ABSTRACT
Coronavirus disease 2019 (COVID-19), which is caused by the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is the most severe emerging infectious disease in the current century. The discovery of SARS-CoV-2-related coronaviruses (SARSr-CoV-2) in bats and pangolins in South Asian countries indicates that SARS-CoV-2 likely originated from wildlife. To date, two SARSr-CoV-2 strains have been isolated from pangolins seized in Guangxi and Guangdong by the customs agency of China, respectively. However, it remains unclear whether these viruses cause disease in animal models and whether they pose a transmission risk to humans. In this study, we investigated the biological features of a SARSr-CoV-2 strain isolated from a smuggled Malayan pangolin (Manis javanica) captured by the Guangxi customs agency, termed MpCoV-GX, in terms of receptor usage, cell tropism, and pathogenicity in wild-type BALB/c mice, human angiotensin-converting enzyme 2 (ACE2)-transgenic mice, and human ACE2 knock-in mice. We found that MpCoV-GX can utilize ACE2 from humans, pangolins, civets, bats, pigs, and mice for cell entry and infect cell lines derived from humans, monkeys, bats, minks, and pigs. The virus could infect three mouse models but showed limited pathogenicity, with mild peribronchial and perivascular inflammatory cell infiltration observed in lungs. Our results suggest that this SARSr-CoV-2 virus from pangolins has the potential for interspecies infection, but its pathogenicity is mild in mice. Future surveillance among these wildlife hosts of SARSr-CoV-2 is needed to monitor variants that may have higher pathogenicity and higher spillover risk.
IMPORTANCE SARS-CoV-2, which likely spilled over from wildlife, is the third highly pathogenic human coronavirus. Being highly transmissible, it is perpetuating a pandemic and continuously posing a severe threat to global public health. Several SARS-CoV-2-related coronaviruses (SARSr-CoV-2) in bats and pangolins have been identified since the SARS-CoV-2 outbreak. It is therefore important to assess their potential of crossing species barriers for better understanding of their risk of future emergence. In this work, we investigated the biological features and pathogenicity of a SARSr-CoV-2 strain isolated from a smuggled Malayan pangolin, named MpCoV-GX. We found that MpCoV-GX can utilize ACE2 from 7 species for cell entry and infect cell lines derived from a variety of mammalian species. MpCoV-GX can infect mice expressing human ACE2 without causing severe disease. These findings suggest the potential of cross-species transmission of MpCoV-GX, and highlight the need of further surveillance of SARSr-CoV-2 in pangolins and other potential animal hosts.
KEYWORDS: MpCoV-GX, SARS-CoV-2-related coronavirus, interspecies infection, pangolin
INTRODUCTION
Coronavirus disease 2019 (COVID-19) emerged at the end of December 2019 and has been the most devastating pandemic in the current century (1, 2). The causative pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), belonging to the species SARS-related coronavirus (SARSr-CoV) in the genus Betacoronavirus of the family Coronaviridae (3), has been identified as the third coronavirus highly pathogenic to humans after the severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) (4–7). It causes viral pneumonia, manifested by fever, cough, chest discomfort, and dyspnea, bilateral lung infiltration, and even death in severe cases (8, 9).
Following the identification of SARS-CoV-2, we and other groups reported the discovery of diverse SARS-CoV-2-related CoVs (SARSr-CoV-2) in bats collected in Southeast Asian countries and China and pangolins seized by the Chinese customs agency during 2010 to 2020 (1, 10–17). The closest SARSr-CoV-2 known to date is BANAL-52, which was identified in 2020 from the rectal swab of a Rhinolophus malayanus bat from Laos, with 96.8% identity to SARS-CoV-2 at the whole-genome level (16). RaTG13, which was detected in a Rhinolophus affinis bat collected in 2013 in Yunnan Province, China, shares 96.2% genome-wide nucleotide identity with SARS-CoV-2 but only 93.1% in the spike (S) gene (1). In addition to these two close relatives of SARS-CoV-2, other coronaviruses similar to SARS-CoV-2 have been identified in various species of Rhinolophus bats from Thailand, Cambodia, China, and Japan sharing 81.5% to 93.3% full-length genome identities and 71.3% to 81.3% S gene sequence identity to SARS-CoV-2 (11, 12, 15, 17). Besides bats, SARSr-CoV-2 strains have been identified in Malayan pangolins smuggled from Southeast Asia (13, 14). These viruses, here named MpCoV-GX and MpCoV-GD, show 85.4% and 92.4% genome-wide and 92.6% and 90.7% S gene nucleotide identity to SARS-CoV-2, respectively (13, 14). Currently, however, only one bat SARSr-CoV-2 has been isolated, which challenges the further characterization of these viruses (16). In contrast, the two pangolin SARSr-CoV-2 (MpCoV-GX and MpCoV-GD) have been successfully isolated (13, 14).
Human angiotensin-converting enzyme 2 (hACE2) is the cell receptor for SARS-CoV, SARS-CoV-2 (1, 18), and some bat SARSr-CoVs (19, 20). Studies have shown that MpCoV-GX and MpCoV-GD efficiently utilize human ACE2 as the receptor, and MpCoV-GX exhibits infection characteristics similar to SARS-CoV-2 and direct contact transmissibility in hamsters (21–23). However, their biological features have not been deeply characterized. Here, we investigated the cellular susceptibility and replication kinetics of MpCoV-GX in vitro and in vivo in wild-type BALB/c mice, human ACE2-transgenic mice, and human ACE2 knock-in mice. Body weight changes, viral genome copies in organ tissues, and the lung histopathology of infected mice were analyzed with the aim of expanding our understanding about the interspecies infection risk of MpCoV-GX. We expect that this information will shed new light on the pathogenesis of wildlife-borne SARSr-CoV-2 and provide reference for the prevention of future emerging infectious diseases caused by this group of viruses.
RESULTS
MpCoV-GX utilizes ACE2s of different origins for cell entry.
