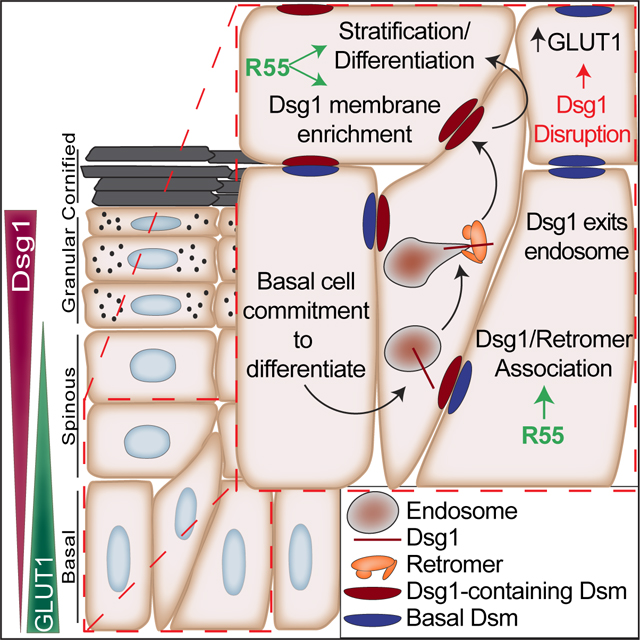
SUMMARY
Sorting transmembrane cargo is essential for tissue development and homeostasis. However, mechanisms of intracellular trafficking in stratified epidermis are poorly understood. Here we identify an interaction between the retromer endosomal trafficking component, VPS35, and the desmosomal cadherin, desmoglein-1 (Dsg1). Dsg1 is specifically expressed in stratified epidermis and, when properly localized on the plasma membrane of basal keratinocytes, promotes stratification. We show that the retromer drives Dsg1 recycling from the endo-lysosomal system to the plasma membrane to support human keratinocyte stratification. The retromer-enhancing chaperone, R55, promotes the membrane localization of Dsg1 and a trafficking-deficient mutant associated with a severe inflammatory skin disorder, enhancing its ability to promote stratification. In the absence of Dsg1, retromer association with and expression of the glucose transporter GLUT1 increases, exposing a potential link between Dsg1 deficiency and epidermal metabolism. Our work provides evidence for retromer function in epidermal regeneration, identifying it as a potential therapeutic target.
Keywords: desmosome, keratinocyte, epidermal differentiation, endosomal trafficking, retromer chaperone, VPS35, SAM syndrome, cadherin, Dsg1, GLUT1
eTOC Blurb
Hegazy et al. identify the endosomal trafficking complex, the retromer, as a regulator of epidermal development and homeostasis through its regulation of the important disease target, a desmosomal cadherin called desmoglein 1. Their work identifies the retromer as possible therapeutic target for a systemic skin disorder.
Graphical Abstract
INTRODUCTION
The epidermis is a regenerating multi-layered tissue composed predominantly of keratinocytes that undergo a highly orchestrated process of differentiation and stratification to create an essential barrier that protects against water loss, pathogens, and allergens.1 An important contributor to the process of stratification is the release of committed basal cells from the basement membrane and transit into the suprabasal layers, a process called delamination.2–5 Restriction of cell spreading and associated junction remodeling promote dynamic changes in cortical tension to drive delamination.6 While the importance of junctional membrane traffic in simple epithelial remodeling is well established, the contribution of specific trafficking machinery to the process of epidermal commitment to differentiation is poorly understood. Further, much of the focus on junction remodeling has been on integrin- and classic cadherin-based adhesions.7,8 Less attention has been paid to the desmosomal cadherin-intermediate filament cytoskeleton in these remodeling processes.
Desmosomes are intercellular junctions required for epidermal integrity and control of cell mechanics in tissues subjected to high mechanical stress such as the skin and heart.9 The desmosomal cadherins are divided into two subclasses, desmogleins and desmocollins, for a total of at least seven protein isoforms. One such isoform, desmoglein-1 (Dsg1), is first expressed during the basal to suprabasal commitment, promoting keratinocyte transit into the next superficial layer on its journey to the skin’s surface.10 Dsg1’s expression gradually increases throughout differentiation, reaching its highest level in the outer living layers of the epidermis.11,12
We recently showed that Dsg1 and intermediate filament connections are required for changes in cell mechanics leading to delamination of committed basal cells.10,13 Dsg1’s function in delamination requires its proper localization on the plasma membrane in detergent insoluble regions segregated from the adherens junction (AJ) protein E-cadherin.10 The importance of Dsg1 trafficking and proper placement on the plasma membrane is also underscored by the existence of human inherited and autoimmune disorders.14 For instance, mutations in the Dsg1 signal sequence that impair Dsg1 trafficking and accumulation on the plasma membrane cause the severe systemic inflammatory disorder, Severe dermatitis, multiple Allergies, and Metabolic wasting (SAM) syndrome.15 While these observations illustrate the importance of Dsg1 localization for normal epidermal homeostasis, there are substantial gaps in our understanding of the mechanisms by which Dsg1 is delivered to the plasma membrane.
A BioID protein interaction screen revealed VPS35, a component of a trafficking complex called the retromer, as a potential Dsg1 interacting protein. The retromer functions in endosomal trafficking including retrograde trafficking from the endosome-to-trans-Golgi network (TGN), polarized transport of membrane proteins, and endosome-to-plasma membrane recycling.16,17 It is composed of a trimer consisting of VPS35-VPS26-VPS29 with a role in cargo selection and a Snx component, which plays a role in promoting the formation of dynamic tubulovesicular structures for protein transport.18 While the importance of the retromer is highlighted by its dysfunction in neuronal diseases such as Parkinson’s, Alzheimer’s and amyotrophic lateral sclerosis (ALS),19 information about the retromer in the epidermis is limited to a role in Human Papilloma Virus (HPV) trafficking.20
Multiple retromer cargo proteins have been identified in vitro and a role for the retromer in trafficking cell surface recycling transporters of solutes such as glucose and glutamine is emerging.21 One of the first solute carriers identified to undergo retromer-mediated endosome-to-plasma membrane recycling is glucose transporter 1 (GLUT1).22 GLUT1 is highly expressed in keratinocytes and known to support basal cell proliferation, where Dsg1 expression is lowest.23 Interestingly, GLUT1 is observed to be upregulated in the skin of psoriasis patients, while conversely, Dsg1 was downregulated in lesional psoriatic skin.23–25 The importance of this reciprocal pattern of Dsg1 and GLUT1 expression in the epidermis has not been explored.
Here we demonstrate that the retromer promotes Dsg1 recycling and accumulation on the plasma membrane to promote human keratinocyte stratification, which is enhanced by a small molecule retromer chaperone called R55. In addition, R55 restores the plasma membrane localization of SAM syndrome-associated Dsg1 (SAM-Dsg1), which has a mutation in its signal sequence. Using a Dsg1 knockout mouse and SAM patient skin, we found Dsg1 disruption is associated with increased GLUT1 plasma membrane localization and retromer association, while Dsg1 overexpression results in GLUT1 cytoplasmic accumulation. These findings raise the possibility that the onset of Dsg1 expression during epidermal differentiation may initiate a switch in retromer cargo trafficking, decreasing GLUT1 plasma membrane localization and activity. These data highlight the retromer’s importance in epidermal regeneration and raise the possibility that compounds that modulate the activity of the retromer may be used clinically in the future to ameliorate symptoms in inflammatory skin disorders.
RESULTS
Endosomal trafficking inhibition disrupts plasma membrane localization of Dsg1, but not E-cadherin.
Dsg1 incorporates into desmosomes that are segregated from E-cadherin in AJs to promote actin cytoskeletal reorganization in cells committing to differentiate.10 This segregation could occur at least in part through utilization of distinct trafficking mechanisms by Dsg1 and E-cadherin. To identify potential mediators of differential trafficking, we carried out parallel BioID screens using Dsg1 and E-cadherin biotin ligase BirA* fusion proteins expressed in differentiated keratinocytes (Figure S1A, B, C; Table S1–S3).26 The retromer component, VPS35 was identified in the Dsg1 screen, but not the E-cadherin screen, which was confirmed with a benchtop BioID (Figure S1D). Identification of VPS35 as a potential binding partner was an indication that Dsg1 may require endosomal-mediated transport for its plasma membrane localization. To test the importance of endosomal trafficking on Dsg1 plasma membrane localization, we treated differentiated keratinocytes with primaquine, a weak base that prevents proteins from exiting the endosome.27 Primaquine treatment significantly diminished endogenous and ectopic Dsg1 levels at cell-cell interfaces with no change to E-cadherin (Figures 1A, B, E and S2A–D, I). In addition, there was a significant accumulation of EEA+ endosomal Dsg1, but not E-cadherin (Figures 1C–E and S2E–H). These data suggest that Dsg1’s localization in the desmosomes of keratinocytes requires distinct endosomal-mediated trafficking steps not used by E-cadherin.
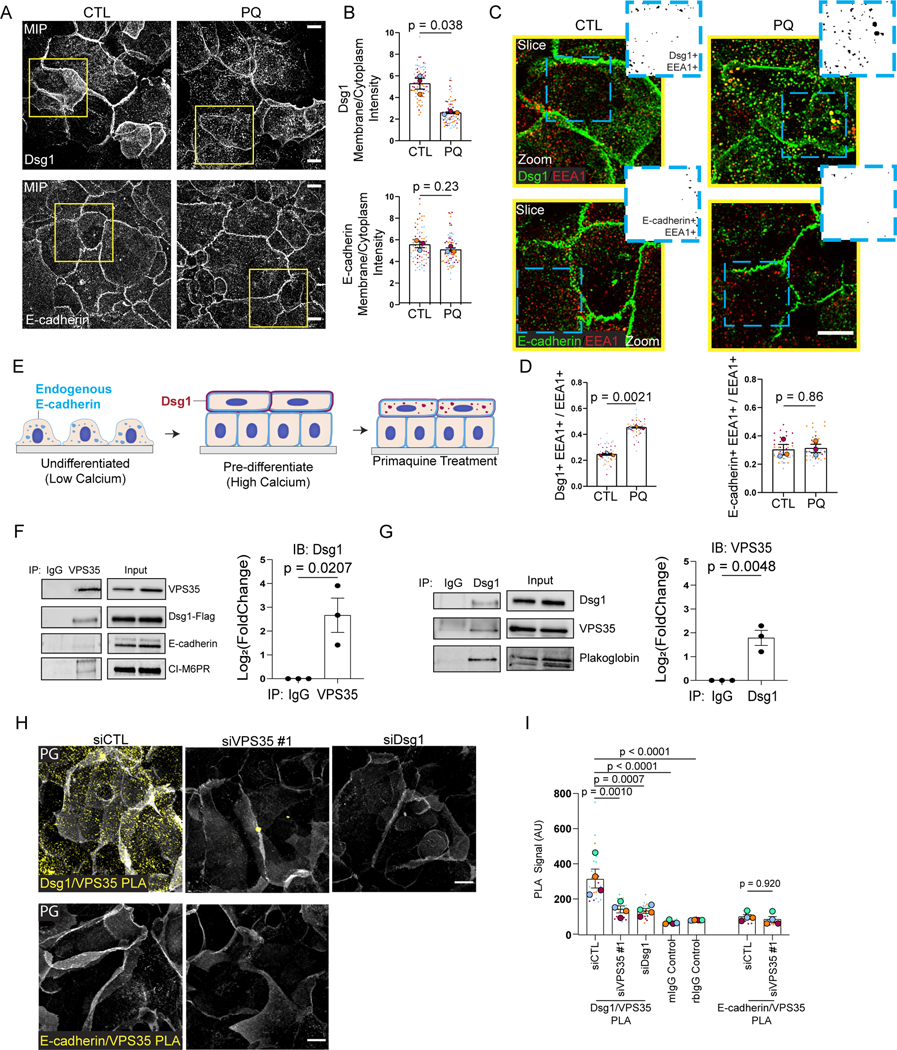
Figure 1. Dsg1 interacts with the endosomal trafficking retromer component, VPS35.
(A) Maximum intensity projection (MIP) of Dsg1 or E-cadherin immunofluorescence following overnight 200μM primaquine (PQ) treatment of pre-differentiated keratinocytes.
(B) Quantification of average Dsg1 and E-cadherin membrane/cytoplasmic ratio following PQ treatment.
(C) 1 μm slice and zoom of confocal images (yellow box in (A)) with dual staining of EEA1 (red) and Dsg1 or E-cadherin (green). Inset: Zoom of the area (blue dashed box). Auto-threshold was implemented using ImageJ; black puncta illustrate the region of overlap between EEA1 signal with Dsg1 or E-cadherin.
(D) Quantification of fraction of EEA1+/Dsg1+ or EEA1+/E-cadherin+ vesicles per field following PQ treatment.
(E) Summary of the impact of PQ treatment on endogenous Dsg1 (red on the plasma membrane in stratified cells and vesicles in PQ treated cells) and E-cadherin (blue in vesicles and at the plasma membrane) localization after the initiation of keratinocyte differentiation.
