ABSTRACT
Little is known about the relationships between symptomatic early severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral load and upper airway mucosal gene expression and immune response. To examine the association of symptomatic SARS-CoV-2 early viral load with upper airway mucosal gene expression, we profiled the host mucosal transcriptome from nasopharyngeal swab samples from 68 adults with symptomatic, mild-to-moderate coronavirus disease 19 (COVID-19). We measured SARS-CoV-2 viral load using reverse transcription-quantitative PCR (RT-qPCR). We then examined the association of SARS-CoV-2 viral load with upper airway mucosal immune response. We detected SARS-CoV-2 in all samples and recovered >80% of the genome from 95% of the samples from symptomatic COVID-19 adults. The respiratory virome was dominated by SARS-CoV-2, with limited codetection of other respiratory viruses, with the human Rhinovirus C being identified in 4 (6%) samples. This limited codetection of other respiratory viral pathogens may be due to the implementation of public health measures, like social distancing and masking practices. We observed a significant positive correlation between SARS-CoV-2 viral load and interferon signaling (OAS2, OAS3, IFIT1, UPS18, ISG15, ISG20, IFITM1, and OASL), chemokine signaling (CXCL10 and CXCL11), and adaptive immune system (IFITM1, CD300E, and SIGLEC1) genes in symptomatic, mild-to-moderate COVID-19 adults, when adjusting for age, sex, and race. Interestingly, the expression levels of most of these genes plateaued at a cycle threshold (CT) value of ~25. Overall, our data show that the early nasal mucosal immune response to SARS-CoV-2 infection is viral load dependent, potentially modifying COVID-19 outcomes.
IMPORTANCE Several prior studies have shown that SARS-CoV-2 viral load can predict the likelihood of disease spread and severity. A higher detectable SARS-CoV-2 plasma viral load was associated with worse respiratory disease severity. However, the relationship between SARS-CoV-2 viral load, airway mucosal gene expression, and immune response remains elusive. We profiled the nasal mucosal transcriptome from nasal samples collected from adults infected with SARS-CoV-2 during spring 2020 with mild-to-moderate symptoms using a comprehensive metatranscriptomics method. We observed a positive correlation between SARS-CoV-2 viral load, interferon signaling, chemokine signaling, and adaptive immune system in adults with COVID-19. Our data suggest that early nasal mucosal immune response to SARS-CoV-2 infection was viral load dependent and may modify COVID-19 outcomes.
KEYWORDS: metatranscriptomics, RNA-seq, mucosal immune response, gene expression, nasal swab, coronavirus, COVID-19, SARS-CoV-2
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection causes coronavirus disease 19 (COVID-19) and is responsible for the 21st century’s most significant pandemic (1). While most SARS-CoV-2 infections are either mild or asymptomatic, in approximately 2% to 10% of cases, it can lead to life-threatening pneumonia and multiple-organ failure (2), resulting in more than 5.5 million official COVID-19 deaths worldwide (3). It has been hypothesized that a dysregulated innate immune activation response induces a cytokine storm that promotes respiratory failure and may lead to acute respiratory distress syndrome (ARDS) (4, 5), which has been the main reason for hospital admission and mortality in COVID-19 patients (5). Several studies suggested that SARS-CoV-2 viral load can predict the likelihood of disease spread and severity (6–8). A higher detectable SARS-CoV-2 plasma viral load was associated with worse respiratory disease severity (8). Recently, we have identified associations between SARS-CoV-2 viral titer and the nasopharyngeal microbiome in adults (9). However, little is known about the relationship between SARS-CoV-2 viral load and airway mucosal immune response. To fill this knowledge gap, we profiled the host mucosal transcriptome from nasopharyngeal samples collected from outpatient adults infected with SARS-CoV-2 during spring 2020 with mild-to-moderate symptoms utilizing a comprehensive metatranscriptomics method we developed recently (10). We examined the association of SARS-CoV-2 viral load with the mucosal immune response.
RESULTS
Patient characteristics and SARS-CoV-2 viral load measurement.
Sixty-eight adults with confirmed, symptomatic, mild-to-moderate COVID-19 (based on criteria from the World Health Organization [11]) were enrolled as part of a clinical trial (12). All patient enrollment and sample collection procedures were done in the early phase of the pandemic (spring of 2020). As described in the Materials and Methods, all research samples were collected early in the infection (within 24 h of clinical diagnosis). The patients’ baseline characteristics are shown in Table 1. The median (interquartile range [IQR]) age was 36 (27 to 57) years. None of the participants had used antibiotics in the prior 2 weeks. Reverse transcription-quantitative PCR (RT-qPCR) was used to quantify the SARS-CoV-2 viral load. Based on the cycle threshold (CT) value, we partitioned the subjects into the following tertiles: lower tertile (CT 14.5 to 21.5, n = 23, hereafter referred to as “high viral load group”), median tertile (CT 21.6 to 25.5, n = 22, hereafter referred to as “medium viral load group”), and upper tertile (CT 25.6 to 36, n = 23, hereafter referred to as “low viral load group”). The three groups had no significant differences in baseline characteristics and underlining comorbidities (Table 1).
TABLE 1.
Demographic characteristics by viral load tertiles
| Characteristic | Value by viral load tertilea |
Test statistic | |||
|---|---|---|---|---|---|
| High viral loadb (N = 23) | Medium viral loadc (N = 22) | Low viral loadd (N = 23) | Combined (N = 68) | ||
| Age | |||||
| Lower quartile, median, and upper quartile | 30, 46, 59 | 24, 30, 45 | 28, 36, 58 | 27, 36, 57 | F2, 65 = 2.1, P = 0.13e |
| Mean ± SD | 44 ± 16 | 35 ± 15 | 41 ± 16 | 40 ± 16 | |
| Male sex | 0.43 (10/23) | 0.45 (10/22) | 0.61 (14/23) | 0.50 (34/68) | χ22 = 1.7, P = 0.44f |
| Race | |||||
| African American | 0.04 (1/23) | 0.05 (1/22) | 0.22 (5/23) | 0.10 (7/68) | χ26 = 11, P = 0.082f |
| Hispanic | 0.00 (0/23) | 0.09 (2/22) | 0.04 (1/23) | 0.04 (3/68) | |
| Unknown | 0.30 (7/23) | 0.09 (2/22) | 0.09 (2/23) | 0.16 (11/68) | |
| White | 0.65 (15/23) | 0.77 (17/22) | 0.65 (15/23) | 0.69 (47/68) | |
| Diabetes | 0.09 (2/23) | 0.09 (2/22) | 0.04 (1/23) | 0.07 (5/68) | χ22 = 0.46, P = 0.79f |
| Hypertension | 0.26 (6/23) | 0.14 (3/22) | 0.17 (4/23) | 0.19 (13/68) | χ22 = 1.2, P = 0.55f |
| Heart disease | 0.04 (1/23) | 0.00 (0/22) | 0.09 (2/23) | 0.04 (3/68) | χ22 = 2, P = 0.36f |
| Pulmonary disease | 0.13 (3/23) | 0.18 (4/22) | 0.09 (2/23) | 0.13 (9/68) | χ22 = 0.88, P = 0.64f |
| Obese | 0.31 (5/16) | 0.26 (5/19) | 0.40 (8/20) | 0.33 (18/55) | χ22 = 0.85, P = 0.65f |
Values are proportion (n) unless otherwise indicated. N = the number of nonmissing values.
Values represent the lower quartile for continuous variables.
Values represent the median for continuous variables.
Values represent the upper quartile for continuous variables.
Values were determined using the Kruskal-Wallis test.
Values were determined using the Pearson test.
Metatranscriptome captured the nasal virome.
Total RNA from the nasopharyngeal swab samples was extracted and further processed as described previously (10). After quality-based trimming and removal of human and bacterial rRNA reads, we had an average of 98,120,791 (median, 740,58,180) human reads and an average of 2,263,290 (median, 848,478) microbiome reads. The microbiome read bin contains viral, bacterial, fungal, and unclassified reads (for details, see Materials and Methods). Following quality control and initial data processing steps, including a taxonomic classification of reads, the reads classified as viruses were used to profile the respiratory virome. The reads mapped to human transcripts were used to analyze the host response to SARS-CoV-2.
With a stringent cutoff of >80% genome coverage, we identified SARS-CoV-2 in 65 (95.5%) out of 68 samples (see Table S3 in the supplemental material). All SARS-CoV-2 genomes identified were placed in the B.1 lineage in the Nextclade tree (13) (see Table S2 in the supplemental material). In addition to SARS-CoV-2, we identified human Rhinovirus C in four samples, and bacteriophage/prophage RNA transcripts were also recovered from many of the samples, albeit with lower genome coverage (Fig. 1A). Among the four samples codetected with human rhinoviruses, three were mapped to human Rhinovirus C reference genome JN798569.1 and one was mapped to human Rhinovirus C reference genome LC004809.1. Human rhinovirus C was spread across three CT value quantile groups; two were in the CT value quantile 1 group (high viral load), one was in CT value quantile 2 (medium viral load), and one was in CT value quantile 3 (low viral load) groups (Table S3). As anticipated, common respiratory viruses were not codetected in these samples, except human Rhinovirus C was codetected in 6% of the samples. This finding may be due to the implementation of public health measures early in the COVID-19 pandemic, such as masking practices and encouragement of social distancing.
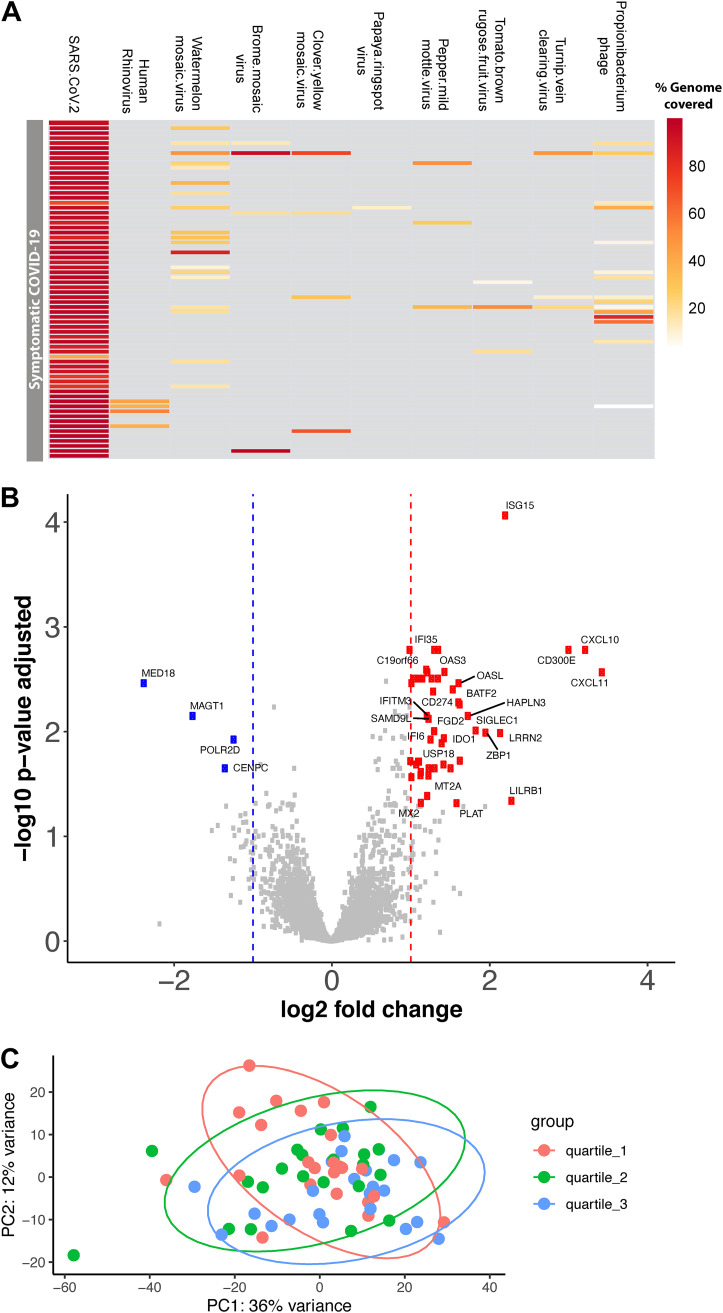
FIG 1.
Differentially expressed genes between high and low viral load SARS-COV-2-infected adults. (A) Heatmap showing the virome profile. Each row represents a sample, and each column represents the percentage of a virus genome recovered. (B) Volcano plot showing log2 fold change and adjusted P value obtained from DESeq2 analyses. Differential expression analysis was conducted using DESeq2 models, including age, sex, and race as covariates. The red squares indicate significantly upregulated genes, and the blue squares indicate significantly downregulated genes in symptomatic SARS-COV-2 adults with high viral load compared with the low viral load group. Only the top 50 most significantly different genes are labeled. (C) Principal-component analysis of the normalized gene-level read counts. Each dot represents a sample, and the samples are colored based on the CT value; we partitioned the subjects into the following tertiles: lower tertile (high viral load group) shown in red, median tertile (medium viral load group) shown in green, and upper tertile (low viral load group) shown in blue color.
Differential host nasal mucosal gene expression based on SARS-CoV-2 viral load.
Differential expression analysis between high and low viral load groups revealed that 48 genes were significantly upregulated and 4 genes were significantly downregulated (based on a threshold of log2 fold change of >|1| and adjusted P < 0.05) in the high viral load subjects (Fig. 1B and C; see Table S1 in the supplemental material). Most of the upregulated genes in the high viral load group are involved in the immune response during viral infection, specifically, genes involved in interferon alpha/beta signaling (IFITM3, IFITM1, RSAD2, MX2, IFI6, ISG15, IFI35, IFIT1, USP18, OASL, BST2, ISG20, OAS1, OAS2, OAS3, IRF7, and XAF1) (Fig. 2A and B) and immunoregulatory interactions between a lymphoid and a nonlymphoid cell (IFITM1, CD300E, LILRB1, and SIGLEC1) (Fig. 2B). Only a few genes were downregulated in the high viral load group, including magnesium transporter 1 (MAGT1), mediator complex subunit 18 (MED18), RNA polymerase II subunit D (POLR2D), and centromere protein C (CENPC) (Fig. 1). There was no significant difference in the gene expression profiles between the high and medium viral load groups and medium and low viral load groups.
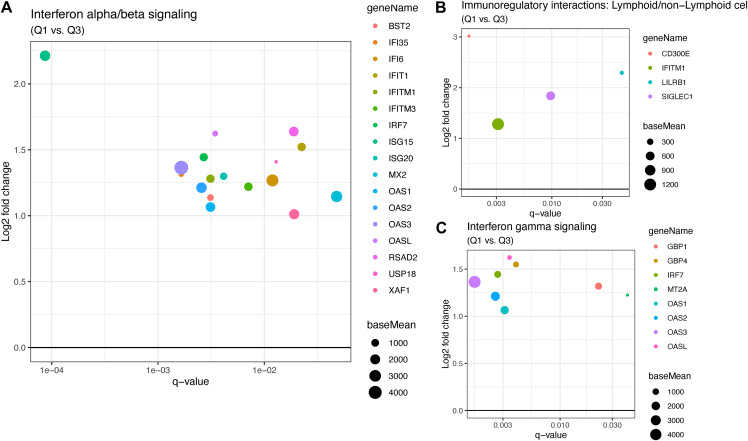
FIG 2.
(A) Interferon alpha/beta signaling pathway genes that are significantly down- or upregulated regulated in symptomatic SARS-COV-2 adults with high viral load compared with those in the low viral load group. On the x axis is displayed the q value for the up- or downregulated genes with q values of <0.05. On the y axis is shown the log2 fold change for those genes. The size of the dots represents “base mean,” which is the mean of normalized counts of all samples. (B) Similar to A, the plot shows the genes involved in immunoregulatory interactions between a lymphoid and a nonlymphoid cell that are upregulated in the high viral load group. (C) Plot showing the genes involved in interferon gamma signaling that are up-regulated in the high viral load group.
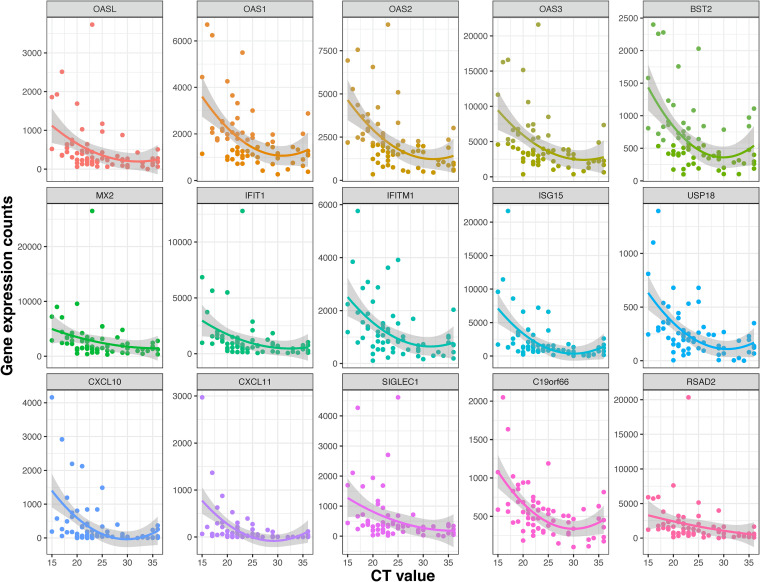
We examined the Spearman correlations between SARS-CoV-2 viral load and the mucosal gene expression levels. The genes involved in mucosal inflammation were inversely correlated with viral loads in terms of the CT values (Fig. 3); a lower CT value corresponds to a high viral load, indicating a higher level of infectiousness. Strikingly, there was a moderate negative correlation (ρ between −0.5 to −0.6; q value of <0.05) between interferon signaling (OAS2, OAS3, IFIT1, UPS18, ISG20, IFITM1, ISG15, and OASL), chemokine receptor signaling (CXCL 10 and CXCL11), and adaptive immune system (IFITM1, CD300E, and SIGLEC1) genes with SARS-CoV-2 CT values in symptomatic, mild-to-moderate COVID-19 adults (Fig. 3). The expression levels of most of these genes were decreased with a lower viral load and seemed to reach the plateau phase at a CT value of ~25 (Fig. 4). The expression of the POLR2D gene, which encodes the fourth largest subunit of RNA polymerase II, responsible for synthesizing mRNA in eukaryotes, showed a weak positive correlation (r = 0.28, q value = 0.02) with SARS-CoV-2 viral load (Fig. 3).
FIG 3.
The Spearman correlation between CT value and mucosal gene expression. The Spearman correlation coefficient was calculated for normalized gene-level expression counts and CT value (viral load). The Benjamini-Hochberg method was used for multiple comparisons and P value adjustments. The gene expression is moderately correlated with viral load. As the adjusted P < 0.05, the correlation is statistically significant.
FIG 4.
Scatterplots showing a correlation between CT value and mucosal gene expression. Normalized expression counts for selected genes of all samples were plotted with the CT values. The expression levels of most of these genes decreased with a lower viral load (i.e., with increased CT values) and reached the plateau phase at a CT value of ~25.
DISCUSSION
High SARS-CoV-2 viral loads during early infection have been associated with severe disease outcomes (14). We did not find an association between disease severity and viral load in our cohort, as all patients had mild-to-moderate symptoms. A multitude of studies have focused on the change in immune response and gene expression in human samples, including lung tissues (15), tracheal aspirates (16), nasal and nasopharyngeal swabs (17–20), bronchoalveolar lavage (BAL) fluid (20), and blood (19, 21–24). Studies using animal models and cell lines have shown that SARS-CoV-2 infection is associated with low type I and type III interferon and elevated interleukin-6 (IL-6) and chemokines that resulted in reduced innate antiviral defense and increased inflammation (25). A metatranscriptomic analysis of human BAL fluid from SARS-CoV-2-infected patients shows robust overexpression of interferon-stimulated genes (ISGs) and other inflammatory genes (20). However, to our knowledge, none of the studies focused on understanding the role of early viral load on the mucosal immune response.
Similar to published data, we found the antiviral protein ISG15 was upregulated upon SARS-CoV-2 infection (26), as it can activate downstream pattern recognition receptor MDA5 to establish an antiviral state. However, this pathway is directly antagonized by SARS-CoV-2 to evade host innate immunity via the overexpression of PLpro, a gene with redundant functionality of ISG-deconjugating enzyme USP18 (a negative regulator of type I and III interferons) (27), as several studies (27, 28) show upregulation of USP18 upon SARS-CoV-2 infection. Here, we show USP18 was upregulated in the nose in a viral-load-dependent manner, which for the first time confirms in vitro findings in clinical samples (Fig. 4). Our findings in the human clinical samples support the hypothesis from Vere et al. (29) that targeting USP18 has strong therapeutic potential for COVID-19.
Similar to our findings, several studies have shown that the ISGs, OAS1, OAS2, OAS3, IFITMs, and OASL are robustly activated upon SARS-CoV-2 infection (30, 31). ISGs are critical in countering other interferon-responsive viruses like SARS-CoV and Middle East respiratory syndrome (MERS)-CoV (32). Supporting this hypothesis, multiple genome-wide association studies have shown that mutations in ISGs are associated with susceptibility to SARS-CoV-2 and the severity of COVID-19. Similarly, we also found interferon alpha and beta signaling genes BST2, IFIT1, OASL, MX2 (known antivirals), IFI35, IFI6, IFITM1, IFIM3, IRF7, ISG15, ISG20, RSAD2, USP18, and XAF1 (Fig. 4) were upregulated in subjects with a higher viral load. These genes have been linked with COVID-19 or other viral infection restrictions. IFIT genes, particularly IFIT1, work in concert with other ISGs to inhibit viral translation by binding viral mRNAs that mimic host mRNAs due to the presence of 5′ cap-1 structures (33, 34), which SARS-CoV-2 contains (35). The IFIT proteins also increase the expression of CXCL10 (36), the 2nd most upregulated gene in our analysis, which induces lymphocyte chemotaxis and may inhibit the replication of infected viruses. Only one study has identified IFIT1 in vitro during COVID-19 infection (37).
CXCL10 and CXCL11 are two T helper type 1 (Th1) chemokines that were upregulated in our analysis, and their gene expression was correlated with viral load. Both chemokines are highly produced in bronchial and alveolar epithelial cells. They have been implicated as predictors of COVID-19 severity in vitro, in vivo in an animal model, and in clinical data (38, 39). Additionally, our findings of upregulation of RSAD2, OAS1-3, IFITM1, MX2, and CD300E (20, 40–42) agree with findings published previously for SARS-CoV-2 infection. We further show that the immune modulatory gene sialic acid-binding Ig-like lectin 1 (Siglec-1/CD169) was upregulated in subjects with high viral load. Lectins, especially Siglec-1/CD169, have been known to mediate the attachment of viruses to antigen-presenting cells (APCs) (43). Recent studies show that blockage of Siglec-1 on monocyte-derived dendritic cells (MDDCs) decreased SARS-CoV-2 viral transfer or trans-infection to bystander target cells (43), and Siglec1 was associated with disease severity (44) and enhanced SARS-CoV-2 infection and influenced neutralizing antibodies (45).
Our study has many strengths, such as the inclusion of an overall young population with no underlining comorbidities (Table 1), no recent use of antibiotics or current use of intranasal medications, and samples were collected early in the infection, i.e., within the first 24 h of SARS-CoV-2 diagnosis. Also, as the patient enrollment and sample collection were done in the early pandemic (spring 2020), none of our patients were vaccinated or potentially infected prior to this infection, which could have confounded immune response and gene expression (9, 12). All the SARS-CoV-2 genotypes in this study were identified as B.1 lineage (Table S2). Our metatranscriptomics method also captured the virome and showed limited coinfection with other respiratory viruses.
We should also acknowledge several limitations. First, our study was cross-sectional; thus, there is a possibility of reverse causation. Second, residual confounding is possible, as we lacked data on the participants’ atopic status or other unknown morbidities that could modify the immune response. Third, though all samples were collected within 24 h after the first diagnosis of COVID-19, it is still impossible to ascertain the days since infection, as the presymptomatic period can vary considerably (3 to 15 days) between patients. Fourth, nasal viral titers drop dramatically after 10 to 15 days of infection (46); thus, prior studies in hospitalized patients have shown blood viral titer to be a better predictor of the disease severity (8). Fifth, we did not have lower respiratory tract samples. However, the upper respiratory tract (URT) is the portal of entry and an active site of replication of SARS-CoV-2, as well as a common harboring site for potential pathogens, and is thus of critical importance in the pathogenesis of this respiratory virus (47). Last, our results cannot be extended to adults with asymptomatic, severe, or critical COVID-19, as only those with symptomatic, mild-to-moderate COVID-19 were included in our study (11). Despite limitations, our study highlights that URT mucosal innate immune response correlates with SARS-CoV-2 viral load.
In summary, we determined that early SARS-CoV-2 viral load was associated with URT gene expression during COVID-19 infection and potentially modified both the innate and adaptive immune response to SARS-CoV-2. Future studies with larger sample sizes, with serial sample collection, and with patients who develop severe disease outcomes will be needed to examine how SARS-CoV-2 interacts with the mucosal gene expression and how these viral-host interactions can impact the clinical progression, severity, and recovery of COVID-19.
MATERIALS AND METHODS
Overview of the study design.
For the current study, we included nonhospitalized patients aged ≥18 years who were diagnosed with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (confirmed by qualitative PCR) at Vanderbilt University Medical Center or one of its affiliated centers in Nashville, Tennessee. These patients were enrolled as part of a clinical trial examining the effect of several types of nasal irrigations on upper respiratory tract (URT) symptoms and viral load during coronavirus disease 2019 (COVID-19). The detailed methods for this clinical trial have been reported previously (12). Exclusion criteria for these patients included current use of nasal saline irrigations or other intranasal medications, inability to perform nasal irrigations or to collect URT samples in a separate house bathroom or away from household contacts, or need for hospitalization related to SARS-CoV-2 infection. Thus, only patients with mild or moderate COVID-19 (based on criteria from the World Health Organization [11]) were included in the clinical trial. Eligible patients were contacted and enrolled in the study within 24 h of initial diagnosis.
Following adequate training, all participants were asked to obtain a midturbinate swab on the day of enrollment (i.e., before any study intervention for those enrolled in the clinical trial) using a self-collection kit (FLOQSwabs; Copan Diagnostics Inc.). These enrollment samples were used for the current study. The sample collection in the present study occurred between April and June 2020. Each adult provided informed consent for their participation. The Institutional Review Board of Vanderbilt University approved this study.
Severe acute respiratory syndrome coronavirus 2 testing by quantitative reverse transcription-PCR.
To measure viral load in SARS-CoV-2-infected patients, we performed quantitative reverse transcription-PCR (RT-qPCR) in the midturbinate swabs. Total RNA was extracted from the swabs using a phenol-chloroform-based method. The swabs were placed in red 1.5-mL RINO screw-cap tubes (NextAdvance) prefilled with RNase-free zirconium oxide beads, and QIAzol lysis reagent (Qiagen) was added. Samples were then homogenized in a Bullet Blender 24 Gold (NextAdvance) system for 3 min at maximum speed. Following homogenization, genomic DNA was eliminated with genomic DNA (gDNA) eliminator columns (Qiagen), and RNA was purified using the RNeasy mini plus kit (Qiagen) following the manufacturer’s protocol. The RNA quality was measured using a 2100 bioanalyzer (Agilent Technologies). The United States Centers for Disease Control and Prevention primers and probes designed for the detection of SARS-CoV-2 (2019-nCoV) were purchased from Integrated DNA Technologies (IDT) (48). Both the SARS-CoV-2 nucleocapsid gene region 1 (N1) and nucleocapsid gene region 2 (N2) were targeted to detect SARS-CoV-2. RNase P was also examined as a measure of RNA quality and quantity. RT-qPCR was performed using the SuperScript III one-step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen) as per manufacturer’s instructions on a CFX96 touch real-time PCR detection system (Bio-Rad). Plasmid controls for 2 SARS-CoV-2 nucleocapsids and RNase P were also ordered from IDT at a concentration of 66,666 copies/reaction. No-template controls and an extraction negative were used as negative controls. Reactions were prepared using 12.5 μL of SuperScript III master mix (ThermoFisher), 1 μL each of 400-nm forward and reverse primer, 1 μL of 400 nM FAM-labeled probe, 1 μL of Platinum Taq polymerase, 3 μL of template RNA, and 7.25 μL of PCR-certified water (Teknova). RNA was reverse transcribed at 50°C for 15 min, and PCR conditions were run on a 95°C denaturation step for 2 min, followed by 40 cycles of 95°C for 15 s and 55°C for 30 s. The cycle threshold (CT) values were captured and calculated by the CFX Maestro (Bio-Rad) software and used as a measure of viral load.
RNA extraction, metatranscriptomic library preparation, and sequencing.
The nasal swab samples in the self-collection kit (FLOQSwabs; Copan Diagnostics Inc.) were vortexed for 2 min, and then an aliquot of the 250-μL nasal swap sample was used for RNA extraction. The total RNA from these samples was extracted as described previously (10). In brief, a 250-μL aliquot from the nasal swab sample was homogenized in 600 μL QIAzol (Qiagen) and 500 μL of 2.0 mm zirconium oxide beads (Next Advance, Inc.; catalog ZROB05) using a Bullet Blender homogenizer (BB24-AU; Next Advance, Inc.). While the samples were homogenizing, the temperature was maintained at or near 4°C by using the dry ice cooling system in the Bullet Blender system. The homogenate was treated with 100 μL of genomic DNA eliminator solution (Qiagen) to remove the genomic DNA. Next, 180 μL of chloroform was added to the samples for phase separation. The total RNA in the aqueous phase was then purified using RNeasy mini spin columns as the Qiagen RNeasy protocol recommended. RNA integrity and quantification were assessed using a bioanalyzer RNA 6000 Nano/Pico chip (Agilent Technologies, Palo Alto, CA). Eukaryotic rRNAs were depleted using the NEBNext rRNA depletion kit (human/mouse/rat; catalog number E6310X). After rRNA depletion, the samples were checked by using the Agilent Bioanalyzer RNA 6000 Nano/Pico chip to ensure depletion of the 18S and 28S ribosomal peaks. Next, Illumina sequencing libraries were made using the NEBNext Ultra II RNA library prep kit (New England BioLabs [NEB]; number E7775). The quality of the libraries was assessed using an Agilent bioanalyzer DNA high-sensitivity chip. The libraries were then sequenced on an Illumina NovaSeq6000 platform (S4 flow cells run) with 2 × 150-base pair reads, with a sequencing depth of ~40 million paired-end reads per sample.
Quantification and statistical analysis.
(i) Preprocessing and quality control of next-generation sequencing (NGS) data. Adapter removal and quality-based trimming of raw reads were performed using Trimmomatic v0.39 (49) using default parameters. Trimmed reads shorter than 50 nucleotides (nt) were discarded. Low-complexity reads were discarded using bbduk from bbtools (50) with entropy set at 0.7 (BBMap, http://sourceforge.net/projects/bbmap/).
(ii) Read binning. Reads were mapped to human rRNA and the human mitochondrial genome using bbmap from bbtools; the mapped reads were discarded. The remaining reads were binned into the human genome, bacterial rRNA, and a bin that contains all microbiome reads, using the seal, from bbtools, with default parameters. The human genome (GRCh38) and SILVA bacterial rRNA database were used as references. Binning resulted in an average of 98,120,791 (median, 740,58,180) reads mapping to the human genome and an average of 2,263,290 (median, 848,478) microbiome reads. The microbiome read bin contains viral, bacterial, fungal, and unclassified reads.
(iii) Taxonomic classification of reads. Reads from the microbiome bin were subjected to taxonomic classification using KrakenUniq (51) with default parameters. The reference NCBI nucleotide database was installed via the kraken2-build script.
(iv) Virome profiling. To produce a high-confidence virome profile, we developed a method that first produces de novo transcriptome assemblies, followed by putative virome identification using BLAST searches, and finally high-confidence virome profiling based on read mapping to reference virus genomes. This workflow was implemented in a bash script. First, reads that were classified as viral by KrakenUniq were extracted using the script krakenuniq-extract-reads, with taxonID 10239 (superkingdom, viruses). If more than 100,000 reads were extracted, they were first normalized to a target depth of 100 using bbnorm from bbtools. Reads were assembled using the metaSPAdes assembler. The resulting contigs were filtered for length, using reformat from bbtools, and only contigs that were at least 300 bp were retained. Nucleotide BLAST (blastn) searches were performed on the resulting contigs against the NCBI nucleotide database with -max_target_seqs and -max_hsps set to 1. From the BLAST results, a list of subjects was compiled, and their genome sequences (fasta) were extracted from the nucleotide BLAST database using the blastdbcmd from BLAST. Each genome sequence was used as a reference, and all the virome reads were mapped using bowtie2. Genome coverage and average read depth statistics were extracted from this mapping using SAMtools (52). The high-confidence virome profile was constructed using these coverage statistics.
(v) SARS-CoV-2 genomes. Full sets of reads (before binning into human, bacterial rRNA, and microbiome) were utilized to produce the SARS-CoV-2 genome sequences. They were mapped to the SARS-CoV-2 reference genome NC_045512 using bowtie2 with default parameters. The consensus sequence was called using SAMtools, and lineages were determined using the Pangolin Web application (https://pangolin.cog-uk.io/). Consensus genome sequences were submitted to Global Initiative on Sharing Avian Influenza Data (GISAID).
(vi) Host response to SARS-CoV-2 infection. The reads identified as originating from human transcripts were mapped to the human genome (hg19) using HISAT2 (53). The read counts for genomic features were quantified using HTSeq (54). The feature counts of all the samples were combined into a single matrix using a custom R script. SARS-CoV-2-positive samples were partitioned into tertiles based on CT values. Differential expression analysis was performed by comparing tertile groups using the DESeq2 package (55). Genes with a significant log2 fold change with an adjusted P value of <0.05 were treated as differentially expressed. The lists of differentially expressed genes for each group were analyzed for enrichment of reactome human pathways using Enrichr (56) and were deemed significant when the false discovery rate (FDR) was <0.05.
Spearman rank correlation.
The Spearman rank correlation coefficient was calculated for gene expression counts and CT value (viral load) using the cor.test() function in R. The Benjamini-Hochberg method was used for multiple comparisons and P value adjustments.
Data availability.
Raw sequence reads are available in the Sequence Read Archive (SRA) BioProject identifier (ID) PRJNA922078.
ACKNOWLEDGMENTS
S.V.R., J.H.T., E.P., S.A.M., and S.R.D. contributed to the study design. K.S.K., M.H.S., B.C.W., V.G., M.H.F., and J.H.T. contributed to the sample collection. M.H.S., B.A.S., H.M.B., and S.R.D. contributed to the sample processing. S.B.P. and S.V.R. contributed to microbiome profiling and data analysis. S.B.P. generated SARS-CoV-2 genome sequences. S.V.R. and M.H.S. contributed to the statistical analysis. J.H.T. and S.R.D. obtained the research funding supporting this study. S.V.R. and S.R.D. wrote the original version of the manuscript, and all authors reviewed and approved the final version.
This work was supported by funds from the Centers For Disease Control and Prevention (CDC; 75D3012110094); National Institute of Allergy and Infectious Diseases (under award numbers R21AI142321-02S1, R21AI142321, R21AI154016, and R21AI149262); the National Heart, Lung, and Blood Institute (under award numbers K23HL148638 and R01HL146401); and the Vanderbilt Technologies for Advanced Genomics Core (grant support from the National Institutes of Health under award numbers UL1RR024975, P30CA68485, P30EY08126, and G20RR030956). The contents are solely the authors’ responsibility and do not necessarily represent the official views of the funding agencies.
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
Contributor Information
Suman R. Das, Email: suman.r.das@vumc.org.
Stacey Schultz-Cherry, St. Jude Children's Research Hospital.
REFERENCES
- 1.Zoumpourlis V, Goulielmaki M, Rizos E, Baliou S, Spandidos DA. 2020. [Comment] The COVID19 pandemic as a scientific and social challenge in the 21st century. Mol Med Rep 22:3035–3048. 10.3892/mmr.2020.11393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaim S, Chong JH, Sankaranarayanan V, Harky A. 2020. COVID-19 and multiorgan response. Curr Probl Cardiol 45:100618. 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam D. 2022. The pandemic’s true death toll: millions more than official counts. Nature 601:312–315. 10.1038/d41586-022-00104-8. [DOI] [PubMed] [Google Scholar]
- 4.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ, Hlh Across Speciality Collaboration UK, HLH Across Speciality Collaboration, UK . 2020. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395:1033–1034. 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. 2020. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8:420–422. 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawasuji H, Morinaga Y, Tani H, Yoshida Y, Takegoshi Y, Kaneda M, Murai Y, Kimoto K, Ueno A, Miyajima Y, Fukui Y, Kimura M, Yamada H, Sakamaki I, Yamamoto Y. 2022. SARS-CoV-2 RNAemia with a higher nasopharyngeal viral load is strongly associated with disease severity and mortality in patients with COVID-19. J Med Virol 94:147–153. 10.1002/jmv.27282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones TC, Biele G, Muhlemann B, Veith T, Schneider J, Beheim-Schwarzbach J, Bleicker T, Tesch J, Schmidt ML, Sander LE, Kurth F, Menzel P, Schwarzer R, Zuchowski M, Hofmann J, Krumbholz A, Stein A, Edelmann A, Corman VM, Drosten C. 2021. Estimating infectiousness throughout SARS-CoV-2 infection course. Science 373:eabi5273. 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fajnzylber J, Regan J, Coxen K, Corry H, Wong C, Rosenthal A, Worrall D, Giguel F, Piechocka-Trocha A, Atyeo C, Fischinger S, Chan A, Flaherty KT, Hall K, Dougan M, Ryan ET, Gillespie E, Chishti R, Li Y, Jilg N, Hanidziar D, Baron RM, Baden L, Tsibris AM, Armstrong KA, Kuritzkes DR, Alter G, Walker BD, Yu X, Li JZ, Abayneh BA, Allen P, Antille D, Balazs A, Bals J, Barbash M, Bartsch Y, Boucau J, Boyce S, Braley J, Branch K, Broderick K, Carney J, Chevalier J, Choudhary MC, Chowdhury N, Cordwell T, Daley G, Davidson S, Desjardins M, The Massachusetts Consortium for Pathogen Readiness, et al. 2020. Massachusetts Consortium for Pathogen R. 2020. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun 11:5493. 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosas-Salazar C, Kimura KS, Shilts MH, Strickland BA, Freeman MH, Wessinger BC, Gupta V, Brown HM, Rajagopala SV, Turner JH, Das SR. 2021. SARS-CoV-2 infection and viral load are associated with the upper respiratory tract microbiome. J Allergy Clin Immunol 147:1226–1233.e2. 10.1016/j.jaci.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajagopala SV, Bakhoum NG, Pakala SB, Shilts MH, Rosas-Salazar C, Mai A, Boone HH, McHenry R, Yooseph S, Halasa N, Das SR. 2021. Metatranscriptomics to characterize respiratory virome, microbiome, and host response directly from clinical samples. Cell Rep Methods 1:100091. 10.1016/j.crmeth.2021.100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization. 2020. Clinical management of COVID-19: interim guidance. World Health Organization, Geneva, Switzerland. 10.15557/PiMR.2020.0004. [DOI] [Google Scholar]
- 12.Kimura KS, Freeman MH, Wessinger BC, Gupta V, Sheng Q, Huang LC, Von Wahlde K, Das S, Chowdhury NI, Turner JH. 2020. Interim analysis of an open-label randomized controlled trial evaluating nasal irrigations in non-hospitalized patients with COVID-19. Int Forum Allergy Rhinol 10:1325–1328. 10.1002/alr.22703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aksamentov I, Roemer C, Hodcroft EB, Neher RA. 2021. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw 6:3773. 10.21105/joss.03773. [DOI] [Google Scholar]
- 14.Argyropoulos KV, Serrano A, Hu J, Black M, Feng X, Shen G, Call M, Kim MJ, Lytle A, Belovarac B, Vougiouklakis T, Lin LH, Moran U, Heguy A, Troxel A, Snuderl M, Osman I, Cotzia P, Jour G. 2020. Association of initial viral load in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) patients with outcome and symptoms. Am J Pathol 190:1881–1887. 10.1016/j.ajpath.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nienhold R, Ciani Y, Koelzer VH, Tzankov A, Haslbauer JD, Menter T, Schwab N, Henkel M, Frank A, Zsikla V, Willi N, Kempf W, Hoyler T, Barbareschi M, Moch H, Tolnay M, Cathomas G, Demichelis F, Junt T, Mertz KD. 2020. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun 11:5086. 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarma A, Christenson SA, Byrne A, Mick E, Pisco AO, DeVoe C, Deiss T, Ghale R, Zha BS, Tsitsiklis A, Jauregui A, Moazed F, Detweiler AM, Spottiswoode N, Sinha P, Neff N, Tan M, Serpa PH, Willmore A, Ansel KM, Wilson JG, Leligdowicz A, Siegel ER, Sirota M, DeRisi JL, Matthay MA, Consortium C, Hendrickson CM, Kangelaris KN, Krummel MF, Woodruff PG, Erle DJ, Calfee CS, Langelier CR, COMET Consortium . 2021. Tracheal aspirate RNA sequencing identifies distinct immunological features of COVID-19 ARDS. Nat Commun 12:5152. 10.1038/s41467-021-25040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mick E, Kamm J, Pisco AO, Ratnasiri K, Babik JM, Castaneda G, DeRisi JL, Detweiler AM, Hao SL, Kangelaris KN, Kumar GR, Li LM, Mann SA, Neff N, Prasad PA, Serpa PH, Shah SJ, Spottiswoode N, Tan M, Calfee CS, Christenson SA, Kistler A, Langelier C. 2020. Upper airway gene expression reveals suppressed immune responses to SARS-CoV-2 compared with other respiratory viruses. Nat Commun 11:5854. 10.1038/s41467-020-19587-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang EH, Willis AL, Romanoski CE, Cusanovich DA, Pouladi N, Li J, Lussier YA, Martinez FD. 2020. Rhinovirus infections in individuals with asthma increase ACE2 expression and cytokine pathways implicated in COVID-19. Am J Respir Crit Care Med 202:753–755. 10.1164/rccm.202004-1343LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng DL, Granados AC, Santos YA, Servellita V, Goldgof GM, Meydan C, Sotomayor-Gonzalez A, Levine AG, Balcerek J, Han LM, Akagi N, Truong K, Neumann NM, Nguyen DN, Bapat SP, Cheng J, Martin CS, Federman S, Foox J, Gopez A, Li T, Chan R, Chu CS, Wabl CA, Gliwa AS, Reyes K, Pan CY, Guevara H, Wadford D, Miller S, Mason CE, Chiu CY. 2021. A diagnostic host response biosignature for COVID-19 from RNA profiling of nasal swabs and blood. Sci Adv 7:eabe5984. 10.1126/sciadv.abe5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, Guo L, Yang J, Wang C, Jiang S, Yang D, Zhang G, Li H, Chen F, Xu Y, Chen M, Gao Z, Yang J, Dong J, Liu B, Zhang X, Wang W, He K, Jin Q, Li M, Wang J. 2020. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 27:883–890.e2. 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thair SA, He YD, Hasin-Brumshtein Y, Sakaram S, Pandya R, Toh J, Rawling D, Remmel M, Coyle S, Dalekos GN, Koutsodimitropoulos I, Vlachogianni G, Gkeka E, Karakike E, Damoraki G, Antonakos N, Khatri P, Giamarellos-Bourboulis EJ, Sweeney TE. 2021. Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections. iScience 24:101947. 10.1016/j.isci.2020.101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Julia A, Bonafonte-Pardas I, Gomez A, Lopez-Lasanta M, Lopez-Corbeto M, Martinez-Mateu SH, Llados J, Rodriguez-Nunez I, Myers RM, Marsal S. 2021. Targeting of the CD80/86 proinflammatory axis as a therapeutic strategy to prevent severe COVID-19. Sci Rep 11:11462. 10.1038/s41598-021-90797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arunachalam PS, Wimmers F, Mok CKP, Perera R, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, Wagh D, Coller J, Pellegrini KL, Kazmin D, Alaaeddine G, Leung WS, Chan JMC, Chik TSH, Choi CYC, Huerta C, Paine McCullough M, Lv H, Anderson E, Edupuganti S, Upadhyay AA, Bosinger SE, Maecker HT, Khatri P, Rouphael N, Peiris M, Pulendran B. 2020. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369:1210–1220. 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overmyer KA, Shishkova E, Miller IJ, Balnis J, Bernstein MN, Peters-Clarke TM, Meyer JG, Quan Q, Muehlbauer LK, Trujillo EA, He Y, Chopra A, Chieng HC, Tiwari A, Judson MA, Paulson B, Brademan DR, Zhu Y, Serrano LR, Linke V, Drake LA, Adam AP, Schwartz BS, Singer HA, Swanson S, Mosher DF, Stewart R, Coon JJ, Jaitovich A. 2021. Large-scale multi-omic analysis of COVID-19 severity. Cell Syst 12:23–40.e7. 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA, tenOever BR. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181:1036–1045.e9. 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honke N, Shaabani N, Zhang DE, Hardt C, Lang KS. 2016. Multiple functions of USP18. Cell Death Dis 7:e2444. 10.1038/cddis.2016.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu G, Lee JH, Parker ZM, Acharya D, Chiang JJ, van Gent M, Riedl W, Davis-Gardner ME, Wies E, Chiang C, Gack MU. 2021. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat Microbiol 6:467–478. 10.1038/s41564-021-00884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munnur D, Teo Q, Eggermont D, Lee HHY, Thery F, Ho J, van Leur SW, Ng WWS, Siu LYL, Beling A, Ploegh H, Pinto-Fernandez A, Damianou A, Kessler B, Impens F, Mok CKP, Sanyal S. 2021. Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat Immunol 22:1416–1427. 10.1038/s41590-021-01035-8. [DOI] [PubMed] [Google Scholar]
- 29.Vere G, Alam MR, Farrar S, Kealy R, Kessler BM, O’Brien DP, Pinto-Fernández A. 2022. Targeting the ubiquitylation and ISGylation machinery for the treatment of COVID-19. Biomolecules 12:300. 10.3390/biom12020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun G, Cui Q, Garcia G, Jr, Wang C, Zhang M, Arumugaswami V, Riggs AD, Shi Y. 2021. Comparative transcriptomic analysis of SARS-CoV-2 infected cell model systems reveals differential innate immune responses. Sci Rep 11:17146. 10.1038/s41598-021-96462-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alqutami F, Senok A, Hachim M. 2021. COVID-19 transcriptomic atlas: a comprehensive analysis of COVID-19 related transcriptomics datasets. Front Genet 12:755222. 10.3389/fgene.2021.755222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang KJ, Su IJ, Theron M, Wu YC, Lai SK, Liu CC, Lei HY. 2005. An interferon-gamma-related cytokine storm in SARS patients. J Med Virol 75:185–194. 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleith RC, Mears HV, Leong XY, Sanford TJ, Emmott E, Graham SC, Mansur DS, Sweeney TR. 2018. IFIT3 and IFIT2/3 promote IFIT1-mediated translation inhibition by enhancing binding to non-self RNA. Nucleic Acids Res 46:5269–5285. 10.1093/nar/gky191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daugherty MD, Schaller AM, Geballe AP, Malik HS. 2016. Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. Elife 5:e14228. 10.7554/eLife.14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viswanathan T, Arya S, Chan SH, Qi S, Dai N, Misra A, Park JG, Oladunni F, Kovalskyy D, Hromas RA, Martinez-Sobrido L, Gupta YK. 2020. Structural basis of RNA cap modification by SARS-CoV-2. Nat Commun 11:3718. 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imaizumi T, Hashimoto S, Sato R, Umetsu H, Aizawa T, Watanabe S, Kawaguchi S, Matsumiya T, Seya K, Ding J, Tanaka H. 2021. IFIT proteins are involved in CXCL10 expression in human glomerular endothelial cells treated with a Toll-like receptor 3 agonist. Kidney Blood Press Res 46:74–83. 10.1159/000511915. [DOI] [PubMed] [Google Scholar]
