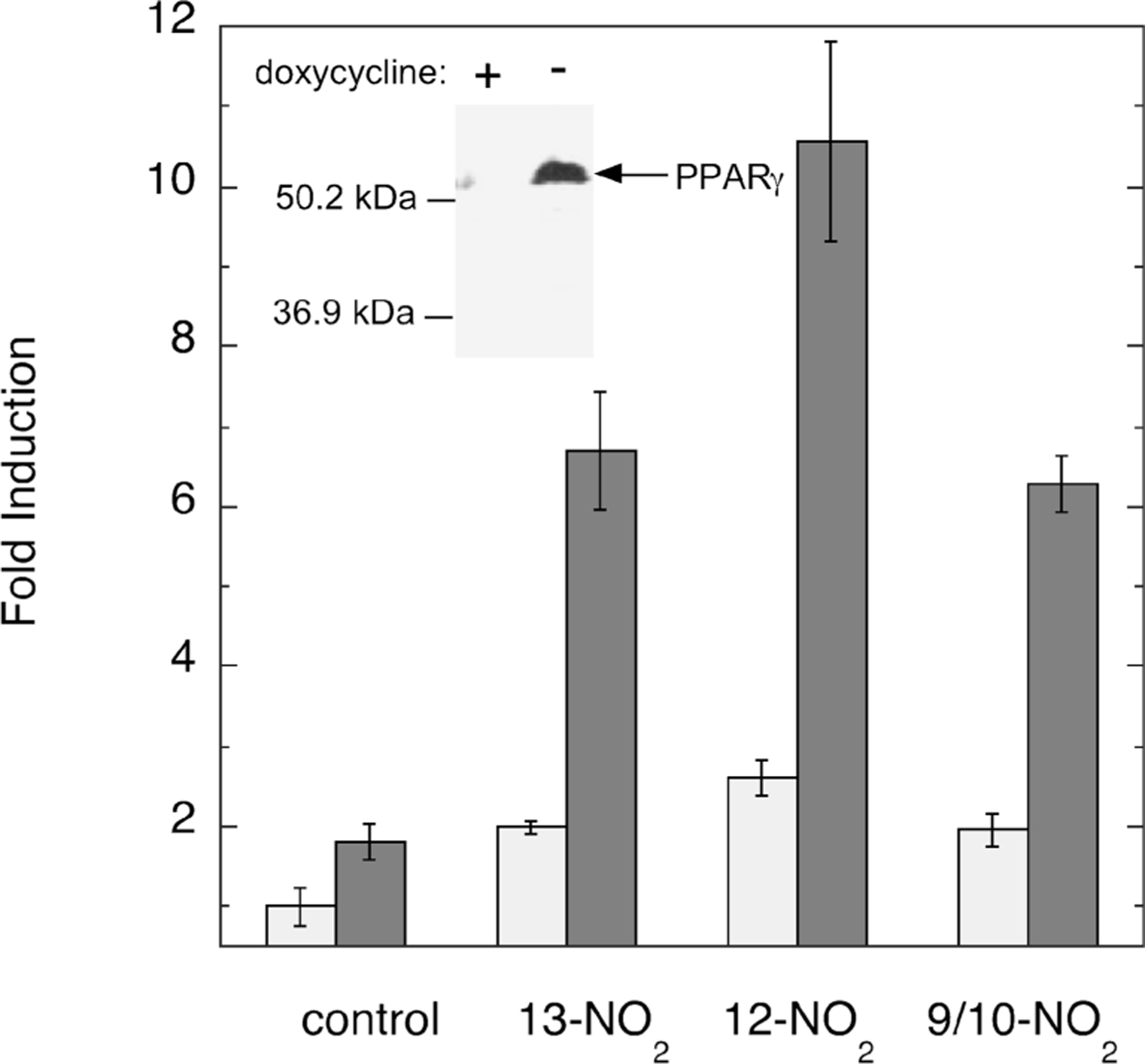
Activation of PPARγ-dependent transcription by regioisomers of NO2-LA., The inset shows a Western blot of PPARγ expression in MCF7/RTO/PPARG1–4ʹ cells grown in the presence (+) or absence (−) of doxycycline. The bar graph shows the induction of PPRE-containing reporter gene expression by NO2-LA isomers in the absence (light gray bars, with doxycycline) or presence (dark gray bars, without doxycycline) of PPARγ transgene expression. Induction was accomplished with 2 µM NO2-LA isomer or vehicle (control) as described in Experimental Procedures. Fold induction is defined as (corrected PPRE reporter activity + inducing agent)/[corrected PPRE reporter activity − inducing agent in doxycycline-treated (−PPARγ) cells]. Bars represent the mean values of triplicate determinations ± one standard deviation.