(F, G) Co-immunoprecipitation using an anti-VPS35 antibody (F) and immunoblotting for Dsg1-FLAG, E-cadherin, and CI-M6PR (positive control) and co-immunoprecipitation using an anti-Dsg1 antibody (G) and immunoblotting for VPS35, and plakoglobin (PG, positive control). Quantification of the Dsg1 and VPS35 to the right of each immunoblot.
(H) Proximity Ligation Assay (PLA) using an anti-VPS35 antibody paired with an anti-Dsg1 or anti-E-cadherin antibody showing VPS35 proximity with endogenous Dsg1 or E-cadherin in situ (yellow). siDsg1, siVPS35 or scramble control (siCTL) demonstrates signal specificity (Additional controls, Figure S3). PG staining was included to visualize cell-cell membranes (white).
(I) Quantification of the Dsg1-VPS35 PLA and E-cadherin-VPS35 PLA compared to IgG control and/or siRNA knockdown controls.
Scale bars are all 20μm. Statistics from at least three biological repeats using unpaired (F, G), paired (B, D) t-test, or one-way ANOVA with Dunnett post-hoc test (I). Error bars are all SEM.
Dsg1 interacts with the essential retromer component VPS35.
The canonical function for VPS35 is cargo recognition,16 therefore to test if Dsg1 is retromer cargo, we immunoprecipitated VPS35 in keratinocytes expressing Dsg1-FLAG and evaluated proteins in the complex. Dsg1-FLAG co-immunoprecipitated with VPS35, whereas E-cadherin was only weakly detectable (Figure 1F). Cation-independent Mannose-6 Phosphate Receptor (CI-M6PR), which undergoes retromer-dependent endosome-to-TGN trafficking, co-immunoprecipitated with VPS35 as expected.28 Similarly, endogenous VPS35 co-immunoprecipitated with endogenous Dsg1 in differentiated keratinocytes (Figure 1G).
To verify that endogenous Dsg1 and VPS35 are in close proximity in situ, a Proximity Ligation Assay (PLA) was performed on differentiated keratinocytes. A Dsg1/VPS35 antibody pairing produced a positive PLA signal in stratified cells where Dsg1 is highly concentrated, when compared with controls (Figures 1H, I and S3A–D; #1 refers to 1 of 2 siVPS35 oligos used in this paper), whereas an E-cadherin/VPS35 antibody pairing did not produce a PLA signal (Figures 1H, I and S3A–D). These data suggest Dsg1 is retromer cargo recognized by the retromer component VPS35 in stratified keratinocytes.
Loss of the retromer results in the accumulation of Dsg1 in the endo-lysosomal system.
The observations that Dsg1 interacts with VPS35 and that endosomal trafficking is necessary for Dsg1 plasma membrane localization suggest that Dsg1 may utilize the retromer for trafficking to the plasma membrane. To test this hypothesis, VPS35 and another essential retromer component, VPS29, were depleted in cells and the amount of plasma membrane localized Dsg1 was quantified. Loss of retromer components resulted in a decrease in surface localized Dsg1 while no change in E-cadherin surface levels was observed (Figures 2A, B and S4A). The retromer is necessary for the tubulation and fission of endosomes to promote cargo exit from the lysosomal degradative pathway.18,29 Live cell imaging of keratinocytes revealed highly dynamic Dsg1-GFP positive tubules extending from vesicles, which became more rounded and less elongated in cells silenced for VPS35 or VPS29 (Figure 2C, D; Movie 1). These data suggest that the retromer is necessary for Dsg1 localization in endosomal tubules and exit out of the endosome.
Figure 2. Retromer component depletion disrupts Dsg1 plasma membrane localization resulting in Dsg1 endo-lysosomal accumulation.
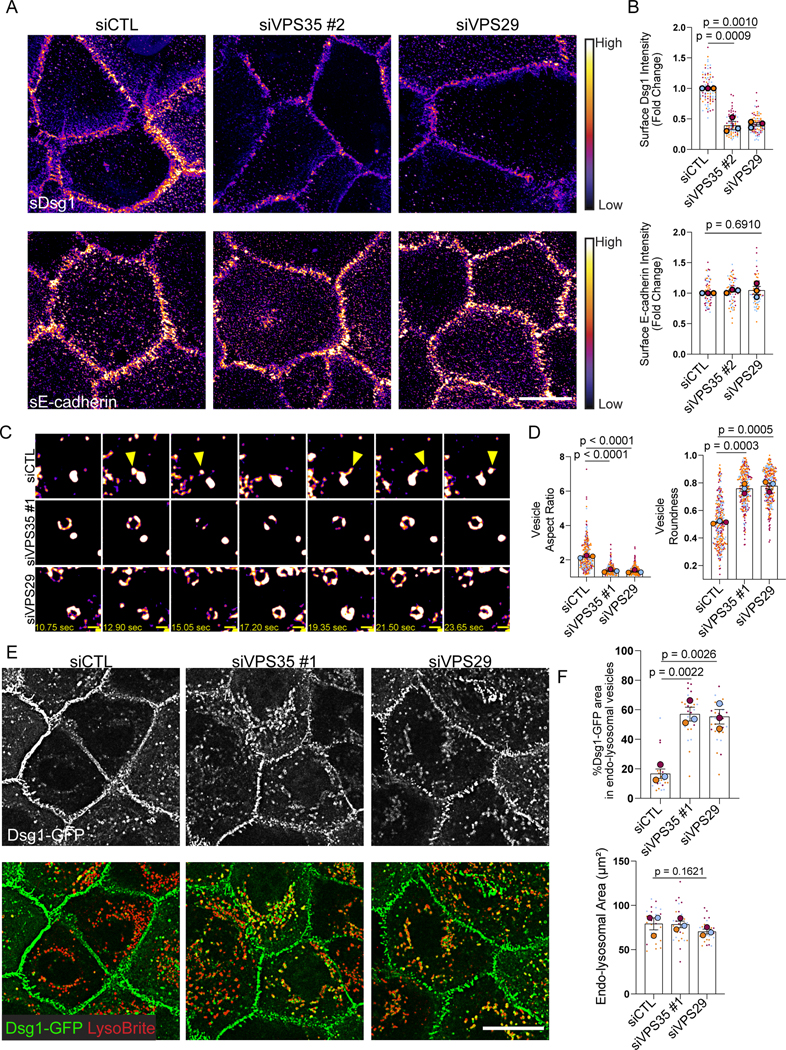
(A) Living cells were surface-labeled for Dsg1 or E-cadherin following siRNA-mediated depletion of VPS35 or VPS29 and compared to scramble transfected control (siCTL). Images were pseudocolored using ImageJ with a Fire look up table. Scale bar, 20 μm.
(B) Quantification of membrane Dsg1 or E-cadherin.
(C) Movie stills from Dsg1-GFP live cell imaging. Yellow arrowheads highlight Dsg1 localization in vesicle tubules and projections. Pseudocolored as in (A). Refer to Supplemental materials to view the movie. Scale bar, 1 μm.
(D) Quantification of Dsg1-GFP vesicle aspect ratio and roundness.
(E) Immunofluorescence of Dsg1 and endo-lysosomal marker, LysoBrite, in live cells following retromer component depletion compared to scramble control (siCTL). Scale bar, 20 μm.
(F) Quantification of the % area of Dsg1 in endo-lysosomal vesicles and the area of LysoBrite labeled endo-lysosomes.
Statistics from three biological repeats using one-way ANOVA with Dunnett post-hoc test. Error bars are all SEM.
To test if Dsg1 becomes trapped in the endo-lysosomal system upon loss of the retromer, LysoBrite was used to label acidic organelles. Consistent with the idea that Dsg1 requires the retromer for its exit from the endosome, depletion of VPS35 or VPS29 resulted in the accumulation of ectopic Dsg1-GFP in the endo-lysosomal system in basal keratinocytes (Figure 2E, F). Similarly, in stratified cells, endogenous Dsg1 displayed enhanced colocalization with the endo-lysosomal marker, LAMP1, following retromer component depletion (Figure S4B, C). Glucose transporter 1 (GLUT1), another retromer cargo, exhibited increased localization in LAMP1-positive vesicles in basal keratinocytes in the absence of VPS35 or VPS29 (Figure S4B, C).
Similar to what has previously been reported, we observed that expression of VPS35 or VPS29 was necessary to maintain protein levels of the other retromer components (Figure S4A, D).30 While it might be predicted that Dsg1 levels would decrease due to sequestration and degradation in lysosomes, we saw such a decrease inconsistently. The fact that loss of the retromer has been reported to reduce lysosomal proteolytic capacity could explain this observation.31 Interestingly, retromer depletion in Dsg1-GFP expressing keratinocytes occasionally resulted in increased extracellular cleavage, which could reflect lysosomal or MMP/ADAM dependent cleavage (Figure S4D). Altogether, these data suggest that Dsg1 requires the retromer for its exit from the endo-lysosomal system and for its plasma membrane localization.
Dsg1 utilizes the retromer for endosome-to-plasma membrane recycling.
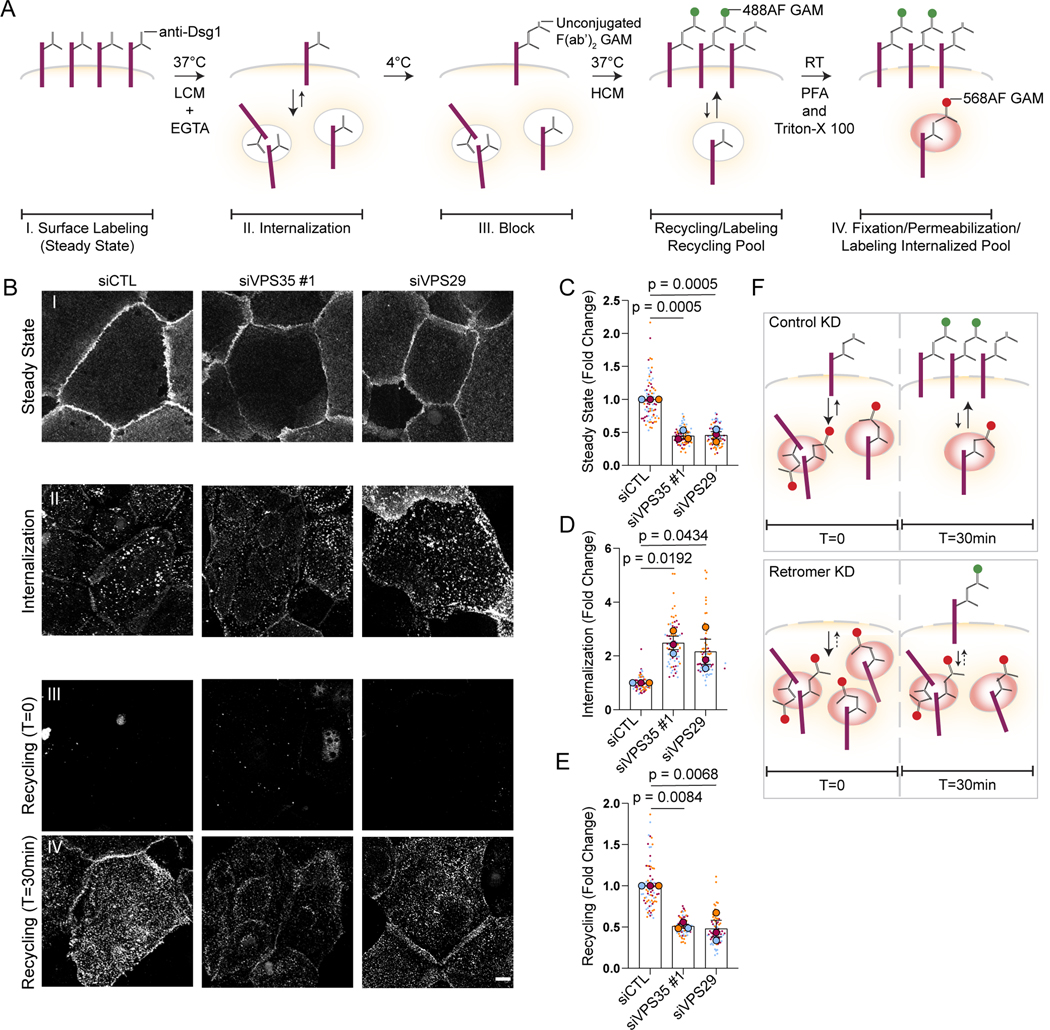
The retromer has been reported to promote endosome-to-plasma membrane recycling of some retromer cargo such as GLUT1.32 Based on the observation that loss of the retromer resulted in decreased levels of plasma membrane Dsg1, we tested the role of the retromer in Dsg1 recycling to the plasma membrane using an antibody-based recycling assay (Figure 3A). After surface labeling cells with an anti-Dsg1 monoclonal antibody (I), internalization was induced using low calcium media for 30min at 37°C (II). Following internalization, remaining surface molecules were blocked using an unconjugated goat anti-mouse F(ab’)2 (III). The cells were then returned to 37°C in high calcium media to induce recycling, and this pool was recognized by surface staining with a Alexa Fluor (AF) 488 conjugated goat anti-mouse secondary antibody (IV). Internalized Dsg1 was visualized with a AF568 conjugated goat anti-mouse secondary antibody after fixation and permeabilization (IV). Surface Dsg1 was reduced in retromer depleted cells at steady state, resulting in a 2-fold higher pool of labeled Dsg1 in control compared with VPS35 or VPS29 knockdown cells (Figure 3B, C). Correspondingly, an increased level of Dsg1 was observed in the internalized pool in retromer depleted cells when normalized to the starting surface pool. This increase might be explained by retention of Dsg1 in vesicles in the absence of the retromer instead of freely cycling back to the plasma membrane (Figure 3B, D). Finally, the plasma membrane associated recycled pool also decreased significantly upon loss of the retromer (Figure 3B, E). These data are consistent with the hypothesis that the retromer is necessary for Dsg1 endosomal recycling to the plasma membrane (Figure 3F).
