ABSTRACT
Viral infections alter host cell metabolism and homeostasis; however, the mechanisms that regulate these processes have only begun to be elucidated. We report here that Zika virus (ZIKV) infection activates the antioxidant nuclear factor erythroid 2-related factor 2 (Nrf2), which precedes oxidative stress. Downregulation of Nrf2 or inhibition of glutathione (GSH) synthesis resulted in significantly increased viral replication. Interestingly, 6-amino-nicotinamide (6-AN), a nicotinamide analog commonly used as an inhibitor of the pentose phosphate pathway (PPP), decreased viral replication by over 1,000-fold. This inhibition was neither recapitulated by the knockdown of PPP enzymes, glucose 6-phosphate dehydrogenase (G6PD), or 6-phosphogluconate dehydrogenase (6PGD), nor prevented by supplementation with ribose 5-phosphate. Instead, our metabolomics and metabolic phenotype studies support a mechanism in which 6-AN depletes cells of NAD(H) and impairs NAD(H)-dependent glycolytic steps resulting in inhibition of viral replication. The inhibitory effect of 6-AN was rescued with precursors of the salvage pathway but not with those of other NAD+ biosynthesis pathways. Inhibition of glycolysis reduced viral protein levels, which were recovered transiently. This transient recovery in viral protein synthesis was prevented when oxidative metabolism was inhibited by blockage of the mitochondrial pyruvate carrier, fatty acid oxidation, or glutaminolysis, demonstrating a compensatory role of mitochondrial metabolism in ZIKV replication. These results establish an antagonistic role for the host cell Nrf2/GSH/NADPH-dependent antioxidant response against ZIKV and demonstrate the dependency of ZIKV replication on NAD(H). Importantly, our work suggests the potential use of NAD(H) antimetabolite therapy against the viral infection.
IMPORTANCE Zika virus (ZIKV) is a major public health concern of international proportions. While the incidence of ZIKV infections has declined substantially in recent years, the potential for the reemergence or reintroduction remains high. Although viral infection alters host cell metabolism and homeostasis to promote its replication, deciphering the mechanism(s) involved in these processes is important for identifying therapeutic targets. The present work reveals the complexities of host cell redox regulation and metabolic dependency of ZIKV replication. An antagonistic effect of the Nrf2/GSH/NADP(H)-dependent antioxidant response against ZIKV infection and an essential role of NAD(H) metabolism and glycolysis for viral replication are established for the first time. These findings highlight the potential use of NAD(H) antimetabolites to counter ZIKV infection and pathogenesis.
KEYWORDS: Zika virus, Nrf2, GSH, 6-AN, oxidative stress, pentose phosphate pathway
INTRODUCTION
Zika virus (ZIKV) is a member of the family Flaviviridae. The ~10.8-kb positive-sense RNA genome of the virus encoding a single open reading frame (ORF) is translated into a polyprotein that is processed by cellular and viral proteases to yield 3 structural proteins (capsid [C], premembrane [prM], and envelope [E]) and 7 nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (1, 2). While the NS proteins are involved in the replication of the genome, assembly of progeny virions, manipulation of cellular signaling pathways, and evasion of host antiviral responses, the structural proteins are mainly responsible for virus attachment and entry into host cells as well as virus particle formation (1, 3).
ZIKV is primarily transmitted by mosquitoes, although other modes of transmission such as sexual and vertical transmission have also been reported. It exhibits broad tissue and cell tropism, including brain and placenta, and the virus can be secreted from body fluids including tears, semen, saliva, urine, and cervical mucus (4). The explosive epidemic in Brazil in 2015 to 2016 was associated with serious congenital abnormalities, including fetal growth restriction and microcephaly (5). While the majority of ZIKV infections in adults are asymptomatic, with only a fraction of the infected individuals showing symptoms of febrile illnesses such as mild fever, rash, and conjunctivitis (5), the viral infection can trigger development of Guillain Barré syndrome (5), and studies have also demonstrated cell death and reduced proliferation of adult stem cells (6).
It is well known that viral infection alters host cell metabolism, but the mechanisms and consequences are still unclear. Host cell metabolism is hijacked and rewired by viruses for the generation of macromolecules, including nucleotides, amino acids, and lipids needed for viral replication, protein synthesis, and virion assembly, which also requires an increase in the utilization of various nutrient sources (7–9). On the other hand, the host cells have evolved complex processes that regulate metabolic engagement and metabolic reconfiguration following viral infections (7, 8, 10). Infections with viruses such as adenoviruses, herpesviruses, influenza virus, hepatitis C virus, and dengue virus have been shown to result in increased glycolysis, while infections with vaccinia virus and adenovirus result in increased glutaminolysis, (7, 8, 10), a process by which glutamine is used to generate tricarboxylic acid (TCA) cycle intermediates in the absence or limited availability of pyruvate. In addition, lipid homeostasis is also altered in cells infected with many other viruses (8). While infection with viruses, including many positive-strand RNA viruses, has been shown to increase fatty acid biosynthesis, fatty acid oxidation (FAO), a process that generates a significant amount of ATP, has also been shown to be up- or down-regulated by viruses (8). Thus, it appears that viruses have evolved to utilize different carbon sources to generate energy and substrates to fuel their replication.
While viruses are known to induce common metabolic changes, unique metabolic perturbations have also been observed for individual virus species (11). Metabolomics studies in virus-infected cells have identified alterations in the metabolites in the pentose phosphate pathway (PPP), which utilizes glucose to eventually generate precursors for nucleotide synthesis (10). Currently, how ZIKV manipulates host cell metabolic pathways to support its replication is not well understood. Only recently, ZIKV infection has been reported to increase glycolysis in human cells to generate TCA cycle intermediates (12). In contrast, a recent study demonstrated that pathogenic ZIKV strains impair glucose metabolism downstream of the PPP toward mitochondrial oxidative phosphorylation, leading to inflammation and apoptosis, which was prevented by pyruvate supplementation (13). Therefore, further studies to delineate ZIKV-induced metabolic changes in cells would help provide a better understanding of replication and pathogenic outcome of the viral infection. Central carbon metabolism, bioenergetics, and redox homeostasis are tightly linked. Increased or disrupted oxidative phosphorylation leads to the leakage of electrons and the formation of reactive oxygen species (ROS) (14). ZIKV infection induces oxidative stress as determined by an increase in the steady-state levels of ROS and accumulation of by-products of oxidative damage to proteins and lipids, which parallels the inhibition of antioxidant responses by the nuclear factor-erythroid factor 2-related factor 2 (Nrf2) (15, 16). Conflicting reports exist regarding the role of Nrf2 on ZIKV replication (17, 18), with some reports demonstrating failure to detect an increase in ROS upon ZIKV infection (19). An increase in carbon flux through the oxidative phase of the PPP is an immediate adaptive cellular response to oxidative stress that would provide reducing equivalents (NADPH) for the recycling of antioxidant systems including glutathione (GSH) (20–22). While an increase in carbon flux from glucose to the PPP was found to be specific for mosquito cells, but not for human foreskin fibroblasts (12), a recent report demonstrated an increase in the PPP in human lung cells infected with ZIKV (13).
In this work, we sought to elucidate the mechanisms by which central carbon metabolism and redox homeostasis regulate ZIKV replication. We report an antagonizing role of the host cell’s antioxidant response by Nrf2 that counters ZIKV replication via GSH synthesis and the PPP. A metabolic profiling study revealed that while the glycolytic output was not altered in cells infected with ZIKV, 6-amino-nicotinamide (6-AN), a nicotinamide adenine dinucleotide (NAD) antimetabolite, depleted cells of intracellular NAD and exerted a potent inhibitory effect on ZIKV replication. These results highlight for the first time the potential use of NAD antimetabolites for therapeutic use.
RESULTS
Nrf2 activation antagonizes ZIKV replication.
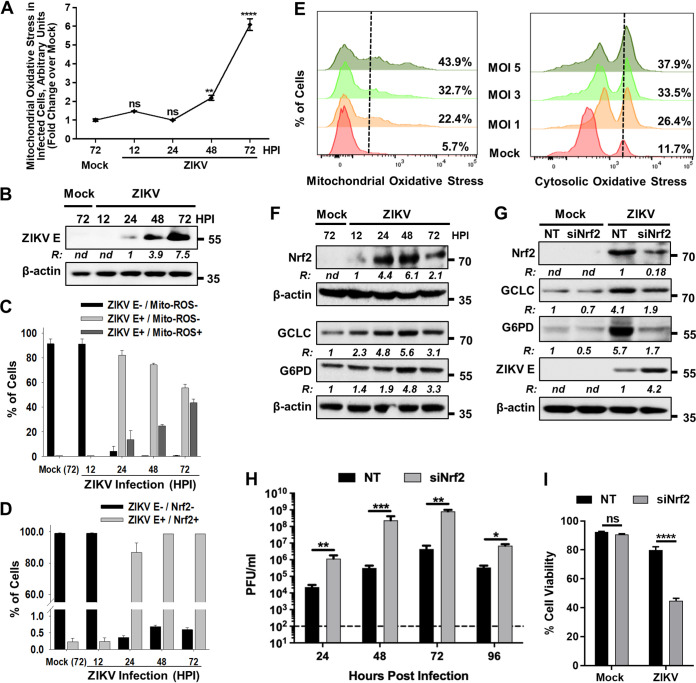
Previous studies have demonstrated that ZIKV induces oxidative stress (15, 16, 19, 23–25), but the status of Nrf2 activation remains underinvestigated. While some studies have shown that silencing of Nrf2 results in attenuation of ZIKV replication (17–19), other reports suggest that Nrf2 is negatively regulated in ZIKV-infected cells (16). Since ZIKV infects astrocytes, oligodendrocyte precursor cells, neural stem cells, and microglia in the brain, we first used A172 human astrocytoma cells and assessed mitochondrial oxidative stress in infected cells by staining with MitoSOX red and performing flow cytometry. Following ZIKV infection, an increase in the steady-state levels of mitochondrial ROS could be detected at 48 h postinfection (hpi), and they were significantly higher by 72 hpi than that in mock-infected cells under similar growth conditions (Fig. 1A). The increase in the steady-state levels of ROS was concomitant with viral replication as demonstrated by an increase in viral E protein accumulation with time (Fig. 1B), as well as an increase in the percentage of infected cells showing ROS as determined by costaining with an antibody against the viral E protein and MitoSOX red (Fig. 1C). In addition, by 48 hpi, >95% of cells infected with ZIKV also costained with an antibody against Nrf2 (Fig. 1D), a key transcription factor detected in cells responding to oxidative stress, indicating that most cells infected with ZIKV exhibit oxidative stress. A viral multiplicity-dependent increase in mitochondrial (Fig. 1E, left panel) as well as cytosolic oxidative stress (Fig. 1E, right panel) was also observed in Lund human mesencephalic (LUHMES) cells, a human embryonic neuronal precursor cell line, suggesting that the induction of oxidative stress in ZIKV-infected cells is not specific to a neuronal cell type.
FIG 1.
Nrf2 activation antagonizes ZIKV replication. (A) A172 cells were mock infected or infected with ZIKV at an MOI of 1, and changes in the steady-state levels of ROS were determined by flow cytometry at the indicated hours postinfection (hpi) following staining of the cells with MitoSOX red. Data from three independent experiments are shown with fold increases in fluorescence intensity over mock-infected sample at 72 hpi. ns, nonsignificant; ****, P ≤ 0.0001. (B) A172 cells were either mock infected or infected with ZIKV at an MOI of 1, and cell lysates prepared at the indicated hpi were subjected to immunoblotting to detect ZIKV E protein and β-actin. A representative Western blot image from three independent experiments is shown with the ratios (R) of the viral E protein to β-actin presented between the blots. nd, not determined. The numbers on right represent the electrophoretic mobility of molecular mass markers in kilodaltons (kDa). (C) Cells mock infected or infected with ZIKV as in panel A were costained at the indicated hpi with ZIKV E antibody and MitoSOX red (Mito-ROS) and subjected to flow cytometry. The percentage of cells showing viral infection and ROS from three independent experiments is presented. Error bars represent the ±SEM. (D) Cells mock infected or infected with ZIKV as in panel A were costained at the indicated hpi with ZIKV E and Nrf2 antibodies and subjected to flow cytometry. The percentage of cells showing viral infection and Nrf2 expression from three independent experiments is presented. Error bars represent the ±SEM. (E) Examination of mitochondrial ROS levels using MitoSOX red (left panel) and cytosolic ROS levels using DHE (right panel) induced in LUHMES cells infected with various MOIs of ZIKV. The percent positive cells exhibiting steady-state levels of ROS at 72 hpi is shown in the panels for each MOI. (F) Detection of Nrf2 in mock- and ZIKV-infected A172 cells at various hpi. Cells were infected as in panel B, and cell lysates were examined by immunoblotting to detect Nrf2, GCLC, G6PD, and β-actin with the respective antibodies. Representative Western blots from three independent experiments are shown, with the ratios (R) of the proteins to β-actin presented at the bottom of the corresponding blots. nd, not determined. The numbers on right represent electrophoretic mobility of molecular mass markers in kDa. (G) A172 cells were transfected with either nontargeting (NT) or Nrf2 siRNA (siNrf2) for 48 h. The cells were subsequently mock infected or infected with ZIKV at an MOI of 1. Cell lysates prepared at 48 hpi were subjected to immunoblotting to detect Nrf2, GCLC, G6PD, ZIKV E protein, and β-actin using the respective antibodies. Representative images from three independent experiments are shown, with the ratios (R) of the proteins to β-actin presented at the bottom of the corresponding blots. nd, not determined. The electrophoretic mobility of molecular mass markers in kDa is shown on the right. (H) Infectious virus titers in NT or Nrf2 siRNA-treated cells at various hpi. Data from four independent experiments are shown. The horizontal dashed line represents the limit of detection. Error bars represent the ±SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (I) Effect of siRNA-mediated depletion of Nrf2 on ZIKV-induced cell viability at 96 hpi. A172 cells were infected as described in panel H, and cell viability was determined as described in Materials and Methods. Data from three independent experiments are shown with error bars representing the ±SEM. ns, nonsignificant; ****, P ≤ 0.0001.
