ABSTRACT
Transcription of herpes simplex virus 1 (HSV-1) immediate early (IE) genes is controlled at multiple levels by the cellular transcriptional coactivator, HCF-1. HCF-1 is complexed with epigenetic factors that prevent silencing of the viral genome upon infection, transcription factors that drive initiation of IE gene expression, and transcription elongation factors required to circumvent RNAPII pausing at IE genes and promote productive IE mRNA synthesis. Significantly, the coactivator is also implicated in the control of viral reactivation from latency in sensory neurons based on studies that demonstrate that HCF-1-associated epigenetic and transcriptional elongation complexes are critical to initiate IE expression and viral reactivation. Here, an HCF-1 conditional knockout mouse model (HCF-1cKO) was derived to probe the role and significance of HCF-1 in the regulation of HSV-1 latency/reactivation in vivo. Upon deletion of HCF-1 in sensory neurons, there is a striking reduction in the number of latently infected neurons that initiate viral reactivation. Importantly, this correlated with a defect in the removal of repressive chromatin associated with latent viral genomes. These data demonstrate that HCF-1 is a critical regulatory factor that governs the initiation of HSV reactivation, in part, by promoting the transition of latent viral genomes from a repressed heterochromatic state.
KEYWORDS: HCF-1, herpes simplex virus, latency, HSV reactivation, viral chromatin
INTRODUCTION
Herpes simplex virus (HSV) is a ubiquitous pathogen with an estimated prevalence of 50 to 90% in the human population. Following an initial primary lytic infection, HSV establishes lifelong persistence or latency in neurons of sensory ganglia. Periodically, this quiescent state is punctuated by reactivation events that result in lytic infection and recurrent disease.
HSV-mediated disease ranges from oral and genital lesions to more significant manifestations including ocular infections that can lead to HSV-mediated blindness, while neonatal infections can result in mortality or developmental and neurological issues (1–3). Importantly, HSV is also a cofactor in acquisition of HIV (4, 5).
The lytic replication cycle is defined by a cascade of viral gene expression where viral immediate early (IE) genes are first transcribed and IE proteins are critical for subsequent waves of gene expression (early and late), for subverting host cell antiviral responses, and for the establishment of a “permissive” state of the host cell. Ultimately productive lytic infection generates progeny virus that can infect sensory neurons proximal to the initial site of lytic infection. Here, the virus establishes a quiescent latency state that is characterized by a lack of significant viral lytic gene expression and the expression of noncoding RNAs (ncRNAs) derived from the viral latency-associated transcript (LAT) locus (1).
During lytic infection, the induction of viral IE gene expression is controlled by complex and cooperative interactions of host cell and viral-encoded transcription factors that promote the assembly of transcription initiation complexes (6) and transcription elongation factors that are required to stimulate the release of paused RNAPII at IE genes to allow for efficient production of IE mRNAs (7, 8). In addition, expression of IE genes is dependent on an initial chromatin dynamic that can result in either heterochromatic suppression of the infecting viral genome or a euchromatic state that ultimately promotes IE gene transcription (9–11).
A central mediator of IE gene expression is the cellular transcriptional coactivator HCF-1. This protein is required for the assembly of an enhancer complex containing the cellular transcription factor OCT-1 and the viral IE activator VP16 on IE gene enhancer core elements (6, 12, 13). HCF-1 also interacts with or potentiates the activity of multiple transcription factors (e.g., SP1, GABP, FHL2, GR) that may synergize with the core complex or stimulate IE expression in the absence of the core complex (6, 12, 14). In addition to interactions that promote initiation of IE gene transcription, HCF-1 also plays a role in transcriptional elongation of IE genes via interactions with the active super elongation complex, a complex that is required to counter RNAPII pausing during IE transcription (7, 8).
Significantly, HCF-1 plays another critical role in IE expression by modulating the character of chromatin associated with IE genes. While HCF-1 interacts with multiple epigenetic factors/complexes, one HCF-1-associated complex consisting of two H3K9-histone demethylases (LSD1, JMJD2s) and an H3K4-methyltransferase (SETD1A, MLLs) is critical for expression of IE genes by limiting the deposition or accumulation of repressive heterochromatin on IE genes and promoting the installation of active histone modifications to stimulate IE transcription (10, 11, 15). Finally, with respect to stimulation of lytic infection, the roles of this coactivator are not restricted to promoting IE genes, as the protein has also been implicated in viral DNA replication by coupling histone chaperones to viral replication proteins, presumably to promote nucleosome remodeling at the replication fork (16).
In addition to control of viral IE gene expression during lytic infection, HCF-1 has also been proposed to be a key regulator of viral reactivation as suggested by studies demonstrating that the protein is (i) uniquely sequestered in the cytoplasm of sensory neurons during latency and is rapidly transported to the nucleus upon stimuli that induce viral reactivation (17, 18) and (ii) recruited to viral IE gene enhancer domains at a very early stage of reactivation (19). In addition, inhibition of the HCF-1-associated histone H3K9 demethylases (LSD1/KDM1A and JMJD2s/KDM4s) suppresses lytic infection, shedding, and spontaneous reactivation in vivo (20, 21). Finally, inhibition of the super elongation complex, found in association with HCF-1, suppressed the initiation of reactivation in the ganglia explant model system of viral latency/reactivation (7).
However, while HCF-1 has been implicated in control of viral reactivation from latency, direct evidence of the significance of the coactivator remains to be assessed in vivo. Here, as HCF-1 is an essential cellular coactivator, a conditional knockout mouse model was developed. In two approaches to delete HCF-1 in sensory neurons in vivo, loss of HCF-1 suppressed the initiation of viral reactivation in the ganglia explant model and upon induction of reactivation in vivo. Inhibition of reactivation in HCF-1 deleted neurons correlated with a defect in chromatin modulation that normally leads to a reduction in the level of repressive H3K9me3 heterochromatin associated with latent viral genomes.
RESULTS
Generation and characterization of a conditional HCF-1-knockout mouse.
To assess the role and impact of the coactivator HCF-1 in vivo, an HCF-1 conditional knockout (HCF-1cKO) mouse strain was derived via homologous recombination of the wild-type (WT) locus with a targeting vector containing a selectable marker (Neo, neomycin phosphotransferase) flanked by FRT sites (flippase recombination targeting sequences) and LoxP sites (Cre recombinase target sequences) flanking HCF-1 exon 2 and the Neo cassette (see Fig. S1A in the supplemental material). FLP-mediated recombination resulted in deletion of the Neo marker to generate the conditional HCF-1cKO strain, while Cre-mediated recombination results in deletion of the HCF-1 exons 2 and 3 and the expression of an mRNA predicted to encode a truncated 64-amino-acid (aa) peptide versus the 2,045-aa WT protein.
Generation and characterization of an HCF-1 conditional knockout mouse. (A) Schematic of the modified HCF-1 locus generated by recombination with a targeting construct containing HCF-1 exon 2 and 3, the selectable marker neomycin phosphotransferase (Neo) flanked by FRT sites, and the loxP sites flanking exon 2 and Neo (HCF-1tm1LVD). FLP-mediated recombination results in deletion of the selectable marker (HCF-1tm1.1LVD), referred to as HCF-1cKO, while Cre-mediated recombination results in deletion of HCF-1 exon 2 and 3 (HCF-1tm1.2LVD). (B and C) Primary fibroblast cells derived from HCF-1cKO or control C57BL/6 mice were infected with the indicated MOI of adenovirus expressing FLPe or Cre. (B) Cre mRNA levels are shown relative to mock-infected cells at 48 h postinfection. Data are from 3 independent experiments (ns, unpaired two-tailed t tests). (C) Controls for Fig. 2B. The levels of control cellular genes SP1 and GAPDH are shown relative to mock-infected cells. Data are from 4 independent experiments (ns, ANOVA with Dunnett’s multiple comparisons test). Download FIG S1, PDF file, 0.4 MB (457.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
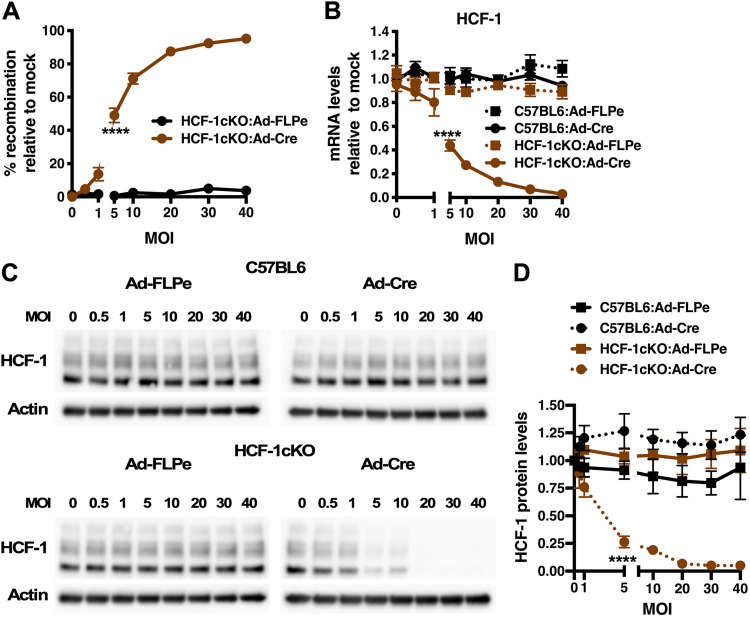
To initially characterize HCF-1 deletion in the conditional strain, primary fibroblast cells were derived from HCF-1cKO and control C57BL/6 mice and infected with adenovirus expressing Cre (Ad-Cre) or control FLP (Ad-FLP) (Fig. S1B). As shown in Fig. 1, infection of HCF-1cKO cells with Ad-Cre resulted in dose-dependent recombination at the HCF-1 locus (Fig. 1A) with a concomitant reduction in HCF-1 mRNA (Fig. 1B) and protein levels (Fig. 1C and D). In contrast, no impact on HCF-1 mRNA or protein levels was observed upon infection of HCF-1cKO cells with Ad-FLP or in control C57BL/6 cells infected with either Ad-Cre or Ad-FLP. Additionally, no impact of Ad-Cre or Ad-FLP was seen on the expression of cellular SP1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes (Fig. S1C).
FIG 1.