The relative potency of NO2-LA isomers in the activation of PPARγ-dependent transcription was examined in dose–response experiments using the PPARγ-expressing (without doxycycline) MCF7 cells. These experiments were conducted in triplicate and were confirmed using at least two independently derived preparations of the NO2-LA isomers. A representative experiment is shown in Figure 3. Values for the EC50 and maximum induction achieved by each isomer and rosiglitazone were calculated as described in Experimental Procedures. These experiments demonstrate that of the NO2-LA isomers, the 12-NO2 derivative is the most potent with an EC50 of 0.045 µM, comparable to that of rosiglitazone (0.067 µM), while 9/10-NO2-LA and 13-NO2-LA are weaker ligands with considerably higher EC50 values of 0.41 and 0.62 µM, respectively (Figure 3 and Table 4). Moreover, the maximum level of induction of PPARγ-dependent reporter gene activity achieved by 12-NO2-LA treatment, while somewhat lower than that by rosiglitazone, is nearly 2-fold higher than the maximum inductions achieved by 9/10- and 13-NO2-LA. In contrast and as shown previously, treatment with the parent un-nitrated compound, linoleic acid, results in negligible PPARγ activation even at concentrations as high as 100 µM (Figure 4).
FIGURE 3:
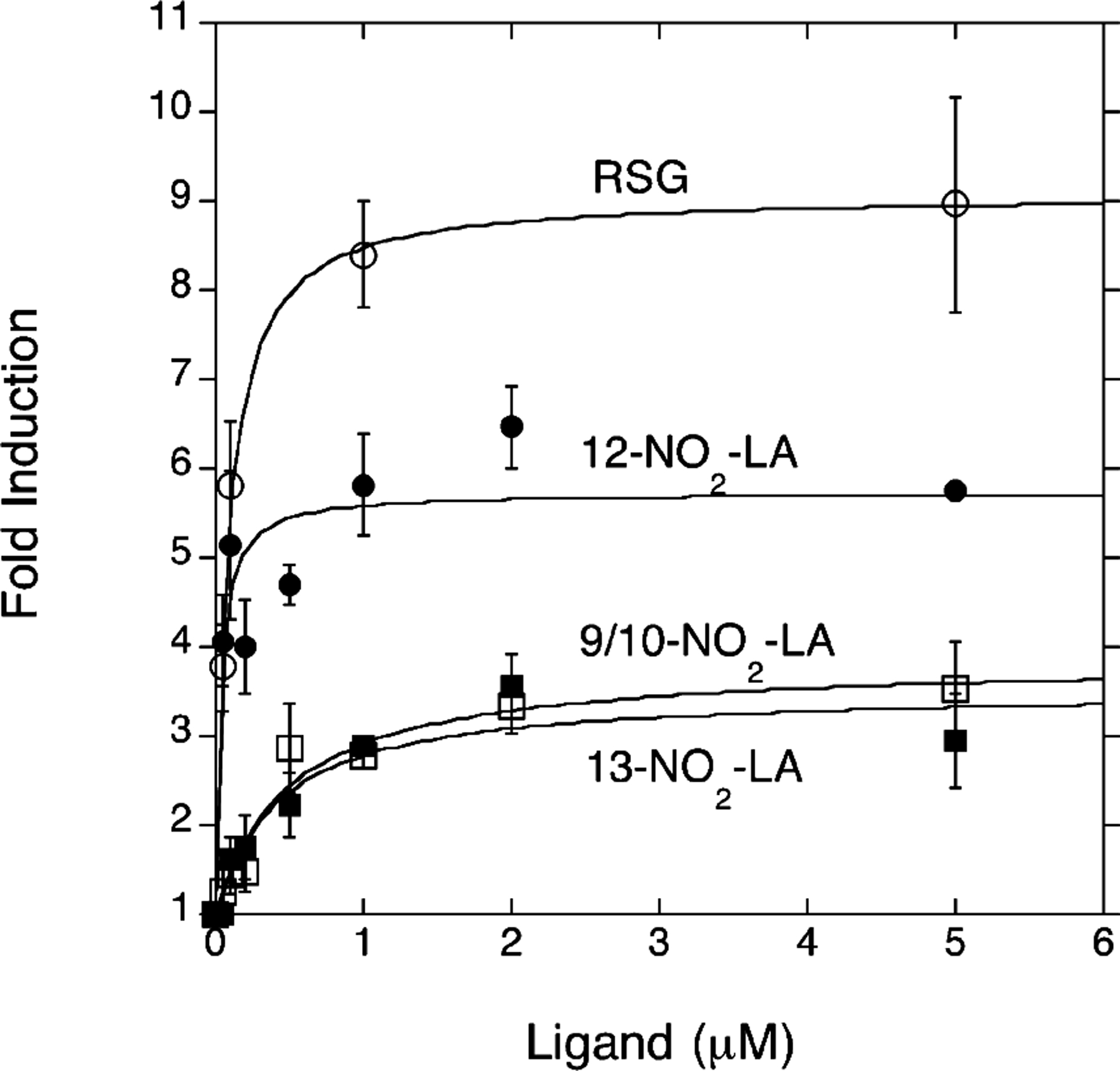
