Abstract
Thirteen psychrophilic sulfate-reducing isolates from two permanently cold fjords of the Arctic island Spitsbergen (Hornsund and Storfjord) were phylogenetically analyzed. They all belonged to the δ subclass of Proteobacteria and were widely distributed within this group, indicating that psychrophily is a polyphyletic property. A new 16S rRNA-directed oligonucleotide probe was designed against the largest coherent cluster of these isolates. The new probe, as well as a set of available probes, was applied in rRNA slot blot hybridization to investigate the composition of the sulfate-reducing bacterial community in the sediments. rRNA related to the new cluster of incompletely oxidizing, psychrophilic isolates made up 1.4 to 20.9% of eubacterial rRNA at Storfjord and 0.6 to 3.5% of eubacterial rRNA at Hornsund. This group was the second-most-abundant group of sulfate reducers at these sites. Denaturing gradient gel electrophoresis and hybridization analysis showed bands identical to those produced by our isolates. The data indicate that the psychrophilic isolates are quantitatively important in Svalbard sediments.
Low environmental temperatures characterize the habitat of many prokaryotes living in marine sediments, since 90% of the sea floor has a temperature of less than 4°C (15). While prokaryotic activity is commonly found to be lower during cold seasons in temperate environments (for a review, see reference 23), the current data suggest that in permanently cold habitats bacterial activity is comparable to that in temperate environments at the respective ambient temperature (2, 9, 23, 27). Arnosti et al. (2) determined the temperature dependence of microbial degradation of organic matter and showed that carbon turnover in the cold Arctic is not intrinsically slower than in temperate environments. Also, Sagemann et al. (27) and Glud et al. (9) found rates of sulfate reduction and benthic carbon mineralization in Arctic sediments to be comparable to those in temperate or even tropical sediments. Optimal temperatures for polysaccharide hydrolysis, oxygen consumption (2), and sulfate reduction (27) in permanently cold sediments were significantly higher than the ambient temperature; however, the relative activity at a low in situ temperature compared to optimum activity was generally higher than in samples from temperate habitats. These observations indicate that the bacterial community in these Arctic sediments is adapted to cold temperature. However, little is known about the diversity and composition of prokaryotic communities in cold marine sediments; only a few cold-adapted psychrophilic isolates from these environments have been studied so far (for a review, see reference 26). In addition, few cultivation-independent studies have been conducted in these habitats (28).
The aims of our project, conducted in the context of the above-mentioned studies, were to characterize the sulfate-reducing bacterial community of permanently cold habitats and to quantify the abundance of psychrophilic sulfate reducers. We chose two sites, off the coast of Spitsbergen (Hornsund and Storfjord), which are never exposed to temperatures higher than 3°C. We concentrated on sulfate-reducing prokaryotes because sulfate reduction is a major process of carbon mineralization in marine sediments (11). A set of probes is available for the main phylogenetic groups of gram-negative mesophilic sulfate reducers (7), and the different phylogenetic groups can be defined by distinct physiological features (35, 36). In a related study, most probable number (MPN) counts were determined and psychrophilic sulfate reducers were isolated to enumerate and identify the sulfate-reducing bacteria (SRB) (12). A new oligonucleotide probe was designed to target the largest cluster of these isolates. This newly developed probe, along with an established set of probes, was applied to quantify sulfate reducer rRNA in the sediment. The presence of the isolates was further evaluated by denaturing gradient gel electrophoresis (DGGE) analysis. The results of this study will be discussed in relation to data from a 16S ribosomal DNA (rDNA) clone library presented in an accompanying paper (22).
MATERIALS AND METHODS
Study site and sampling procedure.
Our study was conducted as part of a research cruise in the Arctic Sea from Tromsø (northern Norway) to Spitsbergen (Arctic Ocean) in September and October of 1995. Sediments from two different stations (Hornsund [76°58.2′N, 15°34.5′E] and Storfjord [77°33.0′N, 19°05.5′E]) were investigated. In situ temperatures and depths were 2.6°C and 155 m for Hornsund and −1.7°C and 175 m for Storfjord. Sediment samples were collected with a multicorer. Samples for MPN dilutions (12) and molecular analysis were taken from the same core. The individual subcores (our replicates A and B) derived from two different multicorer cores. The sediments were anoxic below a depth of approximately 8 mm (9). Five distinct vertical horizons of 2 to 3 cm in thickness were sectioned from the upper 30 cm of each core. The sediment of each section was carefully homogenized, and subsamples of 1 or 2 cm3 were immediately frozen in liquid N2.