Cre-mediated deletion of HCF-1 in cells derived from the HCF-1cKO mouse line. (A) Levels of recombination at the HCF-1 locus in primary fibroblast cells derived from HCF-1cKO mice infected with the indicated MOI of adenovirus expressing enhanced FLP (Ad-FLPe) or Cre (Ad-Cre). Levels are relative to those in mock-infected cells. Data are from 3 independent experiments. (B to D) Primary fibroblast cells derived from HCF-1cKO or control C57BL/6 mice were infected with the indicated MOI of Ad-FLPe or Ad-Cre. (B) HCF-1 mRNA levels are shown relative to those in mock-infected cells. Data are from 4 independent experiments. (C) Representative Western blots of HCF-1 and control Actin protein levels. (D) Quantitation of HCF-1 protein levels normalized to Actin levels. Data are from 3 independent experiments. (A, B, D) Analysis of variance (ANOVA) with Dunnett’s multiple-comparison test.
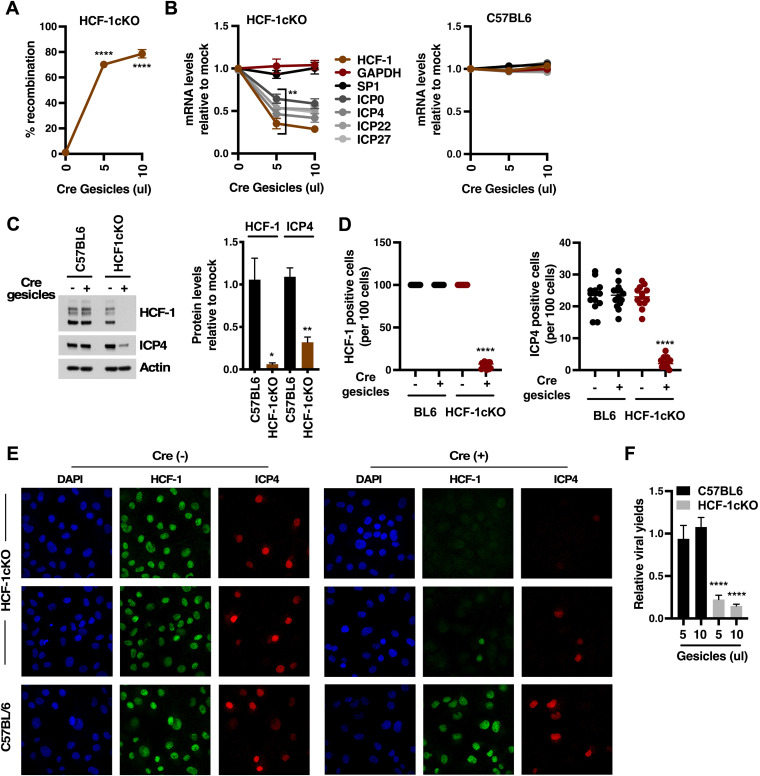
As an alternate approach to supplying Cre, HCF-1cKO and control C57BL/6 cells were transduced with gesicles containing Cre protein (Fig. 2). Transduction of HCF-1cKO cells resulted in efficient recombination at the HCF-1 locus (70 to 80%) (Fig. 2A) and, upon infection, a significant reduction in the levels of viral IE mRNAs (Fig. 2B) that paralleled the reduction in the level of HCF-1 mRNA. In contrast, Cre transduction of C57BL/6 cells had no impact on viral IE expression. As expected, reduction in the levels of HCF-1 and viral mRNAs resulted in a reduction in the levels of HCF-1 and a representative IE protein (ICP4) (Fig. 2C).
FIG 2.
Deletion of HCF-1 suppresses HSV IE gene expression. (A to E) Primary fibroblast cells derived from HCF-1cKO or control C57BL/6 mice were transfected with the indicated amounts of Cre gesicles and 7 days later were infected with HSV-1 ([A to C] MOI, 1; [D to E] MOI, 5). (A) Levels of recombination at the HCF-1 loci. (B) Levels of HSV IE (ICP0, ICP4, ICP22, ICP27) and control (GAPDH, SP1) mRNAs at 1.5 h postinfection relative to mock-transfected cells (3 experiments; 5 samples). (C) Representative Western blot of HCF-1, ICP4, and control Actin protein levels. Quantitation of HCF-1 and ICP4 protein relative to Actin from 3 independent experiments. (D and E) Number and representative images of HCF-1-positive and HSV ICP4-positive cells. Data are represented as the number of positive cells per 100 cells (n ≥ 13 groups of 100 cells). (F) Viral yields from HCF-1cKO or control C57BL/6 primary fibroblast cells transfected with Cre Gesicles and subsequently infected for 12 h (3 experiments; 6 samples). (A, B, F) ANOVA with Dunnett’s multiple-comparison test. (C, D, E) Unpaired two-tailed t test.
Further confirmation of the impact of HCF-1 deletion on IE expression and the initiation of viral infection was obtained by treating HCF-1cKO and control C57BL/6 cells with Cre gesicles, infecting with HSV for 1.5 h, and staining with antibodies to HCF-1 and a marker of early viral transcriptional foci (ICP4). As shown in Fig. 2D and E, reduction in the number of cells positive for HCF-1 staining correlated with a significant reduction in the number of HCF-1cKO-infected cells exhibiting ICP4 transcriptional foci. Finally, Cre gesicle-mediated depletion of HCF-1 in HCF-1cKO cells ultimately led to a significant reduction in viral yields at 12 h postinfection without impact on yields in the control C57BL/6 cells (Fig. 2F).
Overall, the data demonstrate that recombination-mediated deletion of HCF-1 in primary cells derived from the HCF-1cKO mouse model results in suppression of viral IE gene expression and, ultimately, viral yields. These results are consistent with previous studies in which the levels of HCF-1 protein were reduced using small interfering RNA (siRNA)/small hairpin RNA (shRNA) (22) and further affirm the significance of HCF-1 in the initiation of HSV lytic infection.
Specific deletion of HCF-1 in latently infected sensory neurons in vivo suppresses viral reactivation from latency.
HCF-1 plays multiple roles in driving the initiation of viral lytic infection including chromatin modulation, enhancer complex formation, and transcriptional elongation. Previous studies have also suggested that the protein plays a significant role in the initiation of viral reactivation from latency in sensory neurons. However, to directly probe the roles and significance of the coactivation during viral reactivation in vivo, two approaches were used to delete HCF-1 in sensory neurons in the HCF-1cKO mouse model.
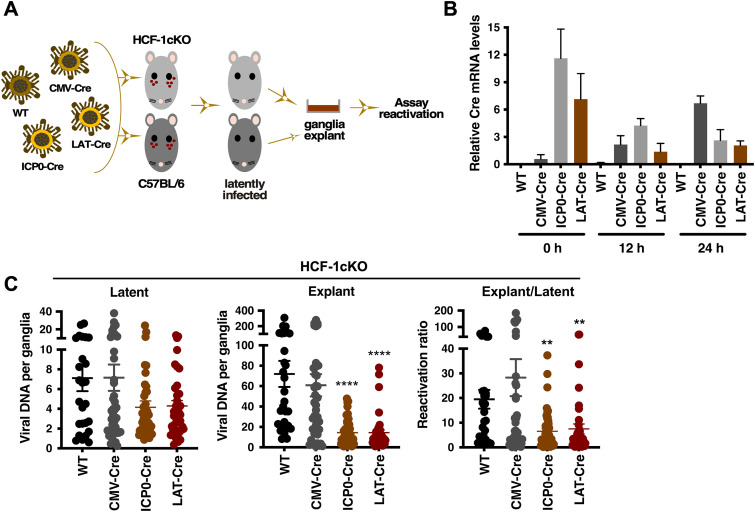
The first approach specifically targeted HCF-1 in latently infected sensory neurons using HSV-1 recombinant viruses that express Cre recombinase under the control of either the human cytomegalovirus (hCMV) IE, HSV IE ICP0, or the HSV LAT promoters (Fig. 3A) (23, 24). Here, HCF-1cKO and control C57BL/6 mice were infected with WT or the HSV-1 recombinants via the ocular route. Following the establishment of latency, trigeminal ganglia were explanted to induce a robust reactivation. As shown in Fig. 3B, both the ICP0-Cre and the LAT-Cre infected mice exhibited readily detectable Cre expression in the latently infected trigeminal ganglia (0 h) relative to the WT or CMV-Cre infected mice. Upon explant induction, Cre mRNA levels derived from the CMV-Cre virus increased, while the levels derived from the ICP0-Cre and LAT-Cre viruses decreased over the course of 24 h. Thus, both ICP0-Cre and LAT-Cre viruses express Cre during latency in contrast to the CMV-Cre recombinant that expresses Cre primarily during lytic reactivation.
FIG 3.
Expression of Cre in latently infected sensory neurons of HCF-1cKO mice results in reduced viral yields upon explant. (A) Schematic representation of the experimental strategy. Pools of HCF-1cKO and control C57BL/6 mice are infected with HSV-1 WT, CMV-Cre, ICP0-Cre, or LAT-Cre. At 30 to 45 days postinfection, ganglia from latently infected mice are explanted to induce reactivation. Reactivation is quantitated by several assays. (B) Cre mRNA levels in ganglia of HCF-1cKO mice latently infected with the indicated HSV strain at 0, 12, and 24 h post explant (2 experiments, pool of 4 ganglia per sample). (C) HSV DNA levels in ganglia of HCF-1cKO mice latently infected with the indicated HSV strain at 0 (latent) and 48 h post explant (Explant). Ratio E/L represents the ratio of DNA levels in explanted to latent ganglia (n ≥ 26 ganglia; Kruskal-Wallis test with Dunnett’s multiple-comparison test).
To assess the impact of HCF-1 deletion on explant-mediated reactivation, ganglia latently infected with WT, CMV-Cre, ICP0-Cre, and LAT-Cre viruses were bisected, and the viral DNA loads in latent and explanted halves were determined for each individual ganglion (Fig. 3C). While there were minor variances in the levels of latent viral genomes between the groups, viral DNA levels in explanted ganglia infected with the ICP0-Cre and LAT-Cre recombinant viruses were significantly lower than those in control WT and CMV-Cre infected ganglia. Furthermore, the ratio of explant to latent viral loads for individual ganglia also reflected the decrease in the levels of viral reactivation in ganglia infected with ICP0-Cre and LAT-Cre viruses relative to the control WT and CMV-Cre infected animals. In contrast no significant differences in the reactivation ratios were seen in control C57BL/6 mice (see Fig. S2 in the supplemental material).