Dose–response analysis of NO2-LA isomer-mediated activation of PPARγ-dependent transcription. Concentration-dependent induction of PPARγ-mediated PPRE reporter gene expression was assessed for the ligands rosiglitazone [RSG (O)], 9/10-NO2-LA (□), 12-NO2-LA (●), and 13-NO2-LA (■). Experiments were conducted using PPARγ-expressing (without doxycycline) MCF7/RTO/PPARG1–4ʹ cells with fold induction defined and curve fitting accomplished as described in Experimental Procedures. Data points represent the mean values of triplicate determinations ± one standard deviation.
Table 4:
Transcriptional Activation of PPARγ by Nitrolinoleic Acid Derivatives and Rosiglitazone
| compound | EC50a (µM) | agonistb |
|---|---|---|
| rosiglitazone | 0.067 ± 0.015 | full (100 ± 4%) |
| 9/10-NO2-linoleic acid | 0.41 ± 0.07 | partial (36 ± 3%) |
| 12-NO2-linoleic acid | 0.045 ± 0.012 | partial (76 ± 9%) |
| 13-NO2-linoleic acid | 0.62 | partial (41 ± 5%) |
EC50 is the concentration of added ligand that yields 50% of the maximum observed PPARγ-dependent reporter gene activation. Values are the means of triplicate determinations ± one standard deviation (rosiglitazone and 9/10-NO2- and 12-NO2-linoleic acid) or the mean of duplicate determinations (13-NO2-linoleic acid).
Rosiglitazone is defined as a full agonist in the PPARγ-dependent reporter gene assay. In parentheses are mean inductions achieved by treatment with the various ligands at 5 µM shown as a percentage of induction achieved by treatment with 0.1 µM rosiglitazone (100%). Values are the means of 6–18 determinations ± one standard error of the mean.
