ABSTRACT
Interferon-inducible protein 16 (IFI16) plays a critical role in antiviral innate immune responses against DNA viruses. Although the acetylation of IFI16 is crucial to its cytoplasmic translocation and downstream signal transduction, the regulation of IFI16 acetylation remains unclear. In this study, we demonstrated that the NAD-dependent deacetylase silent information regulatory 1 (Sirtuin1, Sirt1) interacted with IFI16 and decreased the acetylation of IFI16, resulting in the inhibition of IFI16 cytoplasmic localization and antiviral responses against DNA virus and viral DNA in human cells. Meantime, Sirt1 could not inhibit RNA virus-triggered signal transduction. Interestingly, even p204, the murine ortholog of human IFI16, barely interacted with Sirt1. Thus, Sirt1 could not negatively regulate the acetylation of p204 and subsequent signal transduction upon herpes simplex virus 1 (HSV-1) infection in mouse cells. Taken together, our research work showed a new mechanism by which Sirt1 manipulated IFI16-mediated host defense. Our study also demonstrated a difference in the regulation of antiviral host defense between humans and mice, which might be considered in preclinical studies for antiviral treatment.
IMPORTANCE DNA viruses, such as hepatitis B virus (HBV), human papillomavirus (HPV), human cytomegalovirus (HCMV), Epstein-Barr virus (EBV), and herpes simplex virus (HSV), can cause a wide range of diseases and are considered a global threat to human health. Interferon-inducible protein 16 (IFI16) binds virus DNA and triggers antiviral innate immune responses to restrict viral infection. In this study, we identified that silent information regulatory 1 (Sirtuin1, Sirt1) interacted with IFI16 and regulated IFI16-mediated innate host defense. Therefore, the activator or inhibitor of Sirt1 may have the potential to be used as a novel strategy to treat DNA virus-associated diseases. We also found that Sirt1 barely interacted with p204, the murine ortholog of human IFI16, and could not negatively regulate innate immune responses upon HSV-1 infection in mouse cells. This difference between humans and mice in the regulation of antiviral host defense might be considered in preclinical studies for antiviral treatment.
KEYWORDS: DNA sensor, posttranslational modification, acetylation, antiviral innate immunity, signal transduction, cytoplasmic translocation, IFI16
INTRODUCTION
The initiation of innate immune responses relies on the recognition of pathogen-associated molecular patterns (PAMPs), which are completed by a series of pattern recognition receptors (1). The nucleic acids are relatively conserved in viruses acting as PAMPs for innate immune recognition (2). Recently, four nuclear DNA sensors have been identified to bind virus DNA within the nucleus and play a critical role in antiviral host defense, including interferon (IFN)-inducible protein 16 (IFI16), cyclic GMP-AMP synthase (cGAS), IFIX, and heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1) (3–8).
IFI16, containing an N-terminal pyrin domain (PYD) and two C-terminal HIN domains, was first identified as a DNA sensor critical to antiviral host defense during herpes simplex virus 1 (HSV-1) infection (6). Then, IFI16 was demonstrated to be required in the antiviral immune responses against some other family members of herpesviruses, all of which are DNA viruses, including human cytomegalovirus (HCMV), Kaposi's sarcoma-associated herpesvirus (KSHV), and Epstein-Barr virus (EBV) (9–11). IFI16 recognizes viral DNA in both the cytoplasm and the nucleus (12). During HSV-1 infection, IFI16 redistributes from the nucleolus to the nuclear periphery, where it serves as a scaffold recruiting STING and TANK-binding kinase 1 (TBK1) in the cytoplasm, and activates interferon regulatory factor 3 (IRF3) and nuclear factor kappa-B (NF-κB), leading to the production of type I IFN and proinflammatory cytokines, thus resulting in the induction of host antiviral defense (6, 13). It is established that IFI16 is modified by acetylation at numerous sites. Acetylations occurring at the nuclear localization signal (NLS) of IFI16 prevent the nuclear import of IFI16, thus working as molecular toggles of IFI16 distribution (14). However, the underlying molecular mechanism regulating the acetylation and localization of IFI16 remains unclear.
Silent information regulatory 1 (Sirtuin1, Sirt1), belonging to the Sirtuin family, is an NAD-dependent deacetylase that has been demonstrated to be involved in the regulation of aging, metabolism, apoptosis, cell cycle, and inflammation (15). The role of Sirt1 in antiviral innate immune responses is complicated and differs much according to the type of viruses and host cells. Recently, two reports addressed the role of Sirt1 in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (16, 17). Although both groups revealed the interaction between Sirt1 and viral protein NSP14, Zhang et al. considered that Sirt1/PGC-1α inhibits SARS-CoV-2 replication (16), whereas Marius Walter and his colleagues showed that Sirt1 deficiency or Sirt1 inhibitor Ex527 reduced SARS-CoV-2 viral levels (17). During HSV-1 infection, nutraceutical activators of the AMPK/Sirt1 axis have been reported to inhibit viral production, but the underlying mechanisms are not fully understood (18, 19). Although acetylation is considered an important posttranslational modification to regulate the function of cGAS and IFI16 in innate immunity (14, 20), whether Sirtuin family members are involved in the regulation of acetylation of DNA sensors remains unclear.
In this study, we demonstrated that deacetylase Sirt1 interacted with IFI16 and reduced its acetylation, thus regulating IFI16-mediated antiviral innate immune responses. Sirt1 overexpression inhibited DNA virus- or viral DNA-induced innate immune responses, whereas Sirt1 knockdown or deficiency promoted these antiviral responses in human cells. Consistently, the Sirt1 activator inhibited, and the Sirt1 inhibitor promoted, DNA virus- or viral DNA-triggered innate immune responses. Mechanistic investigation indicated that Sirt1 prevented DNA virus- or viral DNA-induced cytoplasmic translocation of IFI16, by which the association of IFI16 with STING and the following signal transduction were inhibited. However, Sirt1 had only a weak interaction with p204 (the murine ortholog of human IFI16) and could not efficiently reduce the acetylation of p204 in mouse cells. Collectively, our research suggests a novel regulatory role of Sirt1 in innate immunity in human cells and shows the differences between humans and mice in the regulation of antiviral host defense.
RESULTS
Sirt1 interacts with IFI16 and regulates its acetylation.
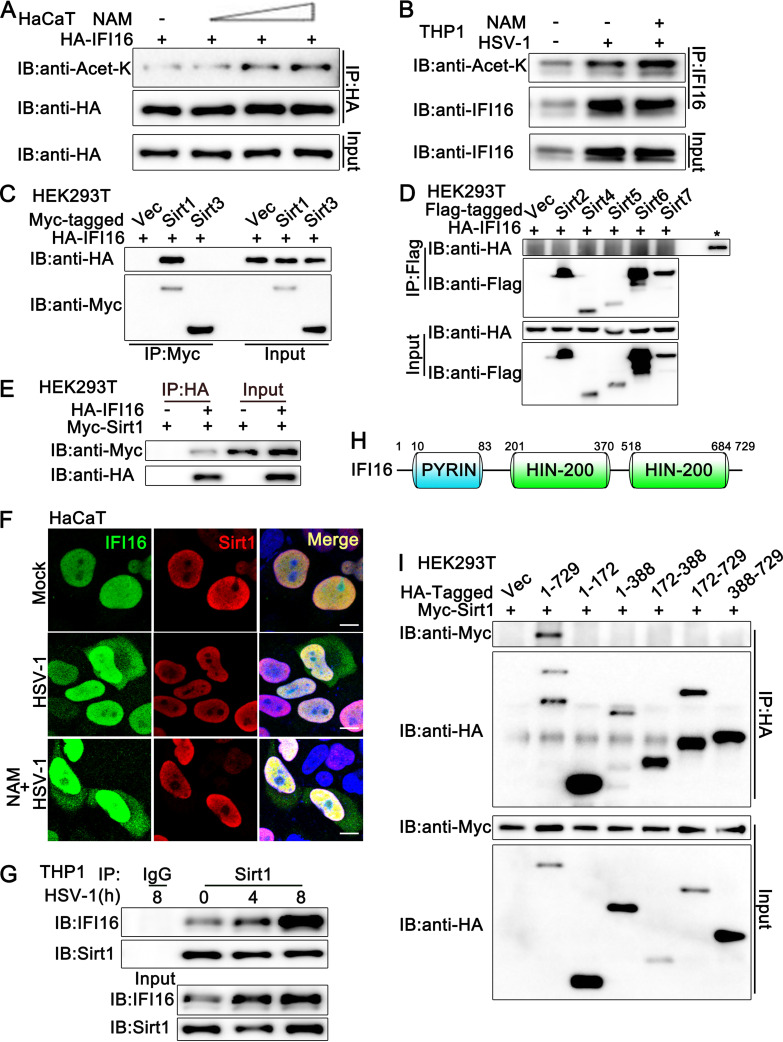
To investigate whether Sirtuin family members play a role in IFI16 function, we transfected the HaCaT keratinocytes with hemagglutinin (HA)-IFI16 and then treated the cells with nicotinamide (NAM), the inhibitor of the Sirtuin family. As shown in Fig. 1A, NAM treatment increased the acetylation of HA-IFI16 in a dose-dependent pattern. Next, we explored the effect of NAM on endogenous IFI16. The acetylation of IFI16 was observed with or without HSV-1 infection, and this acetylation was enhanced by NAM treatment (Fig. 1B), suggesting that the Sirtuin family might be involved in the regulation of IFI16 acetylation during HSV-1 infection. Then, we identified which member of the Sirtuin family was associated with IFI16 and responsible for the deacetylation of IFI16. All seven Sirtuin family members were transfected into HEK293T cells separately, together with an IFI16-expressing plasmid, and coimmunoprecipitation assays indicated that only Sirt1 was associated with IFI16 (Fig. 1C to E). Additionally, confocal microscopy analysis indicated that HA-IFI16 colocalized with Myc-Sirt1 in HaCaT cells with or without HSV-1 infection or NAM treatment (Fig. 1F). This association was confirmed in endogenous coimmunoprecipitation experiments, which indicated that Sirt1 interacted with IFI16 in phorbol myristate acetate-treated THP1 cells (PMA-THP1 cells) (Fig. 1G). Upon HSV-1 infection, the expression level of IFI16 was upregulated and more IFI16 protein was coimmunoprecipitated with Sirt1 (Fig. 1G). Finally, we explored the domain in IFI16 which was responsible for its association with Sirt1. As shown in Fig. 1H, IFI16 contains one pyrin domain and two HIN-200 domains. However, only the full length of IFI16 was found to be associated with Sirt1 in coimmunoprecipitation assays (Fig. 1I). In all, our findings suggest that Sirt1 interacts with IFI16.
FIG 1.
Sirt1 interacts with IFI16. (A) HaCaT keratinocytes were transfected with HA-IFI16 for 24 h and then treated with increasing amounts of nicotinamide (NAM; 0, 2, 5, 10 mM) for another 6 h. Afterward, the cells were lysed and subjected to immunoprecipitation (IP) and immunoblot (IB) analysis. (B) PMA-THP1 cells were infected with HSV-1 (MOI = 1) or left uninfected for 12 h and then treated with NAM (5 mM) for 6 h. Afterward, the cells were lysed and subjected to IP and IB analysis. (C and D) HEK293T cells were transfected with various combinations of plasmids as indicated. At 24 h later, IP and IB analysis were performed. The input of IFI16 is shown in panel D, indicated by a star. (E) HEK293T cells were transfected with Myc-Sirt1, together with HA-IFI16 (+) or control vector (−). At 24 h after transfection, the cell lysates were subjected to IP and IB analysis as indicated. (F) HaCaT keratinocytes were transfected with HA-IFI16 and Myc-Sirt1. At 24 h after transfection, the cells were stimulated with HSV-1 or left uninfected for another 8 h. Then, the cells were treated with or without NAM (5 mM) for 6 h. Immunofluorescence was performed using anti-HA (green) and anti-Myc (red). Nuclei were stained with DAPI. Scale bars, 10 μm. (G) PMA-THP1 cells were infected with HSV-1 (MOI = 1) for the indicated periods and then subjected to IP and IB analysis as indicated. (H) Schematic presentation of full-length IFI16. (I) HEK293T cells were transfected with various combinations of plasmids as indicated. At 24 h later, IP and IB were performed. The data are representative of three independent experiments.
Sirt1 overexpression inhibits HSV-1- or viral DNA-induced innate immune responses.