Nrf2 is a constitutively expressed transcription factor that is negatively regulated in cells by its ubiquitination and degradation via the proteasome in the absence of oxidative stress. However, Nrf2 degradation is interrupted upon oxidative stress, resulting in its stabilization and transcriptional activation (26). Contrary to undetectable levels of ROS observed early in ZIKV infection (Fig. 1A), Nrf2 was detected at 12 hpi, and its levels peaked between 24 and 48 hpi (Fig. 1F). A subsequent decrease in the levels of Nrf2 at 72 hpi (Fig. 1F) coincided with the increase in ROS formation seen in Fig. 1A. Activation of Nrf2 generally results in synthesis of many of its target gene products involved in cellular antioxidant response (27), including glutamate cysteine ligase catalytic subunit (GCLC) and glucose 6-phosphate dehydrogenase (G6PD), two key enzymes that catalyze glutathione synthesis or recycling. We observed that activation of Nrf2 due to ZIKV infection also resulted in increased protein levels of both GCLC and G6PD (Fig. 1F), whose transcription is driven by the antioxidant response element recognized by Nrf2 (28). The temporality of the increased expression of both GCLC and G6PD concurred well with the increased protein accumulation and subsequent activation of Nrf2. Consistent with these results, depletion of Nrf2 by small interfering RNA (siRNA) decreased the elevated levels of GCLC and G6PD (Fig. 1G).
While pharmacological activation of Nrf2 by alkylating agents has been previously shown to inhibit ZIKV genome replication (29), the effect of direct genetic modulation of Nrf2 in ZIKV replication has not been previously studied. siRNA-mediated depletion of Nrf2 increased ZIKV protein levels (Fig. 1G) and infectious progeny production significantly (Fig. 1H). Furthermore, downregulation of Nrf2 levels also enhanced cell death induced by ZIKV (Fig. 1I). These results demonstrate that Nrf2 activation counters ZIKV replication, and its subsequent inactivation facilitates oxidative stress and enhances viral replication.
Glutathione downregulates ZIKV replication.
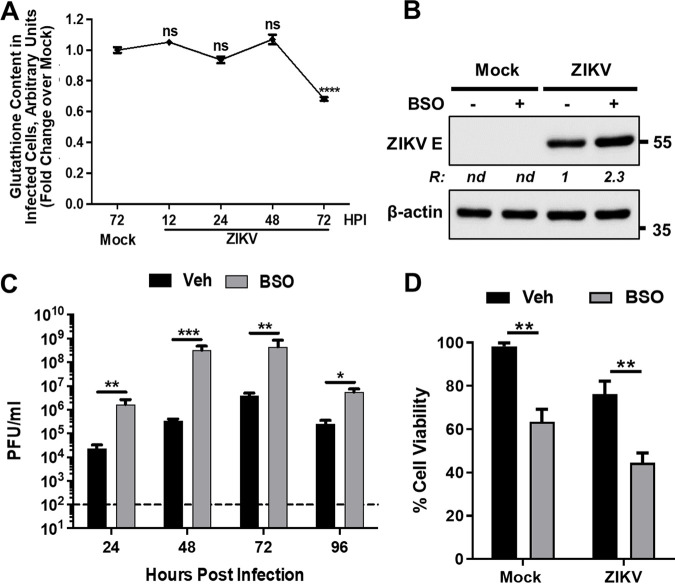
The Nrf2-dependent increase in GCLC observed upon ZIKV infection (Fig. 1F) suggested a role for GSH synthesis in viral infection, as GCLC mediates the rate-limiting step in de novo GSH formation by the synthesis of its precursor γ-glutamylcysteine (30). Examination of GSH content in mock- and ZIKV-infected cells showed significant depletion of GSH levels at 72 hpi compared to that in mock-infected cells (Fig. 2A) that paralleled the downregulation of Nrf2/GCLC at 72 hpi. In the presence of the irreversible GCLC inhibitor, buthionine sulfoximine (BSO), ZIKV protein expression (Fig. 2B) and progeny virus production (Fig. 2C) were enhanced, consistent with the results obtained when Nrf2 was depleted by siRNA (Fig. 1G to H). These results show that inhibition of GSH synthesis leads to increased viral replication, suggesting that GSH downregulates ZIKV replication. BSO treatment significantly reduced cell viability irrespective of the infection status (Fig. 2D), which could be explained by the essential role of GSH in the viability of cells. Overall, these results demonstrate an antiviral role of Nrf2 via GSH synthesis in ZIKV-infected cells.
FIG 2.
Inhibition of glutathione synthesis enhances ZIKV replication. (A) A172 cells were mock infected or infected with ZIKV at an MOI of 1, and cellular GSH content at the indicated hpi was determined by flow cytometry. Data from three independent experiments are shown with the fold change over mock-infected sample at 72 hpi. The error bars show the ±SEM. ns, nonsignificant; ****, P ≤ 0.0001. (B) A172 cells were treated with either vehicle (–) or BSO (+) prior to being mock infected or infected with ZIKV at an MOI of 1 in the continued presence of BSO. Cell lysates prepared at 48 hpi were subjected to immunoblotting to detect ZIKV E protein and β-actin with the respective antibodies. Representative images from three independent experiments are shown. The ratio (R) of the viral E protein to β-actin is shown between the blots. nd, not determined. The electrophoretic mobility of molecular mass markers in kDa is shown on the right. (C) Infectious virus titers in vehicle (veh) or BSO-treated and infected cells at various hpi. Data from four independent experiments are shown. The horizontal dashed line represents the limit of detection. The error bars show the ±SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (D) Effect of BSO on ZIKV-induced cell viability at 96 hpi. The experiment was conducted as described in panel C, and data from three independent experiments are shown with error bars representing the ±SEM. **, P ≤ 0.01.
The PPP inhibits ZIKV replication.
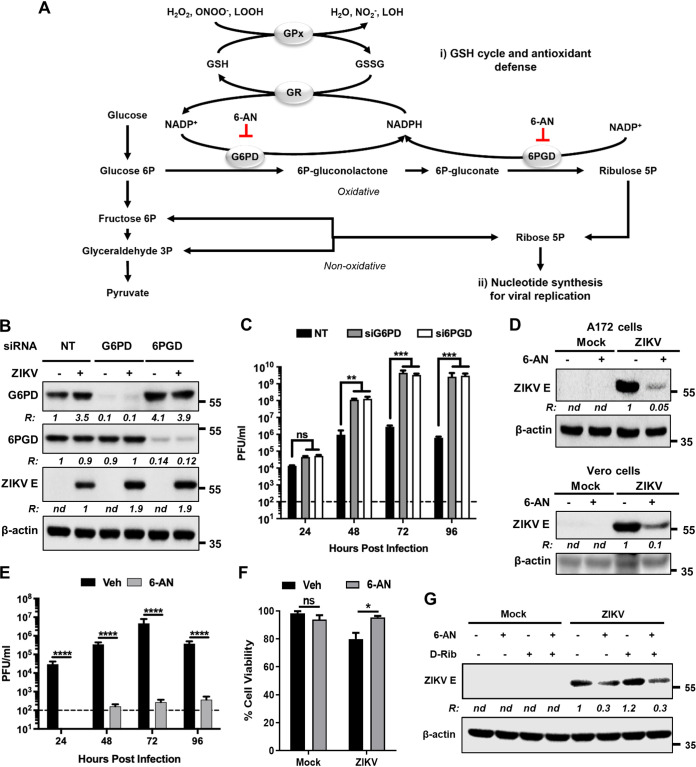
The antioxidant function of GSH is primarily mediated by the activity of GSH peroxidases (GPx), which catalyze the reduction of peroxides (hydrogen peroxide [H2O2], peroxynitrite [ONOO–], or lipid peroxides [LOOH]). The resultant GSH disulfide or oxidized GSH (GSSG) is reduced by GSH reductase (GR) using NADPH as a cofactor (Fig. 3A) (30). G6PD and 6-phosphogluconate dehydrogenase (6PGD) are oxidoreductases within PPP that regenerate NADPH, whose reductive power fuels numerous antioxidant/reductive systems (31). Interestingly, while G6PD levels were transiently upregulated in an Nrf2-dependent manner during ZIKV infection (Fig. 1D), we observed that the 6PGD levels remained unaltered (Fig. 3B). Since our results indicated that GSH synthesis exerted an inhibitory effect on ZIKV replication (Fig. 2B to C), it was expected that G6PD and 6PGD activities would have similar inhibitory effects as well. Consistent with this expectation, downregulation of G6PD or 6PGD using siRNA increased ZIKV protein levels (Fig. 3B) and progeny virus production (Fig. 3C), demonstrating that the PPP antagonizes ZIKV replication.
FIG 3.
The PPP inhibits ZIKV replication. (A) Scheme showing the GSH/NADPH recycling pathway and the PPP enzymatic steps. Formation of 6P-gluconolactone by G6PD and ribulose-5P by 6PGD requires the conversion of NADP+ to NADPH. This provides the reducing power for the conversion of oxidized glutathione (GSSG) to its reduced form (GSH) by glutathione reductase (GR). 6-AN inhibits the activities of G6PD and 6PGD. (B) A172 cells were transfected with either nontargeting (NT) siRNA or siRNAs targeting G6PD or 6PGD for 48 h. The cells were then mock infected (–) or infected with ZIKV (+) at an MOI of 1. Cell lysates prepared at 48 hpi were subjected to immunoblotting to detect G6PD, 6PGD, ZIKV E protein, and β-actin. Representative images from three independent experiments are shown. The ratio (R) of the proteins to β-actin is shown below each blot. nd, not determined. The numbers on the right represent the electrophoretic mobility of molecular mass markers in kDa. (C) Infectious virus production in cells depleted of G6PD or 6PGD. The experiments were conducted as described in panel B; culture supernatants collected at various hpi were assayed for infectious virus by plaque assay. The horizontal discontinuous line represents the limit of detection. Data from three independent experiments are shown. Error bars represent the ±SEM. ns, nonsignificant; **, P ≤ 0.01; ***, P ≤ 0.001. (D) A172 (upper blots) and Vero (lower blots) cells were mock infected or infected with ZIKV at an MOI of 1 and treated without (–) or with (+) 6-AN. Cell lysates prepared at 48 hpi were subjected to immunoblotting to detect ZIKV E protein and β-actin using the corresponding antibodies. Representative images from three independent experiments are shown. The ratio (R) of ZIKV E protein to β-actin is shown between the blots. nd, not determined. The numbers on right represent the electrophoretic mobility of molecular mass markers in kDa. (E) Infectious virus titers in vehicle (veh) or 6-AN-treated cells. The horizontal dashed line represents the limit of detection. Data from four independent experiments are shown with error bars representing the ±SEM. ****, P ≤ 0.0001. (F) Effect of 6-AN on ZIKV-induced cell viability at 96 hpi. A172 cells were infected with ZIKV as described in panel E. Data from three independent experiments are shown with error bars representing the ±SEM. ns, non-significant; *, P ≤ 0.05. (G) A172 cells were mock infected or infected with ZIKV at an MOI of 1 and treated with (+) or without (–) 6-AN along with d-ribose. Cell extracts prepared at 48 hpi were subjected to immunoblotting analysis to examine the levels of ZIKV E protein and β-actin. Representative images from two independent experiments are shown. The ratio (R) of ZIKV E protein to β-actin is shown between the blots. nd, not determined. The numbers on the right represent the electrophoretic mobility of molecular mass markers in kDa.
In contrast to the effects observed with selective knockdown of the PPP enzymes, ZIKV protein levels and viral particle release were significantly inhibited by 6-AN (Fig. 3D to E), a potent antimetabolite of nicotinamide used as a competitive inhibitor of G6PD and 6PGD. This effect was not cell type specific, as it was also observed in Vero cells (Fig. 3D, lower blots). 6-AN also inhibited ZIKV-induced cell death (Fig. 3F). In addition to its role in replenishing NADPH, the PPP generates ribose-5P (R5P), a precursor for the synthesis of nucleotides (Fig. 3A). While the inhibitory effect of 6-AN on viral protein synthesis could be linked to an impairment in nucleotide synthesis required for viral genome replication, supplementation of cells with d-ribose that bypasses the PPP to produce R5P (32) did not reverse the inhibitory effect of 6-AN on viral protein expression (Fig. 3G). These results suggest that although PPP plays a role in antagonizing ZIKV replication, other cellular processes impacted by 6-AN also likely contribute to ZIKV replication.
6-AN alters host cell metabolism, depletes the NAD pool, and inhibits ZIKV replication.
