Abstract
A 16S ribosomal DNA (rDNA) clone library from permanently cold marine sediments was established. Screening 353 clones by dot blot hybridization with group-specific oligonucleotide probes suggested a predominance of sequences related to bacteria of the sulfur cycle (43.4% potential sulfate reducers). Within this fraction, the major cluster (19.0%) was affiliated with Desulfotalea sp. and other closely related psychrophilic sulfate reducers isolated from the same habitat. The cloned sequences showed between 93 and 100% similarity to these bacteria. Two additional groups were frequently encountered: 13% of the clones were related to Desulfuromonas palmitatis, and a second group was affiliated with Myxobacteria spp. and Bdellovibrio spp. Many clones (18.1%) belonged to the γ subclass of the class Proteobacteria and were closest to symbiotic or free-living sulfur oxidizers. Probe target groups were further characterized by amplified rDNA restriction analysis to determine diversity within the groups and within the clone library. Rarefaction analysis suggested that the total diversity assessed by 16S rDNA analysis was very high in these permanently cold sediments and was only partially revealed by screening of 353 clones.
Coastal and shelf sediments play a significant role in the remineralization of organic matter. In shelf areas, an estimated 32 to 46% of the primary production settles to the sea floor (54). While part of it is permanently buried, the majority of this detrital material is reoxidized, mainly through the action of prokaryotes (54). Steep redox gradients provide niches for a wide variety of metabolically diverse microorganisms, and O2, NO3−, manganese and iron oxides, and SO42− have been identified as the most important electron acceptors in marine sediments (3, 19). The various processes of microbial carbon mineralization can be quantified by tracer techniques, and their importance for biogeochemical cycles in the marine environment is recognized; however, little is known about the microbial community responsible for them.
Few cultivation-independent studies of microbial diversity in marine sediments have been conducted (6, 15, 22, 43). The sequences recovered in these studies revealed the presence of mainly unknown organisms only distantly related to known isolates. To further uncover microbial diversity in marine shelf sediments and to identify potentially dominant groups in this habitat, we constructed a 16S ribosomal DNA (rDNA) clone library using general bacterial primers to amplify the almost complete gene.
The screening process was tested by statistical analysis to evaluate whether we had covered total diversity in our clone library by screening 353 clones. Species diversity can be considered to be composed of two components: species richness (the number of species in a community) and species evenness (the distribution of levels of abundance among the species). Two types of analyses have been used to assess diversity. Rarefaction is a statistical technique for different applications in an ecological context and gives an estimation of the decrease in apparent species richness of a community with decreasing subsample size (50). A second approach to evaluate whether diversity within a subsample approaches diversity within a sample of infinite size is to calculate coverage (14). Coverage (C) values are calculated by the equation C = 1 − (n/N) × 100, where n is the number of unique clones and N is the total number of clones examined.
We chose to study permanently cold sediments because 90% of the sea floor has temperatures below 4°C (25). During a cruise to the Arctic Ocean in September and October of 1995, several studies of different aspects of microbial life in this habitat, such as the determination of prokaryotic abundance and the profiling of prokaryotic rRNA (47, 48), were conducted. Temperature dependence was determined, rates of polysaccharide hydrolysis (2) and sulfate reduction were measured (46), and psychrophilic sulfate reducers were enriched (23). Furthermore, benthic exchange and mineralization rates were determined (12, 24). All the above-described studies indicated an active microbial community with metabolic rates comparable to those of temperate habitats. Forty-two percent of total benthic mineralization was due to sulfate reduction at the station sampled for the clone library (46). Here and in the accompanying paper (48), we describe the phylogenetic affiliation and diversity of the prokaryotic community and quantify the contribution of sulfate-reducing bacteria (SRB) to the total microbial community.
MATERIALS AND METHODS
Study site.
Sediment samples were collected at Hornsund off the coast of Spitsbergen, Arctic Ocean, in September and October of 1995. The bottom water temperature was 2.6°C (47), and sediments were anoxic below a depth of approximately 8 mm (12). For a detailed description of the sampling procedure, see the report of Sahm and Berninger (47).
DNA extraction and purification.
Total community DNA was extracted directly from the sediment as described by Zhou et al. (55). The protocol encompassed three cycles of freezing and thawing, chemical lysis in a high-salt extraction buffer (1.5 M NaCl) by heating of the suspension in the presence of sodium dodecyl sulfate (SDS) and hexadecyltrimethylammonium bromide, and a proteinase K step. It was slightly modified by performing only two SDS extraction steps.
Aliquots of 2 g of wet sediment of different sections (0 to 2, 3 to 6, and 8 to 11 cm) from duplicate cores were used for DNA extraction. Extracted DNAs were finally combined. The crude DNA was purified with the WIZARD DNA Clean Up System (Promega, Madison, Wis.). DNA yield was quantified photometrically. Per cubic centimeter of wet sediment, 11.5 μg of DNA was recovered. High-molecular-weight DNA was cut out of an agarose gel and extracted with a GeneClean II Kit (Bio 101 Inc., La Jolla, Calif.) by following the manufacturer’s instructions. Approximately 40% of the crude DNA was recovered after this step.
Cell lysis efficiency.
Cell lysis efficiency of the DNA extraction procedure was checked by enumerating the total number of 4′,6-diamidino-2-phenylindole (DAPI)-stained cells in aliquots of sediments taken before and after cell lysis. Ninety-three percent ± 3.4% of the microorganisms were lysed.
PCR amplification of 16S rDNAs.