Figure 3. The retromer is necessary for Dsg1 endosomal-to-plasma membrane recycling.
(A) Schematic of the antibody-based recycling assay shown in B.
(A, B) I. Dsg1 surface signal at steady state with or without siRNA-mediated depletion of retromer components. II. Dsg1 internalization signal in the 568 channel following fixation and permeabilization after 30min low calcium/EGTA treatment. III. Dsg1 remaining surface signal in the 488 channel after 30min low calcium/EGTA treatment (T=0). IV. Dsg1 recycling signal after 30min high calcium treatment following internalization. Scale bar, 10 μm.
(C-E) Quantification of Dsg1 surface signal at steady state (C), Dsg1 internalization (D), and Dsg1 recycling (E).
(F) Summary of the antibody-based recycling assay results showing that retromer loss impairs Dsg1 surface signal at steady state, disrupting Dsg1 recycling and enhancing Dsg1 internalization due to the lack of constitutive recycling.
Statistics from three biological repeats using one-way ANOVA with Dunnett post-hoc test. Error bars are all SEM.
Loss of the retromer results in abnormal differentiation.
While Dsg1’s function in epidermal differentiation has been established, the retromer’s role in this process has not been addressed. To test if retromer depletion affects epidermal differentiation, the human epidermis was reconstituted using a 3D organotypic epidermal “raft” culture model where primary keratinocytes are lifted to an air-liquid interface on a collagen/fibroblast plug (Figure S5A).33 When retromer components were depleted, H&E staining revealed a disruption in structural features of the rafts including decreased keratohyalin granules and a loss of the superficial cornified layer (Figure 4A). Raft cultures at 3 and 6–7 days after lifting were stained for Dsg1 and Keratin 10 (K10), a suprabasally expressed intermediate filament protein. In day 3 rafts, Dsg1 membrane localization and K10 expression was reduced and staining was frequently absent from the superficial layers where Dsg1 and K10 are typically expressed (Figures 4B–D and S5B, C). The appearance of basal-like cells in the superficial layers that retain the basal markers integrin β1, P-cadherin, and Ki67 was also observed, impairing the regular stacking of the multi-layered tissue, a phenotype not observed in Dsg1 depleted rafts (Figures 4B and S5D, E). The presence of superficial cells with a basal phenotype suggests that loss of the retromer results in the disruption in positional identity or a disconnect between stratification and differentiation signals, independent of Dsg1 loss at the plasma membrane. Retromer depleted day 6–7 rafts exhibited a partial restoration of K10 and a slight increase in Dsg1 membrane levels, as VPS35 and VPS29 expression began to return following knockdown (Figures 4B–D and S5B, C). These data suggest that loss of the retromer impairs epidermal differentiation through Dsg1-dependent and independent pathways.
Figure 4. Retromer depletion disrupts epidermal differentiation and Dsg1-mediated stratification.
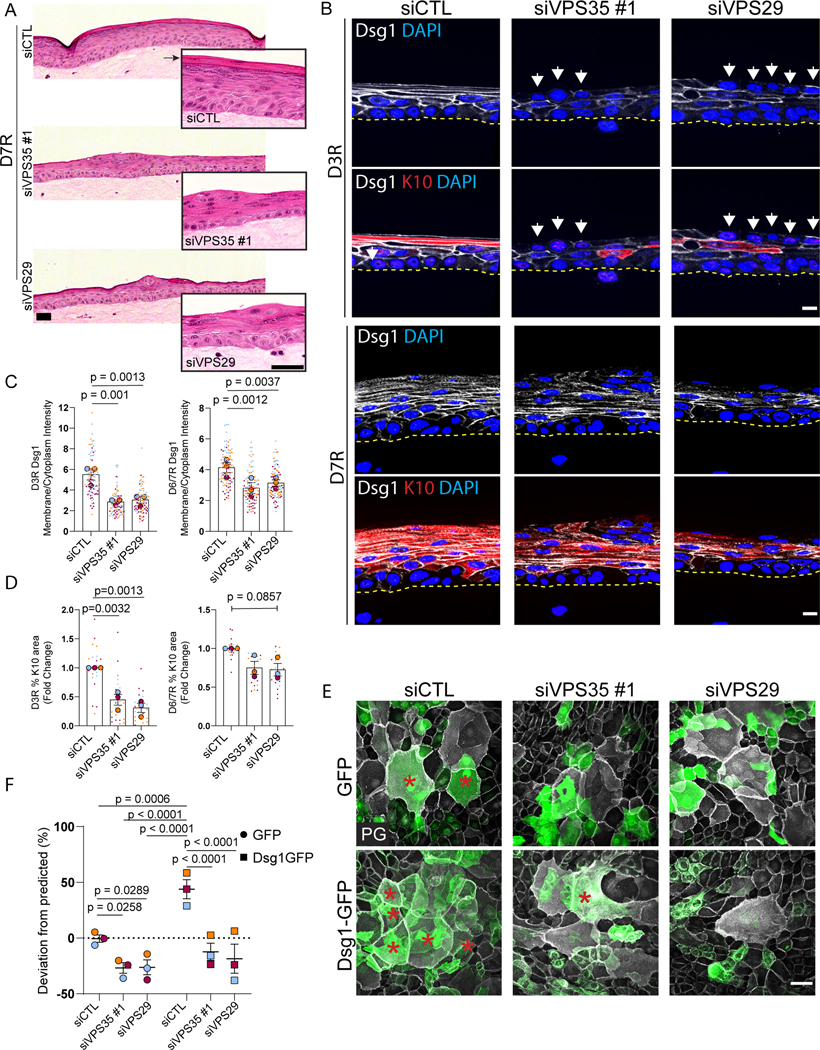
(A) H&E staining of 3D organotypic “raft” cultures harvested 7 days after lifting to an air-liquid interface (D7R). Scale bar, 100 μm. Inset: Representative higher magnification images. Arrow denotes the cornified layer. Scale bar, 50 μm.
(B) Tissue sections from 3D rafts stratified for 3 days (D3R) or 7 days (D7R) stained for Dsg1 and the differentiation marker Keratin 10 (K10). White arrows highlight basal-like cells in the superficial layers. Scale bar, 10 μm.
(C, D) Quantification of average Dsg1 membrane/cytoplasmic ratio (C) or % K10 area per field (D) in rafts harvested 3 days (D3R) and 6/7 days (D6/7R) after lift to air-liquid interface.
(E) GFP- and Dsg1-GFP labeled keratinocytes were transfected with siCTL, siVPS35, or siVPS29, mixed 50:50 with non-transfected, unlabeled keratinocytes and allowed to settle overnight in 0.07mM CaCl2 media. Cells were switched to 1.2mM CaCl2 media for 24hrs then fixed and stained with PG (white). Red asterisks mark GFP positive stratified cells. Scale bar, 50 μm.
(F) Quantification of deviation from predicted % of GFP cells or Dsg1-GFP cells in the stratified layer shown in E.
Statistics from three biological repeats using one-way ANOVA with Dunnett post-hoc test (C, D) or Tukey post-hoc test (F). Error bars are all SEM.
As Dsg1 promotes delamination of basal cells into the next superficial layer, we hypothesized that the observed impairment of differentiation may result in part from a reduction in stratification due to impaired Dsg1 trafficking in retromer-deficient cells.10 To test whether the retromer is required for efficient Dsg1-mediated stratification, we carried out a sorting assay that measures the ability of genetically modified GFP labeled cells to stratify when mixed in a 50:50 ratio with unmodified, unlabeled keratinocytes.13 Depletion of retromer components resulted in a decrease in the number of GFP positive keratinocytes that moved into the superficial layer, compared to the controls (Figures 4E, F and S5F). This could be due to loss of endogenous Dsg1 at the plasma membrane or due to the disruption of another retromer cargo component important for stratification. Ectopic expression of Dsg1-GFP in basal keratinocytes, which we previously showed promotes differentiation and stratification,13 enhanced stratification; however, retromer component depletion blocked this increase. These data support the hypothesis that the retromer is necessary for Dsg1-mediated keratinocyte stratification.
The retromer chaperone R55 promotes VPS35 association with and plasma membrane localization of WT Dsg1 and a disease-associated Dsg1 mutant.
Bi-allelic loss of function mutations in Dsg1 are associated with SAM syndrome, which typically features severe palmoplantar keratoderma and allergies. One of the originally reported mutations, c.49–1G>A, causes skipping of exon 2 in a region with 0.981 probability of encoding a signal peptide, which is important for targeting transmembrane molecules to the endoplasmic reticulum (ER) (Figure S6A, B).34 The c.49–1G>A SAM mutant has a 0.5477 probability of having a signal peptide. This SAM mutant is expected to inefficiently insert into the ER membrane, thus decreasing the pool of mature Dsg1 available to traffic to the plasma membrane. As the SAM mutant retains the predicted propeptide cleavage site RQKR, mutant Dsg1 that gets into the endoplasmic reticulum (ER) is likely to be processed correctly in the Golgi. To test this idea, we first assessed the extent to which Dsg1 missing 12 amino acids encoded by Exon 2 (SAM-Dsg1), which mimics the c.49–1 G>A mutation, accumulated on the plasma membrane. The SAM mutant exhibited decreased plasma membrane localization with a corresponding increase in cytoplasmic staining in primary keratinocytes (Figure 5A, B), recapitulating what has been reported for this mutant in patient tissues.15,35
Figure 5. Retromer function-enhancing chaperone, R55, increases WT- and SAM-Dsg1 plasma membrane localization and proximity to VPS35 in vitro and in vivo.
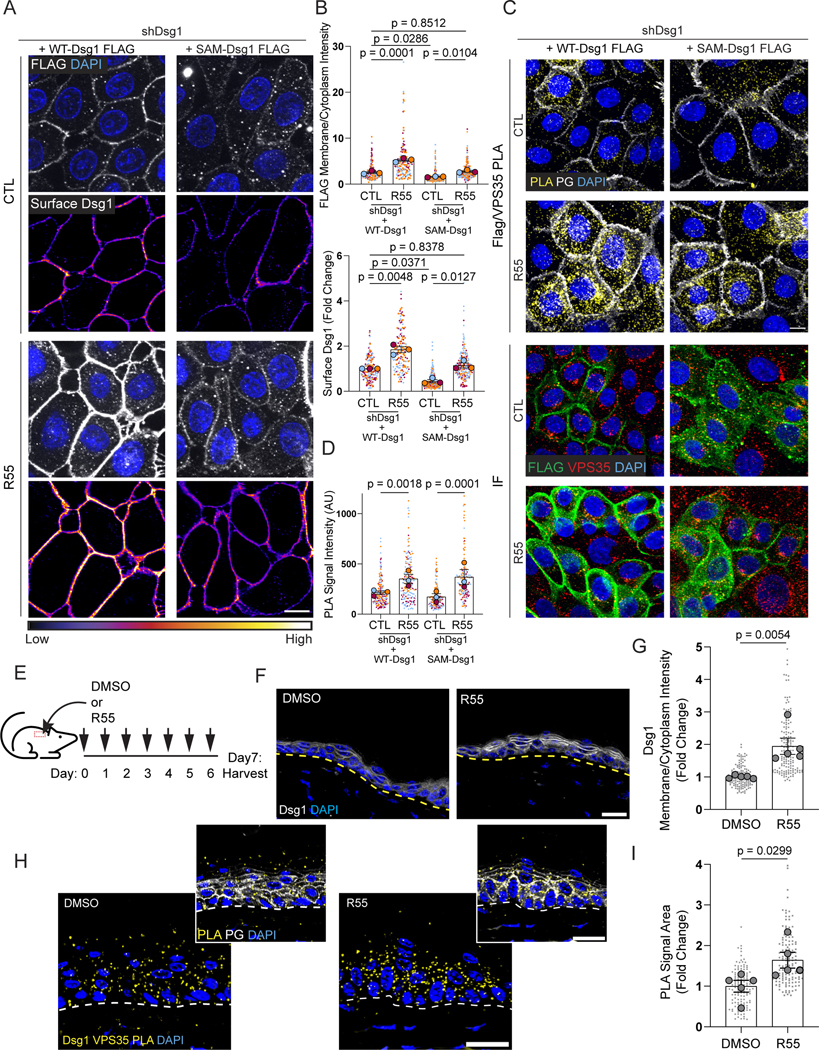
(A) Untreated (CTL) and R55-treated cells expressing FLAG-tagged WT- or SAM-Dsg1 were surface-labeled for Dsg1 in live cells with an anti-Dsg1 antibody. Cells were fixed, permeabilized, and stained with an anti-FLAG antibody. Surface Dsg1 was pseudocolored using ImageJ with a Fire look up table. Scale bar, 10 μm.