- 37.Prasad K, Khatoon F, Rashid S, Ali N, AlAsmari AF, Ahmed MZ, Alqahtani AS, Alqahtani MS, Kumar V. 2020. Targeting hub genes and pathways of innate immune response in COVID-19: a network biology perspective. Int J Biol Macromol 163:1–8. 10.1016/j.ijbiomac.2020.06.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lore NI, De Lorenzo R, Rancoita PMV, Cugnata F, Agresti A, Benedetti F, Bianchi ME, Bonini C, Capobianco A, Conte C, Corti A, Furlan R, Mantegani P, Maugeri N, Sciorati C, Saliu F, Silvestri L, Tresoldi C, Ciceri F, Rovere-Querini P, Di Serio C, Cirillo DM, Manfredi AA, Bio Angels for C-BSG . 2021. CXCL10 levels at hospital admission predict COVID-19 outcome: hierarchical assessment of 53 putative inflammatory biomarkers in an observational study. Mol Med 27:129. 10.1186/s10020-021-00390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callahan V, Hawks S, Crawford MA, Lehman CW, Morrison HA, Ivester HM, Akhrymuk I, Boghdeh N, Flor R, Finkielstein CV, Allen IC, Weger-Lucarelli J, Duggal N, Hughes MA, Kehn-Hall K. 2021. The pro-inflammatory chemokines CXCL9, CXCL10 and CXCL11 are upregulated following SARS-CoV-2 infection in an AKT-dependent manner. Viruses 13:1062. 10.3390/v13061062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou S, Butler-Laporte G, Nakanishi T, Morrison DR, Afilalo J, Afilalo M, Laurent L, Pietzner M, Kerrison N, Zhao K, Brunet-Ratnasingham E, Henry D, Kimchi N, Afrasiabi Z, Rezk N, Bouab M, Petitjean L, Guzman C, Xue X, Tselios C, Vulesevic B, Adeleye O, Abdullah T, Almamlouk N, Chen Y, Chasse M, Durand M, Paterson C, Normark J, Frithiof R, Lipcsey M, Hultstrom M, Greenwood CMT, Zeberg H, Langenberg C, Thysell E, Pollak M, Mooser V, Forgetta V, Kaufmann DE, Richards JB. 2021. A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat Med 27:659–667. 10.1038/s41591-021-01281-1. [DOI] [PubMed] [Google Scholar]
- 41.Bizzotto J, Sanchis P, Abbate M, Lage-Vickers S, Lavignolle R, Toro A, Olszevicki S, Sabater A, Cascardo F, Vazquez E, Cotignola J, Gueron G. 2020. SARS-CoV-2 infection boosts MX1 antiviral effector in COVID-19 patients. iScience 23:101585. 10.1016/j.isci.2020.101585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zenarruzabeitia O, Astarloa-Pando G, Terren I, Orrantia A, Perez-Garay R, Seijas-Betolaza I, Nieto-Arana J, Imaz-Ayo N, Perez-Fernandez S, Arana-Arri E, Borrego F. 2021. T cell activation, highly armed cytotoxic cells and a shift in monocytes CD300 receptors expression is characteristic of patients with severe COVID-19. Front Immunol 12:655934. 10.3389/fimmu.2021.655934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Zsolt D, Munoz-Basagoiti J, Rodon J, Elosua-Bayes M, Raich-Regue D, Risco C, Sachse M, Pino M, Gumber S, Paiardini M, Chojnacki J, Erkizia I, Muniz-Trabudua X, Ballana E, Riveira-Munoz E, Noguera-Julian M, Paredes R, Trinite B, Tarres-Freixas F, Blanco I, Guallar V, Carrillo J, Blanco J, Telenti A, Heyn H, Segales J, Clotet B, Martinez-Picado J, Vergara-Alert J, Izquierdo-Useros N. 2021. SARS-CoV-2 interaction with Siglec-1 mediates trans-infection by dendritic cells. Cell Mol Immunol 18:2676–2678. 10.1038/s41423-021-00794-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doehn JM, Tabeling C, Biesen R, Saccomanno J, Madlung E, Pappe E, Gabriel F, Kurth F, Meisel C, Corman VM, Hanitsch LG, Treskatsch S, Heim K, Stegemann MS, Ruwwe-Glosenkamp C, Muller-Redetzky HC, Uhrig A, Somasundaram R, Spies C, von Bernuth H, Hofmann J, Drosten C, Suttorp N, Witzenrath M, Sander LE, Hubner RH. 2021. CD169/SIGLEC1 is expressed on circulating monocytes in COVID-19 and expression levels are associated with disease severity. Infection 49:757–762. 10.1007/s15010-021-01606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lempp FA, Soriaga LB, Montiel-Ruiz M, Benigni F, Noack J, Park YJ, Bianchi S, Walls AC, Bowen JE, Zhou J, Kaiser H, Joshi A, Agostini M, Meury M, Dellota E, Jr, Jaconi S, Cameroni E, Martinez-Picado J, Vergara-Alert J, Izquierdo-Useros N, Virgin HW, Lanzavecchia A, Veesler D, Purcell LA, Telenti A, Corti D. 2021. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature 598:342–347. 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Zhang L, Sang L, Ye F, Ruan S, Zhong B, Song T, Alshukairi AN, Chen R, Zhang Z, Gan M, Zhu A, Huang Y, Luo L, Mok CKP, Al Gethamy MM, Tan H, Li Z, Huang X, Li F, Sun J, Zhang Y, Wen L, Li Y, Chen Z, Zhuang Z, Zhuo J, Chen C, Kuang L, Wang J, Lv H, Jiang Y, Li M, Lin Y, Deng Y, Tang L, Liang J, Huang J, Perlman S, Zhong N, Zhao J, Malik Peiris JS, Li Y, Zhao J. 2020. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest 130:5235–5244. 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sungnak W, Huang N, Becavin C, Berg M, Queen R, Litvinukova M, Talavera-Lopez C, Maatz H, Reichart D, Sampaziotis F, Worlock KB, Yoshida M, Barnes JL, Network H, HCA Lung Biological Network . 2020. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 26:681–687. 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Centers for Disease Control and Prevention. 2020. CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 49.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bushnell B, Rood J, Singer E. 2017. BBMerge—accurate paired shotgun read merging via overlap. PLoS One 12:e0185056. 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breitwieser FP, Baker DN, Salzberg SL. 2018. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol 19:198. 10.1186/s13059-018-1568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma'ayan A. 2016. Enrichr: a comprehensive gene set enrichment analysis Web server 2016 update. Nucleic Acids Res 44:W90–W97. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jvi.01478-22-s0001.xlsx, XLSX file, 0.03 MB (34.2KB, xlsx)
Table S2. Download jvi.01478-22-s0002.xlsx, XLSX file, 0.01 MB (12.6KB, xlsx)
Table S3. Download jvi.01478-22-s0003.xlsx, XLSX file, 0.02 MB (17.5KB, xlsx)
Data Availability Statement
Raw sequence reads are available in the Sequence Read Archive (SRA) BioProject identifier (ID) PRJNA922078.