FIGURE 4:
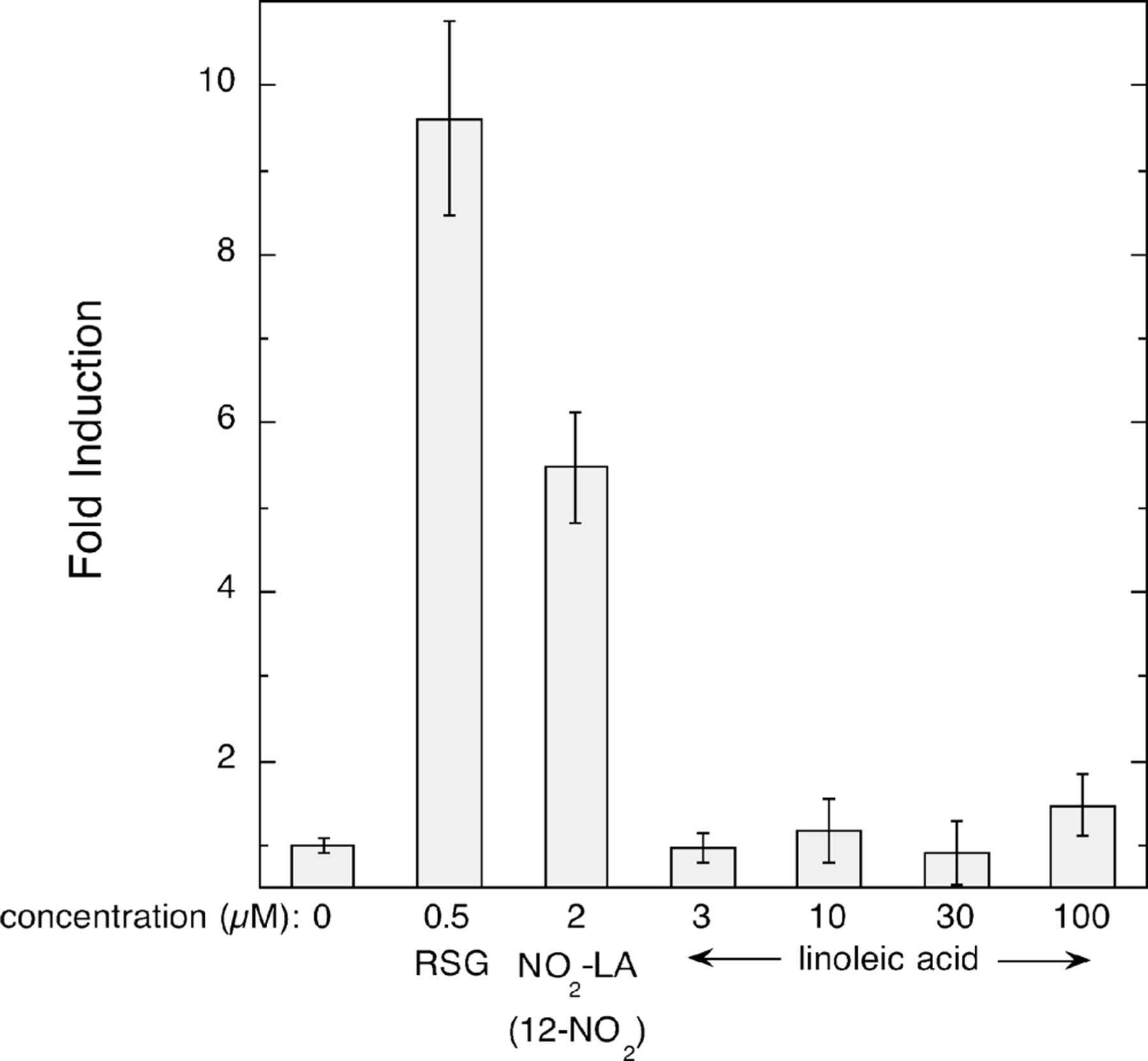
Linoleic acid does not support activation of PPARγ-dependent transcription. Inductions of PPRE reporter gene expression by indicated concentrations of rosiglitazone (RSG), 12-NO2-LA, and linoleic acid were determined in PPARγ-expressing (without doxycycline) MCF7/RTO/PPARG1–4ʹ cells as described in Experimental Procedures. Bars indicate the mean values of triplicate determinations ± one standard deviation.
The dose—response experiments (Figure 3) indicated that, in comparison to rosiglitazone, the weaker ligands, 9/10-NO2-LA and 13-NO2-LA, and to a lesser extent 12-NO2-LA are partial agonists of PPARγ in this cellular system. To verify this, transactivation experiments were conducted in which varying concentrations of NO2-LA isomers were coincubated with a fixed concentration (0.1 µM) of rosiglitazone. As shown in Figure 5, addition of increasing concentrations of 9/10-NO2- and 13-NO2-LA (panels A and C, respectively) significantly reduced the level of induction by rosiglitazone, while inhibition by the stronger agonist, 12-NO2-LA, was less pronounced (panel B). The results of the NO2-LA transcription activation experiments are summarized in Table 4.