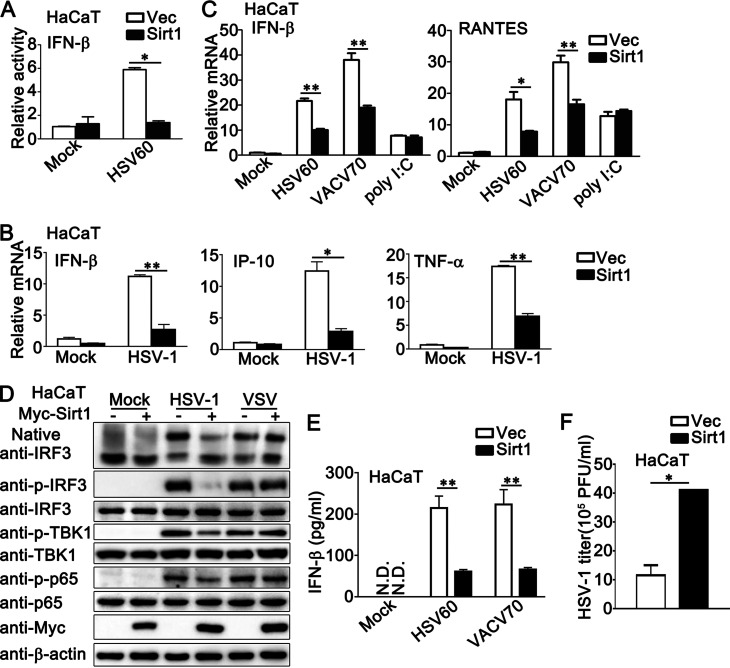
As a DNA sensor, IFI16 plays a critical role in DNA virus- or viral DNA-induced innate immune responses. Therefore, we investigated the role of Sirt1 in the antiviral host defense against HSV-1 infection or exogenous cytosolic DNA stimulation. Luciferase assays with a beta interferon (IFN-β) reporter indicated that Sirt1 overexpression inhibited viral DNA HSV60-triggered activation of IFN-β promoter (Fig. 2A). Upon HSV-1 infection, the inhibition by Sirt1 of the production of IFN-β, IFN-γ-inducible protein-10 (IP-10), and tumor necrosis factor alpha (TNF-α) was observed in the mRNA levels (Fig. 2B). Consistently, Sirt1 overexpression inhibited the production of IFN-β and RANTES triggered by viral DNA, such as HSV60 (a 60-bp HSV-1-derived double-stranded DNA oligonucleotide that stimulates the STING-dependent pathway) and VACV70 (70-mer dsDNA representing the genome of vaccinia virus), but not by poly (I:C) transfection (Fig. 2C). In addition, Sirt1 inhibited the formation of IRF3 dimers and the phosphorylation of IRF3, TBK1, and p65 that was induced by HSV-1 infection but not by RNA virus vesicular stomatitis virus (VSV), suggesting that Sirt1 specifically inhibited DNA virus-triggered antiviral immune responses (Fig. 2D). Interestingly, Sirt1 even caused a slight increase in the formation of IRF3 dimers upon VSV infection (Fig. 2D). Consistently, Sirt1 decreased the secretion of IFN-β upon the stimulation of HSV60 or VACV70 (Fig. 2E). Additionally, Sirt1 overexpression increased the HSV-1 titer, as suggested by the plaque assays (Fig. 2F). Taken together, these findings indicate that the overexpression of Sirt1 inhibited innate immune responses that were triggered by DNA viruses or viral DNA but not by RNA viruses.
FIG 2.
Sirt1 overexpression inhibits HSV-1- or exogenous cytosolic DNA-induced innate immune responses. (A) HaCaT keratinocytes were transfected with an IFN-β luciferase reporter, together with Myc-Sirt1 or vector (Vec). At 24 h later, the cells were transfected with HSV60 (1 μg/mL) for another 24 h and then lysed for the luciferase assay. (B) HaCaT keratinocytes were transfected with Myc-Sirt1 or control vector (Vec). At 24 h after transfection, the cells were infected with HSV-1 (MOI = 1) for 8 h. Afterward, real-time PCR was performed. (C) HaCaT keratinocytes were transfected with Myc-Sirt1 or control vector (Vec). At 24 h after transfection, the cells were transfected with HSV60 (1 μg/mL), VACV70 (1 μg/mL), or poly (I:C) (2.5 μg/mL) for 8 h. Afterward, real-time PCR was performed. (D) HaCaT keratinocytes were transfected with Myc-Sirt1 (+) or control vector (−). At 24 h after transfection, the cells were infected with HSV-1 (MOI = 1) or VSV (MOI = 1) for 8 h. Afterward, the cell lysates were subjected to immunoblot analysis. β-Actin was used as a loading control in the immunoblot assays. (E) HaCaT keratinocytes were transfected with Myc-Sirt1 or control vector (Vec). At 24 h after transfection, the cells were transfected with HSV60 (1 μg/mL) or VACV70 (1 μg/mL) for 24 h. The supernatants were collected and subjected to ELISA analysis. (F) HaCaT keratinocytes were transfected with a control vector (Vec) or the Sirt1 plasmid and then infected with HSV-1 for 24 h. The titers of HSV-1 were determined by a standard plaque assay. The data are representative of three independent experiments and are presented as means ± SDs. *, P < 0.05; **, P < 0.01.
Sirt1 knockdown promotes HSV-1- or viral DNA-induced innate immune responses.
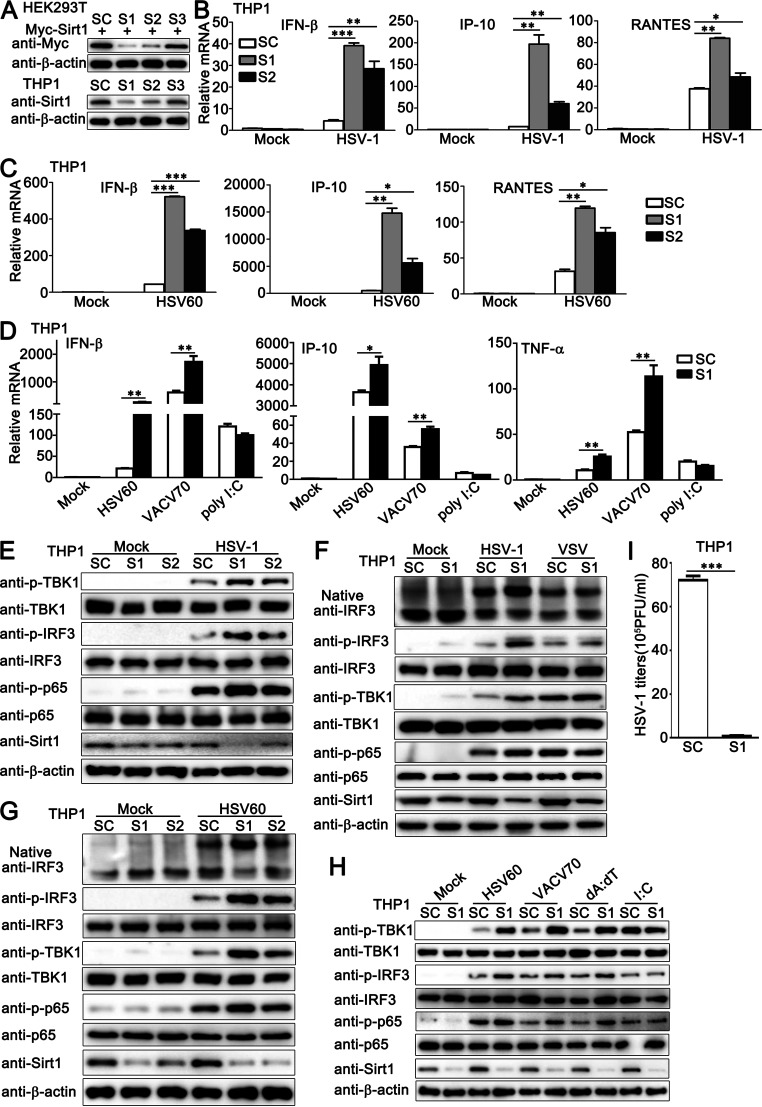
Next, we used a knockdown approach to further address the effect of endogenous Sirt1 on HSV-1- or viral DNA-triggered innate immune responses. Sirt1-transfected HEK293T or PMA-THP1 cells were transfected with control siRNA (SC) or three pairs of small interfering RNA (siRNA) oligonucleotides specific for Sirt1 RNA (S1, S2, S3). As shown in Fig. 3A, both S1 and S2 inhibited exogenous and endogenous Sirt1 expression efficiently, and S1 showed higher efficiency in inhibition than S2. Thus, both S1 and S2 were used in the following experiments. As shown in Fig. 3B and C, both of the siRNAs targeting Sirt1, S1 and S2, upregulated the production of IFN-β, IP-10, and RANTES upon HSV-1 infection or HSV60 transfection in mRNA levels, and the increase by S1 transfection was more significant than that by S2 transfection, consistent with their efficiency in the inhibition of Sirt1 expression (Fig. 3B and C). Similar results were obtained by the stimulation with viral DNA VACV70 but not by poly (I:C) transfection (Fig. 3D). Notably, Sirt1 knockdown did not enhance poly (I:C) transfection-triggered production of IFN-β, IP-10, and TNF-α but caused a trend of inhibition in the expression of these proteins at the mRNA level (Fig. 3D). In immunoblot assays, upon HSV-1 infection or HSV60 stimulation, higher levels of activation of TBK1, IRF3, and p65 were observed in cells transfected with S1 or S2 than in control SC-transfected cells, and cells with S1 transfection exhibited the highest phosphorylation levels of these proteins (Fig. 3E and G). However, Sirt1 knockdown did not show significant differences in signaling activation upon VSV infection or poly (I:C) transfection (Fig. 3F and H). Furthermore, Sirt1 knockdown inhibited the titers of HSV-1 in plaque assays (Fig. 3I). Finally, to exclude the possibility that the effect of Sirt1 knockdown on innate immune responses was limited to immune cells, we further investigated the role of Sirt1 in nonimmune cells, HaCaT cells, and HeLa cells. Consistent with the results obtained in PMA-THP1 cells, in both HaCaT and HeLa cells, upon HSV-1 infection, the knockdown of Sirt1 promoted the production of IFN-β, IP-10, interferon-stimulated gene 56 (ISG56), and TNF-α and the activation of TBK1 and IRF3 (see Fig. S1A to D in the supplemental material). Taken together, these results demonstrate that endogenous Sirt1 negatively regulates innate immune responses triggered by DNA virus HSV-1 or viral DNA but not by RNA virus VSV or poly (I:C) transfection.
FIG 3.
Sirt1 knockdown promotes HSV-1- or exogenous cytosolic DNA- induced innate immune responses. (A) HEK293T cells were transfected with Myc-Sirt1 and then transfected with control siRNA (SC) or Sirt1-specific siRNA (S1, S2, and S3). At 24 h after transfection, the cells were lysed for immunoblot assays (top). PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1, S2, and S3). At 24 h after transfection, the cells were subjected to immunoblot assay (bottom). (B and C) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1 and S2) for 24 h and then stimulated with HSV-1 (MOI = 1) (B) or HSV60 (1 μg/mL) transfection (C) for 8 h. The cells were lysed for real-time PCR analysis. (D) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1) for 24 h and then transfected with HSV60 (1 μg/mL), VACV70 (1 μg/mL), and poly (I:C) (2.5 μg/mL) for 8 h. Then, the cells were lysed for real-time PCR analyses. (E) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1 and S2) for 24 h and then infected with HSV-1 (MOI = 1) for 4 h. The cells were lysed for immunoblot assays. (F) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1) for 24 h and then infected with HSV-1 (MOI = 1) or VSV (MOI = 1) for 4 h. The cells were lysed for native PAGE or SDS-PAGE assays. (G) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1 and S2) for 24 h and then transfected with HSV-60 (1 μg/mL) for 4 h. The cells were lysed for native PAGE or SDS-PAGE assays. (H) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1) for 24 h and then transfected with HSV60 (1 μg/mL), VACV70 (1 μg/mL), poly(dA·dT) (dA:dT; 1 μg/mL), and poly (I:C) (I:C; 2.5 μg/mL) for 4 h. The cells were lysed for immunoblot assays. (I) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1) for 24 h and then infected with HSV-1 for 24 h. The titers of HSV-1 were determined by a standard plaque assay. β-Actin served as a loading control in all the immunoblot assays. The data are representative of three independent experiments and are presented as means ± SDs. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Sirt1 deficiency enhances HSV-1- or viral DNA-triggered innate immune responses in PMA-THP1 cells.