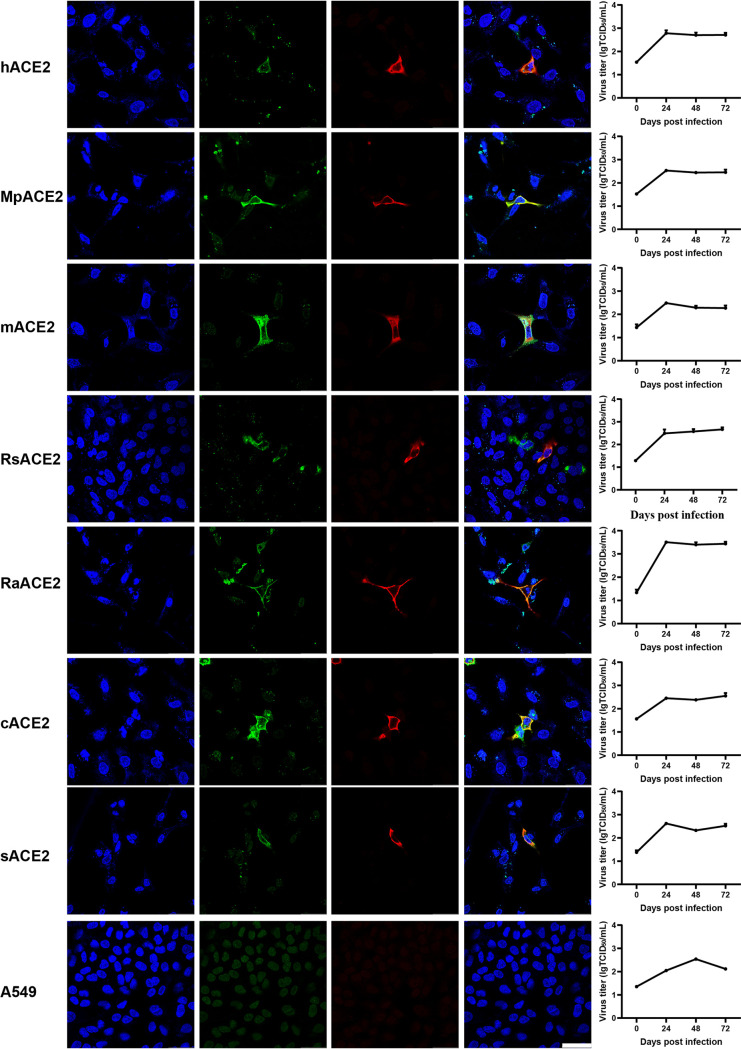
The receptor-binding domain (RBD) of MpCoV-GX has been shown to interact with ACE2 from 18 mammal species by flow cytometry (23). To validate whether MpCoV-GX can also use ACE2 from different species as a cellular entry receptor, we assessed virus infectivity using A549 cells transiently expressing ACE2 derived from pangolin (Manis pentadactyla), human, bat (Rhinolophus sinicus and Rhinolophus affinis), civet, mouse, and swine. We found that MpCoV-GX used all tested ACE2s for the entry and replicated in the ACE2-expressing cells, indicating that the virus has a wide host range in terms of receptor usage. However, quantitative PCR (qPCR) results showed no obvious virus amplification after 24 h of infection, and only a few ACE2-expressing cells were infected. These results suggested that the replication efficiency and infectivity of MpCoV-GX are restricted in A549 cells (Fig. 1).
FIG 1.
Analysis of the ACE2 usage spectrum of MpCoV-GX using immunofluorescence assay and RT-qPCR. A549 cells with and without expression of ACE2 from different species were infected with MpCoV-GX at an MOI of 1.0. ACE2 expression was detected using a mouse anti-S tag monoclonal antibody and an FITC-labeled goat anti-mouse IgG(H+L). At 24 hpi, virus replication was detected using rabbit serum against the SARSr-CoV-Rp3 Np and a Cy3-conjugated goat anti-rabbit IgG. Nuclei were stained with DAPI. hACE2, human ACE2; MpACE2, Manis pentadactyla ACE2; mACE2, mouse ACE2; RsACE2, Rhinolophus sinicus ACE2; RaACE2, Rhinolophus affinis ACE2; sACE2, swine ACE2; cACE2, civet ACE2; A549, no ACE2 expression. The columns (from left to right) show staining of nuclei (blue), ACE2 expression (green), virus replication (red), merged triple-stained images, and RT-qPCR results, respectively (n = 3). Error bars represent the standard error. Staining patterns were examined using a confocal microscope. Bars, 40 μm.
MpCoV-GX RBD binds to human, pangolin, and mouse ACE2 with different efficiencies.
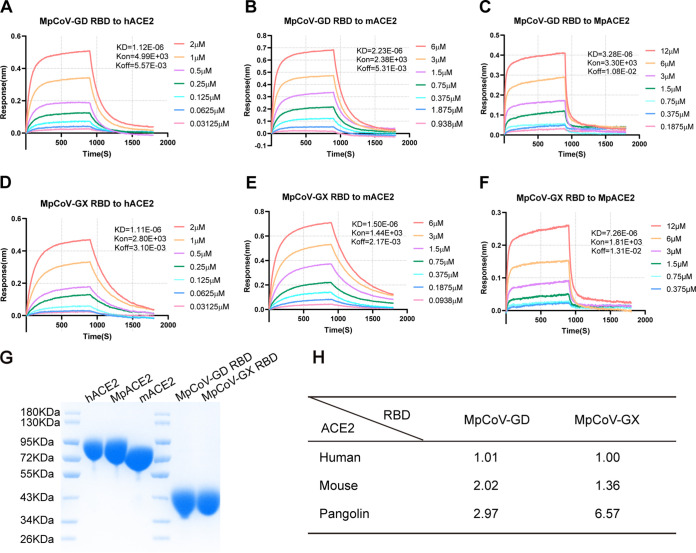
The pangolin coronaviruses from Guangxi and Guangdong were both isolated from Malayan pangolins. Because the live virus of MpCoV-GD was not available, we compared the binding affinities of MpCoV-GX and the MpCoV-GD RBD to human and mouse ACE2. We expressed the RBDs from MpCoV-GX/GD and ACE2s from humans, mice, and Malayan pangolins in HEK293-F cells and then tested the binding affinity between them based on the equilibrium dissociation constant (KD) using biolayer interferometry (Fig. 2G). Both RBDs from MpCoV-GX and -GD bound ACE2s from humans, pangolins, and mice (Fig. 2A to F), with the binding affinity to human ACE2 higher than that to mouse ACE2, and the lowest affinity to pangolin ACE2 (Fig. 2H).
FIG 2.
Binding affinities of MpCoV-GX and -GD RBDs to human, mouse, and pangolin ACE2s. (A to C) Binding of MpCoV-GD RBD protein to human, mouse, and pangolin ACE2s. (D to F) Binding of MpCoV-GX RBD protein to human, mouse, and pangolin ACE2s. (G) Purified ACE2 proteins from humans (hACE2), pangolins (MpACE2), and mice (mACE2) and RBD proteins from MpCoV-GD (MpCoV-GD RBD) and MpCoV-GX (MpCoV-GX RBD). (H) Comparison of dissociation constants (KD) between MpCoV-GD and -GX RBDs and human, mouse, and pangolin ACE2s. Binding was analyzed in comparison to MpCoV-GX RBD binding to human ACE2.
Replication kinetics of MpCoV-GX in different cell lines.