DNA extraction and amplification of 16S rDNA.
After three cycles of freezing and thawing, DNAs were extracted directly by the method of Zhou et al. (38), which is based on lysis with a high-salt extraction buffer and extended heating in the presence of sodium dodecyl sulfate and hexadecyltrimethylammonium bromide. Lysis efficiency was checked by DAPI (4′,6-diamidino-2-phenylindole) staining. In general, at least 90 to 96% of the cells were lysed. The DNA could be used for PCR without further purification. Primers GM5clamp (Escherichia coli positions 341 to 357) and 907R (19) were used to amplify variable regions V3 to V5 of the 16S rDNA in a touchdown PCR, as described by Buchholz-Cleven et al. (5). To amplify the nearly complete 16S rDNA, primers 8F and 1492R (5) were used in a 35-cycle PCR with an annealing temperature of 40°C. Bovine serum albumin (final concentration, 3 mg ml−1) was added routinely to the PCR mixtures to prevent interference by humic acids (24).
16S rDNA sequencing.
PCR products were purified with a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). A Taq DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, Calif.) was used to directly sequence the purified PCR products. Sequencing reactions were analyzed on an Applied Biosystems model 373S DNA sequencer. Both strands of the amplification products were sequenced with primers 8F, 787F, 787R, 1175R, 1099F, and 1492R (5). Primer nomenclature refers to 5′ ends of the respective target sites on the 16S rDNA according to the E. coli numbering of 16S rRNA nucleotides (4).
Phylogenetic analysis.
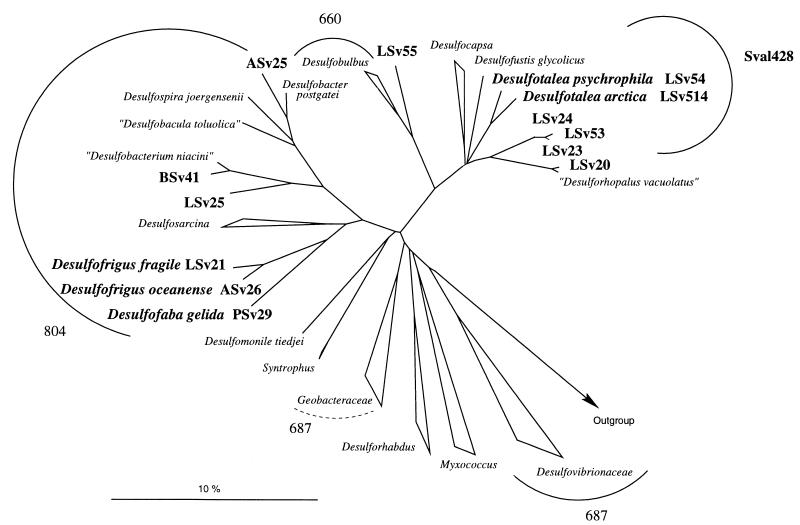
The ARB program package and the ARB database (34) were used for phylogenetic analysis. Sequences were aligned to the 16S rRNA primary structures present in the ARB database by using the automatic aligner tool, and the results were corrected manually where necessary. Pairwise distance matrix analysis was performed with the 16S rRNA sequences by considering only those positions that were present in both sequences. Phylogenetic trees were reconstructed for all available sequences from the δ subclass of Proteobacteria, and a selection of representatives of major groups outside this subclass was used as an outgroup. Only sequences with at least 1,350 nucleotides were used. Tree topology was evaluated by using neighbor-joining, maximum-parsimony, and maximum-likelihood algorithms on the full set of data or on a subset. Furthermore, we applied filters that excluded positions with less than 50% conservation within the δ subclass. Branching orders that were not supported by all methods are shown as multifurcation (Fig. 1).
FIG. 1.