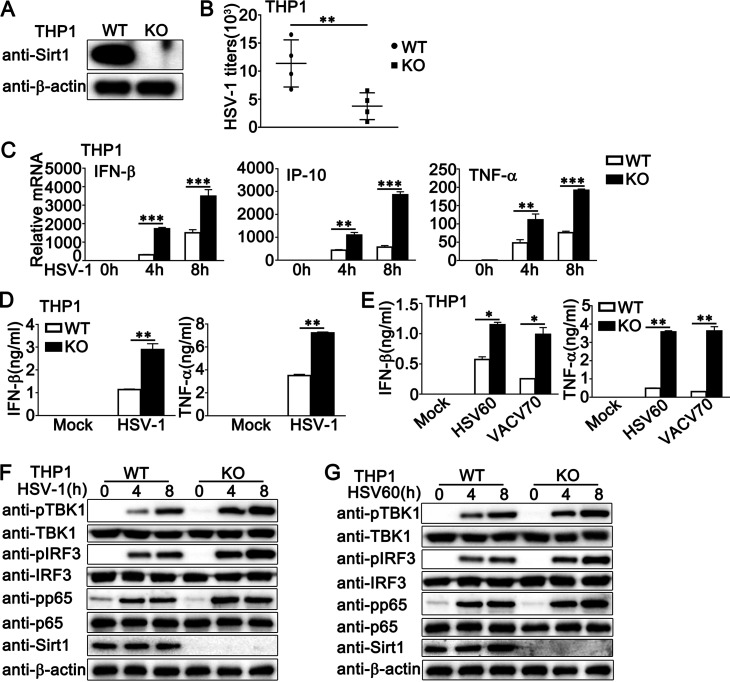
To further explore the role of Sirt1 in antiviral innate immune responses, we generated a Sirt1-deficient THP1 cell line by CRISPR/Cas9 strategy. The expression of Sirt1 could not be detected in Sirt1-deficient PMA-THP1 cells, as suggested by immunoblot assays (Fig. 4A). Compared to wild-type PMA-THP1 cells, Sirt1-deficient cells exhibited decreased HSV-1 titers (Fig. 4B) upon HSV-1 infection. Next, we examined the antiviral innate immune responses in Sirt1-deficient cells. Increased production of IFN-β, IP-10, and TNF-α was observed in mRNA levels in Sirt1-deficient PMA-THP1 cells upon HSV-1 infection (Fig. 4C). Results from enzyme-linked immunosorbent assay (ELISA) analysis indicated that the depletion of Sirt1 promoted HSV-1- or viral DNA-induced production of IFN-β and also TNF-α in protein levels (Fig. 4D and E). Further, Sirt1 depletion increased the phosphorylation of TBK1, IRF3, and p65, which were triggered by HSV-1 infection (Fig. 4F) or HSV60 stimulation (Fig. 4G). Taken together, our findings demonstrate that Sirt1 deficiency enhances antiviral innate immune responses against DNA virus, suggesting the inhibitory role of Sirt1 in DNA virus- or viral DNA-triggered innate immune responses.
FIG 4.
Sirt1 deficiency promotes HSV-1- or exogenous cytosolic DNA-triggered innate immune responses in PMA-THP1 cells. (A) Wild-type (WT) and Sirt1-deficient (knockout [KO]) PMA-THP1 cells were lysed for immunoblot assays. (B) WT and KO PMA-THP1 cells were infected with HSV-1 (MOI = 1) for 24 h. The titers of HSV-1 were determined by standard plaque assay. Data were obtained from four independent experiments. (C) WT and KO PMA-THP1 cells were infected with HSV-1 (MOI = 1) for the indicated periods. Then, the cells were lysed for real-time PCR analyses. (D) WT and KO PMA-THP1 cells were infected with HSV-1 (MOI = 1) for 24 h. The supernatants were collected and subjected to ELISA analysis. (E) WT and KO PMA-THP1 cells were transfected with HSV60 (1 μg/mL) and VACV70 (1 μg/mL) for 24 h. The supernatants were collected and subjected to ELISA analysis. (F and G) WT and KO PMA-THP1 cells were treated with HSV-1 (MOI = 1) (F) or HSV60 (1 μg/mL) transfection (G) for the indicated periods. The cells were lysed for immunoblot assays. β-Actin served as a loading control in all the immunoblot assays. The data are representative of three independent experiments and are presented as means ± SDs. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Sirt1 deficiency enhances the expression of type I IFN, proinflammatory cytokines, and ISGs.
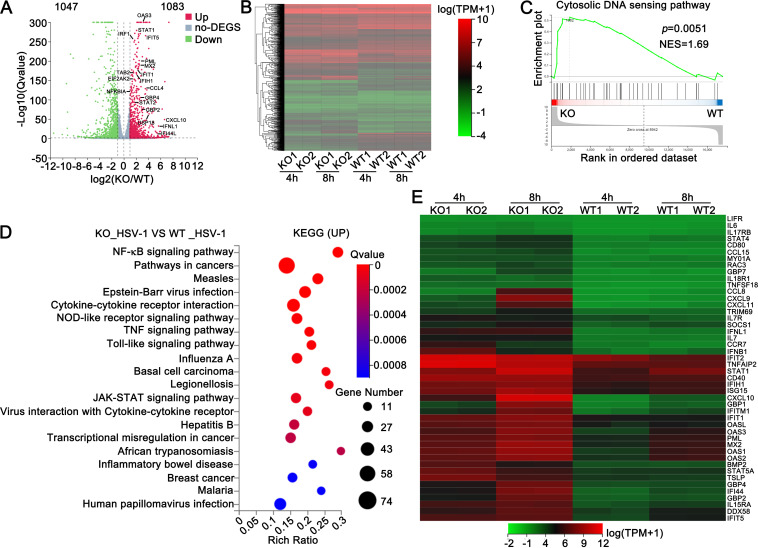
We further performed global RNA sequencing (RNA-seq) analysis to identify genes and pathways regulated by Sirt1. Differential expression analysis identified 1,083 upregulated genes (log2 > 1) and 1,047 downregulated genes (log2 < −1) in response to Sirt1 deficiency in PMA-THP1 cells after HSV-1 infection (Fig. 5A and B). Gene set-enrichment analysis (GSEA) was performed, and the results demonstrated that the core-enriched genes were related to the cytosolic DNA sensing pathway (Fig. 5C). Additionally, gene ontology (GO) analysis further demonstrated that these genes were involved in cellular antiviral immune responses against DNA viruses, including “Epstein-Barr virus infection,” “hepatitis B,” and “human papillomavirus infection” (Fig. 5D). Differential expression analysis further displayed that Sirt1 deficiency resulted in the upregulation of many type I IFN-induced IFN-stimulated genes (ISGs) and proinflammatory cytokines upon HSV-1 infection (log2 > 2) (Fig. 5E). In all, our findings suggest that Sirt1 negatively regulates antivirus responses during viral infection by controlling the expression of type I IFNs, proinflammatory cytokines, and ISGs.
FIG 5.
Sirt1 deficiency enhances the expression of type I IFN, ISGs, and proinflammatory cytokines. (A) Volcano plots comparing gene expression in wild-type (WT) and Sirt1-deficient (KO) PMA-THP1 cells after being stimulated with HSV-1 (MOI = 1) for 4 h. (B) Heat map depiction of differentially expressed genes in WT and KO PMA-THP1 cells. (C) GSEA analysis of cytosolic DNA sensing pathway relative to WT and KO PMA-THP1 cells. Enrichment plots are shown along with the normalized enrichment score and P value. (D) KEGG analysis through Tom (https://biosys.bgi.com/) for genes significantly upregulated (log2 ≥ 1, P < 0.05) after stimulation with HSV-1 (MOI = 1) for 4 h. (E) Heat map view of mRNA variations in type I IFN, ISGs, and proinflammatory cytokines (log2 ≥ 2).
Sirt1 decreases IFI16 acetylation.
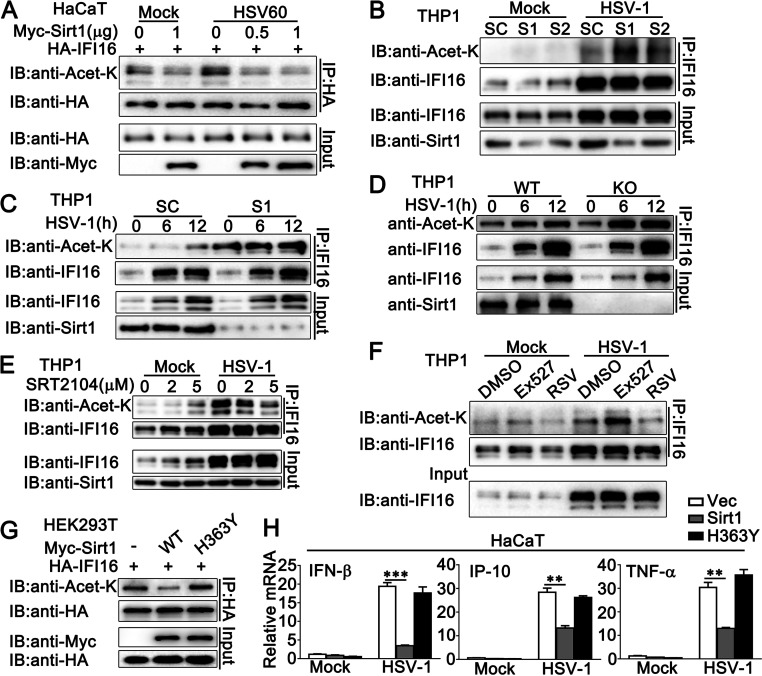
To investigate the mechanism by which Sirt1 regulated IFI16-mediated antiviral immune responses, the effect of Sirt1 on the acetylation of IFI16 was examined. As shown in Fig. 6A, Sirt1 deacetylated IFI16 with or without HSV60 transfection. This deacetylation was more significant after the stimulation of viral DNA HSV60 (Fig. 6A). Consistently, in PMA-THP1 cells, Sirt1 knockdown by S1 or S2 enhanced the acetylation of IFI16 upon HSV-1 infection, and the effects of S1 were more significant than those of S2, which was consistent with their inhibitory efficiency on Sirt1 expression (Fig. 6B and C). Then, the acetylation of IFI16 was examined in Sirt1-deficient PMA-THP1 cells. After HSV-1 infection, Sirt1-deficient cells exhibited a markedly higher level of IFI16 acetylation than that exhibited by wild-type PMA-THP1 cells (Fig. 6D). Then, Sirt1 activators, SRT2104 and resveratrol (RSV), and Sirt1 inhibitor Ex527 were used to investigate the role of Sirt1 in IFI16 acetylation. As expected, both SRT2104 and RSV decreased HSV-1-induced IFI16 acetylation in a dose-dependent pattern, whereas Sirt1 inhibitor Ex527 increased the acetylation of IFI16 significantly (Fig. 6E and F; Fig. S2A). Finally, the H363Y mutant, an enzymatically dead dominant negative mutant of Sirt1, was used to further investigate whether the deacetylase activity of Sirt1 was required for its role in IFI16-mediated innate immune responses. As expected, the exogenously expressed H363Y mutant of Sirt1 could not inhibit the acetylation of IFI16 as efficiently as the wild-type Sirt1 did (Fig. 6G). Upon HSV-1 infection, the H363Y mutant of Sirt1 lost the ability to affect the induction of IFN-β, IP-10, and TNF-α (Fig. 6H). Taken together, our findings suggest that Sirt1 decreases the acetylation of IFI16 and that the deacetylase activity of Sirt1 is required to regulate IFI16-mediated host defense.
FIG 6.
Sirt1 decreases IFI16 acetylation. (A) HaCaT keratinocytes were transfected with the indicated plasmids for 24 h and then transfected with HSV60 (1 μg/mL) for 12 h or left untreated. Afterward, the cells were lysed and subjected to immunoprecipitation (IP) and immunoblot (IB) analysis. (B) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1 and S2) for 24 h and then infected with HSV-1 (MOI = 1) for 12 h. Afterward, the cells were lysed and subjected to IP and IB analysis. (C) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1) for 24 h and then infected with HSV-1 (MOI = 1) for the indicated periods. Afterward, the cells were lysed and subjected to IP and IB analysis. (D) Wild-type (WT) and Sirt1-deficient (KO) PMA-THP1 cells were infected with HSV-1 (MOI = 1) for the indicated periods. The cells were subjected to IP and IB analysis. (E) PMA-THP1 cells were treated with increasing amounts of SRT2104 (0, 2, and 5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for another 12 h. The cells were subjected to IP and IB analysis. (F) PMA-THP1 cells were treated with DMSO, EX527 (5 μM), or resveratrol (RSV) (100 μM) for 12 h and then infected with HSV-1 (MOI = 1) for another 12 h. The cells were subjected to IP and IB analysis. (G) HEK293T cells were transfected with the indicated plasmids. At 24 h after transfection, the cells were subjected to IP and IB analysis. (H) HaCaT keratinocytes were transfected with the indicated plasmids for 24 h and then infected with HSV-1 (MOI = 1) for 8 h. Then, the cells were lysed for real-time PCR analyses. The data are representative of three independent experiments and are presented as means ± SDs. **, P < 0.01; ***, P < 0.001.
