ABSTRACT
P1 is the first protein translated from the genomes of most viruses in the family Potyviridae, and it contains a C-terminal serine-protease domain that cis-cleaves the junction between P1 and HCPro in most cases. Intriguingly, P1 is the most divergent among all mature viral factors, and its roles during viral infection are still far from understood. In this study, we found that telosma mosaic virus (TelMV, genus Potyvirus) in passion fruit, unlike TelMV isolates present in other hosts, has two stretches at the P1 N terminus, named N1 and N2, with N1 harboring a Zn finger motif. Further analysis revealed that at least 14 different potyviruses, mostly belonging to the bean common mosaic virus subgroup, encode a domain equivalent to N1. Using the newly developed TelMV infectious cDNA clones from passion fruit, we demonstrated that N1, but not N2, is crucial for viral infection in both Nicotiana benthamiana and passion fruit. The regulatory effects of N1 domain on P1 cis cleavage, as well as the accumulation and RNA silencing suppression (RSS) activity of its cognate HCPro, were comprehensively investigated. We found that N1 deletion decreases HCPro abundance at the posttranslational level, likely by impairing P1 cis cleavage, thus reducing HCPro-mediated RSS activity. Remarkably, disruption of the Zn finger motif in N1 did not impair P1 cis cleavage and HCPro accumulation but severely debilitated TelMV fitness. Therefore, our results suggest that the Zn finger motif in P1s plays a critical role in viral infection that is independent of P1 protease activity and self-release, as well as HCPro accumulation and silencing suppression.
IMPORTANCE Viruses belonging to the family Potyviridae represent the largest group of plant-infecting RNA viruses, including a variety of agriculturally and economically important viral pathogens. Like all picorna-like viruses, potyvirids employ polyprotein processing as the gene expression strategy. P1, the first protein translated from most potyvirid genomes, is the most variable viral factor and has attracted great scientific interest. Here, we defined a Zn finger motif-encompassing domain (N1) at the N terminus of P1 among diverse potyviruses phylogenetically related to bean common mosaic virus. Using TelMV as a model virus, we demonstrated that the N1 domain is key for viral infection, as it is involved both in regulating the abundance of its cognate HCPro and in an as-yet-undefined key function unrelated to protease processing and RNA silencing suppression. These results advance our knowledge of the hypervariable potyvirid P1s and highlight the importance for infection of a previously unstudied Zn finger domain at the P1 N terminus.
KEYWORDS: Potyviridae, Potyvirus, P1, HCPro, Zn finger motif, RNA silencing suppression, host fitness, telosma mosaic virus
INTRODUCTION
Plant viruses characteristically have small sizes and compact genomes and have evolved varied gene expression strategies to complement their limited coding capacity. Viruses in the family Potyviridae (potyvirids), the largest group of plant-infecting RNA viruses, employ polyprotein processing as a gene expression strategy (1–3). Potyvirids possess one positive-sense, single-stranded RNA genome (~9.7 kb), with the exception of bymoviruses, which have bisegmented genomes (RNA1, 7.2 to 7.6 kb; RNA2, 2.2 to 3.6 kb) (4, 5). The genomes of monopartite potyvirids contain a long, full-genome open reading frame (ORF) and a relatively short ORF (PIPO) embedded in the P3-coding region (1, 6). Exceptionally, sweet potato-infecting potyviruses have an additional small ORF (PISPO) embedded in the P1-coding region (7, 8). Both PIPO and PISPO ORFs become translatable after RNA polymerase slippage during viral replication (9–12). Upon translation, the resulting viral polyproteins are proteolytically processed by virus-encoded proteases into 10 to 12 mature units (2). The central and 3′-proximal parts of potyvirid genomes, encoding proteins from P3 to CP, are relatively conserved (2, 13). In contrast, the 5′-proximal regions, encoding leader proteases, vary greatly among species. A large majority of potyvirids, including all viruses in six (of 12) genera (Potyvirus, Brambyvirus, Rymovirus, Roymovirus, Tritimovirus, and Poacevirus) and two viruses in the genus Ipomovirus, share the same pattern of leader proteases: a chymotrypsin-like serine protease (P1) and a cysteine protease (HCPro) in tandem (2, 4). Their carboxy-proximal halves harbor protease domains that catalyze self cleavage at their own C terminus during polyprotein maturation (14, 15).
Being the first translation products, viral leader cistrons are considered important virulence and pathogenicity factors that can coordinate early infection stages (16). P1 and HCPro represent the most variable potyvirid proteins and are proposed to have evolved diverse roles in different potyvirid-host pathosystems. Potyvirus is the largest genus in the family Potyviridae, including 190 of 235 potyvirid species defined so far (https://ictv.global/report/chapter/potyviridae). Potyviral HCPro is a well-characterized multifunctional protein and has been found to carry out crucial tasks during viral infection cycle, including aphid transmission, RNA silencing suppression (RSS), enhancement of viral particle yield, promotion of viral genome translation, and subversion of RNA decay-mediated defense response (17–22). Among these roles, the molecular mechanism by which HCPro exerts RSS is, perhaps, the most studied: although diverse potyviral HCPros can perturb different steps of the RNA silencing pathway, its capacity to directly interact with, and kidnap, virus-derived small interfering RNAs seems to be the most relevant (13).
Based on phylogeny, chemical features, and functionality, potyvirid P1 proteins are classified as either type A or type B (2, 23, 24). All type B P1s tested so far have RSS activity, whereas those in the type A group do not. In addition, type A P1s, including potyviral P1s, depend on a host cofactor(s) for their proteolytic self cleavage (25–29). Extensive work to understand the biological relevance of P1 protein during viral infection has been performed with two model potyviruses, tobacco etch virus (TEV) and plum pox virus (PPV). Early studies revealed that TEV P1 plays a role in trans as an accessory factor for genome amplification and that the separation of P1 from HCPro is indispensable for viral viability (30, 31). In fact, a nonprocessed PPV P1-HCPro is deficient in RSS activity, providing an important clue to explain the incapacity of potyviruses to infect when the protease activity of P1 is knocked out (28, 29). Accumulated experimental evidences revealed an enhancing effect of P1 on the RSS activity of HCPro when they are coexpressed from the same RNA template (32–36). This regulatory effect of P1 in cis might be indirectly driven by the enhancement of translation when P1 is present (37). An involvement of potyviral P1 in host range definition and adaptation was observed (38, 39). This notion was further reinforced by additional data showing that type A P1 requires a specific host cofactor(s) to promote its self cleavage and the consequent release of mature HCPro (28, 29). Interestingly, the hypervariable amino-proximal moiety seems to block P1 self cleavage unless the host cofactor(s) interacts with it, and hence, removal of this domain allows self cleavage in the absence of the cofactor(s) with the consequent infection of nonpermissive host (28, 40, 41). As a result, it was proposed that this factor-dependent modulation of P1 cis cleavage comprises an elaborate physiological process that aims to optimize viral infection in permissive host (28). Type A P1s are also implicated in other processes of viral infection, such as improvement of translation efficiency of viral genome, cell-to-cell movement, and systemic spread (37, 42, 43).
Telosma mosaic virus (TelMV) is a member of the genus Potyvirus (4, 44). It has been reported to infect Tilia cordata in Vietnam (45), Pogostemon cablin in Indonesia (46), passion fruit (Passiflora edulis Sims) in Thailand (47), and Senna alata in China (48). In recent years, TelMV was found to be highly epidemic in main growing regions of passion fruit in China (49–51). Importantly, TelMV-infected passion fruit exhibits severe small foliage, leaf chlorosis, and mosaic symptoms (52), leading to a serious reduction in yield and quality of passion fruits (49). However, despite this socioeconomical relevance, very little is known about TelMV. In fact, only four complete genome sequences of TelMV isolates are available in the NCBI GenBank database: one from T. cordata and three from passion fruit. Given the industrial importance of passion fruit in tropical and subtropical areas, investigation of the molecular and cellular mechanisms underlying TelMV infection is urgently needed.
In this study, we found that TelMV isolates from passion fruit had two stretches (N1 and N2) at the P1 N terminus, in contrast those from the other known hosts. Here, we aimed to investigate the biological relevance of these protein stretches during viral infection, as well as the underlying molecular mechanism underpinning its function(s). Using newly developed infectious cDNA clones of a TelMV isolate (PasFru) from passion fruit, we demonstrated that the N1 domain, rather than N2, was critical for viral compatible infection in both Nicotiana benthamiana and passion fruit. We further found that the N1 domain in P1 increased the abundance of its cognate HCPro at the posttranslational level, likely via P1 cis cleavage, and thus enhanced HCPro-mediated RSS activity. Moreover, the Zn finger motif encompassed in N1 facilitated viral infection, but it did not function by modulating P1 protease activity and HCPro abundance. The new data obtained enlarge our knowledge of the biological relevance of hypervariable potyvirid P1 proteins.
RESULTS
Two amino-terminal stretches in P1, N1 and N2, are a hallmark of TelMV isolates infecting passion fruit.
To date, TelMV has been reported to infect T. cordata, P. cablin, passion fruit, and S. alata in nature. However, only four isolates with available complete genomic sequences are found in the NCBI public database, including three from passion fruit—TelMV-PasFru (accession no. MG944249), TelMV-Fuzhou (MK340754), and TelMV-Wuyishan (MK340755)—and one from T. cordata, TelMV-Hanoi (NC_009742). To obtain more information about TelMV genome sequences, we searched for the presence of TelMV sequences in available transcriptome sequencing (RNA-seq) libraries (53). Interestingly, we found TelMV not only in samples from passion fruit but also in samples from P. cablin, a host of TelMV from which no whole-genome sequence of this virus has been reported yet. Therefore, with the aim of reconstructing the genome of TelMV from patchouli, we downloaded the RNA-seq data of an infected plant (accession no. SRR12576393) and obtained the transcriptome by using Trinity (54). Yan and colleagues performed the RNA-seq to dissect the molecular mechanism of patchouli alcohol biosynthesis (55). In fact, the sample was found to be coinfected by TelMV, broad bean wilt virus 2 (family Secoviridae) and two different cytorhabdoviruses (one is strawberry cytorhabdovirus 1, and the other is unknown) by the analysis of Serratus Explorer (https://serratus.io/). Fortunately, the full-length genome of TelMV in patchouli was reconstructed with high coverage (see Data Set S1 in the supplemental material). When the corresponding amino acid sequences of TelMV isolates infecting different plant species were aligned, remarkable differences were observed in P1 (Fig. 1). As expected, the C-terminal moieties of P1s representing the serine protease domain were relatively conserved among TelMV P1s. The protease domains are defined with reference to the experimentally proven protease domain of PPV P1 (28). In contrast, N-terminal regions were quite variable (Fig. 1). Notably, TelMV isolates infecting passion fruit (Fuzhou, PasFru, and Wuyishan) have two stretches (referred to here as N1 and N2) at the beginning of P1 in comparison to isolates infecting T. cordata and P. cablin.
FIG 1.