Phylogenetic affiliations of psychrophilic sulfate-reducing isolates and specificities of probes used in this study. The tree shows members of the δ subclass of Proteobacteria and was constructed by using the neighbor-joining algorithm and a 50% conservation filter. The arcs comprise the respective probe target groups. Sval428 is described in this study. Other probes are as described by Devereux et al. (7). Strains isolated from Svalbard sediments are shown in boldface type. Strain names indicate the carbon sources on which the strains were isolated (A, acetate; L, lactate; P, propionate; and B, betaine) and the sampling site (Sv2 indicates Hornsund and Sv5 indicates Storfjord).
Oligonucleotide probes.
Oligonucleotides were purchased from Biometra (Göttingen, Germany). The probe target sequence of Sval428 is 5′ GTAAAATCCTGTCAGATGG 3′ (E. coli positions 428 to 446). Probes used and their specificities are shown in Fig. 1.
RNA extraction and slot blot hybridization.
Nucleic acids were isolated directly by bead beating, phenol extraction, and isopropanol precipitation as described by Sahm and Berniger (28). Between 10 and 100 ng of RNA was blotted on nylon membranes (Magna Charge; Micron Seperations, Westborough, Mass.) in triplicate and probed with radioactively labeled oligonucleotides as described previously (33). Membranes were prehybridized at 40°C and washed at different temperatures depending on the dissociation temperature (Td) of the probe as follows: 54°C (EUB338 [1]), 45°C (687 [7]), 59°C (660 [7]), 46°C (804 [7]), or 52°C (Sval428). The Td for probe Sval428 was determined according to the method of Raskin et al. (21), with rRNA from strain LSv20 being used as one mismatch control. Intensity of hybridization signal was measured with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and quantified as described by Sahm et al. (29) with the program ImageQuant (Molecular Dynamics). rRNA isolated from Desulfovibrio salexigens (DSM 2638), Desulfobulbus elongatus (DSM 2908), Desulfococcus multivorans (DSM 2059), and strain LSv23 served as standards for hybridization with the probes specific for sulfate reducers.
DGGE and Southern hybridization analysis.
DGGE was performed on a D-Gene system (Bio-Rad, Munich, Germany) as described previously (17, 18). PCR products were analyzed directly on a 1-mm-thick 6% polyacrylamide gel containing a denaturing gradient from 20 to 80%. Electrophoresis with 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8]) was performed at 100 V for 20 h. After electrophoresis, the gels were stained in ethidium bromide and photographed on a UV transilluminator. DGGE gels were blotted onto nylon membranes via electroblotting as described by Muyzer et al. (18). Hybridization analysis was performed with probe Sval428 by the protocol described by Santegoeds et al. (30). The probe was end labeled with [γ-32P]ATP, and the membrane was hybridized at 40°C overnight. Stringent washes were performed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% (wt/vol) sodium dodecyl sulfate at the previously determined Td of 56°C. The Td of the probe was determined by the method described by Raskin et al. (21) for DNA and RNA targets. The hybridized membranes were sealed in plastic bags and exposed for 1 to 7 days on an X-ray film or a PhosphorImager screen.
Nucleotide sequence accession numbers.
The 16S rDNA sequences have been deposited in the GenBank database. Accession numbers are AF099054 to AF099065 and AF136008.
RESULTS AND DISCUSSION
Phylogenetic affiliation of isolates.
Psychrophilic sulfate reducers isolated from MPN enrichments (12) were phylogenetically analyzed by 16S rDNA sequencing. All isolates belonged to the δ subclass of the Proteobacteria, as is the case for the majority of mesophilic sulfate reducers (32). Adaptation to cold temperatures as represented by our sulfate-reducing isolates was widely spread within this phylogenetic group (Fig. 1). On the basis of their phylogenetic distance, strains ASv25, BSv41, and LSv20 belong to the existing genera Desulfobacter, Desulfobacterium, and Desulforhopalus (3.6, 1.9, and 0.7% phylogenetic distance, respectively) (Fig. 1). The remaining 10 strains were only distantly related (phylogenetic distance, 6.4 to 11.6%) to known sulfate reducers. Five of these have recently been described as members of three new genera (13). The results suggest that psychrophily is a polyphyletic property within gram-negative sulfate reducers. Furthermore, the wide distribution of our isolates within the δ-Proteobacteria indicates that the diversity among psychrophilic SRB might be as high as among mesophiles.
Probe design.