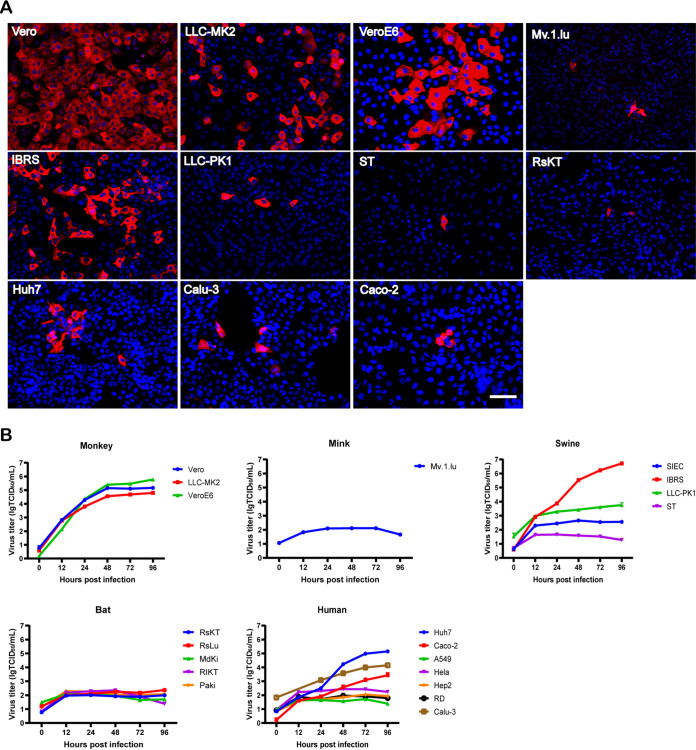
To assess the cross-species infection potential of MpCoV-GX, we inoculated the virus in 30 cell lines derived from different tissues or organs of different host species at a multiplicity of infection (MOI) of 0.1 (Table 1). Of the seven tested human cell lines (Table 1), Caco-2, Calu-3, and Huh7 were susceptible to MpCoV-GX, with the latter two being more supportive. All three cell lines derived from nonhuman primates (Vero, LLC-MK2, and VeroE6) showed high virus replication throughout the 96 h. MpCoV-GX showed stronger replication capacity in Vero cells than in VeroE6 cells from 12 h postinfection (hpi) to 48 hpi. Two (IBRS and LLC-PK1) of the four tested swine cell lines were susceptible to MpCoV-GX infection, with the former being more supportive. In contrast, cells derived from Rhinolophus (RsKT) and mink (Mv.1.lu) showed limited susceptibility to MpCoV-GX infection (Fig. 3A and B).
TABLE 1.
Cell lines derived from different hosts that were tested for susceptibility to MpCoV-GX
| Host | Cell line | Origin of cell line | Expression of N protein at 24 hpia |
|---|---|---|---|
| Human | A549 | Homo sapiens lung | − |
| HeLa | H. sapiens cervix | − | |
| Calu-3 | H. sapiens lung | ++ | |
| Hep2 | H. sapiens larynx | − | |
| Huh7 | H. sapiens liver | ++ | |
| RD | H. sapiens muscle | − | |
| Caco-2 | H. sapiens colon | + | |
| Monkey | Vero | Chlorocebus aethiops kidney | ++++ |
| LLC-MK2 | Macaca mulatta kidney | +++ | |
| VeroE6 | Chlorocebus aethiops kidney | ++++ | |
| Bat | RsKT | Rhinolophus sinicus kidney | + |
| RsLub | R. sinicus lung | − | |
| MdKib | Myotis davidii kidney | − | |
| RlKT | Rousettus leschenaultii kidney | − | |
| PaKi | Pteropus alecto kidney | − | |
| Swine | ST | Sus scrofa testicle | + |
| SIEC | Sus scrofa intestinal | − | |
| IBRS | Sus scrofa kidney | ++++ | |
| LLC-PK1 | Sus scrofa kidney | + | |
| Dog | MDCK | Canis familiaris kidney | − |
| Mink | Mv.1.lu | Mustela vison lung | + |
| Hamster | BHK21 | Mesocricetus auratus kidney | − |
| V79 | Cricetulus griseus lung | − | |
| CHO | Cricetulus griseus ovary | − | |
| Rat | BRL | Rattus norvegicus liver | − |
| H9c2 | Rattus norvegicus heart | − | |
| Mouse | L929 | Mus musculus lung | − |
| MS1 | Mus musculus pancreas | − | |
| SYF | Mus musculus embryo | − | |
| FL83B | Mus musculus liver | − |
Expression efficiency of N protein is defined by Cy3-positive cell ratio: −, 0; +, 1% to 5%; ++, >5% to 25%; +++, >25% to 50%; ++++, >50%.
Primary cell lines.
FIG 3.
Analysis of the host range and replication efficiency of MpCoV-GX. (A) Virus Np was detected at 24 hpi by an immunofluorescence assay using rabbit serum against the SARSr-CoV-Rp3 Np and a Cy3-conjugated goat anti-rabbit IgG. Nuclei were stained with DAPI. Bars, 100 μm. (B) Virus replication was detected by RT-qPCR in supernatants of virus-infected cells. Cell line information is provided in Table 1.
Clinical manifestation and pulmonary pathological features of MpCoV-GX infection in different mouse models.
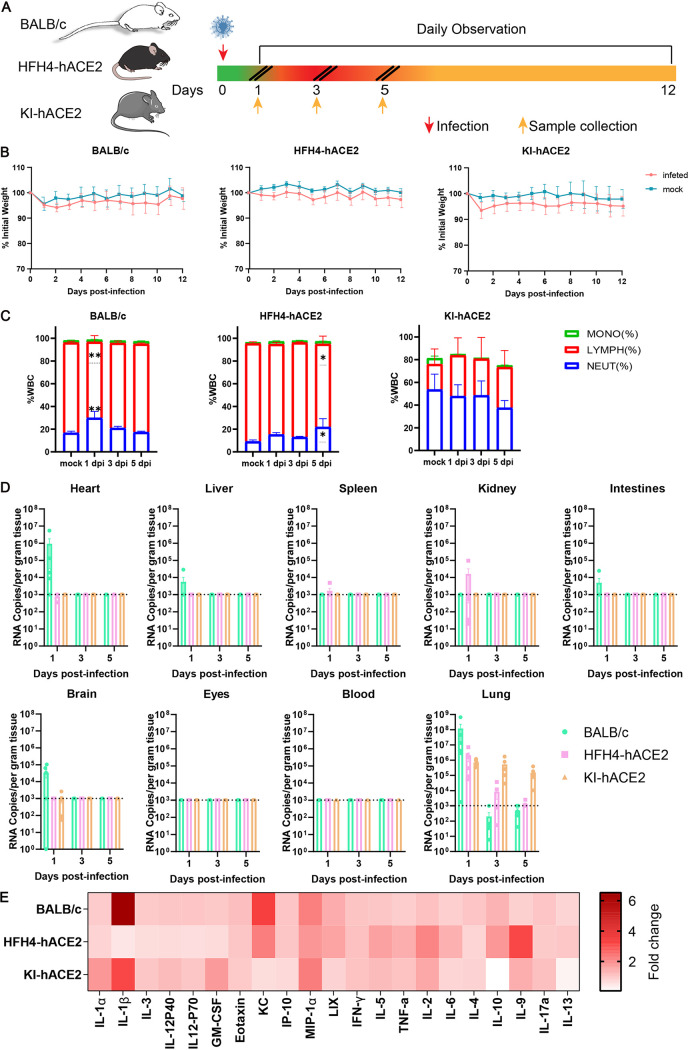
To assess the pathogenicity of MpCoV-GX in mice, wild-type BALB/c mice (8 weeks of age), human ACE2-transgenic mice (8 to 12 weeks of age), and human ACE2 knock-in mice (10 to 12 months of age) were intranasally inoculated with 4 × 105 median tissue culture infectious doses (TCID50) of the virus. Each type of mice was divided into two groups, one for monitoring weight change and the survival rate (n = 8) and one for tissue sampling at various time points after infection (n = 18). In addition, four and six mice were mock infected as corresponding control groups. Six mice (three males and three females) were sacrificed for collection of tissue and blood samples at 1, 3, and 5 days postinfection (dpi) (Fig. 4A). In the survival monitoring groups, the body weight of the infected animals showed a slight decrease from 1 dpi but later recovered in the wild-type and hACE2-KI groups, while there was no obvious body weight change in the HFH4-hACE2 group (Fig. 4B). All infected mice survived to the endpoint without evident symptoms. Mouse peripheral blood was used for routine blood analysis up to 5 dpi. In the infected BALB/c group, neutrophils increased at 1 dpi, decreased at 3 dpi, and recovered to the normal level at 5 dpi, whereas lymphocytes decreased at 1 dpi, increased at 3 dpi, and returned to the normal level at 5 dpi, compared with those in control mice (Fig. 4C). Infected HFH4-hACE2 mice showed the same changes in neutrophils and lymphocytes, except that neutrophils increased and lymphocytes decreased at 5 dpi (Fig. 4C). There were no obvious differences in the blood between infected KI-hACE2 mice and the control group (Fig. 4C).