FIGURE 5:
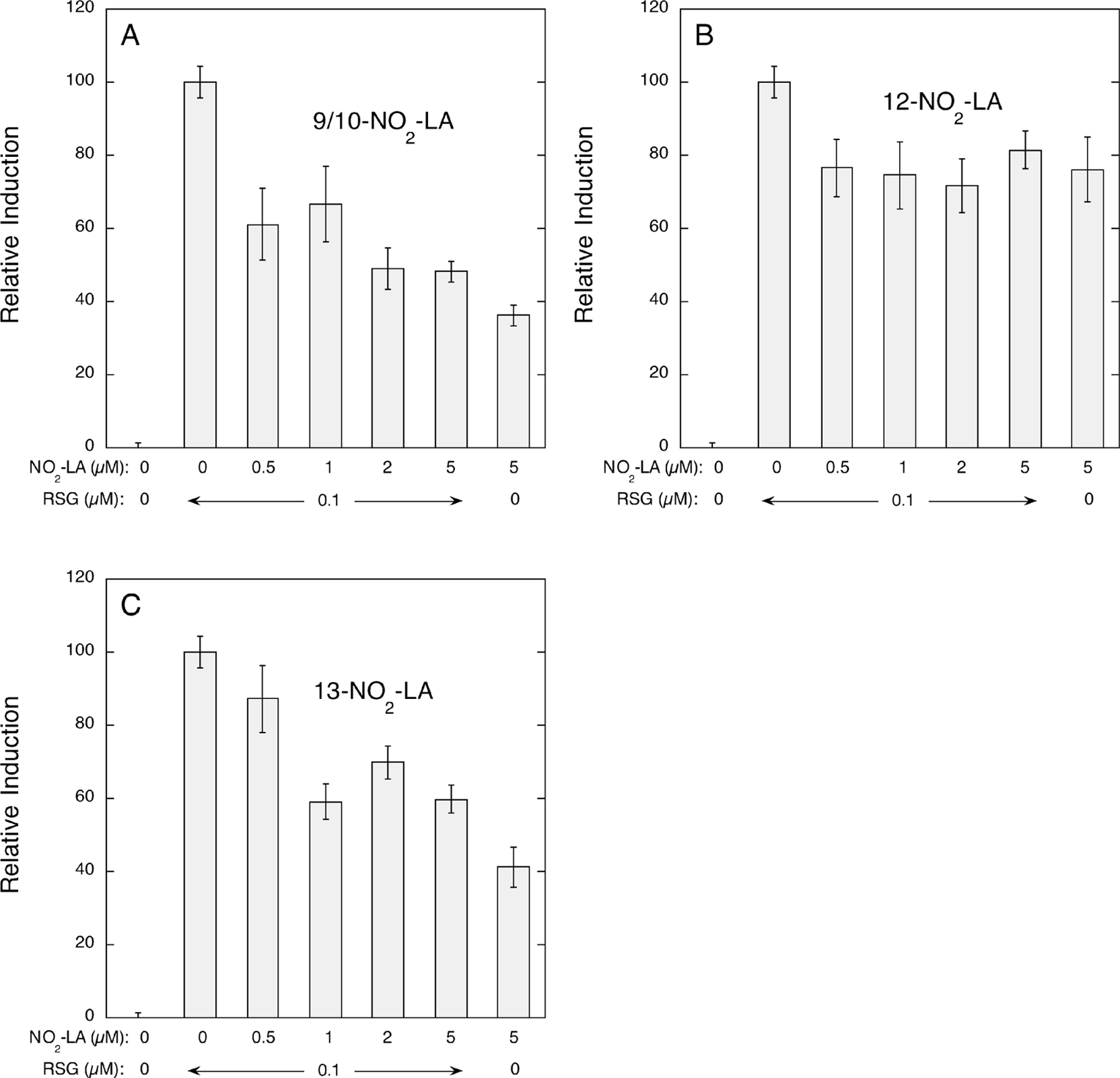
NO2-LA isomers are partial agonists of PPARγ. The effect of adding varying concentrations (0–5 µM) of NO2-LA isomers on induction of PPARγ-dependent reporter gene expression by fixed concentrations (0 or 0.1 µM) of rosiglitazone (RSG) was determined as described in Experimental Procedures and the legends of Figures 3 and 4. The concentrations of NO2-LA [9/10-NO2-LA (A), 12-NO2-LA (B), and 13-NO2-LA (C)] and rosiglitazone used in these inductions are shown under each panel. Relative induction values are derived from fold inductions normalized to 0.1 µM rosiglitazone alone (100%) with no ligand controls defined as 0%. Values are the means of 6–18 determinations ± one standard error of the mean.
