ABSTRACT
Despite active control strategies, including the vaccination program in poultry, H9N2 avian influenza viruses possessing mutations in hemagglutinin (HA) were frequently isolated. In this study, we analyzed the substitutions at HA residue 193 (H3 numbering) of H9N2 and investigated the impact of these mutations on viral properties. Our study indicated that H9N2 circulating in the Chinese poultry have experienced frequent mutations at HA residue 193 since 2013, with viruses that carried asparagine (N) being replaced by those with alanine (A), aspartic acid (D), glutamic acid (E), glycine (G), and serine (S), etc. Our results showed the N193G mutation impeded the multiple cycles of growth of H9N2, and although most of the variant HAs retained the preference for human-like receptors as did the wild-type N193 HA, the N193E mutation altered the preference for both human and avian-like receptors. Furthermore, these mutations substantially altered the antigenicity of H9N2 as measured by both monoclonal antibodies and antisera. In vivo studies further demonstrated that these mutations showed profound impact on viral replication and transmission of H9N2 in chicken. Viruses with D, E, or S at residue 193 acquired the ability to replicate in lungs of the infected chickens, whereas virus with G193 reduced its transmissibility in infected chickens to those in direct contact. Our findings demonstrated that variations at HA residue 193 altered various properties of H9N2, highlighting the significance of the continued surveillance of HA for better understanding of the etiology and effective control of H9N2 in poultry.
IMPORTANCE H9N2 are widespread and have sporadically caused clinical diseases in humans. Extensive vaccinations in poultry helped constrain H9N2; however, they might have facilitated the evolution of the virus. It is therefore of importance to monitor the variation of the circulating H9N2 and evaluate its risk to both veterinary and public health. Here, we found substitutions at position 193 of HA from H9N2 circulated since 2013 and assessed the impact of several mutations on viral properties. Our data showed these mutations resulted in substantial antigenic change. N193E altered the binding preference of HA for human-like to both avian and human-like receptors. More importantly, N193G impaired the growth of H9N2 and its transmission in chickens, whereas mutations from N to D, E, and S enhanced the viral replication in lungs of chickens. Our study enriched the knowledge about H9N2 and may help implement an effective control strategy for H9N2.
KEYWORDS: H9N2 avian influenza virus, HA variation, position 193, receptor, antigenicity, replication, transmission
INTRODUCTION
H9N2 avian influenza virus (AIV) was first isolated from turkeys in Wisconsin, USA in 1966 and has been widespread since the 1990s in Asia, Middle East, and North Africa (1, 2). This virus causes mild illness in poultry; however, it may lead to higher mortality in poultry if co-infects with other pathogens (3, 4). H9N2 AIV also poses a threat to public health as it can be transmitted to humans. The World Health Organization (WHO) has reported 58 confirmed cases of human infection since December 2015. Due to its extensive geographical distribution, H9N2 AIV often co-circulates with other AIV subtypes, donating internal genes to the H5NX, H7N9, and H10N8 viruses through reassortantment (5–8). Meanwhile, H9N2 AIV may acquire gene segments from H7N9 and H10N8 strains (9).
Because of the threat to poultry and public health posed by H9N2 viruses, since the late 1990s China has implemented a vaccination program to control H9N2 infections in poultry (10–13); however, H9N2 variants have often been isolated from vaccinated chicken flocks (14, 15), highlighting immune escape and the antigenic drift of hemagglutinin (HA) of the circulating H9N2 viruses. Vaccines can elicit neutralizing antibodies that target epitopes on viral surface proteins, especially HA, which plays a central role in the viral life cycle through its involvement in receptor recognition, virus attachment, membrane fusion, and virus entry (16). Antigenic drift in HA can directly affect antibody binding, which is the most conventional immune escape mechanism of H9N2 viruses (17, 18).
Each HA monomer contains subunits HA1 and HA2. HA1, located at the globular head, carries the receptor-binding site (RBS) which is responsible for the receptor recognition (19, 20). The structure of the RBS is conserved across all HA subtypes and consists of the 130-loop (residues 135 to 138, H3 number), 190-helix (residues 190 to 198), and 220-loop (residues 221 to 228) (13). The RBS overlaps with the reported antigenic sites, e.g., the 130-loop and 190-helix in the HA of H3N2 virus shared residues in antigenic sites A and B (21, 22), whereas residues 222, 226, and 227 (H3 numbering) in the 220-loop of H9N2 virus were found to be targeted by some murine monoclonal antibodies (MAbs) (23, 24). Amino acid substitutions at RBS have the potential to cause change in both antigenicity and receptor binding (21, 25, 26). In this study, we found that position 193 (H3 numbering) in the HA 190-helix of H9N2 viruses circulating in China has undergone frequent variation since 2013. We demonstrated that amino acid mutations at HA position 193 have profound impact on the biological properties of H9N2 viruses, including the replication efficiency, receptor binding, antigenicity, as well as the in vivo transmissibility in infected chickens.
RESULTS
Amino acid variation at HA position 193 of H9N2 AIVs.
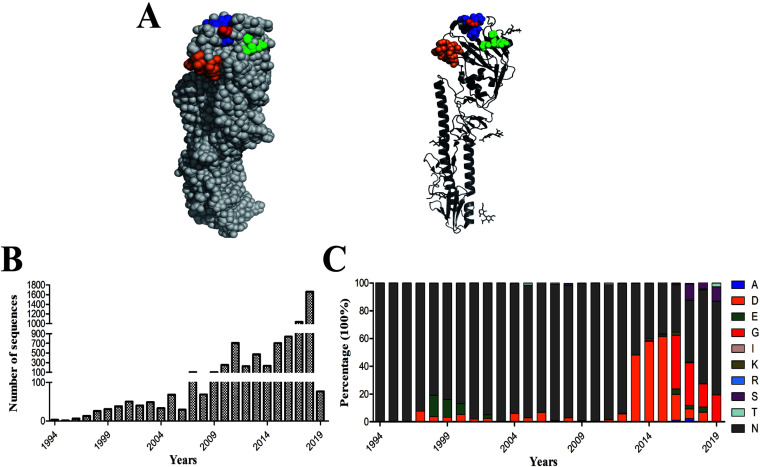
Influenza viruses acquire frequent amino acid changes in HA during the circulation in a host, especially under immune pressure. Some of these changes occurred in the RBS, which is consisted of the 130-1oop, 190-helix, and 220-loop (Fig. 1A). To examine the amino acid variation at HA position 193 of H9N2 viruses (Fig. 1A), belonging to 190 helix, more than 6,800 HA gene sequences of H9N2 viruses isolated from chickens in China since 1994 were analyzed (Fig. 1B). We found that the amino acid at HA position 193 was largely conserved before 2012, with the majority of the H9N2 isolated bearing N and some variants carrying A, D, E, S, and T (Fig. 1C; Table 1). However, since 2013, various mutations occurred at HA position 193 of the circulating H9N2 viruses. Virus containing D193 rapidly surged to be in about 50% of H9N2 strains from 2013 to 2015, and then a relatively high proportion of G193 viruses were isolated between 2016 and 2019. From 2017 to 2019, S193 was present in nearly 10% of H9N2 viruses (Fig. 1C; Table 1). By 2019, viruses with N, S, and G at position 193 dominated the H9N2 strains, whereas a small number of viruses possessed T at HA position 193. Taken together, since 1994, the major amino acid at HA position 193 of H9N2 viruses circulating in China was A, D, E, G, N, and S (>0.7%), while other amino acids (I, K, R, and T) accounted for a small proportion (<0.5%).