FIG 4.
Clinical symptoms, viral replication, and tissue distribution of MpCoV-GX. (A) Mice were mock infected (n = 4) or intranasally infected (n = 8) with 4 × 105 TCID50 of MpCoV-GX and sacrificed for tissue and blood sampling at 1, 3, and 5 dpi. (B) Mouse body weight was monitored until 12 dpi. (C) Peripheral blood was collected from mock-infected (n = 6) and virus-infected mice at 1 (n = 6), 3 (n = 6), and 5 (n = 6) dpi. The white blood cell (WBC) population was measured using a ProCyte Dx hematology analyzer. Error bars indicate the standard error. Statistical significance was measured by two-way analysis of variance in comparison with the mock infection group. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Mice (n = 6) were sacrificed and tissue (heart, liver, spleen, lung, kidney, intestine, brain, and eyes) and blood samples were collected at 1, 3, and 5 dpi, respectively, and viral RNA copy numbers were determined using RT-qPCR. (E) Serum cytokine/chemokine heatmap in infected mice at 1 dpi. GM-CSF, granulocyte-macrophage colony-stimulating factor; IFN-γ, gamma interferon; TNF-a, tumor necrosis factor alpha.
We investigated the replication capability of MpCoV-GX using qPCR. Viral RNA was detected in the heart, liver, lungs, intestines, and brain of wild-type BALB/c mice (Fig. 4D) and in the lungs and kidneys of HFH4-hACE2 mice (Fig. 4D). In infected KI-hACE2 mice, viral RNA copies were detected only in the lung tissue (Fig. 4D). Viral loads were the highest in the lungs in all infected groups. Virus clearance was rapid in BALB/c and HFH4-hACE2 lung tissues; viral RNA was barely detectable at 3 dpi. In the lung tissues of infected KI-hACE2 mice, viral shedding was maintained until 5 dpi (Fig. 4D).
Furthermore, we quantified 21 cytokines/chemokines in sera collected at 1 dpi using a Luminex cytokine assay. The results demonstrated that MpCoV-GX infection altered the cytokine/chemokine profiles. For instance, MIP-1α and interleukin-2 (IL-2) expression was increased in all three models. Notably, IL-1β, a major inflammatory indicator, was highly elevated in BALB/c mice (Fig. 4E).
To monitor viral shedding in the infected animals, oropharyngeal and anal swabs were collected from mice in each survival monitoring group until 5 dpi. Viral RNA was detected in the oropharyngeal swabs of 6 out of 8 BALB/c mice and 4 out of 8 HFH4-hACE2 mice at 1 dpi and in 2 BALB/c mice at 2 dpi (Table 2). No viral RNA was detected in anal swabs in these two groups (Table 2). As no efficient virus replication was detected in oropharyngeal and anal swabs in the BALB/c and HFH4-hACE2 mouse models, we did not assess virus shedding in oropharyngeal or anal swabs from KI-hACE2 mice.
TABLE 2.
Viral shedding by BALB/c and HFH4-hACE2 mice of MpCoV-GXa
| Mouse model | No. of positive mice/total no. |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Oropharyngeal swabb |
Anal swab |
|||||||||
| 1 dpi | 2 dpi | 3 dpi | 4 dpi | 5 dpi | 1 dpi | 2 dpi | 3 dpi | 4 dpi | 5 dpi | |
| BALB/c | 6/8 | 2/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 |
| HFH4-hACE2 | 4/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 |
| KI-hACE2 | — | — | — | — | — | — | — | — | — | — |
Wild-type BALB/c and HFH4-hACE2 mice were intranasally inoculated with 4 × 105 TCID50 of MpCoV-GX.
Oropharyngeal and anal swabs from mice (n = 8) were used for viral RNA detection. —, not determined.
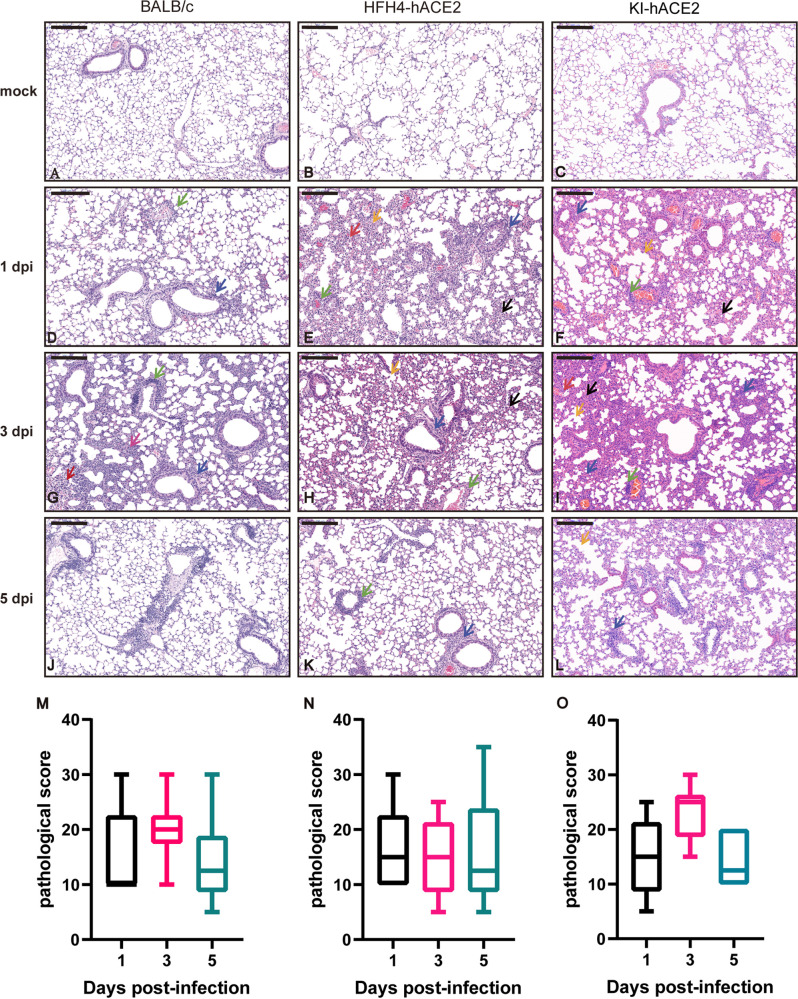
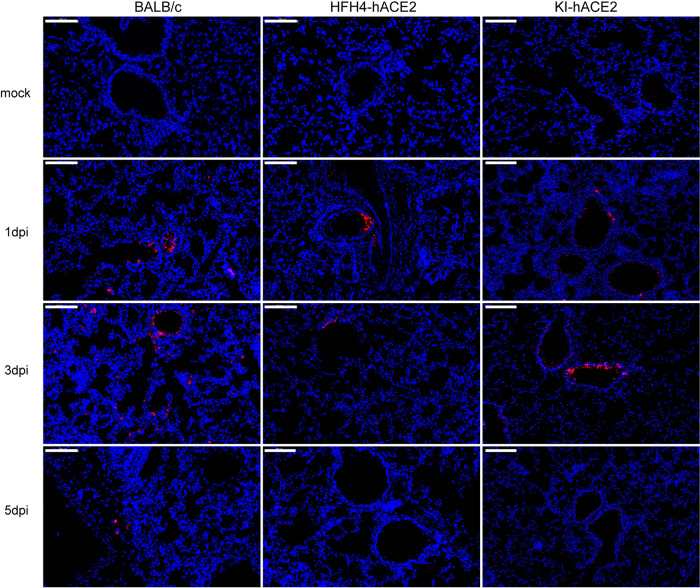
We analyzed lung tissue damage using hematoxylin and eosin (H&E) staining (Fig. 5). The infected mice showed different levels of inflammatory cell infiltration into the lungs during the course of infection, while there was no obvious tissue damage in mock-infected lungs (Fig. 5A to C). At 1 dpi, virus-infected lungs displayed a few lesions of focal interstitial pneumonitis composed of monocytes and lymphocytes, but they were not consistent in all mice. Infected lungs displayed a few thickened alveolar walls with monocyte and lymphocyte infiltration, and increased numbers of macrophages and lymphocytes in some alveolar spaces, with minor fibrin exudation (Fig. 5D to F). Moderate interstitial pneumonia appeared at 3 dpi, including multifocal lesions, increased inflammatory cells (lymphocytes and monocytes) in the peribronchial and perivascular regions, and fibrin exudation and protein-rich edema in some alveoli (Fig. 5G to I). Mild peribronchial and perivascular infiltration was observed in all mice at 5 dpi (Fig. 5J to L). Semiquantitative pathological scoring was in line with the H&E pathological observations in the three mouse models. No pathological changes were observed in the mock-infected mice (Fig. 5M to O). Using an immunofluorescence assay with a cross-reaction polyclonal antibody against a bat SARSr-CoV Rp3 nucleocapsid protein (Np) prepared in-house, viral antigens were detected in the bronchial epithelial and alveolar cells of lungs with higher viral loads (Fig. 6).