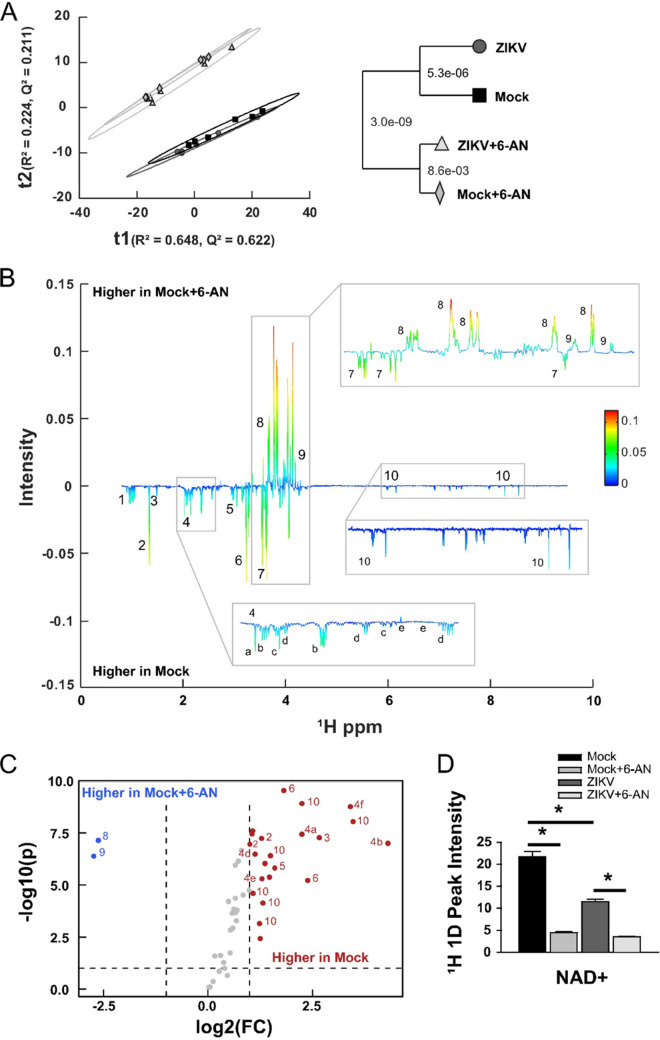
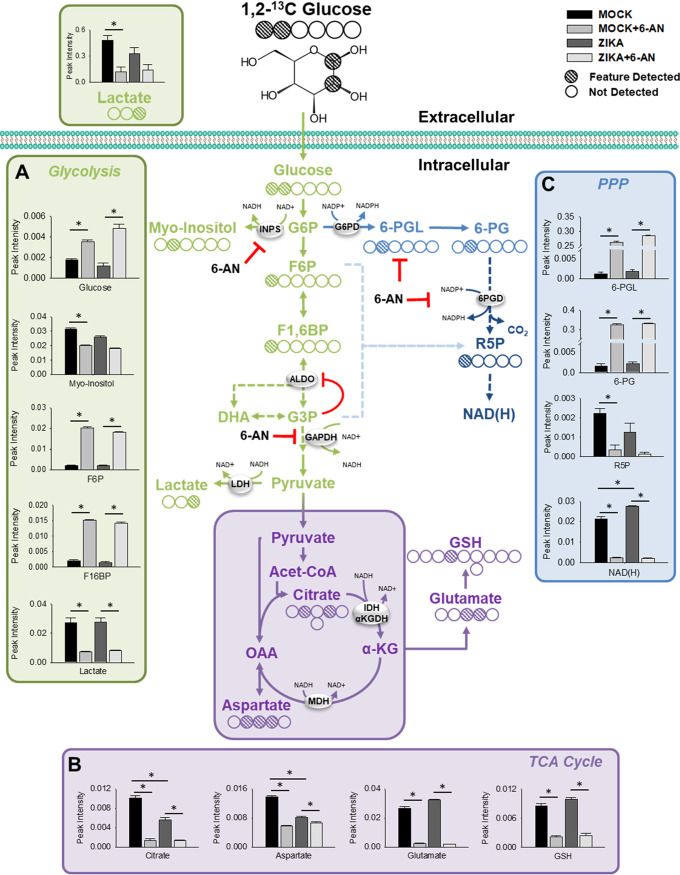
We next sought to identify the mechanisms by which 6-AN exerted its potent inhibitory effect on ZIKV replication. One dimensional (1D) 1H nuclear magnetic resonance (1H-NMR) metabolomic analysis was performed in cells infected with ZIKV for 24 h in the presence or absence of 6-AN to evaluate its effects on cell metabolites. Principal-component analysis (PCA) demonstrated that regardless of ZIKV infection, the most significant axis of separation was a result of 6-AN treatment. This is statistically represented by the dendrogram within the plot showing the Mahalanobis distances displayed as P values (Fig. 4A). When analyzing the groups in a pairwise capacity of mock versus mock plus 6-AN treatment, the results showed significant changes in metabolites within the PPP as well as glycolysis. A back-scaled loading plot from the orthogonal protection to latent structures (OPLS) (Fig. 4B) and a volcano plot univariate analysis of the data (Fig. 4C) demonstrated that 6P-gluconolactone (peak 8) and 6P-gluconate (peak 9) metabolites within the PPP (Fig. 3A) accumulated at elevated levels in mock plus 6-AN-treated cells compared to mock-treated cells. In contrast, metabolites within glycolysis, such as lactate (peak 2), and several other metabolites, including NAD(H) (peak 10), were markedly decreased following 6-AN treatment compared to mock treatment (Fig. 4B and D). Together, the data demonstrate that 6-AN treatment results in a significant buildup of oxidative PPP metabolites.
FIG 4.
1H 1D NMR metabolomics analysis. (A) PCA model of mock (black square), mock plus 6-AN (light gray diamond), ZIKV (dark gray circle), and ZIKV plus 6-AN (cream triangle) samples 48 h after ZIKV exposure and 24 h after 6-AN treatment. R2 = 0.872 and Q2 = 0.833. The model dendrogram depicts the first 4 components with Mahalanobis distances displayed as P values. (B) Back-scaled loading plot comparing mock versus mock plus 6-AN. This plot was generated from a valid OPLS model with a CV-ANOVA P value of 2.6677e-05. Positive values depict metabolites that are higher in mock plus 6-AN samples, while negative values depict metabolites whose concentration is higher in mock samples. (C) Volcano plot for mock versus mock plus 6-AN metabolites with a fold change (FC) threshold (x axis) of log2 and t test threshold (y axis) of 0.1. Blue circles represent metabolites significantly increased in mock plus 6-AN samples, and red circles represent metabolites significantly enriched in mock samples. (D) Bar chart quantifying NAD+ metabolites. Data in graphs are presented as changes in spectrum intensity and are the means ± SEM of n = 6 replicates. *, P ≤ 0.05. Metabolites observed in panels B and C are numbered as follows: (1) branched-chain amino acids (isoleucine, leucine, and valine), (2) threonine and lactate, (3) alanine, (4) TCA-derived metabolites ([a] N-acetylglutamate, [b] glutamate, [c] methionine, [d] glutathione, [e] aspartic acid, [f] glutamine), (5) creatine and phosphocreatine, (6) choline metabolites, (7) myo-inositol, (8) gluconolactone, (9) 6-phosphogluconic acid, (10) energy metabolites (NAD[H], GDP, UDP, and UDPG).
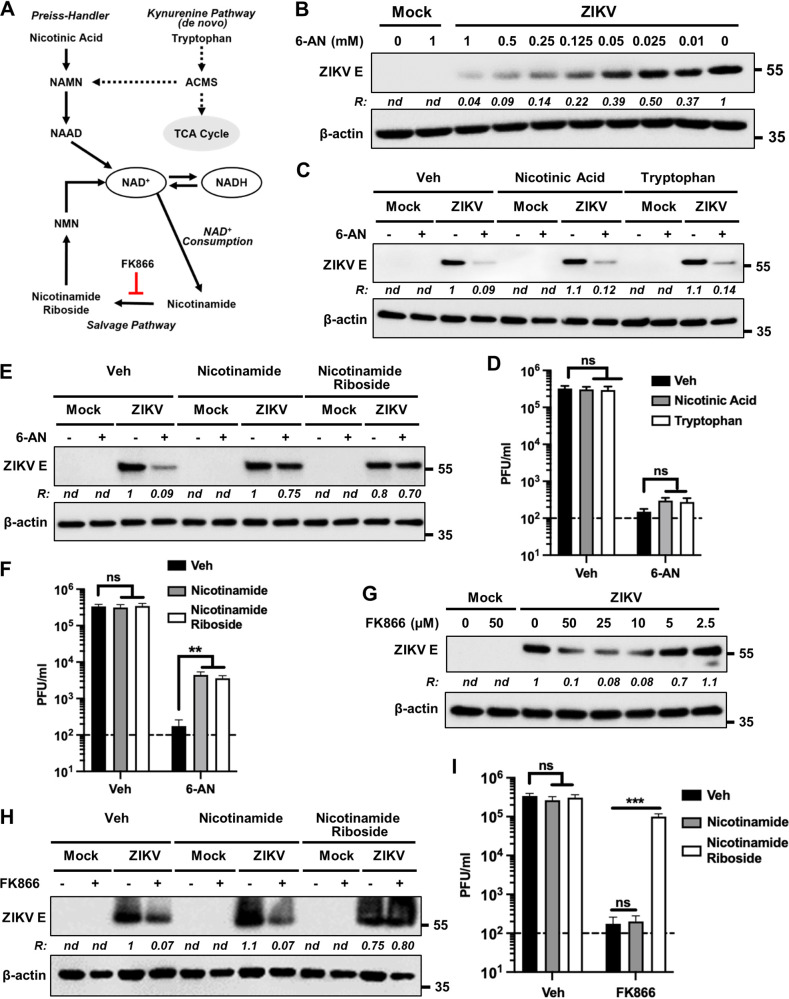
Precursors of NAD+ biosynthesis rescue the inhibitory effects of 6-AN on ZIKV replication.
Since our results suggested that an impairment of NAD(H) metabolism mediates the inhibitory effects of 6-AN on ZIKV replication, we next sought to determine the effects of NAD+ precursors. There are four major precursors for the synthesis of NAD+: tryptophan, nicotinic acid, nicotinamide, and nicotinamide riboside. While NAD+ can be synthesized de novo from tryptophan via the kynurenine pathway through multiple enzymatic steps, it can also be generated from dietary nicotinic acid via the Preiss-Handler pathway (Fig. 5A). In mammals, a major pathway of NAD+ biosynthesis is the salvage pathway from nicotinamide, a by-product of NAD+ consumption, or from extracellular nicotinamide riboside (Fig. 5A) (33).
FIG 5.
NAD precursors rescue the inhibitory effects of 6-AN on ZIKV replication. (A) Schematic showing the different pathways leading to the synthesis and/or recycling of NAD/NADH. FK866 inhibits the conversion of nicotinamide to nicotinamide riboside. ACMS, α-amino-β-carboxymuconate-ε-semialdehyde; NAMN, nicotinic acid mononucleotide; NAAD, nicotinic acid adenine dinucleotide; NMN, nicotinamide mononucleotide. (B) Dose-response inhibition of E protein accumulation in ZIKV-infected cells by 6-AN. A172 cells were either mock infected or infected with ZIKV at an MOI of 1 and treated without or with various concentrations of 6-AN as shown on top of each lane. Cell lysates prepared at 48 hpi were subjected to immunoblotting to detect the viral E protein and β-actin using the corresponding antibodies. The ratio (R) of ZIKV E protein to β-actin is shown between the blots. nd, not determined. The numbers on the right represent the electrophoretic mobility of molecular mass markers in kDa. (C) A172 cells were either mock infected or infected with ZIKV at an MOI of 1 and treated without (–) or with (+) 130 μM 6-AN. Additionally, the cells were treated with either vehicle control (veh), nicotinic acid (300 μM), or tryptophan (300 μM). Cell lysates prepared at 48 hpi were subjected to immunoblotting to detect ZIKV E protein and β-actin using the corresponding antibodies. Representative images from three independent experiments are shown. The ratio (R) of ZIKV E protein to β-actin is shown between the blots. nd, not determined. The numbers on the right represent the electrophoretic mobility of molecular mass markers in kDa. (D) Infectious virus titers in vehicle- or 6-AN-treated cells along with either nicotinic acid or tryptophan. Data from four independent experiments are shown. The horizontal dashed line represents the limit of detection. Error bars represent the ±SEM. ns, non-significant. (E and F) The experiments were conducted as described in panels C and D above except that nicotinamide (300 μM) and nicotinamide riboside (300 μM) were used. Representative Western blot images from three independent experiments are shown in panel E, and data from four independent experiments are shown in panel F. ns, nonsignificant; **, P ≤ 0.01. (G) A172 cells were either mock infected or infected with ZIKV at an MOI 1 and treated with the indicated concentrations of FK866. Cell lysates prepared at 48 hpi were subjected to immunoblotting to detect ZIKV E protein, and β-actin. Representative images from three independent experiments are shown. The ratio (R) of ZIKV E protein to β-actin is shown between the blots. nd, not determined. The numbers on the right represent the electrophoretic mobility of molecular mass markers in kDa. (H) A172 cells were either mock infected or infected with ZIKV at an MOI of 1 and treated without (–) or with (+) a 50-μM concentration of FK866. Cells were also immediately treated without (veh) or with 300-μM concentrations of either nicotinamide or nicotinamide riboside as shown on top of the lanes. Cell lysates prepared at 48 hpi were subjected to immunoblotting to detect ZIKV E protein and β-actin. Representative images are shown. The ratio (R) of ZIKV E protein to β-actin is shown between the blots. nd, not determined. (I) The experiment was conducted as in panel D, and infectious virus yield in the supernatants was assayed at 48 hpi. Data from three independent experiments are shown. The horizontal dashed line represents the limit of detection. Error bars represent the ±SEM. ns, nonsignificant; ***, P ≤ 0.001.
Therefore, we evaluated if impaired NAD(H) synthesis by 6-AN is the mechanism exerting the inhibitory effect on ZIKV replication. In an initial study, we observed that the levels of ZIKV E protein in infected cells were inhibited by 6-AN in a dose-dependent manner (Fig. 5B). Using a nonsaturable concentration of 6-AN (130 μM) that results in substantial inhibition of E protein levels, we then supplemented cells with an excess concentration (300 μM) of NAD+ precursors and examined if they could outcompete and rescue the inhibitory effects of 6-AN. Neither tryptophan nor nicotinic acid rescued the inhibitory effects of 6-AN on ZIKV E protein levels (Fig. 5C) or progeny virus release (Fig. 5D). In contrast, nicotinamide and nicotinamide riboside rescued ZIKV E protein levels (Fig. 5E) and progeny virion production (Fig. 5F), suggesting that it is the impairment of NAD(H) metabolism by which 6-AN inhibits ZIKV replication. This interpretation is further supported by the observation that FK866, an inhibitor of nicotinamide phosphoribosyltransferase (NAMPT) that converts nicotinamide to nicotinamide riboside in the salvage pathway, decreased ZIKV protein levels (Fig. 5G) and infectious progeny production (Fig. 5I). As expected, the viral E protein levels and infectious progeny production in the presence of FK866 could be rescued with nicotinamide riboside but not with nicotinamide (Fig. 5H and I). These results underscore the importance of the salvage pathway of the NAD(H) metabolism in ZIKV replication.
