ABSTRACT
African swine fever (ASF) is a highly contagious infectious disease of domestic pigs and wild boars caused by African swine fever virus (ASFV), with a mortality rate of up to 100%. In order to replicate efficiently in macrophages and monocytes, ASFV has evolved multiple strategies to evade host antiviral responses. However, the underlying molecular mechanisms by which ASFV-encoded proteins execute immune evasion are not fully understood. In this study, we found that ASFV pH240R strongly inhibits transcription, maturation, and secretion of interleukin-1β (IL-1β). Importantly, pH240R not only targeted NF-κB signaling but also impaired NLRP3 inflammasome activation. In this mechanism, pH240R interacted with NF-kappa-B essential modulator (NEMO), a component of inhibitor of kappa B kinase (IKK) complex and subsequently reduced phosphorylation of IκBα and p65. In addition, pH240R bonded to NLRP3 to inhibit NLRP3 inflammasome activation, resulting in reduced IL-1β production. As expected, infection with H240R-deficient ASFV (ASFV-ΔH240R) induced more inflammatory cytokine expression both in vitro and in vivo than its parental ASFV HLJ/18 strain. Consistently, H240R deficiency reduced the viral pathogenicity in pigs compared with its parental strain. These findings reveal that the H240R gene is an essential virulence factor, and deletion of the H240R gene affects the pathogenicity of ASFV HLJ/18 by enhancing antiviral inflammatory responses, which provides insights for ASFV immune evasion mechanisms and development of attenuated live vaccines and drugs for prevention and control of ASF.
IMPORTANCE African swine fever (ASF), caused by African swine fever virus (ASFV), is a highly contagious and acute hemorrhagic viral disease of domestic pigs, with a high mortality approaching 100%. ASFV has spread rapidly worldwide and caused huge economic losses and ecological consequences. However, the pathogenesis and immune evasion mechanisms of ASFV are not fully understood, which limits the development of safe and effective ASF attenuated live vaccines. Therefore, investigations are urgently needed to identify virulence factors that are responsible for escaping the host antiviral innate immune responses and provide a new target for development of ASFV live-attenuated vaccine. In this study, we determined that the H240R gene is an essential virulence factor, and its depletion affects the pathogenicity of ASFV by enhancing NLRP3-mediated inflammatory responses, which provides theoretical support for the development of an ASFV attenuated live vaccine.
KEYWORDS: ASFV, H240R gene, NF-κB signaling, NLRP3 inflammasome, pathogenicity
INTRODUCTION
African swine fever (ASF), caused by African swine fever virus (ASFV), is an acute and lethal hemorrhagic infectious disease of domestic and wild boars, which can cause hemorrhagic necrosis of multiple tissues in infected pigs, with a mortality rate approaching 100% (1). ASF, first described in Kenya in 1921 and then spread to Europe, South American, and Southeast Asia, has caused huge economic losses to the global swine industry (2). The first ASF case was reported in 2018 in China. To date, more than 190 outbreaks have been announced, and several ASFV isolates have been isolated to assess their pathogenicity (3, 4). Unfortunately, since there are currently no commercial vaccines or clinical anti-ASFV drugs for the disease, ASF continues to spread in China, with devastating social-economic consequences for the pig industry and domestic food security (5).
ASFV is a member of the Asfarviridae family, belonging to the nucleoplasmic large DNA virus (NCLDV) group, and it is the only known DNA arbovirus (6). ASFV is a large, enveloped, double-stranded DNA (dsDNA) virus containing a linear DNA genome of 170 to 190 kb with more than 150 open reading frames (ORFs) (7). More than 150 viral proteins are expressed in ASFV-infected macrophages or other cell lines (8), which not only execute functions involved in virus entry, replication, virion assembly, and egress (9), but also regulate host antiviral innate immune responses (10).
The inflammatory responses caused by viral infection play an important role in host antiviral defense, which is beneficial for the host to kill the infected cells and clear invading viruses. The activation of inflammatory responses requires two separate signals: signal 1 is involved in detecting viral nucleic acid or protein through their Toll like receptors (TLRs) to activate inhibitor of kappa B kinase (IKK)-mediated nuclear factor kappa B (NF-κB) signaling, resulting in increased transcription of NOD-like receptor thermal protein domain associated protein 3 (NLRP3), and pro-interleukin-1β (IL-1β), and signal 2 is associated with activation of several inflammasomes (such as NLRP3, NLR family CARD domain-containing protein 4 (NLRC4) and absent in melanoma 2 (AIM2)) to activate pro-caspase-1 (11). Active caspase-1 cleaves pro-IL-1β, pro-IL-18, and gasdermin D (GSDMD), resulting in maturation of IL-1β, IL-18, and pyroptosis, which leads to IL-1β and IL-18 secretion (12).
Previous studies have reported that pigs infected with a virulent ASFV strain promote the release of various inflammatory cytokines, such as IL-1α, IL-1β, IL-6, and tumor necrosis factor-α (TNF-α), which induces an excessive “cytokine storm” (13). Song et al. found that ASFV-encoded I226L, A151R, NP419L, and QP383R proteins can suppress NLRP3-mediated inflammatory responses (14). Our previous studies also showed that pMGF505-7R interacts with the IKK complex to inhibit pro-IL-1β transcription and binds to NLRP3 to inhibit the assembly of the NLRP3 inflammasome, thereby inhibiting ASFV-mediated inflammatory responses (15). Borca et al. noticed through a yeast two-hybrid assay that the ASFV pL83L interacts with IL-1β, suggesting that pL83L may play an important role in regulating IL-1β function (16).
As a capsid protein, ASFV pH240R can be packaged into ASFV virion, where it interacts with the major capsid protein p72 (17, 18). The infectious progeny virus titers of a recombinant ASFV with deletion of the H240R gene (ASFV-ΔH240R) were reduced by approximately 2.0 log units compared with its parental ASFV HLJ/18. Notably, pH240R does affect ASFV assembly, which leads to generation of noninfectious particles in primary porcine alveolar macrophages (PAMs). However, ASFV-ΔH240R infection still induces higher-level expression of inflammatory cytokines in PAMs than its parental ASFV HLJ/18 (18). The underlying mechanisms by which ASFV pH240R regulates inflammatory responses are still unknown.
In this study, we report that pH240R strongly inhibits the transcription of IL-1β by targeting the IKK complex by interacting with IKK-gamma (NEMO). ASFV pH240R also inhibits the maturation and secretion of IL-1β by inhibiting NLRP3 inflammasome formation by interacting with NLRP3. Furthermore, H240R gene deficiency attenuated ASFV pathogenicity in pigs. These findings suggest that H240R is one of the key virulence-related genes of ASFV and that deletion of the H240R gene may serve as a strategy to develop attenuated live vaccines against ASFV.
RESULTS
ASFV pH240R inhibits NF-κB activation although it interacts with IKK complex.
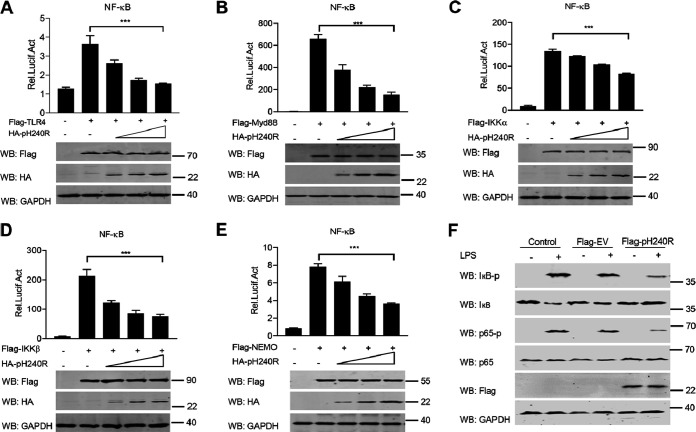
Previous studies reported that ASFV-ΔH240R infection induced higher levels of inflammatory cytokines in PAMs than its parental virus (18), which suggests that ASFV pH240R may inhibit the secretion of inflammatory cytokines. The production of inflammatory cytokines requires activation of two separate signals: NF-κB signaling and assembly of multimolecular inflammasomes (11). To test the effect of pH240R on inflammatory cytokine production, we first examined the effect of pH240R on NF-κB activity. Previous studies demonstrated that TLR4, myeloid differentiation primary response 88 (MyD88), and the components of IκB kinase (IKK) complex (IKKα, IKKβ, and NEMO) play important roles in promoting NF-κB signaling activation (19). Therefore, we first tested the effect of pH240R on the NF-κB signaling triggered by these molecules. The results showed that ectopically expressed pH240R strongly inhibited TLR4-, MyD88-, IKKα-, IKKβ-, and NEMO-induced NF-κB promoter activation in a dose-dependent manner (Fig. 1A to E), indicating that pH240R negatively regulates NF-κB reporter activation. Consistent with this result, we found that overexpression of pH240R significantly inhibited the degradation of IκB and the phosphorylation of IκB and p65 (Fig. 1F).
FIG 1.
ASFV pH240R negatively regulates NF-κB signaling activation. (A to E) Luciferase activity of NF-κB promoter reporter in HEK293T cells overexpressed with TLR4 (A), MyD88 (B), IKKα (C), IKKβ (D), or NEMO (E). HEK293T cells were transfected with an NF-κB reporter, a thymidine kinase (TK) reporter, and increasing amounts of a plasmid (0, 100, 200, 400 ng) encoding pH240R, along with a plasmid encoding Flag-TLR4 (A), MyD88 (B), IKKα (C), IKKβ (D), or NEMO (E). At 24 h posttransfection (hpt), the luciferase activities were measured with a dual-luciferase reporter assay system. The data were normalized to the transfection efficiency by dividing the firefly luciferase activity by the Renilla luciferase activity. The expressions of Flag-tagged TLR4, MyD88, IKKα, IKKβ, and NEMO, HA-tagged pH240R, and GAPDH were detected by Western blotting. (F) Immunoblot analysis of the phosphorylation of IκB and p65 in HEK293 T cells upon pH240R overexpression. HEK293T cells were mock-transfected or transfected with an empty vector or a plasmid expressing pH240R and then mock-treated or treated with LPS for 6 h. The expressions of IκB, p65, pH240R, GAPDH, and phosphorylated IκB and p65 were detected by Western blotting. All assays were independently repeated at least three times. The data are shown as the mean ± SD; n = 3. ***, P < 0.001.