FIG 1.
The spatial location and amino acid diversity of HA position 193 of H9N2 virus. (A) The structure of the HA protein was drawn with PyMoL. Shown is the RBS of 130-loop (orange), 190-helix (blue), and 220-loop (green), and amino acid at position 193 is marked as red. The presence of amino acid mutation at position 193 in the HA of H9N2 circulating in chickens in China from 1994. Shown are (B) the number of HA sequences analyzed by years and (C) the percentage of amino acid at HA position 193.
TABLE 1.
The amino acids at HA position 193 of H9N2 viruses circulating in chickens in China
| Yrs | The amino acids at HA position 193 of H9N2 viruses | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1994–2012 | N (1877/1942) | D (31/1942) | T (41/1942) | E (12/1942) | S (1/1942) | ||||
| 2013 | N (245/474) | D (228/474) | S (1/474) | ||||||
| 2014 | D (137/236) | N (95/236) | G (3/236) | E (1/236) | |||||
| 2015 | D (434/707) | N (260/707) | E (7/707) | G (5/707) | A (1/707) | ||||
| 2016 | G (325/840) | N (287/840) | D (156/840) | E (32/840) | K (18/840) | A (11/840) | S (8/840) | T (2/840) | R (1/840) |
| 2017 | N (464/1034) | S (116/1034) | G (317/1034) | D (73/1034) | E (26/1036) | A (25/1034) | K (5/1034) | R (7/1034) | T (3/1034) |
| 2018 | N (1122/1659) | G (277/1659) | S (168/1659) | E (60/1659) | D (17/1659) | R (12/1659) | K (1/1659) | I (1/1659) | T (1/1659) |
| 2019 | N (52/77) | G (15/77) | S (8/77) | T (2/77) | |||||
To determine the impact of amino acid substitution at HA position 193 on the biological characteristics of H9N2 virus, A/Chicken/Xuzhou/491/2019 (H9N2) (XZ491) containing N at position 193 and belonging to G57 genotype H9N2 virus (with G1-like PB2 and M gens in F/98-like backbone), which has become predominant H9N2 since 2010 in China (18), was selected as a template virus. Wild-type XZ491 and mutant XZ491 viruses, carrying A, D, E, G, or S at this position, were rescued with reverse genetics, and designated rg-XZ491-193N, rg-XZ491-193A, rg-XZ491-193D, rg-XZ491-193E, rg-XZ491-193G, and rg-XZ491-193S, respectively. Each rescued virus was confirmed by Sanger sequencing, and all the rescued viruses carried with the correct amino acid changes at position 193 and without any other mutations. All the rescued viruses grew efficiently in embryonated chicken eggs, whereas rg-XZ491-193G grew to a relatively lower titer of 106 TCID50/mL (Table 2). In addition, effectively agglutinated chicken, goose, turkey, and horse red blood cells (RBCs) at an HA titer of 512 to 2,048, except that rg-XZ491-193G with demonstrated a poor agglutination of all RBCs, with HA titers of 4 (chicken and turkey RBCs) and 32 (goose and horse RBCs), respectively (Table 2).
TABLE 2.
HA titers and TCID50 of H9N2 viruses carrying various residues at HA position 193
| HA titer |
TCID50 (log10/mL)b | ||||
|---|---|---|---|---|---|
| Virus | cRBCa | gRBC | tRBC | hRBC | |
| XZ491-193N | 2,048 | 2,048 | 2,048 | 512 | 8.032 |
| XZ491-193A | 2,048 | 2,048 | 512 | 256 | 8.199 |
| XZ491-193D | 2,048 | 2,048 | 2,048 | 256 | 8.366 |
| XZ491-193E | 2,048 | 2,048 | 512 | 2,048 | 9.199 |
| XZ491-193S | 2,048 | 2,048 | 2,048 | 256 | 7.366 |
| XZ491-193G | 4 | 32 | 4 | 32 | 6.032 |
cRBC, gRBC, tRBC, and hRBC represent chicken, goose, turkey, and horse red blood cells, respectively.
Viruses were titrated in MDCK cells.
HA position 193 impacts replication of H9N2 AIVs in cells.