FIG 5.
Histopathological changes and pathological scoring in the three mouse models. Six euthanized mice were used to examine pathological changes in the lungs at 1, 3, and 5 dpi. (A to C) H&E staining of mock-infected BALB/c, HFH4-hACE2, and KI-hACE2 mouse lungs. (D to F) H&E staining of virus-infected BALB/c and HFH4-hACE2 mouse lungs at 1 dpi showing inflammatory cells in the peribronchial (blue arrows) and perivascular (green arrows) regions. There are some damaged alveoli (orange arrows) and thickened alveoli (black arrows) in HFH4-hACE2 and KI-hACE2 mouse lungs. (G to I) H&E staining of virus-infected BALB/c mouse showing slightly peribronchial (blue arrows) and perivascular (green arrows) infiltration at 1dpi. Moderate peribronchial (blue arrows) and perivascular (green arrows) infiltration and interstitial pneumonia were observed at 3 dpi, with inflammatory cell infiltration (pink arrow), and focal hemorrhage (red arrows). HFH4-hACE2 mouse showing interstitial pneumonia with thickened alveolar septa (black arrows) and damaged alveoli (orange arrows). Moderate peribronchial (blue arrows) and perivascular (green arrows) infiltration and interstitial pneumonia were observed at 3 dpi, with thickened alveolar septa (black arrows) and inflammatory cell infiltration (pink arrow), damaged alveoli (orange arrows), and focal hemorrhage (red arrows). (J to L) H&E staining showing mild peribronchial (blue arrows) and perivascular (green arrow) infiltration at 5 dpi. Moderate peribronchial (blue arrows) and perivascular (green arrows) infiltration and interstitial pneumonia were observed at 1 dpi in KI-hACE2 mouse lung. In addition to confirming cell infiltration, thickened alveoli (black arrows) were also observed at 3 dpi. Damaged alveoli (orange arrows) were observed only in KI-hACE2 mice at 5 dpi. (M to O) Semiquantitative analysis of the pathology of the H&E-stained lung section from infected mice 1 (n = 6), 3 (n = 6), and 5 (n = 6) dpi. Error bars indicate the standard error. Images were collected using a Pannoramic MIDI system. Bars, 200 μm.
FIG 6.
Antigen detection in mice after MpCoV-GX infection. Lung sections of wild-type BALB/c mice, HFH4 mock-infected mice, and KI-hACE2 mice were examined. Viral antigen was detected in lung bronchi and alveoli collected at 1, 3, and 5 dpi, using a SARSr-CoV-Rp3 Np polyclonal antibody. Images were acquired using the Pannoramic MIDI system. Bars, 100 μm.
DISCUSSION
Before the COVID-19 pandemic, we and other groups reported that horseshoe bats carried a large number of SARSr-CoVs with great genetic diversity, some of which used human ACE2 as a receptor, indicating their interspecies transmission potential (24, 25). Infection experiments have demonstrated that some bat SARSr-CoVs can induce pneumonia in human ACE2-transgenic mice but have low pathogenicity compared to SARS-CoV (26, 27). Therefore, a thorough risk assessment of novel SARSr-CoVs and other unknown viruses in wildlife reservoirs based on in vitro and in vivo virus infection data is important for prevention of future disease emergence and preparation of countermeasures.
In this study, we first performed in vitro infection with MpCoV-GX in A549 cells expressing ACE2 from humans, pangolins, Rhinolophus bats, civets, or swine and found that all of them had limited susceptibility to virus replication (Fig. 1). In addition, we demonstrated that some cell lines from humans, nonhuman primates, Chinese horseshoe bats, swine, and minks were susceptible to MpCoV-GX but with different replication efficiencies (Fig. 3). Our results are consistent with previous results showing that both MpCoV-GX and -GD S RBDs have a high binding affinity to human ACE2 (22, 28). Most SARSr-CoVs, including SARS-CoV and SARS-CoV-2, cannot infect or have limited infection potential in wild-type mice (29–32). Therefore, human ACE2-transgenic mice have been widely used to evaluate the pathogenicity of this group of viruses instead (32–35). In this study, we demonstrated that MpCoV-GX successfully infected three mouse models, including wild-type BALB/c mice, human ACE2-transgenic mice, and human ACE2 knock-in mice, though the infected animals presented no obvious clinical symptoms (Fig. 4) and only slight histopathological changes in the lungs (Fig. 5). Taken together, the in vitro and in vivo experimental data suggest that pangolin SARSr-CoV-2 poses a risk of interspecies infection to humans and other animals, though it may not be a highly pathogenic strain.