Metabolomics studies reveal the effects of 6-AN on ZIKV-infected cells.
NAD+ is an essential redox carrier, as it receives hydride from metabolic processes, including glycolysis, the TCA cycle, and fatty acid oxidation (FAO), to form NADH. NADH serves as a central hydride donor to mitochondrial oxidative phosphorylation (33). As glycolysis has been reported to be essential for optimal viral replication (7–9), we evaluated if the antagonistic effect of the NAD(H) antimetabolite, 6-AN, against ZIKV infection involves alterations in central carbon metabolism.
We performed NMR metabolomics using isotope-labeled 1,2-13C2 glucose. A map of the 13C-labeled carbon was generated from the two-dimensional (2D) 1H-13C heteronuclear single quantum coherence (HSQC) NMR spectral data to denote the features detected within each metabolite. We identified three NAD(H)/NADP(H)-dependent nodes within glucose metabolism that were inhibited by 6-AN (Fig. 6), corroborating the impairment in NAD-dependent enzymatic processes. First, 6-AN induced an accumulation of glycolytic metabolites, including glucose, fructose 6-phosphate (F6P), and fructose 1,6-biphosphate (F1,6BP), upstream of glyceraldehyde 3-P (G3P) dehydrogenase (GAPDH) (Fig. 6A). Interestingly, G3P levels were not increased, as it is known to have an inhibitory effect on aldolase (34), an enzyme required for synthesis of F1,6BP. Additionally, previous studies have demonstrated that the levels of F1,6BP are predictive of the rate in glycolysis across GAPDH (35). The levels of metabolic by-products downstream of glycolysis, including lactate (Fig. 6A), and across the TCA cycle (citrate, aspartate, and glutamate) were reduced concurrently (Fig. 6B). Consistent with our previous results, GSH synthesis was also reduced in the presence of 6-AN (Fig. 6B). Second, carbon flux through the oxidative PPP was halted due to the inhibition of 6PGD as evidenced by the accumulation of 6-phosphoglucunolactone (6-PGL) and 6-phosphogluconate (6-PG) along with the concomitant decrease in R5P (Fig. 6C). As expected, because R5P is a precursor for AMP synthesis that can be further metabolized into ATP to be incorporated into NAD+ via nicotinamide mononucleotide adenylyl transferases (NMNATs), NAD(H) synthesis was also impaired by 6-AN (Fig. 6C). Because R5P was primarily found in its C1 labeled form, we consider that the impairment in R5P synthesis occurs mainly via inhibition of the oxidative PPP by 6-AN (36). However, lower levels of G3P could also halt the synthesis of R5P via the nonoxidative PPP. The observation that carbon flux across the PPP did not stop at the G6PD node, as would be expected due to the well-known inhibitory role of 6-AN on this enzyme, correlates with previous observations demonstrating that G6PD is actually dispensable for pentose synthesis (37). A third metabolic node inhibited significantly by 6-AN was the synthesis of myo-inositol only in mock-infected but not in ZIKV-infected cells (Fig. 6A), which could be explained by the impairment in the activity of the NAD+-dependent inositol 1-phosphate synthase (INPS). ZIKV infection, by itself, induced a significant increase in NAD(H) (Fig. 6C) and a reduction in TCA cycle metabolites, citrate, and aspartate (Fig. 6B).
FIG 6.
NMR metabolomics of ZIKV-infected cells in the presence or absence of 6-AN. The 2D 1H-13C HSQC NMR metabolomics were performed on ZIVK-infected cells treated with 6-AN for 24 h. Cells were mock infected or infected with ZIKV at an MOI of 1 and treated with or without 6-AN. Then, 2 hours prior to sample collection, cells were incubated with medium supplemented with 2.5 mM 1,2-13C-glucose. (A to C) The effects of ZIVK infection and 6-AN exposure can be seen by intracellular and extracellular changes in metabolites in (A) glycolysis, (B) TCA cycle, and (C) PPP. The NMR carbon map shows the 1H-13C HSQC bond signal for each carbon in the corresponding metabolite. Data in graphs are presented as changes in spectrum intensity and are means ± SEM of n = 6 replicates. *, P ≤ 0.05.
Glycolytic flux is impaired by 6-AN.
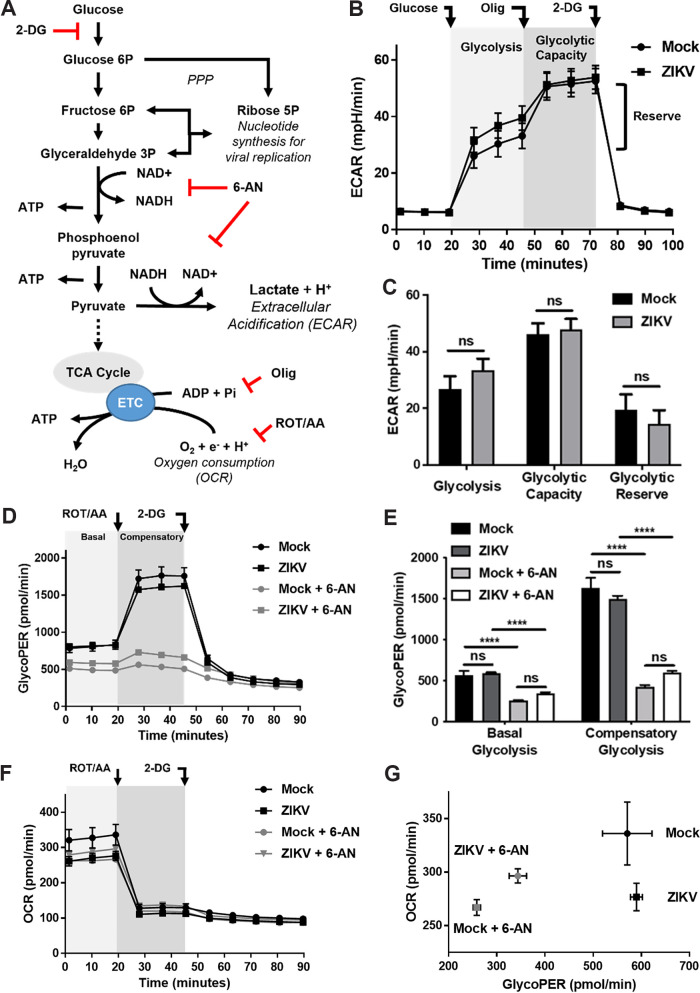
To ascertain the inhibitory effects of 6-AN on glycolysis, we evaluated glycolytic flux by quantification of extracellular medium acidification rates (ECAR) as an indirect proxy of lactate production, the end result of glycolysis (Fig. 7A). No alterations in glycolysis were observed when glycolysis, maximal glycolytic capacity (after inhibition of mitochondrial ATP synthase with oligomycin), and glycolytic reserve (the difference between basal and maximal glycolytic capacity) were assessed in ZIKV-infected cells (Fig. 7B and C). Since mitochondrial-derived CO2 can result in additional extracellular acidification beyond that contributed by glycolysis, we then determined the glycolytic proton efflux rate (glycoPER). Our results revealed that glycoPER was not affected by ZIKV infection (Fig. 7D and E). In contrast, 6-AN decreased basal glycolysis and compensatory glycolysis (after inhibition of the mitochondrial transport chain [ETC] with rotenone [ROT] and antimycin A [AA]) in both mock- and ZIKV-infected cells (Fig. 7D and E). When oxygen consumption rates (OCR) were quantified as a proxy of mitochondrial respiration, we found that while ZIKV infection alone slightly decreased mitochondrial respiration, 6-AN reduced OCR in mock-infected cells but had no major effects on OCR in virus-infected cells (Fig. 7F to G). We conclude that 6-AN impairs glycolysis, which opens the possibility that the inhibitory effect of 6-AN on ZIKV replication also involves glycolysis.
FIG 7.
Glycolytic flux is impaired by 6-AN. (A) Representation of the site of action of the metabolic toxins used to evaluate glycolytic flux. 2-DG blocks the conversion of glucose to glucose 6P; 6-AN inhibits production of NADH from NAD+ as shown; oligomycin (olig) inhibits mitochondrial ATP generation in the electron transport chain (ETC); rotenone (ROT) and antimycin A (AA) inhibit the ETC in the mitochondria. Metabolic flux analysis was conducted in ZIKV-infected cells in the presence or absence of 6-AN (1 mM). (B and C) Glycolytic flux was evaluated by the quantification of extracellular medium acidification rates (ECAR). Glycolysis was induced by the addition of glucose after 1 h of glucose deprivation. Maximal glycolytic capacity is evidenced by the inhibition of mitochondrial ATP synthase with oligomycin (olig, 2 μM). The addition of 2-DG (50 mM) is used to evidence that ECAR is directly related to glycolysis. The data in panel B were plotted in the bar diagram shown in panel C. ns, nonsignificant. (D and E) Determination of glycolytic proton efflux rate (glycoPER). The basal glycolytic rate was evaluated during the first 20 min of incubation in the presence of glucose (10 mM) following glucose starvation, and compensatory glycolysis was elicited by the inhibition of the ETC with ROT and AA (0.5 μM each). The data in panel D were plotted in the bar diagram shown in panel E. The glycoPER values presented in panel E were obtained after subtraction of non-glycolytic acidification values obtained following addition of 2-DG as shown in panel D. ns, nonsignificant; ****, P ≤ 0.0001. (F) Cells were either mock infected or infected with ZIKV at an MOI of 1 and treated without or with 6-AN. ROT/AA and 2-DG were added as shown, and the OCR were measured with time after addition of the inhibitors as described in Materials and Methods. (G) Comparison of changes in OCR versus glycoPER. Please note that the OCR axis in panel G is magnified compared to that shown in panel F. Error bars represent the ±SEM of n = 5 replicates.
Glycolysis and mitochondrial metabolism are important for ZIKV protein expression and glycosylation.
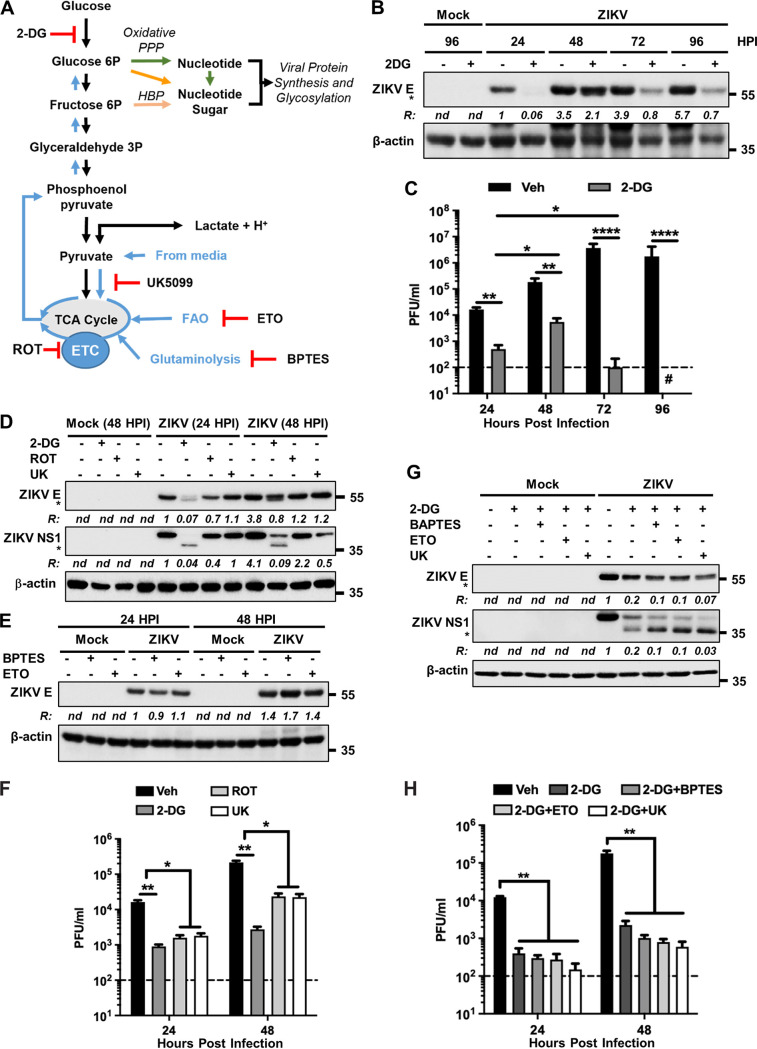
We next examined the importance of glycolysis in ZIKV replication. Hexokinases catalyze the first irreversible step of the glycolytic pathway, wherein glucose is phosphorylated to glucose-6P. Inhibition of hexokinases by 2-deoxy-d-glucose (2-DG) (Fig. 8A) significantly decreased ZIKV E protein levels and viral progeny production (Fig. 8B and C). Interestingly, there was a transient recovery of viral protein levels and infectious progeny production at 48 hpi (Fig. 8B and C), suggesting that additional carbon sources were fueling viral replication. Three primary carbon sources that drive bioenergetics and central carbon metabolism independently from glycolysis are (i) pyruvate, originated by either its reverted synthesis from lactate or uptake from culture media, (ii) acetyl-CoA from FAO, and (iii) glutamate synthesis via glutaminolysis, which is subsequently incorporated into the TCA cycle upon its metabolism to α-ketoglutarate. Progressive oxidation of these carbon sources is coupled to the ETC (Fig. 8A). In addition, reductive carboxylation of pyruvate and glutamine can fuel nucleotide and nucleotide sugar biosynthesis required for viral protein synthesis via gluconeogenesis (Fig. 8A, blue upward arrows) (38). Using inhibitors of ETC complex I (rotenone [ROT]), the mitochondrial pyruvate carrier (UK5099 [UK]), glutaminolysis [bis-2-(5-phenylacetamido-1, 2, 4-thiadiazol-2-yl) ethyl sulfide 3 (BPTES)], and FAO (etomoxir [ETO]), as shown in Fig. 8A, we examined the role of mitochondrial metabolism in ZIKV replication. None of these compounds had any significant inhibitory effect on viral E protein levels (Fig. 8D and E). Only a slight reduction of ZIKV E protein levels was observed with ROT at 24 hpi (Fig. 8D). Examination of the viral NS1 protein revealed a similar trend (Fig. 8D). Infectious progeny yield in the supernatants was inhibited by ROT and UK, but not as significantly as that seen with 2-DG (Fig. 8F). Importantly, the transient recovery of viral protein levels as well as infectious progeny production observed at 48 hpi when glycolysis was blocked by 2-DG (Fig. 8B and C) were prevented by inhibition of mitochondrial pyruvate uptake, FAO, or glutaminolysis (Fig. 8G and H), indicating that carbon sources other than glucose appear to be required for fueling viral replication to some extent.