Expression of Cre in latently infected sensory neurons of control C57BL/6 mice does not affect viral reactivation. The ratio of HSV DNA levels in explanted to latent ganglia (n ≥ 9) of C57BL/6 mice infected with the indicated HSV strain (ns, Kruskal-Wallis test with Dunnett’s multiple comparisons test). Control for Fig. 3C. Download FIG S2, PDF file, 0.1 MB (111.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
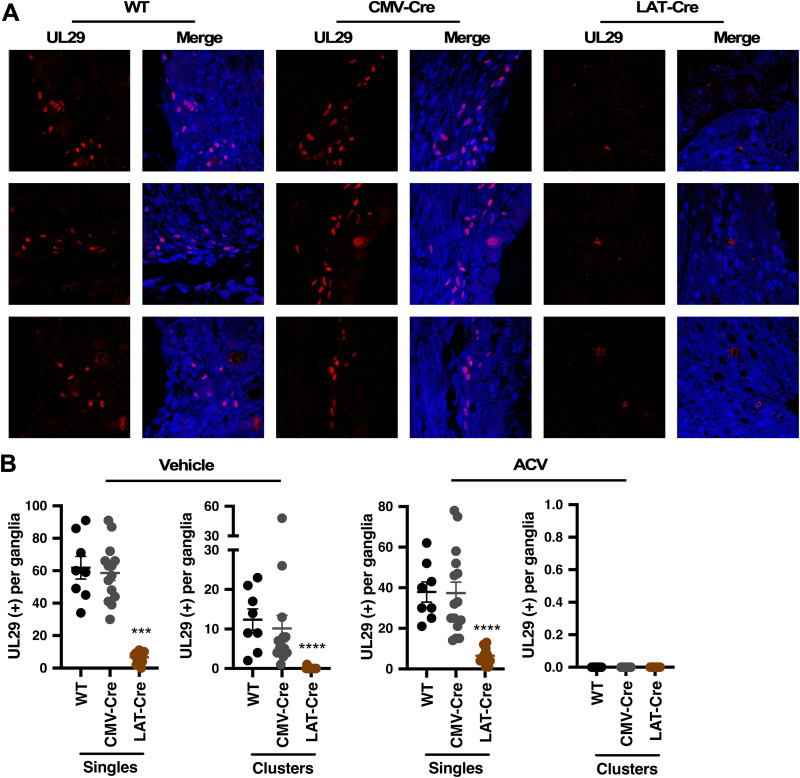
Deletion of HCF-1 suppresses viral reactivation as determined by viral DNA loads. However, to directly assess the impact of HCF-1 deletion on the number of neurons initiating viral reactivation, ganglia from mice latently infected with the WT, CMV-Cre, and LAT-Cre viruses were explanted for 48 h, and sections were stained for the lytic DNA replication protein UL29 (ICP8). As shown in Fig. 4A, ganglia from mice infected with LAT-Cre exhibited dramatically reduced UL29 (+) numbers relative to those infected with WT or CMV-Cre viruses. Quantitation of the number of individual UL29 (+) neurons representing the primary reactivation events (singles) and clusters of UL29 (+) representing lytic spread (Fig. 4B) confirmed that deletion of HCF-1 during latency (LAT-Cre virus) suppressed the number of primary reactivation events and, thus, the number of UL29 (+) clusters (Fig. 4B, vehicle, left panel). To further confirm that the individual UL29 (+) neurons did, in fact, represent primary reactivating neurons, ganglia were explanted in the presence of the DNA replication inhibitor acyclovir (ACV) to inhibit viral spread from the initiating neuron (Fig. 4B, ACV, right panel; see also Fig. S3A in the supplemental material). Similar to the data obtained in the absence of ACV, the number of UL29 (+) singles/initiating neurons was significantly decreased in the LAT-Cre infected ganglia relative to that of the control WT and CMV-Cre infected ganglia. Furthermore, as anticipated based on the level of Cre expression during viral latency in mice infected with the ICP0-Cre virus (Fig. 3B), ganglia infected with this virus also showed a significant reduction in the number of UL29 (+) primary reactivation events (singles) and clusters relative to CMV-Cre infected ganglia (Fig. S3B and C). Thus, deletion of HCF-1 during latency suppresses the initiation of viral reactivation.
FIG 4.
Deletion of HCF-1 reduces primary reactivation events. Ganglia from HCF-1cKO mice that were latently infected with the indicated virus were explanted in the absence or presence of ACV for 48 h. (A) Representative images of ganglia sections stained with anti-UL29 (red) and 4′,6-diamidino-2-phenylindole (DAPI) (blue). See Fig. S3A in the supplemental material for images of sections treated with ACV. (B) Sections were scored for individual UL29+ neurons and clusters. Vehicle, n ≥ 12 ganglia; ACV, n ≥ 8 ganglia; Kruskal-Wallis test with Dunnett’s multiple-comparison test. Representative sections are shown in panel B (vehicle) and Fig. S3A (ACV).
Expression of Cre in latently infected sensory neurons results in reduced numbers of primary neurons undergoing reactivation. (A to C) Ganglia that were latently infected with the indicated virus were explanted for 48 h. Ganglia sections were stained with anti-UL29 (red) and DAPI (blue). (A) Representative sections of ganglia infected with WT, CMV-Cre, and LAT-Cre that were explanted in the presence of ACV for 48 h. Related to Fig. 4A. (B and C) Ganglia sections were scored for individual UL29+ neurons (Single) and clusters (Clusters). (n ≥ 14 ganglia; Kruskal-Wallis test with Dunnett’s multiple comparisons test). Representative sections are shown in panel C. Download FIG S3, PDF file, 2.7 MB (2.8MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
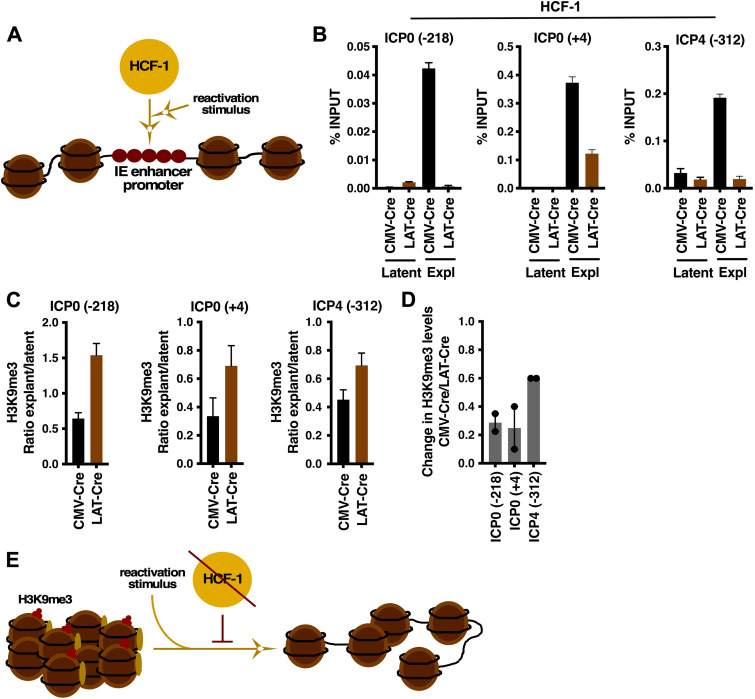
Deletion of HCF-1 in vivo results in a block to the removal of repressive H3K9me3 heterochromatin following stimulation of reactivation.
Latent genomes are characterized by multiple classes of repressive heterochromatin (e.g., H3K9me3, H3K27me3) (20, 25–30). Strikingly, an HCF-1 complex consisting of two histone H3K9 demethylases (LSD1, JMJD2s) and H3K4 histone methyltransferases (SETD1A, MLL1) was identified as being important for driving the initial chromatin dynamic required to promote viral IE gene expression during lytic infection of primary cells in vitro and a mouse model of HSV infection in vivo (10, 11, 21). In addition, inhibition of these histone H3K9 demethylases suppressed viral shedding and reactivation in several animal models of HSV latency/reactivation (20), indicating that removal of this heterochromatin from the latent viral genome is also a key step in the escape from latency. While these data would suggest that HCF-1 epigenetic complexes play critical roles in the initiation of viral reactivation, the direct impact of HCF-1 on the state of viral chromatin during reactivation has not been demonstrated.
To address this, chromatin immunoprecipitation (ChIP) assays were done to assess the occupancy of HSV IE gene regulatory regions by HCF-1 during reactivation (Fig. 5A) and the impact of HCF-1 deletion on the levels of heterochromatin associated with the viral genome following stimulation of viral reactivation. As anticipated, ganglia from mice latently infected with the LAT-Cre or control CMV-Cre did not exhibit significant levels of HCF-1 associated with representative viral IE enhancer/promoters (ICP0, ICP4) of latent viral genomes (Fig. 5B). However, upon explant-mediated induction of reactivation, HCF-1 could readily be detected associated with these IE regions in CMV-Cre-infected ganglia. In contrast, HCF-1 occupancy in ganglia infected with the LAT-Cre virus was not detected or was considerably reduced relative to ganglia infected with the control CMV-Cre.
FIG 5.
Depletion of HCF-1 in vivo blocks the removal of repressive chromatin from latent viral genomes during explant reactivation. (A) Schematic representing rapid recruitment of HCF-1 to viral IE gene enhancer/promoter domains upon stimulation that results in reactivation. (B to D) Ganglia of HCF-1cKO mice latently infected with HSV-CMV-Cre or HSV-LAT-Cre were explanted for 0 h (Latent) or 6 h (Expl). (B) ChIP assays showing HCF-1 occupancy of viral IE gene (ICP0, ICP4) promoter/enhancer regions. Data are from pools of 10 ganglia per IP. (C) Levels of repressive H3K9me3 associated with viral IE gene enhancers (ICP0 −218, ICP4 −312) and promoter (ICP0 +4) regions. Levels are shown as the ratio of explant/latent. Data are from pools of 10 ganglia per IP. (D) Ratio of the change in H3K9me3 levels in HSV-CMV-Cre relative to that of HSV-LAT-Cre. (E) Schematic illustrating that upon stimulation of reactivation, the level of H3K9me3-heterochromatin associated with the viral genome is reduced; a process that is blocked in HCF-1-depleted neurons.
Most significantly, recruitment of HCF-1 to IE enhancer-promoters during reactivation correlated with a substantial decrease in the levels of repressive H3K9me3 associated with these regions in CMV-Cre infected mice relative to LAT-Cre (HCF-1 deleted) mice (Fig. 5C). The differential response is most clearly illustrated by the ratio of change in H3K9me3 levels in mice infected with CMV-Cre relative to LAT-Cre (Fig. 5D). The results are consistent with studies demonstrating that HCF-1-associated histone H3K9-demethylases are required for the initiation of viral reactivation.
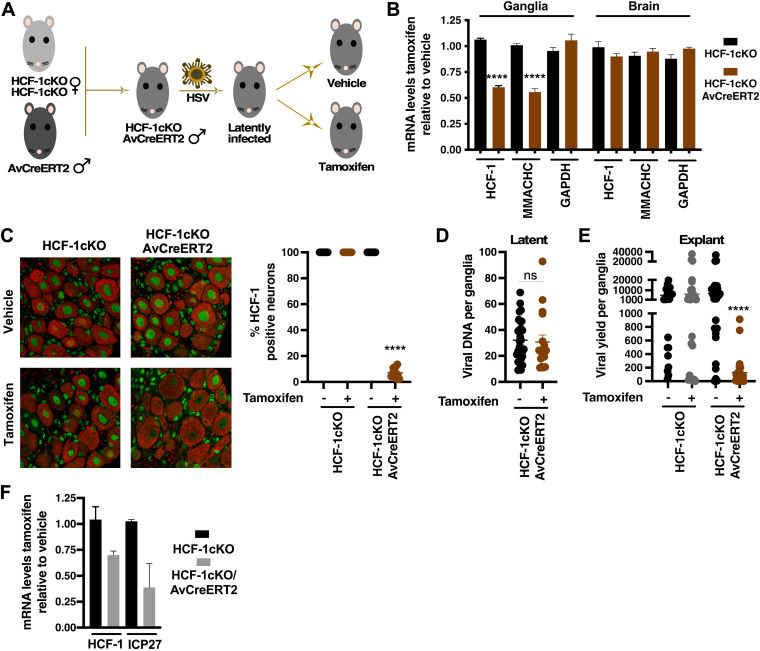
Inducible deletion of HCF-1 in neurons of sensory ganglia in the HCF-1cKO/AvCreERT2 mouse model.
As a second approach to deletion of HCF-1 in sensory neurons that did not depend on expression of Cre from recombinant viruses, the HCF-1cKO strain was mated to a transgenic expressing a tamoxifen-inducible Cre (CreERT2) under the control of the sensory neuron-specific advillin promoter (Fig. 6A) (31). Treatment of the resulting progeny (HCF-1cKO/AvCreERT2) with tamoxifen results in nuclear transport of the CreERT2 fusion protein and Cre-mediated recombination in sensory neurons (see Fig. S4 in the supplemental material). As shown in Fig. 6B, tamoxifen treatment of HCF-1cKO/AvCreERT2 mice resulted in specific reduction in the mRNA levels of HCF-1 and an HCF-1-dependent cellular gene (MMACHC) in trigeminal ganglia. In contrast, no impact of tamoxifen treatment was seen in ganglia of control mice or in brains, livers, or kidneys of either the HCF-1cKO/AvCreERT2 or control strain (Fig. 6B; see also Fig. S5 in the supplemental material). Strikingly, as shown in Fig. 6C, tamoxifen treatment of HCF-1cKO/AvCreERT2 mice resulted in specific reduction in HCF-1 protein in ~90% of neurons in the ganglia.
FIG 6.
Deletion of HCF-1 in sensory neurons reduces viral yields in explanted ganglia. (A) Generation of latently infected HCF-1cKO/AvCreERT2 mice. Homozygous HCF-1cKO females were mated to male Advillin-CreERT2 transgenics to generate hemizygous HCF-1cKO males carrying the Advillin-CreERT2 transgene (HCF-1cKO/AvCreERT2). Animals infected with HSV-1 were allowed to establish latency and subsequently treated with control vehicle or tamoxifen to induce recombination of the HCF-1 locus in sensory neurons. (B) mRNA levels of HCF-1, MMACHC, and GAPDH in trigeminal ganglia and brains of control HCF-1cKO or HCF-1cKO/AvCreERT2 mice. Levels are graphed as those in tamoxifen-treated mice relative to those in vehicle-treated animals. Data are from 4 pools of 6 ganglia/3 individual brains; unpaired two-tailed t tests. (C) Ganglia from HCF-1cKO/AvCreERT2 and control HCF-1cKO mice treated with vehicle or tamoxifen were sectioned and stained for HCF-1 (green) and neurofilament 200 (red). Quantitation is shown as the percent of HCF-1(+) neurons (n ≥ 11 ganglia per group; Mann-Whitney test). (D) Latent viral loads in ganglia from HCF-1cKO/AvCreERT2 mice treated with vehicle or tamoxifen (n ≥ 17; Mann-Whitney test). (E) Viral yields from ganglia explanted from the indicated strain treated with vehicle or tamoxifen (n ≥ 25; Mann-Whitney test). (F) Levels of HCF-1 and a viral IE (ICP27) mRNAs in ganglia explanted from the indicated strain treated with vehicle/JQ1 or tamoxifen/JQ1. Levels are shown in tamoxifen-treated relative to vehicle-treated mice. Data are from 2 experiments; pools of 5 ganglia per cDNA preparation.
Tamoxifen-dependent recombination in sensory ganglia. Latently infected HCF-1cKO/AvCreERT2 and C57BL/6 mice were injected either with tamoxifen or vehicle for 5 days, and DNA was purified from individual trigeminal ganglia (n = 6) at 9 days postinjection. PCR was completed with primers that detect the nonrecombined loci from the first loxP site through exon 2 (P1-P2; 522 bp) and for the detection of recombination (deletion of exon 2 and 3; P1-P6; 287 bp). Download FIG S4, PDF file, 0.3 MB (333.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
mRNA levels in tissues of HCF-1cKO/AvCreERT2 and control C57BL/6 mice. mRNA levels of HCF-1, MMACHC, and GAPDH in kidney and liver of control C57BL/6 and HCF-1cKO/AvCreERT2 mice treated with vehicle or tamoxifen. Levels are shown in tamoxifen-treated relative to vehicle-treated animals. Data are from 2 tissues pooled from each of 2 samples. Download FIG S5, PDF file, 0.2 MB (160KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Importantly, as shown in Fig. 6D, there was no impact of tamoxifen treatment on latent viral loads in HCF-cKO/AvCreERT2 mice compared to that in vehicle-treated animals. However, as determined by viral yields, explant of latently infected ganglia from the tamoxifen-treated mice resulted in a significant decrease in reactivation (mean, 129 PFU) compared to that in vehicle-treated animals (mean, 6,992 PFU) or tamoxifen-treated control HCF-1cKO mice (mean, 6,417 PFU) (Fig. 6E). Reduction in viral yields/reactivation from ganglia of tamoxifen-treated HCF-1cKO/AvCreERT2 mice correlated with reduced mRNA levels of HCF-1 and a representative viral IE (ICP27) (Fig. 6F). It is important to note that while the reduction in HCF-1 mRNA levels in sensory ganglia upon tamoxifen treatment of HCF-1cKO/AvCreERT2 mice appears to be “modest” (Fig. 6B and F), these ganglia are composed of populations of diverse cell types, and neurons are but a smaller fraction of the total population.
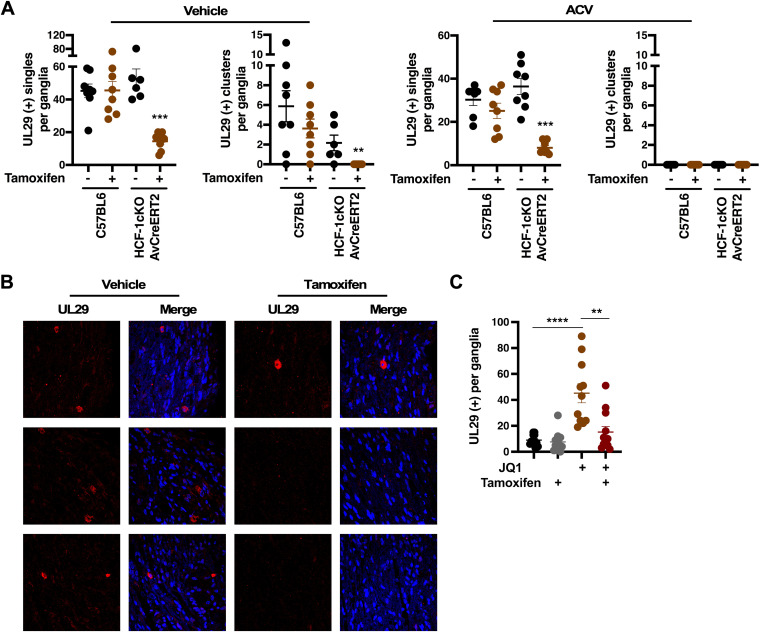
Similar to the results using Cre-expressing recombinant HSV viruses, tamoxifen-mediated deletion of HCF-1 in the HCF-1cKO/AvCreERT2 model resulted in a significant reduction in the number of primary reactivation events (individual/single neurons) and neuron clusters. In contrast, no change in the number of reactivation events was seen in control C57BL/6 mice (Fig. 7; see also Fig. S6 in the supplemental material). The addition of ACV to the ganglia explants further confirmed that the individual UL29 (+) neurons were primary events and not the result of secondary spread in the ganglia.
FIG 7.
Deletion of HCF-1 reduces the number of reactivating neurons. (A and B) Latently infected ganglia from the indicated strain treated with vehicle or tamoxifen were explanted for 48 h in the absence or presence of ACV. Sections were stained with anti-UL29 (red) and DAPI (blue). (A) Sections were scored for UL29+ clusters (Clusters) and individual neurons (Single). (n ≥ 6 ganglia per strain/condition; Mann-Whitney test). (B) Representative sections from HCF-1cKO/AvCreERT2 mice. (C) Latently infected mice were treated with vehicle or tamoxifen and subsequently treated with vehicle or JQ1. Ganglia were removed and fixed, and sections were stained with anti-UL29. The number of UL29 (+) neurons were counted (3 sections each of ≥11 ganglia per group; Mann-Whitney test).
Representative images from explanted ganglia from control C57BL/6 mice. Latently infected ganglia from control C57BL/6 mice were explanted in the presence of vehicle or tamoxifen for 48 h. Sections were stained for UL29 (red) and DAPI (blue). Representative sections are shown. Control for Fig. 7B. Download FIG S6, PDF file, 0.8 MB (891.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Loss of HCF-1 does not result in a significant loss of neurons in sensory ganglia.