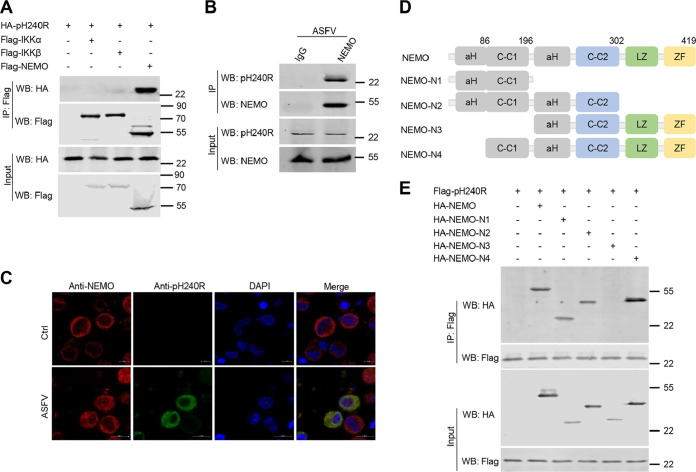
To test whether pH240R targets the IKK complex, we further explored the interaction between pH240R and the components of the IKK complex. Coimmunoprecipitation (co-IP) assays were performed between overexpressed pH240R and IKKα, IKKβ, or NEMO in HEK293T cells. As shown in Fig. 2A, pH240R strongly coprecipitated with NEMO, but not with IKKα and IKKβ. Consistent with this result, pH240R also coprecipitated with endogenous NEMO in PAMs and colocalized with endogenous NEMO upon ASFV infection (Fig. 2B and C). These results indicate that pH240R interacts with the IKK complex component NEMO. NEMO is composed of four functional domains: two coil-coiled domains (CC1 and CC2), a leucine zipper region (LZ), and a zinc finger domain (ZF). To map the domain of NEMO which is required for its interaction with pH240R, four deletion mutants of NEMO (1 to 196 amino acids [aa], 1 to 302 aa, 197 to 419 aa, and 86 to 419 aa) were generated (20), and their interactions with pH240R were evaluated (Fig. 2D). The results showed that pH240R interacts with the CC1 domain (86 to 196 aa) of NEMO (Fig. 2E), which is required for IKKβ binding.
FIG 2.
ASFV pH240R interacts with NEMO. (A) Co-IP analysis of the interaction between overexpressed HA-pH240R and Flag-IKKα, -IKKβ, or -NEMO in HEK293T cells. HEK293T cells were transfected with a plasmid encoding HA-pH240R, along with a plasmid encoding Flag-IKKα, -IKKβ, or -NEMO. At 24 hpt, the cells were collected to perform co-IP with anti-Flag beads. The immunoprecipitants and the whole-cell lysates were detected by Western blotting with the indicated antibodies. (B) Co-IP analysis of the interaction of pH240R and endogenous NEMO in PAMs upon ASFV infection. PAMs were infected with ASFV (multiplicity of infection [MOI], 1) for 24 h, and then the cells were collected to perform co-IP with IgG or anti-NEMO antibody. The immunoprecipitants and the whole-cell lysates were detected by Western blotting with the indicated antibodies. (C) The subcellular colocalization of pH240R and endogenous NEMO upon ASFV infection in PAMs. PAMs were mock-infected or infected with ASFV (MOI, 1); the location of pH240R and NEMO was detected by confocal microscopy at 24 hpi. Scale bars = 5 μm. (D) Schematic of full-length NEMO and its truncated mutants. (E) Co-IP analysis of the interaction between Flag-pH240R and HA-tagged full-length NEMO or its deleted mutants in HEK293T cells. HEK293T cells were transfected with a plasmid expressing Flag-pH240R alone or together with HA-NEMO or its deleted mutants. At 24 hpt, the cells were collected to perform the co-IP analysis with the indicated antibodies.
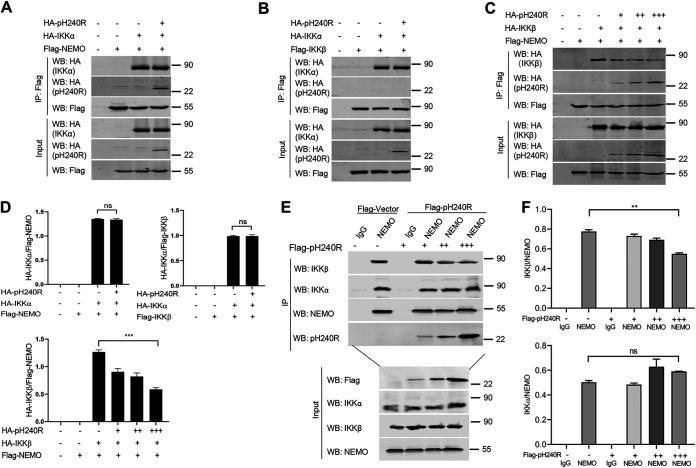
It is well known that NEMO interacts with IKKα and IKKβ to form the IKKα/IKKβ/NEMO complex and activate NF-κB signaling during lipopolysaccharide (LPS) stimulation (21). To explore whether pH240R inhibits the formation of the IKK complex, co-IP assays were performed to evaluate the effect of pH240R on IKKα-NEMO, IKKα-IKKβ, IKKβ-NEMO, and IKKα-IKKβ-NEMO interactions. The results showed that overexpressed pH240R had no obvious effect on IKKα-NEMO and IKKα-IKKβ interactions (Fig. 3A, B, and D). However, pH240R obviously disrupted the IKKβ-NEMO interaction in a dose-dependent manner (Fig. 3C to F).
FIG 3.
ASFV pH240R disrupts the interaction between IKKβ and NEMO. (A) The effect of pH240R on the interaction of HA-IKKα and Flag-NEMO. HEK293T cells were transfected with a plasmid encoding Flag-NEMO alone or together with a plasmid expressing HA-IKKα or a plasmid encoding HA-pH240R as indicated. At 24 hpt, co-IP was performed to test the interaction between IKKα and NEMO with anti-Flag beads. The immunoprecipitants and the whole-cell lysates were detected by Western blotting with the indicated antibodies. (B) The effect of pH240R on the interaction of HA-IKKα and Flag-IKKβ. HEK293T cells were transfected with a plasmid encoding Flag-IKKβ alone or together with a plasmid expressing HA-IKKα or a plasmid encoding HA-pH240R as indicated. At 24 hpt, co-IP was performed to test the interaction between IKKβ and IKKα with anti-Flag beads. The immunoprecipitants and the whole-cell lysates were detected by Western blotting with the indicated antibodies. (C) The effect of pH240R on the interaction of HA-IKKβ and Flag-NEMO. HEK293T cells were transfected with a plasmid encoding Flag-NEMO alone or together with a plasmid expressing HA-IKKβ or increasing amounts of a plasmid encoding HA-pH240R as indicated. At 24 hpt, co-IP was performed to test the interaction between IKKβ and NEMO with anti-Flag beads. The immunoprecipitants and the whole-cell lysates were detected by Western blotting with the indicated antibodies. (D) The relative band intensity analysis of the HA-IKKα/Flag-NEMO, HA-IKKα/Flag-IKKβ, and HA-IKKβ/Flag-NEMO immunoblotting result from panels A to C created using ImageJ. (E) The effect of pH240R on the interaction of endogenous NEMO, IKKα, and IKKβ. CRL-2843 cells were transfected with an empty vector or increasing amounts of a plasmid expressing Flag-pH240R for 24 h and then mock-treated or treated with LPS for another 12 h. The cells were then collected to perform co-IP with IgG or anti-NEMO antibody. The immunoprecipitants and the whole-cell lysates were detected by Western blotting with the indicated antibodies. (F) The relative band intensity analysis of the IKKβ/NEMO and IKKα/NEMO immunoblotting result from panel E produced using ImageJ. All assays were independently repeated at least three times. The data presented are the mean ± SD. ns, not significant; **, P < 0.01, ***, P < 0.001.
Taken together, these results suggest that ASFV pH240R targets the IKK complex component NEMO to suppress the inflammatory response.
ASFV pH240R targets NLRP3 to inhibit IL-1β maturation.
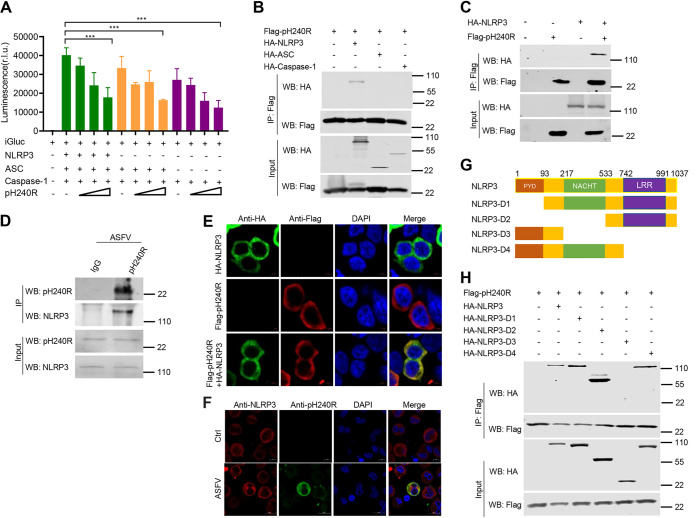
Our previous studies demonstrated that ASFV infection induces IL-1β production through activation of NLRP3 inflammasome, and ASFV pMGF505-7R negatively regulates IL-1β production by inhibiting NLRP3 inflammasome activity (15). Based on our previous research using iGluc reporter, we further investigated whether pH240R affects NLRP3 inflammasome activation. HEK293T cells were cotransfected with plasmids expressing the components of the NLRP3 inflammasome, including NLRP3, apoptosis-associated speck-like protein containing a CARD (ASC), and caspase-1, a plasmid expressing pH240R, along with an pro-interleukin (IL)-1β-gaussia luciferase (iGluc) reporter, and then the luciferase activities were measured. The results showed that pH240R significantly inhibited iGluc reporter activity triggered by the inflammasome activation (Fig. 4A), indicating that pH240R suppresses NLRP3 inflammasome activity. To test whether pH240R interacts with the components of the NLRP3 inflammasome, a co-IP assay was performed between pH240R and NLRP3, ASC, or caspase-1 in HEK293T cells. As shown in Fig. 4B and C, overexpressed pH240R coprecipitated with NLRP3, but not with ASC or caspase-1. Consistent with this result, pH240R also coprecipitated with endogenous NLRP3 in PAMs infected with ASFV (Fig. 4D). Furthermore, we found that both NLRP3 and pH240R were enriched in the cytoplasm of HEK293T cells and PAMs (Fig. 4E and F). These results indicate that pH240R interacts with NLRP3 in the cytoplasm of PAMs following ASFV infection. NLRP3 contains three functionally distinct domains: nucleotide-binding domain (NACHT), leucine-rich-repeat domain (LRR), and pyrin domain (PYD). To map the NLRP3 domain required for its interaction with pH240R, four NLRP3 deletion mutants were constructed, and the related co-IP assay was performed. The results showed that pH240R interacted with the NACHT and LRR domains of NLRP3, but not with the PYD domain (Fig. 4G and H).
FIG 4.
ASFV pH240R inhibits inflammasome activation by interacting with NLRP3. (A) iGLuc activity in HEK293T cells using the iGLuc-based NLRP3 inflammasome system. HEK293T cells were transfected with different doses of a plasmid expressing pH240R in the presence of the components of the iGLuc-based NLRP3 inflammasome system as indicated. At 24 hpt, the supernatants were collected to detect the luciferase activity. (B) Co-IP analysis of interaction between overexpressed pH240R and NLRP3, ASC, or caspase-1 in HEK293T cells. HEK293T cells were transfected with a plasmid encoding pH240R, along with a plasmid encoding HA-tagged NLRP3, ASC, or caspase-1. At 24 hpt, the cells were lysed and the lysates were coimmunoprecipitated with anti-Flag beads. The immunoprecipitants and cell lysates were detected by immunoblotting with the indicated antibodies. (C) Co-IP analysis of the interaction between overexpressed pH240R and NLRP3. HEK293T cells were mock-transfected or transfected with a plasmid expressing HA-NLRP3 or Flag-pH240R alone or both. At 24 hpt, the cells were collected and the co-IP was performed to test the interaction between pH240R and NLRP3. (D) Co-IP analysis of the interaction between pH240R and NLRP3 in PAMs infected with ASFV. PAMs were infected with ASFV (MOI, 1) for 24 h, and then the cells were collected to perform co-IP with anti-pH240R antibody. The immunoprecipitants and the whole-cell lysates were detected by Western blotting with anti-pH240R and anti-NLRP3 antibodies. (E) The subcellular localization of overexpressed pH240R and NLRP3 in HEK293T cells. HEK293T cells were transfected with a plasmid expressing HA-NLRP3 or Flag-pH240R alone or both. At 24 hpt, the cells were fixed and stained with the indicated antibodies for immunofluorescence analysis. Scale bars = 5 μm. (F) The subcellular localization of pH240R and NLRP3 in PAMs infected with ASFV. PAMs were mock infected or infected with ASFV (MOI, 1), and 24 h later, the cells were fixed and stained with the indicated antibodies for immunofluorescence analysis. Scale bars = 5 μm. (G) Schematic representation of the full-length NLRP3 and its truncated mutants. (H) Co-IP analysis of the interaction between HA-NLRP3 and Flag-pH240R or its deletion mutants in HEK293T cells. HEK293T cells were transfected with a plasmid expressing Flag-pH240R alone or together with HA-NLRP3 or its deletion mutants. At 24 hpt, the cells were collected to perform co-IP with anti-Flag beads. The immunoprecipitants and the whole-cell lysates were detected by Western blotting with the indicated antibodies. All assays were independently repeated at least three times. The data presented are the mean ± SD. ***, P < 0.001.
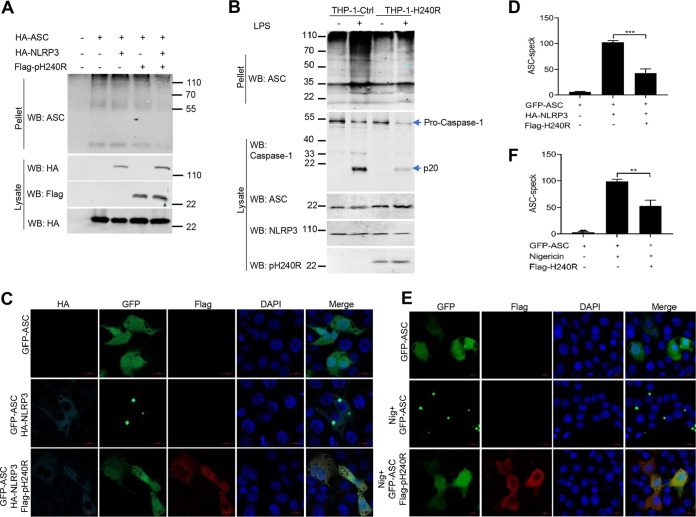
It is well known that formation of the NLRP3 inflammasome complex activates caspase-1, resulting in IL-1β maturation and secretion. Since we found that pH240R binds to NLRP3 and suppresses NLRP3 inflammasome activity, we further tested whether pH240R inhibits NLRP3 inflammasome assembly. Both HEK293T and THP-1 cells were overexpressed with different components of the NLRP3 inflammasome with or without pH240R coexpression, and co-IP was then performed to examine the effect of pH240R on the formation of the NLRP3 inflammasome complex. We found that in the absence of pH240R coexpression, both ASC and caspase-1 could be strongly pulled down by NLRP3, suggesting potent formation of the NLRP3-ASC-caspase-1 complex, while with pH240R coexpression, the amount of ASC and caspase-1 pulled down by NLRP3 were greatly decreased (Fig. 5A to C). In agreement with this result, pH240R inhibited the interaction of endogenous NLRP3 and ASC in a dose-dependent manner in CRL-2843 cells (Fig. 5D and E). In addition, we also noticed that overexpression of pH240R inhibited NLRP3-induced ASC oligomerization in HEK293T and THP-1 cells (Fig. 6A and B). Consistent with this, pH240R strongly suppressed GFP-ASC speck formation in response to NLRP3 overexpression (Fig. 6C and D) and nigericin stimulation (Fig. 6E and F), further indicating that pH240R inhibits NLRP3 inflammasome assembly.
FIG 5.
ASFV pH240R inhibits the formation of NLRP3 inflammasome. (A) The effect of pH240R on the formation of NLRP3 inflammasome in HEK293T cells. HEK293T cells were transfected with a plasmid encoding Flag-NLRP3 alone or together with a plasmid expressing HA-ASC, HA-caspase-1, and HA-pH240R as indicated. At 24 hpt, co-IP was performed with anti-Flag beads. (B) The effect of pH240R on the formation of NLRP3 inflammasome in THP-1 cells. THP-1 or THP-1-pH240R cells were lysed, and then co-IP was performed with IgG or anti-NLRP3 antibody. (C) The relative band intensity analysis of the ASC/NLRP3 and caspase-1/NLRP3 immunoblotting result from panel B using ImageJ. (D) The effect of pH240R on the interaction of endogenous NLRP3 and ASC. CRL-2843 cells were transfected with empty vector or increasing amounts of a plasmid expressing Flag-pH240R and then treated with nigericin (5 μM). Then, 4 h later, the cells were collected to perform co-IP with IgG or anti-NLRP3 antibody. The immunoprecipitants and the whole-cell lysates were detected by Western blotting with the indicated antibodies. (E) The relative band intensity analysis of the ASC/NLRP3 immunoblotting result from panel D using ImageJ. All assays were independently repeated at least three times. The data presented are the mean ± SD. *, P < 0.05; ***, P < 0.001.
FIG 6.
ASFV pH240R inhibits the oligomerization of ASC. (A) Effect of pH240R on the oligomerization of ASC in HEK293T cells. HEK293T cells were transfected with a plasmid encoding HA-ASC alone or together with a plasmid expressing HA-NLRP3 and a plasmid encoding Flag-pH240R as indicated. The cell lysates were prepared to detect the expression of ASC, NLRP3, and pH240R by Western blotting, and the pellets were washed with PBS three times and cross-linked using fresh dextran sulfate sodium (DSS) for detecting the oligomerization of ASC by Western blotting. (B) Effect of pH240R on the oligomerization of ASC in THP-1 cells treated with or without LPS. THP-1 cells and THP-1-pH240R cells were mock-treated or treated with LPS for 6 h, and then the oligomerization of ASC, the cleavage of caspace-1, and the expression of ASC, NLRP3, and pH240R were detected by Western blotting. (C) Effect of pH240R on the ASC specks in CRL-2843 cells with NLRP3 coexpression. CRL-2843 cells were transfected with a plasmid expressing GFP-ASC alone or together with plasmids expressing HA-NLRP3 and Flag-pH240R for 24 h. The cells were then fixed and probed with mouse anti-Flag MAb, rabbit anti-HA MAb, and DAPI (4′,6-diamidino-2-phenylindole) and then observed by confocal microscopy. (D) The number of ASC specks was quantified in three independent visual fields. (E) Effect of pH240R on the ASC specks in CRL-2843 cells with nigericin stimulation. CRL-2843 cells were transfected with a plasmid encoding GFP-ASC alone or together with a plasmid encoding Flag-pH240R. At 24 hpt, cells were stimulated with nigericin (5 μM) for another 4 h. The cells were then fixed and probed with rabbit anti-Flag MAb and DAPI and observed by confocal microscopy. (H) The number of ASC specks was quantified in three independent visual fields. All assays were independently repeated at least three times. The data are shown as the mean ± SD; **, P < 0.01; ***, P < 0.001.
Taken together, these findings suggest that pH240R interacts with NLRP3 to inhibit NLRP3 inflammasome assembly and activation, resulting in reduced IL-1β production.
Deletion of the H240R gene enhances secretion of inflammatory cytokines.
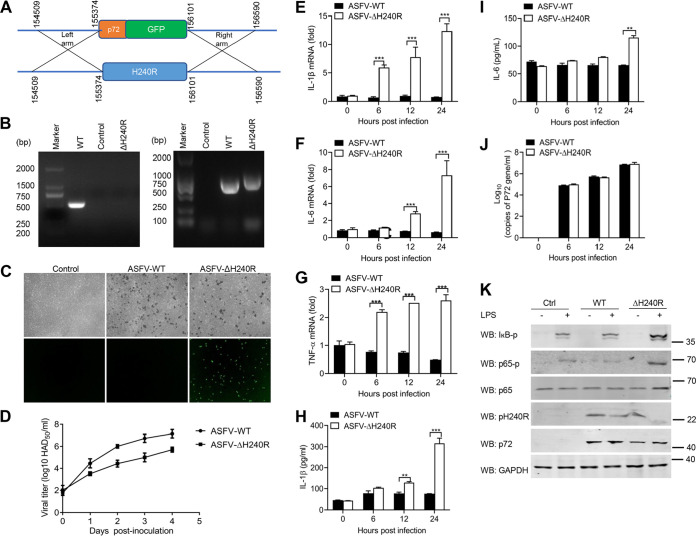
To determine the role of the H240R gene during ASFV infection, a recombinant ASFV lacking the H240R gene (ASFV-ΔH240R) was generated from the highly pathogenic ASFV HLJ/18 strain by homologous recombination. The H240R gene was replaced by a cassette containing the fluorescent gene EGFP under the control of the ASFV B646L gene promoter (named p72) (Fig. 7A). The recombinant ASFV-ΔH240R was obtained after 10 rounds of plaque purification in PAMs based on green fluorescent protein (GFP) expression. To evaluate the accuracy of the genetic modification, the genomic DNA was isolated for PCR amplification of the modified fragment (Fig. 7B) and sequencing, and the results showed that only the H240R gene was successfully replaced by p72-EGFP cassette, and the other DNA sequences in the genome were not affected. GFP expressed successfully under the control of the B646L gene promoter, and deletion of the H240R gene maintains the hemadsorption characteristics of ASFV (Fig. 7C). The growth characteristics of ASFV-ΔH240R in vitro were evaluated in PAMs. As shown in Fig. 7D, the 50% hemadsorption dose (HAD50) of ASFV-ΔH240R is significantly reduced by approximately 2.0 log units compared with that of its parental ASFV HLJ/18 at 3 days postinoculation (dpi).
FIG 7.
ASFV-ΔH240R induces higher levels of mRNA transcription and secretion of inflammatory cytokines. (A) Schematic representation of the experimental design for generation of ASFV-ΔH240R. The H240R gene segment was replaced with the p72-EGFP reporter gene cassette. (B) Verification of the H240R gene deletion in ASFV-ΔH240R. The H240R gene was examined using PCR in both ASFV-WT (lane 1) and ASFV-ΔH240R (lane 3) using primers H240R-F/R, and the p72-EGFP gene was examined using PCR in both ASFV-WT (lane 5) and ASFV-ΔH240R (lane 6) using primers 72EGFP-F/R. Agarose gel (1%) showing the result of the PCR to amplify of the genomic segment containing the targeted gene. (C) Hemadsorption characteristics of ASFV-ΔH240R. PAMs were infected with ASFV-WT or ASFV-ΔH240R. At 24 hpi, the cells were observed by microscope. (D) Growth kinetics of ASFV-ΔH240R and ASFV-WT in PAMs. PAMs were infected with ASFV-ΔH240R or ASFV-WT (MOI, 0.01), and samples were taken from three independent experiments at the indicated time points and titrated. (E to I) Analysis of inflammatory cytokine expression and secretion in PAMs infected with ASFV-WT or ASFV-ΔH240R; PAMs were infected with ASFV or ASFV-ΔH240R at the same number of genome copies (108 for 0, 6, 12, and 24 h. The mRNA levels of Il-1β) (E), Il-6 (F), and Tnf-α (G) in PAMs were detected by qPCR, and the secreted IL-1β (H) and IL-6 (I) in cell supernatants were detected by ELISA. (J) The genomic copy numbers of ASFV were detected by qPCR. (K) Immunoblot analysis of the degradation and phosphorylation of IκB and p65 in PAMs upon ASFV-WT or ASFV-ΔH240R infection. PAMs were mock-infected or infected with ASFV-WT or ASFV-ΔH240R and then mock-treated or treated with LPS for 6 h. The expression of IκB, p65, pH240R, and GAPDH and the phosphorylation of IκB and p65 were detected by Western blotting. All assays were independently repeated at least three times. The data are shown as the mean ± SD; n = 3. **, P < 0.01; ***, P < 0.001.
To detect whether deletion of the H240R gene affects ASFV-induced inflammatory cytokine production, PAMs were infected with same amount of ASFV HLJ/18 or ASFV-ΔH240R (108 genome copies) for 0, 6, 12, and 24 h, and then the levels of expression and secretion of certain inflammatory cytokines were analyzed. Consistent with previous results, we found that ASFV-ΔH240R infection strongly enhanced the mRNA levels of Il-1β, Il-6, and Tnf-α (Fig. 7E to G) and the secretion of IL-1β and IL-6 (Fig. 7H and 7I) compared with its parental virus, while the genomic copy number of the ASFV-wild type (WT) and ASFV-ΔH240R were almost the same at 0, 6, 12, and 24 h postinoculation (hpi) (Fig. 7J), suggesting that loss of H240R greatly compromises ASFV’s ability to block the inflammatory responses. To confirm the potential mechanism, we further explored the effect of ASFV-ΔH240R infection on NF-κB signaling, and we found that compared with the parental virus ASFV-WT, ASFV-ΔH240R strongly promoted the phosphorylation of IκB and p65 upon LPS stimulation (Fig. 7K), demonstrating the higher NF-κB signaling activation.
Deletion of the H240R gene increases inflammatory cytokine production and reduces the pathogenicity of ASFV HLJ/18 in vivo.
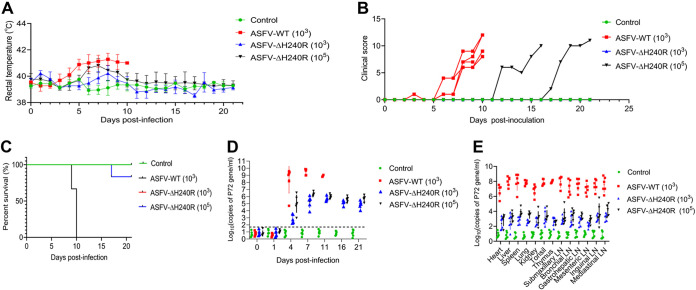
To test whether the H240R gene is related to the pathogenicity of ASFV HLJ/18 (ASFV-WT), specific-pathogen-free (SPF) large Landrace piglets were challenged with ASFV HLJ/18 or ASFV-ΔH240R. As shown in Fig. 8A, the pigs in the ASFV-WT-inoculated group developed fever at 3 dpi, and the body temperature continued to rise to 41°C until death. In comparison, all the pigs in the 103 HAD50 ASFV-ΔH240R-inoculated group had normal body temperature; three pigs in the 105 HAD50 ASFV-ΔH240R-inoculated group developed fever at 6 dpi, and then two of them returned to normal. We also evaluated the clinical scores of the infected pigs and found that all the pigs challenged with 103 HAD50 ASFV-WT developed signs of disease such as mental depression and loss of appetite on the 6th day after challenge until death, while only two pigs in the 105 HAD50 ASFV-ΔH240R-inoculated group showed reduced feed intake and gloomy spirit on the 12th and 17th days after challenge; one died at 18 dpi, and the other one survived with normal temperature (Fig. 8B). In total, only one pig inoculated with 105 HAD50 ASFV-ΔH240R died, and all the remaining 11 pigs challenged with 103 HAD50 or 105 HAD50 ASFV-ΔH240R survived for the duration of the 21-day observation period, while all of the pigs challenged with 103 HAD50 ASFV-WT died within 10 dpi (Fig. 8C).
FIG 8.
Deletion of the H240R gene reduces ASFV pathogenicity in pigs. (A to C) The rectal temperature measurements (A), clinical score (B), and survival rates (C) of ASFV-WT- and ASFV-ΔH240R-challenged pigs. The different groups of pigs were unchallenged (control, n = 4), challenged with 103 HAD50 of parental ASFV-WT (n = 6), 103 HAD50 of ASFV-ΔH240R (n = 6), or 105 HAD50 of ASFV-ΔH240R (n = 6). The rectal temperature measurements (A), clinical scores (B), and survival rates (C) were recorded daily until day 21 postchallenge. (D) ASFV DNA copy numbers in blood samples from ASFV-WT- and ASFV-ΔH240R-challenged pigs. The blood samples were collected at 0, 1, 4, 7, 11, 16, and 21 dpi from unchallenged or ASFV-WT-challenged (103 HAD50) or ASFV-ΔH240R-challenged (103 and 105 HAD50) pigs to detect the viral DNA copy number by qPCR. (E) ASFV DNA copy number in different tissues from ASFV-WT- and ASFV-ΔH240R-challenged pigs. At 21 dpi, the surviving pigs were euthanized, and tissues including the heart, liver, lung, kidney, spleen, tonsil, thymus, and six lymph nodes (inguinal lymph node, submaxillary lymph node, bronchial lymph node, mesenteric lymph node, mediastinal lymph node, and gastrohepatic lymph node) were collected from all pigs for viral DNA quantification by qPCR. For pigs that died before 21 dpi, the indicated tissue samples were collected at the time of death.
Serum samples collected at 0, 1, 4, and 7 dpi and tissue samples including the heart, lung, spleen, tonsil, thymus, and five lymph nodes (intestinal lymph node, inguinal lymph node, submaxillary lymph node, bronchial lymph node, and gastrohepatic lymph node) collected from the dead pigs and surviving pigs before euthanasia were analyzed for viral DNA quantification by quantitative PCR (qPCR). As shown in Fig. 8D, the viral DNA copies in the blood samples of the 103 HAD50-ASFV-ΔH240R group and 105 HAD50-ASFV-ΔH240R-inoculated group were about 5 and 3 log units lower at 4 dpi than that in ASFV-WT inoculated group, and about 4 and 3 log units lower at 7 dpi. The viral DNA copy numbers in the organ and tissue samples in the ASFV-ΔH240R-inoculated group were 4 to 5 log units lower than that in ASFV-WT-inoculated group (Fig. 8E).
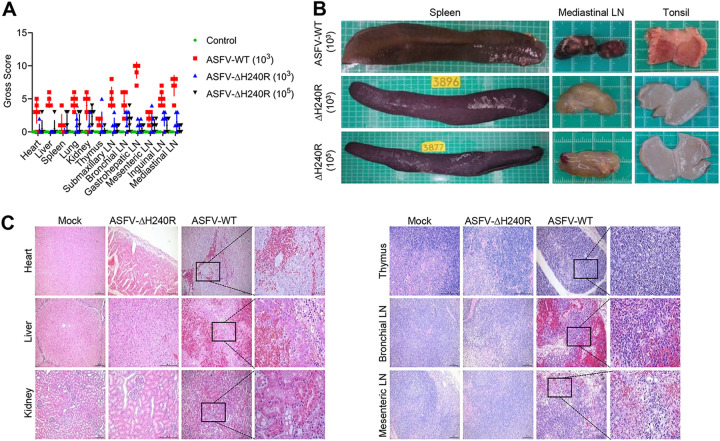
To observe the tissue damage caused by ASFV infection, the organs and tissues including the heart, lung, spleen, tonsil, thymus, and five lymph nodes of all pigs infected with ASFV-WT or ASFV-ΔH240R or those that received mock injection were scored grossly. The gross scores were significantly lower for pigs infected with ASFV-ΔH240R than for those infected with ASFV-WT (Fig. 9A). ASFV-ΔH240R infection caused mild pathological injury in pigs compared with ASFV-WT. The spleens of pigs infected with ASFV-WT showed swelling and congestion; the mediastinal lymph nodes were darkened, which might be due to congestion. The tonsils showed remarkable congestion. In contrast, the spleens, mediastinal lymph nodes, and tonsils of pigs that survived ASFV-ΔH240R infection showed mild pathological changes (Fig. 9B). In pigs infected with ASFV-WT, massive local bleeding and mild muscle cell degeneration were observed in the heart sections; multiple areas of congestion and bleeding and a small amount of inflammatory cell infiltration can be seen in the interlobular connective tissue in the liver section; hepatocyte degeneration and partial necrosis were also observed. In the renal sections, protein tube type, local hyaline degeneration of proximal convoluted tubular epithelial cells, nuclear pyknosis and necrosis of renal tubular epithelial cells could be observed. Extensive lymphocytopenia, nuclear pyknosis, disintegration, and necrosis were observed in the thymus sections. In the bronchial lymph nodes and mesenteric lymph nodes, massive hemorrhage, significant necrosis and reduction of lymphocytes, extensive and massive hemorrhage of medulla, deposition of hemosiderin, and reduction of cortical lymphocytes were also observed. In comparison, we found less pathological damage in the heart, liver, kidney, thymus, bronchial lymph nodes, and mesenteric lymph nodes in pigs infected with ASFV-ΔH240R (Fig. 9C).
FIG 9.
Deletion of the H240R gene attenuates ASFV pathological damage in pigs. (A) Gross scores of the tissues including the heart, liver, lung, kidney, spleen, tonsil, thymus, and six lymph nodes (inguinal lymph node, submaxillary lymph node, bronchial lymph node, mesenteric lymph node, mediastinal lymph node, and gastrohepatic lymph node) of pigs mock-challenged or challenged with 103 HAD50 of parental ASFV-WT (n = 6), 103 HAD50 of ASFV-ΔH240R (n = 6), or 105 HAD50 of ASFV-ΔH240R (n = 6) were recorded and analyzed. (B) Tissue lesions noted in the spleen, mediastinal lymph node, and tonsils of pigs inoculated with ASFV-WT (103 HAD50) or ASFV-ΔH240R (103 HAD50 or 105 HAD50). (C) Hematoxylin and eosin-stained images of heart, liver, kidney, thymus, bronchial lymph nodes, and mesenteric lymph nodes from pigs mock-challenged or challenged with ASFV-WT (103 HAD50) or ASFV-ΔH240R (103 HAD50). Scale bars = 200 μm. The sites of inflammation are magnified in the panes on the right. Scale bars = 50 μm.
Taken together, these results indicate that ASFV-ΔH240R is much less virulent and its infection causes much less pathological damage to organs and tissues of affected pigs than ASFV-WT, suggesting a critical role of pH240R in ASFV virulence and pathogenicity.
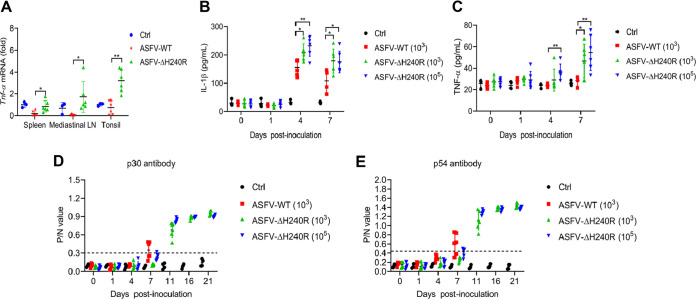
To further compare the inflammatory responses of pigs in response to ASFV-WT and ASFV-ΔH240R challenges, we analyzed the levels of inflammatory cytokines in the affected pigs. The mRNA levels of TNF-α in the immune tissues, including spleen, tonsil, and mediastinal lymph node, were detected by qPCR, and the results showed that the mRNA levels of TNF-α in these tissues from pigs challenged with 103 HAD50 ASFV-ΔH240R were significantly higher than those from pigs challenged with 103 HAD50 ASFV-WT (Fig. 10A). Meanwhile, the secretion levels of IL-1β and TNF-α in peripheral serum of pigs challenged with 103 HAD50 ASFV-WT, 103 HAD50 ASFV-ΔH240R, and 105 HAD50 ASFV-ΔH240R were monitored at 0, 1, 4, and 7 dpi using enzyme-linked immunosorbent assay (ELISA). The results showed that the secretion of IL-1β and TNF-α in the ASFV-ΔH240R-inoculated groups was significantly higher than that in the ASFV-WT-inoculated group at both 4 dpi and 7 dpi (Fig. 10B and C). The serum antibodies against the viral proteins p30 and p54 gradually increased from 4 dpi and peaked at 11 dpi and 14 dpi in the 105-HAD50 ASFV-ΔH240R- and 103-HAD50 ASFV-ΔH240R-inoculated groups, while these antibodies in the ASFV-WT-challenged pigs were very low (Fig. 10D and E). These results indicate stronger inflammatory responses in pigs challenged with ASFV-ΔH240R than with ASFV-WT, suggesting an essential role of pH240R in the ASFV infection-induced inflammatory responses.
FIG 10.
ASFV-ΔH240R infection induces higher inflammatory cytokine production in pigs than its parental ASFV HLJ/18. (A to C) Analysis of inflammatory cytokines in tissues and serum from ASFV-WT- and ASFV-ΔH240R-challenged pigs. Pigs were inoculated with PBS (control [ctrl], n = 4) or challenged intramuscularly with ASFV-ΔH240R (n = 6, 103 and 105 HAD50) or parental ASFV-WT (n = 6, 103 HAD50). At 21 dpi, the surviving pigs were euthanized, and tissues including spleen, tonsil, and mediastinal lymph node were collected from all pigs to detect the Tnf-α mRNA level by qPCR (A). The protein levels of IL-1β (B) and TNF-α (C) in the serum from ASFV-WT- and ASFV-ΔH240R-challenged pigs were measured by ELISA. (D to E) Analysis of antibody in serum from ASFV-WT- and ASFV-ΔH240R-challenged pigs. At 0, 1, 4, 7, 11, 16, and 21 dpi, the antibodies against p30 (D) and p54 (E) in the serum samples from ASFV-WT- and ASFV-ΔH240R-challenged pigs were measured by ELISA. The data presented are the mean ± SD. *, P < 0.05; **, P < 0.01.
DISCUSSION
Pattern recognition receptors (PRRs) and inflammasomes are key parts of the antiviral innate immune system as they help the host detect conserved pathogen-associated molecular patterns (PAMPs) and remove the invading pathogens. The signaling pathways activated by PRRs induce expression of proinflammatory cytokines, which boost the overall immune responses by recruiting immune cells to the site of infection, and mobilize adaptive immunity. Inflammasomes contribute to antiviral responses through the maturation of IL-1β and IL-18 and induction of pyroptotic cell death. The activity of the innate immune system along with the adaptive immune response normally leads to successful virus elimination. Although antiviral innate immune responses provide a powerful first line of defense, ASFV has evolved multiple strategies to counteract host antiviral innate immune responses (22).
Upon DNA virus infection, TLRs in host cells recognize viral genomic nuclear acids to induce production of the inflammatory cytokines, such as IL-1β and IL-18 (23). In signal 1, the IKK-NF-κB axis is a critical step for inflammatory responses. In the resting state, IκBα binds to the NF-κB p50/p65 heterodimer to form an inactive complex. Some viral infections can activate the IKK complex, which phosphorylates IκBα and causes its degradation by the K48-linked ubiquitin-proteasome pathway. Then, the NF-κB p50/p65 heterodimers become activated, enter the nucleus, and induce production of IFN-I and proinflammatory cytokines (such as pro-IL-1β and pro-IL-18) (24, 25). In signal 2, activation of AIM2 or NLRP3 inflammasomes facilitates pro-caspase 1 activity, leading to maturation and secretion of inflammatory cytokines (23). Several DNA viruses have been reported to induce inflammatory responses by activating AIM2 and NLRP3 inflammasomes during infection (26, 27). Previous studies revealed that ASFV infection can induce IL-1β production through activation of the TLR/MyD88 pathway and the NLRP3 inflammasome in PAMs (15), and ASFV-encoded inhibitor of apoptosis (IAP)-like protein/pA224L can induce the activation of NF-κB (28).
To replicate efficiently, ASFV has evolved multiple strategies to antagonize host inflammatory responses by interfering with the activation of both the NF-κB signaling and the NLRP3 inflammasome. For example, ASFV pI215L and pF317L are found to inhibit NF-κB activation to evade host immune responses (29, 30). Transient expression of ASFV pI215L impairs NF-κB and AP-1 transcription factors that act upstream or at the step of IKKβ, leading to reduced p65 nuclear translocation upon cytokine stimulation (30). pF317L interacts with IKKβ and suppresses its phosphorylation, which reduces phosphorylation and ubiquitination of IκBα and results in decreased expression of various proinflammatory cytokines (29). ASFV pMGF505-7R interacts with IKKα to inhibit NF-κB activation, resulting in decreased pro-IL-1β transcription and production. Meanwhile, pMGF505-7R binds to NLRP3 and inhibits NLRP3 inflammasome assembly, resulting in decreased IL-1β maturation and secretion (15). In this study, we found that ASFV pH240R inhibits NF-κB signaling by interacting with NEMO, which is consistent with the results just published (31). Previous studies showed the combination of NEMO with IKKβ through its CC1 domain (32). We found that ASFV pH240R interacted with the CC1 domain of NEMO to inhibit the interaction between IKKβ and NEMO, thereby inhibiting NF-κB activation and IL-1β transcription. Therefore, the NF-κB signaling and the IKK complex have been important targets attacked by ASFV to compromise the host innate immune responses and facilitate its replication.
NLRP3 is an important PRR in the cytoplasm, and it consists of a C-terminal LRR domain that has autoinhibitory functions and signal recognition capabilities, a central NACHT domain that has ATPase activity and mediates self-oligomerization, and an amino-terminal PYD domain that recruits ASC. In response to stimuli, NLRP3 undergoes self-oligomerization via the NACHT domain and then recruits ASC via the PYD domain and induces formation of ASC specks (33). In this study, we found that pH240R interacts with both the NACHT and LRR domains of NLRP3 to suppress the NLRP3 inflammasome assembly and ASC oligomerization and ultimately inhibits IL-1β maturation and secretion. Zhou et al. showed that the interaction of pH240R and the PRR domain inhibits the oligomerization of NLRP3 (31). However, we speculate that the interaction of pH240R and the NACHT domain may inhibit the oligomerization of NLRP3, and the interaction of pH240R and the LRR domain may prevent NLRP3 from sensing stimuli, thereby inhibiting NLRP3-inflammasome activation. Further studies are required to understand the underlying mechanism.
Recently, it was reported that ASFV-ΔH240R virions are mostly aberrant, with a decrease of about 30-fold in the infectious virus-to-particle ratio compared with ASFV-WT (18). Consistent with the study, we also found that ASFV without the H240R gene induces higher mRNA and secretion levels of Il-1β, Il-6, and Tnf-α in PAMs than does ASFV-WT, despite the same numbers of genome copies of ASFV-ΔH240R and ASFV-WT used and detected. These observations indicate that H240R as an important anti-inflammatory factor during ASFV infection.
It has been reported that the ASFV genes that function to antagonize host innate immunity play important roles in determining viral virulence (34, 35). To detect whether the H240R gene is a key virulence gene, the pathogenicity of ASFV-ΔH240R was evaluated in SPF pigs. Consistent with our expectations, deletion of the H240R gene significantly reduces the pathogenicity of ASFV HLJ/18. Only two pigs inoculated with 105 HAD50 ASFV-ΔH240R developed signs including lethargy and reduced feed intake. However, only one pig died, and the other one did not have a fever or high viral load in the tissues, which indicates that the clinical symptoms in tissue samples from pigs in the ASFV-ΔH240R challenge groups were significantly lower than those from the ASFV-WT challenge group, while ASFV-ΔH240R induced more TNF-α and IL-1β than ASFV-WT in peripheral blood. Deletion of the H240R gene reduced the viral load of ASFV in pigs, which may be due to several reasons. On one hand, deletion of the H240R gene reduces ASFV-ΔH240R infectivity due to the production of noninfectious virions (18). On the other hand, increased secretion of TNF-α and IL-1β induced by ASFV-ΔH240R may inhibit ASFV-ΔH240R replication in pigs, as it is reported that inflammatory cytokines can inhibit virus replication (36). In addition, more inflammatory cytokines induced by ASFV-ΔH240R may activate stronger cellular immunity. Of note, ASFV-ΔH240R also induced high levels of antibodies against p30 and p54, which indicates that combined deletion of the H240R gene and other virulence-related genes may serve as a new strategy to develop attenuated live vaccines against ASFV.
Taken together, our findings reveal a novel mechanism by which ASFV pH240R evades host antiviral inflammatory responses. H240R gene is a key ASFV virulence-related gene, and its deletion significantly attenuates the virulence of ASFV HLJ/18, which provides a candidate gene to develop attenuated live vaccines against ASFV.
MATERIALS AND METHODS
Safety protocols.
All experiments involving ASFV HLJ/18 or ASFV-ΔH240R infection were conducted within the enhanced biosafety level 3 (P3+) and level 4 (P4) facilities in the Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (CAAS) approved by the Ministry of Agriculture and Rural Affairs.
Cells and viruses.
HEK293T cells and MA104 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS). CRL-2843 cells were purchased from ATCC and cultured in complete growth RPMI 1640 medium containing 10% FBS and 2 mM l-glutamine. PAMs were isolated from the lung lavage fluid of 4-week-old healthy specific-pathogen-free (SPF) piglets (without ASFV, classical swine fever virus, porcine reproductive and respiratory syndrome virus, pseudorabies virus, and 26 other pathogens) and were cultured in RPMI 1640 medium supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C with 5% CO2. The ASFV HLJ/18 strain was isolated from a pig sample in an ASF outbreak farm in northern China (3).
Plasmids.
To construct plasmids expressing Flag-tagged or hemagglutinin (HA)-tagged proteins in the NF-κB and NLRP3 signaling pathways, the cDNAs corresponding to these swine genes were amplified by standard reverse transcription-PCR (RT-PCR) using total RNA extracted from PAMs as templates and were then cloned into the pCAGGS-Flag or pCAGGS-HA vector, respectively. All constructs were validated by DNA sequencing. The primers used in this study are listed in Table 1.
TABLE 1.
Primers used for PCR in this study
| Plasmids | Primers(5′–3′) |
|---|---|
| pCAGGS-Flag-Myd88 and pCAGGS-HA-Myd88 | F: ATGGCTGCAGGAGGCTCCGGAGC |
| R: TCAGGGCAGGGATAGGGCC | |
| pCAGGS-Flag-IKKα and pCAGGS-HA-IKKα | F: ATGGAGCGGCCCCCGGGGCT |
| R: TCATCCTGTTAACCAACTCCA | |
| pCAGGS-Flag-IKKβ and pCAGGS-HA-IKKβ | F: ATGAGTTCCAGTA TGTGGTATAT |
| R: CTATTCTACAAGTAACAGGTATTC | |
| pCAGGS-Flag-NEMO and pCAGGS-HA-NEMO | F: ATGAGCAGGACCCCCTGGAA |
| R: CTACTCGATACACTCCATGA | |
| pCAGGS-HA-NEMO-N1 | F: ATGAGCAGGACCCCCTGGAA |
| R: ATTAAGATCTGCTAGCTCGAG | |
| pCAGGS-HA-NEMO-N2 | F: ATGAGCAGGACCCCCTGGAA |
| R: ATTAAGATCTGCTAGCTCGAG | |
| pCAGGS-HA-NEMO-N3 | F: GTGCAGGTGGACCAGCTGCGCCTGCAG |
| R: CTACTCGATACACTCCATGA | |
| pCAGGS-HA-NEMO-N4 | F: CAGCGGGAGGAGAAGGAGTTTCTCAT |
| R: CTACTCGATACACTCCATGA |
Reagents and antibodies.
The following antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA): anti-Flag (D6W5B, 14793), anti-HA (C29F4, 3724). Alexa Fluor 633-labeled goat anti-rabbit IgG, and Alexa Fluor 488-labeled goat anti-mouse IgG were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Anti-mouse IgG (H+L) DyLight 800-labeled (042-07-18-06) was purchased from Sera Care (Milford, MA, USA). The IRDye 800CW goat anti-rabbit IgG (H+L) (925-32211) was purchased from LI-COR (Lincoln, NE, USA). IPKine horseradish peroxidase (HRP) goat anti-mouse IgG HCS (A25112) was purchased from Abbkine (Wuhan, China). Mouse anti-IL-1β monoclonal antibodies (MAb) and mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) polyclonal antibodies (pAb) were purchased from Thermo Fisher Scientific. Rabbit anti-swine NLRP3 pAb, anti-swine caspase-1 p20 pAb, and anti-swine ASC pAb were prepared by immunizing the rabbits as previously described (15). The full-length cDNA of the H240R gene was cloned into the pET-21a vector for the pET-21a-H240R construct. The plasmid was then transformed into BL21(DE3) to express recombinant pH240R. Mice were immunized with the purified recombinant pH240R to generate anti-pH240R polyclonal antibody.
iGLuc-based inflammasome reconstruction system.
The iGLuc-based inflammasome reconstruction system was constructed based on Gaussia luciferase with high sensitivity and specificity as previously described (14). Briefly, HEK293T cells plated in 48-well plates were transiently transfected with a total amount of 500 ng of DNA per well using 1 μL of X-treme GENE HP DNA transfection reagent (Roche, Basel, Switzerland). The 500 ng of DNA contained the iGLuc-based NLRP3 inflammasome system (100 ng iGLuc, 10 ng pCAGGS-HA-caspase-1, 10 ng pCAGGS-HA-ASC, and 12.5 ng pCAGGS-HA-NLRP3) and a plasmid expressing a tested protein or an empty vector. After 24 h, the supernatants were collected for GLuc readout.
Confocal microscopy.
CRL-2843 cells were transfected with plasmids expressing HA-tagged and Flag-tagged proteins for 24 h. These cells were then fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.1% Triton X-100. After blocking with 10% FBS, cells were incubated with the indicated anti-Flag and anti-HA antibodies for 1 h, or PAMs were infected with ASFV HLJ/18 or ASFV-ΔH240R for 12 h and then stained with anti-ASC or anti-pH240R antibodies. After washing with 1× cold phosphate-buffered saline (PBS; pH 7.4) three times, the cells were incubated with the indicated secondary antibodies. Samples were visualized with an SP2 confocal system (Leica Microsystems, Wetzlar, Germany).
ASC speck and oligomerization.
CRL-2843 cells were transfected with plasmids encoding GFP-ASC and pH240R. At 24 h posttransfection (hpt), the cells were stimulated with nigericin (5 μM) for an extra 1 h before harvest. HEK293T cells were transfected with different combinations of plasmids encoding GFP-ASC, NLRP3, and pH240R for 24 h before harvest. After harvest, the cells were washed in PBS with 1% fetal calf serum (FCS) and fixed in 2% PFA for 30 min at 4°C and then observed for the ASC specks with a Leica SP2 confocal system.
Generation of H240R gene-deleted ASFV (ASFV-ΔH240R).
A recombinant ASFV without the H240R gene (ASFV-ΔH240R) was generated by homologous recombination in PAMs first infected with ASFV HLJ/18 and then transfected with a recombination transfer vector as described previously (37). Recombinant transfer vectors (p72-EGFP) containing the flanking genomic regions of the targeted genes that mapped approximately 866 bp to the left and 498 bp to the right of the deleted parts, and the reporter gene cassette containing the reporter gene EGFP fused with the ASFV p72 late gene promoter was used as shown in Fig. 7. ASFV-ΔH240R was identified by PCR and confirmed by sequencing.
50% Hemadsorption dose (HAD50) assay.
The HAD50 assay was performed as previously described (3). Briefly, peripheral blood mononuclear cells (PBMC) were seeded in 96-well plates and then infected with 0.1 mL/well of 10-fold serially diluted supernatant in quintuplicate. The quantity of ASFV was determined by identification of the characteristic rosette formation representing hemadsorption of erythrocytes around infected cells. At 7 days postinfection (dpi), the HAD50 was determined using the Reed-Muench method. All data are shown as the means of three independent experiments.
Coimmunoprecipitation (co-IP) and immunoblot analysis.
For co-IP, the cells were collected and lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, and 10% glycerol) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1× protease inhibitor cocktail (Roche, Basel, Switzerland). Then, cell supernatants were incubated with anti-Flag (M2) agarose or with protein G plus agarose immunoprecipitation reagent (Santa Cruz Biotechnology, Santa Cruz, USA) together with 1 μg of the corresponding antibodies at 4°C overnight on a roller. The pellets were washed five times with cell lysis buffer. For Western blot analysis, the cell lysates and immunoprecipitants were resolved by 10 to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Darmstadt, Germany). After incubation with the indicated primary and secondary antibodies, the membranes were visualized by enhanced chemiluminescence (ECL) (Thermo Fisher Scientific, MA, USA) or an Odyssey two-color infrared fluorescence imaging system (LI-COR).
Luciferase reporter gene assay.
Luciferase activities were measured with a dual-luciferase reporter assay system (Promega, Wisconsin, USA) according to the manufacturer’s instructions. The data were normalized to the transfection efficiency by dividing the firefly luciferase activity by the Renilla luciferase activity.
Quantitative real-time PCR (qPCR).
To detect the mRNA levels of Il-1β, Il-6, and Tnf-α, total RNA was extracted using TRIzol reagent (Invitrogen, California, USA), and reverse transcription was performed with a PrimeScript RT reagent kit (TaKaRa, Osaka, Japan). Reverse transcription products were amplified using an Agilent-Strata gene Mx real-time qPCR system with SYBR premix Ex Taq II system (TaKaRa) according to the manufacturer’s instructions. Data were normalized to the level of β-actin expression in each individual sample. For ASFV genomic DNA detection, ASFV genomic DNA was extracted using a QIAamp DNA minikit (Qiagen, Dusseldorf, Germany). qPCR was carried out on a QuantStudio5 system (Applied Biosystems, Waltham, USA) according to The Office of International Education (OIE)-recommended procedure. All the qPCR primers are listed in Table 2.
TABLE 2.
Primers used for qPCR in this study
| Gene name | Sequence (5′–3′) |
|---|---|
| sq-TNF-α | F: ACCACGCTCTTCTGCCTACTGC |
| R: TCCCTCGGCTTTGACATTGGCTAC | |
| sq-β-actin | F: TGAGAACAGCTGCATCCACTT |
| R: CGAAGGCAGCTCGGAGTT | |
| sq-IL-6 | F: CTGCTTCTGGTGATGGCTACTG |
| R: GGCATCACCTTTGGCATCTT | |
| sq-IL-1β | F: CCCAAAAGTTACCCGAAGAGG |
| R: TCTGCTTGAGAGGTGCTGATG | |
| ASFV B646L | F: CTGCTCATGGTATCAATCTTATCGA |
| R: GATACCACAAGATCAGCCGT | |
| Probe: CCACGGGAGGAATACCAACCCAGTG |
Enzyme-linked immunosorbent assay (ELISA).
The protein concentrations of IL-1β and IL-6 in cell culture supernatants and serum from animals were measured by enzyme-linked immunosorbent assay kits according to the manufacturer’s instructions (R&D, Minnesota, USA).
Pathogenicity of ASFV-ΔH240R in domestic pigs.
A total of 22 8-week-old healthy SPF piglets were randomly assigned into four groups (6 piglets inoculated with ASFV-ΔH240R, 103 HAD50/piglet; 6 piglets inoculated with ASFV-ΔH240R, 105 HAD50/piglet; 6 piglets inoculated with ASFV-WT, 103 HAD50/piglet; 4 piglets inoculated with PBS). The piglets were monitored daily for clinical signs prior to feeding, including anorexia, lethargy, fever, and emaciation (38). The blood samples were collected at 0, 1, 4, 7, 11, 16, and 21 dpi to detect virus load, IL-1β, IL-6, anti-p30, and anti-p54 antibodies. ASFV-infected piglets were euthanized in the moribund stage. At 21 dpi, all surviving piglets were euthanized. The tissue samples from heart, liver, lung, spleen, kidney, tonsil, thymus, and six lymph nodes (mesenteric lymph node, mediastinal lymph node, inguinal lymph node, submaxillary lymph node, bronchial lymph node, and gastrohepatic lymph node) were collected to measure ASFV genomic DNA copy numbers, while spleen, mediastinal lymph node, and tonsil were collected to detect Tnf-α mRNA.
Generation of a THP-1-pH240R stable cell line.
The ASFV H240R gene was cloned into the pLVX-ISRE-Puro vector (Clontech). The THP-1 cells were infected with the lentiviral particles expressing pH240R in the presence of 8 mg/mL Polybrene. At 3 days postinfection, the cells were treated with 1 mg/mL puromycin for 2 weeks. ASFV-pH240R overexpression efficiency was assessed by Western blotting, and the cell line was named THP-1-pH240R.
Statistical analysis.
All statistical analyses were performed using one-way analysis of variance (ANOVA) with the SPSS 16.0 software package (version 16.0, SPSS, Inc., Chicago, IL, USA). Data were expressed as the mean ± standard deviation (SD). A P value of <0.05 was considered statistically significant.
Ethics statement.
All animal experiments were performed according to animal protocols approved by the Subcommittee on Research Animal Care at the HVRI and carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China.
ACKNOWLEDGMENTS
This study was supported by the National Key Research and Development Program of China (grant no. 2021YFD1800103), the National Natural Science Foundation of China (grant no. U21A20256, 32270156), and the Natural Science Foundation of Heilongjiang Province of China (grant no. YQ2020C022).
Contributor Information
Zhigao Bu, Email: buzhigao@caas.cn.
Jiangnan Li, Email: lijiangnan520@163.com.
Changjiang Weng, Email: wengchangjiang@caas.cn.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Karger A, Perez-Nunez D, Urquiza J, Hinojar P, Alonso C, Freitas FB, Revilla Y, Le Potier MF, Montoya M. 2019. An update on African swine fever virology. Viruses 11:864. 10.3390/v11090864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costard S, Wieland B, de Glanville W, Jori F, Rowlands R, Vosloo W, Roger F, Pfeiffer DU, Dixon LK. 2009. African swine fever: how can global spread be prevented? Philos Trans R Soc Lond B Biol Sci 364:2683–2696. 10.1098/rstb.2009.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao D, Liu R, Zhang X, Li F, Wang J, Zhang J, Liu X, Wang L, Zhang J, Wu X, Guan Y, Chen W, Wang X, He X, Bu Z. 2019. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg Microbes Infect 8:438–447. 10.1080/22221751.2019.1590128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun E, Zhang Z, Wang Z, He X, Zhang X, Wang L, Wang W, Huang L, Xi F, Huangfu H, Tsegay G, Huo H, Sun J, Tian Z, Xia W, Yu X, Li F, Liu R, Guan Y, Zhao D, Bu Z. 2021. Emergence and prevalence of naturally occurring lower virulent African swine fever viruses in domestic pigs in China in 2020. Sci China Life Sci 64:752–765. 10.1007/s11427-021-1904-4. [DOI] [PubMed] [Google Scholar]
- 5.Gao X, Liu T, Liu Y, Xiao J, Wang H. 2021. Transmission of African swine fever in China through legal trade of live pigs. Transbound Emerg Dis 68:355–360. 10.1111/tbed.13681. [DOI] [PubMed] [Google Scholar]
- 6.Gaudreault NN, Madden DW, Wilson WC, Trujillo JD, Richt JA. 2020. African swine fever virus: an emerging DNA arbovirus. Front Vet Sci 7:215. 10.3389/fvets.2020.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malogolovkin A, Kolbasov D. 2019. Genetic and antigenic diversity of African swine fever virus. Virus Res 271:197673. 10.1016/j.virusres.2019.197673. [DOI] [PubMed] [Google Scholar]
- 8.Cackett G, Matelska D, Sýkora M, Portugal R, Malecki M, Bähler J, Dixon L, Werner F. 2020. The African swine fever virus transcriptome. J Virol 94:e00119-20. 10.1128/JVI.00119-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon LK, Chapman DA, Netherton CL, Upton C. 2013. African swine fever virus replication and genomics. Virus Res 173:3–14. 10.1016/j.virusres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 10.Dixon L, Islam M, Nash R, Reis A. 2019. African swine fever virus evasion of host defences. Virus Res 266:25–33. 10.1016/j.virusres.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spel L, Martinon F. 2021. Detection of viruses by inflammasomes. Curr Opin Virol 46:59–64. 10.1016/j.coviro.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Gao W, Shao F. 2017. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci 42:245–254. 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Zhang J, Zhang Y, Yang J, Wang L, Qi Y, Han X, Zhou X, Miao F, Chen T, Wang Y, Zhang F, Zhang S, Hu R. 2020. Cytokine storm in domestic pigs induced by infection of virulent African swine fever virus. Front Vet Sci 7:601641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song J, Li K, Li T, Zhao G, Zhou S, Li H, Li J, Weng C. 2020. Screening of PRRSV- and ASFV-encoded proteins involved in the inflammatory response using a porcine iGLuc reporter. J Virol Methods 285:113958. 10.1016/j.jviromet.2020.113958. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Song J, Kang L, Huang L, Zhou S, Hu L, Zheng J, Li C, Zhang X, He X, Zhao D, Bu Z, Weng C. 2021. pMGF505-7R determines pathogenicity of African swine fever virus infection by inhibiting IL-1beta and type I IFN production. PLoS Pathog 17:e1009733. 10.1371/journal.ppat.1009733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borca M, O’Donnell V, Holinka L, Ramírez-Medina E, Clark B, Vuono E, Berggren K, Alfano M, Carey L, Richt J, Risatti G, Gladue D. 2018. The L83L ORF of African swine fever virus strain Georgia encodes for a non-essential gene that interacts with the host protein IL-1β. Virus Res 249:116–123. 10.1016/j.virusres.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Wang N, Zhao D, Wang J, Zhang Y, Wang M, Gao Y, Li F, Wang J, Bu Z, Rao Z, Wang X. 2019. Architecture of African swine fever virus and implications for viral assembly. Science 366:640–644. 10.1126/science.aaz1439. [DOI] [PubMed] [Google Scholar]
- 18.Zhou P, Li LF, Zhang K, Wang B, Tang L, Li M, Wang T, Sun Y, Li S, Qiu HJ. 2022. Deletion of the H240R gene of African swine fever virus decreases infectious progeny virus production due to aberrant virion morphogenesis and enhances inflammatory cytokine expression in porcine macrophages. J Virol 96:e0166721. 10.1128/jvi.00308-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinz M, Scheidereit C. 2014. The IkappaB kinase complex in NF-kappaB regulation and beyond. EMBO Rep 15:46–61. 10.1002/embr.201337983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. 2007. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol 8:592–600. 10.1038/ni1465. [DOI] [PubMed] [Google Scholar]
- 21.Rothwarf DM, Zandi E, Natoli G, Karin M. 1998. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature 395:297–300. 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 22.Zheng X, Nie S, Feng WH. 2022. Regulation of antiviral immune response by African swine fever virus (ASFV). Virol Sin 37:157–167. 10.1016/j.virs.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanneganti TD. 2010. Central roles of NLRs and inflammasomes in viral infection. Nat Rev Immunol 10:688–698. 10.1038/nri2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brady G, Bowie AG. 2014. Innate immune activation of NFkappaB and its antagonism by poxviruses. Cytokine Growth Factor Rev 25:611–620. 10.1016/j.cytogfr.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Unterholzner L, Bowie AG. 2008. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem Pharmacol 75:589–602. 10.1016/j.bcp.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 26.Zahid A, Ismail H, Li B, Jin T. 2020. Molecular and structural basis of DNA sensors in antiviral innate immunity. Front Immunol 11:613039. 10.3389/fimmu.2020.613039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Z, Ni G, Damania B. 2018. Innate sensing of DNA virus genomes. Annu Rev Virol 5:341–362. 10.1146/annurev-virology-092917-043244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez CI, Nogal ML, Carrascosa AL, Salas ML, Fresno M, Revilla Y. 2002. African swine fever virus IAP-like protein induces the activation of nuclear factor kappa B. J Virol 76:3936–3942. 10.1128/jvi.76.8.3936-3942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Li S, Feng T, Zhang X, Yang F, Cao W, Chen H, Liu H, Zhang K, Zhu Z, Zheng H. 2021. African swine fever virus F317L protein inhibits NF-kappaB activation to evade host immune response and promote viral replication. mSphere 6:e0065821. 10.1128/mSphere.00658-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrado-Gil L, Del Puerto A, Galindo I, Cuesta-Geijo MA, Garcia-Dorival I, de Motes CM, Alonso C. 2021. African swine fever virus ubiquitin-conjugating enzyme is an immunomodulator targeting NF-kappaB activation. Viruses 13:1160. 10.3390/v13061160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou P, Dai J, Zhang K, Wang T, Li LF, Luo Y, Sun Y, Qiu HJ, Li S. 2022. The H240R protein of African swine fever virus inhibits interleukin 1beta production by inhibiting NEMO expression and NLRP3 oligomerization. J Virol 96:e0095422. 10.1128/jvi.00954-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sebban H, Yamaoka S, Courtois G. 2006. Posttranslational modifications of NEMO and its partners in NF-kappaB signaling. Trends Cell Biol 16:569–577. 10.1016/j.tcb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Lamkanfi M, Dixit VM. 2014. Mechanisms and functions of inflammasomes. Cell 157:1013–1022. 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Hong J, Chi X, Yuan X, Wen F, Rai KR, Wu L, Song Z, Wang S, Guo G, Chen JL. 2022. I226R Protein of African swine fever virus is a suppressor of innate antiviral responses. Viruses 14:575. 10.3390/v14030575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaulagain S, Delhon GA, Khatiwada S, Rock DL. 2021. African swine fever virus CD2v protein induces beta-interferon expression and apoptosis in swine peripheral blood mononuclear cells. Viruses 13:1480. 10.3390/v13081480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koyama S, Ishii KJ, Coban C, Akira S. 2008. Innate immune response to viral infection. Cytokine 43:336–341. 10.1016/j.cyto.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Chen W, Zhao D, He X, Liu R, Wang Z, Zhang X, Li F, Shan D, Chen H, Zhang J, Wang L, Wen Z, Wang X, Guan Y, Liu J, Bu Z. 2020. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci China Life Sci 63:623–634. 10.1007/s11427-020-1657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galindo-Cardiel I, Ballester M, Solanes D, Nofrarias M, Lopez-Soria S, Argilaguet JM, Lacasta A, Accensi F, Rodriguez F, Segales J. 2013. Standardization of pathological investigations in the framework of experimental ASFV infections. Virus Res 173:180–190. 10.1016/j.virusres.2012.12.018. [DOI] [PubMed] [Google Scholar]