FIG 8.
Glycolysis and mitochondrial metabolism are necessary for ZIKV protein expression and glycosylation. (A) Scheme showing glycolysis and mitochondrial metabolism. UK5099 (UK) inhibits the entry of pyruvate into the TCA cycle, ETO inhibits FAO, and BPTES inhibits glutaminolysis. Other inhibitors (2-DG and ROT) with their sites of action are described in the legend to Fig. 7. (B) A172 cells were either mock infected or infected with ZIKV at an MOI of 1 and treated without (–) or with (+) 2-DG. Cell lysates prepared at the indicated hpi were subjected to immunoblotting to detect ZIKV E protein and β-actin. Representative images from three independent experiments are shown. Note that the E protein is seen as a closely migrating doublet in the presence of 2-DG, representing the glycosylated (upper band) and the nonglycosylated (bottom band, identified with an asterisk) species. The ratio (R) of ZIKV E protein (both bands) to β-actin is shown between the blots. nd, not determined. The numbers on the right represent the electrophoretic mobility of molecular mass markers in kDa. (C) Infectious virus titers at various hpi in vehicle (veh)-or 2-DG-treated cells as described in panel B. The horizontal dashed line represents the limit of detection. Data from three independent experiments are shown with error bars representing the ±SEM. *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001. (D) A172 cells were either mock infected or infected with ZIKV at an MOI of 1 and treated with either 2-DG, ROT, or UK. Cell lysates prepared at the indicated hpi were subjected to immunoblotting to detect ZIKV E protein, NS1 protein, and β-actin. Note that the upper bands in the E and NS1 blots represent the glycosylated proteins, whereas their unglycosylated forms (lower bands) are identified with asterisks. Representative images from three independent experiments are shown. The ratio (R) of ZIKV E or NS1 proteins (both bands) to β-actin is shown below the respective blots. nd, not determined. The numbers on the right represent the electrophoretic mobility of molecular mass markers in kDa. (E) The experiment was conducted and the data are presented in a similar manner to that described in panel D but using only BPTES and ETO as inhibitors. (F) Infectious virus titers from cells treated with no drug (veh), 2-DG, ROT, or UK. The horizontal dashed line represents the limit of detection. Data from four independent experiments are shown with error bars representing the ±SEM. *, P ≤ 0.05; **, P ≤ 0.01. (G) A172 cells were either mock infected or infected with ZIKV at an MOI of 1 and treated without (–) or with (+) 2-DG. Additionally, the cells were treated with either BPTES, ETO, or UK. Cell lysates prepared at 48 hpi were subjected to immunoblotting to detect ZIKV E protein, NS1 protein, and β-actin. The unglycosylated forms of E and NS1 are identified with asterisks in the respective blots. Representative images from three independent experiments are shown. The ratio (R) of ZIKV E or NS1 proteins (only the top band) to β-actin is shown below the respective blots. nd, not determined. The numbers on the right represent the electrophoretic mobility of molecular mass markers in kDa. (H) Infectious virus titers from cells treated with vehicle control or with 2-DG and either BPTES, ETO, or UK. The horizontal dashed line represents the limit of detection. Data from four independent experiments are shown with error bars representing the ±SEM. **, P ≤ 0.01.
We and others have previously examined the N-glycosylation of ZIKV E and NS1 proteins and its importance for viral assembly, infectivity, and pathogenicity (39–42). A recent study demonstrated that the major proportion of N-glycan on ZIKV E contains N-acetylglucosamine (GlcNAc) and glucose among others (43). G6P and fructose 6P are precursors of the nucleotide-activated sugar donors UDP-glucose and UDP-GlcNAc in the hexosamine biosynthetic pathway (HBP), respectively, that mediate protein N-glycosylation (Fig. 8A, orange arrows). Inhibition of glycolysis by 2-DG reduced ZIKV E and NS1 protein glycosylation (Fig. 8B, D, and G). Under this inhibitory condition, inhibition of mitochondrial pyruvate uptake by UK, FAO by ETO, or glutaminolysis by BPTES further reduced E and NS1 glycosylation (Fig. 8G). Together, these results show that viral protein levels and glycosylation depend on glycolysis and suggest that mitochondrial metabolism via gluconeogenesis can partially compensate for glucose deficiency.
DISCUSSION
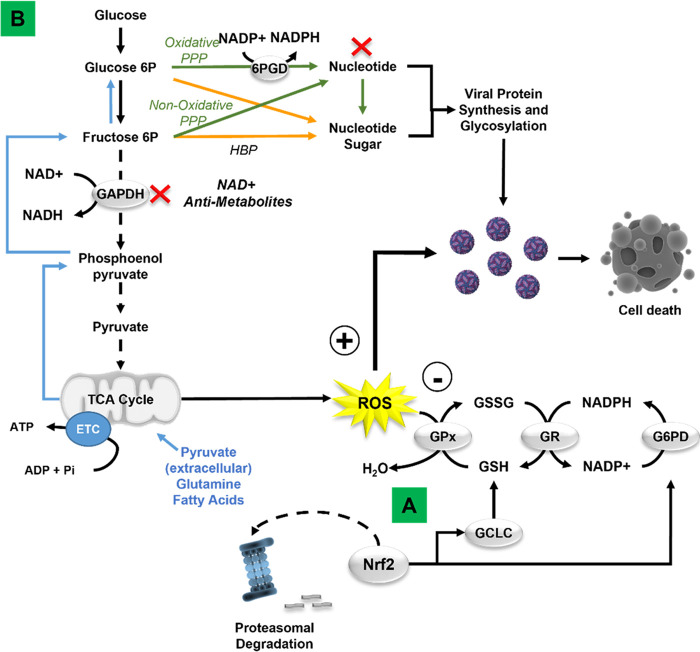
Viruses hijack cell-host metabolism to facilitate their replication. Mechanistic studies geared toward understanding the metabolic reliance of viral replication and the mechanisms involved in rewiring host cell metabolism open new avenues for antiviral drug development. Here, we report on the mechanisms by which redox homeostasis and central carbon metabolism regulate ZIKV replication. We first demonstrated that during viral replication, the host cell’s antioxidant response mediated by Nrf2 counters viral replication (Fig. 9A). Then, while examining the role of the PPP, we serendipitously discovered that 6-AN, an NAD(H) antimetabolite, also counters viral replication significantly. The NAD(H)-dependent mechanisms involved in viral replication appear to depend, at least partially, on central carbon metabolism via glycolysis (Fig. 9B). These novel findings uncover the potential for NAD(H) antimetabolites and Nrf2 activators as antiviral strategies against ZIKV infection.
FIG 9.
Schematic illustration of the redox regulation and metabolic dependency of Zika virus replication. (A) ZIKV induces a transient stabilization and activation of the antioxidant transcription factor Nrf2. Pharmacological inhibition of de novo GSH synthesis and knockdown of Nrf2, and the NADPH regenerating enzymes G6PD and 6PGD (not depicted), enhance viral replication and cell death, demonstrating an antiviral role (–) of Nrf2 signaling and GSH redox cycle, while ROS seem to facilitate viral replication (+). (B) Metabolomic studies and metabolic flux assays suggest that the antiviral effects of the NAD antimetabolite 6-AN likely involve multiple mechanisms, including NAD depletion and interference with NAD-dependent metabolic processes. 6-AN inhibits glycolysis and the PPP (red X), which are required for energy production and synthesis of nucleotides. Mitochondrial metabolism (blue lines) exerts a compensatory role in viral replication when glycolysis is inhibited.
Oxidative stress has been reported in cells infected with viruses, including members of the Flaviviridae family (44–47). Contradictory results exist regarding whether ZIKV infection induces oxidative stress or not (15, 16, 19). In our study, we clearly observed an increase in mitochondrial and cytosolic ROS levels and a concomitant decrease in GSH content in both A172 glial and LUHMES neuroblast cells infected with the virus. Importantly, Nrf2 activation as demonstrated by its increased accumulation as well as that of its target gene products (GCLC and G6PD) preceded ROS detection. The observation that Nrf2 activation could be readily seen prior to significant ROS detection in infected cells suggests that during ZIKV infection, ROS levels are kept in check early by the activation of the Nrf2-depedent antioxidant responses and that these ROS only increase once Nrf2-mediated antioxidant responses are suppressed. Discrepancy with other studies could be explained by the time differences postinfection when ROS levels were evaluated (19), which could also explain why previous studies failed to report the activation of Nrf2 signaling in ZIKV-infected cells (15, 16).
The mechanism by which ZIKV downregulates Nrf2 is unclear but likely involves degradation in a Kelch-like ECH-associated protein 1 (KEAP1)-proteasomal- or p62-autophagy-independent manner, as KEAP1 is expected to remain inactivated by high oxidative stress at 72 hpi, and our previous studies demonstrated that ZIKV inhibits autophagy during the same time frame (48). Glycogen synthase kinase 3 β (GSK3-β)-dependent Nrf2 degradation via the recruitment of the 1-Cullin1-Rbx1/β-transducin repeat-containing protein (SCF/β-TrCP) E3 ubiquitin ligase complex (49, 50) is a possibility, as GSK3-β is commonly activated by viruses, including ZIKV (51, 52). The NS2B3 protease complex in dengue virus has been recently reported to degrade Nrf2 (44). It is also possible that the same complex of ZIKV could be involved as well in downregulation of Nrf2.
Nrf2 activators have been reported to decrease viral replication (17, 29, 53). Our results presented here corroborate these findings by demonstrating that knockdown of Nrf2 enhances ZIKV replication. In addition, we have demonstrated that the antioxidant Nrf2 response involving de novo GSH synthesis and redox cycle antagonize virus replication (Fig. 9A). This was evident by the downregulation of Nrf2 and the NADPH synthesizing enzymes G6PD and 6PGD, as well as the inhibition of GSH synthesis, which had an enhancing effect on viral replication. These findings underscore the antiviral role of Nrf2 signaling and antioxidant defenses (54).
In addition to antioxidant genes, Nrf2 regulates the transcription of genes involved in cell metabolism, immune, and inflammatory responses. Although the exact mechanism by which Nrf2/GSH antagonize ZIKV remains to be elucidated, it clearly involves the GSH redox cycle, as ZIKV replication was enhanced when the NADPH-synthesizing enzymes G6PD and 6PGD were downregulated. Another possible mechanism by which Nrf2 exerts its antiviral effects on ZIKV could be through ROS scavenging mediated by heme oxygenase 1 (HO-1). HO-1 is known to have antioxidant effects via the degradation of heme (55, 56) and has also been shown to mediate antiviral effects on dengue virus (57, 58) and ZIKV (17). Additionally, ROS have been demonstrated to benefit viral replication through modulation of host pathways and covalent changes in viral components (44, 45, 59, 60). Furthermore, a recent study reported that the antiviral effect of a free-form amino acid formulation containing cysteine, glycine, and glutamate is mediated by increased GSH formation (61). Therefore, it appears that multiple mechanisms involving several pathways might be operating for Nrf2’s antiviral effect on ZIKV.
While studying the role of the NADPH recycling enzymes on ZIKV replication, we discovered that 6-AN, a commonly used inhibitor of G6PD and 6PGD, significantly blocked viral protein expression and progeny production, which contrasted with the stimulatory effect of downregulating these enzymes by siRNA. A recent study reported that 6-AN reduces hepatitis B virus replication, but the mechanisms involved were not identified (62). 6-AN was first described as a potent antagonist of nicotinamide (63) and was subsequently demonstrated to form an abnormal NAD+ analog that inhibits NAD(H)-dependent enzymes, particularly GAPDH (64). Our metabolomic analysis demonstrated that 6-AN depleted cells of NAD(H) and that it indeed inhibited glycolysis by blocking carbon flux upstream of GAPDH (Fig. 6). Downstream metabolic pathways, including the TCA cycle, were also inhibited by 6-AN, resulting in decreased glutamate and, consequently, GSH levels. The inhibitory effects of 6-AN on ZIKV replication were reversed by supplementation with precursors of the NAD+ salvage pathway and mimicked by inhibition of NAMPT with FK866, the rate-limiting enzyme in NAD+ salvage, demonstrating that NAD antimetabolites in general could inhibit ZIKV replication. To our knowledge, this is the first study demonstrating the role of the NAD salvage pathway in viral replication and the potential of NAD-antimetabolites as antiviral agents.