Sirt1 inhibitors and activators regulate DNA virus- or viral DNA-triggered innate immune responses.
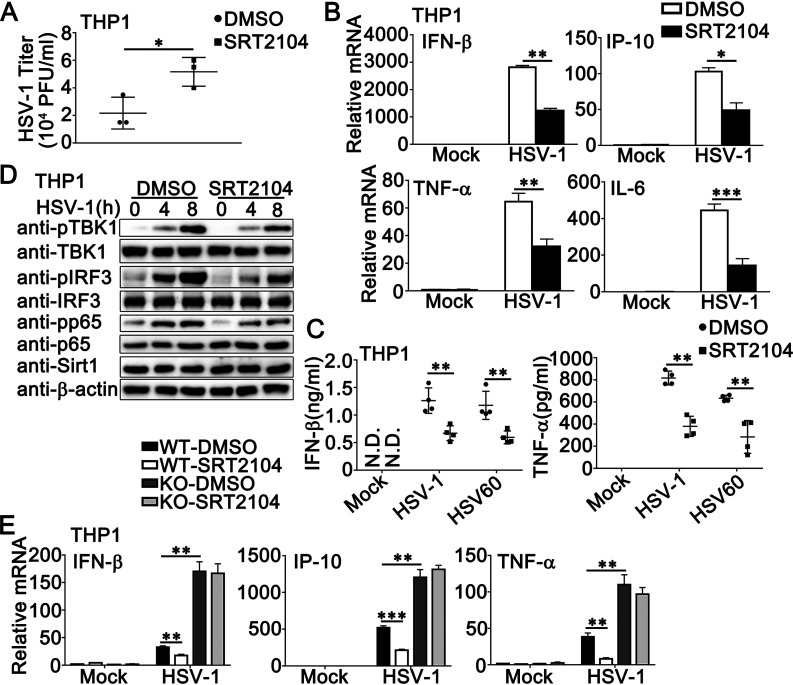
Given our findings that the deacetylase activity of Sirt1 was essential to its regulatory role in antiviral responses, we used the activator or inhibitor of Sirt1 to regulate its deacetylase activity and then confirmed the effect of Sirt1 on the innate immune responses that were induced by DNA virus or viral DNA. As shown in Fig. 7A, Sirt1 activator SRT2104 increased the titers of HSV-1 in PMA-THP1 cells. The treatment with SRT2104 inhibited HSV-1- or HSV-60-triggered antiviral innate immune responses, including the production of IFN-β, IP-10, IL-6, and TNF-α in mRNA levels (Fig. 7B), the induction of IFN-β and TNF-α in protein levels (Fig. 7C), and the phosphorylation of TBK1, IRF3, and p65 (Fig. 7D), whereas SRT2104 treatment did not affect the expression of Sirt1 (Fig. 7D), suggesting that the antiviral innate immune responses against DNA virus or viral DNA were regulated by the deacetylase activity of Sirt1. Similar results were obtained by the treatment of RSV in PMA-THP1 cells (Fig. S2B to D). Finally, we examined whether the effect of SRT2104 on innate immune responses was dependent on Sirt1. Wild-type or Sirt1-deficient PMA-THP1 cells were treated with or without SRT2104 and then infected with HSV-1. As shown in Fig. 7E, the antiviral immune responses were inhibited by SRT2104 in wild-type PMA-THP1 cells but not in Sirt1-deficient PMA-THP1 cells, indicating that the role of SRT2104 in antiviral signaling relied on the presence of Sirt1.
FIG 7.
Sirt1 activator SRT2104 inhibits HSV-1- or exogenous cytosolic DNA-induced innate immune responses. (A) PMA-THP1 cells were treated with SRT2104 (5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for 24 h. The titers of HSV-1 were determined by a standard plaque assay. Data were obtained from three independent experiments. (B) PMA-THP1 cells were treated with SRT2104 (5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for 8 h. Afterward, real-time PCR analysis was performed. (C) PMA-THP1 cells were treated with SRT2104 (5 μM) for 12 h and then stimulated with HSV-1 (MOI = 1) or HSV60 (1 μg/mL) transfection. The supernatants were collected and subjected to ELISA analysis. Data were obtained from four independent experiments. (D) PMA-THP1 cells were treated with SRT2104 (5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for the indicated periods. Afterward, the cells were lysed for immunoblot analysis. (E) Wild-type (WT) and Sirt1-deficient (KO) PMA-THP1 cells were treated with control DMSO or SRT2104 (5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for 4 h. Afterward, real-time PCR analysis was performed. β-Actin served as a loading control in all the immunoblot assays. The data are representative of three independent experiments and are presented as means ± SDs. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
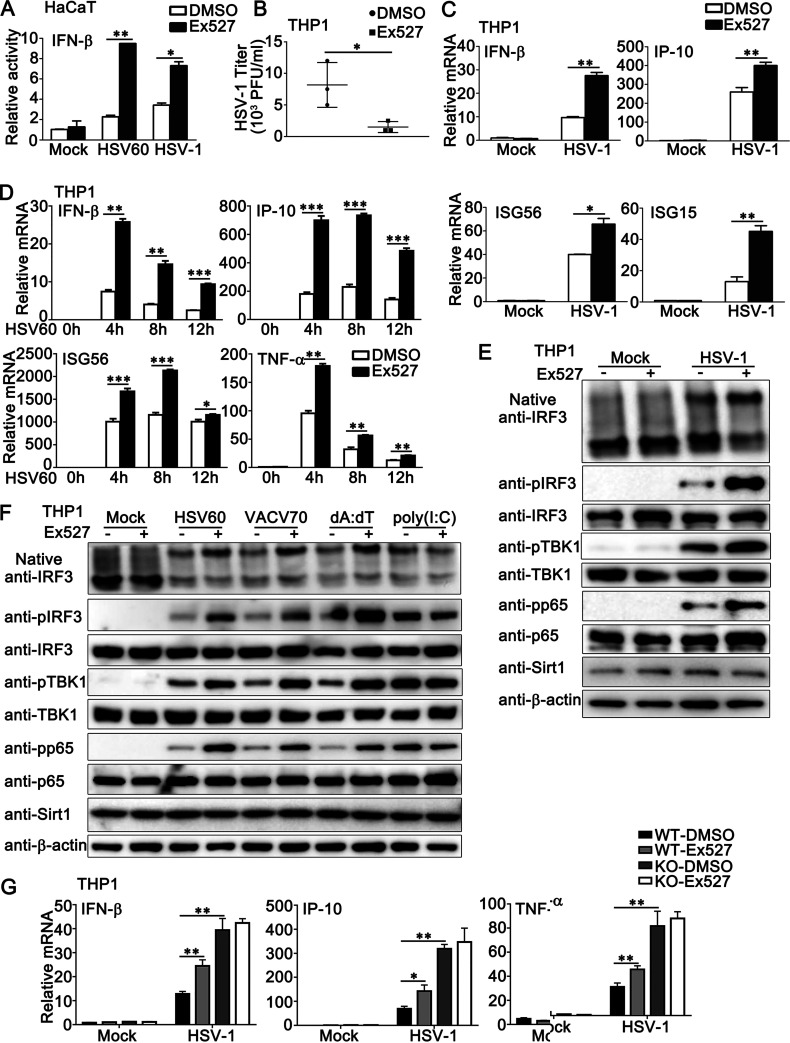
Next, we investigated the regulation of antiviral innate immune responses by Ex527, the inhibitor of Sirt1. In HaCaT cells with the transfection of an IFN-β luciferase reporter, Ex527 treatment enhanced the IFN-β reporter activity induced by HSV-1 infection or HSV60 transfection (Fig. 8A). In PMA-THP1 cells, Ex527 treatment decreased HSV-1 titers, as shown by plaque assays (Fig. 8B). Real-time PCR results indicated that the induction of IFN-β, IP-10, ISG56, and ISG15 by HSV-1 infection was promoted by Ex527 treatment in PMA-THP1 cells (Fig. 8C). Similar results were obtained when PMA-THP1 cells were treated with Ex527 and then stimulated by HSV60 (Fig. 8D). In addition, Ex527 treatment enhanced the formation of IRF3 dimers and the phosphorylation of IRF3, TBK1, and p65 that was triggered by HSV-1 infection (Fig. 8E) or exogenous cytosolic DNA transfection but not by poly (I:C) transfection (Fig. 8F). However, Ex527 could not regulate the antiviral innate immune responses against HSV-1 in the absence of Sirt1, indicating that Ex527 regulated DNA virus-triggered innate immune responses via Sirt1 (Fig. 8G). Taken together, our results demonstrate that Sirt1 inhibitors and activators regulate DNA virus- or viral DNA-triggered innate immune responses.
FIG 8.
Sirt1 inhibitor Ex527 promotes HSV-1- or exogenous cytosolic DNA-induced innate immune responses. (A) HaCaT keratinocytes were transfected with an IFN-β luciferase reporter. At 24 h later, the cells were treated with Ex527 (5 μM) for another 12 h. Then, the cells were stimulated with HSV60 (1 μg/mL) transfection or HSV-1 (MOI = 1) for 24 h and subjected to a luciferase assay. (B) PMA-THP1 cells were treated with Ex527 (5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for 24 h. The titers of HSV-1 were determined by a standard plaque assay. Data were obtained from three independent experiments. (C and D) PMA-THP1 cells were treated with Ex527 (5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for 4 h (C) or transfected with HSV60 (1 μg/mL) for the indicated periods (D). Afterward, real-time PCR analysis was performed. (E and F) PMA-THP1 cells were treated with Ex527 (5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for 8 h (E) or transfected with HSV60 (1 μg/mL), VACV70 (1 μg/mL), poly(d·dT) (dA:dT; 1 μg/mL), or poly (I:C) (2.5 μg/mL) (F) for 8 h. Then the cells were lysed for native PAGE and SDS-PAGE assays. (G) Wild-type (WT) and Sirt1-deficient (KO) PMA-THP1 cells were treated with control DMSO or Ex527 (5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for 4 h. Afterward, real-time PCR analysis was performed. β-Actin served as a loading control in all the immunoblot assays. The data are representative of three independent experiments and are presented as means ± SDs. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Sirt1 prevents DNA virus- or viral DNA-induced cytoplasmic translocation of IFI16.
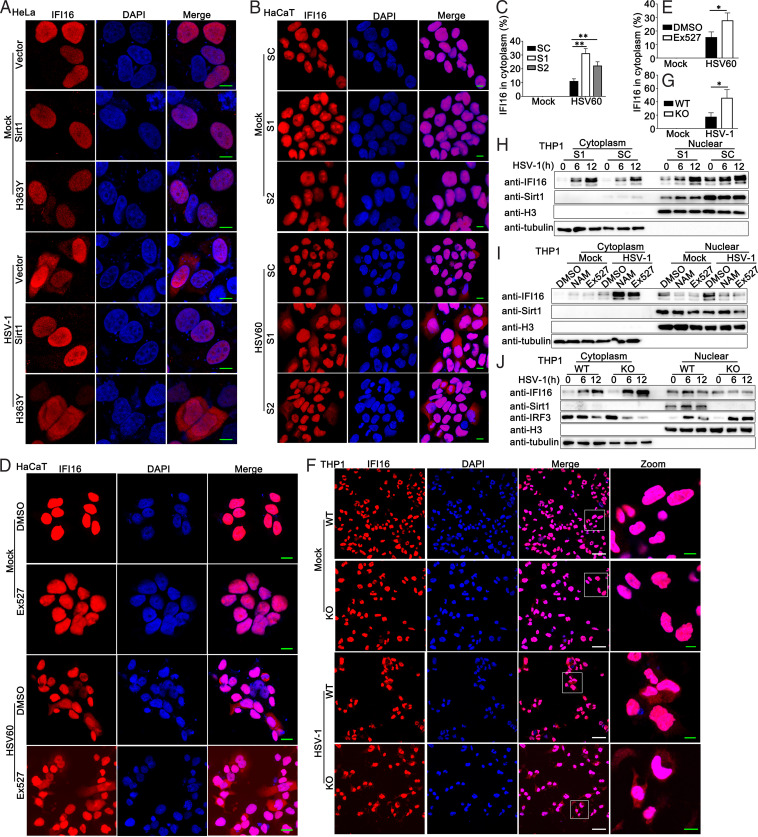
It has been reported that herpesvirus genome recognition induced acetylation of nuclear IFI16, which is essential for the cytoplasmic translocation of IFI16 and IFN-β responses (14). Therefore, we examined the role of Sirt1 in the redistribution of IFI16 upon DNA virus or viral DNA stimulation. As shown in Fig. 9A, in HeLa cells, Sirt1 overexpression inhibited HSV-1-induced cytoplasmic translocation of IFI16, whereas the H363Y mutant of Sirt1 transfection did not exhibit the inhibitory function under fluorescence microscopy. In HaCaT cells with HSV60 stimulation, Sirt1 knockdown or Ex527 treatment promoted the cytoplasmic translocation of IFI16 (Fig. 9B to E). Consistently, increased cytoplasmic IFI16 localization was detected in Sirt1-deficient cells (Fig. 9F and G). Next, we tried to confirm these findings about IFI16 redistribution by immunoblot assays. PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1 siRNA (S1) and then infected with HSV-1. The cells were fractionated into nuclear and cytoplasmic components. The purity of the nuclear and cytoplasmic fractions was monitored by the presence of histone H3 protein and tubulin, respectively (Fig. 9H). HSV-1 infection induced the cytoplasmic translocation of IFI16, which was enhanced by Sirt1 knockdown (Fig. 9H). In addition, both Sirtuin family inhibitor NAM and Sirt1 inhibitor Ex527 promoted cytoplasmic translocation of IFI16 (Fig. 9I). Compared to wild-type PMA-THP1 cells, Sirt1-deficient PMA-THP1 cells exhibited increased accumulation of IFI16 in the cytoplasm upon HSV-1 infection (Fig. 9J). In all, these results suggest that Sirt1 prevents DNA virus- or viral DNA-induced cytoplasmic translocation of IFI16.