Since cultivation is inherently selective and often leads to the isolation of quantitatively less-important microbial groups, we estimated the abundance of these isolates by a cultivation-independent method, i.e., rRNA slot blot hybridization. Besides LSv55, all strains that were related to the Desulfobacteriaceae were targeted by an already existing probe (probe 804) designed by Devereux et al. (7). Of the remaining strains, six incomplete oxidizers formed a cluster related to Desulfocapsa sp., Desulfofustis glycolicus, and the only other known moderately psychrophilic sulfate reducer, “Desulforhopalus vacuolatus” ltk10 (10) (Fig. 1). Two strains of this cluster have recently been described as species of the new genus Desulfotalea (13). The cluster will in what follows be referred to as the Desulfotalea cluster. This cluster was chosen for the design of a new oligonucleotide probe (Sval428). In addition to the two Desulfotalea strains and strains LSv23, LSv24, and LSv53, one of the mesophilic sulfate reducers, Desulfofustis glycolicus (8), is targeted by the probe (Fig. 1). It has one mismatch to LSv20 and “Desulforhopalus vacuolatus” ltk10 (10). LSv55 and LSv20 are the only strains isolated from Spitsbergen sediments not targeted by the set of probes used.
rRNA quantification of sulfate reducers.
rRNA slot blot hybridization revealed that the concentration of SRB rRNA for all target groups generally decreased with depth (Table 1), following the trend of total prokaryotic rRNA (28) and MPN counts (12). The relative contribution of SRB rRNA to total eubacterial rRNA (pooled signals from all probes), however, increased from 3.8% in core A and 4.0% in core B at the surface to 10.5% in core A and 17% in core B at a depth of 15 to 18 cm in Hornsund and from 10.5% in core A and 7% in core B to 58.6% in core A and 36.4% in core B at a depth of 27 to 30 cm in Storfjord (Table 2).
TABLE 1.
Recovered SRB rRNAs
| Site | Depth interval (cm) | Total SRB rRNA recovered (ng per cm3 of sediment) from core:
|
|
|---|---|---|---|
| A | B | ||
| Hornsund | 0–2 | 113 | 134 |
| 3–6 | 71 | 81 | |
| 8–11 | 26 | 55 | |
| 15–18 | 8 | 43 | |
| 25–28 | 4 | 8 | |
| Storfjord | 0–3 | 87 | 36 |
| 3–6 | 82 | 25 | |
| 7–10 | 21 | 17 | |
| 17–20 | 15 | 13 | |
| 27–30 | 10 | 12 | |
TABLE 2.
Relative contributions of different probe target groups to the total eubacterial rRNA
| Site | Depth interval (cm) | % of total bacterial rRNA (core A/core B)
|
% of Sval428-specific SRB (mean) | |||
|---|---|---|---|---|---|---|
| Desulfotalea sp. (Sval428 specific) | Desulfovibrionaceae (687 specific) | Desulfobulbus sp. (660 specific) | Sum of detected SRB | |||
| Hornsund | 0–2 | 0.6/1.1 | 2.5/2.3 | 0.7/0.6 | 3.8/4.0 | 22 |
| 3–6 | 0.7/2.5 | 4.6/3.6 | 1.2/1.5 | 6.5/7.6 | 22 | |
| 8–11 | 0.6/3.5 | 6.4/10.1 | 1.6/1.4 | 8.6/15.0 | 15 | |
| 15–18 | 0.3/2.6 | 8.6/12.9 | 1.6/1.5 | 10.5/17.0 | 9 | |
| 25–28 | 0.0/1.9 | 6.0/11.1 | 1.2/0.5 | 7.2/13.5 | 13 | |
| Storfjord | 0–3 | 1.6/1.4 | 8.1/5.0 | 0.8/0.6 | 10.5/7 | 18 |
| 3–6 | 2.6/3.5 | 14.9/6.5 | 2.7/0.8 | 20.2/10.8 | 23 | |
| 7–10 | 3.9/4.4 | 10.4/6.7 | 1.4/0.7 | 15.7/11.8 | 31 | |
| 17–20 | 11.5/9.2 | 20.7/14.1 | 1.5/0.9 | 33.7/24.2 | 36 | |
| 27–30 | 20.9/15.0 | 36.0/20.4 | 1.7/1.0 | 58.6/36.4 | 38 | |
The high concentration of detectable SRB rRNA in the first 2 to 3 cm of the sediment is in striking contrast to the very low sulfate reduction rates measured within this layer in parallel cores (12, 27). The high abundance of SRB in the zone where low sulfate reduction rates are measured can also be seen in MPN counts (12) and was also observed in a coastal sediment of the Baltic Sea by RNA slot blot hybridization (29). These findings suggest that SRB might use electron acceptors other than sulfate in the oxidized zone of the sediment. In this respect it is interesting that four of the five isolated strains tested for iron reduction were able to grow on Fe(III) (13). In addition Kostka et al. (14) showed that in Storfjord sediments Fe(III) reduction accounted for almost 10% of the total carbon oxidation but that it was insignificant at Hornsund. Other potential electron acceptors like NO3− or Mn(IV) were of minor importance in the investigated sediments (14).