NAD+ and its metabolites are critical regulators of numerous physiological processes. NAD(H) and NADP(H) are cofactors of metabolic pathways involved in transferring hydrogen in oxidation-reduction reactions. Postsynthesis, NAD+-dependent chemical modifications of proteins, RNA, and DNA regulate signal transduction, enzymatic activity, DNA ligation and repair, RNA processing, and epigenetics. NAD+ also serves as a precursor for the synthesis of second messengers (33). NAD+-dependent deacetylases/mono-ADP ribosyltransferases (sirtuins) function as viral restriction factors, inhibiting replication of DNA and RNA viruses (65, 66). Poly(ADP-ribose) polymerases (PARPs) elicit direct antiviral activity, targeting certain steps of the virus replication cycle, or indirect antiviral activity via modulation of the innate immune response (67). Interestingly, sirtuin inhibitors have also been shown to be highly effective host-targeting antivirals by preventing accumulation of viral RNA and proteins in cells infected with arboviruses, including ZIKV (68, 69). PARP11/12-mediated ADP-ribosylation of nonstructural NS1 and NS3 proteins prompts their proteasomal degradation, resulting in inhibition of ZIKV replication (70, 71). In contrast, NS3-mediated activation of PARP1 mediates ZIKV-induced cell death (72). A recent investigation reported that supplementation with NAD(H) or its precursors protected against microcephaly and improved survival of ZIKV-infected mice. However, NAD(H) supplementation per se had no effect on ZIKV replication (73), a result consistent with the findings presented here. Our findings along with previously published data suggest that ZIKV hijacks NAD(H) metabolism for its replication, consequently impairing NAD+-dependent antiviral responses of the host. Consistent with this interpretation, conditions in which cells depleted of NAD+ by NAD+-utilizing enzymes possessing antiviral activities such as PARPs and sirtuins have been linked to poor pathogenic outcomes in viral infections (74–76).
Glycolysis has been shown to participate in viral replication (77–81). It is noteworthy that discrepancy exists regarding the effect of ZIKV on glycolysis, where some studies report an increase (82), decrease (13), or no effect (83) on glycolytic flux, which could be ascribed to the time postinfection wherein these measurements were taken. Although we did not detect any major changes in glycolytic flux in ZIKV-infected cells as measured by NMR metabolomics or extracellular flux analysis experiments, blocking the activity of hexokinase—the first rate-limiting enzyme in glycolysis—with 2-DG significantly inhibited ZIKV replication. The decreased levels of viral proteins and glycosylation by 2-DG were further exacerbated by inhibiting the mitochondrial carbon inputs from pyruvate, FAO, or glutaminolysis. Thus, our results expand the role of central carbon metabolism to mitochondria in ZIKV infection. A recent report demonstrated that high glucose in the medium reduces ZIKV replication in kidney cells, but these effects could be ascribed to the toxicity due to hyperglycemic conditions (84).
The mechanisms by which inhibition of glycolysis and/or mitochondrial metabolism impair viral replication were not explored in detail but likely involve a decrease in ATP and reduced synthesis of building blocks required for viral macromolecular synthesis and virion assembly. While the potent inhibitory effects of 6-AN can be attributed to impaired glycolysis, our metabolomic analysis also corroborated its strong inhibitory effect on the PPP. Thus, the antiviral effects of 6-AN likely involve multiple mechanisms, including (i) NAD depletion by interference with NAD synthesis, (ii) inhibition of glycolysis, and (iii) inhibition of the PPP. This is supported by the stronger inhibitory effect of 6-AN on ZIKV virus replication compared to the direct inhibition of glycolysis by 2-DG. While knockdown of the PPP enzymes 6PGD and G6PD alone had a stimulatory effect on viral replication, our results reconcile these observations with their involvement in antiviral effects of the Nrf2/GSH antioxidant response via supply of NADPH. In contrast, the inhibitory effect of the downregulation of G6PD or 6PGD within the oxidative branch of the PPP on the synthesis of nucleotides could be compensated by the nonoxidative branch, which is known to be involved in nucleotide metabolism (85). Our metabolomics results suggest that 6-AN has the potential to inhibit both the oxidative and nonoxidative branch simultaneously by its inhibitory effects on 6PGD and GAPDH.
Overall, our results report for the first time the antiviral role of the Nrf2/GSH antioxidant response against ZIKV replication. In addition, they reveal the potential of NAD(H)-antimetabolites against viral replication. A fine balance between the NAD(H)-dependent host antiviral responses and NAD(H)-dependent proviral replication processes likely exists in infected cells. Future studies addressing the mechanisms that regulate this balance will facilitate development of antiviral strategies against ZIKV.
MATERIALS AND METHODS
Cells and viruses.
A172 (ATCC CRL-1620), Vero E6 (ATCC CCL-81) cells, and nondifferentiated Lund Human Mesencephalic (LUHMES) cells (ATCC CRL-2927) were maintained as described before (48). Briefly, A172 and Vero cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1× antibiotics (100 U/mL penicillin and 20 U/mL streptomycin) at 37°C and 5% CO2 atmosphere. LUHMES cells were maintained similarly in culture-wares precoated with 50 μg/mL poly-l-ornithine (Sigma-Aldrich, St. Louis, MO) and 1 μg/mL fibronectin (BD Biosciences). A stock of the infectious clone-derived MR766 ZIKV was prepared in Vero E6 cells as described previously (39). For quantitation of virus yield, 100-μL aliquots of culture supernatants collected at the indicated hpi were used to determine virus titers by plaque assay (86).
Antibodies and other reagents.
Anti-ZIKV E (catalog [cat.] no. GTX1333325 for Western blotting and no. GTX634155 for flow cytometry) and anti-ZIKV NS1 (cat. no. GTX133307) antibodies were obtained from GeneTex. Anti-GCLC (cat. no. ab53179), anti-G6PD (cat. no. ab129199), and anti-Nrf2 (cat. no. EP1808Y and ab62352) antibodies were obtained from Abcam. Anti-β-actin (cat. no. A2228), goat anti-rabbit immunoglobulin (IgG)-horseradish peroxidase (HRP)-conjugated (cat. no. A6154) and goat anti-mouse IgG-HRP-conjugated (cat. no. A4416) antibodies were obtained from Sigma-Aldrich. l-buthionine sulfoximine (BSO), monochlorobimane (mBCl), dihydro ethidium (DHE), MitoSOX red, FK866, nicotinamide, nicotinic acid, l-tryptophan, 2-deoxy-d-glucose (2-DG), and d-ribose were obtained from Thermo Scientific. Nicotinamide riboside was obtained from Combi-Blocks (San Diego, CA). 6-Aminonicotinamide (6-AN) was obtained from Alfa Aesar (Haverhill, MA). Etomoxir (ETO), bis-2-(5-phenylacetamido-1, 2, 4-thiadiazol-2-yl) ethyl sulfide 3 (BPTES), and UK5099 (UK) were obtained from Cayman Chemical (Ann Arbor, MI). Oligomycin (olig), rotenone (ROT), and antimycin A (AA) were obtained with the Seahorse kit from Agilent (Santa Clara, CA). Propidium iodide (PI) was procured from Sigma-Aldrich. Unless otherwise described in the figures or the figure legends, the final concentrations of various drugs and inhibitors used in these studies were as follows: mBCl, 10 μM; DHE, 5 μM; MitoSOX red, 5 μM; BSO, 500 μM; 6-AN, 1 mM; 2-DG, 50 mM; PI, 1 μg/mL; oligomycin, 2 μM; AA, 0.5 μM; BPTES, 10 μM; UK5099, 5 μM; ROT, 0.5 μM; ETO, 50 μM; d-(-)-ribose, 25 mM; nicotinamide riboside, 300 μM; tryptophan, 300 μM; nicotinic acid, 300 μM; nicotinamide, 300 μM. BSO, oligomycin, 2-DG, ROT, ETO, d-(-)-ribose, nicotinamide riboside, tryptophan, nicotinic acid, and nicotinamide were dissolved in water; 6-AN, BPTES, and UK were dissolved in dimethyl sulfoxide (DMSO).
siRNA-mediated protein depletion.
Nrf2, G6PD, and 6PGD depletion was performed using small interfering RNA (siRNA) pools targeting Nrf2 (cat. no. L-003755-00-0005), G6PD (cat. no. JL-008181-02-0005), or 6PGD (cat. no. L-008371-00-0005), respectively, obtained from Horizon Discovery (Waterbeach, UK). Cells were transfected with siRNAs at a final concentration of 80 nM using Lipofectamine RNAiMax (Invitrogen) following the manufacturer’s recommendations. A nontargeting (NT) siRNA (Qiagen, Hilden, Germany; cat. no. 1027281) was used as a negative control. At 24 h posttransfection, the cells were replenished with DMEM containing 2% FBS and 1× antibiotic solution and were incubated for a further 24 h prior to virus infection.
Cell lysate preparation and Western blotting.
Cell lysates were prepared as described previously (48). Briefly, cells washed with cold phosphate-buffered saline (PBS) were recovered from the culture dishes following trypsin treatment and pelleting at 500 × g for 5 min at 4°C. Cell pellets were resuspended in lysis buffer (48), and 25 μg of total protein from the clarified lysates was used for detection of the desired proteins by immunoblot analysis. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting was performed as described previously (48).
Flow cytometry and data analysis.
Cell viability was examined by determining PI uptake, as a marker of plasma membrane integrity loss, and changes in the forward scatter properties of the cell (cell size) using flow cytometry (fluorescence-activated cell sorting). Changes in intracellular GSH content were examined by staining with mBCl. ROS production was determined using DHE. ROS production in the mitochondria was assessed using MitoSOX red, a derivative of DHE with a cationic triphenylphosphonium substituent responsible for the electrophoretic uptake into actively respiring mitochondria. To determine the percentage of ZIKV-infected A172 cells exhibiting oxidative stress and/or Nrf2 expression, cells seeded in 6-well plates were infected with ZIKV at a multiplicity of infection (MOI) of 1, and at the indicated hpi, they were stained with either DHE (5 μM) or MitoSOX red (5 μM) for 20 min and harvested by trypsinization. The cells were washed with PBS and fixed for 20 min at 4°C with 4% paraformaldehyde. The fixed cells were pelleted and washed twice with staining buffer (PBS, 1% FBS, 0.1% sodium azide, and 0.1% saponin). Cell pellets were incubated with primary antibodies against ZIKV E protein (GeneTex, cat. no. GTX634155) and Nrf2 (Abcam, cat. no. ab62352) diluted 1:100 in staining buffer and incubated at room temperature for 1 h, following which they were incubated in the appropriate secondary antibodies (goat anti-mouse-Alexa 488 for ZIKV E protein, goat anti-rabbit-Alexa 647 for Nrf2) diluted 1:1,000 in staining buffer. The cells were incubated in the dark at room temperature for 1 h, washed twice with staining buffer, and finally, resuspended in staining buffer prior to data acquisition using a Cytek flow cytometer (BD Biosciences). To determine the percentage of LUHMES cells showing mitochondrial or cytosolic oxidative stress at various MOIs of ZIKV, infected cells were stained with the appropriate dyes at 72 hpi and subjected to flow cytometry as described above. Data analysis was performed using FlowJo software as described previously (87, 88).
Metabolomics studies.
After treatment, cells were washed twice with PBS. Two hours prior to collection, cells were incubated in culture medium supplemented with 2.5 mM 1,2-13C glucose (Sigma-Aldrich). Cells were washed with PBS, and metabolites were extracted with cold methanol (–80°C), followed by 80%/20% methanol/water. Culture media and cell extracts from ~5 × 106 cells were utilized for 1D 1H- and 2D 1H-13C HSQC NMR experiments. NMR data were collected and analyzed as described previously (89). Briefly, all data were collected on a Bruker Avance III-HD 700-MHz spectrometer equipped with a 5-mm quadruple resonance QCI-P cryoprobe (1H, 13C, 15N, 31P with 2H lock and decoupling) and a SampleJet automated sample changer. The 1D 1H NMR data were collected at 298K with 128 scans and 4 dummy scans and a spectral width of 9,090 Hz using an excitation sculpting pulse sequence. The 2D 1H-13C HSQC spectra were collected at 298K with 1K data points and a spectral width of 11,160 Hz in the direct dimension and 64 data points with a spectral width of 29,059 Hz in the indirect dimension. The 1D 1H metabolites were identified and quantified with Chenomx NMR Suite 8.0 (http://www.chenomx.com/), while the 2D metabolites were identified with HMDB (90) and BMRB (91) metabolite libraries.
Extracellular flux analysis.
The extracellular acidification rate (ECAR, mpH/min) and oxygen consumption rates (OCR, pmol/min) were determined using the XFe24 analyzer (Seahorse Biosciences/Agilent) as described before (92). All kits for the studies were obtained from Agilent, and the studies were conducted as per the manufacturer’s recommendations. After treatment, cells were incubated for 1 h in DMEM supplemented with 2 mM l-glutamine (HyClone, SH30034), 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, and 1 mM pyruvate (HyClone, SH30239) prior to OCR and ECAR measurements. When indicated, media were also supplemented with 10 mM glucose (Thermo Fisher Scientific, D16500). The pH of DMEM was adjusted to 7.4 with sodium hydroxide. Both ECAR and OCR measurements were used to determine the glycolytic proton efflux rate (glycoPER) of the cells.
Statistical analysis.
Experimental replicas were independent and performed on separate days. The data were analyzed by using one-way or two-way analysis of variance (ANOVA), and the appropriate post hoc test using the GraphPad Prism package. When ANOVA assumptions were not met (normality [Shapiro Wilk test] or equal variance [Kruskal-Wallis]), one-way ANOVA on ranks or data transformation (two-way ANOVA) were performed on the data collected. Data were plotted as the mean ± standard error of the mean (SEM) using the same package for statistical analysis. Multivariate analysis of the 1D 1H NMR metabolomics data set was obtained using PCA and OPLS model analyses in the MVAPACK software suite (http://bionmr.unl.edu/mvapack.php) (93). Briefly, the 1D 1H data were Fourier transformed, automatically phase corrected, and binned via the adaptive intelligent binning in the MVAPACK software (94). The data were normalized by standard normal variate normalization, and the data were univariately scaled for PCA analysis. Dendrograms were generated from the PCA score plot using the PCA-PLS-DA utilities package as previously described (95). OPLS models were generated from full data following normalization and removal of solvent peak (4.5 to 5.0 ppm). The 2D NMR spectra were processed in NMRPipe (96) and NMRViewJ as previously described (89). The 2D metabolites were assigned using 0.08 1H and 0.25 13C ppm chemical shift errors. Metabolites were identified to metabolomics standards initiatives (MSI) level 2 standards.