DISCUSSION
The nitroalkenes have been shown to influence many signaling pathways (1–7). Their activities include suppression of inflammation and induction of cellular defense responses, relaxation of the vasculature, and changes in gene expression and differentiation via receptor-mediated processes. The physiological importance of nitroalkenes is suggested by reports indicating that nitrated oleic (NO2-OA) and linoleic (NO2-LA) acids are present in human plasma and blood cells at aggregate concentrations in the micromolar range, concentrations in excess of those necessary to mediate the bioactivities attributed to them (9). Although the mechanisms that underlie some of these activities are poorly or incompletely understood, many depend upon the electrophilic reactivity of the nitroalkenes. These mechanisms include the ability of nitroalkenes to form adducts with accessible protein thiols, resulting, for example, in the inhibition of NFkB (6) and glyceraldehyde-3-phosphate dehydrogenase (18). In addition, nitroalkenes can, by depleting intracellular glutathione, perturb redox homeostasis, and they can, by disrupting Keap1-mediated negative regulation of Nrf2, activate ARE-dependent transcription of genes involved in protecting cells from redox or xenobiotic stress (7). These activities are not unique to nitroalkenes but are shared by many electrophiles. Their mechanisms are not necessarily expected to be selective for particular nitroalkene species or isomers. In contrast, receptor-mediated processes, such as PPARγ-dependent transcription activation, are dependent upon precise interactions between the ligand and the ligand-binding pocket. For such interactions, the position of nitration on the nitroalkene ligand could have a profound effect on PPARγ binding and induced conformational changes associated with transcription activation. Hence, we hypothesized that regioisomers of NO2-LA differ in their PPARγ activating abilities.
Results presented herein confirm this expectation. The 12-NO2-LA isomer was the most potent with the highest maximum level of activation of PPARγ-dependent transcription and the lowest EC50 (Figure 3 and Table 4). The 9/10-NO2 and 13-NO2 derivatives were considerably less potent. While the 12-NO2 derivative had nearly full agonist activity relative to rosiglitazone, the 9/10-NO2 and 13-NO2 isomers were partial agonists with maximum levels of PPARγ activation that were only ~50% of that of the 12-NO2 isomer. Moreover, the EC50 values for 9/10-NO2-LA and 13-NO2-LA were more than 10-fold higher than for 12-NO2-LA.
A recent X-ray crystallography study reporting structures determined for the PPARγ ligand-binding domain in complex with NO2-LA provides considerable insight into the specificity of NO2-LA-PPARγ interactions, especially the role of NO2 in stabilizing the complex and inducing conformational changes in PPARγ that are associated with its transcriptional activation (19). The selectivity for NO2-LA is indicated by specific noncovalent interactions within the ligand-binding pocket between Arg288 or Glu343 and the nitro groups of NO2-LA. Interactions between these moieties stabilize PPARγ-NO2-LA complexes, and, from mutational analyses, these interactions appear to be required for optimal PPARγ activation by NO2-LA. Importantly, though, Arg288 interacts with the 10-NO2 group, whereas Glu343 interacts with the 12-NO2 group, suggesting that regioisomer-specific interactions occur between NO2-LA and PPARγ, perhaps providing a clue about the structural basis of our PPARγ activation observations. Indeed, it is tempting to speculate that the superior activity of 12-NO2-LA in our reporter gene system may be because of important interactions with Glu343 that, among the four NO2-LA isomers, is unique to this regioisomer. By similar reasoning, alternative interactions between the nitro groups and the ligand-binding pocket may ultimately explain the partial agonist activities observed for the 9/10- and 13-NO2 isomers.
An interesting unresolved finding deserves some additional discussion. It was surprising that the EC50 for 12-NO2-LA-mediated activation of PPARγ activation, which was more than 10-fold lower than those of 9/10-NO2-LA and 13-NO2-LA (Table 4), was not reflected by a significantly lower binding affinity [IC50 (Table 3)]. While it is possible that there are two binding configurations for 12-NO2-LA (one a low-affinity, kinetically favored, configuration comparable to those of the other isomers and a second high-affinity, kinetically unfavorable, configuration), it is not likely that the putative high-affinity configuration would have been missed by the radioligand competition assay used: this method involves a prolonged, 16 h, incubation period which should be sufficient to approach equilibrium for all but the slowest ligand-receptor interactions. A plausible explanation for the discrepancy between the relative EC50 and IC50 values includes the possibility that the 12-NO2-LA isomer is relatively more readily taken up by the cells or is more stable within the intracellular environment. With regard to the latter possibility, a recent report indicates that the position of nitration can have a significant effect on the stability of the NO2-LA isomer in aqueous solution (20). Indeed, this study found that the regioisomer nitrated at the terminal C-9 position of the 1,4-pentadiene moiety of LA was markedly less stable than the isomer nitrated at the internal C-10 position. While such differential stabilities of internal versus terminal nitrated derivatives of LA would not explain the higher maximum induction achieved with 12-NO2-LA (discussed above), it could help explain why 12-NO2-LA, an internally nitrated isomer, has a lower EC50 than either 13-NO2-LA or 9/10-NO2-LA. Alternatively, the 12-NO2-LA isomer may be metabolized, including the formation of adducts with small molecule nucleophiles such as glutathione, to high-affinity PPARγ ligands intracellularly. According to X-ray crystallography data (19), NO2-LA occupies only ~40% of the ligand-binding pocket, leaving ample room for accommodation of such additional moieties attached to the nitrated ligand.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants CA70338 (C.S.M.) and HL62198 (S.B.K.).