Two universal bacterial primers, EUB008 (17) and EUB1492 (20), were used to amplify 16S rDNAs from the extracted and purified chromosomal DNAs. PCR was performed with a model PHC-3 Temperature Cycler (Techne, Cambridge, United Kingdom) as follows: 50 pmol of each primer, 2.5 μmol of each deoxyribonucleoside triphosphate, 300 μg of bovine serum albumin, 1× PCR buffer, and 1 U of Super Taq DNA polymerase (HT Biotechnology, Cambridge, United Kingdom) were adjusted to a final volume of 100 μl with sterile water. Template DNA (80 to 500 ng) was added to the reaction mixture (preheated to 70°C) to avoid nonspecific annealing of the primers to nontarget DNA. The cycles used were as follows: 1 cycle at 70°C for 1 min; 33 cycles at 95°C for 1 min, 40°C for 1 min, and 72°C for 3 min; and 1 final cycle at 95°C for 1 min, 40°C for 1 min, and 72°C for 10 min. The number of amplification cycles during PCR was reduced as much as possible to reduce PCR biases (52), chimera formations (53), and Taq polymerase error rates; however, 34 cycles were needed to yield sufficient product.
Clone library construction.
Products of three parallel PCRs were combined and precipitated to concentrate the DNAs for cloning. DNA was ligated in the pGEM-T-Easy vector by using the protocol of the manufacturer (Promega). Ligation reaction mixtures were purified and used for electroporation of Escherichia coli XL1 Blue (Stratagene GmbH, Heidelberg, Germany) or E. coli JM-109 cells (Promega) as described by Flohr (8). Recombinant transformants were selected by blue and white screening.
Dot blot hybridization.
Plasmid DNA was prepared from overnight cultures with a WIZARD Mini Prep Purification Kit (Promega) by following the manufacturer’s recommendations. Plasmids were checked for insert presence on agarose gels. All plasmids known to contain the correctly sized insert of 1.5 kb were used for dot blot hybridization.
For blotting, plasmid DNA was denatured for 5 min at 95°C and cooled immediately on ice. Aliquots of 100 to 400 ng of DNA were spotted onto a prewetted nylon membrane (Hybond-N+; Amersham, Little Chalfont, Buckinghamshire, United Kingdom) with a Bio-Rad (Munich, Germany) dot blot apparatus. For additional denaturation, blots were placed on filter paper soaked with 0.4 M NaOH–0.6 M NaCl for 15 min. Finally, membranes were equilibrated with 2× SSC (0.3 M NaCl, 0.03 M sodium citrate [pH 7.0]) for 10 min. For immobilization of the DNA, the membrane was baked at 80°C for 2 h. Oligonucleotide probes were 5′ end labeled with [γ-32P]ATP by using T4 polynucleotide kinase according to the recommendation of the manufacturer (New England Biolabs, Schwalbach, Germany). The unincorporated [γ-32P]ATP was removed from the labeled probes by using Sephadex columns (NAP columns; Pharmacia Biotech, Freiburg, Germany) according to the manufacturer’s protocol.
Membranes were prehybridized for 1 h at 40°C in hybridization solution (10× Denhardt solution, 4× SSC, 0.1% SDS, 2 mM EDTA [pH 8.0], 50 μg of salmon sperm DNA per ml [32]) before 32P-labeled probes were added. Hybridization was carried out at 40°C (except with membranes for hybridization with a gram-positive probe, for which the temperature was 30°C) for 14 to 16 h. Thereafter, the membranes were washed twice for 30 min with washing buffer (2× SSC, 0.1% SDS) at hybridization temperature. To eliminate nonspecific binding, the membranes were washed two more times for 15 min at the dissociation temperature (Td), which had been determined according to the method of Raskin et al. (39). Probes and Tds used in this study are given in Table 1. Control 16S rDNAs different in sequence from each particular probe by one nucleotide were also spotted on membranes and hybridized as well to check the stringency of washing conditions. Hybridization signals were analyzed with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
TABLE 1.
Oligonucleotide probes used in this study
| Probe | Specificity | Sequence (5′→3′) | Positiona | Td (°C) | Reference |
|---|---|---|---|---|---|
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 338–355 | 57 | 1 |
| ALF968 | α subclass of the Proteobacteria, several members of the δ subclass of Proteobacteria, and most Pelobacter-Geobacter spp. | GGTAAGGTTCTGCGCGTT | 968–985 | 58 | 35 |
| 687 | Desulfovibrio and some species of Geobacteriaceae | TACGGATTTCACTCCT | 687–702 | 48 | 5 |
| 660 | Desulfobulbus | GAATTCCACTTTCCCCTCTG | 660–679 | 56 | 5 |
| 804 | Desulfobacterium, Desulfobacter, Desulfobotulus, Desulfosarcina, Desulfococcus | CAACGTTTACTGCGTGGA | 804–821 | 52 | 5 |
| Sval428 | Desulfotalea, Desulfofustis | CCATCTGACAGGATTTTAC | 428–446 | 56 | 48 |
| GP | Most gram-positive bacteria | Unpublished | 41 | 28 | |
| CF319a | Cytophaga-Flavobacterium cluster | TGGTCCGTGTCTCAGTAC | 319–336 | 58 | 30 |
| Gamma598 | 16S rDNA clone sequences affiliated with endosymbionts and some other species in the γ subclass of Proteobacteria | CGGATGTGAAAGCCCTGG | 598–615 | 58 | This study |
Position in the 16S rRNA of E. coli.
ARDRA and rarefaction analysis.
Amplified rDNA restriction analysis (ARDRA) was performed to analyze the diversity of clones within each group defined by dot blot hybridization. Isolated plasmid DNAs of 16S rDNA clones were used as templates for insert amplification. The PCR was performed as described above, except that the primer annealing temperature was higher (44°C). PCR products were purified, and aliquots of 200 to 400 ng of the amplified insert were digested with 7.5 U of the restriction endonuclease HaeIII (Promega) for 3 h at 37°C. The resulting fragments were analyzed on an 8% polyacrylamide gel, and restriction patterns within each group were compared. Diversity of the clone library was further investigated by rarefaction analysis (16, 18, 50). Rarefaction curves were produced by using the analytical approximation algorithm of Hurlbert (18) and 95% confidence intervals estimated as described by Heck et al. (16). Calculations were performed on a personal computer with the freeware program aRarefactWin (17a).