(B) Quantification of average surface Dsg1 and FLAG membrane/cytoplasmic ratio following R55 treatment.
(C) Top: Tandem PLA showing VPS35 proximity to FLAG-tagged WT- and SAM-Dsg1 with and without R55 treatment. PG staining labels cell-cell membranes (white). Bottom: Immunofluorescence staining (IF) using FLAG and VPS35 antibodies to confirm antibody recognition of the proteins of interest. Scale bar, 10 μm.
(D) Quantification of FLAG-VPS35 PLA +/− R55.
(E) Schematic of daily topical application of R55 or vehicle on mouse dorsal skin for 7 days.
(F) DMSO or R55 treated dorsal skin was stained with an anti-Dsg1 antibody and DAPI to label nuclei. Scale bar, 20 μm.
(G) Quantification of Dsg1 membrane/cytoplasmic ratio following R55 treatment of mice shown in (F).
(H) Tandem PLA showing Dsg1 proximity to VPS35 after R55 treatment compared to vehicle. Scale bar, 20 μm.
(I) Quantification of Dsg1-VPS35 PLA following R55 treatment of mice shown in (H) Statistics from at least three biological repeats (N=5 in mouse experiments) using one-way ANOVA with Sidak post-hoc test (B, D) or using unpaired t-test (G, I). Error bars are all SEM.
Because the retromer is necessary for Dsg1 exit from the endo-lysosomal system, we tested if enhancing retromer function with R55, a small molecule chaperone developed to stabilize and increase retromer function, could increase SAM-Dsg1 plasma membrane localization.36 Although there was no noticeable change in Dsg1 total protein levels with the addition of R55, the chaperone enhanced WT- and SAM-Dsg1 localization on the plasma membrane such that the level of SAM-Dsg1 on the plasma membrane was now comparable with untreated WT-Dsg1 (Figures 5A, B and S6C, D). This membrane enrichment by R55 corresponded with increased VPS35-Dsg1 association in keratinocytes expressing FLAG-tagged WT-Dsg1 or the SAM-Dsg1 mutant, as assessed using PLA with a VPS35-FLAG antibody pair (Figure 5C, D). These data illustrate that the SAM mutant can undergo retromer-mediated trafficking, and that R55 is sufficient to enhance WT- and SAM-Dsg1 on the plasma membrane in vitro.
We next tested the effect of topical R55 treatment on Dsg1 localization in vivo, in mouse epidermis (Figure 5E–G). Consistent with our in vitro results, immunofluorescence revealed an enrichment of plasma membrane localized Dsg1 in R55-treated epidermis that was accompanied by an increase in Dsg1/VPS35 PLA signal (Figure 5F–I). The retromer cargo, GLUT1’s membrane localization was enhanced with R55 treatment as expected, however E-cadherin localization did not change (Figure S6E, F). These data suggest that Dsg1 utilizes the retromer in vivo and that topical application of R55 is sufficient to enhance Dsg1 and GLUT1 at the plasma membrane.
The retromer chaperone R55 enhances Dsg1-dependent keratinocyte stratification.
Considering R55’s ability to increase cell surface Dsg1, we hypothesized that treatment with the retromer chaperone would enhance keratinocyte stratification. As expected, shRNA-mediated depletion of Dsg1 resulted in decreased stratification in vitro (Figures 6A, B and S6G). Addition of R55 enhanced the stratification of control GFP-labeled keratinocytes, but not Dsg1-depleted keratinocytes (Figures 6A, B and S6G). Ectopic expression of WT-Dsg1, which forces Dsg1 expression earlier than expression of endogenous Dsg1 in the unlabeled cells, rescued stratification of Dsg1-depleted keratinocytes above normal levels. R55 did not further enhance the number of stratifying cells under this condition, likely as a threshold was reached in cells ectopically expressing WT-Dsg1. Importantly, while the SAM-Dsg1 mutant rescued stratification to a certain extent in untreated cells, R55 significantly increased stratification up to or beyond levels induced by WT-Dsg1. Importantly, while PLA indicated that the desmosomal cadherins Dsg3 and Dsc2 are also in close proximity to VPS35 (Figure S7A, B), R55 promoted stratification only when Dsg1 was present (Figures 6C and S7C). These data reveal a nonredundant role for Dsg1 and its trafficking-deficient variant in epidermal stratification that depends on retromer function.
Figure 6. R55 enhances keratinocyte stratification in cells expressing Dsg1 or SAM-Dsg1.
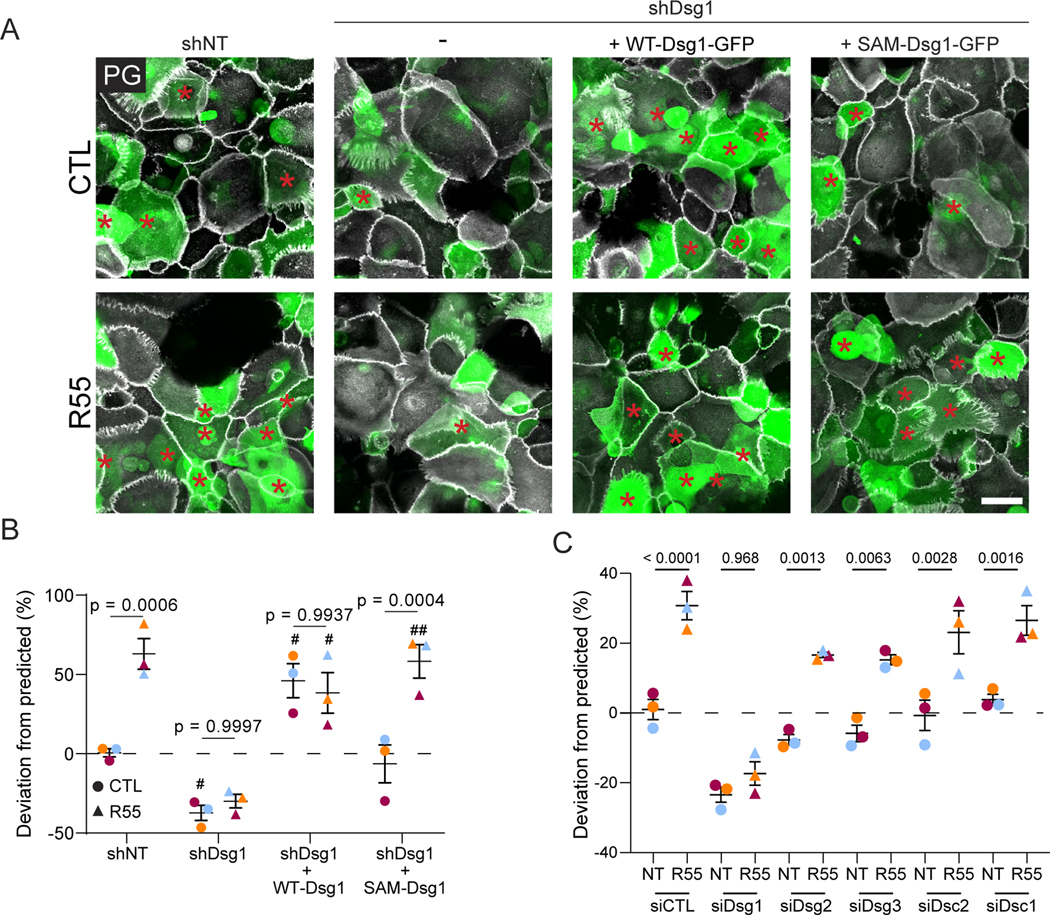
(A) Keratinocytes expressing non-targeting shRNA (shNT), shDsg1, shDsg1+ WT-Dsg1-GFP, or shDsg1+SAM-Dsg1-GFP were pretreated +/− R55 for 24hrs, mixed 50:50 with non-transfected, unlabeled cells and allowed to settle overnight in 0.07mM CaCl2 media +/− R55. Cells were switched to 1.2mM CaCl2 media +/− R55 for 24hrs then fixed and stained with PG (white). Red asterisks mark GFP positive stratified cells. Scale bar, 50 μm.
(B) Quantification of deviation from predicted % of GFP cells in the stratified layer. When comparing non-treated (CTL) and R55 treatments for each condition, p-values are shown. When comparing shDsg1 +/− WT-Dsg1 or SAM-Dsg1 to the control (shNT), significant pvalue <0.05 or <0.01 denoted with # or ##, respectively.
(C) Keratinocytes transfected with siCTL, siDsg1, siDsg2, siDsg3, siDsc2, siDsc1, were treated as described in (A). Shown is the quantification of deviation from predicted % of GFP cells in the stratified layer.
Statistics from three biological repeats using one-way ANOVA with Tukey post-hoc test. Error bars are all SEM.
Dsg1 expression in keratinocytes negatively regulates the retromer cargo, GLUT1.
The work described here exposes a previously unappreciated role for the retromer in promoting the stratification necessary to maintain a multi-layered epidermis. On the other hand, the retromer cargo GLUT1 is reported to regulate the proliferation of basal keratinocytes and exhibits a reciprocal expression pattern to Dsg1 in the skin of mice (Figures 5F, 7A and S6E).23 In agreement with this, Dsg1-VPS35 PLA shows a strong signal in suprabasal layers of the mouse epidermis, while GLUT1 staining is greatest in the basal layer (Figure 7A–C). Interestingly, GLUT1 membrane localization in the more superficial layers of E18.5 Dsg1 knockout epidermis was enhanced, accompanied by increased GLUT1-VPS35 PLA signal. GLUT1 expression has been shown to be upregulated in psoriasis while Dsg1 expression is downregulated.23,25 This raises the possibility that GLUT1 membrane localization and retromer association occurs as a result of an increase in retromer availability upon loss of Dsg1 in differentiating keratinocytes. To test this idea, we expressed Dsg1-GFP in primary basal keratinocytes and analyzed GLUT1 expression. Expression of Dsg1-GFP in basal keratinocytes resulted in increased cytoplasmic accumulation of GLUT1 (Figure 7D, E), suggesting that Dsg1 competes with GLUT1 for retromer association. In support of this hypothesis, immunohistochemistry of skin from 3 SAM syndrome patients harboring the c.49–1 G>A Dsg1 mutation demonstrated an increase in GLUT1 expression and retromer association in the suprabasal layers of the epidermis, consistent with the expression pattern observed in the Dsg1 knockout mouse (Figure 7F–H).15,35 These data support our observations that altering Dsg1 expression changes GLUT1 localization and retromer association in the epidermis, which may be associated with increased inflammation in skin diseases like SAM syndrome or the common disorder psoriasis.37
Figure 7. Dsg1 expression regulates retromer association with and localization of GLUT1.
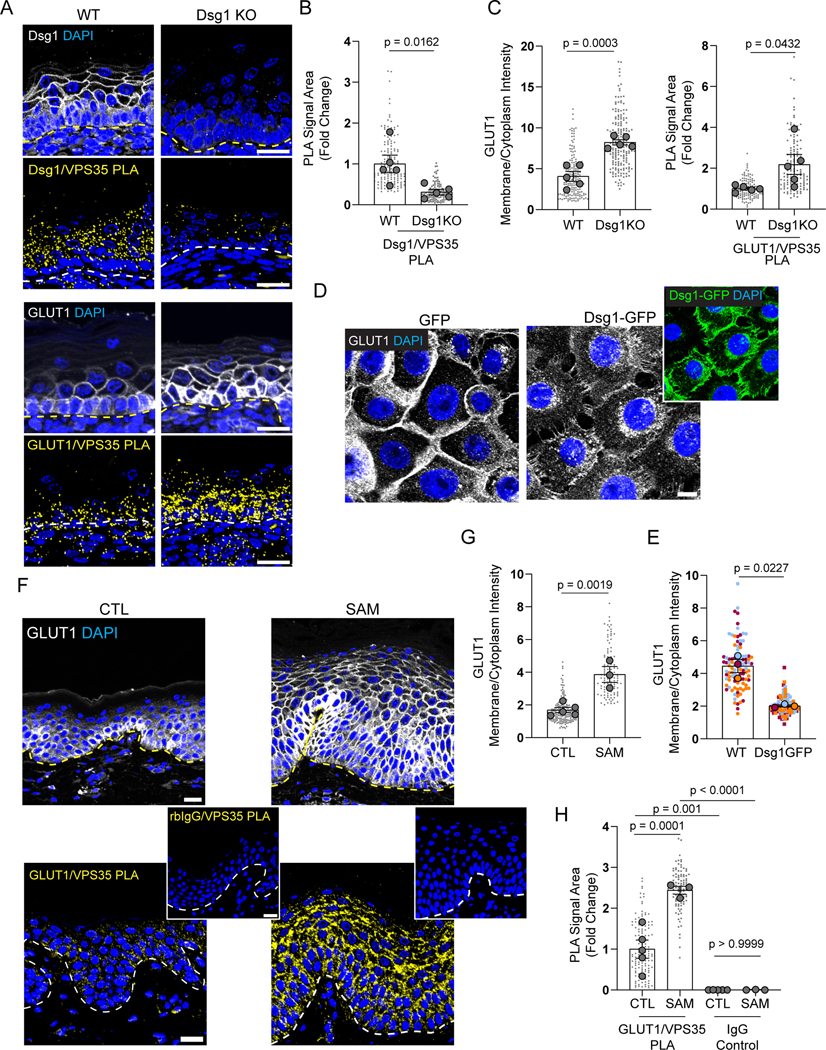
(A) Top: Immunofluorescence staining (IF) using an anti-Dsg1 antibody or an anti-GLUT1 antibody in wild type (WT) and Dsg1 knockout (KO) E18.5 epidermis. Bottom: Tandem PLA demonstrating Dsg1-VPS35 and GLUT1-VPS35 proximity. Scale bar, 20 μm
(B) Quantification of Dsg1-VPS35 PLA shown in (A) in WT and Dsg1 KO mice.