HCF-1 is an important cellular transcriptional regulator. Therefore, to ensure that upon deletion of HCF-1, a loss of neurons was not a contributing factor to the observed decrease in viral reactivation, latently infected ganglia from vehicle-treated and tamoxifen-treated HCF-1cKO/AvCreERT2 and control C57BL/6 mice were assessed for the presence of apoptotic neurons by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (see Fig. S7A and B in the supplemental material). Control treatment of sections of ganglia of HCF-1cKO/AvCreERT2 mice with DNase I clearly showed a robust TUNEL signal in cells throughout the ganglia. In contrast, no TUNEL signal was detected in sections of ganglia derived from either HCF-1cKO/AvCreERT2 or the control C57BL/6 mice upon either vehicle or tamoxifen treatment. In addition, visual counts of neurons in ganglia sections did not detect any significant differences in the numbers of neurons from HCF-1cKO/AvCreERT2 or control C57BL/6 mice treated with tamoxifen compared to those treated with vehicle. The data indicate that neuronal integrity is not a significant factor in the apparent decrease in viral reactivation in HCF-1-deleted ganglia.
Deletion of HCF-1 does not result in loss of neurons or induction of neuronal apoptosis. Latently infected HCF-1cKO/AvCreERT2 or control C57BL/6 mice were treated with either vehicle or tamoxifen. (A and B) TUNEL assays were done on ganglia sections, which were counterstained with DAPI (blue) and neurofilament-200 (red). DNaseI treated HCF-1cKO/AvCreERT2 sections were used as positive TUNEL controls. Representative sections are shown (1× and 3× zoom) for each condition. (C) Numbers of neurons per ganglia (2 sections were counted for each of 8 ganglia per group) of HCF-1cKO/AdCreERT2 and control C57BL/6 mice treated with either vehicle or tamoxifen (Mann-Whitney test). Download FIG S7, PDF file, 2.4 MB (2.5MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Deletion of HCF-1 suppresses induction of viral reactivation in vivo.
The data above were derived using the ganglia explant model to induce robust viral reactivation. To demonstrate that deletion of HCF-1 in sensory neurons reduces viral reactivation following an in vivo reactivation stimulus, HCF-1cKO/AvCreERT2 mice were treated with vehicle or tamoxifen followed by treatment with the BET inhibitor JQ1. As previously demonstrated, JQ1 induces the expression of HSV IE genes by promoting transcriptional elongation, a key rate-limiting step in the initiation of viral reactivation (7, 8). As shown in Fig. 7C, injection of JQ1 resulted in significant induction of viral reactivation as determined by counts of UL29 (+) primary reactivation events while deletion of HCF-1 clearly reduced the number of neurons undergoing in vivo reactivation. Interestingly, in contrast to explanted ganglia, there was no evidence of spread beyond the initial reactivating neurons as only single UL29 (+) neurons were seen. This is perhaps reflective of a more restrictive control of the spread of lytic infection beyond the primary initiating neuron in the context of the ganglia in vivo.
DISCUSSION
Much of what has been learned with respect to latency and reactivation has been elucidated using animal model systems. However, reductionist approaches utilizing primary rodent neuronal cell culture systems and neuronal cultures derived from differentiation of human induced pluripotent stem cells (iPSCs) or human embryonic neuronal precursors have also contributed to understanding aspects of the control of latency and reactivation (32–39). Importantly, these systems have allowed for the elucidation of neuronal signaling pathways that either support the maintenance of latency or promote reactivation (17, 40–45). Most significantly, both animal models and in vitro neuronal cultures have contributed to a dynamic model of viral latency that may reflect various populations of latent genomes in distinct chromatin states and/or stages of transcriptional potential/activity (7, 32, 46, 47).
However, despite advances in understanding parameters of viral latency/reactivation, the identification and characterization of many of the components and molecular mechanisms governing this key phase of the viral life cycle remain to be determined. Of particular interest are those that control the induction of viral IE gene transcription during the initial stage of reactivation. One key factor that is central to the viral IE gene regulatory paradigm is the cellular transcriptional coactivator HCF-1. With respect to IE expression during lytic infection, HCF-1 mediates chromatin modulation/dynamics via preventing accumulation of repressive H3K9-heterochromatin on the infecting viral genome, complexes with multiple transcription factors involved in initiation of viral IE transcription, and promotes the critical rate-limiting stage of IE gene transcriptional elongation via interactions with cellular elongation factors including the super elongation complex.
HCF-1 has also been proposed to play multiple roles in the initiation of viral reactivation. The protein can be detected associated with viral IE gene enhancer domains at a very early stage of reactivation, although the factors involved in recruitment of HCF-1 at this point in reactivation remain unknown. Furthermore, studies utilizing inhibitors of HCF-1-associated H3K9-histone demethylases (LSD1, JMJD2s) or transcriptional elongation components (super elongation complex) have indirectly implicated HCF-1 in both chromatin modulation of the latent viral genome required for IE expression and in mediating the critical rate-limiting step of IE mRNA transcriptional elongation. Given these data, a model was proposed in which multiple populations of genomes ranging from strictly repressed to transcriptionally poised are acted on by HCF-1 complexes to modulate viral chromatin state and drive transcriptional initiation and elongation of IE genes during the initiation of viral reactivation (7).
These studies/observations have implicated HCF-1 as a critical regulatory factor that promotes both lytic infection and reactivation from latency. To investigate the role of this protein in control of viral reactivation in vivo, two approaches were used to selectively delete it in a conditional knockout mouse model. It should be noted that the first approach using Cre-expressing recombinant viruses to selectively delete HCF-1 in infected neurons was more challenging due to the requirement to establish consistent and comparable latent infections with the different viruses. Additionally, not all latently infected neurons express LAT and, therefore, by extension, Cre driven by the LAT promoter. However, similar results were also obtained using a second recombinant (ICP0-Cre) expressing Cre during latency (Fig. 3B) (48, 49).
In contrast to the first approach, which was dependent on comparisons between different Cre-expressing HSV recombinants, the second approach was based upon transgenic expression of Cre in the total population of neurons in the sensory ganglia. Irrespective of approach, the data clearly demonstrate that loss of HCF-1 in latently infected sensory neurons suppressed the number of neurons initiating viral reactivation in the trigeminal ganglia explant model. Most significantly, using the HCF-1cKO/AvCreERT2 model, depletion of HCF-1 resulted in a significant reduction in the number of primary reactivating neurons following in vivo induction of reactivation.
The reduction in the number of neurons undergoing reactivation upon loss of HCF-1 is reflective of a block in the very early stage(s) of reactivation as indicated by (i) the reduction in the mRNA levels of an IE gene (ICP27) and (ii) a block in the removal repressive H3K9me3-heterochromatin associated with viral IE gene promoter/regulatory domains, a chromatin remodeling process that is expected to be required early in reactivation. Importantly, the data indicate that HCF-1 is critical for at least one, if not more, steps in the early initiation of reactivation in vivo. However, given this early block, it is not possible to determine potential impacts of the loss of HCF-1 on later stages of viral reactivation (e.g., DNA replication) (16). It is important to note that in the experiments described in this study, the impacts of HCF-1 depletion on viral IE gene expression during lytic infection and on the initiation of viral reactivation are not absolute. This may be due to a lack of complete efficiency of recombination at the HCF-1 locus and depletion of the HCF-1 protein. Regardless, deletion of HCF-1 in the conditional knockout mouse model demonstrates the significance of the coactivator in control of viral reactivation.
With respect to induction of reactivation in vivo, the BET inhibitor JQ1 has been shown to promote transcriptional elongation of IE genes in vitro by enhancing the levels of the active super elongation complex-P-TEFb associated with viral IE gene promoter regions (8). In addition, inhibition of the super elongation complex blocked expression of viral IE genes and the initiation of viral reactivation in the ganglia explant model (7). Interestingly, the observed stimulation of viral reactivation by JQ1 in vivo is consistent with the proposed model that a population of latent viral genomes may be “poised” for reactivation and that transcriptional elongation is a critical rate-limiting or determining factor in reactivation of these genomes.
HCF-1 is an important cellular transcriptional regulator modulating gene expression through interactions with multiple transcription factors (e.g., Sp1, EGR2, THAP family members, Myc, YY1, ZNF143, GABP, E2F family members, insulin receptor) (50–59) and epigenetic complexes (e.g., SET/MLLs, BAP1, NSL, Sin3A) (58, 60–65). These interactions ultimately impact cellular processes, such as cell cycle progression and differentiation (50, 58, 66, 67). Despite its significance in regulation of the expression of a large set of cellular genes in replicating cells, deletion in sensory neurons did not impact neuronal viability within the time frame of these experiments. The lack of impact may be due to the nonreplicating and highly differentiated nature of sensory neurons. The unique cytoplasmic localization of HCF-1 and signal-mediated nuclear transport in these cells may also reflect a more limited role in sensory neuronal gene expression where its function may be particularly important in or restricted to signal-mediated regulation of transcription.
It is apparent that the chromatin state of viral genomes is a key parameter in the establishment and maintenance of viral latency and that modulation of that chromatin state is critical for the initiation of viral reactivation. During latency, viral lytic genes are associated with repressive heterochromatin (e.g., H3K9me3, H3K27me3) (20, 25–30), although the contribution of these different repressive marks and whether they represent distinct populations of latent genomes remain to be determined. In contrast to latency, reactivation is characterized by a reduction in the levels of viral associated heterochromatin and the induction of lytic gene expression. Importantly, deletion of HCF-1 in vivo resulted in a defect in the modulation of the levels of viral repressive heterochromatin (H3K9me3) that is normally seen upon initiation of reactivation. This is likely due to the loss of targeting of HCF-1-associated epigenetic factors, such as the histone H3K9 demethylases LSD1 and JMJD2s to latent viral genomes. However, HCF-1 is associated with multiple components/complexes that govern chromatin modulation, transcription initiation, and transcription elongation. It is, therefore, possible that the reduction in reactivation events in HCF-1-deleted neurons may reflect blocks in multiple HCF-1-dependent roles in the reactivation of distinct populations of latent viral genomes that may exist in other chromatin states or in various stages of “poised” transcriptional activation. The HCF-1cKO/AvCreERT2 model will allow for further investigation into the roles of HCF-1 in both HSV and neuronal biology.
MATERIALS AND METHODS
Generation and breeding of an HCF-1 conditional KO mouse.