Sequencing and phylogenetic analysis.
Representatives of all major ARDRA pattern groups were chosen for sequencing. Plasmid DNAs from selected 16S rDNA clones were sequenced (partially or in full) by Taq Cycle Sequencing with universal rRNA-specific primers with a model ABI377 (Applied Biosystems, Inc.) or a Li-Cor (MWG Biotech, Ebersberg, Germany) sequencer. A total of 116 clones were sequenced partially or fully. All sequences were checked for chimera formation with the CHECK_CHIMERA software of the Ribosomal Database Project (29), and the phylogenetic affiliations of their 5′ and 3′ ends were compared. By this procedure seven potential chimeras (6.0%) were detected. This figure probably underestimates the real chimera fraction because it is more difficult to detect chimera formation of two closely related sequences (53). Potential chimeras were eliminated before phylogenetic trees were constructed.
Sequence data were analyzed with the ARB software package (51). Phylogenetic trees were calculated by parsimony, neighbor-joining, and maximum-likelihood analysis with different sets of filters. For tree reconstruction, only full-length sequences were considered.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper will appear in the EMBL, GenBank, and DDBJ nucleotide sequence databases under the accession no. AJ240966 to AJ241022. Only sequences of more than 1,000 bases in length were submitted.
RESULTS
Initial clone library analysis.
A sample of 30 clones was initially selected for sequencing and phylogenetic analysis to get a first overview of the quality of and the diversity in the 16S rDNA clone library. Within the 30 clones, we detected 21 different sequences. Two major groups became evident: approximately 50% of the 16S rDNA clone sequences were related to gram-negative SRB and other members of the δ subclass of the class Proteobacteria, and approximately 40% were affiliated with the γ subclass of Proteobacteria, most closely with sulfur-oxidizing bacteria. Additional sequences were related to Cytophaga spp. (one sequence) and gram-positive bacteria (two sequences).
Grouping of clones by dot blot hybridization and ARDRA.
On the basis of the initial sequence analysis, we developed a new probe (Gamma598) and used this probe, in addition to others, for dot blot hybridization (for an overview of probes used, see Table 1).
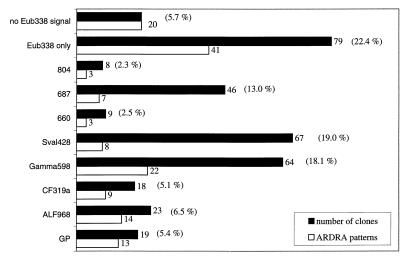
Of the screened clones, 94.3% hybridized with the bacterial probe EUB338 and contained a 16S rDNA insert. The remaining 5.7% had no hybridization signal with EUB338 (Fig. 1), although all of them had a correctly sized insert of 1.5 kb. ARDRA of this group resulted in 20 different patterns, with each pattern being represented by a single clone (see Fig. 2J). Fourteen of these clones were sequenced, and 12 fell into the division Planctomycetes-Verrucomicrobiales. This result is in agreement with the work of Neef et al., who showed that Planctomycetales spp. have at least one nucleotide that is different from the sequence of probe EUB338 (36). The other clones were distantly related to low-G+C-content gram-positive and green nonsulfur bacteria. Of the EUB-positive clones, 71.9% bound one of the group-specific probes used.
FIG. 1.
Dot blot hybridization and ARDRA of 16S rDNA clones. Three hundred fifty-three clones were screened by dot blot hybridization with different probes. The diversity within each group was further investigated by ARDRA with one restriction endonuclease (HaeIII). The filled bars represent the numbers of clones detected with specific probes, and the open bars show the numbers of different ARDRA patterns after digestion with HaeIII. Probe GP is specific for gram-positive bacteria (28), ALF968 is specific for members of the α subclass of Proteobacteria (35), CF319 is specific for the Cytophaga-Flavobacterium group (30), Gamma598 targets three gene clusters affiliated with sulfur-oxidizing bacteria in the γ subclass of Proteobacteria, Sval428 is specific for psychrophilic sulfate reducers isolated from the same site (48), probe 660 targets Desulfobulbus species (5), 687 is specific for Desulfovibrio and some species of Geobacteraceae (5), and 804 targets Desulfobacter, Desulfobacterium, and Desulfobotulus species (5). “EUB338 only” indicates the clones which hybridized only with the universal eubacterial probe (1). No EUB338 signal describes clones with a correctly sized insert of 1.5 kb but no hybridization signal at all.
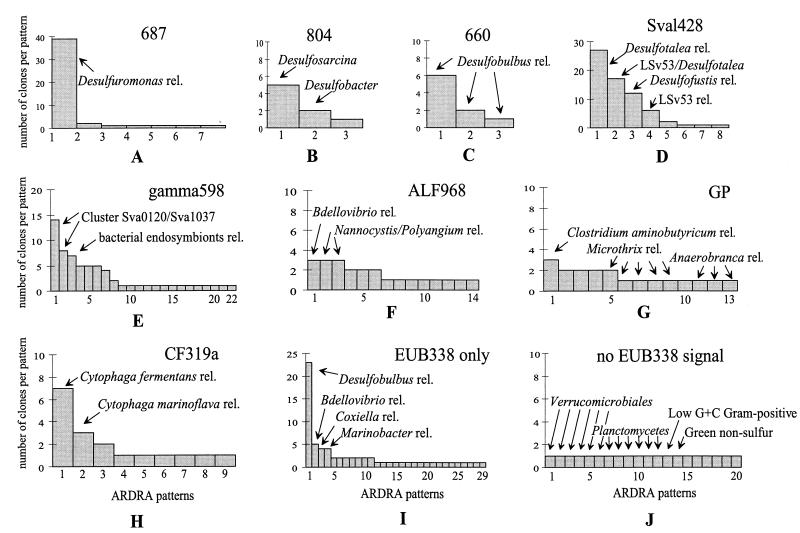
FIG. 2.