Abbreviations:
- ARE
antioxidant (or electrophile) responsive element
- COSY
correlation spectroscopy
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- HMBC
heteronuclear multiple-bond correlation
- HMQC
heteronuclear multiple-quantum coherence
- LA
linoleic acid [(9Z,12Z)-octadeca-9,12-dienoic acid]
- NO2-LA
nitrolinoleic acid (nitrooctadeca-9,12-dienoic acid)
- 9/10-NO2-LA
1:1 mixture of (9E,12Z)-9-nitrooctadeca-9,12-dienoic and (9E,12Z)-10-nitrooctadeca-9,12-dienoic acid
- 12-NO2-LA
(9Z,12E)-12-nitrooctadeca-9,12-dienoic acid
- 13-NO2-LA
(9Z,12E)-13-nitrooctadeca-9,12-dienoic acid
- NO2-OA
nitrooleic acid (nitrooctdeca-9-enoic acid)
- PPAR
peroxisomal proliferator-activated receptor
- PPRE
PPAR responsive element
- TOCSY
total correlation spectroscopy
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional NMR data and a discussion of the structural assignments made for the regioisomers of NO2-LA. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Freeman BA, Baker PRS, Schopfer FJ, Woodcock SR, Napolitano A, and d’Ischia M (2008) Nitro-fatty Acid Formation and Signaling. J. Biol. Chem 283, 15515–15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coles B, Bloodsworth A, Clark SR, Lewis MJ, Cross AR, Freeman BA, and O’Donnell VB (2002) Nitrolinoleate Inhibits Superoxide Generation, Degranulation, and Integrin Expression by Human Neutrophils: Novel Antiinflammatory Properties of Nitric Oxide-Derived Reactive Species in Vascular Cells. Circ. Res 91, 375–381. [DOI] [PubMed] [Google Scholar]
- 3.Lima ES, Bonini MG, Augusto O, Barbeiro HV, Souza HP, and Abdalla DS (2005) Nitrated lipids decompose to nitric oxide and lipid radicals and cause vasorelaxation. Free Radical Biol. Med 39, 532–539. [DOI] [PubMed] [Google Scholar]
- 4.Schopfer FJ, Lin Y, Baker PRS, Cui T, Garcia-Barrio M, Zhang J, Chen K, Chen YE, and Freeman BA (2005) Nitrolinoleic acid: An endogenous peroxisome proliferator-activated receptor γ ligand. Proc. Natl. Acad. Sci. U.S.A 102, 2340–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright MM, Schopfer FJ, Baker PRS, Vidyasagar V, Powell P, Chumley P, Iles KE, Freeman BA, and Agarwal, (2006) Fatty acid transduction of nitric oxide signaling: Nitrolinoleic acid potently activates endothelial heme oxygenase 1 expression. Proc. Natl. Acad. Sci. U.S.A 103, 4299–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui T, Schopfer FJ, Zhang J, Chen K, Ichikawa T, Baker PRS, Batthyany C, Chacko BK, Feng X, Patel RP, Agarwal A, Freeman BA, and Chen YE (2006) Nitrated Fatty Acids: Endogenous Anti-inflammatory Signaling Mediators. J. Biol. Chem 281, 35686–35698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villacorta L, Zhang J, Garcia-Barrio MT, Chen X. l., Freeman BA, Chen YE, and Cui T (2007) Nitro-linoleic acid inhibits vascular smooth muscle cell proliferation via the Keap1/Nrf2 signaling pathway. Am. J. Physiol 293, H770–H776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker PRS, Lin Y, Schopfer FJ, Woodcock SR, Groeger L, Batthyany C, Sweeney S, Long MH, Iles KE, Baker LMS, Branchaud BP, Chen YE, and Freeman BA (2005) Fatty Acid Transduction of Nitric Oxide Signaling: Multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J. Biol. Chem 280, 42464–42475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker PRS, Schopfer FJ, Sweeney S, and Freeman BA (2004) Red cell membrane and plasma linoleic acid nitration products: Synthesis, clinical identification, and quantitation. Proc. Natl. Acad. Sci. U.S.A 101, 11577–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexander RL, Bates DJ, Wright MW, King SB, and Morrow CS (2006) Modulation of Nitrated Lipid Signaling by Multidrug Resistance Protein 1 (MRP1): Glutathione Conjugation and MRP1-Mediated Efflux Inhibit Nitrolinoleic Acid-Induced, PPARγ-Dependent Transcription Activation. Biochemistry 45, 7889–7896. [DOI] [PubMed] [Google Scholar]