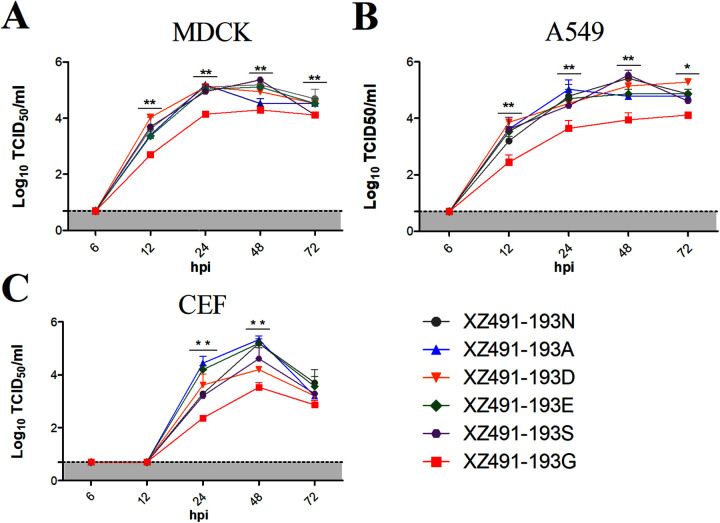
To investigate whether the variation in amino acids at HA position 193 impacts viral replication, we examined the growth kinetics of the six rescued H9N2 viruses in both mammalian and avian cells. Madin-Darby canine kidney (MDCK), human pulmonary epithelial A549, and chicken embryo fibroblast (CEF) cells were infected with each virus at a multiple of infection (MOI) of 0.001, and supernatants were collected at different time points for viral titration. All of these viruses grew efficiently in MDCK, A549, and CEF cells (Fig. 2), reaching peak titers at 24 or 48 h postinfection (hpi); however, compared with the mammalian cells, all viruses replicated slower in CEF cells, and no virus was detected in CEF cells at 12 days postinfection (dpi). The rg-XZ491-193G virus had evidently lower replication efficiency than other viruses as it grew to lower titers at all of the time points examined (P values < 0.05 or 0.01). These data demonstrate that amino acid mutations at HA position 193 of H9N2 viruses may alter the multiple cycles of replication in mammalian cells.
FIG 2.
Growth kinetics of the H9N2 viruses with various amino acids at HA position 193. Viral growth kinetics were determined in MDCK (A), A549 (B), and CEF (C) cells. Confluent monolayers of MDCK, A549, or CEF cells were infected with the H9N2 viruses at an MOI of 0.001, and supernatant samples were collected at 6, 12, 24, 48, and 72 hpi. The virus titers were measured in MDCK or A549 cells by TCID50. The statistical analysis was performed with Student's t test. *, **, and *** indicate P values of less than 0.05, 0.01, and 0.001, respectively.
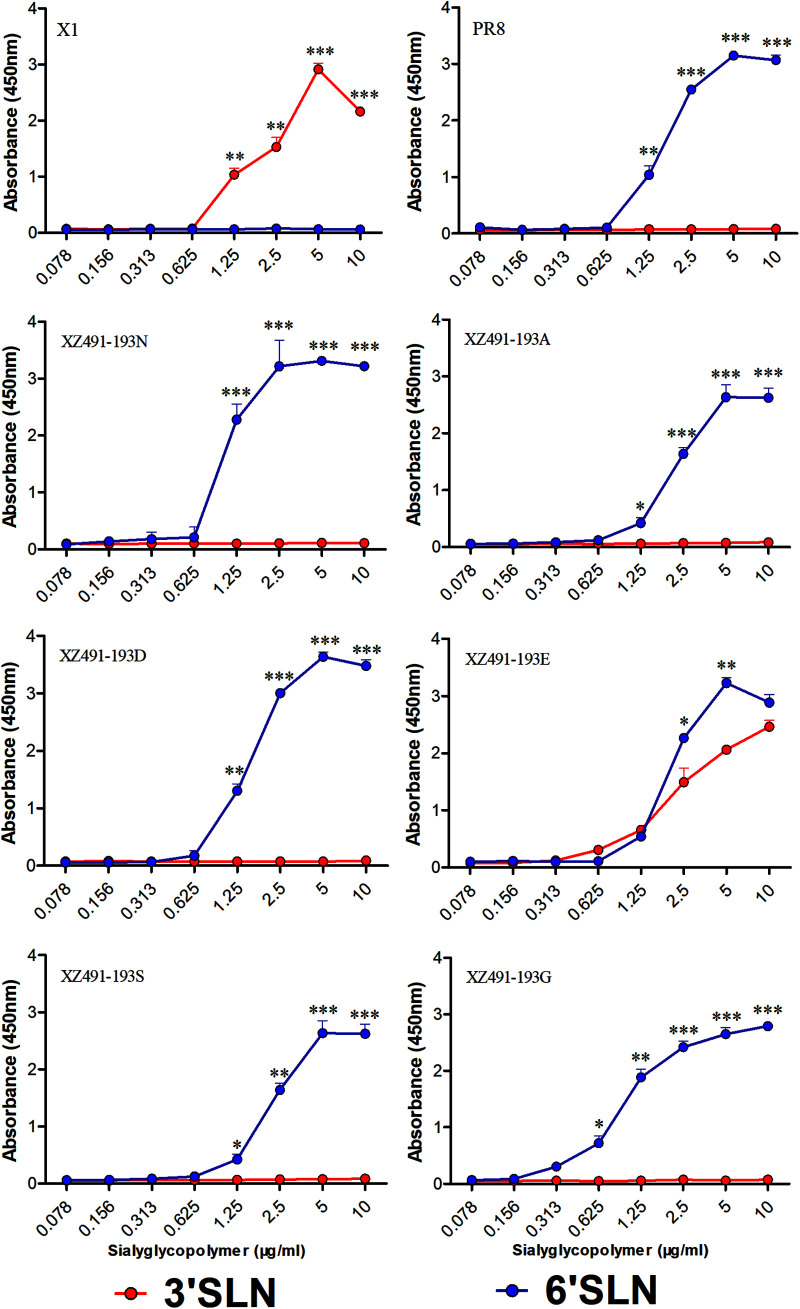
HA position 193 affects receptor specificity of H9N2 AIVs.
Receptor specificity plays a critical role in the transmission of influenza virus between avian and mammals. The rescued H9N2 viruses were tested with two receptor analogs, i.e., Neu5Acα2-3Galb1-4GlcNAcb (3’SLN) and Neu5Acα2-6Galb1-4GlcNAcb (6’SLN), for receptor preference. A/chicken/Jiangsu/X1/2004 (X1, H9N2) and A/Puerto Rico/8/1934 (PR8, H1N1) were included as controls for ɑ-2,3 sialic acid (SA) and ɑ-2,6 SA, respectively (Fig. 3). Five of the viruses, rg-XZ491-193N, rg-XZ491-193A, rg-XZ491-193D, rg-XZ491-193G, and rg-XZ491-193S, mainly bound to α-2,6 SA; however, rg-XZ491-193E efficiently bound to both α-2,3 SA and α-2,6 SA (Fig. 3). These data demonstrate that the variation at HA position 193 influences the receptor preference of H9N2 viruses.
FIG 3.
Receptor-binding properties of the H9N2 viruses with various amino acids at HA position 193. The binding of the H9N2 viruses with sialic acids were determined using various concentrations of sialic acids conjugated to biotinylated sialylglycopolymers (3’SLN and 6’SLN) via direct solid-phase binding assays. The statistical analysis was performed with Student's t test. *, **, and *** indicate P values of less than 0.05, 0.01, and 0.001, respectively.
Variation in HA position 193 alters antigenicity of H9N2 AIVs.
The HA RBS overlaps with antigenic sites, and thus, amino acid substitutions around the RBS may contribute to the antigenic drift of influenza viruses (23, 24, 27, 28). To assess the effect of amino acid variation at position 193 on HA antigenicity, the rescued H9N2 viruses were first tested with H9-specific MAbs in hemagglutination-inhibition (HI) assays. Due to the low HA titer of rg-XZ491-193G virus with chicken RBCs, goose RBCs were used to test rg-XZ491-193G in all HI assays. All MAbs well inhibited rg-XZ491-193N, with HI titers of 10,240 and 20,480, and all viruses were inhibited by MAbs 2G10 and 3F2 at similar HI titers. However, viruses with N mutated to other residues exhibited dramatic reduced inhibition by some of the tested MAbs (Table 3). For instance, compared to rg-XZ491-193N, HI titer of MAb 6E6 against rg-XZ491-193A virus was 32-fold lower, the inhibition of rg-XZ491-193D virus by MAbs 5B4, 6A10, 6B6, and 6E6 was also significantly decreased, whereas rg-XZ491-193E virus was poorly inhibited by these four antibodies as well as MAb 6A5 (Table 3).
TABLE 3.
Inhibition of H9N2 viruses carrying various residues at HA position 193 by H9-specific MAbs in HI assay
| Virus | HI titer |
|||||||
|---|---|---|---|---|---|---|---|---|
| 2G4a | 3F2 | 2G10 | 5B4 | 6A5 | 6A10 | 6B6 | 6E6 | |
| XZ491-193N | 20,480 | 20,480 | 10,240 | 20,480 | 20,480 | 20,480 | 20,480 | 20,480 |
| XZ491-193A | 20,480 | 20,480 | 20,480 | 20,480 | 20,480 | 10,240 | 10,240 | 640 |
| XZ491-193D | 20,480 | 20,480 | 20,480 | 1,280 | 20,480 | 320 | 320 | 640 |
| XZ491-193E | 20,480 | 20,480 | 20,480 | 640 | 640 | 640 | 640 | 160 |
| XZ491-193S | 20,480 | 10,240 | 20,480 | 5,120 | 10,240 | 10,240 | 10,240 | 1,280 |
| XZ491-193G | 5,120 | 10,240 | 20,480 | 2,560 | 20,480 | 10,240 | 10,240 | 2,560 |
MAbs used in this study were untreated mouse ascitic fluid of hybridomas.
To further evaluate the antigenic change, chicken antisera against these H9N2 viruses were generated and cross inhibition of each virus was examined with HI assays. Compared to the homologous HI titers, some of the antisera exhibited 4- or 8-fold reduction in the titers with viruses with a different residue at HA position 193 (Table 4). The difference in antigenicity of these viruses was also evident when tested with chicken sera raised against a commercial vaccine containing H9N2 virus with N at HA position 193 (Table 4). For instance, serum#5 inhibited rg-XZ491-193N virus at an HI titer of 5,120, whereas the titers with other viruses were either 4- or 8-fold lower (Table 4). Taken together, HI assay using both murine MAbs and chicken antisera demonstrated the variation in antigenicity of viruses with different residues at HA position 193.
TABLE 4.
Inhibition of H9N2 viruses carrying various residues at HA position 193 by chicken antisera in HI assay
| Virus | Antisera against rg-XZ491 virusesa |
Antisera against vaccineb |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 193N | 193A | 193D | 193E | 193S | 193G | #1 | # 2 | # 3 | #4 | #5 | |
| XZ491-193N | 2,560 | 1,280 | 1,280 | 1,280 | 2,560 | 1,280 | 5,120 | 2,560 | 5,120 | 5,120 | 5,120 |
| XZ491-193A | 1,280 | 2,560 | 1,280 | 2,560 | 5,120 | 5,120 | 5,120 | 2,560 | 2,560 | 2,560 | 1,280 |
| XZ491-193D | 5,120 | 2,560 | 10,240 | 1,280 | 10,240 | 2,560 | 1,280 | 1,280 | 1,280 | 1,280 | 640 |
| XZ491-193E | 5,120 | 640 | 2,560 | 5,120 | 5,120 | 2,560 | 2,560 | 1,280 | 1,280 | 2,560 | 640 |
| XZ491-193S | 10,240 | 2,560 | 5,120 | 5,120 | 10,240 | 2,560 | 2,560 | 1,280 | 2,560 | 2,560 | 1,280 |
| XZ491-193G | 1,280 | 1,280 | 2,560 | 1,280 | 2,560 | 10,240 | 2,560 | 1,280 | 1,280 | 1,280 | 640 |
Antisera collected from chickens immunized with H9N2 viruses XZ491-193N (193N), XZ491-193A (193A), XZ491-193D (193D), XZ491-193E (193E), XZ491-193S (193S), and XZ491-193G (193G), respectively.
Antisera collected from chickens immunized with a commercial vaccine, which contains an H9N2 virus bearing N193 in the HA.
Mutations at HA position 193 influences the transmission of H9N2 AIVs in chickens.
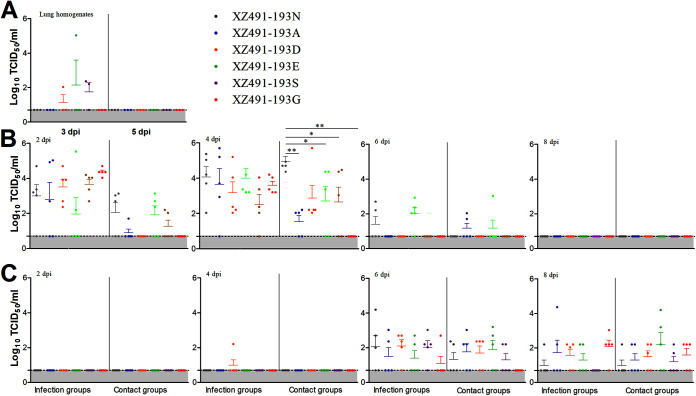
To determine whether amino acid changes at HA position 193 affects in vivo viral shedding and transmissibility, chickens were infected with each of the six H9N2 viruses and uninfected chickens were introduced as the direct-contact. At days 3 and 5 postinfection, three chickens per infection group were sacrificed and lungs were collected for viral detection with MDCK cells. No clinical signs or mortality were observed for any group during 14-day experiment. As shown in Fig. 4A, no virus was detected in lungs from chickens infected with rg-XZ491-193N, rg-XZ491-193A, or rg-XZ491-193G; however, low titers of virus in lungs from some of the chickens infected with rg-XZ491-193D (1/3), rg-XZ491-193E (1/3), and rg-XZ491-193S (2/3) were detected on 3 dpi. To further examine the virus shedding and transmission, oropharyngeal and cloacal swabs were collected from both the infected and direct-contact chickens at 2, 4, 6, and 8 dpi and titrated for virus. All infected chickens had virus shedding in oropharyngeal at 2 and 4 dpi (Fig. 4B). Chickens infected with rg-XZ491-193N or rg-XZ491-193E had detectable virus shedding at 6 dpi, whereas no virus shedding was detected in any group by 8 dpi. Birds that were in direct contact with chickens infected with rg-XZ491-193N, rg-XZ491-193A, rg-XZ491-193E, or rg-XZ491-193S had virus shedding in oropharyngeal at 2 and 4 dpi; however, by 6 dpi, only those contacted with chickens infected with rg-XZ491-193D or rg-XZ491-193E had virus shedding (Fig. 4B). Interestingly, no virus shedding was detected in the oropharyngeal from birds in contact with rg-XZ491-193G virus infected chickens. The statistical analysis showed that the chickens in the contact group of rg-XZ491-193N had evidently higher virus shedding than those in the contact group of rg-XZ491-193A, rg-XZ491-193D, and rg-XZ491-193G at 2 dpi. Moreover, the chickens in the contact group of rgXZ491-193N had much higher virus shedding than those in other contact groups of rg-XZ491-193A, rg-XZ491-193E, rg-XZ491-193S, and rg-XZ491-193G (P values < 0.05 or 0.01) at 4 dpi (Fig. 4B). Virus shedding in cloaca of the infected chickens was later than in oropharyngeal (Fig. 4C). At 2 dpi, no virus shedding was detected, and only one cloacal swab of from the rg-XZ491-193D infected chickens had low titer at 4 dpi. However, by 6 dpi, all infected groups had virus shedding in cloaca (Fig. 4C). Virus was also detected from the cloaca swabs collected the direct-contact groups on 6 and 8 dpi.
FIG 4.
Replication and transmission of the H9N2 viruses with various amino acids at HA position 193 in chickens. Groups of 2-week-old chickens were infected intranasally with 105 TCID50 of each virus and five uninfected chickens were introduced as direct-contact group. At 3 and 5 dpi, three chickens each infected group were sacrificed and lung samples were collected to determine virus titers (A). At 2, 4, 6, 8, and 10 dpi, oropharyngeal (B) and cloaca (C) swabs from infected and contact groups were collected to determine the virus titers. The statistical analysis was performed with Mann-Whitney U test. *, **, and *** indicate P values of less than 0.05, 0.01, and 0.001, respectively.
To assess antibody response to virus infection, the remaining chickens in each group were bled at 7, 14, and 21 dpi and sera were collected for measuring antibody titers by HI assays. Chickens from each infection group mounted efficient antibody response by 7 dpi and had increased HI antibody titers by 14 and 21 dpi (Fig. 5). The statistical analysis showed that HI antibody titers of rg-XZ491-193N group was significantly higher than those of rg-XZ491-193A group at 14 dpi (Fig. 5B). For direct-contact groups, at 7 dpi, all birds had low serum HI titers, and although the HI titers increased by 14 and 21 dpi, they were generally lower than those observed for the infected groups. Notably, sera from chickens in contact with the rg-XZ491-193G-infected birds, had substantially low HI antibody titers compared to other groups (Fig. 5). To confirm the results of animal study, the four viruses (rg-XZ491-193N, rg-XZ491-193D, rg-XZ491-193E, and rg-XZ491-193G) were selected to repeated animal study. The results of this independent repeated animal experiment (Fig. S1 and S2) were similar to the above.
FIG 5.
Antibody response in chickens to H9N2 viruses with various amino acids at HA position 193. Shown were HI titers of sera collected at 7, 14, and 21 dpi from chickens in infected and direct-contact groups. The statistical analysis was performed with Mann-Whitney U test. *, **, and *** indicate P values of less than 0.05, 0.01, and 0.001, respectively.
DISCUSSION
H9N2 virus is the predominant AIV subtype in poultry, especially in Asia (29, 30). Due to the threat posed by this virus to poultry and public health, a vaccination strategy has been implemented in China since 1998 (12). However, H9N2 variants have been isolated from the vaccinated chicken flocks (31), highlighting the continuous evolution of H9N2 virus under the immune pressure and the significance of assessment on the impact of amino acid changes in viral surface proteins, especially HA. In this study, more than 6,800 HA sequences of H9N2 viruses, almost isolated from the vaccinated chickens in China, were analyzed. The results revealed that the amino acid at HA position 193 of H9N2 viruses has undergone frequent substitutions since 2013, with the majority of the circulating strains carry N, A, D, E, G, and S at this position. The phylogenetic analysis further showed that HA genes of H9N2 viruses circulating in China could be clustered into multiples sublineages; however, the amino acid at position 193 was not association with the classification of the sublineages (data not shown). Moreover, the amino acid at HA position 193 was highly conserved in other parts of world, with the majority of H9N2 isolates bearing N (about 95%). These data suggest that the frequent variation of amino acid at HA position 193 of H9N2 viruses circulating in chickens in China may be related to the immune pressure.