ACKNOWLEDGMENTS
The cell sorting was performed at the Flow Cytometry Service Center of the Center for Biotechnology, University of Nebraska-Lincoln.
The research was supported in part by funds from the University of Nebraska-Lincoln and National Institutes of Health (NIH) grant GM131701-01 (to O.K.). We acknowledge support from the Nebraska Center for Integrated Biomolecular Communication, which is funded by the National Institute of General Medical Sciences (P20 GM113126), NIH. A.D.T.-G. was funded by a grant (DBI-1757951) from the National Science Foundation.
Contributor Information
Rodrigo Franco, Email: rodrigo.franco@unl.edu.
Asit K. Pattnaik, Email: apattnaik2@unl.edu.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Lindenbach BD, Murray CL, Thiel RJ, Rice CM. 2013. Flaviviridae. p 712–746. In Knipe DM, Howley PM (ed), Fields Virology, 6th ed. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 2.van Hemert F, Berkhout B. 2016. Nucleotide composition of the Zika virus RNA genome and its codon usage. Virol J 13:95. 10.1186/s12985-016-0551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serman TM, Gack MU. 2019. Evasion of innate and intrinsic antiviral pathways by the Zika virus. Viruses 11:970. 10.3390/v11100970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miner JJ, Diamond MS. 2017. Zika virus pathogenesis and tissue tropism. Cell Host Microbe 21:134–142. 10.1016/j.chom.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazear HM, Diamond MS. 2016. Zika virus: new clinical syndromes and its emergence in the Western Hemisphere. J Virol 90:4864–4875. 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Saucedo-Cuevas L, Regla-Nava JA, Chai G, Sheets N, Tang W, Terskikh AV, Shresta S, Gleeson JG. 2016. Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell 19:593–598. 10.1016/j.stem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thaker SK, Ch’ng J, Christofk HR. 2019. Viral hijacking of cellular metabolism. BMC Biol 17:59. 10.1186/s12915-019-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girdhar K, Powis A, Raisingani A, Chrudinova M, Huang R, Tran T, Sevgi K, Dogus Dogru Y, Altindis E. 2021. Viruses and metabolism: the effects of viral infections and viral insulins on host metabolism. Annu Rev Virol 8:373–391. 10.1146/annurev-virology-091919-102416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer KA, Stockl J, Zlabinger GJ, Gualdoni GA. 2019. Hijacking the supplies: metabolism as a novel facet of virus-host interaction. Front Immunol 10:1533. 10.3389/fimmu.2019.01533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer CS. 2022. Innate metabolic responses against viral infections. Nat Metab 4:1245–1259. 10.1038/s42255-022-00652-3. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, He G, Filipowicz NA, Randall G, Belov GA, Kopek BG, Wang X. 2019. Host lipids in positive-strand RNA virus genome replication. Front Microbiol 10:286. 10.3389/fmicb.2019.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thaker SK, Chapa T, Garcia G, Jr., Gong D, Schmid EW, Arumugaswami V, Sun R, Christofk HR. 2019. Differential metabolic reprogramming by Zika virus promotes cell death in human versus mosquito cells. Cell Metab 29:1206–1216.e4. 10.1016/j.cmet.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yau C, Low JZH, Gan ES, Kwek SS, Cui L, Tan HC, Mok DZL, Chan CYY, Sessions OM, Watanabe S, Vasudevan SG, Lee YH, Chan KR, Ooi EE. 2021. Dysregulated metabolism underpins Zika-virus-infection-associated impairment in fetal development. Cell Rep 37:110118. 10.1016/j.celrep.2021.110118. [DOI] [PubMed] [Google Scholar]
- 14.Tirichen H, Yaigoub H, Xu W, Wu C, Li R, Li Y. 2021. Mitochondrial reactive oxygen species and their contribution in chronic kidney disease progression through oxidative stress. Front Physiol 12:627837. 10.3389/fphys.2021.627837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ledur PF, Karmirian K, Pedrosa C, Souza LRQ, Assis-de-Lemos G, Martins TM, Ferreira J, de Azevedo Reis GF, Silva ES, Silva D, Salerno JA, Ornelas IM, Devalle S, Madeiro da Costa RF, Goto-Silva L, Higa LM, Melo A, Tanuri A, Chimelli L, Murata MM, Garcez PP, Filippi-Chiela EC, Galina A, Borges HL, Rehen SK. 2020. Zika virus infection leads to mitochondrial failure, oxidative stress and DNA damage in human iPSC-derived astrocytes. Sci Rep 10:1218. 10.1038/s41598-020-57914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almeida LT, Ferraz AC, da Silva Caetano CC, da Silva Menegatto MB, Dos Santos Pereira Andrade AC, Lima RLS, Camini FC, Pereira SH, da Silva Pereira KY, de Mello Silva B, Perucci LO, Talvani A, de Magalhaes JC, de Brito Magalhaes CL. 2020. Zika virus induces oxidative stress and decreases antioxidant enzyme activities in vitro and in vivo. Virus Res 286:198084. 10.1016/j.virusres.2020.198084. [DOI] [PubMed] [Google Scholar]
- 17.Huang H, Falgout B, Takeda K, Yamada KM, Dhawan S. 2017. Nrf2-dependent induction of innate host defense via heme oxygenase-1 inhibits Zika virus replication. Virology 503:1–5. 10.1016/j.virol.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bottino-Rojas V, Talyuli OAC, Carrara L, Martins AJ, James AA, Oliveira PL, Paiva-Silva GO. 2018. The redox-sensing gene Nrf2 affects intestinal homeostasis, insecticide resistance, and Zika virus susceptibility in the mosquito Aedes aegypti. J Biol Chem 293:9053–9063. 10.1074/jbc.RA117.001589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendonca-Vieira LR, Anibal-Silva CE, Menezes-Neto A, Azevedo EAN, Zanluqui NG, Peron JPS, Franca RFO. 2021. Reactive oxygen species (ROS) are not a key determinant for Zika virus-induced apoptosis in SH-SY5Y neuroblastoma cells. Viruses 13:2111. 10.3390/v13112111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuehne A, Emmert H, Soehle J, Winnefeld M, Fischer F, Wenck H, Gallinat S, Terstegen L, Lucius R, Hildebrand J, Zamboni N. 2015. Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol Cell 59:359–371. 10.1016/j.molcel.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 21.Christodoulou D, Kuehne A, Estermann A, Fuhrer T, Lang P, Sauer U. 2019. Reserve flux capacity in the pentose phosphate pathway by NADPH binding is conserved across kingdoms. iScience 19:1133–1144. 10.1016/j.isci.2019.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filosa S, Fico A, Paglialunga F, Balestrieri M, Crooke A, Verde P, Abrescia P, Bautista JM, Martini G. 2003. Failure to increase glucose consumption through the pentose-phosphate pathway results in the death of glucose-6-phosphate dehydrogenase gene-deleted mouse embryonic stem cells subjected to oxidative stress. Biochem J 370:935–943. 10.1042/bj20021614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simanjuntak Y, Liang JJ, Chen SY, Li JK, Lee YL, Wu HC, Lin YL. 2018. Ebselen alleviates testicular pathology in mice with Zika virus infection and prevents its sexual transmission. PLoS Pathog 14:e1006854. 10.1371/journal.ppat.1006854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G, Poulsen M, Fenyvuesvolgyi C, Yashiroda Y, Yoshida M, Simard JM, Gallo RC, Zhao RY. 2017. Characterization of cytopathic factors through genome-wide analysis of the Zika viral proteins in fission yeast. Proc Natl Acad Sci USA 114:E376–E385. 10.1073/pnas.1619735114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Retallack H, Di Lullo E, Arias C, Knopp KA, Laurie MT, Sandoval-Espinosa C, Mancia Leon WR, Krencik R, Ullian EM, Spatazza J, Pollen AA, Mandel-Brehm C, Nowakowski TJ, Kriegstein AR, DeRisi JL. 2016. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc Natl Acad Sci USA 113:14408–14413. 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen T, Nioi P, Pickett CB. 2009. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284:13291–13295. 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nezu M, Suzuki N. 2020. Roles of Nrf2 in protecting the kidney from oxidative damage. Int J Mol Sci 21:2951. 10.3390/ijms21082951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322. 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 29.Olagnier D, Farahani E, Thyrsted J, Blay-Cadanet J, Herengt A, Idorn M, Hait A, Hernaez B, Knudsen A, Iversen MB, Schilling M, Jorgensen SE, Thomsen M, Reinert LS, Lappe M, Hoang HD, Gilchrist VH, Hansen AL, Ottosen R, Nielsen CG, Moller C, van der Horst D, Peri S, Balachandran S, Huang J, Jakobsen M, Svenningsen EB, Poulsen TB, Bartsch L, Thielke AL, Luo Y, Alain T, Rehwinkel J, Alcami A, Hiscott J, Mogensen TH, Paludan SR, Holm CK. 2020. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat Commun 11:4938. 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franco R, Cidlowski JA. 2012. Glutathione efflux and cell death. Antioxid Redox Signal 17:1694–1713. 10.1089/ars.2012.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanton RC. 2012. Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life 64:362–369. 10.1002/iub.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauly DF, Pepine CJ. 2000. d-Ribose as a supplement for cardiac energy metabolism. J Cardiovasc Pharmacol Ther 5:249–258. 10.1054/JCPT.2000.18011. [DOI] [PubMed] [Google Scholar]
- 33.Xie N, Zhang L, Gao W, Huang C, Huber PE, Zhou X, Li C, Shen G, Zou B. 2020. NAD(+) metabolism: pathophysiologic mechanisms and therapeutic potential. Signal Transduct Target Ther 5:227. 10.1038/s41392-020-00311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose IA, O’Connell EL. 1969. Studies on the interaction of aldolase with substrate analogues. J Biol Chem 244:126–134. 10.1016/S0021-9258(19)78201-5. [DOI] [PubMed] [Google Scholar]
- 35.Shestov AA, Liu X, Ser Z, Cluntun AA, Hung YP, Huang L, Kim D, Le A, Yellen G, Albeck JG, Locasale JW. 2014. Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. Elife 3:03342. 10.7554/eLife.03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrigan JB, Reed MA, Ludwig C, Khanim FL, Bunce CM, Günther UL. 2016. Tracer-based metabolic NMR-based flux analysis in a leukaemia cell line. Chempluschem 81:453–459. 10.1002/cplu.201500549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandolfi PP, Sonati F, Rivi R, Mason P, Grosveld F, Luzzatto L. 1995. Targeted disruption of the housekeeping gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is dispensable for pentose synthesis but essential for defense against oxidative stress. EMBO J 14:5209–5215. 10.1002/j.1460-2075.1995.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yip J, Geng X, Shen J, Ding Y. 2016. Cerebral gluconeogenesis and diseases. Front Pharmacol 7:521. 10.3389/fphar.2016.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Annamalai AS, Pattnaik A, Sahoo BR, Muthukrishnan E, Natarajan SK, Steffen D, Vu HLX, Delhon G, Osorio FA, Petro TM, Xiang SH, Pattnaik AK. 2017. Zika virus encoding nonglycosylated envelope protein is attenuated and defective in neuroinvasion. J Virol 91:e01348-17. 10.1128/JVI.01348-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fontes-Garfias CR, Shan C, Luo H, Muruato AE, Medeiros DBA, Mays E, Xie X, Zou J, Roundy CM, Wakamiya M, Rossi SL, Wang T, Weaver SC, Shi PY. 2017. Functional analysis of glycosylation of Zika virus envelope protein. Cell Rep 21:1180–1190. 10.1016/j.celrep.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carbaugh DL, Baric RS, Lazear HM. 2019. Envelope protein glycosylation mediates Zika virus pathogenesis. J Virol 93:e00113-19. 10.1128/JVI.00113-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Annamalai AS, Pattnaik A, Sahoo BR, Guinn ZP, Bullard BL, Weaver EA, Steffen D, Natarajan SK, Petro TM, Pattnaik AK. 2019. An attenuated Zika virus encoding non-glycosylated envelope (E) and non-structural protein 1 (NS1) confers complete protection against lethal challenge in a mouse model. Vaccines (Basel) 7:112. 10.3390/vaccines7030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Routhu NK, Lehoux SD, Rouse EA, Bidokhti MRM, Giron LB, Anzurez A, Reid SP, Abdel-Mohsen M, Cummings RD, Byrareddy SN. 2019. Glycosylation of Zika virus is important in host-virus interaction and pathogenic potential. Int J Mol Sci 20:5206. 10.3390/ijms20205206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrari M, Zevini A, Palermo E, Muscolini M, Alexandridi M, Etna MP, Coccia EM, Fernandez-Sesma A, Coyne C, Zhang DD, Marques ETA, Olagnier D, Hiscott J. 2020. Dengue virus targets Nrf2 for NS2B3-mediated degradation leading to enhanced oxidative stress and viral replication. J Virol 94:e01551-20. 10.1128/JVI.01551-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng L, Liang Y, Ariffianto A, Matsui C, Abe T, Muramatsu M, Wakita T, Maki M, Shibata H, Shoji I. 2022. Hepatitis C virus-induced ROS/JNK signaling pathway activates the E3 ubiquitin ligase itch to promote the release of HCV particles via polyubiquitylation of VPS4A. J Virol 96:e0181121. 10.1128/JVI.01811-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olagnier D, Peri S, Steel C, van Montfoort N, Chiang C, Beljanski V, Slifker M, He Z, Nichols CN, Lin R, Balachandran S, Hiscott J. 2014. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog 10:e1004566. 10.1371/journal.ppat.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Boehning DF, Qian T, Popov VL, Weinman SA. 2007. Hepatitis C virus core protein increases mitochondrial ROS production by stimulation of Ca2+ uniporter activity. FASEB J 21:2474–2485. 10.1096/fj.06-7345com. [DOI] [PubMed] [Google Scholar]