- 11.Gurnell M, Wentworth JM, Agostini M, Adams M, Collingwood TN, Provenzano C, Browne PO, Rajanayagam O, Burris TP, Schwabe JW, Lazar MA, and Chatterjee VKK (2000) A Dominant-negative Peroxisome Proliferator-activated Receptor γ (PPARγ) Mutant Is a Constitutive Repressor and Inhibits PPARγ-mediated Adipogenesis. J. Biol. Chem 275, 5754–5759. [DOI] [PubMed] [Google Scholar]
- 12.Elbrecht A, Chen Y, Adams A, Berger J, Griffin P, Klatt T, Zhang B, Menke J, Zhou G, Smith RG, and Moller DE (1999) L-764406 Is a Partial Agonist of Human Peroxisome Proliferator-activated Receptor γ. The Role of Cys313 in ligand binding. J. Biol. Chem 274, 7913–7922. [DOI] [PubMed] [Google Scholar]
- 13.Paumi CM, Wright M, Townsend AJ, and Morrow CS (2003) Multidrug Resistance Protein (MRP) 1 and MRP3 Attenuate Cytotoxic and Transactivating Effects of the Cyclopentenone Prostaglandin, 15-Deoxy-∆(12,14)Prostaglandin J(2) in MCF7 Breast Cancer Cells. Biochemistry 42, 5429–5437. [DOI] [PubMed] [Google Scholar]
- 14.Smitherman PK, Townsend AJ, Kute TE, and Morrow CS (2004) Role of Multidrug Resistance Protein 2 (MRP2, ABCC2) in Alkylating Agent Detoxification: MRP2 Potentiates Glutathione S-Transferase A1–1-Mediated Resistance to Chlorambucil Cytotoxicity. J. Pharmacol. Exp. Ther 308, 260–267. [DOI] [PubMed] [Google Scholar]
- 15.Berger JP, Petro AE, Macnaul KL, Kelly LJ, Zhang BB, Richards K, Elbrecht A, Johnson BA, Zhou G, Doebber TW, Biswas C, Parikh M, Sharma N, Tanen MR, Thompson GM, Ventre J, Adams AD, Mosley R, Surwit RS, and Moller DE (2003) Distinct Properties and Advantages of a Novel Peroxisome Proliferator-Activated Protein γ Selective Modulator. Mol. Endocrinol 17, 662–676. [DOI] [PubMed] [Google Scholar]
- 16.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, and Evans RM (1995) 15-Deoxy-∆12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR γ. Cell 83, 803–812. [DOI] [PubMed] [Google Scholar]
- 17.Sibhatu MB, Smitherman PK, Townsend AJ, and Morrow CS (2008) Expression of MRP1 and GSTP1–1 modulate the acute cellular response to treatment with the chemopreventive isothiocyanate, sulforaphane. Carcinogenesis 29, 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batthyany C, Schopfer FJ, Baker PRS, Duran R, Baker LMS, Huang Y, Cervenansky C, Branchaud BP, and Freeman BA (2006) Reversible Post-translational Modification of Proteins by Nitrated Fatty Acids in Vivo. J. Biol. Chem 281, 20450–20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Zhang J, Schopfer FJ, Martynowski D, Garcia-Barrio MT, Kovach A, Suino-Powell K, Baker PR, Freeman BA, Chen YE, and Xu HE (2008) Molecular recognition of nitrated fatty acids by PPARγ. Nat. Struct. Mol. Biol , doi: 10.1038/nsmb.1447. [DOI] [PMC free article] [PubMed]
- 20.Manini P, Capelli L, Reale S, Arzillo M, Crescenzi O, Napolitano A, Barone V, and d’Ischia M (2008) Chemistry of nitrated lipids: Remarkable instability of 9-nitrolinoleic acid in neutral aqueous medium and a novel nitronitrate ester product by concurrent autoxidation/nitric oxide-release pathways. J. Org. Chem 73, 7517–7525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