In this study, we found that rg-XZ491-193G virus, bearing an N193G mutation, showed lower binding ability to RBCs (Table 2) and poor replication efficiency in mammalian and avian cells (Fig. 2). Moreover, rg-XZ491-193G had reduced transmissibility in chickens, as evidenced by the lack of viral shedding in oropharyngeal swabs and the low serum HI titers from chickens in direct-contact group (Fig. 4 and 5). A previous report showed that H9N2 virus with 193G in HA cannot be rescued with PR8 backbone (32). These findings suggest that H9 HA with 193G is less functional. Interestingly, a large number of H9N2 viruses containing 193G in HA position 193 were isolated from 2016 to 2019. To investigate whether there are correlated mutations in HA with 193G, we used the Fastcov to analyze the HA sequences. The result showed that the 193G had a correlated mutation at position 95 in HA (H3 numbering, N95H). However, whether N95H has a compensatory effect need further investigation.
The antigenic variation of HA of influenza viruses is generally considered to be determined by the alterations of multiple amino acids at the five major antigenic sites in the globular head (21, 33). In this study, compared with rg-XZ491-193N, all variant viruses have reduced reactivity with chicken sera raised against a commercial H9N2 vaccine, demonstrating 2- to 8-fold lower HI titer, especially the variant virus of rg-XZ491-193N (Table 4). It is noteworthy that the proportion of H9N2 viruses circulating in China carrying D at HA position 193 were second largest (Fig. 1). These results suggest that N193D mutation may help H9N2 to escape from the vaccine protection (Table 4). Here, we also found amino acid mutations at HA position 193 can decrease the HI titers of some H9-specific MAbs, although the target sites of these MAbs are not position 193 (Table 3). For examples, 6A5 targeting position 160, 6A10 targeting position 156, 6B6 targeting positions 188 and 199, and 6E6 targeting 159 (27) significantly decreased HI titers with some variant viruses (Table 3), which proved that the amino acid change at position 193 may lead to the conformational change of HA, then reducing the reactivity with the antibodies.
Receptor binding is an important determinant of host specificity, and modification of receptor-binding property is often a critical step in cross-species viral transmission. It has been reported that more and more circulating H9N2 viruses can bind to α-2,6 SA (34). Multiple molecular determinants have been shown to influence the receptor-binding profiles of H9N2 viruses. The Q226L substitution alone renders H9N2 virus to replicate efficiently in human epithelial airway cells (32, 35, 36). Substitutions at 155, 184, 188, 190, 196, 225, and 227 have also been shown to play an important role in receptor-binding in some H9N2 strains (26, 34, 37, 38). In our study, we found that 193N, 193A, 193D, 193G, and 193 S mainly bound to α-2,6 SA. This finding is consistent with reports about the human-like receptor preference by viruses with R, G, and N at HA position 193 (26, 32). Interestingly, we confirmed that virus with 193E in HA can efficiently bind to both α-2,3 SA and α-2,6 SA (Fig. 3). The impact of this change in receptor preference deserves further investigation.
In conclusion, the results from this study demonstrated that amino acid substitutions at HA position 193 of H9N2 virus affect the viral replication, antigenicity, and receptor binding in vitro, as well as the transmission and antibody response in chickens. Our findings highlight the importance of continuous monitoring on the evolution of H9N2 virus, which may facilitate the comprehensive control of this virus in both poultry and humans.
MATERIALS AND METHODS
Viruses and cells.
The H9N2 AIV of XZ491 used in this study was isolated from sick chickens in Xuzhou, Jiangsu Province, China in 2019. The virus was propagated in 10-day-old specific pathogen free (SPF) embryonated chicken eggs and stocked in −80°C before use. MDCK, A549, CEF, and 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM), containing 10% fetal calf serum (FCS).
Site-directed mutagenesis.
Nucleotide changes corresponding to single amino acid mutation at position 193 were introduced into HA gene in plasmid pDP2000 with the Mut Express II Fast Mutagenesis Kit V2 (Vazyme Biotech Co., Ltd.), and the plasmids were sequenced to verify the presence of introduce mutations.
Reverse genetics.
The H9N2 viruses with the wild-type or mutant HA of XZ491 were generated as previously described (39). Briefly, eight plasmids (1 μg each plasmid) were co-transfected into a mixture of 293T and MDCK cells. At 6 h posttransfection, the medium was replaced with 1 mL of opti-MEM. After 24 h, the medium was replaced with 2 mL of opti-MEM with 1 μg/mL TPCK-Trypsin. At 72 h posttransfection, the culture supernatant was collected and inoculated into 10-day-old SPF embryonated chicken eggs for preparing of viral stocks.
HI assay.
The antigenic phenotypes of the H9N2 viruses were determined by performing HI assay. Briefly, chicken serum and H9-specific MAbs were diluted with PBS to 1:10 and subsequently serially 2-fold diluted and mixed with 8 hemagglutination units (HAUs) in 96-well plate and incubated at room temperature for 30 min. The HI activity was visualized by adding 0.5% chicken RBCs or 0.5% goose RBCs to the virus-antibody mixtures and incubated at room temperature for 30 min before reading. All HI assays in this study were performed in duplicates, and data shown as the average values of two independent experiments.
Viral growth kinetics.
Confluent MDCK, A549, and CEF cells were infected with the H9N2 viruses at an MOI of 0.001, and then maintained in opti-MEM containing 1 μg/mL TPCK-Trypsin. The culture supernatants were collected at 6, 12, 24, 48, and 72 hpi, and the viral titers were determined in MDCK, A549, and CEF cells by TCID50 by immunofluorescence assay (IFA) with MAb 2G10 against HA protein of H9N2 virus (40).
Solid-phase binding assay.
The receptor binding property of the H9N2 viruses was analyzed by solid-phase binding assay as previously described (41). Briefly, the Perce streptavidin high binding capacity coated 96-well plates (Thermo Fisher Scientific, USA) were coated serially 2-fold diluted Neu5Aca2-3Galb1-4GlcNAcb-PAA-biotin and Neu5Aca2-6Galb1-4GlcNAcb-PAA-biotin (Glycitech, USA) and incubated overnight at 4°C. After blocked with 5% skim milk in PBST, the plates were incubated at 4°C overnight with 64 HAUs of the H9N2 viruses. After three washes with PBST, the plates were incubated with H9-specific MAb 2G10 at 4°C for 2 h (40). The plates were washed three times with PBST and incubated with a horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody for 2 h at 4°C and then washed three times with PBST again. Then, 100 μL TMB solution was added to each well and incubated 10 min. The reaction was stopped by adding 2 M H2SO4, and the OD450 values were measured.
Animal study.
Two-week-old SPF chickens were randomly distributed into six groups. Eleven 2-week-old SPF chickens per group were infected with each H9N2 virus at 105 TCID50 in a volume of 0.2 mL by intranasal inoculation, respectively. In addition, five uninfected chickens per virus were used as the direct-contact group. At 3 and 5 dpi, three infected chicken per group were sacrificed and lungs were collected to determine viral titers in MDCK cells. At 2, 4, 6, and 8 dpi, oropharyngeal and cloaca swabs from the infection and direct-contact groups were collected and titrated in MDCK cells. At 7, 14, and 21 dpi, the serum samples of all chickens were collected and used to evaluate antibodies by HI assay.
Statistical analysis.
All the results are presented as means ± standard deviations. The statistical analysis in this study was performed with a Student's t test or Mann-Whitney U test using GraphPad 5 software. A P value of below 0.05 was considered significant. *, **, and *** indicate P values of less than 0.05, 0.01, and 0.001, respectively.
Analysis of HA sequences.
HA sequences of H9N2 viruses, which isolated from chickens in China, were obtained from the GISAID Initiative (accessed from 1994) and cleaned as described previously (42). The sequences were aligned using MAFF (version 7.2.1). The duplicated and highly similar sequences were removed by BioAider (v1.423) (43). Then the amino acid at position 193 was analyzed.
Ethics statement.
The animal study was performed in accordance with the institutional animal guidelines approved by the Animal Care Committee at Yangzhou University, China. The ethical permission code (no. NSFC2020-SYXY-10, licensed on March 23, 2020) was provided by the Animal Committee of Yangzhou University.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (32002262), Basic Research Program of Jiangsu Province (BK20200922), the 111 Project D18007, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Footnotes
Supplemental material is available online only.
Contributor Information
Zhimin Wan, Email: wanzm@yzu.edu.cn.
Aijian Qin, Email: aijian@yzu.edu.cn.
Jianqiang Ye, Email: jqye@yzu.edu.cn.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Homme PJ, Easterday BC. 1970. Avian influenza virus infections. I. Characteristics of influenza A-turkey-Wisconsin-1966 virus. Avian Dis 14:66–74. 10.2307/1588557. [DOI] [PubMed] [Google Scholar]
- 2.Alexander DJ. 2007. An overview of the epidemiology of avian influenza. Vaccine 25:5637–5644. 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 3.Kim JA, Cho SH, Kim HS, Seo SH. 2006. H9N2 influenza viruses isolated from poultry in Korean live bird markets continuously evolve and cause the severe clinical signs in layers. Vet Microbiol 118:169–176. 10.1016/j.vetmic.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Aamir UB, Wernery U, Ilyushina N, Webster RG. 2007. Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology 361:45–55. 10.1016/j.virol.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Zhang Y, Li H, Gong T, Shi Y, Ni X, Li J, Zhou J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. 2014. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383:714–721. 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 6.Deng G, Shi J, Wang J, Kong H, Cui P, Zhang F, Tan D, Suzuki Y, Liu L, Jiang Y, Guan Y, Chen H. 2015. Genetics, receptor binding, and virulence in mice of H10N8 influenza viruses isolated from ducks and chickens in live poultry markets in China. J Virol 89:6506–6510. 10.1128/JVI.00017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, Lu SH, Yang YD, Fang Q, Shen YZ, Xi XM, Gu Q, Zhou XM, Qu HP, Yan Z, Li FM, Zhao W, Gao ZC, Wang GF, Ruan LX, Wang WH, Ye J, Cao HF, Li XW, Zhang WH, Fang XC, He J, Liang WF, Xie J, Zeng M, Wu XZ, Li J, Xia Q, Jin ZC, Chen Q, Tang C, Zhang ZY, Hou BM, Feng ZX, Sheng JF, Zhong NS, Li LJ. 2013. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med 368:2277–2285. 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- 8.Guan Y, Shortridge KF, Krauss S, Webster RG. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci USA 96:9363–9367. 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye G, Liang CH, Hua DG, Song LY, Xiang YG, Guang C, Lan CH, Ping HY. 2016. Phylogenetic analysis and pathogenicity assessment of two strains of avian influenza virus subtype H9N2 isolated from migratory birds: high homology of internal genes with human H10N8 virus. Front Microbiol 7:57. 10.3389/fmicb.2016.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banet-Noach C, Perk S, Simanov L, Grebenyuk N, Rozenblut E, Pokamunski S, Pirak M, Tendler Y, Panshin A. 2007. H9N2 influenza viruses from Israeli poultry: a five-year outbreak. Avian Dis 51:290–296. 10.1637/7590-040206R1.1. [DOI] [PubMed] [Google Scholar]
- 11.Choi JG, Lee YJ, Kim YJ, Lee EK, Jeong OM, Sung HW, Kim JH, Kwon JH. 2008. An inactivated vaccine to control the current H9N2 low pathogenic avian influenza in Korea. J Vet Sci 9:67–74. 10.4142/jvs.2008.9.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Yu K, Tian G, Yu D, Liu L, Jing B, Ping J, Chen H. 2005. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology 340:70–83. 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69:531–569. 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 14.Park KJ, Kwon HI, Song MS, Pascua PN, Baek YH, Lee JH, Jang HL, Lim JY, Mo IP, Moon HJ, Kim CJ, Choi YK. 2011. Rapid evolution of low-pathogenic H9N2 avian influenza viruses following poultry vaccination programmes. J Gen Virol 92:36–50. 10.1099/vir.0.024992-0. [DOI] [PubMed] [Google Scholar]
- 15.Zhang P, Tang Y, Liu X, Peng D, Liu W, Liu H, Lu S, Liu X. 2008. Characterization of H9N2 influenza viruses isolated from vaccinated flocks in an integrated broiler chicken operation in eastern China during a 5 year period (1998–2002). J Gen Virol 89:3102–3112. 10.1099/vir.0.2008/005652-0. [DOI] [PubMed] [Google Scholar]
- 16.de Graaf M, Fouchier RA. 2014. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J 33:823–841. 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Pu J, Jiang Z, Guan T, Xia Y, Xu Q, Liu L, Ma B, Tian F, Brown EG, Liu J. 2010. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Vet Microbiol 146:215–225. 10.1016/j.vetmic.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C, Wei Y, Wang D, Zhu B, Lemmon G, Jiao Y, Duan S, Wang Q, Du Q, Sun M, Bao J, Sun Y, Zhao J, Zhang H, Wu G, Liu J, Webster RG. 2015. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci USA 112:548–553. 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. 1988. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 333:426–431. 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 20.Skehel JJ, Bayley PM, Brown EB, Martin SR, Waterfield MD, White JM, Wilson IA, Wiley DC. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci USA 79:968–972. 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376. 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 22.Grgic H, Costa M, Friendship RM, Carman S, Nagy E, Wideman G, Weese S, Poljak Z. 2014. Molecular characterization of H3N2 influenza A viruses isolated from Ontario swine in 2011 and 2012. Virol J 11:194. 10.1186/s12985-014-0194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaverin NV, Rudneva IA, Ilyushina NA, Lipatov AS, Krauss S, Webster RG. 2004. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 escape mutants. J Virol 78:240–249. 10.1128/jvi.78.1.240-249.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peacock T, Reddy K, James J, Adamiak B, Barclay W, Shelton H, Iqbal M. 2016. Antigenic mapping of an H9N2 avian influenza virus reveals two discrete antigenic sites and a novel mechanism of immune escape. Sci Rep 6:18745. 10.1038/srep18745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers GN, D'Souza BL. 1989. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173:317–322. 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 26.Peacock TP, Sealy JE, Harvey WT, Benton DJ, Reeve R, Iqbal M. 2021. Genetic determinants of receptor-binding preference and zoonotic potential of H9N2 avian influenza viruses. J Virol 95. 10.1128/JVI.01651-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan Z, Ye J, Xu L, Shao H, Jin W, Qian K, Wan H, Qin A. 2014. Antigenic mapping of the hemagglutinin of an H9N2 avian influenza virus reveals novel critical amino acid positions in antigenic sites. J Virol 88:3898–3901. 10.1128/JVI.03440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, Skepner E, Lewis NS, Spronken MI, Russell CA, Eropkin MY, Hurt AC, Barr IG, de Jong JC, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Smith DJ. 2013. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342:976–979. 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 29.Gu M, Xu L, Wang X, Liu X. 2017. Current situation of H9N2 subtype avian influenza in China. Vet Res 48:49. 10.1186/s13567-017-0453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carnaccini S, Perez DR. 2020. H9 influenza viruses: an emerging challenge. Cold Spring Harb Perspect Med 10:a038588. 10.1101/cshperspect.a038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. 1999. Predicting the evolution of human influenza A. Science 286:1921–1925. 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 32.Fan M, Liang B, Zhao Y, Zhang Y, Liu Q, Tian M, Zheng Y, Xia H, Suzuki Y, Chen H, Ping J. 2022. Mutations of 127, 183 and 212 residues on the HA globular head affect the antigenicity, replication and pathogenicity of H9N2 avian influenza virus. Transbounding Emerging Dis 69. 10.1111/tbed.14363. [DOI] [PubMed] [Google Scholar]
- 33.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. 2001. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci USA 98:11181–11186. 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Shi J, Guo J, Deng G, Zhang Q, Wang J, He X, Wang K, Chen J, Li Y, Fan J, Kong H, Gu C, Guan Y, Suzuki Y, Kawaoka Y, Liu L, Jiang Y, Tian G, Li Y, Bu Z, Chen H. 2014. Genetics, receptor binding property, and transmissibility in mammals of naturally isolated H9N2 avian influenza viruses. PLoS Pathog 10:e1004508. 10.1371/journal.ppat.1004508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan H, Perez DR. 2007. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol 81:5181–5191. 10.1128/JVI.02827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Li S, Sun H, Pan L, Cui X, Zhu X, Feng Y, Li M, Yu Y, Wu M, Lin J, Xu F, Yuan S, Huang S, Sun H, Liao M. 2020. Variation and molecular basis for enhancement of receptor binding of H9N2 avian influenza viruses in China isolates. Front Microbiol 11:602124. 10.3389/fmicb.2020.602124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sealy JE, Yaqub T, Peacock TP, Chang P, Ermetal B, Clements A, Sadeyen JR, Mehboob A, Shelton H, Bryant JE, Daniels RS, McCauley JW, Iqbal M. 2018. Association of increased receptor-binding avidity of influenza A(H9N2) viruses with escape from antibody-based immunity and enhanced zoonotic potential. Emerg Infect Dis 25:63–72. 10.3201/eid2501.180616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng Q, Xu D, Shen W, Liu Q, Rong G, Li X, Yan L, Yang J, Chen H, Yu H, Ma W, Li Z. 2016. A single mutation at position 190 in hemagglutinin enhances binding affinity for human type sialic acid receptor and replication of H9N2 avian influenza virus in mice. J Virol 90:9806–9825. 10.1128/JVI.01141-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA 97:6108–6113. 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin F, Dong X, Wan Z, Ren D, Liu M, Geng T, Zhang J, Gao W, Shao H, Qin A, Ye J. 2019. A single mutation N166D in hemagglutinin affects antigenicity and pathogenesis of H9N2 avian influenza virus. Viruses 11:709. 10.3390/v11080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu K, Gao R, Gu M, Li J, Shi L, Sun W, Liu D, Gao Z, Wang X, Hu J, Liu X, Hu S, Chen S, Gao S, Peng D, Jiao XA, Liu X. 2018. Genetic and biological characterization of two reassortant H5N2 avian influenza A viruses isolated from waterfowl in China in 2016. Vet Microbiol 224:8–16. 10.1016/j.vetmic.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 42.He D, Gu J, Gu M, Wu H, Li J, Zhan T, Chen Y, Xu N, Ge Z, Wang G, Hao X, Wang X, Hu J, Hu Z, Hu S, Liu X, Liu X. 2021. Genetic and antigenic diversity of H7N9 highly pathogenic avian influenza virus in China. Infect Genet Evol 93:104993. 10.1016/j.meegid.2021.104993. [DOI] [PubMed] [Google Scholar]
- 43.Zhou ZJ, Qiu Y, Pu Y, Huang X, Ge XY. 2020. BioAider: an efficient tool for viral genome analysis and its application in tracing SARS-CoV-2 transmission. Sustain Cities Soc 63:102466. 10.1016/j.scs.2020.102466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2. Download jvi.01379-22-s0001.pdf, PDF file, 0.4 MB (456KB, pdf)