- 48.Sahoo BR, Pattnaik A, Annamalai AS, Franco R, Pattnaik AK. 2020. Mechanistic target of rapamycin signaling activation antagonizes autophagy to facilitate Zika virus replication. J Virol 94:e01575-20. 10.1128/JVI.01575-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. 2011. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31:1121–1133. 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dodson M, de la Vega MR, Cholanians AB, Schmidlin CJ, Chapman E, Zhang DD. 2019. Modulating NRF2 in disease: timing is everything. Annu Rev Pharmacol Toxicol 59:555–575. 10.1146/annurev-pharmtox-010818-021856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee SE, Choi H, Shin N, Kong D, Kim NG, Kim HY, Kim MJ, Choi SW, Kim YB, Kang KS. 2022. Zika virus infection accelerates Alzheimer’s disease phenotypes in brain organoids. Cell Death Discov 8:153. 10.1038/s41420-022-00958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alfhili MA, Alsughayyir J, McCubrey JA, Akula SM. 2020. GSK-3-associated signaling is crucial to virus infection of cells. Biochim Biophys Acta Mol Cell Res 1867:118767. 10.1016/j.bbamcr.2020.118767. [DOI] [PubMed] [Google Scholar]
- 53.Pereira SH, Almeida LT, Ferraz AC, Ladeira MS, Ladeira LO, Magalhaes CLB, Silva BM. 2021. Antioxidant and antiviral activity of fullerol against Zika virus. Acta Trop 224:106135. 10.1016/j.actatropica.2021.106135. [DOI] [PubMed] [Google Scholar]
- 54.Herengt A, Thyrsted J, Holm CK. 2021. NRF2 in viral infection. Antioxidants (Basel) 10:1491. 10.3390/antiox10091491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Araujo JA, Zhang M, Yin F. 2012. Heme oxygenase-1, oxidation, inflammation, and atherosclerosis. Front Pharmacol 3:119. 10.3389/fphar.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waza AA, Hamid Z, Ali S, Bhat SA, Bhat MA. 2018. A review on heme oxygenase-1 induction: is it a necessary evil. Inflamm Res 67:579–588. 10.1007/s00011-018-1151-x. [DOI] [PubMed] [Google Scholar]
- 57.Tseng CK, Lin CK, Wu YH, Chen YH, Chen WC, Young KC, Lee JC. 2016. Human heme oxygenase 1 is a potential host cell factor against dengue virus replication. Sci Rep 6:32176. 10.1038/srep32176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu YH, Chen WC, Tseng CK, Chen YH, Lin CK, Lee JC. 2022. Heme oxygenase-1 inhibits DENV-induced endothelial hyperpermeability and serves as a potential target against dengue hemorrhagic fever. FASEB J 36:e22110. 10.1096/fj.202100688RRR. [DOI] [PubMed] [Google Scholar]
- 59.Foo J, Bellot G, Pervaiz S, Alonso S. 2022. Mitochondria-mediated oxidative stress during viral infection. Trends Microbiol 30:679–692. 10.1016/j.tim.2021.12.011. [DOI] [PubMed] [Google Scholar]
- 60.Tao L, Lemoff A, Wang G, Zarek C, Lowe A, Yan N, Reese TA. 2020. Reactive oxygen species oxidize STING and suppress interferon production. Elife 9:57837. 10.7554/eLife.57837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vasireddi M, Crum A, May H, Katz D, Hilliard J. 2019. A novel antiviral inhibits Zika virus infection while increasing intracellular glutathione biosynthesis in distinct cell culture models. Antiviral Res 161:46–52. 10.1016/j.antiviral.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Ren F, Yang X, Hu ZW, Wong VKW, Xu HY, Ren JH, Zhong S, Jia XJ, Jiang H, Hu JL, Cai XF, Zhang WL, Yao FL, Yu HB, Cheng ST, Zhou HZ, Huang AL, Law BYK, Chen J. 2019. Niacin analogue, 6-Aminonicotinamide, a novel inhibitor of hepatitis B virus replication and HBsAg production. EBioMedicine 49:232–246. 10.1016/j.ebiom.2019.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson WJ, McColl JD. 1955. 6-Aminonicotinamide: a potent nicotinamide antagonist. Science 122:834. 10.1126/science.122.3174.834. [DOI] [PubMed] [Google Scholar]
- 64.Dietrich LS, Friedland IM, Kaplan LA. 1958. Pyridine nucleotide metabolism: mechanism of action of the niacin antagonist, 6-aminonicotinamide. J Biol Chem 233:964–968. 10.1016/S0021-9258(18)64687-3. [DOI] [PubMed] [Google Scholar]
- 65.Koyuncu E, Budayeva HG, Miteva YV, Ricci DP, Silhavy TJ, Shenk T, Cristea IM. 2014. Sirtuins are evolutionarily conserved viral restriction factors. mBio 5:e02249-14. 10.1128/mBio.02249-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Budayeva HG, Rowland EA, Cristea IM. 2016. Intricate roles of mammalian sirtuins in defense against viral pathogens. J Virol 90:5–8. 10.1128/JVI.03220-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malgras M, Garcia M, Jousselin C, Bodet C, Leveque N. 2021. The antiviral activities of poly-ADP-ribose polymerases. Viruses 13:582. 10.3390/v13040582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hackett BA, Dittmar M, Segrist E, Pittenger N, To J, Griesman T, Gordesky-Gold B, Schultz DC, Cherry S. 2019. Sirtuin inhibitors are broadly antiviral against arboviruses. mBio 10:e01446-19. 10.1128/mBio.01446-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rausch K, Hackett BA, Weinbren NL, Reeder SM, Sadovsky Y, Hunter CA, Schultz DC, Coyne CB, Cherry S. 2017. Screening bioactives reveals nanchangmycin as a broad spectrum antiviral active against Zika virus. Cell Rep 18:804–815. 10.1016/j.celrep.2016.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li L, Shi Y, Li S, Liu J, Zu S, Xu X, Gao M, Sun N, Pan C, Peng L, Yang H, Cheng G. 2021. ADP-ribosyltransferase PARP11 suppresses Zika virus in synergy with PARP12. Cell Biosci 11:116. 10.1186/s13578-021-00628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L, Zhao H, Liu P, Li C, Quanquin N, Ji X, Sun N, Du P, Qin CF, Lu N, Cheng G. 2018. PARP12 suppresses Zika virus infection through PARP-dependent degradation of NS1 and NS3 viral proteins. Sci Signal 11:aas9332. 10.1126/scisignal.aas9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu G, Li S, Liu X, Gao P, Chen X, Wang H, Zhang M, Yang Y, Gao GF, Zhang F. 2019. PARP-1 mediated cell death is directly activated by ZIKV infection. Virology 537:254–262. 10.1016/j.virol.2019.08.024. [DOI] [PubMed] [Google Scholar]
- 73.Pang H, Jiang Y, Li J, Wang Y, Nie M, Xiao N, Wang S, Song Z, Ji F, Chang Y, Zheng Y, Yao K, Yao L, Li S, Li P, Song L, Lan X, Xu Z, Hu Z. 2021. Aberrant NAD(+) metabolism underlies Zika virus-induced microcephaly. Nat Metab 3:1109–1124. 10.1038/s42255-021-00437-0. [DOI] [PubMed] [Google Scholar]
- 74.Zheng M, Schultz MB, Sinclair DA. 2022. NAD(+) in COVID-19 and viral infections. Trends Immunol 43:283–295. 10.1016/j.it.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shang J, Smith MR, Anmangandla A, Lin H. 2021. NAD+-consuming enzymes in immune defense against viral infection. Biochem J 478:4071–4092. 10.1042/BCJ20210181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan A, Doig CL. 2021. NAD(+) degrading enzymes, evidence for roles during infection. Front Mol Biosci 8:697359. 10.3389/fmolb.2021.697359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Codo AC, Davanzo GG, Monteiro LB, de Souza GF, Muraro SP, Virgilio-da-Silva JV, Prodonoff JS, Carregari VC, de Biagi Junior CAO, Crunfli F, Jimenez Restrepo JL, Vendramini PH, Reis-de-Oliveira G, Bispo Dos Santos K, Toledo-Teixeira DA, Parise PL, Martini MC, Marques RE, Carmo HR, Borin A, Coimbra LD, Boldrini VO, Brunetti NS, Vieira AS, Mansour E, Ulaf RG, Bernardes AF, Nunes TA, Ribeiro LC, Palma AC, Agrela MV, Moretti ML, Sposito AC, Pereira FB, Velloso LA, Vinolo MAR, Damasio A, Proenca-Modena JL, Carvalho RF, Mori MA, Martins-de-Souza D, Nakaya HI, Farias AS, Moraes-Vieira PM. 2020. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1alpha/glycolysis-dependent axis. Cell Metab 32:437–446.e5. 10.1016/j.cmet.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Passalacqua KD, Lu J, Goodfellow I, Kolawole AO, Arche JR, Maddox RJ, Carnahan KE, O’Riordan MXD, Wobus CE. 2019. Glycolysis is an intrinsic factor for optimal replication of a norovirus. mBio 10:e02175-18. 10.1128/mBio.02175-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boodhoo N, Kamble N, Sharif S, Behboudi S. 2020. Glutaminolysis and glycolysis are essential for optimal replication of Marek’s disease virus. J Virol 94:e01680-19. 10.1128/JVI.01680-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu X, Liu X, Bai J, Gao Y, Song Z, Nauwynck H, Wang X, Yang Y, Jiang P. 2021. Glyceraldehyde-3-phosphate dehydrogenase restricted in cytoplasmic location by viral GP5 facilitates porcine reproductive and respiratory syndrome virus replication via its glycolytic activity. J Virol 95:e0021021. 10.1128/JVI.00210-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu YH, Yang Y, Chen CH, Hsiao CJ, Li TN, Liao KJ, Watashi K, Chen BS, Wang LH. 2021. Aerobic glycolysis supports hepatitis B virus protein synthesis through interaction between viral surface antigen and pyruvate kinase isoform M2. PLoS Pathog 17:e1008866. 10.1371/journal.ppat.1008866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh S, Singh PK, Suhail H, Arumugaswami V, Pellett PE, Giri S, Kumar A. 2020. AMP-activated protein kinase restricts Zika virus replication in endothelial cells by potentiating innate antiviral responses and inhibiting glycolysis. J Immunol 204:1810–1824. 10.4049/jimmunol.1901310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Q, Gouilly J, Ferrat YJ, Espino A, Glaziou Q, Cartron G, El Costa H, Al-Daccak R, Jabrane-Ferrat N. 2020. Metabolic reprogramming by Zika virus provokes inflammation in human placenta. Nat Commun 11:2967. 10.1038/s41467-020-16754-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reslan A, Haddad JG, Moukendza KL, Despres P, Bascands JL, Gadea G. 2021. Zika virus growth in human kidney cells is restricted by an elevated glucose level. Int J Mol Sci 22:2495. 10.3390/ijms22052495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Q, Qin T, Bi Z, Hong H, Ding L, Chen J, Wu W, Lin X, Fu W, Zheng F, Yao Y, Luo ML, Saw PE, Wulf GM, Xu X, Song E, Yao H, Hu H. 2020. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat Commun 11:1456. 10.1038/s41467-020-15308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pattnaik A, Palermo N, Sahoo BR, Yuan Z, Hu D, Annamalai AS, Vu HLX, Correas I, Prathipati PK, Destache CJ, Li Q, Osorio FA, Pattnaik AK, Xiang SH. 2018. Discovery of a non-nucleoside RNA polymerase inhibitor for blocking Zika virus replication through in silico screening. Antiviral Res 151:78–86. 10.1016/j.antiviral.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 87.Garcia-Garcia A, Anandhan A, Burns M, Chen H, Zhou Y, Franco R. 2013. Impairment of Atg5-dependent autophagic flux promotes paraquat- and MPP(+)-induced apoptosis but not rotenone or 6-hydroxydopamine toxicity. Toxicol Sci 136:166–182. 10.1093/toxsci/kft188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rodriguez-Rocha H, Garcia-Garcia A, Pickett C, Li S, Jones J, Chen H, Webb B, Choi J, Zhou Y, Zimmerman MC, Franco R. 2013. Compartmentalized oxidative stress in dopaminergic cell death induced by pesticides and complex I inhibitors: distinct roles of superoxide anion and superoxide dismutases. Free Radic Biol Med 61:370–383. 10.1016/j.freeradbiomed.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhinderwala F, Lei S, Woods J, Rose J, Marshall DD, Riekeberg E, Leite AL, Morton M, Dodds ED, Franco R, Powers R. 2019. Metabolomics analyses from tissues in Parkinson’s disease. Methods Mol Biol 1996:217–257. 10.1007/978-1-4939-9488-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. 2013. HMDB 3.0: The Human Metabolome Database in 2013. Nucleic Acids Res 41:D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. 2008. BioMagResBank. Nucleic Acids Res 36:D402–D408. 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Montesinos-Cruz V, Rose J, Pappa A, Panayiotidis MI, De Vizcaya-Ruiz A, Franco R. 2020. Survival mechanisms and xenobiotic susceptibility of keratinocytes exposed to metal-derived nanoparticles. Chem Res Toxicol 33:536–552. 10.1021/acs.chemrestox.9b00398. [DOI] [PubMed] [Google Scholar]
- 93.Worley B, Powers R. 2014. MVAPACK: a complete data handling package for NMR metabolomics. ACS Chem Biol 9:1138–1144. 10.1021/cb4008937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Worley B, Powers R. 2015. Generalized adaptive intelligent binning of multiway data. Chemometr Intell Lab Syst 146:42–46. 10.1016/j.chemolab.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Worley B, Halouska S, Powers R. 2013. Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal Biochem 433:102–104. 10.1016/j.ab.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]