(C) Left: Quantification of GLUT1 membrane/cytoplasmic ratio in the spinous layer of WT and Dsg1 KO mouse epidermis. Right: Quantification of GLUT1-VPS35 PLA in WT and Dsg1 KO mice.
(D) Keratinocytes transduced with Dsg1-GFP were differentiated in 1.2mM CaCl2 media for 6hrs to over represent Dsg1 expression in basal cells. Cells were fixed and stained for GLUT1 and DAPI. Scale bar, 10 μm.
(E) Quantification of GLUT1 membrane/cytoplasmic ratio in basal keratinocyte +/− Dsg1-GFP.
(F) Normal control (CTL) and c.49–1 G>A SAM syndrome patient skin sections were stained with an anti-GLUT1 antibody and DAPI to mark nuclei (Top). Tandem PLA using antiGLUT1 anti-body paired with an anti-VPS35 antibody in normal control (CTL) and c.49–1 G>A SAM patient skin (Bottom). Scale bar, 20 μm.
(G, H) Quantification of (G) GLUT1 membrane/cytoplasmic ratio and (H) GLUT1-VPS35 PLA in normal control (CTL) and c.49–1 G>A SAM patient skin.
Statistics from at least three biological repeats (N=5 in mouse experiments) using unpaired (B, C, G, H) or paired (E) t-test. Error bars are all SEM.
DISCUSSION
Intracellular trafficking coordinates with changes in the cytoskeleton and cell polarity machinery to regulate developmental and morphogenetic processes as well as tissue homeostasis.38 Alterations in protein trafficking and consequent defects in cell polarity are known to impair barrier and transport functions in simple epithelia. Interfering with basolateral sorting, endocytosis, recycling and transcytosis can lead to diseases such as cystic fibrosis, Wilson disease, familial hypercholesterolemia and autosomal dominant polycystic kidney disease.39 Less is known about the importance of intracellular trafficking in polarized multilayered epithelia such as the epidermis. Depletion of Beclin 1 and SNAP29, which have roles in endocytic recycling, result in disrupted epidermal barrier and postnatal lethality in mice, phenotypes also exhibited by Dsg1 knockout animals.25,40–42 However, mechanisms for regulation of cell-cell junction proteins are not well-characterized in the epidermis. Here, we identify a role for the retromer trafficking machinery in regeneration of the multi-layered epidermis. We show that the retromer functions at least in part by its association with and recycling of desmosomal cadherin Dsg1 to drive epidermal stratification and differentiation. We show that the retromer can be exploited to enhance delivery and consequently function of a Dsg1 mutant associated with SAM syndrome.
The retromer was originally discovered in yeast to be required for retrieval of a sorting receptor called Vps10 from the endosome to the TGN,43 and is now well-established to be more broadly involved in processes of endosomal trafficking and recycling in metazoans.16,18,21 Since its discovery, the retromer has been shown to play essential roles in specific aspects of development, such as wing morphogenesis in flies, and establishing anterior/posterior neuronal polarity in C. elegans.44,45 While a study in HeLa cells identified ~150 retromer cargoes,22 in vivo, the retromer seems to be involved in selected pathways, for instance the Wnt, but not Hh, Notch or BMP pathways in Drosophila wing development.46–48 In mammals, depletion of retromer components affects specific aspects of early development, leading to embryonic lethality in early embryos, including delayed development of the embryonic ectoderm in VPS26a deficient animals.49,50 In humans, specific roles for the mammalian retromer are emerging due to the association of the complex with diseases such as ALS, Alzheimer’s and Parkinson’s disease.19 In complex epithelia, previous work established that the retromer is necessary for HPV infection in keratinocytes and identified a HPV specific peptide that inhibits the association of the retromer with the cellular cargo DMT1-II.19,20,51 However, until now its role in normal homeostasis of complex epithelia such as the epidermis has not been addressed.
Our data show that the retromer complex promotes endosomal trafficking of the stratified tissue-specific desmosomal cadherin Dsg1, but not the AJ cadherin E-cadherin, in epidermal keratinocytes. Although retromer-mediated trafficking seems to be specific to Dsg1 when compared to E-cadherin in keratinocytes, it has been reported that the retromer component Snx1 affected bulk-internalized E-cadherin recycling in simple epithelial cells.52 However, the authors were unable to find an interaction between E-cadherin and Snx1 and suggested that the effect on E-cadherin internalization is indirect, consistent with our finding that E-cadherin does not require retromer-mediated trafficking under steady state conditions. The specificity of the retromer machinery for Dsg1 and not E-cadherin could help explain how these proteins are segregated into different domains during keratinocyte differentiation to promote actin reorganization and tension redistribution required for delamination.10 Delamination is an important contributor to the process of stratification in conjunction with the asymmetric division of basal cells to establish and maintain the epidermal layer.2,3,10 The dynein light chain Tctex-1 was also found to be necessary for Dsg1’s proper localization on the insoluble plasma membrane, segregated from E-cadherin.10 As dynein has been shown to interact with the retromer to promote endosome-to-TGN trafficking,53,54 it seems plausible that the retromer works with dynein/Tctex-1 to tailor Dsg1-specific trafficking to promote proper plasma membrane localization required for Dsg1’s functions in morphogenesis.
Collectively, our observations provide an alternative perspective to the textbook view of desmosomes as stable, adhesive spot welds, supporting the emerging concept that these intercellular junctions function as dynamic complexes that undergo constant remodeling during epidermal regeneration.14,55,56 While not addressed in this study, it is also possible that the retromer contributes to patterning of cadherins in the epidermis by regulating the exchange of suprabasal desmosomal cadherins into desmosomes containing basal cadherins during a cell’s transit into superficial layers.57 The mechanism of this exchange is not clear but is likely important for establishing the patterned distribution of desmosomal cadherins. The data presented here suggest that the onset of Dsg1 expression results in a change in retromer cargo. Interestingly, depletion of Dsg1 in mice is associated with an increase in Dsg3, which in part may be due to transcriptional changes, but considering our data demonstrating that Dsg3 is in close proximity with the retromer suggests that there may be an increased association between Dsg3 and the retromer in the absence of Dsg1.25 The patterned distribution of desmosomal cadherins is functionally relevant.14 For example, during epidermal morphogenesis, the stratified tissue-specific Dsg1 regulates multiple processes important for maintaining epidermal homeostasis, including patterning the function of EGFR and ErbB2 to promote the basal to suprabasal transition while maintaining the tight junction barrier in the superficial layers.13,58
Our data suggest that Dsg1 promotes changes in retromer cargo either through release of retromer occupancy or via changes in cellular signaling. Dsg1 responds acutely to environmental stresses to provide keratinocytes a mechanism for adaptive responses,14 which may include increased transport of retromer cargo associated with metabolism and nutrient intake.59 In this report we demonstrate that in the absence of Dsg1 membrane trafficking, the glucose transporter GLUT1 increases at the membrane along with an increased association with the retromer. Further studies are needed to address the mechanism of retromer cargo distribution and the contributions that Dsg1 loss and increased GLUT1 association with the retromer make to the pathomechanisms of inflammatory skin diseases.23
Lastly, this work identified the retromer as a target for therapeutic intervention in skin disease. Most investigation into the retromer’s role in human health has been focused on neurological diseases and tauopathies.19 Furthermore, treatment of neurons in vitro and animal models for Alzheimer’s disease and ALS with the small molecule retromer chaperones have been shown to ameliorate common pathological features in neurological diseases.36,60–62 Here, we illustrate a role for the retromer as a therapeutic target for epidermal diseases, such as SAM syndrome. Targeting stress-enriched retromer cargo may be another method of treatment for inflammatory skin diseases. One such target could be GLUT1, which was found to play a role in UV-mediated stress response and psoriasiform hyperplasia in a psoriasis mouse model.23 Additionally, as Dsg1 is also disrupted in common disorders such as psoriasis, squamous cell carcinoma, and eosinophilic esophagitis, retromer therapy may have broad benefit beyond rare genetic disorders.14 This study reveals the importance of understanding Dsg1 trafficking mechanisms for determining underlying molecular mechanisms, and potentially treating, disorders of stratified epithelia.
Limitations of the Study
This study identifies the retromer as an important endocytic trafficking complex for Dsg1 plasma membrane localization in vitro and in vivo. A detailed analysis of epidermal-specific VPS35-depleted mice (or embryos if the phenotype is lethal) could elucidate Dsg1-dependant and independent retromer functions in epidermal differentiation. Furthermore, in vitro studies were performed using neonatal human keratinocytes isolated from foreskin, differences in differentiation and stratification based on age and sex have not been investigated.
STAR METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kathleen J. Green (kgreen@northwestern.edu).
Materials availability
Requests and further details for newly generated materials should be directed to the lead contact.