Relative abundance of SRB and physiological considerations.
SRB can, in general, be divided into two major groups: those that oxidize the carbon source completely to CO2 and those that oxidize the carbon source incompletely to acetate. rRNA hybridization revealed a predominance of incompletely oxidizing SRB in these sediments. We detected incompletely oxidizing groups targeted by probes 687 (Desulfovibrionaceae but also some Geobacteraceae [16]), 660 (Desulfobulbus sp.), and Sval428 (Desulfotalea cluster), while the completely oxidizing genera Desulfococcus, Desulfosarcina, Desulfobacterium, and Desulfobacter targeted by probe 804 were below the detection limit (Table 2). This result is in agreement with cultivation-dependent data, since cell numbers of complete oxidizers on acetate were 10- to 100-fold lower than those of lactate oxidizers (12). If this finding reflects the actual relation between complete and incomplete oxidizers, the RNA concentration for acetate oxidizers would be at or below the detection limit of slot blot hybridization. Another possible explanation for not detecting RNA of complete oxidizers is that these organisms were not targeted by the probe used; all completely oxidizing strains isolated in this study, however, had the target sequence of probe 804.
The low abundance of complete oxidizers contrasts with results from estuarine, coastal, and vegetated salt marsh sediments (6, 25, 29), where the 804 target group was reported to be one of the major groups of SRB. Dominant sulfate reducers in these studies were species of the nutritionally versatile genera Desulfobacterium, Desulfococcus, and Desulfosarcina. The occurrence of these groups may reflect the input of a wide variety of carbon sources in coastal zone habitats close to the mainland. Our study, in contrast, was conducted in a remote, sparsely populated region. Data on the amount and type of biologically available carbon sources would be necessary to determine the relationship between substrates and the occurrence of specific groups of SRB.
A second group of complete oxidizers, namely, those that grow readily on acetate, is of special interest in sediments. These bacteria belong to the genus Desulfobacter, like strain ASv25, or to the gram-positive genus Desulfotomaculum. Although acetate is hypothesized to be one of the major carbon sources for SRB in marine sediments (20, 31), we could not detect Desulfobacter sp. rRNA in our samples. Boschker et al. (3) showed recently that addition of [13C]acetate to an intertidal sediment led to an incorporation of label in polar-lipid-derived fatty acids typical of Desulfotomaculum acetoxidans. An rRNA probe for Desulfotomaculum sp. is not yet available. Neither cultivation (12) nor clone library (22) data indicate the presence of Desulfotomaculum sp., but the use of a specific probe is needed to further investigate their role in acetate oxidation in marine sediments.
The major group of SRB was the Desulfovibrionaceae or Geobacteraceae cluster (probe 687), which in the deeper zones accounted for up to 8.6 and 12.9% of the RNA at Hornsund (15 to 18 cm) and for 36.0 and 20.4% at Storfjord (27 to 30 cm) (Table 2); however, no Desulfovibrionaceae were isolated from MPN cultures (12). We took this as an indication that the detected RNA might be coming from organisms of the Geobacteraceae group. A clone library established for Hornsund sediment samples (see the accompanying publication [22]) further supported this theory. Of all clones screened, 46 (13%) gave a positive signal with probe 687. Diversity within this group was very low, with one phylotype being represented by 39 clones and six additional phylotypes being represented by only one or two clones (22). All belonged to the family Geobacteraceae and were most closely related to Desulfuromonas palmitatis. Species of the genus Desulfuromonas belong to the δ subclass of Proteobacteria and are able to completely oxidize acetate via reduction of sulfur (37). Since completely oxidizing genera of sulfate reducers (804 target group) were below the detection limit, the high abundance of sequences related to Desulfuromonas sp. in the clone library might indicate that acetate is mineralized by sulfur reducers in these sediments; however, the phylogenetic distance between the clones and Desulfuromonas palmitatis is so large (6.3%) that we can only speculate on the possible physiological properties of this group until pure cultures have been isolated. Sequence information derived from the clone library will enable us now to design a specific probe for this 687-positive clone group and investigate its actual abundance. Furthermore, the phylogenetic affiliation might help to choose selective culture conditions.
RNA related to the genus Desulfobulbus (probe 660) was present in small amounts in both stations, with relative contributions varying between 0.5 and 2.7%. Clone library data suggest at least one additional group of SRB not targeted by our probes. This group is related to Desulfobulbus sp. and to isolate LSv55 and represents 6.5% of the clone library (22). A new specific probe is being developed to investigate the abundance of these isolates in the sediment.
Occurrence of the new psychrophilic isolates.
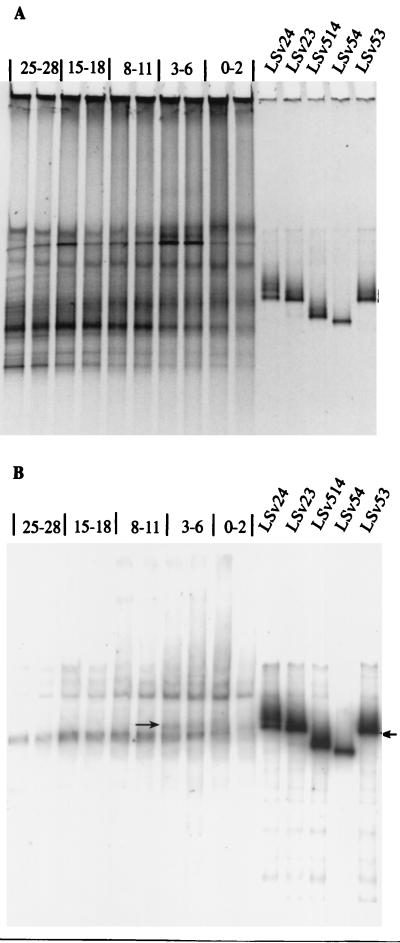
The Desulfotalea cluster (probe 428), containing many psychrophilic isolates, was the second largest group among the detected sulfate reducers. In Storfjord, up to 15.0 and 20.9% (27 to 30 cm) of the eubacterial RNA was related to our isolates (Table 2). To estimate whether potentially dominant strains of the target group had been isolated, DGGE and Southern hybridization with probe Sval428 were performed on community DNAs. Positions of the hybridization signals within the community pattern were compared to the positions of amplified 16S rDNAs from isolates belonging to this group (Fig. 2). In both stations, we could detect several positive bands. One of them had the same position as isolates LSv23, LSv24, and LSv53 (Fig. 2). These isolates are closely related, showing three to four bases’ difference within the amplified DNA fragment, and cannot be distinguished by DGGE. The presence of additional positive bands showed that there are at least three additional RNA species of this group present in Hornsund (Fig. 2) and one in Storfjord (data not shown) sediments. The high abundance of Desulfotalea-related rRNAs and the identification of bands corresponding to isolates in the community DNA profile demonstrate that a quantitatively important group of sulfate reducers was isolated from Svalbard sediment. In addition, this group was the second most abundant of the detected SRB (Table 2). Since it is doubtful that the most abundant SRB group, target group 687, is really a group of sulfate reducers, the Desulfotalea cluster, containing mainly psychrophilic strains, may even be the most abundant of SRB. This observation relates to the question of whether the bacterial community in Arctic sediments consists of cold-adapted prokaryotes. Our results show that a major group of SRB in this habitat is psychrophilic.
FIG. 2.
DGGE-hybridization analysis of community 16S rDNA patterns with probe Sval428 for Hornsund sediments. (A) DGGE results; (B) corresponding hybridization results. Numbers give the depth ranges from which target DNAs were extracted. For each depth, two cores have been investigated. “LSv” followed by a number indicates a DGGE fragment of a psychrophilic sulfate-reducing isolate targeted by probe Sval428. The arrow indicates corresponding bands between the community profile and isolates.
In the present study, we showed that psychrophilic SRB are related to mesophilic strains and are probably as phylogenetically diverse as the mesophiles. A new group of psychrophilic sulfate-reducing isolates is abundant in their source sediments. Their abundance suggests that they may play a previously unrecognized role in the sulfur cycle of marine sediments.
ACKNOWLEDGMENTS
We are grateful to Birgit Rattunde for excellent technical assistance. We also thank the cruise leader Donald E. Canfield and the crew of the RV Jan Mayen for a successful cruise. We acknowledge two anonymous reviewers for helpful comments on the manuscript.
This work was supported by the Max-Planck-Society.
REFERENCES
- 1.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnosti C, Sagemann J, Jørgensen B B, Thamdrup B. Temperature dependence of microbial degradation of organic matter in marine sediments: polysaccharide hydrolysis, oxygen consumption, and sulfate reduction. Mar Ecol Prog Ser. 1998;165:59–70. [Google Scholar]
- 3.Boschker H T S, Nold S C, Wellsbury P, Bos D, de Graaf W, Pel R, Parkes R J, Cappenberg T E. Direct linking of microbial population to specific biogeochemical processes by 13C-labelling of biomarkers. Nature. 1998;392:801–805. [Google Scholar]
- 4.Brosius J, Dull T, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 5.Buchholz-Cleven B E E, Rattunde B, Straub K L. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst Appl Microbiol. 1997;20:301–309. [Google Scholar]
- 6.Devereux R, Hines M E, Stahl D A. S cycling: characterization of natural communities of sulfate-reducing bacteria by 16S rRNA sequence comparisons. Microb Ecol. 1996;32:283–292. doi: 10.1007/BF00183063. [DOI] [PubMed] [Google Scholar]
- 7.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 8.Friedrich M, Springer N, Ludwig W, Schink B. Phylogenetic positions of Desulfofustis glycolicus gen. nov., sp. nov., and Syntrophobotulus glycolicus gen. nov., sp. nov., two new strict anaerobes growing with glycolic acid. Int J Syst Bacteriol. 1996;46:1065–1069. doi: 10.1099/00207713-46-4-1065. [DOI] [PubMed] [Google Scholar]
- 9.Glud R N, Holby O, Hoffmann F, Canfield D E. Benthic mineralization and exchange in Arctic sediments (Svalbard, Norway) Mar Ecol Prog Ser. 1998;173:237–251. [Google Scholar]
- 10.Isaksen M F, Teske A. Desulforhopalus vacuolatus gen. nov., sp. nov., a new moderately psychrophilic sulfate-reducing bacterium with gas vacuoles isolated from a temperate estuary. Arch Microbiol. 1996;166:160–168. [Google Scholar]
- 11.Jørgensen B B. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature. 1982;296:643–645. [Google Scholar]
- 12.Knoblauch C, Jørgensen B B, Harder J. Community size and metabolic rates of psychrophilic sulfate-reducing bacteria in Arctic marine sediments. Appl Environ Microbiol. 1999;65:4230–4233. doi: 10.1128/aem.65.9.4230-4233.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoblauch, C., K. Sahm, and B. B. Jørgensen. Psychrophilic sulfate-reducing bacteria isolated from permanently cold Arctic marine sediments: description of Desulfofrigus oceanense gen. nov., sp. nov., Desulfofrigus fragile sp. nov., Desulfofaba gelida gen. nov., sp. nov., Desulfotalea psychrophila gen. nov., sp. nov., and Desulfotalea arctica sp. nov. Int. J. Syst. Bacteriol., in press. [DOI] [PubMed]
- 14.Kostka J E, Thamdrup B, Glud R N, Canfield D. Rates and pathways of carbon oxidation in permanently cold Arctic sediments. Mar Ecol Prog Ser. 1999;180:2–21. [Google Scholar]
- 15.Levitus S, Boyer T. World ocean atlas. Vol. 4. Washington, D.C: Temperature. U.S. Department of Commerce; 1994. [Google Scholar]
- 16.Lonergan D J, Jenter H L, Coates J D, Phillips E J P, Schmidt T M, Lovley D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Waver C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 3rd ed. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 3.4.4.1–3.4.4.23. [Google Scholar]