Distribution of 16S rDNA clone sequences in different ARDRA patterns. The profiles are based on ARDRA and sequence analysis. The closest cultivated relatives (rel.) for the individual ARDRA groups are indicated.
δ-Proteobacteria.
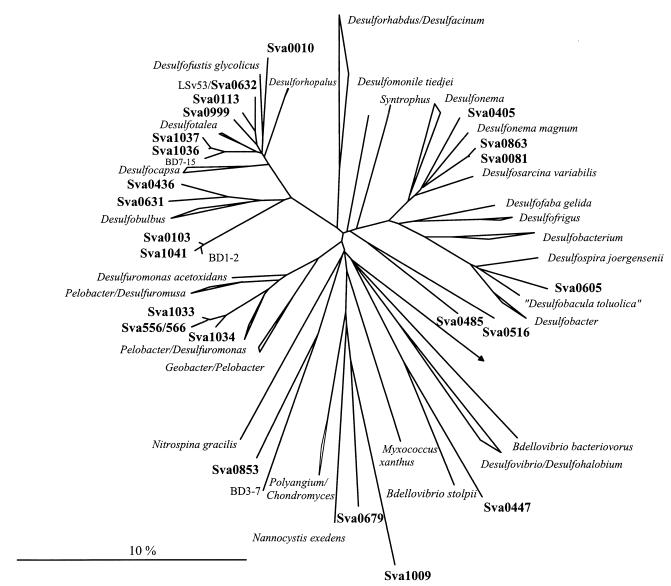
The most abundant group of clones was affiliated with the δ subclass of Proteobacteria. A total of 36.8% of the clones hybridized with different probes specific for SRB. The majority of these clones were targeted by probe Sval428 (Fig. 1). Sixty-seven clones (19.0%) hybridized with the probe specific for SRB first isolated from the same habitat. ARDRA of this fraction resulted in eight different restriction patterns, subsequently referred to as phylotypes (Fig. 2D). Sequence analysis revealed that the dominant phylotypes were almost all closely related (95 to 100%) to SRB isolates from the same cruise (strains with the prefix “LSv,” Desulfotalea [23, 48]). The most abundant pattern was found in 27 clones (e.g., Sva1036 and Sva1037), their sequences showing 95% similarity to the 16S rDNA sequence of LSv20 and Desulforhopalus vacuolatus (Fig. 3). The sequences of the second-most-dominant pattern, represented by 17 clones, were affiliated with Desulfotalea sp. (Sva0999) and LSv23 or LSv53 (Sva0632). Twelve of these clones were identical to the 16S rDNA of LSv53, a psychrophilic SRB isolated by Knoblauch et al. (23) during the same cruise but from a different sampling station (Storfjord). A third dominant phylotype in the Sval428-positive fraction (12 clones, e.g., Sva0010) was phylogenetically related to Desulfocapsa sp. (93% similarity) (Fig. 3), while a fourth phylotype (6 clones, e.g., Sva0113) was affiliated with LSv23 or LSv53 (97% similarity). Remaining patterns were represented by only one or two clones.
FIG. 3.
Phylogenetic tree showing the affiliations of 16S rDNA clone sequences to selected reference sequences of the δ subclass of Proteobacteria. The tree was calculated by neighbor-joining analysis and corrected with filters which considered only 50% conserved regions of the 16S rRNA of δ-Proteobacteria. 16S rDNA clone sequences are in boldface type. The bar represents 10% estimated sequence divergence.
The second-largest fraction (13.0%) of SRB-related 16S rDNA clone sequences was targeted by probe 687, specific for Desulfovibrio and some species of the Geobacteraceae (Fig. 1). Diversity in this group was very low (Fig. 2A). Of the seven different ARDRA patterns, one was represented by 39 clones and the remaining six patterns were each represented by one or two clones only. Phylogenetic analysis revealed that the major group (e.g., Sva1033 and Sva0566) was related to Desulfuromonas sp. (Fig. 3). The highest similarity was 93.7% (to Desulfuromonas palmitatis). Sequencing of clones representing the other patterns also placed them with Desulfuromonas sp.
Relatively few 16S rDNA clones were affiliated with Desulfobulbus sp. by probe 660 (2.5%) or with Desulfobacter sp., Desulfobacterium sp., or Desulfobotulus sp. by probe 804 (2.3%) (Fig. 1). The diversity in these two groups was also low (Fig. 2B and C). Only three different ARDRA patterns per group were found. The three different clusters that were detected with probe 660 were all, as expected, phylogenetically related to Desulfobulbus sp. (e.g., Sva0436 and Sva0631). Five of the clones targeted by probe 804 (e.g., Sva0081 and Sva0863) were affiliated with Desulfosarcina sp. (Fig. 3). The two other groups were represented by two clones (e.g., Sva0605) and one clone (Sva0405) only. Although clone Sva0605 hybridized with probe 804, its 16S rDNA sequence was most closely related to Desulfobacula toluolica (93.1% similarity).
γ-Proteobacteria.
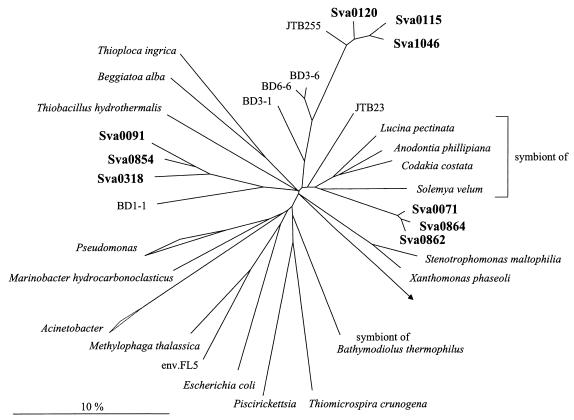
In addition to the SRB, there was a second dominant group in the clone library represented by 16S rDNA clone sequences which fell in the γ subclass of Proteobacteria. They were only distantly related to known bacteria (between 85.6 and 92.1%), being related most closely to sulfur-oxidizing bacteria. This group was detected by dot blot hybridization with probe Gamma598. Sixty-four clones (18.0%) hybridized with this new probe developed on the basis of preliminary screening of 30 clones (see above). After restriction endonuclease digestion, 22 different ARDRA patterns became evident (Fig. 1 and 2E). Phylogenetically, the clones formed three distinct clusters (Fig. 4). Clones Sva0071 and Sva0864 belonged to a cluster that was affiliated with sulfur-oxidizing endosymbiotic bacteria such as the gill symbionts Solemya velum (92.0%) and Codakia costata (92.1%). The second cluster, containing clones Sva1046, Sva0115, and Sva0120, was most closely related to other clone sequences published by Kato and Li (21), derived from deep-sea sediments (97.9% highest similarity). The third cluster (containing, e.g., Sva0091, Sva0854, and Sva0318) could not be assigned stably. Different tree reconstructions affiliated the sequences with sulfur-oxidizing endosymbionts, the Beggiatoa-Thioploca group, or with separate groups. To no members of the above-named groups did they show more than 89.5% 16S rDNA similarity. The phylogenetic position in Fig. 4 was consequently indicated by a multifurcation.
FIG. 4.
Phylogenetic tree showing the affiliations of 16S rDNA clone sequences with selected reference sequences of the γ subclass of Proteobacteria. The tree was calculated by neighbor-joining analysis and corrected with filters which considered only 50% conserved regions of the 16S rRNAs of γ-Proteobacteria. Sva0862 and Sva0854 are not full-length sequences (1,000 bp) and have therefore been added to the existing tree, by a special algorithm included in the ARB software, without allowing for changes of the tree topology based on almost complete sequences. 16S rDNA clone sequences are in boldface type. The bar represents 10% estimated sequence divergence.
Other probe target groups.
Probe ALF968 was designed to target the α subclass of Proteobacteria. This probe is known to also target some members of the δ subclass of Proteobacteria (35). In our study, 59 of the 82 clones hybridizing with probe ALF968 could be assigned to SRB by hybridization with probe Sval428 and sequencing. Consequently, we investigated only the diversity of the remaining 23 clones, which displayed 14 different ARDRA patterns (Fig. 1). The three most abundant patterns were each represented by three clones. The sequences were most closely related to Bdellovibrio and Nannocystis or Polyangium sp., i.e., genera of the δ subclass of Proteobacteria (Fig. 2F). The remaining patterns were represented by one or two clones only. Sequencing of 11 of 14 phylotypes hybridizing with ALF968 showed that only one phylotype, represented by two clones, was indeed affiliated with the α subclass of Proteobacteria (Rhodobacter spp.).
Probe GP, specific for gram-positive bacteria, hybridized with 19 clones (5.4%). Diversity within this group was very high since it contained 13 different patterns (Fig. 2G). The sequences were fairly distantly related to Clostridium sp. (89.8%), Microthrix parvicella (85.6%), and Anaerobranca sp. (86%).
Eighteen clones were assigned to the Cytophaga-Flavobacterium cluster by probe CF319a (Fig. 1). We found nine different phylotypes by ARDRA. The most abundant phylotype was represented by seven clones which were most similar (89.1%) to Cytophaga fermentans (Fig. 2H).
A large number of clones (37 clones) hybridized only with probe EUB338. We found 29 patterns in this fraction. Only four of these patterns were represented by more than two clones (Fig. 2I). The most dominant pattern (23 clones) was represented by sequences (e.g., Sva0103 and Sva1041) that were most closely related to Desulfobulbus sp. relatives (90.1%) (Fig. 3). These sequences had three nucleotide differences from probe 660, specific for Desulfobulbus sp., and were, therefore, not detected by this probe. Other phylotypes that were represented by more than two clones were related to Bdellovibrio sp. or Nitrospina sp. (82.7%; δ-Proteobacteria), Coxiella sp., and Marinobacter sp. (91.5 and 89.8%; both γ-Proteobacteria). The less frequent patterns had sequences related to the γ subclass of Proteobacteria (e.g., Marinobacter, Methylophaga, and Coxiella relatives), to the δ subclass of Proteobacteria (Desulfobacula, Desulfosarcina, Nitrospina, and Bdellovibrio relatives), and to the newly described phylum Holophaga-Acidobacterium (27).
Rarefaction analysis.
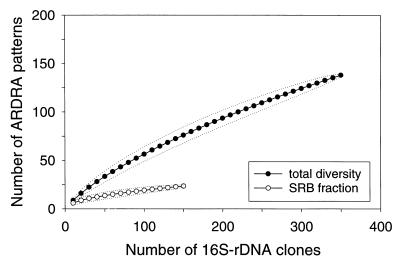
We applied rarefaction analysis to evaluate whether screening of 353 clones was sufficient to estimate diversity within the clone library. The expected number of different ARDRA patterns was plotted versus the number of 16S rDNA clones in the clone library. ARDRA of 353 clones resulted in 140 different patterns. The calculated rarefaction curves did not reach a clear saturation, indicating that analysis of an increasing number of clones would have revealed further diversity (Fig. 5).
FIG. 5.
Rarefaction curves for the different ARDRA patterns of 16S rDNA clones. Rarefaction curves were calculated by using the analytical approximation algorithm described by Hurlbert (18) and 95% confidence intervals estimated as described by Heck et al. (16). The number of different ARDRA patterns in the clone library was determined after digestion with one restriction endonuclease. The expected number of ARDRA patterns (●) is plotted versus the number of clones. Rarefaction curves were also calculated for the fraction of SRB (○). The dotted lines represent 95% confidence intervals.
We did the same rarefaction analysis with the fraction of clones representing 16S rDNAs of SRB (including Desulfobulbus relatives detected by EUB338 only). Twenty-two different ARDRA patterns were represented by 155 clones. The calculated rarefaction curve approached saturation, indicating that the diversity of SRB in the clone library was almost covered.
DISCUSSION
Diversity.
The sediments investigated in this study are never exposed to temperatures higher than 3°C and might, therefore, be regarded as extreme environments. Studies of the bacterial communities of extreme environments such as a saltern (31) and a low-pH hydrothermal vent system (33) have indicated low bacterial diversity in these habitats. By applying rarefaction analysis to restriction fragment length polymorphism patterns of their 16S rDNA clone library, Moyer et al. (33) could demonstrate that screening of 48 bacterial clones was enough to detect the majority of taxa in the clone library of a hydrothermal vent microbial mat. One aim of our study was to assess bacterial diversity, which was expected to be limited considering the extremely low environmental temperatures; however, rarefaction analysis revealed that by screening 353 clones, the actual diversity in our clone library was only partially covered. It is unlikely that new major groups will be discovered by analyzing additional clone sequences, since the major groups were the same after 30 and 353 clones were screened. Total phylotype richness, i.e., the number of phylotypes present, on the other hand, might reflect the potential within a microbial community to respond to changes in environmental conditions. At a different time point, those phylotypes not detected or represented by only one clone might play an important role in this habitat.
Another approach that has been used to assess completeness of a clone library analysis is to calculate coverage. In our case, coverage was 71.95%, indicating that almost three-quarters of total diversity in the clone library was detected; however, since coverage is based only on the number of unique clones relative to total richness, not taking evenness into account, it should be regarded only as a rough estimate of diversity within a sample of infinite size.
The 16S rDNA inserts of the clones were digested with one tetrameric restriction enzyme. Use of a second enzyme resulted in an increased number of patterns (data not shown). In a study of sulfate-reducing isolates, Rooney-Varga et al. (44) demonstrated that use of four enzymes was necessary to differentiate between sequences having more than 95% similarity. However, since our study was aimed towards an overview of diversity, we concentrated on the differences revealed by one enzyme. The results presented here should therefore be regarded as indicating minimal diversity.
As can be seen in Fig. 2, diversity within each probe target group varied greatly. In particular, group 687 showed very little evenness (distribution of the number of clones per pattern), with one phylotype making up 85% of the 687 positive clones; however, since different probe target groups represent different phylogenetic depths (probe 660, e.g., is specific for one genus and probe 804 is specific for a group of different genera), we refrain from comparing levels of diversity among the different groups. The data might, however, serve as a basis in future analyses for comparing levels of diversity of the same target group in different environmental samples.
Methodological considerations.
Clone libraries of 16S rDNAs have been widely used to investigate in a cultivation-independent approach the microbial communities of different, mainly pelagic or terrestrial habitats (4, 7, 9, 11, 33, 37, 38, 49). They have helped to elucidate common features within the microbial communities of specific habitats such as marine pelagic environments (9, 38) and have provided additional sequence information for the design and evaluation of probes. However, this experimental approach suffers from specific limitations that potentially confer selectivity via differential cell lysis, variable nucleic acid extraction efficiencies, or biased amplification in the PCR. The high lysis efficiency (93% ± 3.4%) and the high overall diversity in the clone library presented here suggest that our analysis was based on a substantial fraction of the bacterial community from Hornsund sediments; however, it is difficult to assess the potential bias introduced during amplification of the 16S rDNA. These biases are due to primer selectivity or erroneous product ratios caused by product saturation in the later cycles of amplification (52). Furthermore, oligonucleotides specific for a very general phylogenetic group, such as the bacterial 16S rDNA primers we used in PCR, are ultimately bound to miss some members of the community, which, in turn, leads to an underestimation of diversity.
Despite the caveats that clone abundance in the library does not necessarily reflect bacterial abundance at the site and that diversity might not be fully covered, the correlations between results of the clone library and results of completely different approaches such as 16S rRNA quantification analysis (see the accompanying paper [48]) and most probable number counts (23) are encouraging. The largest group of clones (19%) was detected by a probe designed especially for SRB isolated on the same cruise (23). While some of these isolates came from the same sampling site on the west coast of Spitsbergen (Hornsund), others were obtained from sediments sampled off the east coast (Storfjord). The clones were closely related to these isolates, with one phylotype even showing 100% sequence identity to strain LSv53, which was isolated from the east coast station (Fig. 3). This phylotype was represented by 12 of 353 clones. The same phylotype was also detected in a denaturing gradient gel electrophoresis-Southern blot analysis described in the accompanying paper; however, as expected, quantitative representation of the phylotypes in the clone library corresponds only weakly to the results from 16S rRNA slot blot hybridization (see the accompanying paper [48]).
Phylogenetic composition of the clone library.
The clone library was dominated by sequences related to δ-Proteobacteria. Even within the clones targeted by the general EUB338 probe only, we could detect one additional phylotype affiliated with the δ subclass of Proteobacteria, loosely related to Desulfobulbus. Twenty-three clones (6.5%) belonged to this phylotype not targeted by any of the specific SRB probes. Phylogenetic affiliation makes it likely that they are also sulfate reducers. The design and application of a new probe specific for this group and its employment in quantitative rRNA slot blot and in situ hybridizations will show the extent to which it contributes to the bacterial community.
Detailed analysis of the clones targeted by probe 687 showed that all 16S rDNA inserts were affiliated with Desulfuromonas palmitatis, whereas no Desulfovibrio was detected. Desulfuromonas palmitatis is known to reduce sulfur or thiosulfate and iron or to employ a fermentative metabolism; however, the phylogenetic distance between Desulfuromonas and the clones is so large (6.3%) that we cannot determine whether these clones represent sulfur or sulfate reducers. All clones had one nucleotide that was different from the probe sequence but gave a clearly positive signal in dot blot hybridization. A one-mismatch control also included in the hybridization analysis did show a distinguishable weaker signal. This example serves as a reminder that discrimination by one nucleotide might not always be possible.
A second dominant group of 16S rDNA clones was distantly related to sulfur-oxidizing symbiotic or free-living bacteria of the γ subclass of Proteobacteria, with a similarity value of 92 or 86%. Since no pure culture representatives for this group have been isolated, we can only speculate that they might indeed be involved in the oxidative part of the sulfur cycle. Selective cultivation of sulfur oxidizers from the same habitat is under way.
When investigating Wadden Sea sediments by fluorescence in situ hybridization, Llobet-Brossa et al. (26) found members of the Cytophaga-Flavobacterium cluster to be even more abundant than δ-Proteobacteria. This cluster has also been found in marine aggregates (4, 40); Cytophagales, in general, are known for their ability to associate and glide on surfaces and to degrade a wide variety of polymeric substances (42). They were also a significant constituent of our clone library (5.1%), indicating that Cytophagales might be a common member of marine sediment microbial communities.
Since sedimentation regularly brings in organic matter from the water column, we expected to find evidence of allochthonous input in the sediment. Groups that are commonly found in planktonic communities, like some genera of α-Proteobacteria (13, 34), were not abundant in the clone library; only 2 of 353 clones belonged to the α-Proteobacteria. Furthermore, we did not detect any cyanobacterial sequence and detected only one plastid sequence; however, the presence of allochthonous microorganisms is probably dependent on the time of sampling, with higher abundances expected after a phytoplankton bloom.
Comparison with other clone libraries.
Open ocean and coastal planktonic communities are well-studied ecosystems with regard to clone libraries (10, 11, 34, 38, 49). Although the bacterial communities of these habitats are phylogenetically diverse, distinct phylogenetic clusters are repeatedly detected. These results are in line with the idea that in similar climate zones, a limited number of phylotypes account for a substantial fraction of the bacterioplankton at certain times (34). It is still an open question whether the same applies to benthic environments, since only limited data are available on marine sediments. Devereux and Mundfrom (6) established a clone library from a sandy marine sediment, selectively amplifying partial 16S rDNAs of SRB. Gray and Herwig (15) set up a general 16S rDNA clone library, examining 22 clones. Kato and Li (21) investigated clones from deep-sea sediments off Japan. A comparison is difficult, in particular because in many cases only partial sequences are available, but some trends are noteworthy. Sequences related to the Desulfotalea-Desulforhopalus cluster were frequently recovered (Fig. 3). The highest similarity values among clones with almost complete sequences were between 99.4 and 97.2%. Furthermore, all these clone libraries also contained sequences related to Myxobacteria and Bdellovibrio (Fig. 3; see below). Myxobacteria have been known mainly as terrestrial organisms (41); their isolation from coastal marine sediments has been attributed to resting cells because of their low salt tolerance. Bdellovibrio, on the other hand, has been repeatedly isolated from marine sediments (45). Considering the fact that related known pure cultures are almost all micropredators (41), they might play a role in the control of bacterial abundance. Within the sequences of the γ-Proteobacteria, similar congruencies occurred. Both Kato and Li (21) and Gray and Herwig (15) found sequences from the symbiont cluster (Fig. 4) (highest similarities among clones, between 98 and 92%).
More data on the prokaryotic diversity of marine benthic habitats are needed to identify common benthic features. Furthermore, the actual abundance of these conspicuous groups has to be determined via in situ and rRNA slot blot hybridization to evaluate their roles in the bacterial community of marine sediments.
ACKNOWLEDGMENTS
We thank Ulrich Nübel for inspiring discussions and Birgit Rattunde for technical assistance. We acknowledge Steven M. Holland for providing the freeware program aRarefactWin.
This work was supported by the Max-Planck-Society.
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnosti C, Sagemann J, Jørgensen B B, Thamdrup B. Temperature dependence of microbial degradation of organic matter in marine sediments: polysaccharide hydrolysis, oxygen consumption, and sulfate reduction. Mar Ecol Prog Ser. 1998;165:59–70. [Google Scholar]
- 3.Canfield D E, Jørgensen B B, Fossing H, Glud R, Gundersen J, Ramsing N B, Thamdrup B, Hansen J W, Nielsen L P, Hall P O J. Pathways of organic carbon oxidation in three continental margin sediments. Mar Geol. 1993;113:27–40. doi: 10.1016/0025-3227(93)90147-n. [DOI] [PubMed] [Google Scholar]
- 4.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 5.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 6.Devereux R, Mundfrom G W. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in sandy marine sediment. Appl Environ Microbiol. 1994;60:3437–3439. doi: 10.1128/aem.60.9.3437-3439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felske A, Wolterink A, Van Lis R, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequence in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flohr T. Molekulare Klonierung und funktionale Charakterisierung Interferon-induzierbarer Gene: Tryptophanyl-tRNA-Synthetase und Cytokeratin K17. Ph.D. thesis. Hannover, Germany: Universität Hannover; 1993. [Google Scholar]
- 9.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial groups from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 10.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbioal communites from the Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 12.Glud R N, Holby O, Hoffmann F, Canfield D E. Benthic mineralization and exchange in Arctic sediments (Svalbard, Norway) Mar Ecol Prog Ser. 1998;173:237–251. [Google Scholar]
- 13.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Good I J. The population frequencies of species and the estimation to the population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- 15.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heck K L, Jr, Van Belle G, Simberloff D. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology. 1975;56:1459–1461. [Google Scholar]
- 17.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;68:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Holland S. posting date. [Online.] aRarefactWin program. Athens: University of Georgia; August 1998. http://www.uga.edu/∼strata/AnRareReadme.html . 8 December 1998, last date accessed.] [Google Scholar]
- 18.Hurlbert S H. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 19.Jørgensen B B. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature. 1982;296:643–645. [Google Scholar]
- 20.Kane M D, Poulsen L K, Stahl D A. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato C, Li L. Bacterial diversity in the deep-sea sediments from different depths. 1998. DDBJ, EMBL, and GenBank accession no. AB015248, AB015254, AB015514, AB015515, AB015547, AB015548, AB015576, and AB015588. [Google Scholar]
- 22.Kato C, Li L, Tamaoka J, Horikoshi K. Molecular analyses of the sediment of the 11000-m deep Mariana Trench. Extremophiles. 1997;1:117–123. doi: 10.1007/s007920050024. [DOI] [PubMed] [Google Scholar]
- 23.Knoblauch C, Jørgensen B B, Harder J. Community size and metabolic rates of psychrophilic sulfate-reducing bacteria in Arctic marine sediments. Appl Environ Microbiol. 1999;65:4230–4233. doi: 10.1128/aem.65.9.4230-4233.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostka J E, Thamdrup B, Glud R N, Canfield D. Rates and pathways of carbon oxidation in permanently cold Arctic sediments. Mar Ecol Prog Ser. 1999;180:2–21. [Google Scholar]
- 25.Levitus S, Boyer T. World ocean atlas. Vol. 4. Washington, D.C: Temperature. U.S. Department of Commerce; 1994. [Google Scholar]
- 26.Llobet-Brossa H, Rosselló-Mora R, Amann R I. Microbial community composition of Wadden Sea sediments as revealed by fluorescent in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 28.MacGregor, B. J., S. Toze, R. Sharp, C. J. Ziemer, and D. A. Stahl. Unpublished data. [DOI] [PubMed]
- 29.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 31.Martínez-Murcia A J, Acinas S G, Rodriguez-Valera F. Evaluation of prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol Ecol. 1995;17:247–256. [Google Scholar]
- 32.Martinez-Picado J, Blanch A R. Rapid detection and identification of Vibrio anguillarum by using a specific oligonucleotide probe complementary to 16S rRNA. Appl Environ Microbiol. 1994;60:732–737. doi: 10.1128/aem.60.2.732-737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullins T C, Britschgi T B, Krest R L, Gionvannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 35.Neef A. Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populations-Analyse in komplexen mikrobiellen Biozönosen. Ph.D. thesis. Munich, Germany: Technische Universität München; 1997. [Google Scholar]
- 36.Neef A, Amann R, Schlesner H, Schleifer K H. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology. 1998;144:3257–3266. doi: 10.1099/00221287-144-12-3257. [DOI] [PubMed] [Google Scholar]
- 37.Nüsslein K, Tiedje J M. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl Environ Microbiol. 1998;64:1283–1289. doi: 10.1128/aem.64.4.1283-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 39.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rath J, Wu K Y, Herndl G J, DeLong E F. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol. 1998;14:261–269. [Google Scholar]
- 41.Reichenbach H, Dworkin M. The myxobacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Vol. 4. New York, N.Y: Springer-Verlag; 1992. pp. 3416–3487. [Google Scholar]
- 42.Reichenbach H, Dworkin M. The order Cytophagales. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Vol. 4. New York, N.Y: Springer-Verlag; 1992. pp. 3631–3687. [Google Scholar]
- 43.Rochelle P A, Cragg B A, Fry J C, Parkes R J, Weightman A J. Effect of sample handling on estimation of bacterial diversity in marine sediments by 16S rRNA gene sequence analysis. FEMS Microbiol Ecol. 1994;15:215–226. [Google Scholar]
- 44.Rooney-Varga J N, Sharak Genthner B R, Devereux R, Willis S G, Friedman S D, Hines M E. Phylogenetic and physiological diversity of sulphate-reducing bacteria isolated from a salt marsh sediment. Syst Appl Microbiol. 1998;21:557–568. doi: 10.1016/s0723-2020(98)80068-4. [DOI] [PubMed] [Google Scholar]
- 45.Ruby E G. The genus Bdellovibrio. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Vol. 4. New York, N.Y: Springer-Verlag; 1992. pp. 3400–3415. [Google Scholar]
- 46.Sagemann J, Jørgensen B B, Greeff O. Temperature dependence and rates of sulfate reduction in cold sediments of Svalbard, Arctic Ocean. Geomicrobiol J. 1998;15:85–100. [Google Scholar]
- 47.Sahm K, Berninger U-G. Abundance, vertical distribution, and community structure of benthic prokaryotes from permanently cold marine sediments (Svalbard, Arctic Ocean) Mar Ecol Prog Ser. 1998;165:71–80. [Google Scholar]
- 48.Sahm, K., C. Knoblauch, and R. Amann. Phylogenetic affiliation and quantification of psychrophilic sulfate-reducing isolates in marine arctic sediments. Appl. Environ. Microbiol. 65:3976–3981. [DOI] [PMC free article] [PubMed]
- 49.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simberloff D. Use of rarefaction and related methods in ecology. In: Dickson K L, Cairns J J, Livingston R J, editors. Biological data in water pollution assessment: quantitative and statistical analyses. Philadelphia, Pa: American Society for Testing and Materials; 1978. pp. 150–165. [Google Scholar]
- 51.Strunk O, Gross O, Reichel B, May M, Hermann S, Stuckman N, Nonhoff B, Lenke M, Ginhart A, Vilbig A, Ludwig T, Bode A, Schleifer K-H, Ludwig W. ARB: a software environment for sequence data. Munich, Germany: Department of Microbiology, Technische Universität München; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang G C Y, Wang Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology. 1996;142:1107–1114. doi: 10.1099/13500872-142-5-1107. [DOI] [PubMed] [Google Scholar]
- 54.Wollast R. The coastal organic carbon cycle: fluxes, sources, and sinks. In: Mantoura R F C, Martin J-M, Wollast R, editors. Ocean margin processes in global change. New York, N.Y: John Wiley & Sons; 1991. pp. 365–381. [Google Scholar]
- 55.Zhou J, Brunns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]