Pangolin SARSr-CoV-2 viruses are more distantly related to SARS-CoV-2 than RaTG13 and the cluster of BANAL viruses from bats in Laos. However, studies based on protein interaction and pseudovirus infection have shown that MpCoV-GX and -GD exhibit higher RBD binding affinity to human ACE2 and higher infectivity in SARS-CoV-2-permissive cell lines than RaTG13, which showed almost no infectivity in various cell lines (22, 36). Furthermore, the S proteins of the pangolin coronaviruses can use ACE2 receptors from a broader range of host species than can the S protein of RaTG13 (23, 36). These findings, along with the results in this study, suggest that despite their lower genetic similarity to SARS-CoV-2, the interspecies infection risk of pangolin coronaviruses cannot be underestimated. Besides MpCoV-GX characterized in this study, special attention should be paid to MpCoV-GD because of its wide range of ACE2 usage and high infectivity in cell lines from a wide variety of animals and humans (36). Our study highlights the need for continued surveillance and characterization of coronaviruses among pangolins and animals or humans exposed to pangolins in order to identify any novel variants of SARSr-CoV-2 and understand their risk of interspecies transmission. Besides bats and humans, coronaviruses can infect a wide range of domestic and wild animals, including pigs, birds, cats, ferrets, and dogs (37–41). We propose that for better prevention of and precautions against future emerging coronaviruses, extensive coronavirus surveillance studies in animal hosts need to be pursued worldwide. Such surveillance should target not only free-ranging wildlife but also captured or smuggled species such as pangolins, as well as domestic animals with potential exposure to wildlife. In addition, wildlife managers, customs officers, and quarantine-related personnel should be given priority in concern because these people have higher chances of contact with animals and are more likely to be infected.
In this century, human society has encountered several emerging infectious diseases unpreparedly, including the SARS and COVID-19 pandemics, suffering tremendous losses to public health and the global economy. The lesson of these pandemics has convinced us of the necessity of preparing prophylactic and therapeutic methods against viruses that may cause new diseases but have yet to emerge. Considering the spillover potential of MpCoV-GX, we conducted preliminary antiviral drug screening for this novel coronavirus and identified six monomeric compounds that exhibited efficient anti-MpCoV-GX and anti-SARS-CoV-2 activities from the Anti-COVID-19 Traditional Chinese Medicine Compound Library, including artemether, artesunate, arteannuin B, echinatin, licochalcone B, and andrographolide (42). Similar researches aimed at identifying and developing effective antiviral agents, as well as antibodies and vaccines against novel SARSr-CoVs of animal origin, should be advocated in the future, as the technical reserve of these countermeasures would be highly valuable for preparedness for the next coronavirus pandemic.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Thirty cell lines (Table 1) derived from different host species, including human (A549, HeLa, Hep2, Huh7, RD, Calu-3, Caco-2), monkey (Vero, LLC-MK2, VeroE6), bat (RsKT, RsLu, MdKi, RlKT, PaKi), swine (ST, SIEC, IBRS, LLC-PK1), dog (MDCK), mink (Mv.1.lu), hamster (BHK21, V79, CHO), rat (BRL, H9c2), and mouse (L929, MS1, SYF, FL83B), were tested for susceptibility and viral replication efficiency. All bat cells were developed in-house. RsKT and RlKT cells derived from R. sinicus and Rousettus, respectively, and PaKi cells derived from Pteropus were kindly provided by Linfa Wang (Duke-NUS Medical School, Singapore). These are continuous cell lines that stably express the large T antigen of simian virus 40. ST cells were kindly provided by Shaobo Xiao (College of Veterinary Medicine, Huazhong Agricultural University). Other cell lines were obtained from the National Virus Resource Center of the Wuhan Institute of Virology (Wuhan, China). Depending on the cell type, cells were maintained in minimum Roswell Park Memorial Institute 1640, Dulbecco’s modified Eagle’s medium (DMEM), or DMEM–F-12 (all from Gibco), according to the supplier’s instructions. The cell lines were routinely tested for mycoplasma and were maintained mycoplasma free. The cells were maintained at 37°C in the presence of 5% CO2. MpCoV-GX (GenBank accession no. MT040334.1) was isolated in 2019 (14). A rabbit polyclonal antibody against SARSr-CoV Rp3 Np, which shares >90% amino acid identity with SARS-CoV-2, was developed in-house.
Expression constructs, protein expression, and purification.
Codon-optimized S RBDs of MpCoV-GD (S residues: amino acids [aa] 326 to 579, accession no. MT121216) and MpCoV-GX (S residues: aa 330 to 583, accession no. MT040334.1) were synthesized at Sangon Biotech (Shanghai, China) and cloned into an expression vector with an N-terminal signal peptide and a C-terminal S tag, as previously described (28). The ectodomains of human ACE2 (aa 19 to 615, accession no. AB046569), Malayan pangolin ACE2 (aa 19 to 615, accession no. XM_017650263.2), and mouse ACE2 (aa 19 to 615, accession no. NM_001130513.1) were amplified or synthesized and cloned into the expression vector with an N-terminal signal peptide and a C-terminal S tag.
The RBD and ACE2 proteins used in the biolayer interferometry-based binding assay were produced in HEK293-F cells. The cells were transiently transfected with expression plasmids using PEI Max transfection-grade linear polyethylenimine hydrochloride (catalog no. 24765-1; Life Technologies) and cultured at 37°C in a humidified 5% CO2 incubator. Supernatants were harvested when the cell viability was 95% and centrifuged at 4,000 × g and 4°C for 10 min. The clarified supernatants were purified using S tag agarose beads and eluted with 3 M MgCl2. The purified protein was buffered with phosphate-buffered saline (PBS), quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA), and stored at −80°C until use.
Biolayer interferometry binding assays.
The RBD-ACE2 binding assays were performed using the Octet Red system (ForteBio, Menlo Park, CA, USA) in 96-well microplates at 30°C with shaking at 1,000 rpm. The RBD was biotinylated using EZ-Link NHS-LC-LC-biotin (Thermo Fisher Scientific). Streptavidin biosensors were activated for 200 s before coupling with 40 μg/mL biotinylated RBD proteins for 600 s. A baseline was collected in kinetic buffer (1 M NaCl, 0.1% bovine serum albumin [BSA], 0.02% Tween 20; pH 6.5) for 200 s before immersing the sensors in 1:2 serially diluted ACE2 protein for 900 s and was then dissociated in the same kinetic buffer for another 900 s. Data analysis included reference subtraction. Interstep correction and Y-alignment were used to minimize tip-dependent variability. Curve fitting was performed in a 1:1 model using Octet data analysis software v7.1 (ForteBio). Mean Kon and Koff values were determined by applying a global fit to all data. The coefficient of determination (R2) for the interactions was close to 1.0, as reported previously (28).
Testing of the ACE2 receptor usage for MpCoV-GX infection.
The ACE2-expressing plasmids were maintained in our laboratory. A549 cells transiently expressing ACE2 were prepared using FuGENE HD (Promega, USA) in a confocal dish, and vector-transfected cells were used as controls. MpCoV-GX cultured in VeroE6 cells was used for infection at an MOI of 1. After adsorption at 37°C, the supernatant was removed and the cells were washed twice with PBS and incubated in medium. At 24 h postinfection (hpi), the cells were washed with PBS and fixed with 4% paraformaldehyde at room temperature for 30 min. ACE2 expression was detected using a mouse anti-S tag monoclonal antibody (made in-house) and a fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG(H+L) (ab96879; Abcam). Viral replication was detected using a rabbit antibody against the Rp3 Np (1:2,000) and a Cy3-conjugated goat anti-rabbit IgG (1:200, ab6939; Abcam). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime). The staining patterns were examined by confocal microscopy using a Stellaris 8 WLL microscope (Leica).