Data and code availability
BioID proteomics data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) partner repository: PXD036646. Accession number is also listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-VPS35 | Abcam | Cat# ab157220, RRID:AB_2636885 |
| rabbit anti-VPS35 | Life Technologies | Cat# PA5-21898, RRID:AB_11153540 |
| mouse anti-VPS35 | Santa Cruz Biotechnology | Cat# sc-374372, RRID:AB_10988942 |
| mouse anti-VPS29 | Santa Cruz Biotechnology | Cat# sc-398874, RRID:AB_2885036 |
| mouse anti-Dsg1 | PROGEN | Cat# 651111, RRID:AB_1541107 |
| mouse anti-Dsg1 | R&D Systems | Cat# MAB944, RRID:AB_2093296 |
| mouse anti-Dsg1 | Thermo Fisher Scientific | Cat# 32-6000, RRID:AB_2533088 |
| rabbit anti-Dsg1 | Abcam | Cat# ab124798, RRID:AB_10974963 |
| mouse anti- FLAG | Sigma-Aldrich | Cat# F1804, RRID:AB_262044 |
| chicken anti-plakoglobin | Aves Laboratories | 1407 and 1408 |
| mouse anti-EEA1 | BD Bioscience | Cat# 610456, RRID:AB_397829 |
| mouse anti-LAMP1 | Santa Cruz Biotechnology | Cat# sc-20011, RRID:AB_626853 |
| mouse anti-GFP | Takara | Cat# 632381, RRID: AB_2313808 |
| mouse anti-E-cadherin | M. Takeichi and O. Abe, Riken Center for Developmental Biology | HecD1 |
| mouse anti-E-cadherin | BD Biosciences | Cat# 61081, RRID:AB_397580 |
| rabbit anti-E-cadherin | R. Marsh and R. Brackenbury, University of Cincinnati | 795 |
| goat anti-E-cadherin | R&D Systems | Cat# AF748, RRID:AB_355568 |
| rabbit anti-β-catenin | Sigma | Cat# C2206, RRID:AB_476831 |
| rabbit anti-K10 | J. Segre, National Human Genome Research Institute | N/A |
| rabbit anti-M6PR (cation independent) | Abcam | Cat# ab124767, RRID:AB_10974087 |
| rabbit anti-GLUT1 | Abcam | Cat# ab115730, RRID:AB_10903230 |
| mouse anti-GLUT1 | Novus Biologicals | Cat# NBP2-75785, RRID:AB_2923123 |
| mouse anti-Dsg2 (6D8) and anti-Dsc2 (7G6) | J. Wahl III, University of Nebraska Medical Center | N/A |
| mouse anti-Dsg3 | Millipore Sigma | Cat# MABT335, RRID:AB_2923124 |
| mouse anti-Dsc1 | PROGEN | Cat# 61092, RRID:AB_1541100 |
| mouse anti-Integrin β1 | BD Biosciences | Cat# 610467, RRID:AB_2128060 |
| rabbit anti-Ki67 | EMD Millipore | Cat# AB9260, RRID:AB_2142366 |
| mouse anti-P-cadherin | M. Wheelock, University of Nebraska Medical Center | N/A |
| rabbit anti-HA-tag | Cell Signaling | Cat# 3724S, RRID:AB_1549585 |
| rabbit anti-GAPDH | Sigma-Aldrich | Cat# G9545, RRID: AB_796208 |
| mouse anti-tubulin | Developmental Hybridoma Studies Bank | 12G10, RRID: AB_1157911 |
| mouse anti-actin | Millipore Sigma | Cat# MAB1501, RRID:AB_2223041 |
| mIgG isotype control | Abcam | Cat# ab37355, RRID:AB_2665484 |
| rbIgG isotype control | Abcam | Cat# ab27478, RRID:AB_2616600 |
| Secondary goat anti-mouse, -rabbit, and -chicken HRP | SeraCare Life Sciences | N/A |
| Secondary goat anti-mouse conjugated with IRDye 680RD | LI-COR Biosciences | Cat# 926-68070, RRID:AB_10956588 |
| Secondary goat anti-mouse conjugated with IRDye 800CW | LI-COR Biosciences | Cat# 925-32210, RRID:AB_2687825 |
| Secondary goat anti-rabbit conjugated with IRDye 680RD | LI-COR Biosciences | Cat# 926-68071, RRID:AB_10956166 |
| Secondary goat anti-rabbit conjugated with IRDye 800CW | LI-COR Biosciences | Cat# 926-32211, RRID:AB_621843 |
| Secondary goat anti-mouse IgG1 isotype conjugated with IRDye 680LT | LI-COR Biosciences | Cat# 926-68050, RRID:AB_2783642 |
| Secondary goat anti-mouse IgG2a isotype conjugated with IRDye 800 CW | LI-COR Biosciences | Cat# 926-32351, RRID:AB_2782998 |
| Secondary goat anti-mouse IgG2a isotype conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-21131; RRID: AB_2535771 |
| Secondary goat anti-mouse IgG2a isotype conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21124; RRID: AB_2535766 |
| Secondary goat anti-mouse IgG1 isotype conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-21121; RRID: AB_2535764 |
| Secondary goat anti-mouse conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-11029; RRID: AB_2534088 |
| Secondary goat anti-mouse conjugated with Alexa Fluor-568 | Thermo Fisher Scientific | Cat# A-11004; RRID: AB_2534072 |
| Secondary goat anti-mouse conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-32728; RRID: AB_2633277 |
| Secondary goat anti-rabbit conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-11008; RRID: AB_143165 |
| Secondary goat anti-rabbit conjugated with Alexa Fluor-568 | Thermo Fisher Scientific | Cat# A-11011; RRID: AB_143157 |
| Secondary goat anti-rabbit conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21245; RRID: AB_2535813 |
| Secondary goat anti-chicken conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21449; RRID: AB_2535866 |
| Secondary donkey anti-goat conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-11055, RRID:AB_2534102 |
| Secondary donkey anti-goat conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21447, RRID:AB_2535864 |
| Secondary donkey anti-mouse conjugated with Alexa Fluor-568 | Thermo Fisher Scientific | Cat# A10037, RRID:AB_2534013 |
| Secondary goat anti-mouse AffiniPure F(ab’)2 Fragment unconjugated | Jackson Laboratory | Cat# 115-006-003, RRID:AB_2338466 |
| Secondary goat anti-mouse AffiniPure F(ab’)2 Fragment conjugated with Alexa Fluor-488 | Jackson Laboratory | Cat# 715-546-150, RRID:AB_2340849 |
| Bacterial and virus strains | ||
| pLZRS-GFP | Getsios et al.58 | N/A |
| pLZRS-WT-Dsg1-FLAG | Getsios et al.58 | N/A |
| pLZRS-shDsg1 | Getsios et al.58 | N/A |
| pLZRS-SAM Dsg1-FLAG | Cohen-Barak et al.35 | N/A |
| pLZRS-shNT (Non-targeting) | Arnette et al.63 | N/A |
| pLZRS-E-cadherin-mCherry | Godsel et al.64 | N/A |
| pLZRS-Dsg1-BirA* | This manuscript | N/A |
| pLZRS-E-cadherin-BirA* | This manuscript | N/A |
| pLZRS-WT-Dsg1-FLAG/GFP (miRNA Resistant) | This manuscript | N/A |
| pLZRS-SAM Dsg1-FLAG/GFP (miRNA Resistant) | This manuscript | N/A |
| Biological samples | ||
| Neonatal human foreskin | NU SBDRC | N/A |
| Control adult human skin biopsies | NU SBDRC | N/A |
| SAM syndrome patient c.49-1 G>A skin biopsies | Samuelov et al.15 and Cohen-Barak et al.35 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 4’,6-Diamido-2-Phenylindole (DAPI) | Sigma-Aldrich | Cat# D9542 |
| Dispase II | Roche | Cat# 04942078001 |
| Goat serum | Sigma-Aldrich | Cat# G9023 |
| LysoBrite NIR | AAT Bioquest | Cat# 22641 |
| R55 | Sigma-Aldrich | Cat# 531084 |
| R55 | MedKoo Biosciences | Cat # 504210 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat# D8418 |
| Primaquine bisphosphate | Sigma-Aldrich | Cat # 160393 |
| Ingenio Electroporation Solution | Mirus Bio | Cat# MIR 50117 |
| Polybrene | Sigma-Aldrich | Cat# H9268 |
| Albumin from bovine serum (BSA) | Sigma-Aldrich | Cat# A7906 |
| 16% Formaldehyde (w/v), Methanol-free | Thermo Fisher Scientific | Cat# PI28908 |
| Urea | Thermo Fisher Scientific | Cat# BP169-212 |
| Sodium Dodecyl Sulfate (SDS) | Thermo Fisher Scientific | Cat# BP166-500 |
| Tris Hydrochloride | Thermo Fisher Scientific | Cat# BP153-500 |
| Pyronin-Y | Sigma-Aldrich | Cat# P7017 |
| β-Mercaptoethanol | Sigma-Aldrich | Cat# M6250 |
| Ethylenediaminetetraacetic acid (EDTA) | Thermo Fisher Scientific | Cat# BP120-1 |
| Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) | Sigma-Aldrich | Cat# E4378 |
| Acrylamide/Bis-acrylamide, 30% solution | Sigma-Aldrich | Cat# A3574 |
| Formalin solution, neutral buffered, 10% | Sigma-Aldrich | Cat# HT501320 |
| Xylene | Thermo Fisher Scientific | Cat# X3P |
| Citric acid | Sigma-Aldrich | Cat# C0759 |
| Glycine | Sigma-Aldrich | Cat# G8790 |
| Methanol | Sigma-Aldrich | Cat# 179337 |
| Glycerol | Thermo Fisher Scientific | Cat# BP229 |
| Ethyl Alcohol, 200 proof | Thermo Fisher Scientific | Cat# A4094 |
| Triton X-100 | Sigma-Aldrich | Cat# X100 |
| Tween20 | Sigma-Aldrich | Cat# P1379 |
| ProLong Gold antifade reagent | Thermo Fisher Scientific | Cat# P36930 |
| PBS | Thermo Fisher Scientific | Cat# 21040CV |
| Opti-MEM I Reduced Serum Medium | Thermo Fisher Scientific | Cat# 11058021 |
| Intercept (PBS) Blocking Buffer | LI-COR Biosciences | Cat# 927-70001 |
| Sodium Chloride (NaCl) | Sigma-Aldrich | Cat# S9625 |
| Empigen | Sigma-Aldrich | Cat# 30326 |
| HEPES | Sigma-Aldrich | Cat# H4034 |
| Medium 154CF, Kit (M154): With gentamicin/amphotericin B solution and CaCl2 | Thermo Fisher Scientific | Cat# M154CF500 |
| Human Keratinocyte Growth Supplement (HKGS) Kit: | Thermo Fisher Scientific | Cat# S001K |
| FluoroBrite DMEM | Thermo Fisher | Cat# A1896701 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher | Cat# 11668019 |
| Streptavidin conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# S11223 |
| Streptavidin conjugated with HRP | Cell Signaling Technology | Cat# 3999 |
| Pierce Streptavidin Magnetic Beads | Thermo Fisher | Cat# 88816 |
| Dynabeads Protein G | Thermo Fisher | Cat# 10004D |
| Critical commercial assays | ||
| Duolink® In Situ PLA® Probe Anti-Rabbit PLUS | Sigma-Aldrich | Cat# DUO92002 |
| Duolink® In Situ PLA® Probe Anti-Mouse MINUS | Sigma-Aldrich | Cat# DUO92004 |
| Duolink® In Situ Detection Reagents Red | Sigma-Aldrich | Cat# DUO92008 |
| Duolink® In Situ Detection Reagents Far Red | Sigma-Aldrich | Cat# DUO92013 |
| Deposited data | ||
| Dsg1 and E-cadherin BioID | This manuscript | PXD036646 |
| Experimental models: Cell lines | ||
| Neonatal human epidermal keratinocytes (NHEKs) | N/A | Isolated from neonatal human foreskin |
| Phoenix cells | G. Nolan, Stanford University | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: wild-type C57BL/6 | The Jackson Laboratory | Strain #:000664 |
| Mouse: Dsg1−/− C57BL/6 | Godsel et al. 25 | N/A |
| Oligonucleotides | ||
| siGENOME non-targeting siRNA pool #2 | Dharmacon | cat# D-001206-14 |
| Silencer Select VPS35 siRNA (siVPS35 #1): 5’- CCATGGATTTTGTACTGCTTT-3’ | Sigma | Product # s31375 |
| VPS35 siRNA (siVPS35 #2): 5’- GAGATATTCAATAAGCTCAACCTTG-3’ | Integrated DNA Technologies | N/A |
| VPS29 siRNA: 5’- ATGATGTGAAAGTAGAACGAATCGA-3’ | Integrated DNA Technologies | N/A |
| Dsg1 siRNA: 5′-CCATTAGAGAGTGGCAATAGGATGA-3′ | Integrated DNA Technologies | N/A |
| Dsg2 siRNA: 5′- CCTGGAAGCAGAGACAGTGTGGTCCTT −3′ | Integrated DNA Technologies | N/A |
| siGENOME SMARTpool Dsg3 siRNA | Dharmacon | cat# M-011646-01-0005 |
| siGENOME SMARTpool Dsc1 siRNA | Dharmacon | cat# M-011995-01-0005 |
| Dsc2 siRNA: 5′-AGAGAGACACTATAAACAAGTACAC-3′ | Integrated DNA Technologies | N/A |
| Software and algorithms | ||
| LI-COR Image Studio (Version 5.2) | LI-COR Biosciences | RRID:SCR_015795 |
| ZEN 2.3 software | Carl Zeiss | RRID: SCR_013672 |
| NIS-Elements | Nikon | RRID: SCR_014329 |
| Leica Application Suite | Leica | RRID:SCR_016555 |
| ImageJ | NIH | RRID: SCR_003070 |
| Prism | GraphPad | RRID: SCR_002798 |
| Microsoft Excel | Microsoft | RRID: SCR_016137 |
| Photoshop | Adobe | RRID: SCR_014199 |
| Illustrator | Adobe | RRID: SCR_010279 |
| MaxQuant (Version 2.1.3.0) | Max Planck Institute of Biochemistry | RRID:SCR_014485 |
| Other | ||
| Amersham™ Protran® Western blotting membranes, nitrocellulose (0.2 μm pore size) | Cytiva | Cat# GE10600001 |
| Amersham™ Protran® Western blotting membranes, nitrocellulose (0.45 μm pore size) | Cytiva | Cat# GE10600002 |
| 4-Chamber 35mm glass bottom dish with 20 mm microwell, #1.5 cover glass | CellVis | Cat# D35C4-20-1.5-N |
| Nucleofector 2b Device | Lonza | Cat# AAB-1001 |
| Amicon Ultra-15 Centrifugal Filter Unit | Sigma-Aldrich | Cat# UFC9030 |
Experimental model and subject details
Neonatal human epidermal keratinocytes
Primary keratinocytes were isolated as previously described from human neonatal foreskin obtained from Northwestern University’s Skin Biology and Disease Resource-based Center (IRB Protocol #STU00009443).33 Undifferentiated keratinocytes were cultured in M154 media (Thermo Fisher) supplemented with 0.07mM CaCl2, human keratinocyte growth supplement (HKGS), gentamicin, and amphotericin B. To induce differentiation in 2D cultures, the calcium concentration of the growth medium was raised to 1.2mM CaCl2. 3D organotypic epidermal raft cultures using primary keratinocytes were generated as previously described.33 Drug treatments were performed with 200μM Primaquine (Sigma-Aldrich) for 6hr-overnight or 10μM R55 (Sigma-Aldrich) for 48–72hrs. Experiment with pre-differentiation, keratinocytes were differentiated for 1.5 days before being treated with 200μM primaquine overnight. During timelapse imaging, keratinocytes were cultured with FluoroBrite DMEM (Thermo Fisher).
Animals
All housing care and use of animals was handled according to Northwestern University Institutional Animal Care and Use Committee Protocols (ID IS00001419). Mice on a C57BL/6 background were housed in a barrier facility in a temperature-controlled room with a 12-hour light cycle and given ad libitum access to food and water. Adult C57BL/6 mice were singly housed and treated topically with 2% DMSO or 0.23mg/day (~10mg/kg/day) R55 (MedKoo) diluted in 2% DMSO on the shaved back for 7 days before harvesting dorsal skin for formalin fixation and paraffin embedding. Generation of the Dsg1 knockout mouse was described previously 25. Mice were mated overnight and separated the following morning to generate timed pregnancies. Embryos were harvested on day E18.5 and dorsal skin of wild-type and Dsg1 knockout embryos was harvested, formalin-fixed and paraffin-embedded (FFPE). Sections from dorsal skin were processed for immunohistochemistry as described below.
Patient Samples
SAM syndrome c.49–1 G>A patients were originally identified and described 15,35 and recruited from the Department of Dermatology of Emek Medical Center (EMC), Afula, Israel from 2016–2020 (IRB Protocol #0086–15) following informed consent according to a protocol approved by the EMC Institutional Review Board (no. 0086–15) and by the Israel National Committee for Human Genetic Studies in adherence to the Helsinki guidelines. Control skin biopsies were provided by the Skin Biology and Diseases Resource-Based Center of Northwestern University (IRB Protocol #STU00024696) following written informed consent and de-identification.
Method details
siRNA-mediated transfection
Keratinocytes were transiently transfected using the Amaxa Nucleofector System (Lonza) for electroporation of cells according to manufacturer’s instructions with siRNA listed in the key resources table. Following trypsinization, 5×105–2×106 keratinocytes were pelleted and resuspended in 100μl Ingenio Electroporation Solution with 5μl siRNA per 1×106 keratinocytes (20μM stock concentration) and electroporated using program X-001.
Plasmids and viral transduction
pLZRS constructs (listed in the key resources table) were transiently transfected into Phoenix cells (provided by G. Nolan, Stanford University, Stanford CA) using Lipofectamine 2000.
Phoenix cells successfully transfected with the plasmid were selected using 1 μg/ml puromycin. Following a 24–48hr incubation at 32°C, retrovirus-containing Phoenix cell supernatant was collected and concentrated using Amicon Ultra-15 Centrifugal Filter Unit (EMD Millipore). Keratinocytes were infected with concentrated retrovirus diluted in growth medium and 4μg/ml polybrene at 20% confluency for 1.5 hours at 32°C.
Immunoblotting and immunoprecipitation
Keratinocytes were lysed using Urea Sample Buffer (8 M deionized urea, 1% SDS, 60 mM Tris, pH 6.8, 10% glycerol, 0.1% pyronin-Y, 5% β-mercaptoethanol). Proteins were separated using SDS-PAGE electrophoresis before transfer to a nitrocellulose membrane (0.2–0.45 μm pore size). Membranes were blocked with either 5% Milk or LICOR Intercept (PBS) Blocking Buffer, (LI-COR). Primary and secondary antibodies were diluted in blocking buffer. Immunoreactive proteins were visualized using NIR fluorescence or chemiluminescence using the Odyssey FC Imaging system (LI-COR). When probing for VPS29, lysates were run on a gradient gel. Following electrophoresis, the gel was cut in with the upper half subjected to a normal transfer protocol and the lower half undergoing a quick transfer (80V for 40min) on nitrocellulose membrane (0.2 μm pore size).
For immunoprecipitation of VPS35, keratinocytes infected with WT-Dsg1 FLAG virus were grown to confluency before being differentiated overnight with 1.2mM Ca2+ media. Cells were then lysed and scraped in ice cold Triton buffer (50mM Tris-HCl pH 7.5, 1mM EDTA pH 8, 0.5% Triton X-100, 150mM NaCl, and 10% glycerol) and transferred to 1.5 ml tubes. For immunoprecipitation of Dsg1, confluent keratinocytes were differentiated for two days in 1.2mM CaCl2 before being lysed and collected in ice cold Triton/Empigen buffer (40mM HEPES pH 7.5, 120mM NaCl, 1mM EDTA, 1% Triton, 2% Empigen, 10% glycerol). Following a 1min vortex, the lysate was allowed to sit on ice for 10 min followed by centrifugation at 4°C for 30 min at 21,130 x g. 5% volume of the supernatant was saved as the input, the remaining supernatant incubated with either rabbit-IgG primary antibody (negative control) and VPS35 antibody (Life Technologies) or mouse-IgG primary antibody (negative control) and Dsg1 antibody (R&D Systems). Following primary antibody incubation, Dynabeads Protein G (Life Technologies) were used per manufacturer’s protocol. VPS35 IP samples were eluted using 3X Laemmli sample buffer (30% glycerol, 3% SDS, 188mM Tris pH 6.8, 5% BME) and boiled before proceeding to standard immunoblotting protocol. Dsg1 IP samples were eluted using Urea Sample Buffer before proceeding to standard immunoblotting protocol.
Immunofluorescence and histology
Primary keratinocytes were cultured on 12–15mm glass coverslips. Cells were fixed with either 4% paraformaldehyde solution for 10–15min room temperature or 100% ice-cold anhydrous methanol for 2min on ice. For live cell staining of surface proteins, the primary antibody was added before fixation for 1–2hr(s) on ice. To stain for cytoplasmic domains of transmembrane proteins or cytoplasmic proteins in PFA-fixed cells, 0.2% Triton X-100 was used to permeabilize the cells for 10 min at room temperature. Cells were then blocked with 1% BSA solution for 30min at room temperature. 5% normal goat serum was added to the blocking solution when primary antibodies did not include goat IgG. Primary antibodies and secondary antibodies were diluted in block solution and added to cells for either 1–2hrs at 37°C or overnight at 4°C. Paraffin-embedded tissue sections of 3D organotypic rafts, adult and E18.5 mouse dorsal skin, and control and SAM patient skin were baked at 60°C overnight. Xylene/ethanol was used to deparaffinize the tissue section and 0.5% Triton X-100 permeabilized the tissue. 0.01M citrate buffer pH 6.0 was used for antigen retrieval. When using a mouse antibody to stain mouse tissue, unconjugated goat anti-mouse F(ab’)2 antibody was used to block endogenous mIgG before addition of primary antibody. 2D, 3D, and tissue samples were mounted using ProLong Gold antifade reagent to glass slides. 3D raft tissue sections were also processed for hematoxylin and eosin (H&E) staining using standard methods.
Proximity ligation assay
For the proximity ligation assay (PLA) protocol, the immunofluorescence protocol described above for 2D and tissue samples was followed through the addition of primary antibody. Following primary antibody incubation, the PLA protocol for 2D and tissue samples was performed according to manufacturer’s instructions. Additional information on troubleshooting, image acquisition, and analysis is detailed in Hegazy et al.65
Image acquisition and analysis
Apotome images were acquired with an epifluorescence microscope (Axio Imager Z2, Carl Zeiss) equipped with an Apotome.2 slide module, Axiocam 503 Mono digital camera, X-Cite 120 LED Boost System, and Plan-Apochromat 40x/1.4 and 63x/1.4 objective. H&E images were obtained with a microscope (DMR, Leica) equipped with 10x and 40x objective a digital camera (Leica DFC295) using Leica Application Suite software. Confocal images were acquired with a Nikon A1R+ confocal with GaAsP detectors and Nikon W1 Spinning Disk Confocal using NIS Elements software (Nikon) and equipped with 95B prime Photometrics camera, Plan-Apochromat 60x/1.4 objective and Plan-Apochromat 40x/1.3 objective. Time lapse imaging with Nikon A1R+ was performed using Plan-Apochromat TIRF 100x/1.49 objective and captured with no delay. To improve image quality, background in A1R confocal images was subtracted using rolling ball background subtraction followed by 2D deconvolution using NIS Elements software (Nikon). 3D reconstruction of A1R confocal z-stack images was implemented with Volume Viewer’s Depth Coded Alpha Blending (rainbow contrast look up table). ImageJ was also used to pseudocolor images using look up tables such as Fire.
Antibody-based recycling assay
Keratinocytes transfected with siCTL, siVPS35, or siVPS29 were each plated on three glass coverslips. All three coverslips were incubated with an anti-Dsg1 monoclonal antibody for 1hr on ice. Following PBS washes, two coverslips were switched to 0.07mM CaCl2 (low calcium) media with 3mM EGTA (calcium chelator) for 30min at 37°C to promote endocytosis while the third coverslip remained on ice (Steady State). Following internalization and subsequent PBS washes, unconjugated goat anti-mouse secondary was added to the two coverslips on ice to block remaining surface labeled Dsg1. 1.2mM CaCl2 (high calcium) media was added to one of the PBS-washed coverslips and incubated in 37ºC for 30min to induce recycling while the second coverslip remained on ice (Block/Internalization). Following a 30min incubation, the last coverslip was PBS washed on ice (Recycle). To stain for surface Dsg1 (Steady State), residual Dsg1 (Block/Internalization), and recycled Dsg1 (Recycle), AF488-conjugated Goat anti-mouse secondary antibody was added the live cells for 30min before cells were fixed with 4% PFA and permeabilized with 0.1–0.2% Triton-X 100 for 10min. To stain for the internalized pool AF568conguated goat anti-mouse was added following permeabilization. Internalization of Dsg1 was quantified using the 568 channels in the Block/Internalization coverslip and normalized to intensity of Dsg1 at Steady State. Residual Dsg1 was quantified using the intensity of 488 channel of the Block/Internalization coverslip and normalized to the internalization pool in the same coverslip. The recycling pool of Dsg1 was quantified using the intensity of 488 channel of the Recycle coverslip and normalized for both internalized signal in the same coverslip and residual Dsg1.
Stratification assay
Stratification assays and analysis were performed as previously described for Dsg1-GFP stratification studies allowing GFP/Dsg1-GFP labeled keratinocytes mixed with unlabeled keratinocyte to stratify for 24hrs.13 For retromer depletion, VPS35 and VPS29 siRNA were transfected into undifferentiated GFP/Dsg1-GFP expressing keratinocytes before mixing with non-transfected and unlabeled keratinocytes in a known ratio. For R55 stratification experiments, GFP-labeled keratinocytes expressing shNT, shDsg1, shDsg1+WT-Dsg1, and shDsg1+SAM-Dsg1 or expressing siCTL, siDsg1, siDsg2, siDsg3, siDsc1, and siDsc2, were pretreated with R55 for 24hrs before mixing with unlabeled keratinocytes and cultured with continued R55 treatment. Following 24hrs of stratification, 4% paraformaldehyde-fixed cells were processed as described above and stained for plakoglobin (PG) and DAPI. Confocal images with 0.5μm z-stacks were obtained to calculate the ratio of stratified cells that are GFP or Dsg1-GFP positive. The percentage of GFP-positive stratified cells per condition was compared to the predicted percentage of cells that express GFP after mixing with unlabeled cells to obtain the percent deviation from predicted.
BioID screen
Preliminary BioID screens for Dsg1-BirA* and E-cadherin-BirA* expressing keratinocytes were performed as previously described for EphA2-BirA*.66 Briefly, confluent keratinocytes were differentiated with 1.2mM CaCl2 media for 24hrs before incubating the cells in the presence of 50μM biotin for 24hrs. Biotinylated proteins were pulled down with Streptavidin magnetic beads (Pierce) and underwent on-bead trypsin digestion before being sent to Northwestern University Proteomics Center of Excellence for mass spectrometry analysis using an LTQ Orbitrap Velos MS instrumentation (Thermo Scientific). Raw MS data files were analyzed using MaxQuant v2.1.3.0 with built-in search engine Andromeda against Human Uniprot proteome UP000005640 database with isoforms, downloaded 07112022.67 The false discovery rate (FDR) for peptide and protein identification was set to 0.05. A protein was considered a “hit” the number of spectra was 1.5 fold higher compared to control or the presence of at least two unique peptides and no spectra in control (see Table S1). Hits were also filtered using Crapome 2.0 database,68 a repository for negative control MS data, with the requirement of being present in less than 40% of negative control experiments (see Table S2 for Dsg1 Hits and Table S3 for E-cadherin Hits).
QUANTIFICATION AND STATISTICAL ANALYSIS
A minimum of three independent experiments (biological repeats) was performed for each experiment. Keratinocyte clones isolated from distinct foreskin are treated as biological repeats. For immunofluorescence studies, population analysis of each independent experiment was performed with a minimum of 15–30 cells spanning at least 3 areas on the coverslip. All images in each experiment were captured using the same parameters. Fluorescence intensity, area, aspect ratio, and roundness were analyzed using FIJI/ImageJ software (NIH). Quantification of aspect ratio and roundness was performed for at least 100 vesicles spanning at least 5 movies for each biological repeat. For biochemical assays, bands on the immunoblots were quantified using LICOR Image Studio (Version 5.2) software. Graphing and statistical analyses were done on GraphPad Prism (version 8.0) software. Specific tests are indicated in the figure legends. P-value < 0.05 was considered statistically significant. The graphs show the mean of the independent experiments represented with large circles as well as the small individual data points with colors corresponding to each independent experiment containing paired samples when applicable. The error bars represent standard error of the mean (SEM).
Supplementary Material
Representative time-lapse images of Dsg1-GFP in keratinocytes that were differentiated overnight in cells transfected with siCTL, siVPS35, or siVPS29. Yellow arrowheads illustrate Dsg1 localization in vesicle tubules and projections. Images were pseudocolored based on signal intensity using the Fire lookup table (see Figure 2A). Interval time between frames is 2.15 seconds. Movie corresponds with images in Figure 2C. Scale bars, 10 μm, 1μm.
Excel document includes gene name, spectra count, number of unique peptides, q-value, score, peptide sequence, which are output from MaxQuant/Andromeda search. Fold change of Dsg1 and E-cadherin sample spectra compared to control spectra is included. In the absence of spectra in the control sample, the number of unique peptides for the Dsg1/E-cadherin samples were listed. Proteins were considered potential protein interactors (“Hits”) if there is a 1.5 fold increase over control or there is an absence in spectra in the control with >2 unique peptides.
Excel document of Dsg1 hits (refer to Table S1 for full list of the BioID screen). Potential hits were further filtered for their % appearance in the Crapome. Hits with a Crapome % less than 40% were listed in Figure S1A.
Excel document of E-cadherin hits (refer to Table S1 for full list of the BioID screen). Potential hits were further filtered for their % appearance in the Crapome. Hits with a Crapome % less than 40% were listed in Figure S1A.
Highlights.
Desmosomal cadherin, Desmoglein 1, requires the retromer for endosomal trafficking
The retromer is necessary for epidermal differentiation and stratification
Retromer chaperone enhances the function of Dsg1 and a disease-associated mutant
Retromer cargo GLUT1 is upregulated in Dsg1-deficent epidermis
ACKNOWLEDGMENTS
We thank Dr. David Sullivan, Dr. Bethany Perez-White, Dr. Gregory Petsko, Dr. Scott Small, Dr. Pierfausto Seneci, Dr. Neil Kelleher, Dr. Rafael Melani, and all Green laboratory members for technical advice and useful discussion.
Research was supported by NIH/NIAMS P30 AR075049 awarded to Northwestern University Skin Biology & Diseases Resource-Based Center. Imaging work was performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. Histology services were provided by the Northwestern University Mouse Histology and Phenotyping Laboratory which is supported by NCI P30-CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. Proteomics services were performed by the Northwestern Proteomics Core Facility, generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center, instrumentation award (S10OD025194) from NIH Office of Director, and the National Resource for Translational and Developmental Proteomics supported by P41 GM108569. This work was supported by NIH/NIAMS R01 AR041836, NIH/NIAMS R01 AR043380, and NIH/NCI R01 CA228196 with partial support from J.L. Mayberry Endowment to KJG. MH was supported by NIH/NCI T32 CA009560 and NIH/NIAMS F31 AR076188. JAB was supported by NIH/NIAMS K01 AR075087 and NIH/NIAMS T32 AR060710.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Proksch E, Brandner JM, and Jensen JM (2008). The skin: an indispensable barrier. Exp Dermatol 17, 1063–1072. 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- 2.Williams SE, Ratliff LA, Postiglione MP, Knoblich JA, and Fuchs E. (2014). Par3-mInsc and Galphai3 cooperate to promote oriented epidermal cell divisions through LGN. Nat Cell Biol 16, 758–769. 10.1038/ncb3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Damen M, Wirtz L, Soroka E, Khatif H, Kukat C, Simons BD, and Bazzi H. (2021). High proliferation and delamination during skin epidermal stratification. Nat Commun 12, 3227. 10.1038/s41467-021-23386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miroshnikova YA., Le HQ., Schneider D., Thalheim T., Rubsam M., Bremicker N., Polleux J., Kamprad N., Tarantola M., Wang I., et al. (2018). Adhesion forces and cortical tension couple cell proliferation and differentiation to drive epidermal stratification. Nat Cell Biol 20, 69–80. 10.1038/s41556-017-0005-z. [DOI] [PubMed] [Google Scholar]
- 5.Watt FM, and Green H. (1982). Stratification and terminal differentiation of cultured epidermal cells. Nature 295, 434–436. 10.1038/295434a0. [DOI] [PubMed] [Google Scholar]
- 6.Biggs LC, Kim CS, Miroshnikova YA, and Wickstrom SA (2020). Mechanical Forces in the Skin: Roles in Tissue Architecture, Stability, and Function. J Invest Dermatol 140, 284–290. 10.1016/j.jid.2019.06.137. [DOI] [PubMed] [Google Scholar]
- 7.Bruser L, and Bogdan S. (2017). Adherens Junctions on the Move-Membrane Trafficking of E-Cadherin. Cold Spring Harb Perspect Biol 9. 10.1101/cshperspect.a029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno-Layseca P, Icha J, Hamidi H, and Ivaska J. (2019). Integrin trafficking in cells and tissues. Nat Cell Biol 21, 122–132. 10.1038/s41556-018-0223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmer SE, and Kowalczyk AP (2020). The desmosome as a model for lipid raft driven membrane domain organization. Biochim Biophys Acta Biomembr 1862, 183329. 10.1016/j.bbamem.2020.183329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nekrasova O, Harmon RM, Broussard JA, Koetsier JL, Godsel LM, Fitz GN, Gardel ML, and Green KJ (2018). Desmosomal cadherin association with Tctex-1 and cortactin-Arp2/3 drives perijunctional actin polymerization to promote keratinocyte delamination. Nat Commun 9, 1053. 10.1038/s41467-018-03414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rübsam M, Broussard JA, Wickström SA, Nekrasova O, Green KJ, and Niessen CM (2018). Adherens Junctions and Desmosomes Coordinate Mechanics and Signaling to Orchestrate Tissue Morphogenesis and Function: An Evolutionary Perspective. Cold Spring Harb Perspect Biol 10, a029207. 10.1101/cshperspect.a029207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green KJ, Jaiganesh A, and Broussard JA (2019). Desmosomes: Essential contributors to an integrated intercellular junction network. F1000Res 8, F1000 Faculty Rev-2150. 10.12688/f1000research.20942.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broussard JA, Koetsier JL, Hegazy M, and Green KJ (2021). Desmosomes polarize and integrate chemical and mechanical signaling to govern epidermal tissue form and function. Curr Biol 31, 3275–3291 e3275. 10.1016/j.cub.2021.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegazy M, Perl AL, Svoboda SA, and Green KJ (2022). Desmosomal Cadherins in Health and Disease. Annu Rev Pathol 17, 47–72. 10.1146/annurev-pathol-042320-092912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samuelov L., Sarig O., Harmon RM., Rapaport D., Ishida-Yamamoto A., Isakov O., Koetsier JL., Gat A., Goldberg I., Bergman R., et al. (2013). Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet 45, 1244–1248. 10.1038/ng.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seaman MN (2012). The retromer complex - endosomal protein recycling and beyond. J Cell Sci 125, 4693–4702. 10.1242/jcs.103440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonifacino JS, and Rojas R. (2006). Retrograde transport from endosomes to the transGolgi network. Nat Rev Mol Cell Biol 7, 568–579. 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 18.Burd C, and Cullen PJ (2014). Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol 6, a016774. 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carosi JM, Denton D, Kumar S, and Sargeant TJ (2021). Retromer dysfunction at the nexus of tauopathies. Cell Death Differ 28, 884–899. 10.1038/s41418-020-00727-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CD, Atwood WJ, and DiMaio D. (2013). Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A 110, 7452–7457. 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandra M, Kendall AK, and Jackson LP (2021). Toward Understanding the Molecular Role of SNX27/Retromer in Human Health and Disease. Front Cell Dev Biol 9, 642378. 10.3389/fcell.2021.642378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Heesom KJ, Tavare JM, and Cullen PJ (2013). A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat Cell Biol 15, 461–471. 10.1038/ncb2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Zi Z, Lee EE, Zhao J, Contreras DC, South AP, Abel ED, Chong BF, Vandergriff T, Hosler GA, et al. (2018). Differential glucose requirement in skin homeostasis and injury identifies a therapeutic target for psoriasis. Nat Med 24, 617–627. 10.1038/s41591-018-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao J, Yang J, Wang L, Li Y, Liu YQ, Dong J, Li L, Wen X, Shen GX, and Tu YT (2008). Expression of GLUT-1 in psoriasis and the relationship between GLUT1 upregulation induced by hypoxia and proliferation of keratinocyte growth. J Dermatol Sci 51, 203–207. 10.1016/j.jdermsci.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Godsel LM, Roth-Carter QR, Koetsier JL, Tsoi LC, Huffine AL, Broussard JA, Fitz GN, Lloyd SM, Kweon J, Burks HE, et al. (2022). Translational implications of Th17-skewed inflammation due to genetic deficiency of a cadherin stress sensor. J Clin Invest 132. 10.1172/JCI144363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roux KJ, Kim DI, Raida M, and Burke B. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810. 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Weert AW., Geuze HJ., Groothuis B., and Stoorvogel W. (2000). Primaquine interferes with membrane recycling from endosomes to the plasma membrane through a direct interaction with endosomes which does not involve neutralisation of endosomal pH nor osmotic swelling of endosomes. Eur J Cell Biol 79, 394–399. 10.1078/0171-9335-00062. [DOI] [PubMed] [Google Scholar]
- 28.Arighi CN, Hartnell LM, Aguilar RC, Haft CR, and Bonifacino JS (2004). Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol 165, 123–133. 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Weering JR, Verkade P, and Cullen PJ (2012). SNX-BAR-mediated endosome tubulation is co-ordinated with endosome maturation. Traffic 13, 94–107. 10.1111/j.16000854.2011.01297.x. [DOI] [PubMed] [Google Scholar]
- 30.Fuse A, Furuya N, Kakuta S, Inose A, Sato M, Koike M, Saiki S, and Hattori N. (2015). VPS29-VPS35 intermediate of retromer is stable and may be involved in the retromer complex assembly process. FEBS Lett 589, 1430–1436. 10.1016/j.febslet.2015.04.040. [DOI] [PubMed] [Google Scholar]
- 31.Cui Y, Carosi JM, Yang Z, Ariotti N, Kerr MC, Parton RG, Sargeant TJ, and Teasdale RD (2019). Retromer has a selective function in cargo sorting via endosome transport carriers. J Cell Biol 218, 615–631. 10.1083/jcb.201806153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seaman MNJ (2021). The Retromer Complex: From Genesis to Revelations. Trends Biochem Sci 46, 608–620. 10.1016/j.tibs.2020.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Arnette C, Koetsier JL, Hoover P, Getsios S, and Green KJ (2016). In Vitro Model of the Epidermis: Connecting Protein Function to 3D Structure. Methods Enzymol 569, 287–308. 10.1016/bs.mie.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almagro Armenteros JJ, Salvatore M, Emanuelsson O, Winther O, von Heijne G, Elofsson A, and Nielsen H. (2019). Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance 2, e201900429. 10.26508/lsa.201900429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen-Barak E, Godsel LM, Koetsier JL, Hegazy M, Kushnir-Grinbaum D, Hammad H, Danial-Farran N, Harmon R, Khayat M, Bochner R, et al. (2020). The Role of Desmoglein 1 in Gap Junction Turnover Revealed through the Study of SAM Syndrome. J Invest Dermatol 140, 556–567 e559. 10.1016/j.jid.2019.08.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mecozzi VJ, Berman DE, Simoes S, Vetanovetz C, Awal MR, Patel VM, Schneider RT, Petsko GA, Ringe D, and Small SA (2014). Pharmacological chaperones stabilize retromer to limit APP processing. Nat Chem Biol 10, 443–449. 10.1038/nchembio.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cibrian D, de la Fuente H, and Sanchez-Madrid F. (2020). Metabolic Pathways That Control Skin Homeostasis and Inflammation. Trends Mol Med 26, 975–986. 10.1016/j.molmed.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Xie Y, Miao H, and Blankenship JT (2018). Membrane trafficking in morphogenesis and planar polarity. Traffic 19, 679–689. 10.1111/tra.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stein M, Wandinger-Ness A, and Roitbak T. (2002). Altered trafficking and epithelial cell polarity in disease. Trends Cell Biol 12, 374–381. 10.1016/s0962-8924(02)02331-0. [DOI] [PubMed] [Google Scholar]
- 40.Schiller SA, Seebode C, Wieser GL, Goebbels S, Mobius W, Horowitz M, Sarig O., Spreche E., and Emmer S. (2016). