Abstract
Lactobacillus johnsonii La1, a probiotic bacterium with demonstrated health effects, grows in milk, where it ferments lactose to d- and l-lactate in a 60:40% ratio. The d-lactate dehydrogenase (D-LDH) gene (ldhD) of this strain was isolated, and an in vitro-truncated copy of that gene was used to inactivate the genomic copy in two strains, La1 and N312, by gene replacement. For that, an 8-bp deletion was generated within the cloned ldhD gene to inactivate its function. The plasmid containing the altered ldhD was transferred to L. johnsonii via conjugative comobilization with Lactococcus lactis carrying pAMβ1. Crossover integrations of the plasmid at the genomic ldhD site were selected, and appropriate resolution of the cointegrate structures resulted in mutants that had lost the plasmid and in which the original ldhD was replaced by the truncated copy. These mutants completely lacked D-LDH activity. Nevertheless, the lower remaining L-LDH activity of the cells was sufficient to reroute most of the accumulating pyruvate to l-lactate. Only a marginal increase in production of the secondary end products acetaldehyde, diacetyl, and acetoin was observed. It can be concluded that in L. johnsonii D- and L-LDH are present in substantial excess for their role to eliminate pyruvate and regenerate NAD+ and that accumulated pyruvate is therefore not easily redirected in high amounts to secondary metabolic routes.
Lactobacillus johnsonii La1 is a probiotic lactic acid bacterium which has been intensively investigated in clinical and nutritional studies for its beneficial health effects (8). Upon ingestion, La1 survives the gastrointestinal transit, where it is responsible for an immunomodulating effect of the host (29, 38, 39). La1 grows on different sugars as a carbon source and can be cultured in milk, where it ferments lactose to a racemic mixture of d(−)- and l(+)-lactic acid in a ratio of ca. 60:40%. Lactic acid is formed via reduction of pyruvate by lactate dehydrogenase (LDH) for the regeneration of NAD+. Thereby, the two isomeric forms of lactate, d(−) and l(+), are formed by distinct stereospecific NAD-dependent LDHs.
Today, the La1 strain is commercialized in many countries as part of a mixed cultured fermented milk product and in 1997 alone more than 1018 live La1 cells were consumed worldwide. As in most standard yogurt products, the presence of d-lactate in the La1-containing end product and the capacity of the strain to produce d-lactate after ingestion do not pose an adverse effect for the vast majority of the adult population. Nevertheless, it has been reported that after the consumption of food, d-lactic acid can accumulate in the blood of patients suffering from short-bowel syndrome and intestinal failure, leading to a manifestation of d-lactic acidosis and encephalopathy (24, 25, 34, 40). Thus, it was determined that d-lactate-producing colonizing intestinal lactobacilli were the main factor in the pathogenesis (6, 37). In view of the probiotic and immune-defense-stimulating effects of L. johnsonii La1, a non-d-lactate-producing variant of this strain would be most helpful as a food supplement to improve the nutritional value of clinical and parenteral diets for patients with intestinal failures and to reconstitute their intestinal microflora after antibiotic treatment.
Furthermore, newborn infants may fail to completely metabolize ingested or by intestinal microorganisms produced d-lactate because of liver immaturity (15, 16). Hence, food products containing these ingredients are not recommended for nutrition to infants and young children to the age of 3 years (17). Here again, a non-d-lactate-producing variant of La1 would be beneficial in the nutrition for young children for building up and regulating their intestinal microflora. In particular, the immunostimulating effects of a probiotic Lactobacillus strain, e.g., La1, may help to mitigate bacterium- or virus-induced diarrhea in infants (4).
Recently, the genes encoding the d-LDH (D-LDH) (ldhD) of Lactobacillus plantarum (42), Lactobacillus bulgaricus (3, 26), and Lactobacillus helveticus (28) have been cloned and sequenced. Furthermore, successful gene inactivations of the ldhD in L. helveticus and L. plantarum have been reported (5, 18). For the L. johnsonii species only a little genetic work has been reported so far (1, 13, 44). In this study, we report the first isolation of the ldhD gene of L. johnsonii. In addition, the genomically encoded ldhD gene was replaced with an in vitro-truncated copy in two different strains of L. johnsonii, and the resulting effects on the production of primary and secondary end products of the lactose metabolism were analyzed.
MATERIALS AND METHODS
Bacterial strains, media, plasmids, and growth conditions.
L. johnsonii La1 and N312 are from the Nestec Culture Collection. They are both resistant to bile salt and gastric juice and survive the transit through the stomach and gut (31, 38). They were maintained in MRS broth (Difco, Detroit, Mich.) and were grown either in MRS at 37 or at 42°C in supplemented milk (SM) which is ultrahigh-temperature-treated (UHT) full fat milk supplemented with 2.5% skimmed-milk powder, 1% yeast extract (Difco), and 0.25% proteose peptone number 3 (Difco). Growth curves in SM were made by the rapid automated bacterial impedance technique (RABIT; Don Whitley, Scientific Ltd.). L. lactis MG1363 (20) was grown at 30°C in M17 broth (Difco) supplemented with 0.5% glucose (GM17). Escherichia coli XL1-Blue was obtained from Stratagene (La Jolla, Calif.) and grown in 2×-concentrated YT broth (35) at 37°C under shaking conditions. Plasmids pGEMT (Promega, Madison, Wis.) and pUC19 (46) were maintained in E. coli XL1-Blue by the addition of 100 μg of ampicillin per ml. The conjugative plasmid pAMβ1 (9) was maintained in L. lactis by the addition of 5 μg of erythromycin per ml. Plasmid pMD14 is a derivative of pBluescript SK(+) carrying the cloned chloramphenicol resistance gene (cat) from pNZ12 (12) and a ca. 800-bp fragment cloned from the region upstream of the erythromycin resistance gene of pUC-838 (32), which is homologous to pAMβ1. pMD14 was maintained in L. lactis and in E. coli by the addition of 14 and 30 μg of chloramphenicol per ml, respectively.
Construction of a truncated ldhD (ldhD*) in pGEMT.
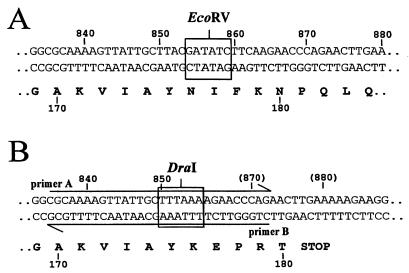
Site-directed mutagenesis was performed by PCR (Gold Taq polymerase from Perkin-Elmer) by using the method of gene splicing by overlap extension (23). The ldhD gene from La1 was amplified from genomic DNA as two fragments in two separate PCRs with the primer pairs A-C and B-D, respectively (Table 1). The two amplified fragments were agarose gel purified, mixed together, and reamplified by PCR with primers C and D. The resulting 1,460-bp fragment was cloned into pGEMT in E. coli XL1-Blue, and single plasmid isolates were tested by DraI and EcoRV digestions. Primer A, containing a DraI site, and primer B, with the same sequence but in the reverse orientation, introduced a deletion of eight nucleotides in the middle of the ldhD gene (ldhD*) (Fig. 1). This resulted in the conversion of the EcoRV site at position 854 into a DraI restriction site at position 849 and the formation of a translational stop signal 16 nucleotides downstream of the newly created DraI site. A correct clone was verified by DNA sequencing and retained as plasmid pLL83.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| A | CGCAAAAGTTATTGCTTTAAAAGAACCCAG |
| B | CTGGGTTCTTTTAAAGCAATAACTTTTGCG |
| C | TGATTTTCAGAGCGTGC |
| D | GCTCTCCTGAAATTTGC |
| E | GTAGTGTAGAAGGCGGTGTGTGG |
| F | GCTTACGCTATTCGAAAAGACG |
| G | GGTACTGGTCACATCGG |
| H | CATCTTTACGAATAGCG |
FIG. 1.
Site of mutagenesis within the ldhD gene. The wild-type (A) and mutated ldhD∗ (B) sequences are shown. Numbers refer to the nucleotide and amino acid sequence positions. Nucleotide restriction sites EcoRV and DraI are boxed. Primers A and B are indicated by arrows.
Introduction of the ldhD* into L. johnsonii.
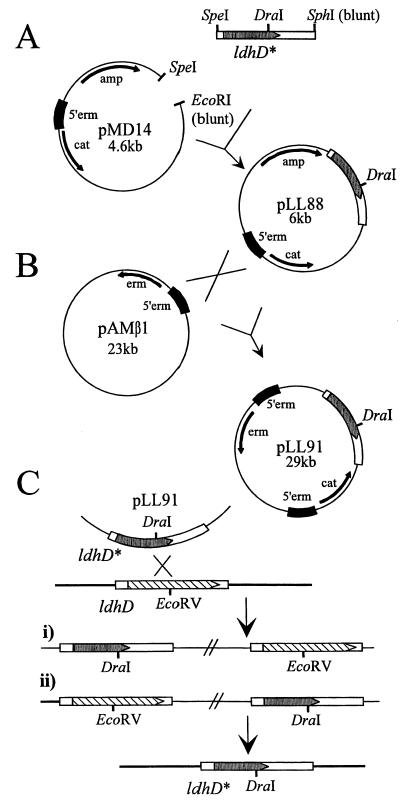
In the first step, ldhD* was isolated from pLL83 as a SpeI-SphI fragment and then cloned into pMD14 in E. coli XL1-Blue. The resulting plasmid was named pLL88 (Fig. 2A). Plasmid pLL88 was transformed by electroporation into L. lactis MG1363(pAMβ1), plated onto GM17 agar containing 14 μg of chloramphenicol per ml, and incubated at 30°C. Since pLL88 cannot autonomously replicate in L. lactis, only transformed cells containing pLL88 inserted via homologous recombination into pAMβ1 were isolated (Fig. 2B). The correct formation of the cointegrate (pLL91) was verified by PCR analysis. Plasmid pLL91 was transferred to L. johnsonii via conjugation on solid agar plates (Fig. 2C). For that, L. lactis MG1363(pLL91) and L. johnsonii were grown separately in appropriate media until the late logarithmic phase and then mixed in a 1:10 ratio, centrifuged, and spread onto LCMG (14) agar plates containing 0.5% polyethylene glycol. Plates were overlaid with LCMG top agar (0.7% agar) and incubated overnight at 37°C, and the cells were then resuspended in LCMG broth. Serial dilutions of the suspensions were plated on MRS agar containing 100 μg of phosphomycin and 14 μg of chloramphenicol per ml. Plates were incubated anaerobically (GasPack; BBL) at 37°C and chloramphenicol-resistant L. johnsonii colonies were recovered after 2 days with a frequency of 10−5 transconjugants per recipient cells. The presence of pLL91 in L. johnsonii was verified by Southern blot analysis.
FIG. 2.
Strategy for ldhD gene replacement in La1. (A) Cloning steps in E. coli XL1-Blue. (B) Vector cointegrate formation in L. lactis MG1363. (C) Gene replacement in L. johnsonii. Antibiotic resistance genes are indicated by arrows; the ldhD and ldhD* genes are boxed. The dark box labeled 5′erm represents the 800-bp fragment cloned from the region upstream of the erythromycin resistance gene of pAMβ1. The figure is not drawn to scale.
DNA preparation.
All DNA manipulations and gel electrophoresis procedures were carried out as recommended by the suppliers or by standard methods (35). Plasmid DNA was isolated from E. coli by the alkaline lysis method. Plasmid pAMβ1 was prepared by the method for isolating large plasmid DNA (2). Lactobacillus DNA was obtained as described previously (10), except that the cells were incubated for 1 h at 37°C with a mixture of mutanolysin (120 μg/ml) and lysozyme (1 mg/ml). When necessary, the digested DNA was blunt ended with T4 DNA polymerase (Boehringer GmbH, Mannheim, Germany) for 5 min at 37°C.
Transformations of E. coli and L. lactis.
Electrotransformation of E. coli was performed according to standard procedures (35). Competent cells and transformations of L. lactis were realized according to the method of Holo and Nes (22).
Southern hybridizations.
Chromosomal DNA was digested with EcoRV and DraI (Boehringer) and submitted to electrophoresis on 1.0% (wt/vol) agarose gels (1× Tris-acetate-EDTA buffer, pH 8.0). Southern blot hybridizations and washings were performed under stringent conditions. Fragments were radiolabelled with [α-32P]dCTP (Amersham, Sunnyvale, Calif.) by using the random primer labeling kit of Boehringer.
DNA sequencing.
DNA sequencing was performed by the dideoxy chain termination method (36) with [γ-33P]dATP (Isotopchim, Peyruis, France). Sequencing reactions were carried out on plasmid or directly on PCR-amplified chromosomal DNA with the VISTRA Thermosequenase kit (Amersham). Sequences were analyzed with the help of the University of Wisconsin Genetics Computer Group (GCG) computer software package (11).
Enzyme assays.
Concentrations of d- and l-lactate in SM and in MRS cultures were determined as follows. Bacterial cultures were diluted in 100 mM Tris-HCl (pH 8.0)–150 mM NaCl, kept on ice for 10 to 30 min, and centrifuged at maximal speed for 10 min. When necessary, supernatants were kept frozen at −20°C until use. Supernatants were further diluted in the same buffer, mixed with 1 ml of mixture reaction (100 mM Tris-HCl, pH 9.0; 2 mM EDTA; 3% hydrazine; 1 mM NAD+) in the presence of D-LDH or L-LDH (20 U/ml each; D-LDH was from Leuconostoc mesenteroides Sigma L-2395, and L-LDH was from rabbit muscle Sigma L-2500, St. Louis, Mo.) and incubated for 90 min at room temperature. NADH accumulation was monitored by measuring the optical density at 340 nm. For the quantification of intracellular D-LDH and L-LDH, cells were washed with a solution containing 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 2 mM EDTA, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride, centrifuged, concentrated five times in the same buffer containing 0.5 mg of lysozyme and 50 μg of mutanolysin per ml, and incubated for 20 min at 37°C. The solution of lysed cells was cleared by centrifugation, and the D- and L-LDH activities were determined in 100 mM Tris-HCl (pH 8.0), 15 mM NAD+, and 500 mM d-lactate (Fluka 71716) or l-lactate (Fluka 71718), respectively, by monitoring the optical density at 342 nm. The total LDH activity was measured under the same conditions but in the presence of 20 mM pyruvate and 0.15 mM NADH in 100 mM potassium buffer at pH 7.5. Protein concentrations were determined as described previously (7), with bovine serum albumin as a standard.
GC analysis of secondary end products.
To determine the production of secondary end products, cells were grown in SM, and the volatile compounds were trapped on a Tenax column and subsequently analyzed by gas chromatography (GC) on a DB wax fused silica capillary column as previously described (33). For that, aliquots of 25-g samples were placed into the sample space of a headspace cell and equilibrated for 2 h at 30°C in a water bath. The headspace of the cell (160 ml) was then passed through a trap containing 250 mg of Tenax with a flow rate of 40 ml/min. Volatile compounds were thermally desorbed from Tenax at 300°C for 15 min by using an ATD400 thermal desorber (Perkin-Elmer Corp., Norwalk, Conn.). They were then refocused on an internal cold Tenax trap (−30°C) and desorbed at 300°C for 3 min into an HP5890 GC (Hewlett-Packard, Avondale, Pa.) equipped with a 60-m-long, 0.53-mm (inner diameter), 1.00-μm-phase-thickness DBwax column (J&W Scientific, Folsom, Calif.). Helium was used as carrier gas at a 10.6-ml/min flow rate. The column was kept at 20°C for 5 min, increased at a rate of 4°C/min to 200°C, and then maintained for 10 min at 200°C. After each sampling, the cell was cleaned in a vacuum oven at 50°C under 104 Pa for at least 1 h. Tenax sampling tubes were cleaned before use by heating for 1 h at 300°C under helium flow (50 ml/min).
Nucleotide sequence accession number.
The ldhD nucleotide sequence reported here has been deposited in the GenBank database under nucleotide accession number AF071558.
RESULTS AND DISCUSSION
Isolation and analysis of the ldhD gene of L. johnsonii La1.
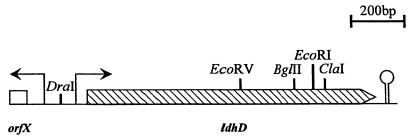
Based on the L. helveticus ldhD gene sequence (28), the degenerated oligonucleotides, primers E and F (Table 1), were used to amplify an 890-bp fragment from genomic La1 DNA. This fragment was cloned in pGEMT and used as probe to screen a BglII genomic library of La1 in pUC19 in E. coli XL1-Blue. A 3-kb BglII fragment was isolated, and its DNA sequence was determined. The sequence analysis revealed the presence of two open reading frames oriented in opposite directions. One of the open reading frames demonstrated significant sequence homology to the L. helveticus ldhD gene and was tentatively designated ldhD. The C-terminal end of this ldhD gene, which was not present on the cloned 3-kb BglII fragment, was isolated by inverted PCR (43) on SpeI-digested DNA with primers G and H. The sequences were assembled, and a map of this region is given in Fig. 3.
FIG. 3.
Physical map of ldhD of La1. Arrows above the map indicate putative promoters; the hairpin represents a putative rho-independent termination signal.
The ldhD gene is 1,014 nucleotides long and encodes the putative 338-amino-acid D-LDH. It shows significant homologies with the known D-LDH enzymes from other lactobacilli, i.e., amino acid sequence identities of 94% with L. helveticus (28), 86.7% with L. bulgaricus (3, 26), and 51.7% with L. plantarum (42). Catalytic domains common to several dehydrogenases were also present in the D-LDH of L. johnsonii. The NADH-binding domain G-X-G-X-X-G17-D is located at amino acid positions 152 to 175, corresponding to the location of the equivalent domain in the other D-LDHs. The histidine essential for catalysis is located at position 296, the arginine involved in substrate binding is located at position 235, and the glutamic acid modulating pH dependence (27) is located at position 264. The presence of a putative promoter sequence and ribosome binding site upstream, as well as a potential rho-independent termination signal at the end of the gene, suggests that the identified ldhD is independently transcribed as a functional gene.
Integration of pLL91 into the La1 genome.
Preliminary attempts to transform La1 by electroporation (22, 30, 45) were unsuccessful. We therefore introduced the vector for gene replacement, pLL91, via conjugative comobilization from L. lactis MG1363(pAMβ1) (9) to L. johnsonii La1. Six independent La1(pLL91) transconjugant colonies were isolated and subcultivated in MRS broth with 14 μg of chloramphenicol per ml for 20 to 30 generations at 37°C. The pAMβ1 replicon is functional in L. johnsonii at a lower growth temperature (37°C), but is inactive at 45°C. To select for genomic integrations of pLL91, 5-ml portions of the six cultures were used to subsequently inoculate six 100-ml volumes of fresh MRS broth, which were incubated at 45°C until saturated growth was reached. The cultures were diluted and grown on MRS agar plates with 10 μg of chloramphenicol per ml. One colony of each transconjugant was isolated, purified, and maintained in MRS-chloramphenicol broth.
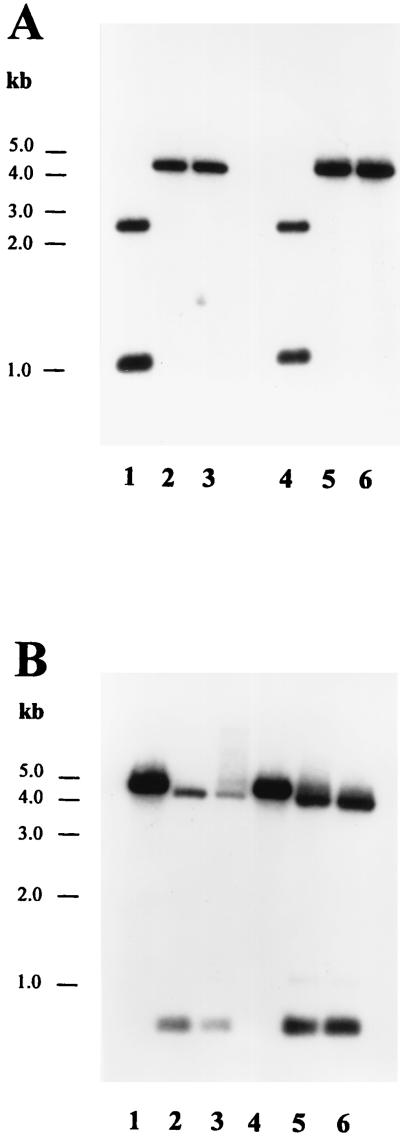
Genomic DNA of the six resulting cultures was prepared and analyzed by Southern blot hybridizations with ldhD and vector-based DNA probes. For all six independent transconjugants, the results revealed targeted integration of pLL91 into the genomic ldhD locus, as well as a duplication of the ldhD signal, indicating cointegrate formation as shown in Fig. 2C. EcoRV and DraI restriction enzyme analysis was used to differentiate between the two types of cointegrates formed following homologous integration either upstream or downstream of the generated mutation in ldhD, respectively. Five of the six integrants recombined as shown in Fig. 2Ci, and one recombined as shown in Fig. 2Cii. One culture per cointegrate structure was retained for further studies.
The two different cointegrate cultures were grown for 100 generations in nonselective MRS broth supplemented with 0.05% d-lactate (Fluka, Buchs, Switzerland) at 37°C. d-Lactate was added in case the ldhD mutant strain would need this isomer for cell wall synthesis, as shown previously for L. plantarum (18). The cultures were then diluted on MRS agar plates containing d-lactate and grown at 37°C. Fifty colonies from each culture were picked and tested on MRS-chloramphenicol (10 μg/ml) and MRS-erythromycin (5 μg/ml) plates for their antibiotic resistance pattern. With one of the cointegrate cultures, 2 of the 50 isolates were sensitive to both antibiotics tested. Genomic DNA of these two isolates was prepared and Southern blot analysis confirmed that plasmid pLL91 was excised from the genome and lost from the cells. Furthermore, Southern blot analysis with EcoRV and DraI revealed that, by chance, in both cases the wild-type ldhD allele was replaced by the ldhD* gene (Fig. 4). Mutants were called La1 ldhD-1 and ldhD-2.
FIG. 4.
Analysis of replacement of the ldhD gene of L. johnsonii La1 and N312 by Southern hybridization. The DNA probe is the complete ldhD gene. (A) Chromosomal DNA digested with EcoRV. (B) Chromosomal DNA digested with DraI. Lane 1, La1; lane 2, La1 ldhD-1; lane 3, La1 ldhD-2; lane 4, N312; lane 5, N312 ldhD-1; lane 6, N312 ldhD-2.
Identical ldhD mutants were constructed for L. johnsonii N312 with the same strategy as that described for La1. Two mutants lacking D-LDH activity were selected and called N312 ldhD-1 and ldhD-2. All mutants were verified by Southern blot analysis of EcoRV and DraI digests to confirm the replacement of the ldhD wild type by the mutated ldhD* gene (Fig. 4). The probe used comprised the entire ldhD gene obtained by PCR with the oligonucleotides C and D (Table 1). After resequencing of the complete gene, all of these strains were retained for further physiological characterization.
Genetic and physiological stability of the mutation.
The stability of the introduced mutation was determined by monitoring the D-LDH activity over many generations. Mutant strains were subcultured 30 times (ca. 200 generations) in 10 ml of MRS broth at 37°C. Production of d- and l-lactate was measured in the culture supernatant, and no d-lactate could be detected. From the last subculture, 200 colonies of each mutant were isolated and then grown in 10 ml of MRS broth, and the occurrence of d-lactate was determined. All strains produced exclusively l-lactate. The mutation was thus considered genetically stable. Furthermore, the sensitivity of La1, N312, and the mutant strains toward the antibiotic vancomycin was determined. For that, small discs treated with 5 μg of vancomycin per ml were put onto inoculated MRS agar plates and incubated for 24 h at 37°C. The sensitivities of both wild types and their mutants toward the antibiotic were identical, showing that, in contrast to L. plantarum (18), the peptidoglycan synthesis pathway of L. johnsonii is not affected by the absence of d-lactate.
Physiological characteristics.
Growth rates of the different strains were compared in SM. Thereby, it was observed that the ldhD mutant strains grew as well as their respective wild-type strains. After a 2-h lag phase, cells grew exponentially for 3 h to reach a final plateau after 5 h of incubation. After a 24-h incubation, production of d- and l-lactate was measured in all supernatants. La1 and N312 produced d- and l-lactate in a ratio of about 60:40 (Table 2). On the contrary, no accumulation of d-lactate was observed in the ldhD mutants, and instead most of the metabolic carbon flux was deviated to the production of l-lactate, which revealed that, in contrast to other lactobacilli (21, 41), L. johnsonii does not possess a dl-lactate racemase. However, the total amount of lactate produced by the mutant strains did not reach the same levels as in the wild-type strains. Lactate production was also measured in the supernatant of cells grown in MRS broth (Table 2). The same characteristics were found for La1, N312, and their mutants: i.e., lactate was produced in a d-lactate/l-lactate ratio of ca. 70:30, and no d-lactate was found in the mutants. However, cells grew much better in milk and, hence, the total production of lactate for all strains was clearly superior in milk than in MRS.
TABLE 2.
Concentration of lactate in the supernatant of overnight cultures grown in SM medium and in MRS broth
| L. johnsonii strains | Concna of lactate in:
|
|||||
|---|---|---|---|---|---|---|
| SM medium
|
MRS broth
|
|||||
| d-Lactate (g/liter) | l-Lactate (g/liter) | Total lactate (%) | d-Lactate (g/liter) | l-Lactate (g/liter) | Total lactate (%) | |
| La1 | 18.0 ± 0.4 (58.1) | 13.0 ± 1.5 (41.9) | 100 | 13.7 ± 0.4 (70.3) | 5.8 ± 0.2 (29.7) | 100 |
| La1 ldhD-1 | <0.1 | 24.3 ± 2.6 | 78 | <0.1 | 16.3 ± 1.2 | 84 |
| La1 ldhD-2 | <0.1 | 24.5 ± 2.4 | 79 | <0.1 | 17.3 ± 0.6 | 89 |
| N312 | 17.0 ± 0.45 (54.7) | 14.1 ± 4.8 (45.3) | 100 | 14.2 ± 0.5 (70.0) | 6.1 ± 0.5 (30.0) | 100 |
| N312 ldhD-1 | 0.2 ± 0.3 | 22.2 ± 2.4 | 71 | <0.1 | 16.4 ± 1.5 | 81 |
| N312 ldhD-2 | 0.3 ± 0.2 | 23.2 ± 3.3 | 75 | <0.1 | 15.5 ± 1.5 | 76 |
The percentage of d- or l-lactate per total lactate concentration is given in parentheses.
Cells grown in MRS were harvested, washed, and used for the determination of intracellular D-LDH, L-LDH, and total LDH activities in the presence of d- or l-lactate and either NAD+ or pyruvate-NADH, respectively. The results showed no detectable D-LDH activity for all mutant strains tested (Table 3). For La1 and its mutants, the L-LDH activity was constant, whereas for N312 a slight increase in this activity was observed for the mutant strains. It was not investigated whether this increase was due to an increased transcriptional or translational activity. Furthermore, it was interesting that, in both wild-type strains, the endogenous D-LDH activity was higher than the L-LDH activity. Nevertheless, upon ldhD inactivation, the ca.-four-times-lower activity of the remaining L-LDH was sufficient to compensate for most of the lost D-LDH activity without significantly affecting the L-LDH expression or inducing a reduction of the bacterial growth rate.
TABLE 3.
Internal LDH activities of cells grown in MRS broth
| L. johnsonii strains | LDH activity
|
||
|---|---|---|---|
| D-LDH (U/mg) | L-LDH (U/mg) | Total LDH (U/mg) | |
| La1 | 1.18 ± 0.04 | 0.32 ± 0.01 | 15.58 ± 0.83 |
| La1 ldhD-1 | <0.01 | 0.32 ± 0.01 | 1.88 ± 0.07 |
| La1 ldhD-2 | <0.01 | 0.35 ± 0.01 | 1.95 ± 0.05 |
| N312 | 1.88 ± 0.04 | 0.39 ± 0.03 | 20.68 ± 1.54 |
| N312 ldhD-1 | <0.01 | 0.61 ± 0.01 | 3.88 ± 0.14 |
| N312 ldhD-2 | <0.01 | 0.57 ± 0.02 | 2.78 ± 0.08 |
Impact of the ldhD* mutation on the production of secondary end products.
L. johnsonii is a strict homofermentative bacterium and does not produce organic acids as end products other than lactate. This was verified for La1, N312, and their ldhD-deficient mutants in SM cultures by high-pressure liquid chromatography (HPLC) analysis (data not shown). In particular, no accumulation of acetic and succinic acid was detectable in L. johnsonii as was observed for L. plantarum (19). However, inactivation of the ldhD gene resulted in an increased production of some of the secondary end products, in particular acetaldehyde, acetoin, and two unidentified compounds with retention times of 38 and 39 min. Interestingly, the results were different for the La1 and N312 ldhD-deficient mutants (Table 4). The mutant strains of N312 produced up to 25 times more acetaldehyde than the parent strain compared to La1, where the increase was by a factor of only two. On the other hand, production of diacetyl and acetoin was slightly increased for the La1 PdhD mutants. These results indicated that the natural metabolic pathways leading to these flavor metabolites are not equally equilibrated between the two strains. However, it has to be noted that the increased production of these metabolites in quantitative terms is very low, i.e., an increase from ca. 1 to 10 ppm for the production of acetaldehyde by the N312 PdhD mutants (quantities were determined by static headspace gas chromatography [data not shown]). Hence, the increase observed in secondary end products cannot fully account for the lower total production of the lactic acid detected. A slight undetectable reduction in the fermentation yield could explain this difference between the wild-type strain and the mutants. It seems that accumulated pyruvate cannot easily be redirected in large amounts to secondary metabolic routes and that it is essential for L. johnsonii to almost stoichiometrically reduce pyruvate to lactate in order to regenerate NAD+.
TABLE 4.
Headspace GC analysis of volatiles produced by strains grown in SM medium
| L. johnsonii strains | Relative peak areas of:
|
|||
|---|---|---|---|---|
| Acetaldehyde | Diacetyl | Acetoin | Acetone | |
| La1 | 305 | 186 | 75 | 2,782 |
| La1 ldhD-1 | 530 | 250 | 227 | 3,014 |
| La1 ldhD-2 | 509 | 254 | 263 | 2,500 |
| N312 | 170 | 153 | 117 | 2,489 |
| N312 ldhD-1 | 4,291 | 188 | 261 | 2,974 |
| N312 ldhD-2 | 4,222 | 192 | 235 | 2,964 |
Perspectives.
Functional foods containing probiotic microorganisms with scientifically supported health benefits to the consumer constitute a growing market. Thereby, selected lactobacilli play an important role because they contribute to an improved health status, mainly through their beneficial effects in the small intestine. In today’s probiotic products, the Lactobacillus strains used are all natural strains, mainly isolated from human or animal intestinal sources, and are selected in screening experiments and validated in clinical studies. The findings presented here show a new approach for improving the health benefit and applicability of already-established probiotic strains by targeted genetic engineering, which may lead to a new generation of probiotic products once they have been approved by the consumers and by legislative bodies. This approach allows us to build new probiotic microorganisms on already-selected and clinically validated strains without going again through new screening procedures either on new isolates or with spontaneous mutants of established strains. Moreover, targeted genetic engineering prevents the accumulation of numerous uncharacterized mutations in the bacterial genome, which may occur in spontaneous mutation and screening experiments. This adds to the safety aspect of this new technology and assures the preservation of the desired health-beneficial character of the new strain. Nevertheless, it is clear that also in this case all newly developed strains have to run through a vigorous safety assessment before an application in food can be envisaged.
ACKNOWLEDGMENTS
We thank David Pridmore for his help in the construction of pLL88, Sunil Kochhar for his advice in the LDH enzyme assays, Aline Mamin for her assistance with the Rapid Automated Bacterial Impedance Technique, and Michel Richard for the HPLC analysis. We thank Elaine Vaughan and Aude Bourniquel for critical reading of the manuscript. We are also grateful to Willem De Vos for the gift of L. lactis MG1363 containing pAMβ1.
REFERENCES
- 1.Allison G E, Klaenhammer T R. Functional analysis of the gene encoding immunity to lactacin F, lafI, and its use as a Lactobacillus-specific, food-grade genetic marker. Appl Environ Microbiol. 1996;62:4450–4460. doi: 10.1128/aem.62.12.4450-4460.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson D, McKay L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard N, Ferain T, Garmyn D, Hols P, Delcour J. Cloning of the d-lactate dehydrogenase gene from Lactobacillus delbrueckii subsp. bulgaricus by complementation in Escherichia coli. FEBS Lett. 1991;290:61–64. doi: 10.1016/0014-5793(91)81226-x. [DOI] [PubMed] [Google Scholar]
- 4.Bernet M F, Brassart D, Neeser J R, Servin A. Lactobacillus acidophilus La1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut. 1994;35:483–489. doi: 10.1136/gut.35.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhowmik T, Fernandez L, Steele J L. Gene replacement in Lactobacillus helveticus. J Bacteriol. 1993;175:6341–6344. doi: 10.1128/jb.175.19.6341-6344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bongaerts G, Tolboom J, Naber T, Bakkeren J, Severijnen R, Willems H. d-Lactic acidemia and aciduria in pediatric and adult patients with short bowel syndrome. Clin Chem. 1995;41:107–110. [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Brassart D, Schiffrin E J. The use of probiotics to reinforce mucosal defence mechanisms. Trends Food Sci Technol. 1997;8:321–326. [Google Scholar]
- 9.Clewell D B, Yagi Y, Dunny G M, Schultz S K. Characterization of three plasmid DNA molecules in a strain of Streptococcus faecalis: identification of a plasmid determined erythromycin resistance. J Bacteriol. 1974;117:283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delley M, Mollet B, Hottinger H. DNA probe for Lactobacillus delbrueckii. Appl Environ Microbiol. 1990;56:1967–1970. doi: 10.1128/aem.56.6.1967-1970.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Sharpe E. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Vos W. Gene cloning and expression in lactic streptococci. FEMS Microbiol Rev. 1987;46:281–295. [Google Scholar]
- 13.Djordjevic G M, Klaenhammer T R. Positive selection, cloning vectors for gram-positive bacteria based on a restriction endonuclease cassette. Plasmid. 1996;35:37–45. doi: 10.1006/plas.1996.0004. [DOI] [PubMed] [Google Scholar]
- 14.Efthymiou C, Hansen P A. An antigenic analysis of Lactobacillus acidophilus. J Infect Dis. 1962;110:258–267. doi: 10.1093/infdis/110.3.258. [DOI] [PubMed] [Google Scholar]
- 15.Food and Agriculture Organization/World Health Organization. Specifications for the identity and purity of food additives and their toxicological evaluation: some emulsifiers and stabilizers and certain other substances. Tenth report of the Joint FAO-WHO Expert Committee on Food Additives. WHO Tech Rep Ser. 1967;373:5–47. [PubMed] [Google Scholar]
- 16.Food and Agriculture Organization/World Health Organization. Toxicological evaluation of certain food additives with a review of general principles and of specifications. Seventeenth report of the Joint FAO-WHO Expert Committee on Food Additives. WHO Tech Rep Ser. 1974;539:1–40. [PubMed] [Google Scholar]
- 17.Food and Agriculture Organization/World Health Organization. Codex Alimentarius. Foods for special dietary uses (including foods for infants and children) 2nd ed. Rome, Italy: F.A.O./World Health Organization; 1994. [Google Scholar]
- 18.Ferain T, Hobbs J R, Richardson J, Bernard N, Garmyn D, Hols P, Allen N E, Delcour J. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J Bacteriol. 1996;178:5431–5437. doi: 10.1128/jb.178.18.5431-5437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferain T, Schanck A N, Delcour J. 13C nuclear magnetic resonance analysis of glucose and citrate end products in an ldhL-ldhD double-knockout strain of Lactobacillus plantarum. J Bacteriol. 1996;178:7311–7315. doi: 10.1128/jb.178.24.7311-7315.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasson M D, Davies F L. Conjugal transfer of the drug resistance plasmid pAMβ1 in the lactic streptococci. FEMS Microbiol Lett. 1980;7:51–53. [Google Scholar]
- 21.Hiyama T, Fukui S, Kitahara K. Purification and properties of lactate racemase from Lactobacillus sake. J Biochem. 1968;64:99–107. doi: 10.1093/oxfordjournals.jbchem.a128870. [DOI] [PubMed] [Google Scholar]
- 22.Holo M Y, Nes I F. High frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton R M. PCR-mediated recombination and mutagenesis. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- 24.Hudson M, Pocknee R, Mowat N A. d-Lactic acidosis in short bowel syndrome—an examination of possible mechanisms. Q J Med. 1990;74:157–163. [PubMed] [Google Scholar]
- 25.Karton M, Rettmer R L, Lipkin E W. Effect of parenteral nutrition and enteral feeding on d-lactic acidosis in a patient with short bowel. J Parenter Enteral Nutr. 1987;11:586–589. doi: 10.1177/0148607187011006586. [DOI] [PubMed] [Google Scholar]
- 26.Kochhar S, Chuard N, Hottinger H. Cloning and overexpression of the Lactobacillus bulgaricus NAD+-dependent d-lactate dehydrogenase gene in Escherichia coli: purification and characterization of the recombinant enzyme. Biochem Biophys Res Commun. 1992;185:705–712. doi: 10.1016/0006-291x(92)91683-h. [DOI] [PubMed] [Google Scholar]
- 27.Kochhar S, Chuard N, Hottinger H. Glutamate 264 modulates the pH dependence of the NAD-dependent d-lactate dehydrogenase. J Biol Chem. 1992;267:20298–20301. [PubMed] [Google Scholar]
- 28.Kochhar S, Hottinger H, Chuard N, Taylor G, Atkinson T, Scawen M D, Nichols D L. Cloning and overexpression of Lactobacillus helveticusd-lactate dehydrogenase gene in Escherichia coli. Eur J Biochem. 1992;208:799–805. doi: 10.1111/j.1432-1033.1992.tb17250.x. [DOI] [PubMed] [Google Scholar]
- 29.Link-Amster H, Rochat F, Saudan K Y, Mignot O, Aeschlimann J M. Modulation of a specific humoral immune response and changes in intestinal flora mediated through fermented milk intake. FEMS Immun Med Microbiol. 1994;10:55–63. doi: 10.1111/j.1574-695X.1994.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 30.Luchansky J B, Muriana P M, Klaenhammer T R. Application of electroporation for transfer of plasmid DNA to Lactobacillus, Lactococcus, Listeria, Pediococcus, Bacillus, Staphylococcus, Enterococcus and Propionibacterium. Mol Microbiol. 1988;2:637–646. doi: 10.1111/j.1365-2958.1988.tb00072.x. [DOI] [PubMed] [Google Scholar]
- 31.Marteau P, Pochart P, Bouhnik Y, Rambaud J C. The fate and effects of transiting, non-pathogenic microorganisms in the human intestine. World Rev Nutr Diet. 1993;74:1–21. doi: 10.1159/000422599. [DOI] [PubMed] [Google Scholar]
- 32.Mollet B, Knol J, Poolman B, Marciset O, Delley M. Directed genomic integration, gene replacement, and integrative gene expression in Streptococcus thermophilus. J Bacteriol. 1993;175:4315–4324. doi: 10.1128/jb.175.14.4315-4324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott A, Fay L B, Chaintreau A. Headspace determination and origin of the aroma impact compounds of yogurt flavor. J Agric Food Chem. 1997;45:850–858. [Google Scholar]
- 34.Rosenthal P, Pesce M. Long-term monitoring of d-lactic acidosis in a child. J Pediatr Gastroenterol Nutr. 1985;4:674–676. [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satoh T, Narisawa K, Konno T, Katoh T, Fujiyama J, Tomoe A, Metoki K, Hayasaka K, Tada K, Ishibashi M, Yamane N, Mitsuoka T, Benno Y. d-Lactic acidosis in two patients with short bowel syndrome: bacteriological analyses of the fecal flora. Eur J Pediatr. 1982;138:324–326. doi: 10.1007/BF00442509. [DOI] [PubMed] [Google Scholar]
- 38.Schiffrin E J, Rochat F, Link-Amster H, Aeschlimann J M, Donnet-Hugues A. Immunomodulation of human blood cells following the ingestion of lactic acid bacteria. J Dairy Sci. 1995;78:491–497. doi: 10.3168/jds.S0022-0302(95)76659-0. [DOI] [PubMed] [Google Scholar]
- 39.Schiffrin E J, Brassart D, Servin A L, Rochat F, Donnet-Hughes A. Immune modulation of blood leukocytes in humans by lactic acid bacteria: criteria for strain selection. Am J Clin Nutr. 1997;66:515S–520S. doi: 10.1093/ajcn/66.2.515S. [DOI] [PubMed] [Google Scholar]
- 40.Scully T B, Kraft S C, Carr W C, Harig J M. d-Lactate-associated encephalopathy after massive small-bowel resection. J Clin Gastroenterol. 1989;11:448–451. doi: 10.1097/00004836-198908000-00020. [DOI] [PubMed] [Google Scholar]
- 41.Stetter K O, Kandler O. Formation of dl-lactic acid by lactobacilli and characterization of a lactic acid racemase from several streptobacteria. Arch Mikrobiol. 1973;94:221–247. [PubMed] [Google Scholar]
- 42.Taguchi H, Ohta T. d-Lactate dehydrogenase is a member of the d-isomer specific 2-hydroxyacid dehydrogenase family: cloning, sequencing and expression in Escherichia coli of the d-lactate dehydrogenase gene of Lactobacillus plantarum. J Biol Chem. 1991;266:12588–12594. [PubMed] [Google Scholar]
- 43.Triflia T, Peterson M G, Kemp D J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker D C, Klaenhammer T R. Isolation of a novel IS3 group insertion element and construction of an integration vector for Lactobacillus spp. J Bacteriol. 1994;176:5330–5340. doi: 10.1128/jb.176.17.5330-5340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei M Q, Rush C M, Norman J M, Hafner L M, Epping R J, Timms P. An improved method for the transformation of Lactobacillus strains using electroporation. J Microbiol Methods. 1995;21:97–109. [Google Scholar]
- 46.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]