FIG 9.
Sirt1 prevents HSV-1- or exogenous cytosolic DNA-induced cytoplasmic translocation of IFI16. (A) HeLa cells were transfected with a control vector, Sirt1 (WT), or H363Y mutant of Sirt1. At 24 h after transfection, the cells were infected with HSV-1 (MOI = 1) or left uninfected for another 8 h. Immunofluorescence was performed using anti-IFI16 (red). Nuclei were stained with DAPI. Scale bars, 10 μm. (B) HaCaT keratinocytes were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1 and S2). At 24 h after transfection, the cells were transfected with HSV60 (1 μg/mL) for another 8 h. Immunofluorescence was performed using anti-IFI16 (red). Nuclei were stained with DAPI. Scale bars, 10 μm. (C) Quantification of the cytoplasmic IFI16 redistribution in panel B. (D) HaCaT keratinocytes were treated with Ex527 (5 μM) for 12 h and then transfected with HSV60 (1 μg/mL) for 8 h. Immunofluorescence was performed using anti-IFI16 (red). Nuclei were stained with DAPI. Scale bars, 10 μm. (E) Quantification of the cytoplasmic IFI16 redistribution in panel D. (F) Wild-type (WT) and Sirt1-deficient (KO) PMA-THP1 cells were infected with HSV-1 (MOI = 1) or left uninfected for 8 h. Immunofluorescence was performed using anti-IFI16 (red). Nuclei were stained with DAPI. Scale bars in merge or zoom images are 50 μm or 10 μm, respectively. (G) Quantification of the cytoplasmic IFI16 redistribution in panel F. (H) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1). At 24 h after transfection, the cells were infected with HSV-1 (MOI = 1) for the indicated periods and then fractionated into cytosolic and nuclear subfractions. The immunoblot assays were performed as indicated. (I) PMA-THP1 cells were treated with control DMSO, NAM (10 mM) for 6 h, or Ex527 (5 μM) for 12 h. Afterward, the cells were infected with HSV-1 (MOI = 1) or left uninfected for another 12 h and then fractionated into cytosolic and nuclear subfractions. The immunoblot assays were performed as indicated. (J) Wild-type (WT) and Sirt1-deficient (KO) PMA-THP1 cells were infected with HSV-1 (MOI = 1) for the indicated periods and then fractionated into cytosolic and nuclear subfractions. The immunoblot assays were performed as indicated. The data are representative of three independent experiments.
Sirt1 prevents the association of IFI16 with STING.
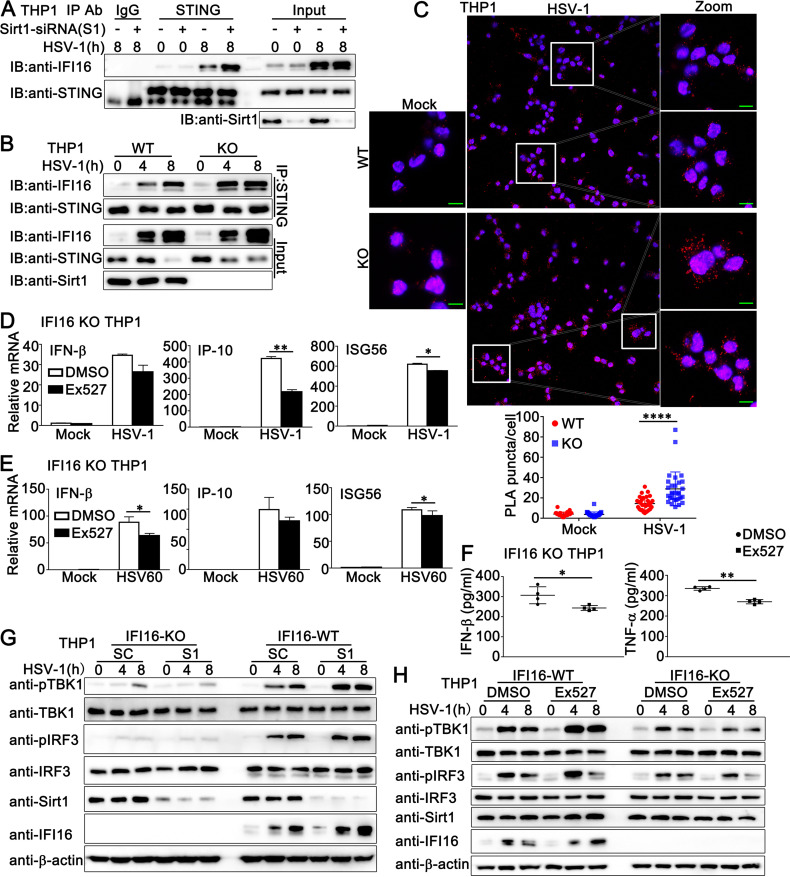
IFI16 acetylation has been reported to be required for its cytoplasmic interaction with STING and subsequent IFN-β production (21). Therefore, we examined the role of Sirt1 in the IFI16-STING interaction during DNA virus infection. Coimmunoprecipitation reactions with an anti-STING antibody revealed that Sirt1 knockdown or deficiency promoted IFI16-STING interaction (Fig. 10A and B). Further, Ex527 treatment enhanced IFI16-STING interaction, suggesting that Sirt1 regulates the association of IFI16 with STING by manipulating acetylation (Fig. S3A). Next, we performed an in situ proximity ligation assay (PLA) to validate the effect of Sirt1 on IFI16-STING interaction. Wild-type or Sirt1-deficient PMA-THP1 cells were infected with HSV-1 or left uninfected and then subjected to PLAs using anti-IFI16 and anti-STING antibodies. The detected red dots depicted the colocalization of IFI16 and STING. As shown in Fig. 10C, with HSV-1 infection, more red dots were detected in Sirt1-deficient PMA-THP1 cells than in wild-type PMA-THP1 cells, suggesting that Sirt1 deficiency promoted the IFI16-STING interaction. Finally, confocal microscope analysis demonstrated that Sirt1 deficiency or Ex527 treatment resulted in more cytoplasmic IFI16 distribution, leading to more IFI16-STING colocalization, suggesting that Sirt1 inhibited the IFI16 interaction with STING by regulating IFI16 distribution (Fig. S3B and C). Taken together, these results suggest that Sirt1 prevents the IFI16-STING interaction.
FIG 10.
Sirt1 regulates DNA virus- or viral DNA-induced signal transduction in an IFI16-dependent pattern. (A) PMA-THP1 cells were transfected with control siRNA (−) or Sirt1-specific siRNA (S1) (+). At 24 h after transfection, the cells were infected with HSV-1 (MOI = 1) for 8 h or left uninfected and then lysed for immunoprecipitation (IP) and immunoblot (IB) analysis. (B) Wild-type (WT) and Sirt1-deficient (KO) PMA-THP1 cells were infected with HSV-1 (MOI = 1) for the indicated periods and then lysed for IP and IB analysis. (C) WT and KO PMA-THP1 cells were infected with HSV-1 (MOI = 1) for 8 h or left uninfected. In situ PLAs were performed to examine the colocalization of IFI16 and STING. IFI16-STING complex, red; nuclei, blue. Scale bars, 10 μm. PLA puncta quantification is shown in the bottom panel. Quantitative data are presented as means ± SDs of triplicate samples (50 cells per sample). (D) IFI16-deficient PMA-THP1 cells were treated with control DMSO or Ex527 (5 μM) for 12 h. Then, the cells were infected with HSV-1 (MOI = 1) or left uninfected for 8 h and lysed for real-time analysis. (E) IFI16-deficient PMA-THP1 cells were treated with DMSO or Ex527 (5 μM) for 12 h. Then, the cells were transfected with HSV60 (1 μg/mL) for 8 h and lysed for real-time analysis. (F) IFI16-deficient PMA-THP1 cells were treated with DMSO or Ex527 (5 μM) for 12 h. Then, the cells were infected with HSV-1 (MOI = 1) for 24 h and the supernatants were subjected to ELISA analysis. Data were obtained from four independent experiments. (G) Wild-type (WT) and IFI16-deficient (KO) PMA-THP1 cells were transfected with control siRNA (SC) or Sirt1-specific siRNA (S1). At 24 h after transfection, the cells were infected with HSV-1 (MOI = 1) for the indicated periods and then lysed for immunoblot analysis. (H) Wild-type (WT) and IFI16-deficient (KO) PMA-THP1 cells were treated with control DMSO or Ex527 (5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for the indicated periods. Afterward, the cells were lysed for immunoblot assay. The data are representative of three independent experiments and are presented as means ± SDs. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Sirt1 inhibits DNA virus- or viral DNA-triggered innate immune responses in an IFI16-dependent pattern.
To explore whether the inhibitory role of Sirt1 in HSV-1-induced antiviral signaling was dependent on IFI16, we first examined the function of Sirt1 in IFI16-silenced HaCaT cells. HaCaT cells were transfected with control siRNA (SC) or IFI16 siRNA (Si-IFI16) and then stimulated them with SRT2104 or EX527, followed by HSV-1 infection. As shown in Fig. S4A and B, upon HSV-1 infection, Ex527 increased antiviral innate immune responses in control HaCaT cells but not in IFI16-silenced HaCaT cells. Similarly, SRT2104 could not inhibit HSV-1-triggered antiviral responses after IFI16 knockdown in HaCaT cells (Fig. S4A and B). Next, we investigated the role of Sirt1 in IFI16-deficient cells. Wild-type or IFI16-deficient PMA-THP1 cells were treated with Sirt1 inhibitor Ex527 and then stimulated with HSV-1 infection or HSV60 transfection. As shown in Fig. 10D, in contrast to the findings in wild-type PMA-THP1 cells (Fig. 8C), in IFI16-deficient cells, Ex527 caused a slight drop in IFN-β production and a significant decrease in the production of IP-10 and ISG56 in response to HSV-1 infection. Similar results were observed in IFI16-deficient PMA-THP1 cells upon HSV60 transfection (Fig. 10E). Consistently, ELISA analysis indicated that Ex527 treatment caused lowered protein levels of IFN-β and TNF-α in IFI16-deficient PMA-THP1 cells after HSV-1 infection (Fig. 10F). In addition, after HSV-1 infection, whereas Sirt1 knockdown or Ex527 treatment resulted in enhanced activation of TBK1 and IRF3 in wild-type PMA-THP1 cells, Sirt1 knockdown or Ex527 treatment resulted in no elevation in the phosphorylation of TBK1 in IFI16-deficient PMA-THP1 cells (Fig. 10G and H). We even observed that Ex527 treatment caused a drop in HSV-1-triggered phosphorylation of IRF3 in IFI16-deficient PMA-THP1 cells (Fig. 10H).
Considering that cGAS is an important DNA sensor involved in HSV-1 infection, we wondered whether cGAS was involved in the regulation of antiviral immune responses by Sirt1. First, we examined the interaction between cGAS and Sirt1 by coimmunoprecipitation assays. As shown in Fig. S4C, Sirt1 did not interact with cGAS. Then, we transfected cGAS and STING into HEK293T cells and explored the effect of Sirt1 on type I IFN production. As shown in Fig. S4D, Sirt1 could not affect cGAS/STING-triggered activation of IFN-β or interferon-sensitive response element (ISRE) reporter in luciferase assays. Consistently, we did not observe a significant effect of Sirt1 on cGAS/STING-induced phosphorylation of TBK1 and IRF3 (Fig. S4E). Next, we investigated the role of Sirt1 in the transfected cyclic GMP-AMP (cGAMP)-induced signaling pathway. Wild-type and Sirt1-deficient PMA-THP1 cells were stimulated with transfected cGAMP, and the innate immune responses were evaluated. Compared to wild-type PMA-THP1 cells, upon cGAMP stimulation, Sirt1-deficient cells exhibited reduced production of IFN-β and ISG56, with decreased activation of TBK1 and IRF3, suggesting that Sirt1 played different roles in the regulation of signaling pathways in response to cGAMP and HSV-1 (Fig. S4F to H).