An HCF-1 conditional knockout mouse was generated by Ozgene Pty Ltd. The core HCF-1 targeting cassette contained a 5′ loxP site, HCF-1 exons 2 and 3, an FRT flanked selectable marker (PGK promoter-neomycin phosphotransferase [Neo]) and a second 3′ loxP site. The core was flanked by 5′ (~6.1 kb) and 3′ (~3.8 kb) HCF-1 genomic sequences to produce the complete targeting vector. Recombination in Ozgene C57BL/6 Bruce 4 ES cells resulted in cells used to generate chimeric mice that were subsequently bred to isolate offspring exhibiting germ line transmission of the modified HCF-1 allele (HCF-1tm1LVD). A similar approach to generate an HCF-1 conditional line was previously described (68). Removal of the Neo marker was done via mating of HCF-1tm1LVD mice to an Flp transgenic (OzFlp, Ubic-Flpe; Gt(ROSA)26Sortm1.1(UBC-flpe)Ozg) to generate HCF-1tm1.1LVD/Flp, followed by subsequent breeding to WT C57BL/6J mice to remove the Flp transgene (HCF-1tm1.1LVD). After backcrossing to C57BL/6J for 4 generations, hemizygous males were mated to heterozygous females to generate the hemizygous males and homozygous females (hereafter referred to as HCF-1cKO mice) that were used in these studies. Genotyping was done by Southern blotting (Ozgene Pty Ltd.) or sequencing of PCR products from genomic DNA isolated from ear or tail samples according to standard procedures. PCR primer sequences are listed in Table S1 in the supplemental material. Animal care and handling were done in accordance with the U.S. National Institutes of Health Animal Care and Use Guidelines and as approved by the NIAID Animal Care and Use Committee.
Primers and reagents. Reagents and sequences of primers used in this study. Download Table S1, PDF file, 0.05 MB (52.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Tamoxifen-dependent recombination-depletion of HCF-1 in sensory ganglia.
Homozygous HCF-1cKO females were bred to Tg(Advil-iCre/ERT2)AJwo males (referred to as AvCreERT2; Jax catalog number 032027) (31). Male progeny carrying the HCF-1cKO allele were screened for the presence of the AvCreERT2 transgene (HCF-1cKO/AvCreERT2) by PCR using primers listed in Table S1 or were genotyped by Transnetyx, Inc. To delete HCF-1, groups of male HCF-1cKO/AvCreERT2 or control mice (HCF-1cKO or C57BL/6) were injected intraperitoneally with 2 mg tamoxifen (in corn oil, according to standard protocol) or vehicle control once per day for 5 days. Mice were maintained for 7 to 9 days postinjection before use. Detection of Cre-mediated recombination at the HCF-1 loci in sensory neurons was done by PCR amplification using primer sets to detect the nonrecombined (P1-P2) and recombined (P1-P6) forms. Primer locations are illustrated in Fig. S5 in the supplemental material, and the sequences are listed in Table S1.
Cell cultures and viral infections.
Vero cells were cultured and maintained according to standard procedures. Primary fibroblast cultures derived from HCF-1cKO and C57BL/6 mice were prepared by incubation of minced ear tissue in Dulbecco modified Eagle medium (DMEM) with Liberase TM (0.14 u/mL; Sigma) at 37°C for 75 min. Pelleted material was washed once in complete DMEM (10% fetal bovine serum [FBS], non-essential amino acids (NEAA), Pen-Strep, Glutamax, 25 μg/mL Fungin), resuspended, plated, and grown in complete DMEM at 37°C/5% CO2. HSV-1 infections were done by incubating cells with HSV-1 (strain 17) at the indicated multiplicity of infection (MOI) for 1 h at 37°C in DMEM containing 1% FBS. Following absorption, the inoculum was removed, and cells were incubated in DMEM containing 10% FBS for the indicated time. For adenovirus infections, primary ear fibroblast cultures were trypsinized, washed, and 4 × 105 cells were resuspended in 0.2 mL DMEM containing 1% FBS and the indicated MOI of adenovirus-Cre-nls (Ad-Cre) (ViraQuest Inc.; number 041405) or control adenovirus-FLPe (Ad-FLPe) (ViraQuest Inc.; number 1760) in 1.5-mL microcentrifuge tubes. Tubes were rotated at 37°C for 4 h, brought to 2 mL with DMEM/10% FBS, plated, and maintained for 3.5 days prior to use. Intracellular delivery of Cre protein was done by treating primary fibroblast cultures with Cre-gesicles (TaKaRa Bio USA) according to the manufacturer’s instructions. Cre-mediated recombination at the HCF-1 loci in cells derived from HCF-1cKO mice was assessed by quantitative PCR (qPCR) using primers listed in Table S1.
Mouse infections and reactivation in explanted trigeminal ganglia.
Groups of randomized HCF-1cKO, HCF-1cKO/AvCreERT2, or C57BL/6 mice were infected with the indicated WT HSV-1 strains [HSV-1(F), HSV-1(SC16)] or HSV-1 SC16-derived recombinant viruses (HSV-CMV-Cre, HSV-ICP0-Cre, and HSV-LAT-Cre) (23, 24) via the ocular route. After establishment of latency (30 to 45 days), trigeminal ganglia were removed and were explanted into DMEM/10% FBS/Pen-Strep. Viral yields were determined at 48 h post explant by titering of ganglia homogenates on Vero cells. Where indicated, latently infected ganglia were bisected, and latent viral loads were determined from one half while viral yields were determined from the other half following explant for 48 h. mRNA levels of viral and cellular mRNAs were determined at the indicated time post explant. Primer sequences are listed in Table S1. Animal care and handling were done in accordance with the National Institutes of Health Animal Care and Use Guidelines and as approved by the NIAID Animal Care and Use Committee (T.M.K.; Animal Protocol LVD40E). For detection of viral reactivation in neurons of trigeminal ganglia by immunofluorescence, ganglia were explanted for 48 h in the presence or absence of acyclovir (ACV), fixed in 4% paraformaldehyde for 12 h, and embedded in paraffin. Deparaffinized sections were treated with citric acid for antigen retrieval, stained with the indicated antibodies (Table S1), and visualized using a Leica TSC SP8X confocal microscope.
JQ1-mediated viral reactivation in vivo.
Latently infected HCF-1cKO/AvCreERT2 mice were injected with vehicle or tamoxifen as noted above. Nine days following the last tamoxifen injection, the mice were injected intraperitoneally with vehicle or JQ1 (50 mg/kg in 10% hydroxy-β-cyclodextrin/phosphate-buffered saline [PBS]) every 12 h for 48 h as previously described (7). Tissue sections of trigeminal ganglia were prepared at 48 h following the last JQ1 injection and were stained with anti-UL29 to detect neurons undergoing viral reactivation.
RNA isolation and quantitation.
For quantitation of RNA levels, trigeminal ganglia, brains, kidneys, and livers of HCF-1cKO, HCF-1cKO/AvCreERT2, and C57BL/6 mice were homogenized in TriPure reagent (Roche; catalog number 11667165001) with Lysing Matrix D (MP Biomedicals; catalog number 6913-100) using a FastPrep-24 instrument (MP Biomedicals). Total RNAs from tissue homogenates or from primary cells derived from these mice were isolated using a NucleoSpin RNA kit (Macherey-Nagel; catalog no. 740955.250). cDNAs were synthesized using qScript cDNA (Quant Biosciences; catalog no. 95047-500) or Maxima First Strand cDNA (Thermo Scientific; catalog no. K1672) synthesis kits and quantitated in triplicate by qPCR with SYBR green master mix (Roche; catalog no. 04913914001) or TaqMan Universal Master Mix II, no UNG (Applied Biosystems; catalog no. 4427788) and a QuantStudio 3 (Applied Biosystems; QuantStudio V1.4 software). The sequence of primers and probes are listed in Table S1.
Western blotting.
Fibroblast cultures derived from HCF-1cKO or control C57BL/6 mice were mock infected or infected with Ad-Cre or Ad-FLPe. At 3.5 days postinfection, cells were lysed, and protein extracts were prepared using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 1 mM EDTA, 1% NP40, 1% sodium deoxycholate (DOC), 0.1% SDS, cOmplete protease inhibitors, 2 mM NaV04, 1 mM NaF, 10 mM B-glycerophosphate). Western blots were done using antibodies to HCF-1 and control actin (Table S1), visualized with WesternBright Quantum (Advansta; catalog number K-12042-D20), and bands were quantitated using Syngene G:Box Chemi XT4 (Syngene; GeneTools 4.03.02.0v software).
Chromatin immunoprecipitations.
Trigeminal ganglia from latently infected mice (45 to 60 days postinfection) were isolated and were flash frozen in liquid nitrogen or were explanted into culture media for 6 h at 37°C. Ganglia were fixed in 1% formaldehyde for 10 min at room temperature (RT), quenched with 125 mM glycine for 10 min at RT. Cross-linked ganglia were homogenized in sonication buffer (0.25% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], cOmplete protease inhibitors, and phosphatase inhibitors), and chromatin was fragmented using a Covaris M220 sonicator (15 min 10% duty cycle, 250 burst per cycle, 75 W peak incident power) to obtain DNA fragments of 200 to 800 bp. Extracts were centrifuged at 12,000 rpm for 10 min at 4°C, and the supernatant diluted 1:2.5 with ChIP dilution buffer (1.6% Triton X-100, 250 mM NaCl, cOmplete protease inhibitors, and phosphatase inhibitors). After preclearing with Dynabeads Protein G (ThermoFisher Scientific; catalog number 10009D) for 2 h at 4°C, immunoprecipitations (IPs) were carried out overnight at 4°C using chromatin from 10 ganglia per IP and 6 μg of rabbit IgG, HCF-1, or H3K9me3 antibodies (see Table S1). Immunoprecipitates were washed twice with low-salt wash buffer (0.1% SDS, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100), once with high-salt buffer (0.1% SDS, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1% Triton X-100), once with LiCl buffer (1 mM EDTA, 10 mM Tris-HCl [pH 8.0], 250 mM LiCl, 1% NP40), and once with Tris-EDTA (10 mM Tris-HCl [pH 8.0], 1.2 mM EDTA). After the final wash, the IPed material was eluted with 1% SDS and 0.1 M NaHCO3 for 30 min at 65°C, and cross-links were reversed overnight at 65°C with the addition of 250 mM NaCl. Following treatment with RNase and proteinase K, the DNA was purified using the ChIP DNA Clean and Concentrator kit (Zymo Research; catalog number D5205). Precipitated DNA was assessed by reverse transcriptase quantitative PCR (RT-qPCR) using the QuantStudio 3 real-time PCR system (Applied Biosystems) and TaqMan Universal Master Mix II, no UNG (Applied Biosystems; catalog number 4427788) in triplicate. RT-qPCR signals were normalized to input DNA. The location and sequence of primers and probe sets used are shown in Table S1.
TUNEL assays.
To identify DNA breaks, characteristic of apoptotic sensory neurons, paraffin-embedded ganglia from latently infected mice were deparaffinized and treated with citric acid for permeabilization and antigen retrieval. Sections were subjected to TUNEL assay (Roche; number 11684795910) according to the manufacturer’s recommendations and were costained with anti-N200 (neurofilament 200) antibody. Ganglia sections treated with 1,000 U/mL DNase I at 37°C for 15 min prior to the TUNEL reaction served as positive controls.
Confocal microscopy.
Immunofluorescent microscopy was done according to standard protocols using antibodies listed in Table S1. Stained cells and trigeminal ganglia sections were visualized with a Leica TCS-SP8 laser scanning confocal microscope using a ×63 oil immersion objective. Deconvolution was completed with Huygens Essential (Scientific Volume Imaging; 21.04.0p6), and sequential z-sections were assembled with Imaris software (Bitplane AG, 9.6).
Statistical analyses.
Results are presented as means ± standard error of the means (SEM). Analyses were done using GraphPad Prism 8. Details are presented in the appropriate figure legends and in Table S2 in the supplemental material. Asterisks indicate statistical significance as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Statistics. Relevant statistical tests and P values of data described in this study. Download Table S2, PDF file, 0.1 MB (55.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
We thank the NIAID Building 33 Vivarium staff for excellent technical support.
This study was funded by the Intramural Research Program of the NIH, NIAID.
Footnotes
This article is a direct contribution from Thomas M. Kristie, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by David Bloom, University of Florida College of Medicine, and Luis Schang, Cornell University College of Veterinary Medicine.
Contributor Information
Thomas M. Kristie, Email: tkristie@niaid.nih.gov.
Nancie M. Archin, University of North Carolina at Chapel Hill
REFERENCES
- 1.Roizman B, Knipe DM, Whitley RJ. 2013. Herpes simplex viruses, p 2501–2601. In Knipe DM, Howley PM (ed), Fields virology, vol 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Wald A, Corey L. 2007. HSV: persistence in the population: epidemiology, transmission, p 656–671. In Arvin R (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 3.Whitley R, Kimberlin DW, Prober CG. 2007. HSV-1 and 2: pathogenesis and disease, p 589–601. In Arvin R, Whitley R (ed), Human herpesviruses biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, UK. [PubMed] [Google Scholar]
- 4.Freeman EE, Weiss HA, Glynn JR, Cross PL, Whitworth JA, Hayes RJ. 2006. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS 20:73–83. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- 5.Wald A, Link K. 2002. Risk of human immunodeficiency virus infection in herpes simplex virus type 2-seropositive persons: a meta-analysis. J Infect Dis 185:45–52. doi: 10.1086/338231. [DOI] [PubMed] [Google Scholar]
- 6.Vogel JL, Kristie TM. 2013. The dynamics of HCF-1 modulation of herpes simplex virus chromatin during initiation of infection. Viruses 5:1272–1291. doi: 10.3390/v5051272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfonso-Dunn R, Arbuckle JH, Vogel JL, Kristie TM. 2020. Inhibition of the super elongation complex suppresses herpes simplex virus immediate early gene expression, lytic infection, and reactivation from latency. mBio 11:e01216-20. doi: 10.1128/mBio.01216-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alfonso-Dunn R, Turner AW, Jean Beltran PM, Arbuckle JH, Budayeva HG, Cristea IM, Kristie TM. 2017. Transcriptional elongation of HSV immediate early genes by the super elongation complex drives lytic infection and reactivation from latency. Cell Host Microbe 21:507–517. doi: 10.1016/j.chom.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JS, Raja P, Knipe DM. 2016. Herpesviral ICP0 protein promotes two waves of heterochromatin removal on an early viral promoter during lytic infection. mBio 7:e02007-15. doi: 10.1128/mBio.02007-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang Y, Vogel JL, Arbuckle JH, Rai G, Jadhav A, Simeonov A, Maloney DJ, Kristie TM. 2013. Targeting the JMJD2 histone demethylases to epigenetically control herpesvirus infection and reactivation from latency. Sci Transl Med 5:167ra5. doi: 10.1126/scitranslmed.3005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med 15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristie TM, Liang Y, Vogel JL. 2010. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim Biophys Acta 1799:257–265. doi: 10.1016/j.bbagrm.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wysocka J, Herr W. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem Sci 28:294–304. doi: 10.1016/S0968-0004(03)00088-4. [DOI] [PubMed] [Google Scholar]
- 14.Sawant L, Kook I, Vogel JL, Kristie TM, Jones C. 2018. The cellular coactivator HCF-1 is required for glucocorticoid receptor-mediated transcription of bovine herpesvirus 1 immediate early genes. J Virol 92:e00987-18. doi: 10.1128/JVI.00987-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narayanan A, Ruyechan WT, Kristie TM. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci USA 104:10835–10840. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng H, Nogueira ML, Vogel JL, Kristie TM. 2010. Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc Natl Acad Sci USA 107:2461–2466. doi: 10.1073/pnas.0911128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JY, Mandarino A, Chao MV, Mohr I, Wilson AC. 2012. Transient reversal of episome silencing precedes VP16-dependent transcription during reactivation of latent HSV-1 in neurons. PLoS Pathog 8:e1002540. doi: 10.1371/journal.ppat.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristie TM, Vogel JL, Sears AE. 1999. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc Natl Acad Sci USA 96:1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitlow Z, Kristie TM. 2009. Recruitment of the transcriptional coactivator HCF-1 to viral immediate-early promoters during initiation of reactivation from latency of herpes simplex virus type 1. J Virol 83:9591–9595. doi: 10.1128/JVI.01115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, Foster TP, Bosch-Marce M, Raja P, Lee JS, Bernstein DI, Krause PR, Knipe DM, Kristie TM. 2014. Inhibition of LSD1 reduces herpesvirus infection, shedding, and recurrence by promoting epigenetic suppression of viral genomes. Sci Transl Med 6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y, Quenelle D, Vogel JL, Mascaro C, Ortega A, Kristie TM. 2013. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio 4:e00558-12. doi: 10.1128/mBio.00558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narayanan A, Nogueira ML, Ruyechan WT, Kristie TM. 2005. Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J Biol Chem 280:1369–1375. doi: 10.1074/jbc.M410178200. [DOI] [PubMed] [Google Scholar]
- 23.Proença JT, Coleman HM, Connor V, Winton DJ, Efstathiou S. 2008. A historical analysis of herpes simplex virus promoter activation in vivo reveals distinct populations of latently infected neurones. J Gen Virol 89:2965–2974. doi: 10.1099/vir.0.2008/005066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proenca JT, Nelson D, Nicoll MP, Connor V, Efstathiou S. 2016. Analyses of herpes simplex virus type 1 latency and reactivation at the single cell level using fluorescent reporter mice. J Gen Virol 97:767–777. doi: 10.1099/jgv.0.000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta 1799:246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cliffe AR, Coen DM, Knipe DM. 2013. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. mBio 4:e00590-12. doi: 10.1128/mBio.00590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol 6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 28.Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J Virol 83:8173–8181. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci USA 102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson Z, Dhummakupt A, Messer H, Phelan D, Bloom D. 2013. Role of polycomb proteins in regulating HSV-1 latency. Viruses 5:1740–1757. doi: 10.3390/v5071740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau J, Minett MS, Zhao J, Dennehy U, Wang F, Wood JN, Bogdanov YD. 2011. Temporal control of gene deletion in sensory ganglia using a tamoxifen-inducible Advillin-Cre-ERT2 recombinant mouse. Mol Pain 7:1744-8069-7-100. doi: 10.1186/1744-8069-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloom DC. 2016. Alphaherpesvirus latency: a dynamic state of transcription and reactivation. Adv Virus Res 94:53–80. doi: 10.1016/bs.aivir.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Colgin MA, Smith RL, Wilcox CL. 2001. Inducible cyclic AMP early repressor produces reactivation of latent herpes simplex virus type 1 in neurons in vitro. J Virol 75:2912–2920. doi: 10.1128/JVI.75.6.2912-2920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards TG, Bloom DC. 2019. Lund human mesencephalic (LUHMES) neuronal cell line supports herpes simplex virus 1 latency in vitro. J Virol 93:e02210-18. doi: 10.1128/JVI.02210-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pourchet A, Modrek AS, Placantonakis DG, Mohr I, Wilson AC. 2017. Modeling HSV-1 latency in human embryonic stem cell-derived neurons. Pathogens 6:24. doi: 10.3390/pathogens6020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith RL, Pizer LI, Johnson EM Jr, Wilcox CL. 1992. Activation of second-messenger pathways reactivates latent herpes simplex virus in neuronal cultures. Virology 188:311–318. doi: 10.1016/0042-6822(92)90760-m. [DOI] [PubMed] [Google Scholar]
- 37.Wilcox CL, Johnson EM Jr. 1987. Nerve growth factor deprivation results in the reactivation of latent herpes simplex virus in vitro. J Virol 61:2311–2315. doi: 10.1128/JVI.61.7.2311-2315.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson AC, Mohr I. 2012. A cultured affair: HSV latency and reactivation in neurons. Trends Microbiol 20:604–611. doi: 10.1016/j.tim.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grams TR, Edwards TG, Bloom DC. 2020. Herpes simplex virus 1 strains 17syn(+) and KOS(M) differ greatly in their ability to reactivate from human neurons in vitro. J Virol 94:e00796-20. doi: 10.1128/JVI.00796-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camarena V, Kobayashi M, Kim JY, Roehm P, Perez R, Gardner J, Wilson AC, Mohr I, Chao MV. 2010. Nature and duration of growth factor signaling through receptor tyrosine kinases regulates HSV-1 latency in neurons. Cell Host Microbe 8:320–330. doi: 10.1016/j.chom.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cliffe AR, Arbuckle JH, Vogel JL, Geden MJ, Rothbart SB, Cusack CL, Strahl BD, Kristie TM, Deshmukh M. 2015. Neuronal stress pathway mediating a histone methyl/phospho switch is required for herpes simplex virus reactivation. Cell Host Microbe 18:649–658. doi: 10.1016/j.chom.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuddy SR, Schinlever AR, Dochnal S, Seegren PV, Suzich J, Kundu P, Downs TK, Farah M, Desai BN, Boutell C, Cliffe AR. 2020. Neuronal hyperexcitability is a DLK-dependent trigger of herpes simplex virus reactivation that can be induced by IL-1. Elife 9:e58037. doi: 10.7554/eLife.58037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu HL, Shiflett LA, Kobayashi M, Chao MV, Wilson AC, Mohr I, Huang TT. 2019. TOP2beta-dependent nuclear DNA damage shapes extracellular growth factor responses via dynamic AKT phosphorylation to control virus latency. Mol Cell 74:466–480. doi: 10.1016/j.molcel.2019.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi M, Wilson AC, Chao MV, Mohr I. 2012. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translation repressor. Genes Dev 26:1527–1532. doi: 10.1101/gad.190157.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzich JB, Cuddy SR, Baidas H, Dochnal S, Ke E, Schinlever AR, Babnis A, Boutell C, Cliffe AR. 2021. PML-NB-dependent type I interferon memory results in a restricted form of HSV latency. EMBO Rep 22:e52547. doi: 10.15252/embr.202152547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JS, Raja P, Pan D, Pesola JM, Coen DM, Knipe DM. 2018. CCCTC-binding factor acts as a heterochromatin barrier on herpes simplex viral latent chromatin and contributes to poised latent infection. mBio 9:e02372-17. doi: 10.1128/mBio.02372-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh N, Tscharke DC. 2020. Herpes simplex virus latency is noisier the closer we look. J Virol 94:e01701-19. doi: 10.1128/JVI.01701-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raja P, Lee JS, Pan D, Pesola JM, Coen DM, Knipe DM. 2016. A herpesviral lytic protein regulates the structure of latent viral chromatin. mBio 7:e00633-16. doi: 10.1128/mBio.00633-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maillet S, Naas T, Crepin S, Roque-Afonso AM, Lafay F, Efstathiou S, Labetoulle M. 2006. Herpes simplex virus type 1 latently infected neurons differentially express latency-associated and ICP0 transcripts. J Virol 80:9310–9321. doi: 10.1128/JVI.02615-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dejosez M, Levine SS, Frampton GM, Whyte WA, Stratton SA, Barton MC, Gunaratne PH, Young RA, Zwaka TP. 2010. Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev 24:1479–1484. doi: 10.1101/gad.1935210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gunther M, Laithier M, Brison O. 2000. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol Cell Biochem 210:131–142. doi: 10.1023/A:1007177623283. [DOI] [PubMed] [Google Scholar]
- 52.Luciano RL, Wilson AC. 2003. HCF-1 functions as a coactivator for the zinc finger protein Krox20. J Biol Chem 278:51116–51124. doi: 10.1074/jbc.M303470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mazars R, Gonzalez-de-Peredo A, Cayrol C, Lavigne AC, Vogel JL, Ortega N, Lacroix C, Gautier V, Huet G, Ray A, Monsarrat B, Kristie TM, Girard JP. 2010. The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: a link between DYT6 and DYT3 dystonias. J Biol Chem 285:13364–13371. doi: 10.1074/jbc.M109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michaud J, Praz V, James Faresse N, Jnbaptiste CK, Tyagi S, Schutz F, Herr W. 2013. HCFC1 is a common component of active human CpG-island promoters and coincides with ZNF143, THAP11, YY1, and GABP transcription factor occupancy. Genome Res 23:907–916. doi: 10.1101/gr.150078.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker JB, Yin H, Vinckevicius A, Chakravarti D. 2014. Host cell factor-1 recruitment to E2F-bound and cell-cycle-control genes is mediated by THAP11 and ZNF143. Cell Rep 9:967–982. doi: 10.1016/j.celrep.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Popay TM, Wang J, Adams CM, Howard GC, Codreanu SG, Sherrod SD, McLean JA, Thomas LR, Lorey SL, Machida YJ, Weissmiller AM, Eischen CM, Liu Q, Tansey WP. 2021. MYC regulates ribosome biogenesis and mitochondrial gene expression programs through its interaction with host cell factor-1. Elife 10:e60191. doi: 10.7554/eLife.60191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas LR, Foshage AM, Weissmiller AM, Popay TM, Grieb BC, Qualls SJ, Ng V, Carboneau B, Lorey S, Eischen CM, Tansey WP. 2016. Interaction of MYC with host cell factor-1 is mediated by the evolutionarily conserved Myc box IV motif. Oncogene 35:3613–3618. doi: 10.1038/onc.2015.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyagi S, Chabes AL, Wysocka J, Herr W. 2007. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell 27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 59.Vogel JL, Kristie TM. 2000. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J 19:683–690. doi: 10.1093/emboj/19.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. 2010. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem 285:4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hancock ML, Meyer RC, Mistry M, Khetani RS, Wagschal A, Shin T, Sui SJH, Naar AM, Flanagan JG. 2019. Insulin receptor associates with promoters genome-wide and regulates gene expression. Cell 177:722–736. doi: 10.1016/j.cell.2019.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. 2009. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem 284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Nuland R, Smits AH, Pallaki P, Jansen PW, Vermeulen M, Timmers HT. 2013. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol Cell Biol 33:2067–2077. doi: 10.1128/MCB.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev 17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, Hart GW, Rauscher FJ, Drobetsky E, Milot E, Shi Y, Affar EB. 2010. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol 30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP. 2008. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell 133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zargar Z, Tyagi S. 2012. Role of host cell factor-1 in cell cycle regulation. Transcription 3:187–192. doi: 10.4161/trns.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Minocha S, Sung TL, Villeneuve D, Lammers F, Herr W. 2016. Compensatory embryonic response to allele-specific inactivation of the murine X-linked gene Hcfc1. Dev Biol 412:1–17. doi: 10.1016/j.ydbio.2016.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation and characterization of an HCF-1 conditional knockout mouse. (A) Schematic of the modified HCF-1 locus generated by recombination with a targeting construct containing HCF-1 exon 2 and 3, the selectable marker neomycin phosphotransferase (Neo) flanked by FRT sites, and the loxP sites flanking exon 2 and Neo (HCF-1tm1LVD). FLP-mediated recombination results in deletion of the selectable marker (HCF-1tm1.1LVD), referred to as HCF-1cKO, while Cre-mediated recombination results in deletion of HCF-1 exon 2 and 3 (HCF-1tm1.2LVD). (B and C) Primary fibroblast cells derived from HCF-1cKO or control C57BL/6 mice were infected with the indicated MOI of adenovirus expressing FLPe or Cre. (B) Cre mRNA levels are shown relative to mock-infected cells at 48 h postinfection. Data are from 3 independent experiments (ns, unpaired two-tailed t tests). (C) Controls for Fig. 2B. The levels of control cellular genes SP1 and GAPDH are shown relative to mock-infected cells. Data are from 4 independent experiments (ns, ANOVA with Dunnett’s multiple comparisons test). Download FIG S1, PDF file, 0.4 MB (457.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression of Cre in latently infected sensory neurons of control C57BL/6 mice does not affect viral reactivation. The ratio of HSV DNA levels in explanted to latent ganglia (n ≥ 9) of C57BL/6 mice infected with the indicated HSV strain (ns, Kruskal-Wallis test with Dunnett’s multiple comparisons test). Control for Fig. 3C. Download FIG S2, PDF file, 0.1 MB (111.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression of Cre in latently infected sensory neurons results in reduced numbers of primary neurons undergoing reactivation. (A to C) Ganglia that were latently infected with the indicated virus were explanted for 48 h. Ganglia sections were stained with anti-UL29 (red) and DAPI (blue). (A) Representative sections of ganglia infected with WT, CMV-Cre, and LAT-Cre that were explanted in the presence of ACV for 48 h. Related to Fig. 4A. (B and C) Ganglia sections were scored for individual UL29+ neurons (Single) and clusters (Clusters). (n ≥ 14 ganglia; Kruskal-Wallis test with Dunnett’s multiple comparisons test). Representative sections are shown in panel C. Download FIG S3, PDF file, 2.7 MB (2.8MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Tamoxifen-dependent recombination in sensory ganglia. Latently infected HCF-1cKO/AvCreERT2 and C57BL/6 mice were injected either with tamoxifen or vehicle for 5 days, and DNA was purified from individual trigeminal ganglia (n = 6) at 9 days postinjection. PCR was completed with primers that detect the nonrecombined loci from the first loxP site through exon 2 (P1-P2; 522 bp) and for the detection of recombination (deletion of exon 2 and 3; P1-P6; 287 bp). Download FIG S4, PDF file, 0.3 MB (333.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
mRNA levels in tissues of HCF-1cKO/AvCreERT2 and control C57BL/6 mice. mRNA levels of HCF-1, MMACHC, and GAPDH in kidney and liver of control C57BL/6 and HCF-1cKO/AvCreERT2 mice treated with vehicle or tamoxifen. Levels are shown in tamoxifen-treated relative to vehicle-treated animals. Data are from 2 tissues pooled from each of 2 samples. Download FIG S5, PDF file, 0.2 MB (160KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Representative images from explanted ganglia from control C57BL/6 mice. Latently infected ganglia from control C57BL/6 mice were explanted in the presence of vehicle or tamoxifen for 48 h. Sections were stained for UL29 (red) and DAPI (blue). Representative sections are shown. Control for Fig. 7B. Download FIG S6, PDF file, 0.8 MB (891.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Deletion of HCF-1 does not result in loss of neurons or induction of neuronal apoptosis. Latently infected HCF-1cKO/AvCreERT2 or control C57BL/6 mice were treated with either vehicle or tamoxifen. (A and B) TUNEL assays were done on ganglia sections, which were counterstained with DAPI (blue) and neurofilament-200 (red). DNaseI treated HCF-1cKO/AvCreERT2 sections were used as positive TUNEL controls. Representative sections are shown (1× and 3× zoom) for each condition. (C) Numbers of neurons per ganglia (2 sections were counted for each of 8 ganglia per group) of HCF-1cKO/AdCreERT2 and control C57BL/6 mice treated with either vehicle or tamoxifen (Mann-Whitney test). Download FIG S7, PDF file, 2.4 MB (2.5MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Primers and reagents. Reagents and sequences of primers used in this study. Download Table S1, PDF file, 0.05 MB (52.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistics. Relevant statistical tests and P values of data described in this study. Download Table S2, PDF file, 0.1 MB (55.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.