Multiple alignment of amino acid sequences of TelMV P1s. The serine protease domain and amino-terminal stretches N1 and N2 in P1 are shown. A triad of amino acid residues (His-Gly-Ser) as putative catalytic activity sites are indicated by asterisks. These sequences were retrieved from NCBI GenBank database, and their accession numbers are MG944249 (TelMV-PasFru), MK340754 (TelMV-Fuzhou), MK340755 (TelMV-Wuyishan), and NC_009742 (TelMV-Hanoi). The genome of TelMV-Patchouli from patchouli was reconstructed from RNA-seq data in this study (Data Set S1).
The N1 domain, uniquely shared by a subset of potyviral P1s, encompasses a putative Zn finger motif.
A BLAST search using TelMV N1 as a query revealed that a total of 14 potyviruses encoded the N1 domain (Fig. 2 and Fig. S1). The amino acid sequence identities of N1 when comparing TelMV and the other potyviruses range from 30.84% to 85.98% with 58% to 100% coverage. Remarkably, the N1 domain among these viruses was much more conserved than the serine protease domain (26.17% to 90.65% versus 15.94% to 79.72%) (Fig. 2 and Tables S1 and S2). Notably, a putative Zn finger motif characterized by the sequence CX3CX25CX2C (where X is any amino acid) was identified in the N1 domain (Fig. 2). Given that the N2 stretch among these viruses shared less conservation and was found to not affect TelMV infectivity (see below), this stretch was not further analyzed. Furthermore, a total of 136 definitive potyviral species whose P1 sequences are available (https://ictv.global/report/chapter/potyviridae) were subjected to phylogenetic analysis. The results showed that 11 of the 14 viruses with an N1 domain belong to the bean common mosaic virus (BCMV) subgroup in the genus Potyvirus (Fig. 3). The remaining viruses are classified into three subgroups represented by daphne mosaic virus (DapMV), papaya ringspot virus (PRSV), and bean yellow mosaic virus (BYMV) (Fig. 3).
FIG 2.
Multiple amino acid sequence alignment of N1-containing potyviral P1s. Identical residues are marked with asterisks, and the catalytic triad residues His-Glu/Asp-Ser in the serine protease domain are indicated by stars. Alignment quality is shown as a bar graph below the alignment. The accession numbers of these sequences in GenBank database are MH286883 for passion fruit Vietnam virus (PVNV), KC845322 for soybean mosaic virus (SMV), MK217416 for watermelon mosaic virus (WMV), MK069986 for bean common mosaic virus (BCMV), LC377302 for East Asian Passiflora distortion virus (EAPDV), KT724930 for East Asian Passiflora virus (EAPV), MG197985 for papaya leaf distortion mosaic virus (PLDMV), LC228573 for gomphocarpus mosaic virus (GoMV), MK241979 for dendrobium chlorotic mosaic virus (DeCMV), EU410442 for Algerian watermelon mosaic virus (AWMV), MN124782 for cowpea aphid-borne mosaic virus (CABMV), MH376747 for pleione flower breaking virus (PlFBV), and MN549985 for paris virus 1 (ParV1).
FIG 3.
Phylogenetic relationships of 136 virus species within the genus Potyvirus. The amino acid sequence alignment of complete polyprotein of these virus species was retrieved from the ICTV webpage (https://ictv.global/report/chapter/potyviridae/potyviridae/resources) (5). The pairwise unrooted maximum-likelihood tree was generated using PhyML v.3.0 (93) and visualized using iTOL (https://itol.embl.de/). The numbers at the branch nodes indicate bootstrap support (100 replicates), and values below 70% are not shown. The bar represents 0.5 substitution per site. The subgroups represented by corresponding viruses are defined with reference to a recent publication (5). A subset of potyviruses having the Zn finger-encompassing N1 domain are indicated with red dots.
Development of full-length infectious cDNA clones of a TelMV isolate from passion fruit, TelMV-PasFru.
Previously, we determined and annotated the entire genome of the TelMV-PasFru isolate, which infects passion fruit (52) (Fig. 4A). Four portions covering the entire genome of TelMV-PasFru (from the 5′ terminus: Nf1, Nf2, M, and C in Fig. 4B) were integrated individually into a low-copy-number mini-binary T-DNA vector (pCB301) to generate the full-length cDNA clone of TelMV-PasFru, named pPasFru (Fig. 4C). N. benthamiana seedlings (n = 10) were inoculated with pPasFru via Agrobacterium infiltration. Fifteen days later, each of the infiltrated plants displayed chlorosis and mottle symptoms (Fig. 4D). Electron microscopy revealed the presence of flexuous filament particles in diseased plants (Fig. 4E), having a size (14 by 780 nm) similar to those previously observed in passion fruit (52). Further, TelMV was detected by RT-PCR in systemic leaves of these diseased plants (Fig. 4F). To monitor viral infection in planta, a complete green fluorescent protein (GFP)-coding sequence was engineered into pPasFru at the NIb-CP junction to create a GFP-tagged TelMV clone, designated pPasFru-G (Fig. 4C). All eight N. benthamiana seedlings infiltrated with pPasFru-G exhibited obvious GFP signals along veins of newly expanded leaves at 5 days postinfiltration (dpi) and systemic signals at 10 dpi (Fig. 4G). Protein samples from upper noninoculated leaves of diseased plants were subjected to Western blotting. As expected, a major band corresponding to the predicted size for the recombinant GFP (27.8 kDa) was detected, indicating that this protein was efficiently processed and released from the viral polyprotein (Fig. 4H).
FIG 4.
Development of infections cDNA clones of TelMV-PasFru. (A) Diagram representing the genomic organization of TelMV-PasFru. “VPg” and “(A)n” at the 5′ and 3′ ends, respectively, denote the genome-linked viral protein VPg and poly(A). The 5′ and 3′ untranslated regions are indicated by two short horizontal lines. The large rectangle and short bar indicate the corresponding long and small ORFs. (B) Schematic representation of the full-length cDNA clone of TelMV-PasFru (pPasFru). The full-length cDNA of TelMV-PasFru was placed between the 35S promoter and Nos terminator in a modified T-DNA vector. Nucleotides belonging to pCB301 backbone and virus are in lowercase and uppercase, respectively. The MluI site, downstream of the poly(A) tail, is shown in italic type. (C) Schematic representation of pPasFru and pPasFru-G. (D) Infectivity test of pPasFru in N. benthamiana. Representative photographs of infiltrated plants were taken at 15 dpi. The leaves indicated by the box are enlarged in the lower images. Mock, empty vector control. (E) Transmission electron micrograph of TelMV particle. Bar, 200 nm. (F) RT-PCR detection of TelMV. The systemic leaves of N. benthamiana plants at 15 dpi (top) and passion fruit seedlings at 20 dpri (bottom) were sampled. (G) Infectivity test of pPasFru-G in N. benthamiana. Representative photographs were taken under a UV lamp at 10 dpi. Mock, empty vector control. (H) Western blot detection of GFP in upper noninoculated leaves of N. benthamiana (at 10 dpi) and passion fruit (at 20 dpri). (I and J) Infectivity test of virus progeny derived from pPasFru and pPasFru-G in passion fruit.
The infectivity of pPasFru and pPasFru-G was also tested in the natural host, passion fruit. In several experiments, Agrobacterium infiltration of both clones (with and without GFP) produced no infection (data not shown). Therefore, a sap rub inoculation assay was used to examine their infectivity. The homogenates were prepared from upper noninoculated leaves of N. benthamiana plants infected with either TelMV or TelMV-GFP (Fig. 4D and G). As a result, all passion fruit seedlings (n = 20) rub inoculated with TelMV displayed dwarfism, small foliage, and leaf chlorosis and distortion symptoms at 20 days post-rub inoculation (dpri) (Fig. 4I). Consistently, strong GFP signals were observed in all seedlings (n = 10) inoculated with TelMV-GFP at 9 dpri (Fig. 4J). The presence of TelMV or TelMV-GFP in diseased plants was confirmed by reverse transcription-PCR (RT-PCR) and Western blotting (Fig. 4F and H). From these results, we conclude that (i) the newly developed full-length cDNA clones of TelMV are highly infectious in N. benthamiana and (ii) the viral progeny is able to efficiently infect the natural host, passion fruit, via sap rub inoculation.
N1, but not N2, is critical for viral systemic infection in both N. benthamiana and passion fruit.
To examine the biological significance of by the two P1 stretches during viral infection, we constructed two single-deletion mutants, pPasFru-G(ΔN1) and pPasFru-G(ΔN2), and one double-deletion mutant, pPasFru-G(ΔN1N2) (Fig. 5A). N. benthamiana seedlings (n = 10 per clone) were inoculated with pCB301 (negative control), pPasFru-G (positive control), or deletion derivatives via Agrobacterium infiltration. At 12 dpi, all plants inoculated with pPasFru-G(ΔN2) displayed infection symptoms similar to those of plants with the wild-type pPasFru-G (data not shown). Consistently, the distribution pattern and signal intensity of GFP in systemically infected leaves were equivalently observed in all plants infected with these two variants (Fig. 5B). In contrast, pPasFru-G(ΔN1) and pPasFru-G(ΔN1N2) failed to systemically infect N. benthamiana plants, as observed under UV light and by RT-PCR assay (Fig. 5B and C), even until 60 dpi.
FIG 5.
Infectivity test of N1 and N2 deletion TelMV clones in N. benthamiana and passion fruit. (A) Schematic diagrams of pPasFru-G and its truncated mutants. The ΔN1 and ΔN2 deletions are represented with labeled black bars. (B) Infectivity test of the indicated virus clones in N. benthamiana and passion fruit. Representative photographs were taken under a UV lamp at 12 dpi for N. benthamiana and at 18 dpri for passion fruit. (C) RT-PCR detection of virus progeny derived from the indicated virus clones. The upper noninoculated leaves of N. benthamiana (at 12 dpi) and passion fruit (at 18 dpri) were sampled for the assay. (D) Immunoblot detection of GFP in upper noninoculated leaves of N. benthamiana (at 12 dpi) and passion fruit (at 18 dpri).
Further, the infectivity of these deletion mutants was evaluated in passion fruit. Leaf tissues (N. benthamiana) infiltrated with the deletion mutants and control clones were sampled at 3 dpi, and each resulting homogenate was inoculated into passion fruit seedlings (n = 8 per extract). Similar to that observed in N. benthamiana, pPasFru-G(ΔN2) behaved like pPasFru-G in terms of GFP distribution pattern or accumulation (Fig. 5B and D). All plants treated with pPasFru-G(ΔN1N2) and six of eight plants inoculated with pPasFru-G(ΔN1) were not systemically infected (Fig. 5B to D), even until 90 dpri. Notably, the remaining two plants infiltrated with pPasFru-G(ΔN1) displayed scattered fluorescence spots in only one newly expanded leaf at 18 dpri (Fig. 5B), and indeed, the leaf was found to be virus infected via RT-PCR assay (Fig. 5C). The genomic region covering the partial 5′ untranslated region (UTR) (72 nucleotides [nt]), complete P1ΔN1 (1,011 nt), and partial HCPro (168 nt) was sequenced for viral progeny derived from pPasFru-G(ΔN1), and no compensatory nucleotide changes were observed. The two plants were monitored until 90 dpri and did not display any symptoms or viral accumulation in the remaining expanded leaves (data not shown). Similar results were obtained in two additional independent experiments using a total of 16 seedlings for each clone: all plants treated with either pPasFru-G(ΔN2) or pPasFru-G were systemically infected at 18 dpri, whereas those with pPasFru-G(ΔN1ΔN2) were not; only three of 16 plants with pPasFru-G(ΔN1) showed scattered fluorescent spots in a single upper noninoculated leaf.
Taken together, our results show that the short stretch N2 in P1 is dispensable for TelMV infectivity and spread, but the N1 domain is required for the virus to produce a successful infection in both N. benthamiana and passion fruit.
N1 deletion significantly attenuates viral local infection.
Local infection of N. benthamiana leaves by the N1 deletion mutant was further examined. First, we determined whether N1 deletion affects viral intercellular movement. Each agrobacterial culture harboring pPasFru-G(ΔN1), pPasFru-G, and pPasFru-G(ΔGDD) (a replication-null deletion mutant that lacks the well-conserved GDD motif of the viral replicase [56, 57]) was highly diluted to an optical density at 600 nm (OD600) of 0.0005 to initiate viral infection from single cells when subjected to infiltration into N. benthamiana leaves. Time course observation of viral intercellular movement was performed (Fig. 6A). At 60 h postinfection (hpi), single cells displaying obvious fluorescent signals representing primarily transfected cells were observed for all tested clones. Clear viral intercellular movement from primarily transfected to neighboring cells was observed at 72 hpi or later for both pPasFru-G and pPasFru-G(ΔN1), but not in the case of pPasFru-G(ΔGDD). Importantly, the spreading area for the N1 deletion virus was significantly reduced at both 72 and 96 hpi in comparison with that of the wild-type virus (Fig. 6A and B). To test the effects of N1 deletion on viral replication, N. benthamiana leaves inoculated with these clones (OD600 of 0.3) were sampled for RT-qPCR analysis at 60 hpi as the viral intercellular movement did not occur at this time point (Fig. 6A). As a result, a significant reduction of either the viral RNA or protein level was observed for N1 deletion mutant in comparison to the wild-type virus (Fig. 6C and D). All in all, these results support the idea that the N1 deletion in TelMV greatly impairs viral replication and, presumably as a consequence, intercellular movement.
FIG 6.
Effects of N1 deletion in TelMV clone on viral intercellular movement and replication. (A) Time course observation of viral intercellular movement for the indicated virus clones. The viral intercellular movement from single primarily infected cells was monitored at 48, 60, 72, and 96 hpi. Bars, 100 nm. (B) Statistic analysis of spreading area of viral infection foci at 72 hpi. Error bars denote standard errors for at least 25 infection foci for each clone from three independent experiments. *, 0.01 < P < 0.05. (C and D) Effects of N1 deletion in TelMV clone on viral genomic RNA and protein accumulations. Viral genomic RNA accumulations were determined by real-time RT-qPCR (C), and GFP accumulation by immunoblot assay (D). Error bars denote the standard errors from three biological replicates. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01.
N1 deletion in P1 markedly decreases HCPro abundance at the posttranslational level.
For PPV, the P1 N-terminal moiety (upstream of the serine-protease domain) modulates its cis cleavage activity and contributes to the yield of the cognate HCPro (28, 41). Based on this observation, we investigated whether the N1 deletion decreased the abundance of cognate HCPro in planta. To test this idea, two T-DNA constructs for ectopically expressing P1-HCProMyc and P1ΔN1-HCProMyc with a Myc tag fused at the C terminus of HCPro were generated (Fig. 7A). Agrobacterial cultures harboring either pCaM-P1-HCProMyc or pCaM-P1ΔN1-HCProMyc were adjusted to an OD600 of 0.3 and infiltrated into N. benthamiana leaves. Western blot assays showed that the abundance of HCProMyc was markedly reduced at 2 dpi or 3 dpi when it was produced from P1ΔN1-HCProMyc, relative to that from P1-HCProMyc (Fig. 7B and C). Further, real-time quantitative PCR (RT-qPCR) revealed that the abundances of the corresponding mRNA transcripts produced by pCaM-P1-HCProMyc and pCaM-P1ΔN1-HCProMyc were not significantly different (Fig. 7D), ruling out the possibility that the lower production of HCPro in the case of pCaM-P1ΔN1-HCProMyc is due to differences in mRNA accumulation.
FIG 7.
Effects of N1 deletion in P1 on the abundance of its cognate HCPro. (A) Schematic diagrams of P1-HCProMyc, P1ΔN1-HCProMyc, P1-MycHCPro-GFP, and P1ΔN1-MycHCPro-GFP. The black bar labeled “ΔN1” represents N1 deletion. Red arrows denote cis cleavage sites of P1 or HCPro. (B) Immunoblot detection of HCProMyc abundance in N. benthamiana leaves. (C) Quantitative analysis of HCProMyc signals in panel B at 2 dpi. The signal intensity values are presented as means and standard deviations (SD) (n = 3). The average value for P1-HCProMyc was designated 100% to normalize the data. *, 0.01 < P < 0.05. (D) Real-time RT-qPCR assay of the mRNA transcripts from N. benthamiana leaves infiltrated with pCaM-P1-HCProMyc and pCaM-P1ΔN1-HCProMyc. Agrobacterial cultures harboring relevant plasmids were adjusted to an OD600 of 0.3 and infiltrated into N. benthamiana leaves. The infiltrated leaves were sampled at 48 hpi for real-time RT-qPCR assay. Error bars denote the standard errors from three biological replicates. NS, no significant difference. (E) Immunoblot detection of MycHCPro accumulation levels. Agrobacterial cultures harboring relevant plasmids were adjusted to an OD600 of 0.3 for infiltration into N. benthamiana leaves, and the treated leaves were sampled at 48 hpi for Western blot assay. (F) Quantitative analysis of MycHCPro signals in panel E. The signal intensity values are presented as means and SD (n = 3). The average values for P1-MycHCPro-GFP were designated 100% to normalize the data. *, 0.01 < P < 0.05. (G) Immunoblot detection of GFP accumulation levels. (H) Quantitative analysis of GFP signals in panel G. NS, no significant difference.
TEV P1 is implicated in stimulation of viral genome translation via interaction with cytoplasmic ribosomes (43), and a Kozak-like mRNA motif in potato virus Y (PVY) P1 seems to enhance viral genome translation (37). The Kozak-like motif in TelMV is close to Kozak plant consensus (AGCAAUGGC versus AACAAUGGC). Based on those findings, we hypothesized that N1 deletion might disturb the mRNA translation, leading to the decreased abundance of HCPro. To test this idea, two additional constructs were generated to express P1-MycHCPro-GFP and P1ΔN1- MycHCPro-GFP with a Myc tag at the N terminus of HCPro (Fig. 7A). For each of them, two conserved peptides, VDVHHY/SL and KHYRVG/GD, cleaved by P1 or P1ΔN1 and HCPro in cis, respectively, are preserved. It is worth noting that GFP was translated in frame with P1-HCPro or P1ΔN1-HCPro and further released by HCPro cis cleavage at its C terminus. The accumulation levels of GFP reflect the translational dynamic of corresponding mRNAs. In line with the results above, the HCPro abundance was significantly reduced when the N1 was deleted from P1-HCPro (Fig. 7E and F). However, the accumulation levels of GFP were comparable between P1-HCProMyc-GFP and P1ΔN1-HCProMyc-GFP (Fig. 7G and H), indicating that N1 deletion does not affect the translation efficiency. In conclusion, the N1 in P1 positively regulates the abundance of downstream HCPro at the posttranslational level when the module P1-HCPro is expressed in planta.
Taken together, our results support the idea that the N1 domain in P1 facilitates the accumulation of its cognate HCPro at the posttranslational stage.
N1 domain facilitates P1 cis cleavage and consequently contributes to HCPro abundance.
Subsequently, we investigated whether N1 domain was involved in P1 cis cleavage, thus contributing to the accumulation of free HCPro. To test this hypothesis, two constructs were generated for ectopically expressing P1(S/A)-HCProMyc and P1ΔN1(S/A)-HCProMyc in which the catalytic active serine in the protease domain was mutated to alanine (Fig. 8A). Moreover, two additional plasmids were built to express P1(S/A-CS)-HCProMyc and P1ΔN1(S/A-CS)-HCProMyc, in which the NIa-Pro cleavage site GESVSLQ/SGK (from the TelMV NIb-CP junction) was engineered between P1(S/A) or P1ΔN1(S/A) and HCProMyc (Fig. 8A). Both P1(S/A-CS)-HCProMyc and P1ΔN1(S/A-CS)-HCProMyc could be cleaved by NIa-Pro in trans. Each of these constructs, together with NIa-expressing plasmid, was coinoculated into N. benthamiana leaves, followed by Western blotting analysis of HCProMyc abundance at 2 dpi. As expected, free HCProMyc was not detected from either P1(S/A)-HCProMyc or P1ΔN1(S/A)-HCProMyc due to the inactivated serine-protease domain (Fig. 8B). Moreover, signals corresponding to unprocessed P1(S/A)-HCProMyc and P1ΔN1(S/A)-HCProMyc were not detected (Fig. 8B), supporting the idea that the P1-HCPro complex is not stable, as previously reported for other potyviruses (28). Remarkably, P1(S/A-CS)-HCProMyc and P1ΔN1(S/A-CS)-HCProMyc produced almost equivalent amount of free HCProMyc via NIa-Pro trans cleavage, in contrast with the significant difference observed when comparing P1-HCProMyc versus P1ΔN1-HCProMyc (Fig. 8B). A real-time RT-qPCR assay showed that there is no significant difference in the mRNA transcript abundance between P1-HCProMyc and its modified versions (Fig. 8C). These results indicate that the N1 domain positively promotes P1 cis cleavage and thus contributes to the accumulation of free HCPro. However, a lower abundance of free HCPro when this protein is produced from P1(S/A-CS)-HCProMyc and P1ΔN1(S/A-CS)-HCProMyc in comparison to P1-HCProMyc (Fig. 8B) might be due to a potentially low-efficiency processing of the complex by NIa-Pro in trans.
FIG 8.
The N1 domain facilitates P1 cis cleavage and thus enhances the abundance of HCPro. (A) Schematic diagrams of P1-HCProMyc, P1ΔN1-HCProMyc, P1(S/A)-HCProMyc, P1ΔN1(S/A)-HCProMyc, P1(S/A-CS)-HCProMyc, and P1ΔN1(S/A-CS)-HCProMyc. (B) Immunoblot analysis of HCProMyc abundance from N. benthamiana leaves coexpressing TelMV NIa and the indicated proteins. Mock, empty vector control. Quantitative analysis of HCProMyc signals is shown (bottom). The signal intensity values are presented as means and SD (n = 3). The average value for NIa+P1-HCProMyc was designated 100% to normalize the data. **, 0.001 < P < 0.01. (C) Real-time RT-qPCR assay of mRNA transcripts of the indicated proteins. The coinfiltrated N. benthamiana leaves were sampled at 48 hpi for the assay. Error bars denote standard errors from three biological replicates. NS, no significant difference.
N1 domain consistently maintains HCPro abundance in the presence of other viral proteins, and thus enhances its RSS activity.
The regulatory effects of N1 domain on HCPro abundance in the presence of other viral proteins (from P3 to CP) were further examined. The construct pP3-CP-GFP was generated for the transient expression of remaining viral proteins P3-to-CP (Fig. 9A). The other separate expression cassettes for P1-HCProMyc, P1ΔN1-HCProMyc, and HCProMyc were each engineered into pP3-CP-GFP to generate pP3-CP-GFP//P1-HCProMyc, pP3-CP-GFP//P1ΔN1-HCProMyc, and pP3-CP-GFP//HCProMyc, respectively (Fig. 9A). In this case, the dual-expression cassettes would be codelivered into the same cells via Agrobacterium infiltration. Agrobacterial cultures harboring relevant plasmids at an OD600 of 0.3 were individually infiltrated into N. benthamiana leaves. As an indication of P3-to-CP expression, mRNA and GFP accumulations were assessed in infiltrated patches by RT-PCR and fluorescence microscopy, respectively (Fig. 9B). Western blot assays revealed that both pP3-CP//P1ΔN1-HCProMyc and pP3-CP//HCProMyc produced nearly undetectable HCPro accumulation (Fig. 9C and D). In contrast, the HCPro abundance was significantly increased for pP3-CP//P1-HCProMyc (Fig. 9C and D), indicating that the N1 domain in P1 maintains HCPro abundance in the presence of other viral proteins.
FIG 9.
The N1 domain in P1 modulates the abundance of HCPro and its RSS activity in the presence of other viral proteins (P3 to CP). (A) Schematic diagrams of four T-DNA constructs (pP3-CP-GFP, pP3-CP-GFP//P1-HCProMyc, pP3-CP-GFP//P1ΔN1-HCProMyc, and pP3-CP-GFP//HCProMyc). (B) Expression analysis of P3 to CP in N. benthamiana leaves. The mRNA transcripts of P3 to CP were detected by RT-PCR. The intracellular GFP signals were monitored by fluorescence microscopy. GFP was translated along with the truncated viral genome and released from polyprotein by proteolytic cleavage of NIa-Pro in trans. Lanes 1 and 2, empty vector control; lanes 3 and 4, pP3-CP-GFP. (C) Immunoblot analysis of HCProMyc accumulation in N. benthamiana leaves infiltrated with the indicated plasmids. (D) Quantitative analysis of HCProMyc signals in panel C. The signal intensity values are presented as means and SD (n = 3). The average value for pP3-CP-GFP//P1ΔN1-HCProMyc was designated 100% to normalize the data. *, 0.01 < P < 0.05. (E) RSS activity test of P1-HCProMyc, P1ΔN1-HCProMyc, and HCProMyc in the presence of viral proteins P3 to CP. Representative photographs of coinfiltrated N. benthamiana leaves were taken at 2 dpi. The patch design for coinfiltration was as follows: a GFP-expressing plasmid, together with a candidate protein-expressing construct (indicated by 3), P19-expressing construct (positive control, indicated by 1), or empty vector pCaMterX (negative control, indicated by 2). (F) Immunoblot detection of GFP accumulation from coinfiltrated leaf patches at 2 dpi. Coomassie blue staining of the large subunit of RubisCO was used as the loading control. Quantitative analysis of GFP signals is shown (bottom). The signal intensity values are presented as means and SD (n = 3). The average value for P3-CP//P1-HCProMyc was designated 100% to normalize the data. *, 0.01 < P < 0.05. (G) Northern blot analysis of GFP mRNA accumulation in coinfiltrated leaf patches at 2 dpi. GelRed staining of rRNA served as the loading control. (H) Real-time RT-qPCR assay of GFP mRNA transcripts from coinfiltrated N. benthamiana leaves. The samples were collected at 48 hpi for the assay. Error bars denote the standard errors from three biological replicates. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01.
Next, we examined the RSS activity of HCProMyc, P1ΔN1-HCProMyc, and P1-HCProMyc in the presence of other viral proteins. For this, three additional constructs, pP3-CP//HCProMyc, pP3-CP//P1ΔN1-HCProMyc, and pP3-CP//P1-HCProMyc, were generated (Fig. S2) and used in the RSS test via coinfiltration with the GFP-expressing plasmid. The leaf patches expressing P3-CP//P1-HCProMyc displayed strong fluorescence signals under UV light, whereas no obvious signals were observed on leaf patches expressing either P3-CP//P1ΔN1-HCProMyc or P3-CP//HCProMyc, similar to the negative control (Fig. 9E). Supporting this, a higher abundance of GFP at both the protein and RNA levels was observed in leaf patches expressing P3-CP//P1-HCProMyc (Fig. 9F to H). These results indicate that only the module P1-HCProMyc confers sufficient RSS activity in the presence of other viral proteins.
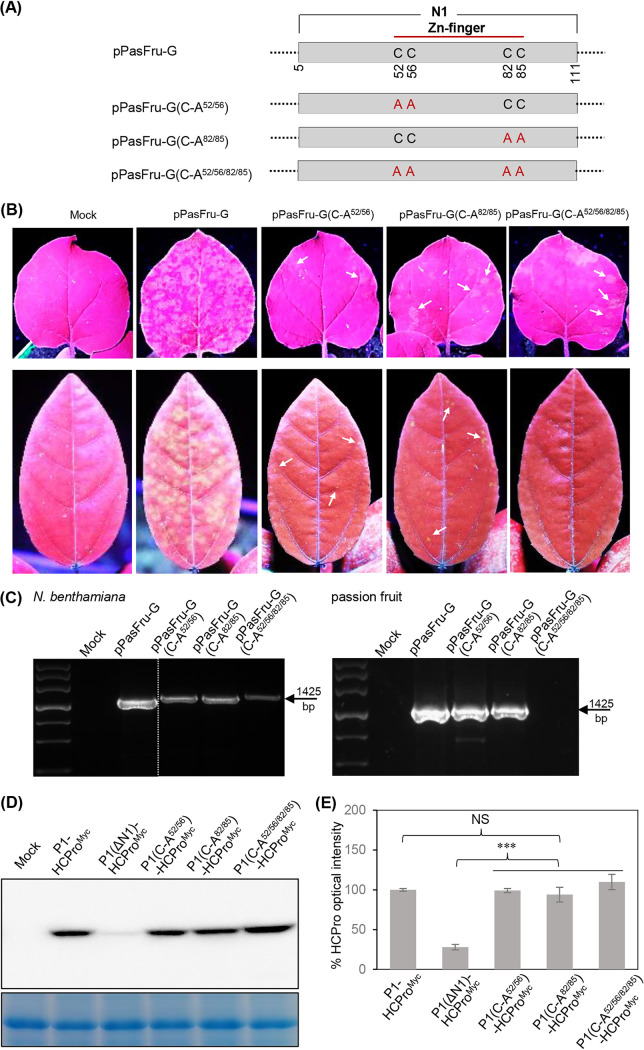
Disruption of the Zn finger motif in N1 does not impact HCPro abundance but severely debilitates viral infectivity.
As stated above, a putative Zn finger motif is encompassed in the N1 domain and strictly conserved among a subset of potyviruses mostly belonging to the BCMV subgroup. This prompted us to associate the Zn finger motif with N1-mediated HCPro abundance and the consequent effect on viral infection. Three mutated virus clones, pPasFru-G(C-A52/56), pPasFru-G(C-A82/85), and pPasFru-G(C-A52/56/82/85), with artificial disruption of the Zn finger motif were constructed (Fig. 10A). Each of these mutated virus clones was inoculated into N. benthamiana seedlings (n = 7 per clone) via Agrobacterium infiltration. Plants inoculated with pPasFru-G exhibited obvious fluorescence signals in newly expanded leaves under UV light at 15 dpi; however, only scattered weak fluorescence spots were observed in new leaves from four, six, and three plants inoculated with pPasFru-G(C-A52/56), pPasFru-G(C-A82/85), and pPasFru-G(C-A52/56/82/85), respectively (Fig. 10B). To examine their infectivity in the natural host, these virus clones were each mechanically inoculated into eight passion fruit seedlings. As expected, all eight plants inoculated with pPasFru-G were systemically infected at 18 dpri (Fig. 10B and C). However, only one, two, and one passion fruit plants inoculated with pPasFru-G(C-A52/56), pPasFru-G(C-A82/85), and pPasFru-G(C-A52/56/82/85), respectively, displayed scattered weak fluorescence spots in upper leaves, indicative of attenuated viral infectivity; the remaining plants were not infected (Fig. 10B and C). The genomic regions surrounding the mutated sites (approximately 170- and 250-nt sequences upstream and downstream of the mutated sites, respectively) for the progeny derived from these mutated clones were sequenced, and no nucleotide reversions were found. Altogether, disruption of the Zn finger motif in the N1 domain greatly impaired viral infectivity in both N. benthamiana and passion fruit plants.
FIG 10.
Disruption of Zn finger motif in N1 debilitates viral compatible infection but does not affect P1 cis cleavage and HCPro accumulation. (A) Schematic diagrams of pPasFru-G and its mutated versions. (B) Infectivity test of the indicated virus clones in N. benthamiana and passion fruit. Representative photographs of upper noninoculated leaves were taken at 15 dpi for N. benthamiana and 18 dpri for passion fruit. The scattered weak fluorescence spots are indicated by arrows. (C) RT-PCR detection of virus progeny derived from the indicated virus clones. The upper noninoculated leaves of N. benthamiana at 15 dpi (left) and passion fruit at 18 dpri (right) were sampled. (D) Immunoblot detection of HCProMyc from N. benthamiana leaves inoculated with the relevant plasmids. (E) Quantitative analysis of HCProMyc signals in panel D. The signal intensity values are presented as means and SD (n = 3). The average value for P1-HCProMyc was designated 100% to normalize the data. ***, P < 0.001; NS, no significant difference.
To examine whether the Zn finger motif in P1 is associated with P1 cis cleavage and HCPro abundance, three T-DNA plasmids for transiently expressing P1(C-A52/56)-HCProMyc, P1(C-A82/85)-HCProMyc, and P1(C-A52/56/82/85)-HCProMyc were generated (Fig. S3). Western blot assays showed that all mutant combinations yielded comparable amounts of HCProMyc, which was almost equivalent to that from P1-HCProMyc but significantly higher than that from P1(ΔN1)-HCProMyc (Fig. 10D and E). These results indicate that the Zn finger motif in N1 is not involved in P1 cis cleavage and HCPro accumulation and suggest that this motif plays a yet-unknown critical role during viral infection.
DISCUSSION
P1, the most variable potyvirid factor, has long been deemed a mysterious protein during viral infection, and it has attracted great scientific interest. During the past decade, it has been reported that type A P1s are implicated in host range definition and host adaption. In this study, we characterized and defined a highly conserved Zn finger motif-encompassing domain (N1) at the N terminus of type A P1, which is shared by at least 14 potyviruses. Using TelMV as a model, we demonstrated that the N1 domain in P1 promoted compatible viral infection in both N. benthamiana (model host) and passion fruit (natural host). It was further revealed that (i) the N1 domain in P1 facilitates the accumulation of its cognate HCPro, likely by promoting P1 cis cleavage, and thus enhances the HCPro-mediated RSS activity and (ii) the Zn finger motif mediates another crucial function(s) of P1, independently of modulating P1 cis cleavage and HCPro abundance.
The coding sequences of potyvirid P1s, in particular that coding for their N-terminal regions, were subjected to many recombination events among both intra- and intergenus viruses (23, 58–60). Equivalently to TelMV, all soybean mosaic virus (SMV) isolates belonging to the SC7 strain have a stretch at the N terminus of P1, which is homolog of TelMV N1, quite likely as a result of recombination between an SMV variant without N1 and BCMV (59, 61–64). Moreover, watermelon mosaic virus (WMV) isolates might arise from a recombination between BCMV and SMV-related potyvirus, as previously proposed (58, 65), and consequently, WMV also encodes the N1 domain in P1. We found in this study that at least 14 potyviruses contain the Zn finger domain. Remarkably, the vast majority of them belong to the BCMV group in the genus Potyvirus. Thus, a more likely evolutionary scenario is that a common ancestor of potyviruses in the BCMV group already had the Zn finger domain, which was further maintained by some of them. If this hypothesis is correct, the potyviruses encoding N1 but not belonging to the BCMV group would have obtained this domain by interspecies recombination. Certainly, the possibility could not be ruled out that a loss-of-N1 virus in this group might reobtain the domain through recombination with a phylogenetically closely related virus carrying the N1 domain during coinfection in the common hosts.
Diverse groups of plant viruses encode Zn finger proteins, such as carlaviral p12, geminiviral TrAP, begomoviral C2, and potyviral HCPro (66–69). Zn finger motifs in these proteins have been found to correlate with key biological activities, such as DNA/RNA binding, transcription activation, and RNA silencing suppression (70–74). Notably, a conserved region characterized by LX2AX6Y plus a Zn finger motif is present in all type B P1s and exhibits enough RSS activity in a context of viral infection (75). Therefore, the LX2AX6Y+Zn finger domain as an RNA silencing suppression module for type B P1s was postulated (75). In a previous review, the Cys and His residues that may form part of the Zn finger structures at the N terminus of type A P1s were detected in several potyviruses (23). This study defined the Zn finger motif-encompassing domain and revealed its specific presence in at least 14 different potyviruses. The Zn finger domains have no obvious sequence homology with the LX2AX6Y+Zn finger modules. Nevertheless, the origins of both domains await investigation.
Recent experimental data revealed extreme adaptation capacity of potyviral P1s, accompanied by various recombination events during viral replication, which probably contributes to the extraordinary success of potyvirids as wide-host-range colonizers (75). Our study demonstrated that N1 domain in TelMV P1 facilitates viral successful infection in both N. benthamiana and passion fruit, supporting the notion that the specific N1 domain might be associated with the host range and fitness of TelMV. To corroborate this speculation, a comprehensive investigation of the correlation between host species and TelMV isolates carrying the N1 domain needs to be performed. Given the enhancement effect of the N1 domain on viral infectivity either by increasing the abundance of its cognate HCPro or by carrying out a Zn finger motif-mediated crucial function, a likely explanation for TelMV isolates from other hosts lacking the N1 domain might be that these hosts are more susceptible to TelMV infection, and the isolates lacking N1 and exhibiting attenuated virulence evolved to better adapt to these hosts.
PPV, a well-studied model virus in the Potyvirus genus, does not contain the N1 domain in P1. Whereas the P1 N-terminal moiety (upstream of the serine protease domain) modulates its cis cleavage activity, the complete inactivation of P1 serine-protease activity leads to the generation of nonprocessed P1-HCPro with a consequent inactivation of HCPro-mediated RSS (28, 29, 41). Removal of the antagonistic element or the complete P1 region accelerates early amplification of PPV and elicits severe hypersensitive response-like symptoms, leading to reduced viral load (28). Therefore, it is proposed that the autoinhibited P1 self-cleavage plays an immune evasion role, which modulates viral multiplication to optimize viral host adaptation by controlling the release of the functional silencing suppressor HCPro (16, 76). Supporting this, two natural SMV isolates, NB and GZL, with the former containing the N1 homolog, differentially elicit salicylic acid signaling pathway (64), although it is unknown whether the contrasting immune response is associated with N1. Different from the observations from PPV, two TelMV mutants with deletion of the N-terminal moiety (upstream of the serine-protease domain) and the complete P1 region and the N1 deletion mutant all failed to produce successful infection (Fig. S4). In line with this, HCPro was almost undetectable when it was coexpressed with viral proteins P3 to CP or coproduced with truncated P1 versions (P1ΔN1, P1ΔN, and ΔP1); however, its abundance could be greatly recovered once it was preceded by a complete P1 sequence (Fig. 9 and Fig. S5). Conclusively, TelMV P1, in particular its N1 domain, positively regulates compatible viral infection, which is different from the antagonistic effects of PPV P1 on viral multiplication to evade the host immune response.
In recent years, intracellular hydrolase and degradation pathways were demonstrated to have regulatory effects on potyviral HCPro abundance during viral infection (77–79). In the case of turnip mosaic virus (TuMV), the autophagy receptor NBR1 targets and degrades HCPro, whereas viral VPg and 6K2 proteins antagonize this NBR1-dependent degradation (78). We propose that similar degradation events might also take place for TelMV HCPro, because it is hardly detectable when it is coexpressed with other viral P3 to CP proteins in planta (Fig. 9). However, the abundance of HCPro is significantly rescued when it is preceded by complete P1 (Fig. 9 and Fig. S5), implying that the N1 domain in P1 might contribute to HCPro abundance by enhancing P1 cis cleavage and/or interfering with potential HCPro-related degradation events, as in the case of VPg and 6K2 in TuMV. Our current working model in this regard is presented in Fig. 11.
FIG 11.
Working model depicting the involvement of the N1 domain of TelMV P1 in viral host fitness.
As the first expressed viral protein, potyviral P1 has long been deemed an accessary factor during viral infection. In recent years, extensive studies revealed the involvement of P1 in viral host-range definition and host adaption. However, few reports have been dedicated to unveiling the direct functional roles of P1 (besides self-cleavage). A recent report, for instance, revealed that TuMV P1 is able to promote the degradation of cpSRP54 via the 26S proteasome system, preventing the delivery of allene oxide cyclases to the thylakoid membrane and the further synthesis of jasmonic acid, to then facilitate viral infection (80). Our study supports the idea that TelMV P1, through its Zn finger domain, plays a crucial role during infection that, as in the case of TuMV P1, is also independent of self-cleavage. Another intriguing study reported the modulatory effect of WMV P1 (N1 containing) on an unrelated VSR protein of a cucurbit-infecting crinivirus, suggesting its involvement in the complex virus-virus interactions during mixed infections (81). The actual role of this Zn finger domain in TelMV, as well as in other potyviruses carrying this motif, awaits further investigation.
MATERIALS AND METHODS
Virus isolate and plant materials.
In this study, a TelMV PasFru isolate that we previously characterized from passion fruit (52) was used for the generation of TelMV-derived cDNA clones. N. benthamiana and yellow passion fruit seedlings were maintained in a growth cabinet set to 16 h of light at 25°C and 8 h of darkness at 23°C, with 70% relative humidity.
Construction of TelMV-derived cDNA clones.
A low-copy-number mini-binary T-DNA vector, pCB301 (82), was used as the backbone to construct the full-length cDNA clone of TelMV-PasFru. In brief, a modified pCB301 with desired multiple cloning sites (StuI, SalI, BamHI, SpeI, and MluI) was amplified from pVPH-GFP/mCherry (57) using the primer set PCB-F/PCB-R (Table S3). Four pairs of primers (N1-F/N1-R, N2-F/N2-R, M-F/M-R, and C-F/C-R) (Table S3) were designed to amplify corresponding portions in the TelMV genome (from the 5′ terminus: N1f, N2f, M, and C), which were separated with SalI, BamHI, and SpeI sites. The fragments M, N1f, C, and N2f were engineered, one by one, into the modified pCB301 vector by utilization of the multiple cloning sites to generate the full-length cDNA clone of TelMV-PasFru (pPasFru).
A complete GFP-coding sequence was inserted at the NIb/CP intercistronic site to create a GFP-tagged TelMV clone, pPasFru-G. For this clone, the original cleavage sequence ESVSLQ/SG at NIb/CP junction (proteolytically processed by NIa-Pro) was engineered into NIb/GFP and GFP/CP junctions for the release of GFP during viral infection. In case the GFP-coding region was removed via potential recombination events during viral replication, the corresponding nucleotide sequences of the two cleavage peptides differ in eight nucleotides (GAATCAGTCTCGCTCCAA/TCCGGC versus GAGTCGGTTTCACTTCAG/TCTGGA). Standard DNA manipulation technologies, including overlapping PCR, were used to construct pPasFru-G. Briefly, the GFP coding region was amplified from pVPH-GFP/mCherry (57) using the primer set SOE-2-F/GFP-2-R (Table S3). Two other fragments upstream and downstream of NIb/CP cleavage site were amplified with the primer sets C-F/SOE-1-R and H-3-F/C-R, respectively (Table S3). Then, overlapping PCR with the primer set C-F/C-R was performed using a mixture of the above fragments as the template. The resulting fragment was inserted back into pPasFru by using SpeI/MluI sites to generate pPasFru-G. A replication-defective virus clone, named pPasFru-G(ΔGDD), was created via the removal of the highly conserved GDD motif in the viral RNA polymerase, NIb. Two fragments upstream and downstream of the GDD motif in pPasFru-G were amplified with the corresponding primer sets C-F/T-d-GDD-F and T-d-GDD-F/C-R (Table S3) and then mixed as the template for overlapping PCR with the primer set C-F/C-R (Table S3). The resulting fragment was inserted back into SpeI/MluI-treated pPasFru-G to generate pPasFru-G(ΔGDD).
A series of P1-truncated and -mutated TelMV clones, including pPasFru-G(ΔN1), pPasFru-G(ΔN2), pPasFru-G(ΔN1N2), pPasFru-G(ΔN), pPasFru-G(ΔP1), pPasFru-G(C-A52/56), pPasFru-G(C-A82/85), and pPasFru-G(C-A52/56/82/85), were constructed by using pPasFru-G as the backbone. A similar strategy was employed to construct these clones, and the detailed description for the creation of pPasFru-G(ΔN1), in which the N1 stretch in TelMV was deleted, serves as an example here. Two PCRs with pPasFru-G as the template were performed with the primer sets PCB301-1F/T-A-1-R and T-A-2-F/T-1216-R (Table S3). A mixture of the resulting PCR products was used as the template for overlapping PCR with the primer set PCB301-F/T-1216-R (Table S3). The obtained fragment was inserted back into pPasFru-G by using the PmeI/PstI sites to generate pPasFru-G(ΔN1).
pP3-CP-G was generated via the deletion of the entire coding region of P1-HCPro in pPasFru-G. Briefly, two fragments upstream and downstream of P1-HCPro in pPasFru-G were amplified with the primer sets PCB301-1F/T-dPH-R and T-dPH-F/N2-R, respectively (Table S3), followed by overlapping PCR with primer set PCB301-1F/N2-R. The resulting fragment was inserted back into pPasFru-G by using the PmeI/BamHI sites to generate pP3-CP-G. To examine the regulatory effects of the N1 domain on HCPro abundance in the presence of other viral proteins (P3 to CP), three separate expression cassettes for P1-MycHCPro, P1ΔN1-MycHCPro, and MycHCPro driven by the 35S promoter were each placed in the backbone pP3-CP-G (adjacent to the P3-CP-G cassette). The resulting constructs were designated pP3-CP-G//P1-HCProMyc, pP3-CP-G//P1ΔN1-HCProMyc, and pP3-CP-G//HCProMyc. Here, the detailed information for the construction of pP3-CP-G//P1-MycHCPro is presented as an example. The Nos-35S region was amplified from pVPH-GFP//mCherry (57) with the primer set T-trans-N-F/T-35S-PH-R and the P1-HCProMyc fragment from pPasFru-G with T-transPH-F/T-trans-C-R (Table S3). A mixture of the resulting PCR products was used as the template for overlapping PCR with the primer set T-trans-N-F/T-trans-C-R (Table S3). The fragment obtained was inserted into pP3-CP-G by using a single MluI site to generate pP3-CP-G//P1-MycHCPro.
All these constructs were verified by Sanger sequencing.
Construction of binary plant-expression vectors.
A binary plant-expression vector pCaMterX (83) was employed as the backbone for the transient expression of genes of interest in planta. Two plasmids, pCaM-P1-HCProMyc and pCaM-P1ΔN1-HCProMyc, were constructed to test the effect of N1 deletion on HCPro accumulation in planta. The coding region of P1-HCProMyc was amplified from pPasFru-G with the primer set P1HC-F/P1HC-myc-R, and P1ΔN1-HCProMyc from pPasFru-G(ΔN1) with the set P1HC(dA)-F/P1HC-myc-R (Table S3). The products obtained were integrated into pCaMterX by BamHI/KpnI sites. Using a similar strategy, the constructs pCaM-P1(C-A52/56)-HCProMyc, pCaM-P1(C-A82/85)-HCProMyc, pCaM-P1(C-A52/56/82/85)-HCProMyc, pCaM-P1(S/A)-HCProMyc, pCaM-P1ΔN1(S/A)-HCProMyc, pCaM-P1(S/A-CS)-HCProMyc, and pCaM-P1ΔN1(S/A-CS)-HCProMyc were generated for testing the effects of P1 mutations on HCPro accumulation in planta.
The plasmids pCaM-P1-MycHCPro-GFP and pCaM-P1ΔN1-MycHCPro-GFP were constructed by overlapping PCR to determine the effect of N1 deletion from the module P1-HCPro on translation efficiency. For these, the cleavage sites VDVHHY/SL in P1 (or P1ΔN1) and KHYRVG/GD in HCPro were kept for the dissociation of the precursor proteins. To construct pCaM-P1-MycHCPro-GFP, the coding regions for P1, MycHCPro, and GFP were amplified from pPasFru-G with the corresponding primer sets P1HC-F/mycHC-1-R, mycHC-2-F/HG-1-R, and HG-2-F/HG-2-R, and the products obtained were mixed as the template for overlapping PCR with the primer set P1HC-F/HG-2-R (Table S3). The resulting fragment was inserted into the pCaMterX by using the BamHI/KpnI sites. Similarly, pCaM-P1ΔN1-MycHCPro-GFP was generated. Besides pCaM-MycHCPro, three more plasmids, pCaM-P1-MycHCPro, pCaM-P1ΔN1-MycHCPro, and pCaM-P1ΔN-MycHCPro, were constructed to examine the effects of different domains in P1 on the abundance of its cognate HCPro. Here, the detailed description for the generation of pCaM-P1-MycHCPro serves as an example. The coding regions of P1 and MycHCPro with the corresponding primer sets P1HC-F/mycHC-1-R and mycHC-2-F/T-H-T-R (Table S3) were amplified from pPasFru-G, and the products obtained were mixed as the template for overlapping PCR with the primer set P1HC-F/T-H-T-R. The resulting fragment was inserted into pCaMterX by using BamHI/KpnI sites to generate pCaM-P1-MycHCPro.
All these plasmids were verified by Sanger sequencing.
Agrobacterium infiltration and sap rub inoculation.
Agrobacterium (strain GV3101)-mediated transformation in N. benthamiana was performed essentially as previously described (84, 85). Fully developed leaves were inoculated with the relevant plasmids via Agrobacterium infiltration. Seedlings at the 3- to 5-leaf stage were used for an infectivity test of TelMV-derived cDNA clones, and the 6- to 8-leaf stage was used for transient expression of genes of interest. A sap rub inoculation assay was employed to test the infectivity of TelMV-derived cDNA clones in passion fruit, following a previously described protocol (85). Briefly, leaf tissues of N. benthamiana were collected at the indicated days and ground in phosphate buffer (KH2PO4, 0.01 M, Na2HPO4, 0.01 M; pH 6.8) in a ratio of 1:10 (grams per milliliter). The resulting homogenate as an inoculum source was immediately rub inoculated into the newly developed leaves of passion fruit seedlings (approximately 8 to 10 cm in height). Each leaf was rubbed twice from the leaf bottom to the top using an inoculum-dipped forefinger.
Electron microscopy and fluorescence microscopy.
A transmission electron microscope (Hitachi HT-7700) was employed to examine the presence of viral particles in upper noninoculated leaves of N. benthamiana plants infiltrated with pPasFru by following a previously described protocol (86). To monitor viral cell-to-cell movement, an inverted fluorescence microscope (BX53F; Olympus) was employed to cellularly examine GFP signals in N. benthamiana leaves inoculated with relevant TelMV clones. The spreading area of GFP signals (representing viral infection foci) was calculated by ImageJ software (87).
UV observation.
A LUYOR-3410 hand-held UV lamp (LUYOR) was used to monitor GFP fluorescence signals in N. benthamiana and passion fruit plants with the stated treatment. Representative photographs were taken in a dark room at the indicated time points.
RNA extraction, RT-PCR, and RT-qPCR.
For RT-PCR and real-time RT-qPCR, total RNAs were extracted from leaf tissues of N. benthamiana or passion fruit using TRNzol universal reagents (Tiangen), followed by treatment with DNase I (Thermo Fisher Scientific). The first-strand cDNAs were synthesized by reverse transcription reactions using a RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific) with random hexamer primers. The PCRs with primers TelMV-8200-F and TelMV-8900-R (52), homologous to and complementary to NIb- and CP-coding regions, respectively, were performed to detect the presence of TelMV or its derivatives. RT-qPCR with SuperReal PreMix Plus (SYBR green) (Tiangen) was performed, and results were analyzed with qTOWER3 real-time PCR thermal cycler (Analytic Jena AG) following the manufacturer’s instructions. The primer set T-CP-F/T-CP-R (Table S3) was designed for real-time qPCR analysis of viral genomic RNA accumulation. The primer set HCpro-F/HCPro-R (Table S3) was used for the relative quantitative analysis of mRNA transcripts of P1-HCProMyc and its derivatives. The primer set GFP-q-F/GFP-q-R (Table S3) was used to relatively quantify the abundance of GFP mRNA transcripts. The actin transcripts obtained with the primer set Actin-145F/Actin-145R (57) were used as an internal control to normalize the data.
RNA silencing suppression assay.
A classic two-component transient coexpression via Agrobacterium infiltration of the GFP-expressing construct pCHF3-35S-GFP (88, 89) together with another plasmid expressing the gene of interest was performed to test the single-stranded-RNA-mediated silencing suppression. Coexpression of the GFP plasmid with P19 from tobacco bushy stunt virus and a negative empty vector (pCaMterX) was used as the positive and negative controls, respectively. The accumulations of GFP in both RNA and protein levels were determined by Northern blotting/RT-qPCR and Western blot assays, respectively, essentially as previously described (90).
Immunoblotting and antibodies.
An immunoblotting assay was performed essentially as previously described (90). An anti-GFP polyclonal antibody (Abcam) or anti-Myc tag monoclonal antibody (Abcam) was used in immune detection. Goat anti-rabbit immunoglobulin antibody (Abcam) conjugated to horseradish peroxidase was used as the secondary antibody. The hybridization signals in the blotted membranes were detected with the substrates of enhanced chemiluminescence detection reagents (Thermo Fisher Scientific), visualized using an ImageQuant LAS 4000 mini-biomolecular imager (GE Healthcare), and quantitatively analyzed with ImageJ software (87).
Sequence analysis and statistical analysis.
Nucleotide sequence analysis, such as the deduction of protein sequences and molecular weights, was performed using the Lasergene software package version 7.1. Multiple alignment of amino acid sequences was performed using ClustalX 1.8.1. The resulting files were analyzed with ESPript 3.0 (91) or Jalview version 2.11.1.4 (92). Statistically significant differences were determined by an unpaired two-tailed Student's t test (SAS software).
ACKNOWLEDGMENTS
This work is supported by grants from the Hainan Provincial National Science Foundation (grant nos. 2019RC010 and 322CXTD505), Sanya Yazhou Bay Sci-Tech City (SYND-2022-32 and SYND-2022-02), the National Natural Science Foundation of China (32060603), and the Central Public Interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (19CXTD-33).
We thank Fangfang Li (Chinese Academy of Agricultural Sciences) for providing pCHF3-35S-GFP and P19-expressing plasmid.
H.C., A.A.V., B.G., Z.D., and L.Q. conceived and designed the project. B.G., Z.D., L.Q., and M.R. carried out experiments. H.C. supervised the work. All authors analyzed and discussed the data. H.C., B.G., Z.D., L.Q., and A.A.V. wrote the manuscript. All authors reviewed and approved the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Adrián A. Valli, Email: avalli@cnb.csic.es.
Hongguang Cui, Email: hongguang.cui@hainanu.edu.cn.
W. Allen Miller, Iowa State University.
REFERENCES
- 1.Revers F, García JA. 2015. Molecular biology of potyviruses. Adv Virus Res 92:101–199. 10.1016/bs.aivir.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Cui H, Wang A. 2019. The biological impact of the hypervariable N-terminal region of potyviral genomes. Annu Rev Virol 6:255–274. 10.1146/annurev-virology-092818-015843. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Li Y, Wang A. 2021. Research advances in potyviruses: from the laboratory bench to the field. Annu Rev Phytopathol 59:1–29. 10.1146/annurev-phyto-020620-114550. [DOI] [PubMed] [Google Scholar]
- 4.Wylie SJ, Adams M, Chalam C, Kreuze J, López-Moya JJ, Ohshima K, Praveen S, Rabenstein F, Stenger D, Wang A, Zerbini FM, ICTV Report Consortium . 2017. ICTV virus taxonomy profile: Potyviridae. J Gen Virol 98:352–354. 10.1099/jgv.0.000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inoue-Nagata AK, Jordan R, Kreuze J, Li F, López-Moya JJ, Mäkinen K, Ohshima K, Wylie SJ, ICTV Report Consortium . 2022. ICTV virus taxonomy profile: Potyviridae 2022. J Gen Virol 103:e001738. 10.1099/jgv.0.001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung BYW, Miller WA, Atkins JF, Firth AE. 2008. An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci USA 105:5897–5902. 10.1073/pnas.0800468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark CA, Davis JA, Abad JA, Cuellar WJ, Fuentes S, Kreuze JF, Gibson RW, Mukasa SB, Tugume AK, Tairo FD, Valkonen JP. 2012. Sweetpotato viruses: 15 years of progress on understanding and managing complex diseases. Plant Dis 96:168–185. 10.1094/PDIS-07-11-0550. [DOI] [PubMed] [Google Scholar]
- 8.Li F, Xu D, Abad J, Li R. 2012. Phylogenetic relationships of closely related potyviruses infecting sweet potato determined by genomic characterization of sweet potato virus G and sweet potato virus 2. Virus Genes 45:118–125. 10.1007/s11262-012-0749-2. [DOI] [PubMed] [Google Scholar]
- 9.Olspert A, Chung BY, Atkins JF, Carr JP, Firth AE. 2015. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep 16:995–1004. 10.15252/embr.201540509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodamilans B, Valli A, Mingot A, San León D, Baulcombe D, López-Moya JJ, García JA. 2015. RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J Virol 89:6965–6967. 10.1128/JVI.00337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mingot A, Valli A, Rodamilans B, San León D, Baulcombe DC, García JA, López-Moya JJ. 2016. The P1N-PISPO trans-frame gene of sweet potato feathery mottle potyvirus is produced during virus infection and functions as an RNA silencing suppressor. J Virol 90:3543–3557. 10.1128/JVI.02360-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Untiveros M, Olspert A, Artola K, Firth AE, Kreuze JF, Valkonen JP. 2016. A novel sweet potato potyvirus open reading frame (ORF) is expressed via polymerase slippage and suppresses RNA silencing. Mol Plant Pathol 17:1111–1123. 10.1111/mpp.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valli AA, Gallo A, Rodamilans B, López-Moya JJ, García JA. 2018. The HCPro from the Potyviridae family: an enviable multitasking helper component that every virus would like to have. Mol Plant Pathol 19:744–763. 10.1111/mpp.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams MJ, Antoniw JF, Beaudoin F. 2005. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol Plant Pathol 6:471–487. 10.1111/j.1364-3703.2005.00296.x. [DOI] [PubMed] [Google Scholar]
- 15.Rodamilans B, Shan H, Pasin F, García JA. 2018. Plant viral proteases: beyond the role of peptide cutters. Front Plant Sci 9:666. 10.3389/fpls.2018.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasin F, Daròs JA, Tzanetakis IE. 2022. Proteome expansion in the Potyviridae evolutionary radiation. FEMS Microbiol Rev 46:fuac011. 10.1093/femsre/fuac011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hafrén A, Eskelin K, Mäkinen K. 2013. Ribosomal protein P0 promotes potato virus A infection and functions in viral translation together with VPg and eIF(iso)4E. J Virol 87:4302–4312. 10.1128/JVI.03198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafrén A, Lõhmus A, Mäkinen K. 2015. Formation of potato virus A-induced RNA granules and viral translation are interrelated processes required for optimal virus accumulation. PLoS Pathog 11:e1005314. 10.1371/journal.ppat.1005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, Wang A. 2018. RNA decay is an antiviral defense in plants that is counteracted by viral RNA silencing suppressors. PLoS Pathog 14:e1007228. 10.1371/journal.ppat.1007228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valli AA, Gallo A, Calvo MV, Pérez JJ, García JA. 2014. A novel role of the potyviral helper component proteinase contributes to enhance the yield of viral particles. J Virol 88:9808–9818. 10.1128/JVI.01010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De S, Pollari M, Varjosalo M, Mäkinen K. 2020. Association of host protein VARICOSE with HCPro within a multiprotein complex is crucial for RNA silencing suppression, translation, encapsidation and systemic spread of potato virus A infection. PLoS Pathog 16:e1008956. 10.1371/journal.ppat.1008956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollari M, De S, Wang A, Mäkinen K. 2020. The potyviral silencing suppressor HCPro recruits and employs host ARGONAUTE1 in pro-viral functions. PLoS Pathog 16:e1008965. 10.1371/journal.ppat.1008965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valli A, Lopez-Moya JJ, Garcia JA. 2007. Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J Gen Virol 88:1016–1028. 10.1099/vir.0.82402-0. [DOI] [PubMed] [Google Scholar]
- 24.Rodamilans B, Valli A, García JA. 2013. Mechanistic divergence between P1 proteases of the family Potyviridae. J Gen Virol 94:1407–1414. 10.1099/vir.0.050781-0. [DOI] [PubMed] [Google Scholar]
- 25.Carrington JC, Freed DD, Oh CS. 1990. Expression of potyviral polyproteins in transgenic plants reveals three proteolytic activities required for complete processing. EMBO J 9:1347–1353. 10.1002/j.1460-2075.1990.tb08249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verchot J, Herndon KL, Carrington JC. 1992. Mutational analysis of the tobacco etch potyviral 35-kDa proteinase: identification of essential residues and requirements for autoproteolysis. Virology 190:298–306. 10.1016/0042-6822(92)91216-h. [DOI] [PubMed] [Google Scholar]
- 27.Thornbury DW, Van den Heuvel JFJM, Lesnaw JA, Pirone TP. 1993. Expression of potyvirus proteins in insect cells infected with a recombinant baculovirus. J Gen Virol 74:2731–2735. 10.1099/0022-1317-74-12-2731. [DOI] [PubMed] [Google Scholar]
- 28.Pasin F, Simón-Mateo C, García JA. 2014. The hypervariable amino-terminus of P1 protease modulates potyviral replication and host defense responses. PLoS Pathog 10:e1003985. 10.1371/journal.ppat.1003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shan H, Pasin F, Valli A, Castillo C, Rajulu C, Carbonell A, Simón-Mateo C, García JA, Rodamilans B. 2015. The Potyviridae P1a leader protease contributes to host range specificity. Virology 476:264–270. 10.1016/j.virol.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 30.Verchot J, Carrington JC. 1995. Debilitation of plant potyvirus infectivity by P1 proteinase-inactivating mutations and restoration by second-site modifications. J Virol 69:1582–1590. 10.1128/JVI.69.3.1582-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verchot J, Carrington JC. 1995. Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification. J Virol 69:3668–3674. 10.1128/JVI.69.6.3668-3674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruss G, Ge X, Shi XM, Carrington JC, Bowman Vance V. 1997. Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9:859–868. 10.1105/tpc.9.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB. 1998. A viral suppressor of gene silencing in plants. Proc Natl Acad Sci USA 95:13079–13084. 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasschau KD, Carrington JC. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461–470. 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 35.Rajamäki ML, Kelloniemi J, Alminaite A, Kekarainen T, Rabenstein F, Valkonen JP. 2005. A novel insertion site inside the potyvirus P1 cistron allows expression of heterologous proteins and suggests some P1 functions. Virology 342:88–101. 10.1016/j.virol.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 36.Valli A, Martín-Hernández AM, López-Moya JJ, García JA. 2006. RNA silencing suppression by a second copy of the P1 serine protease of cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro. J Virol 80:10055–10063. 10.1128/JVI.00985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tena Fernandez F, González I, Doblas P, Rodríguez C, Sahana N, Kaur H, Tenllado F, Praveen S, Canto T. 2013. The influence of cis-acting P1 protein and translational elements on the expression of potato virus Y helper-component proteinase (HCPro) in heterologous systems and its suppression of silencing activity. Mol Plant Pathol 14:530–541. 10.1111/mpp.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salvador B, Saenz P, Yangüez E, Quiot JB, Quiot L, Delgadillo MO, García JA, Simón-Mateo C. 2008. Host-specific effect of P1 exchange between two potyviruses. Mol Plant Pathol 9:147–155. 10.1111/j.1364-3703.2007.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maliogka VI, Salvador B, Carbonell A, Saenz P, León DS, Oliveros JC, Delgadillo MO, García JA, Simón-Mateo C. 2012. Virus variants with differences in the P1 protein coexist in a plum pox virus population and display particular host-dependent pathogenicity features. Mol Plant Pathol 13:877–886. 10.1111/j.1364-3703.2012.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carbonell A, Dujovny G, García JA, Valli A. 2012. The cucumber vein yellowing virus silencing suppressor P1b can functionally replace HCPro in plum pox virus infection in a host-specific manner. Mol Plant Microbe Interact 25:151–164. 10.1094/MPMI-08-11-0216. [DOI] [PubMed] [Google Scholar]
- 41.Shan H, Pasin F, Tzanetakis IE, Simón-Mateo C, García JA, Rodamilans B. 2018. Truncation of a P1 leader proteinase facilitates potyvirus replication in a non-permissive host. Mol Plant Pathol 19:1504–1510. 10.1111/mpp.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohožková J, Navrátil M. 2011. P1 peptidase–a mysterious protein of family Potyviridae. J Biosci 36:189–200. 10.1007/s12038-011-9020-6. [DOI] [PubMed] [Google Scholar]
- 43.Martínez F, Daròs JA. 2014. Tobacco etch virus protein P1 traffics to the nucleolus and associates with the host 60S ribosomal subunits during infection. J Virol 88:10725–10737. 10.1128/JVI.00928-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. 2018. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res 46:D708–D717. 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ha C, Coombs S, Revill PA, Harding RM, Vu M, Dale JL. 2008. Design and application of two novel degenerate primer pairs for the detection and complete genomic characterization of potyviruses. Arch Virol 153:25–36. 10.1007/s00705-007-1053-7. [DOI] [PubMed] [Google Scholar]
- 46.Noveriza R, Suastika G, Hidayat SH, Kartosuwondo U. 2012. Potyvirus associated with mosaic disease on patchouli (Pogostemon cablin (Blanco) Benth.) plants in Indonesia. J Int Soc Southeast Asian Agric Sci 18:131–146. [Google Scholar]
- 47.Chiemsombat P, Prammanee S, Pipattanawong N. 2014. Occurrence of telosma mosaic virus causing passion fruit severe mosaic disease in Thailand and immunostrip test for rapid virus detection. Crop Prot 63:41–47. 10.1016/j.cropro.2014.04.023. [DOI] [Google Scholar]
- 48.Yao LZ, Li XQ, Wang JG, Chen SY, Wang XJ. 2019. First report of telosma mosaic virus infecting emperor’s candlesticks (Senna alata) in China. Plant Dis 103:594–594. 10.1094/PDIS-04-18-0706-PDN. [DOI] [Google Scholar]
- 49.Xie L, Zhang X, Zheng S, Zhang L, Li T. 2017. Molecular identification and specific detection of telosma mosaic virus infecting passion fruit. Sci Agric Sin 50:4725–4734. [Google Scholar]
- 50.Chen S, Yu N, Yang S, Zhong B, Lan H. 2018. Identification of telosma mosaic virus infection in Passiflora edulis and its impact on phytochemical contents. Virol J 15:168. 10.1186/s12985-018-1084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu C, Lian Q, Lin H, Chen L, Lu Y, Zhai Y, Han X, Du Z, Gao F, Wu Z. 2021. A clade of telosma mosaic virus from Thailand is undergoing geographical expansion and genetic differentiation in passionfruit of Vietnam and China. Phytopathol Res 3:24. 10.1186/s42483-021-00101-1. [DOI] [Google Scholar]
- 52.Yang K, Yan H, Song L, Jin P, Miao W, Cui H. 2018. Analysis of the complete genome sequence of a potyvirus from passion fruit suggests its taxonomic classification as a member of a new species. Arch Virol 163:2583–2586. 10.1007/s00705-018-3885-8. [DOI] [PubMed] [Google Scholar]
- 53.Edgar RC, Taylor J, Lin V, Altman T, Barbera P, Meleshko D, Lohr D, Novakovsky G, Buchfink B, Al-Shayeb B, Banfield JF, de la Peña M, Korobeynikov A, Chikhi R, Babaian A. 2022. Petabase-scale sequence alignment catalyses viral discovery. Nature 602:142–147. 10.1038/s41586-021-04332-2. [DOI] [PubMed] [Google Scholar]
- 54.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29:644–652. 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan W, Ye Z, Cao S, Yao G, Yu J, Yang D, Chen P, Zhang J, Wu Y. 2021. Transcriptome analysis of two Pogostemon cablin chemotypes reveals genes related to patchouli alcohol biosynthesis. Peer J 9:e12025. 10.7717/peerj.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng P, Wu Z, Wang A. 2015. The multifunctional protein CI of potyviruses plays interlinked and distinct roles in viral genome replication and intercellular movement. Virol J 12:141. 10.1186/s12985-015-0369-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui H, Wang A. 2016. Plum pox virus 6K1 protein is required for viral replication and targets the viral replication complex at the early stage of infection. J Virol 90:5119–5131. 10.1128/JVI.00024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desbiez C, Lecoq H. 2004. The nucleotide sequence of watermelon mosaic virus (WMV, Potyvirus) reveals interspecific recombination between two related potyviruses in the 5′ part of the genome. Arch Virol 149:1619–1632. 10.1007/s00705-004-0340-9. [DOI] [PubMed] [Google Scholar]
- 59.Yang Y, Gong J, Li H, Li C, Wang D, Li K, Zhi H. 2011. Identification of a novel soybean mosaic virus isolate in China that contains a unique 5′ terminus sharing high sequence homology with bean common mosaic virus. Virus Res 157:13–18. 10.1016/j.virusres.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 60.Desbiez C, Wipf-Scheibel C, Millot P, Verdin E, Dafalla G, Lecoq H. 2017. New species in the papaya ringspot virus cluster: insights into the evolution of the PRSV lineage. Virus Res 241:88–94. 10.1016/j.virusres.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 61.Chen J, Zheng HY, Lin L, Adams MJ, Antoniw JF, Zhao MF, Shang YF, Chen JP. 2004. A virus related to soybean mosaic virus from Pinellia ternata in China and its comparison with local soybean SMV isolates. Arch Virol 149:349–363. 10.1007/s00705-003-0184-8. [DOI] [PubMed] [Google Scholar]
- 62.Yang Y, Lin J, Zheng G, Zhang M, Zhi H. 2014. Recombinant soybean mosaic virus is prevalent in Chinese soybean fields. Arch Virol 159:1793–1796. 10.1007/s00705-014-1980-z. [DOI] [PubMed] [Google Scholar]
- 63.Chen YX, Wu M, Ma FF, Chen JQ, Wang B. 2017. Complete nucleotide sequences of seven soybean mosaic viruses (SMV), isolated from wild soybeans (Glycine soja) in China. Arch Virol 162:901–904. 10.1007/s00705-016-3163-6. [DOI] [PubMed] [Google Scholar]
- 64.Mao C, Shan S, Huang Y, Jiang C, Zhang H, Li Y, Chen J, Wei Z, Sun Z. 2022. The hypervariable N-terminal of soybean mosaic virus P1 protein influences its pathogenicity and host defense responses. Phytopathol Res 4:10. 10.1186/s42483-022-00115-3. [DOI] [Google Scholar]
- 65.Ali A, Natsuaki T, Okuda S. 2006. The complete nucleotide sequence of a Pakistani isolate of watermelon mosaic virus provides further insights into the taxonomic status in the bean common mosaic virus subgroup. Virus Genes 32:307–311. 10.1007/s11262-005-6915-z. [DOI] [PubMed] [Google Scholar]
- 66.Noris E, Jupin I, Accotto GP, Gronenborn B. 1996. DNA-binding activity of the C2 protein of tomato yellow leaf curl geminivirus. Virology 217:607–612. 10.1006/viro.1996.0157. [DOI] [PubMed] [Google Scholar]
- 67.Hartitz MD, Sunter G, Bisaro DM. 1999. The tomato golden mosaic virus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 263:1–14. 10.1006/viro.1999.9925. [DOI] [PubMed] [Google Scholar]
- 68.Koonin EV, Boyko VP, Dolja VV. 1991. Small cysteine-rich proteins of different groups of plant RNA viruses are related to different families of nucleic acid-binding proteins. Virology 181:395–398. 10.1016/0042-6822(91)90512-a. [DOI] [PubMed] [Google Scholar]
- 69.Robaglia C, Durand-Tardif M, Tronchet M, Boudazin G, Astier-Manifacier S, Casse-Delbart F. 1989. Nucleotide sequence of potato virus Y (N strain) genomic RNA. J Gen Virol 70:935–947. 10.1099/0022-1317-70-4-935. [DOI] [PubMed] [Google Scholar]
- 70.Lukhovitskaya NI, Ignatovich IV, Savenkov EI, Schiemann J, Morozov SY, Solovyev AG. 2009. Role of the zinc finger and basic motifs of chrysanthemum virus B p12 protein in nucleic acid binding, protein localization and induction of a hypersensitive response upon expression from a viral vector. J Gen Virol 90:723–733. 10.1099/vir.0.005025-0. [DOI] [PubMed] [Google Scholar]
- 71.Lukhovitskaya NI, Solovieva AD, Boddeti SK, Thaduri S, Solovyev AG, Savenkov EI. 2013. An RNA virus-encoded zinc finger protein acts as a plant transcription factor and induces a regulator of cell size and proliferation in two tobacco species. Plant Cell 25:960–973. 10.1105/tpc.112.106476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Voinnet O, Pinto YM, Baulcombe DC. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA 96:14147–14152. 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Wezel R, Dong X, Liu H, Tien P, Stanley J, Hong Y. 2002. Mutation of three cysteine residues in tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene-silencing suppression. Mol Plant Microbe Interact 15:203–208. 10.1094/MPMI.2002.15.3.203. [DOI] [PubMed] [Google Scholar]
- 74.Xu XJ, Zhu Q, Jiang SY, Yan ZY, Geng C, Tian YP, Li XD. 2021. Development and evaluation of stable sugarcane mosaic virus mild mutants for cross-protection against infection by severe strain. Front Plant Sci 12:788963. 10.3389/fpls.2021.788963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodamilans B, Casillas A, García JA. 2021. P1 of sweet potato feathery mottle virus shows strong adaptation capacity, replacing P1-HCPro in a chimeric plum pox virus. J Virol 95:e00150-21. 10.1128/JVI.00150-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pasin F, Shan H, García B, Müller M, San León D, Ludman M, Fresno DH, Fátyol K, Munné-Bosch S, Rodrigo G, García JA. 2020. Abscisic acid connects phytohormone signaling with RNA metabolic pathways and promotes an antiviral response that is evaded by a self-controlled RNA virus. Plant Commun 1:100099. 10.1016/j.xplc.2020.100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakahara KS, Masuta C, Yamada S, Shimura H, Kashihara Y, Wada TS, Meguro A, Goto K, Tadamura K, Sueda K, Sekiguchi T, Shao J, Itchoda N, Matsumura T, Igarashi M, Ito K, Carthew RW, Uyeda I. 2012. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc Natl Acad Sci USA 109:10113–10118. 10.1073/pnas.1201628109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hafrén A, Üstün S, Hochmuth A, Svenning S, Johansen T, Hofius D. 2018. Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol 176:649–662. 10.1104/pp.17.01198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu XJ, Geng C, Jiang SY, Zhu Q, Yan ZY, Tian YP, Li XD. 2022. A maize triacylglycerol lipase inhibits sugarcane mosaic virus infection. Plant Physiol 189:754–771. 10.1093/plphys/kiac126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ji M, Zhao J, Han K, Cui W, Wu X, Chen B, Lu Y, Peng J, Zheng H, Rao S, Wu G, Chen J, Yan F. 2021. Turnip mosaic virus P1 suppresses JA biosynthesis by degrading cpSRP54 that delivers AOCs onto the thylakoid membrane to facilitate viral infection. PLoS Pathog 17:e1010108. 10.1371/journal.ppat.1010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Domingo-Calap ML, Chase O, Estapé M, Moreno AB, López-Moya JJ. 2021. The P1 protein of watermelon mosaic virus compromises the activity as RNA silencing suppressor of the P25 protein of cucurbit yellow stunting disorder virus. Front Microbiol 12:645530. 10.3389/fmicb.2021.645530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. 1999. A mini binary vector series for plant transformation. Plant Mol Biol 40:711–717. 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- 83.Joensuu JJ, Conley AJ, Lienemann M, Brandle JE, Linder MB, Menassa R. 2010. Hydrophobin fusions for high-level transient protein expression and purification in Nicotiana benthamiana. Plant Physiol 152:622–633. 10.1104/pp.109.149021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cui H, Wang A. 2017. An efficient viral vector for functional genomic studies of Prunus fruit trees and its induced resistance to plum pox virus via silencing of a host factor gene. Plant Biotechnol J 15:344–356. 10.1111/pbi.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y, Shen W, Dai Z, Gou B, Liu H, Hu W, Qin L, Li Z, Tuo D, Cui H. 2021. Biological and molecular characterization of two closely-related arepaviruses and their antagonistic interaction in Nicotiana benthamiana. Front Microbiol 12:755156. 10.3389/fmicb.2021.755156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang K, Shen W, Li Y, Li Z, Miao W, Wang A, Cui H. 2019. Areca palm necrotic ringspot virus, classified within a recently proposed genus Arepavirus of the family Potyviridae, is associated with necrotic ringspot disease in areca palm. Phytopathology 109:887–894. 10.1094/PHYTO-06-18-0200-R. [DOI] [PubMed] [Google Scholar]
- 87.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li F, Huang C, Li Z, Zhou X. 2014. Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog 10:e1003921. 10.1371/journal.ppat.1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li F, Zhao N, Li Z, Xu X, Wang Y, Yang X, Liu S, Wang A, Zhou X. 2017. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog 13:e1006213. 10.1371/journal.ppat.1006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qin L, Shen W, Tang Z, Hu W, Shangguan L, Wang Y, Tuo D, Li Z, Miao W, Valli AA, Wang A, Cui H. 2020. A newly identified virus in the family Potyviridae encodes two leader cysteine proteases in tandem that evolved contrasting RNA silencing suppression functions. J Virol 95:e01414-20. 10.1128/JVI.01414-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3, Fig. S1 to S5, and Data S1. Download jvi.01444-22-s0001.pdf, PDF file, 1.5 MB (1.5MB, pdf)