Immunofluorescence assay.
Cell susceptibility was determined by an immunofluorescence assay targeting Rp3 Np in virus-incubated cells. Briefly, cells in 24-well plates at 80% confluence were inoculated with MpCoV-GX (MOI = 0.1). After 1 h, the infected cells were washed with PBS and maintained in DMEM or DMEM–F-12 (Gibco) supplemented with 2% fetal bovine serum. At 24 hpi, the cells were fixed with 4% paraformaldehyde at room temperature for 30 min and permeabilized with 0.1% Triton X-100. The cells were blocked with bovine serum albumin (5%) diluted in PBS for 1 h and then incubated with rabbit anti-Rp3 Np polyclonal antibody for 1 h. After three washes with PBS, the cells were incubated with Cy3-conjugated anti-rabbit IgG for 1 h, followed by DAPI staining for 15 min. After three washes with PBS, fluorescence images were acquired and examined using an AMF4300 Evos FL cell imaging system (Life Technologies, Carlsbad, CA, USA).
Animal infection experiments.
All animals infected with MpCoV-GX were handled in biosafety level 3 animal facilities in accordance with the recommendations for animal care and use of the Institutional Review Board of the Wuhan Institute of Virology of the Chinese Academy of Sciences (CAS; study approval number WIVA05202014). Mice were inoculated with the virus under proper anesthesia, and efforts were made to minimize any potential pain and distress. Eight- to 12-week-old wild-type BALB/c mice (Laboratory Animal Centre of the Wuhan Institute of Virology, CAS), HFH4-hACE2 mice (kindly provided by Ralph Baric from the University of North Carolina), and KI-hACE2 mice (generated at Shanghai Model Organisms Center Inc. and bred in the Laboratory Animal Centre of the Wuhan Institute of Virology, CAS) were randomly divided into two groups: (i) a survival monitoring group comprising 12 mice for body weight and survival monitoring, in which 4 males and 4 females were inoculated with the virus and 2 males and 2 females were inoculated with DMEM as a control, and (ii) a pathology progression group comprising 24 mice for testing of virus replication and observation of pathological changes, in which 9 males and 9 females were inoculated with the virus and 3 males and 3 females were inoculated with DMEM as a control. Mice were anesthetized with 250 mg avertin (Sigma-Aldrich, St. Louis, MO, USA) per kg of body weight before they were inoculated intranasally with 4 × 105 TCID50 of MpCoV-GX or DMEM as a mock control. Clinical symptoms and body weight in the survival monitoring group were recorded daily for up to 12 days. Virus replication was determined in tissues collected from 6 mice at 1, 3, and 5 dpi.
RNA extraction and RT-qPCR.
Cell lines derived from different hosts in 6-well plates were inoculated with the virus at an MOI of 0.1. Two hundred microliters of supernatant was collected at 0, 12, 24, 48, 72, and 96 hpi. RNA was extracted using a QIAcube HT instrument (Qiagen, Hilden, Germany). Mouse organs were homogenized in DMEM, and viral RNA was extracted using the QIAamp viral RNA minikit (Qiagen). A one-step quantitative reverse transcription-PCR (RT-qPCR) assay targeting the RNA-dependent RNA polymerase (RdRp) gene was used to detect MpCoV-GX using AgPath-ID one-step RT-PCR reagent (Applied Biosystems, USA). Specific primers for MpCoV-GX RdRp (forward, 5′-CAAAATGTGATAGAGCCATGCC-3′; reverse, 5′-GAAACGATGTGACAAACTACAGC-3′) and a fluorescein-labeled probe (5′-6-carboxyfluorescein [FAM]-TGCGAGCAAGAACAAGAGAAGCCA-BHQ1-3′) were designed according to the reference sequence (accession no. MT072864.1). Reaction mixtures were incubated at 50°C for 10 min, followed by 95°C for 5 min, and then underwent thermal cycling at 95°C for 15 s and 55°C for 40 s for 45 cycles. RNA from diluted live virus of the purified MpCoV-GX stock and a plasmid containing the RdRp gene of the virus were used as standards.
Histological analysis.
Lung samples were collected in formalin and embedded in paraffin for pathological analysis. Fixed samples were used for H&E staining, and immunofluorescence assay was performed for MpCoV-GX antigen detection. Optimized protocols were used in the H&E and immunohistochemical staining as previously described (33).
ACKNOWLEDGMENTS
We thank Fan Zhang and Xue-Fang An from the Animal Center of the Wuhan Institute of Virology for transgenic mouse breeding. We thank Tao Du and Jin Xiong from the Center for Biosafety Mega-Science for essential support. We thank Ding Gao of the WIV Core Facility and Technical Support for help with the Octet Red technology.
This study was supported by the Strategic Priority Research Program of the CAS (XDB29010101), the Key Program of the CAS (KJZD-SW-L11) (to Z.-L.S.), and the National Natural Science Foundation of China (32070180 to X.-L.Y.).
Contributor Information
Yi-Gang Tong, Email: tongyigang@mail.buct.edu.cn.