Taken together, our findings suggest that the inhibitory role of Sirt1 in DNA virus- or viral DNA-triggered innate immune responses is dependent on IFI16.
Sirt1 does not inhibit HSV-1 or viral DNA-induced signaling pathways in mouse cells.
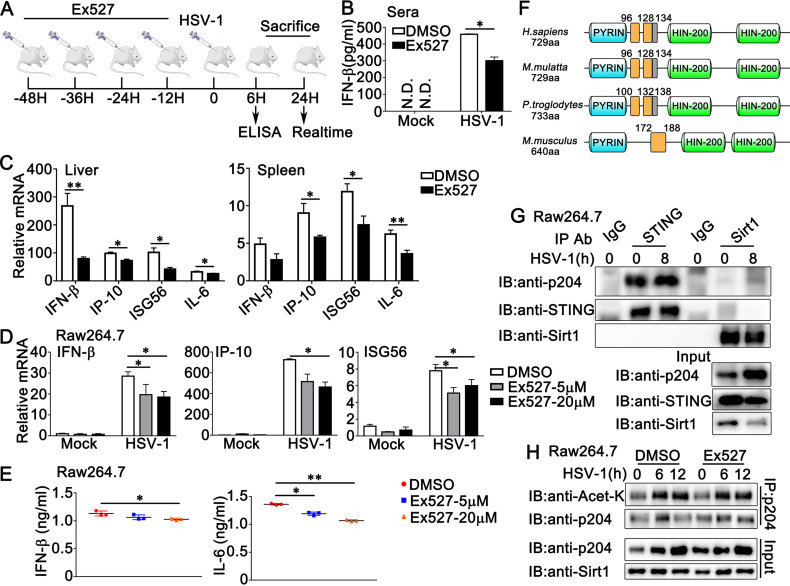
To further investigate the role of Sirt1 in DNA virus infection in vivo, we injected mice with Sirt1 inhibitor Ex527 or DMSO as a control every 12 h. At 48 h after the first injection, the mice were infected with HSV-1. The mice were sacrificed and subjected to ELISA and real-time PCR assays after the indicated periods, as shown in Fig. 11A. Surprisingly, mice injected with Ex527 displayed lower IFN-β expression levels in serum than the control group (Fig. 11B). Additionally, in the liver and spleen, Ex527 treatment inhibited HSV-1-triggered antiviral immune responses, as suggested by the generation of IFN-β, IP-10, ISG56, and interleukin 6 (IL-6) (Fig. 11C). Confused by these findings, we wondered whether the effect of Sirt1 on antiviral host defense in mice might be different from its role in humans. To investigate this possibility, we examined the role of Sirt1 in HSV-1-triggered signal transduction in mouse cells. Consistent with the results from the mouse model, in mouse cell line RAW264.7, real-time PCR analysis suggested that Ex527 treatment resulted in a drop in the production of IFN-β, IP-10, and ISG56 in response to HSV-1 infection (Fig. 11D). Further, Ex527 treatment caused a significant decrease in the secretion of IFN-β and IL-6 in RAW264.7 cells upon HSV-1 infection (Fig. 11E). Considering the constructive difference between IFI16 and its mouse ortholog p204 (Fig. 11F), we proposed the hypothesis that Sirt1 might not be associated with p204 and thus could not inhibit the HSV-1-induced signaling pathway in mouse cells. To confirm our hypothesis, coimmunoprecipitation was performed in RAW264.7 cells, and the results indicated that Sirt1 barely interacted with p204 with or without HSV-1 infection (Fig. 11G). Consistently, Ex527 treatment did not show a significant effect on the acetylation of p204 in RAW264.7 cells (Fig. 11H). Taken together, our findings suggest that Sirt1 barely interacts with p204 and thus could not play a negative regulatory role in HSV-1-induced signal transduction in mouse cells.
FIG 11.
Sirt1 does not inhibit the HSV-1- or viral DNA-induced signaling pathway in mouse cells. (A) Diagram of the experiment illustrated in panels B and C. (B) Sex and age-matched wild-type mice were injected intraperitoneally (i.p.) with Ex527 (10 mg/kg) 48 h before HSV-1 (1 × 107 PFU) infection. At 6 h after HSV-1 infection, ELISA analysis was performed with sera from the mice. (C) Wild-type mice were injected intraperitoneally (i.p.) with DMSO or Ex527 48 h before HSV-1 (1 × 107 PFU) infection. At 24 h after HSV-1 infection, liver and spleen sections were analyzed by real-time PCR. (D) Raw264.7 cells were treated with DMSO or Ex527 (5 μM or 20 μM) for 12 h. Then, the cells were infected with HSV-1 (MOI = 1) for 8 h and the cells were lysed and subjected to real-time PCR analysis. (E) Raw264.7 cells were treated with DMSO or Ex527 (5 μM or 20 μM) for 12 h. Then, the cells were infected with HSV-1 (MOI = 1) for 24 h and the supernatants were collected and subjected to ELISA analysis. Data were obtained from three independent experiments. (F) Distributions of the domains and multipartite NLS motifs within IFI16 homologs. The predicted NLS motifs of IFI16 homologs are represented by orange bars (complete motifs) or gray bars (partial motifs) within sequence schematics. The position of the first amino acid of each motif is shown. (G) Raw264.7 cells were infected with HSV-1 (MOI = 1) for 8 h or left untreated. Afterward, the cells were lysed and subjected to immunoprecipitation (IP) and immunoblot (IB) analysis. (H) Raw264.7 cells were treated with DMSO or Ex527 (5 μM) for 12 h and then infected with HSV-1 (MOI = 1) for the periods indicated. The cells were subjected to IP and IB analysis. The data are representative of three independent experiments and are presented as means ± SDs. *, P < 0.05; **, P < 0.01.
DISCUSSION
Sirt1 has many substrates with diverse functions as a deacetylase (22). In antiviral host defense, the role of Sirt1 is complicated and controversial and is dependent on a wide range of conditions, including host cell type, virus type, virus strain, and infection status. Recently, emerging evidence has demonstrated the role of Sirt1 in herpesvirus infection (23–25). However, these studies focus mainly on the gammaherpesvirus KSHV. The role of Sirt1 in HSV-1 infection remains unclear. For the first time, our research proposed the hypothesis that Sirt1 negatively regulated HSV-1-induced antiviral innate immune responses. To confirm our hypothesis, we investigated the effects of Sirt1 on HSV-1-triggered innate immune responses by using a series of methods to regulate Sirt1 expression or activity as deacetylase, including Sirt1 overexpression, Sirt1 knockdown, and knockout in cells and by using Sirt1-specific activators (SRT2104 and RSV) and inhibitor (Ex527), in both immune cells (human macrophage-like PMA-THP1 cells) and nonimmune cells (human keratinocyte cell line HaCaT). In all the assays we performed, such as the IFN-β luciferase assay, real-time PCR assays for the production of IFN-β, IP-10, TNF-α, or RANTES, ELISA analysis for the secretion of IFN-β and TNF-α, immunoblot assays for the phosphorylation of TBK1, IRF3, and p65, and plaque assays for the titers of HSV-1, consistent results were obtained, suggesting that Sirt1 promoted HSV-1 infection and inhibited HSV-1-induced host innate immune responses. Additionally, a similar phenomenon was observed in cells that were stimulated by exogenous DNA, including HSV60, VACV70, and poly(dA:dT). Further, our RNA-seq analysis on wild-type and Sirt1-deficient PMA-THP1 cells revealed that Sirt1 deficiency in PMA-THP1 cells resulted in the upregulation of a variety of ISGs and proinflammatory cytokines in response to HSV-1 infection. The GSEA analysis demonstrated a close association between Sirt1 and the cytosolic DNA sensing pathway. Considering all these consistent results, we concluded that Sirt1 served as a negative regulator of DNA virus or exogenous DNA-triggered innate immune responses in human cells.
When we explored the potential mechanism by which Sirt1 regulated antiviral host defense, we noticed three facts. First, it has been reported that IFI16 is acetylated during HSV-1 infection and that the acetylation of IFI16 promotes its cytoplasmic localization (14). Second, our findings indicated that Sirt1 interacted with IFI16 and deacetylated IFI16. Third, we found that Sirt1 knockdown or Sirt1 inhibitor promoted the HSV-1-induced innate immune responses only in the wild-type PMA-THP1 cells, not in the IFI16-deficient PMA-THP1 cells, suggesting that the effect of Sirt1 on HSV-1 infection was dependent on the existence of IFI16. Based on these facts, we proposed the hypothesis that Sirt1 negatively regulated the HSV-1-triggered antiviral signaling pathway by decreasing the acetylation of IFI16 and its cytoplasmic distribution. Although IFI16 detects viral DNA in both the cytoplasm and the nucleus (12), recognition of herpesvirus genomes in the nucleus, such as with KSHV, EBV, and HSV-1, results in IFI16 acetylation and cytoplasmic translocation (21). Then, in the cytoplasm, IFI16 interacts with STING, leading to TBK1 and IRF3 phosphorylation and downstream signal transduction (21). Thus, cytoplasmic translocation of IFI16 upon viral DNA sensing is critical to its function in innate immune signal transduction. In this study, we demonstrated that Sirt1 inhibited the cytoplasmic translocation of IFI16 in response to HSV-1 infection or viral DNA stimulation, as suggested by both confocal microscopy and immunoblot assays. Further, Sirt1 knockdown or deficiency increased the IFI16-STING interaction. Thus, we concluded that Sirt1 negatively regulates the DNA virus- or exogenous DNA-triggered innate immune signaling pathway by inhibiting the cytoplasmic translocation of IFI16.
Based on the results that we obtained in human cells, we further explored the effect of Sirt1 on HSV-1 infection in animal models. Surprisingly, injection of Sirt1 inhibitor Ex527 into mice resulted in decreased production of IFN-β and proinflammatory cytokines, suggesting a positive role of Sirt1 in HSV-1-induced antiviral host defense in mice. Confused by these findings, we noticed that although p204 is considered the murine ortholog of human IFI16, the distributions of the NLS motifs in IFI16 and p204 are different (14). Therefore, we examined whether Sirt1 interacted with p204 and affected p204-mediated host defense. In our study, we found that Sirt1 barely interacted with p204 in mouse cell line RAW264.7. In addition, Sirt1 inhibitor Ex527 could not affect the acetylation of p204 upon HSV-1 infection in RAW 264.7 cells. Further, Ex527 treatment in HSV-1-infected RAW264.7 cells caused a drop in the production of IFN-β and proinflammatory cytokines, different from the findings observed in human cells. These results suggested that upon HSV-1 infection, Sirt1 inhibited IFI16-mediated host defense in human cells but could not inhibit p204-mediated innate immune responses. Thus, our findings demonstrate a difference in the regulation of antiviral host defense between humans and mice, which might expand our knowledge in understanding viral infection and host defense in different species.
When we were preparing the manuscript, Qin et al. reported a mechanism by which Sirt1 positively regulated IFN production by affecting IRF3/IRF7 liquid-liquid phase separation in innate antiviral immunity in mice and RAW 264.7 cells (26). Consistent with their findings, our research also suggested that Ex527 caused a drop in HSV-1-triggered production of IFN-β in mouse models, mouse cells, and IFI16-deficient human cells. In addition, we observed that Sirt1 slightly promoted the VSV-induced formation of IRF3 dimers, which was not mediated by IFI16. This phenomenon was also explained well by their publication. However, we also observed the regulation of proinflammatory cytokines by Sirt1 in HSV-1-infected mouse cells, which could not be explained by its effect on IRF3/IRF7 liquid-liquid phase separation. Thus, further investigation on the role of Sirt1 in virus-triggered proinflammatory cytokine production may be needed.
It is worth noticing that IFI16 also plays an important role in inflammasome. Upon binding viral DNA in the nucleus, IFI16 recruited the adaptor molecule ASC and procaspase-1 to form a functional IFI16–ASC–procaspase-1 inflammasome in the nucleus or perinuclear space (12). The activation of IFI16 inflammasome has been reported to be detected in infection by HSV-1/2, KSHV, EBV, human papillomavirus (HPV), HCMV, bovine herpesvirus 1 (BoHV-1), and HIV (12). In this research, we focused mainly on the effect of Sirt1 on IFI16-induced STING-TBK1-IRF3/NF-κB signaling pathways. It should be meaningful to investigate the role of Sirt1 in IFI16 inflammasome and IFI16 inflammasome-involved viral infection in further studies.
Besides its role in type I IFN production and inflammasome, IFI16 also plays an important role in cell proliferation, cellular senescence, and cell survival (27). Thus, IFI16 is associated with the development of various human diseases, including chronic hepatitis B and hepatitis B virus (HBV)-associated acute-on-chronic liver failure, HSV-1-associated severe corneal inflammatory herpetic disease, systemic lupus erythematosus, inflammatory bowel disease, aging-associated inflammatory diseases, and cancer (27–32). Considering that Sirt1 has several highly selective activators and inhibitors, our research on the Sirt1-IFI16 interaction may shed some light on the treatment of these diseases targeting IFI16. Given that Sirt1 has many substrates and broad cellular functions, including metabolic homeostasis, stress response, tumorigenesis, and autophagy, further study may be needed to characterize the role of Sirt1 and IFI16 in these diseases to help with the generation of new therapeutic strategies.
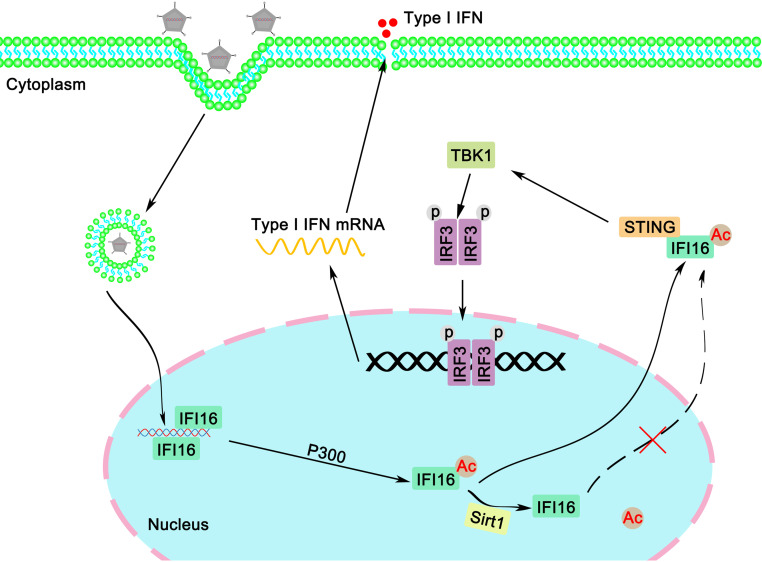
Considering these findings together, in this study, we demonstrated that Sirt1 negatively regulated host innate immune responses against DNA virus and exogenous cytosolic DNA by inhibiting the acetylation and the cytoplasmic translocation of IFI16, thereby affecting DNA virus infection in human cells (Fig. 12). Our findings suggest Sirt1 as a new regulator in IFI16-mediated antiviral signaling pathways in human cells. This research may expand our knowledge about host antiviral defense and help to understand the difference between humans and mice in host antiviral responses.
FIG 12.
Proposed model for the regulation of the IFI16-mediated response by Sirt1. IFI16 locates predominantly in the nucleus. Upon binding with double-stranded DNA derived from viruses, IFI16 is acetylated, leading to the accumulation of IFI16 in the cytoplasm, where IFI16 recruits STING and activates downstream host defense. In the presence of Sirt1, the acetylation and cytoplasmic distribution of IFI16 were inhibited, leading to impaired antiviral immune responses.
MATERIALS AND METHODS
cDNA constructs and reagents.
Human IFI16 and its deletion mutants were amplified by PCR using cDNA from THP1 cells and were subsequently cloned into a pcDNA3 vector (Invitrogen). All IFI16 deletion mutants were constructed by PCR and also subcloned into a pcDNA3 vector. Human Myc-Sirt1, Flag-Sirt2, Myc-Sirt3, Flag-Sirt4, and Flag-Sirt5 were amplified by PCR and subsequently cloned into a pcDNA3 vector (Invitrogen). Flag-Sirt6 and Flag-Sirt7 were obtained from Vigene Biosciences. pIFN-β-Luc was obtained as described previously (33).
The following antibodies were used for immunoblot analysis or immunoprecipitation: anti-Flag (F3165; Sigma-Aldrich), anti-HA (901515; Biolend), anti-Myc (66004-1-Ig; Proteintech), anti-IFI16 (sc-8023; Santa Cruz), anti-Sirt1 (2493; Cell Signaling Technology), anti-p-TBK1 (5483T; Cell Signaling Technology), anti-TBK1 (CSB-PA024154LA01HU; Flarbio), anti-p-IRF3 (4947; Cell Signaling Technology), anti-IRF3 (11312-1-AP; Proteintech), anti-p-p65 (3033; Cell Signaling Technology), anti-p65 (10745-1-AP; Proteintech), anti-acetylated-lysine (9441; Cell Signaling Technology), anti-p204 (NBP2-27153; Novus), anti-STING (19851-1-AP; Proteintech), anti-β-tubulin (10068-1-AP; Proteintech), anti-histone H3 (CSB-PA010109LA01HU; Flarbio), and anti-β-actin (60008-1; Proteintech).
Phorbol myristate acetate (PMA; S1819) was purchased from Beyotime Biotechnology. Ex527 (S1541) and SRT2104 (S7792) were obtained from Selleck. Resveratrol (R5010) was purchased from Sigma-Aldrich. The poly(d·dT) (catalog code tlrl-patn), HSV60 (tlrl-hsv60n), and poly (I:C) (tlrl-picw) were obtained from InvivoGen. VACV70 was synthesized by Sangon Biotech. The sequence was as follows: 5′-CCATCAGAAAGAGGTTTAATATTTTTGTGAGACCATGGAAGAGAGAAAGA GATAAAACTTTTTTACGACT-3′.
Mice.
C57BL/6 mice were purchased from the Beijing Vital River Laboratory Animal Technology Co., Ltd. All mice were bred and kept in specific-pathogen-free conditions in the Xinxiang Medical University. All animal care and use protocols were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People's Republic of China. The animal experiments were approved by the committee on animal care at Xinxiang Medical University (approval number XYLL-2020167).
Cell culture, transfection, and stimulation.
Human embryonic kidney (HEK) 293T cells and Tohoku Hospital Pediatrics 1 (THP1) cells were kindly provided by the Stem Cell Bank, Chinese Academy of Sciences. HaCaT keratinocytes were purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, China). IFI16-deficient THP1 cells (thpd-koifi16) were purchased from InvivoGen. HaCaT and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium. THP1 cells were grown in RPMI 1640. PMA-THP1 cells are THP1 cells that were pretreated with 100 ng/mL PMA for 24 h. All cells were supplemented with 10% fetal bovine serum (Gibco), 4 mM l-glutamine, 100 μg/mL penicillin, and 100 U/mL streptomycin under humidified conditions with 5% CO2 at 37°C. Transfection of HaCaT, HEK293T, and THP1 cells and mouse embryo fibroblasts (MEFs) was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Preparations of MEFs.
The procedure for generating MEFs has been described previously (34).
Immunoprecipitation and immunoblot analysis.
Immunoprecipitation and immunoblot analysis were performed as described previously (35).
IRF3 dimerization assay.
Cell extracts were prepared in native lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 1% protease inhibitor cocktail [Roche], and 1% orthovanadate) for 30 min at 4°C. Lysates were loaded onto a native PAGE gel (without SDS), which was prerun in electrophoresis buffer (25 mM Tris-HCl [pH 8.4] and 192 mM glycine with and without 0.2% deoxycholate in the cathode and anode chamber, respectively) for 60 min at 40 mA and electrophoresed for 60 min at 25 mA. Then, the native gel was analyzed by immunoblotting as described above.
Real-time PCR.
Total RNA was extracted from the cultured cells with TRIzol reagent (Invitrogen) as described by the manufacturer. All gene transcripts were quantified by real-time PCR with SYBR green qPCR master mix using a 7500 fast real-time PCR system (Applied Biosystems). The relative fold induction was calculated using the 2−ΔΔCT method. The primers used for real-time PCR were described previously (34, 36).
ELISA.
THP1 cells or HaCaT keratinocytes were infected with viruses or transfected with synthetic nucleic acids for 24 h. The supernatants were collected for measurement of IFN-β (KE00187; Proteintech) and TNF-α (88-7346-88; Thermo Fisher Scientific). Eight-week-old wild-type mice were treated with Ex527 before infection with HSV-1 for 6 h, and then the sera of mice were collected for measurement of IFN-β (R&D) by ELISA.
RNA interference (RNAi).
Sirt1 Stealth-RNAi siRNA was designed by the Invitrogen BLOCKiT RNAi designer. The siRNA sequences used were as follows: for S1, forward, 5′-CCCAUGAAGUGCCUCAGAUAUUAAU-3′, and reverse, 5′-AUUAAUAUCUGAGGCACUUCAUGGG-3′; for S2, forward, 5′-CCAAACUUUGCUGUAACCCUGUAAA-3′, and reverse, 5′-UUUACAGGGUUACAGCAAAGUUUGG-3′; for S3, forward, 5′-UGGGACAUGCCAGAGUCCAAGUUUA-3′, and reverse, 5′-UAAACUUGGACUCUGGCAUGUCCCA-3′.
The Silencer Select negative-control siRNA was purchased from Invitrogen (catalog no. 4390843). PMA-THP1 or HaCaT cells were transfected with siRNA using Lipofectamine 2000 according to the manufacturer’s instructions. At 24 h after transfection, the cells were used for further experiments.
Generation of Sirt1-deficient THP1 cell line.
A Sirt1-deficient THP1 cell line was generated by the CRISPR-Cas9 system. The following oligonucleotides specifically targeting the Sirt1 gene were used: oligonucleotide 1, 5′-TTCTACCATTTGCTTGATAC-3′; oligonucleotide 2, 5′-TACCCAGAACATAGACACGC-3′; and oligonucleotide 3, 5′-ATGGTTAGTAAACTTCAGAG-3′.
Viruses and infection.
Cells were infected with HSV-1 (KOS strain, multiplicity of infection [MOI] of 1) for 1.5 h. Then, the cells were washed with phosphate-buffered saline (PBS) and cultured in fresh medium. For the in vivo study, age- and sex-matched groups of mice were intravenously infected with HSV-1. HSV-1 viral titer was determined by the plaque-forming assay on Vero cells.
The infection of VSV was performed as described previously (37).
Luciferase reporter gene assay.
Luciferase reporter gene assays were performed as described previously (38).
Confocal microscopy.
After treatment, THP1 cells and HaCaT keratinocytes were fixed with 4% paraformaldehyde (PFA) in PBS, permeabilized with Triton X-100, and then blocked with 1% bovine serum albumin (BSA) in PBS. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Images were taken using Nikon A1R microscopy.
In situ PLA.
In situ PLA was studied using a Duolink PLA kit (Sigma). THP1 cells were seeded on Teflon-coated glasses and cultured overnight. After HSV-1 infection for 8 h, cells were fixed in 4% PFA in PBS for 15 min and washed with PBS three times. After that, cells were permeabilized with 0.1% Triton X-100 on ice for 5 min, washed in PBS, blocked in 5% BSA in PBS for 1 h at 37°C, and incubated in primary antibodies overnight at 4°C. The next day, the cells were washed with the wash buffer (Sigma; DUO82049) three times and incubated with the secondary antibodies with PLA probes (Sigma; DUO92001 and DUO92005) for 2 h at 37°C, followed by three washes with the wash buffer. Cells were incubated with the Duolink in situ detection reagents red (Sigma; DUO92008) for 2 h at 37°C. Finally, nuclei were stained with DAPI and covered on slides. Images were taken using Nikon A1R microscopy, and the Image Pro Plus 6.0 was used for quantitative analyses.
RNA-seq analysis.
After HSV-1 stimulation for the indicated periods, total RNA was extracted from control and Sirt1-deficient PMA-THP1 cells using the TRIzol reagent. Library preparation and sequencing were performed by BGI (Shenzhen, China). To gain insight into the change of phenotype, GO (http://www.geneontology.org/) and KEGG (https://www.kegg.jp/) enrichment analyses of annotated differently expressed genes were performed by Dr. Tom (https://biosys.bgi.com/). The significant levels of terms and pathways were corrected by Q value (false discovery rate, FDR) with a rigorous threshold (Q value ≤ 0.05) by Benjamini - Hochberg.
Nuclear extracts.
The nuclear extracts were prepared as described previously (39).
Statistics.
The data are presented as means ± standard deviations (SDs) from at least three independent experiments. The statistical comparisons between the different treatments were performed using the unpaired Student t test, and a P of <0.05 was considered statistically significant.
Data availability.
All RNA-seq data have been deposited in the NCBI database under accession number PRJNA882382 and are publicly available.
ACKNOWLEDGMENTS
We thank Yinming Liang (Xinxiang Medical University) for providing Sirt1-deficient THP1 cells and all the members of Henan Key Laboratory of immunology and targeted drug for sharing valuable material and research support.
This work was supported by the National Natural Science Foundation of China (grant no. 31970847, 32070949, U2004103, and 32170871), the Natural Science Foundation of Henan Province (grant no. 212300410065 and 222300420064), and the Program for Science & Technology Innovation Talents in Higher Education of Henan Province (grant no. 23HASTIT050).
B.Y., J.W., H.W., and Z.X. supervised the project. B.Y., J.W., X.Q., and Y.H. designed the experiments. J.W., X.Q., Y.H., G.Z., Y.W., Y.L., Y.C., J.P., S.M., Z.S., and Y.L. performed cell experiments, qPCR analysis, ELISA, and Western blotting. B.Y. acquired images using Nikon A1R microscopy. J.W., X.Q., and Y.H. performed the immunoprecipitation. J.W., X.Q., Y.H., and B.Y. analyzed the data. J.W. and B.Y. wrote the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Jie Wang, Email: jiewang618@xxmu.edu.cn.
Xiaofei Zhu, Email: zhuxf@xxmu.edu.cn.
Hui Wang, Email: wanghui@xxmu.edu.cn.
Bo Yang, Email: byang94@xxmu.edu.cn.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Tenthorey JL, Emerman M, Malik HS. 2022. Evolutionary landscapes of host-virus arms races. Annu Rev Immunol 40:271–294. 10.1146/annurev-immunol-072621-084422. [DOI] [PubMed] [Google Scholar]
- 2.Webb LG, Fernandez-Sesma A. 2022. RNA viruses and the cGAS-STING pathway: reframing our understanding of innate immune sensing. Curr Opin Virol 53:101206. 10.1016/j.coviro.2022.101206. [DOI] [PubMed] [Google Scholar]
- 3.Zhang F, Yuan Y, Ma F. 2020. Function and regulation of nuclear DNA sensors during viral infection and tumorigenesis. Front Immunol 11:624556. 10.3389/fimmu.2020.624556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diner BA, Lum KK, Cristea IM. 2015. The emerging role of nuclear viral DNA sensors. J Biol Chem 290:26412–26421. 10.1074/jbc.R115.652289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341:903–906. 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Wen M, Cao X. 2019. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 365:eaav0758. 10.1126/science.aav0758. [DOI] [PubMed] [Google Scholar]
- 8.Diner BA, Li T, Greco TM, Crow MS, Fuesler JA, Wang J, Cristea IM. 2015. The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol Syst Biol 11:787. 10.15252/msb.20145808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gariano GR, Dell'Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. 2012. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog 8:e1002498. 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy A, Dutta D, Iqbal J, Pisano G, Gjyshi O, Ansari MA, Kumar B, Chandran B. 2016. Nuclear innate immune DNA sensor IFI16 is degraded during lytic reactivation of Kaposi's sarcoma-associated herpesvirus (KSHV): role of IFI16 in maintenance of KSHV latency. J Virol 90:8822–8841. 10.1128/JVI.01003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu H, Li X, Rousseau BA, Akinyemi IA, Frey TR, Zhou K, Droske LE, Mitchell JA, McIntosh MT, Bhaduri-McIntosh S. 2022. IFI16 partners with KAP1 to maintain Epstein-Barr virus latency. J Virol 96:e0102822. 10.1128/jvi.01028-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupfer C, Malik A, Kanneganti TD. 2015. Inflammasome control of viral infection. Curr Opin Virol 12:38–46. 10.1016/j.coviro.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiliveru S, Rahbek SH, Jensen SK, Jorgensen SE, Nissen SK, Christiansen SH, Mogensen TH, Jakobsen MR, Iversen L, Johansen C, Paludan SR. 2014. Inflammatory cytokines break down intrinsic immunological tolerance of human primary keratinocytes to cytosolic DNA. J Immunol 192:2395–2404. 10.4049/jimmunol.1302120. [DOI] [PubMed] [Google Scholar]
- 14.Li T, Diner BA, Chen J, Cristea IM. 2012. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci USA 109:10558–10563. 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koyuncu E, Budayeva HG, Miteva YV, Ricci DP, Silhavy TJ, Shenk T, Cristea IM. 2014. Sirtuins are evolutionarily conserved viral restriction factors. mBio 5:e02249-14. 10.1128/mBio.02249-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Wang J, Wang L, Aliyari S, Cheng G. 2022. SARS-CoV-2 virus NSP14 impairs NRF2/HMOX1 activation by targeting Sirtuin 1. Cell Mol Immunol 19:872–882. 10.1038/s41423-022-00887-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter M, Chen IP, Vallejo-Gracia A, Kim IJ, Bielska O, Lam VL, Hayashi JM, Cruz A, Shah S, Soveg FW, Gross JD, Krogan NJ, Jerome KR, Schilling B, Ott M, Verdin E. 2022. SIRT5 is a proviral factor that interacts with SARS-CoV-2 Nsp14 protein. PLoS Pathog 18:e1010811. 10.1371/journal.ppat.1010811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leyton L, Hott M, Acuna F, Caroca J, Nunez M, Martin C, Zambrano A, Concha MI, Otth C. 2015. Nutraceutical activators of AMPK/Sirt1 axis inhibit viral production and protect neurons from neurodegenerative events triggered during HSV-1 infection. Virus Res 205:63–72. 10.1016/j.virusres.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 19.Martin C, Leyton L, Arancibia Y, Cuevas A, Zambrano A, Concha MI, Otth C. 2014. Modulation of the AMPK/Sirt1 axis during neuronal infection by herpes simplex virus type 1. J Alzheimers Dis 42:301–312. 10.3233/JAD-140237. [DOI] [PubMed] [Google Scholar]
- 20.Dai J, Huang YJ, He X, Zhao M, Wang X, Liu ZS, Xue W, Cai H, Zhan XY, Huang SY, He K, Wang H, Wang N, Sang Z, Li T, Han QY, Mao J, Diao X, Song N, Chen Y, Li WH, Man JH, Li AL, Zhou T, Liu ZG, Zhang XM, Li T. 2019. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell 176:1447–1460.e14. 10.1016/j.cell.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iqbal J, Ansari MA, Kumar B, Dutta D, Roy A, Chikoti L, Pisano G, Dutta S, Vahedi S, Veettil MV, Chandran B. 2016. Histone H2B-IFI16 recognition of nuclear herpesviral genome induces cytoplasmic interferon-beta responses. PLoS Pathog 12:e1005967. 10.1371/journal.ppat.1005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Zhao W, Yang JS, Cheng Z, Luo H, Lu Z, Tan M, Gu W, Zhao Y. 2012. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol Cell Proteomics 11:1048–1062. 10.1074/mcp.M112.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, He M, Zhou F, Ye F, Gao SJ. 2014. Activation of Kaposi's sarcoma-associated herpesvirus (KSHV) by inhibitors of class III histone deacetylases: identification of sirtuin 1 as a regulator of the KSHV life cycle. J Virol 88:6355–6367. 10.1128/JVI.00219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye F, Zeng Y, Sha J, Jones T, Kuhne K, Wood C, Gao SJ. 2016. High glucose induces reactivation of latent Kaposi's sarcoma-associated herpesvirus. J Virol 90:9654–9663. 10.1128/JVI.01049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He M, Yuan H, Tan B, Bai R, Kim HS, Bae S, Che L, Kim JS, Gao SJ. 2016. SIRT1-mediated downregulation of p27Kip1 is essential for overcoming contact inhibition of Kaposi's sarcoma-associated herpesvirus transformed cells. Oncotarget 7:75698–75711. 10.18632/oncotarget.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin Z, Fang X, Sun W, Ma Z, Dai T, Wang S, Zong Z, Huang H, Ru H, Lu H, Yang B, Lin S, Zhou F, Zhang L. 2022. Deactylation by SIRT1 enables liquid-liquid phase separation of IRF3/IRF7 in innate antiviral immunity. Nat Immunol 23:1193–1207. 10.1038/s41590-022-01269-0. [DOI] [PubMed] [Google Scholar]
- 27.Choubey D, Deka R, Ho SM. 2008. Interferon-inducible IFI16 protein in human cancers and autoimmune diseases. Front Biosci 13:598–608. 10.2741/2705. [DOI] [PubMed] [Google Scholar]
- 28.Choubey D, Panchanathan R. 2016. IFI16, an amplifier of DNA-damage response: role in cellular senescence and aging-associated inflammatory diseases. Ageing Res Rev 28:27–36. 10.1016/j.arr.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Caneparo V, Cena T, De Andrea M, Dell'oste V, Stratta P, Quaglia M, Tincani A, Andreoli L, Ceffa S, Taraborelli M, Magnani C, Landolfo S, Gariglio M. 2013. Anti-IFI16 antibodies and their relation to disease characteristics in systemic lupus erythematosus. Lupus 22:607–613. 10.1177/0961203313484978. [DOI] [PubMed] [Google Scholar]
- 30.Vanhove W, Peeters PM, Staelens D, Schraenen A, Van der Goten J, Cleynen I, De Schepper S, Van Lommel L, Reynaert NL, Schuit F, Van Assche G, Ferrante M, De Hertogh G, Wouters EF, Rutgeerts P, Vermeire S, Nys K, Arijs I. 2015. Strong upregulation of AIM2 and IFI16 inflammasomes in the mucosa of patients with active inflammatory bowel disease. Inflamm Bowel Dis 21:2673–2682. 10.1097/MIB.0000000000000535. [DOI] [PubMed] [Google Scholar]
- 31.Pang X, Li X, Mo Z, Huang J, Deng H, Lei Z, Zheng X, Feng Z, Xie D, Gao Z. 2018. IFI16 is involved in HBV-associated acute-on-chronic liver failure inflammation. BMC Gastroenterol 18:61. 10.1186/s12876-018-0791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coulon PG, Dhanushkodi N, Prakash S, Srivastava R, Roy S, Alomari NI, Nguyen AM, Warsi WR, Ye C, Carlos-Cruz EA, Mai UT, Cruel AC, Ekmekciyan KM, Pearlman E, BenMohamed L. 2019. NLRP3, NLRP12, and IFI16 inflammasomes induction and caspase-1 activation triggered by virulent HSV-1 strains are associated with severe corneal inflammatory herpetic disease. Front Immunol 10:1631. 10.3389/fimmu.2019.01631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang B, Wang J, Wang Y, Zhou H, Wu X, Tian Z, Sun B. 2013. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol 190:3613–3619. 10.4049/jimmunol.1202507. [DOI] [PubMed] [Google Scholar]
- 34.Yang B, Liu Y, Cui Y, Song D, Zhang G, Ma S, Liu Y, Chen M, Chen F, Wang H, Wang J. 2020. RNF90 negatively regulates cellular antiviral responses by targeting MITA for degradation. PLoS Pathog 16:e1008387. 10.1371/journal.ppat.1008387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Kang L, Song D, Liu L, Yang S, Ma L, Guo Z, Ding H, Wang H, Yang B. 2017. Ku70 senses HTLV-1 DNA and modulates HTLV-1 replication. J Immunol 199:2475–2482. 10.4049/jimmunol.1700111. [DOI] [PubMed] [Google Scholar]
- 36.Yang B, Song D, Liu Y, Cui Y, Lu G, Di W, Xing H, Ma L, Guo Z, Guan Y, Wang H, Wang J. 2018. IFI16 regulates HTLV-1 replication through promoting HTLV-1 RTI-induced innate immune responses. FEBS Lett 592:1693–1704. 10.1002/1873-3468.13077. [DOI] [PubMed] [Google Scholar]
- 37.Yang B, Zhang G, Qin X, Huang Y, Ren X, Sun J, Ma S, Liu Y, Song D, Liu Y, Cui Y, Wang H, Wang J. 2021. Negative regulation of RNF90 on RNA virus-triggered antiviral immune responses targeting MAVS. Front Immunol 12:730483. 10.3389/fimmu.2021.730483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Yang S, Liu L, Wang H, Yang B. 2017. HTLV-1 Tax impairs K63-linked ubiquitination of STING to evade host innate immunity. Virus Res 232:13–21. 10.1016/j.virusres.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Yang B, Hu Y, Zheng Y, Zhou H, Wang Y, Ma Y, Mao K, Yang L, Lin G, Ji Y, Wu X, Sun B. 2013. Negative regulation of Nmi on virus-triggered type I IFN production by targeting IRF7. J Immunol 191:3393–3399. 10.4049/jimmunol.1300740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download jvi.01975-22-s0001.pdf, PDF file, 1.8 MB (1.8MB, pdf)
Data Availability Statement
All RNA-seq data have been deposited in the NCBI database under accession number PRJNA882382 and are publicly available.