- 18.Muyzer G, Hottenträger S, Teske A, Waver C. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—a new molecular approach to analyse the genetic diversity of mixed microbial communities. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 2nd ed. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 3.4.4.1–3.4.4.23. [Google Scholar]
- 19.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 20.Parkes R J, Gibson G R, Mueller-Harvey I, Buckingham W J, Herbert R A. Determination of the substrates for sulphate-reducing bacteria within marine and estuarine sediments with different rates of sulphate reduction. J Gen Microbiol. 1989;135:175–187. [Google Scholar]
- 21.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravenschlag K, Sahm K, Pernthaler J, Amann R. High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol. 1999;65:3982–3989. doi: 10.1128/aem.65.9.3982-3989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivkin R B, Anderson M R, Lajzerowicz C. Microbial processes in cold oceans. I. Relationship between temperature and bacterial growth rate. Aquat Microb Ecol. 1996;10:243–254. [Google Scholar]
- 24.Romanowski G, Lorenz M G, Wackernagel W. Use of polymerase chain reaction and electroporation of Escherichia coli to monitor the persistence of extracellular plasmid DNA introduced into natural soils. Appl Environ Microbiol. 1993;59:3438–3446. doi: 10.1128/aem.59.10.3438-3446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rooney-Varga J N, Devereux R, Evans R S, Hines M E. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl Environ Microbiol. 1997;63:3895–3901. doi: 10.1128/aem.63.10.3895-3901.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russel N J, Hamamoto T. Psychrophiles. In: Horikoshi K, Grant W D, editors. Extremophiles: microbial life in extreme environments. New York, N.Y: John Wiley & Sons; 1998. pp. 25–45. [Google Scholar]
- 27.Sagemann J, Jørgensen B B, Greeff O. Temperature dependence and rates of sulfate reduction in cold sediments of Svalbard, Arctic Ocean. Geomicrobiol J. 1998;15:85–100. [Google Scholar]
- 28.Sahm K, Berninger U-G. Abundance, vertical distribution, and community structure of benthic prokaryotes from permanently cold marine sediments (Svalbard, Arctic Ocean) Mar Ecol Prog Ser. 1998;165:71–80. [Google Scholar]
- 29.Sahm K, MacGregor B J, Jørgensen B B, Stahl D A. Sulphate reduction and vertical distribution of sulphate-reducing bacteria quantified by rRNA slot-blot hybridization in a coastal marine sediment. Environ Microbiol. 1999;1:65–74. doi: 10.1046/j.1462-2920.1999.00007.x. [DOI] [PubMed] [Google Scholar]
- 30.Santegoeds C M, Ferdelman T G, Muyzer G, de Beer D. Structural and functional dynamics of sulfate-reducing populations in bacterial biofilms. Appl Environ Microbiol. 1998;64:3731–3739. doi: 10.1128/aem.64.10.3731-3739.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sørensen J, Christensen D, Jørgensen B B. Volatile fatty acids and hydrogen as substrates for sulfate-reducing bacteria in anaerobic marine sediments. Appl Environ Microbiol. 1981;42:5–11. doi: 10.1128/aem.42.1.5-11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stackebrandt E, Stahl D A, Devereux R. Taxonomic relationships. In: Barton L L, editor. Sulfate-reducing bacteria. New York, N.Y: Plenum Press; 1995. pp. 49–87. [Google Scholar]
- 33.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strunk O, Gross O, Reichel B, May M, Hermann S, Stuckman N, Nonhoff B, Lenke M, Ginhart A, Vilbig A, Ludwig T, Bode A, Schleifer K-H, Ludwig W. ARB: a software environment for sequence data. Munich, Germany: Department of Microbiology, Technische Universität München; 1998. [Google Scholar]
- 35.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]
- 36.Widdel F, Hansen T. The dissimilatory sulfate- and sulfur-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 583–624. [Google Scholar]
- 37.Widdel F, Pfennig N. The genus Desulfuromonas and other Gram-negative sulfur-reducing eubacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. Vol. 4. New York, N.Y: Springer-Verlag; 1992. pp. 3379–3389. [Google Scholar]
- 38.Zhou J, Brunns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]