Zheng-Li Shi, Email: zlshi@wh.iov.cn.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. 2020. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group . 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 6.Zhong NS, Zheng BJ, Li YM, Poon, Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ, Xu J, Li DX, Yuen KY, Peiris, Guan Y. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362:1353–1358. 10.1016/s0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 8.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu B, Guo H, Zhou P, Shi ZL. 2021. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 19:141–154. 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC, Hughes AC, Bi Y, Shi W. 2020. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol 30:2196–2203. 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, Kaewpom T, Chia WN, Ampoot W, Lim BL, Worachotsueptrakun K, Chen VC, Sirichan N, Ruchisrisarod C, Rodpan A, Noradechanon K, Phaichana T, Jantarat N, Thongnumchaima B, Tu C, Crameri G, Stokes MM, Hemachudha T, Wang LF. 2021. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun 12:972. 10.1038/s41467-021-21240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami S, Kitamura T, Suzuki J, Sato R, Aoi T, Fujii M, Matsugo H, Kamiki H, Ishida H, Takenaka-Uema A, Shimojima M, Horimoto T. 2020. Detection and characterization of bat sarbecovirus phylogenetically related to SARS-CoV-2, Japan. Emerg Infect Dis 26:3025–3029. 10.3201/eid2612.203386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, Li N, Guo Y, Li X, Shen X, Zhang Z, Shu F, Huang W, Li Y, Zhang Z, Chen RA, Wu YJ, Peng SM, Huang M, Xie WJ, Cai QH, Hou FH, Chen W, Xiao L, Shen Y. 2020. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 583:286–289. 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 14.Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, Li WJ, Jiang BG, Wei W, Yuan TT, Zheng K, Cui XM, Li J, Pei GQ, Qiang X, Cheung WY, Li LF, Sun FF, Qin S, Huang JC, Leung GM, Holmes EC, Hu YL, Guan Y, Cao WC. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583:282–285. 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 15.Delaune D, Hul V, Karlsson EA, Hassanin A, Ou TP, Baidaliuk A, Gambaro F, Prot M, Tu VT, Chea S, Keatts L, Mazet J, Johnson CK, Buchy P, Dussart P, Goldstein T, Simon-Loriere E, Duong V. 2021. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat Commun 12:6563. 10.1038/s41467-021-26809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chretien D, Sanamxay D, Xayaphet V, Paphaphanh P, Lacoste V, Somlor S, Lakeomany K, Phommavanh N, Perot P, Dehan O, Amara F, Donati F, Bigot T, Nilges M, Rey FA, van der Werf S, Brey PT, Eloit M. 2022. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 604:330–336. 10.1038/s41586-022-04532-4. [DOI] [PubMed] [Google Scholar]
- 17.Li LL, Wang JL, Ma XH, Sun XM, Li JS, Yang XF, Shi WF, Duan ZJ. 2021. A novel SARS-CoV-2 related coronavirus with complex recombination isolated from bats in Yunnan province, China. Emerg Microbes Infect 10:1683–1690. 10.1080/22221751.2021.1964925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li WH, Moore MJ, Vasilieva N, Sui JH, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang YJ, Luo CM, Tan B, Wang N, Zhu Y, Crameri G, Zhang SY, Wang LF, Daszak P, Shi ZL. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503:535–538. 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang XL, Hu B, Wang B, Wang MN, Zhang Q, Zhang W, Wu LJ, Ge XY, Zhang YZ, Daszak P, Wang LF, Shi ZL. 2015. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J Virol 90:3253–3256. 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wrobel AG, Benton DJ, Xu P, Calder LJ, Borg A, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. 2021. Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2. Nat Commun 12:837. 10.1038/s41467-021-21006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Qiao S, Yu J, Zeng J, Shan S, Tian L, Lan J, Zhang L, Wang X. 2021. Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution. Nat Commun 12:1607. 10.1038/s41467-021-21767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu S, Wang J, Bai B, Wu L, Zheng A, Chen Q, Du P, Han P, Zhang Y, Jia Y, Qiao C, Qi J, Tian WX, Wang HW, Wang Q, Gao GF. 2021. Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin. EMBO J 40:e107786. 10.15252/embj.2021107786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, Xie JZ, Shen XR, Zhang YZ, Wang N, Luo DS, Zheng XS, Wang MN, Daszak P, Wang LF, Cui J, Shi ZL. 2017. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog 13:e1006698. 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui J, Li F, Shi ZL. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192. 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menachery VD, Yount BL, Jr, Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge XY, Donaldson EF, Randell SH, Lanzavecchia A, Marasco WA, Shi ZL, Baric RS. 2015. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21:1508–1513. 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menachery VD, Yount BL, Jr, Sims AC, Debbink K, Agnihothram SS, Gralinski LE, Graham RL, Scobey T, Plante JA, Royal SR, Swanstrom J, Sheahan TP, Pickles RJ, Corti D, Randell SH, Lanzavecchia A, Marasco WA, Baric RS. 2016. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci USA 113:3048–3053. 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo H, Hu B, Si HR, Zhu Y, Zhang W, Li B, Li A, Geng R, Lin HF, Yang XL, Zhou P, Shi ZL. 2021. Identification of a novel lineage bat SARS-related coronaviruses that use bat ACE2 receptor. Emerg Microbes Infect 10:1507–1514. 10.1080/22221751.2021.1956373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY, Liu SS, Zhang NN, Li XF, Xiong R, Guo Y, Deng YQ, Huang WJ, Liu Q, Liu QM, Shen YL, Zhou Y, Yang X, Zhao TY, Fan CF, Zhou YS, Qin CF, Wang YC. 2020. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28:124–133.e4. 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glass WG, Subbarao K, Murphy B, Murphy PM. 2004. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J Immunol 173:4030–4039. 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- 31.Frieman M, Yount B, Agnihothram S, Page C, Donaldson E, Roberts A, Vogel L, Woodruff B, Scorpio D, Subbarao K, Baric RS. 2012. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J Virol 86:884–897. 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asaka MN, Utsumi D, Kamada H, Nagata S, Nakachi Y, Yamaguchi T, Kawaoka Y, Kuba K, Yasutomi Y. 2021. Highly susceptible SARS-CoV-2 model in CAG promoter-driven hACE2-transgenic mice. JCI Insight 6:e152529. 10.1172/jci.insight.152529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang RD, Liu MQ, Chen Y, Shan C, Zhou YW, Shen XR, Li Q, Zhang L, Zhu Y, Si HR, Wang Q, Min J, Wang X, Zhang W, Li B, Zhang HJ, Baric RS, Zhou P, Yang XL, Shi ZL. 2020. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182:50–58.e8. 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bao L, Deng W, Huang B, Gao H, Liu J, Ren L, Wei Q, Yu P, Xu Y, Qi F, Qu Y, Li F, Lv Q, Wang W, Xue J, Gong S, Liu M, Wang G, Wang S, Song Z, Zhao L, Liu P, Zhao L, Ye F, Wang H, Zhou W, Zhu N, Zhen W, Yu H, Zhang X, Guo L, Chen L, Wang C, Wang Y, Wang X, Xiao Y, Sun Q, Liu H, Zhu F, Ma C, Yan L, Yang M, Han J, Xu W, Tan W, Peng X, Jin Q, Wu G, Qin C. 2020. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583:830–833. 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 35.Lee KS, Wong TY, Russ BP, Horspool AM, Miller OA, Rader NA, Givi JP, Winters MT, Wong ZYA, Cyphert HA, Denvir J, Stoilov P, Barbier M, Roan NR, Amin MS, Martinez I, Bevere JR, Damron FH. 2022. SARS-CoV-2 Delta variant induces enhanced pathology and inflammatory responses in K18-hACE2 mice. PLoS One 17:e0273430. 10.1371/journal.pone.0273430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nie J, Li Q, Zhang L, Cao Y, Zhang Y, Li T, Wu J, Liu S, Zhang M, Zhao C, Liu H, Nie L, Qin H, Wang M, Lu Q, Li X, Liu J, Liang H, Jiang T, Duan K, Yang X, Shen Y, Huang W, Wang Y. 2021. Functional comparison of SARS-CoV-2 with closely related pangolin and bat coronaviruses. Cell Discov 7:21. 10.1038/s41421-021-00256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XL, Wang YY, Xing SZ, Chen YS, Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG, Ma JY. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martina BE, Haagmans BL, Kuiken T, Fouchier RA, Rimmelzwaan GF, Van Amerongen G, Peiris JS, Lim W, Osterhaus AD. 2003. Virology: SARS virus infection of cats and ferrets. Nature 425:915. 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan JF, To KK, Tse H, Jin DY, Yuen KY. 2013. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol 21:544–555. 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z. 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368:1016–1020. 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. 2016. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 24:490–502. 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, Liu M, Qin H, Lin H, An X, Shi Z, Song L, Yang X, Fan H, Tong Y. 2021. Artemether, artesunate, arteannuin B, echinatin, licochalcone B and andrographolide effectively inhibit SARS-CoV-2 and related viruses in vitro. Front Cell Infect Microbiol 11:680127. 10.3389/fcimb.2021.680127. [DOI] [PMC free article] [PubMed] [Google Scholar]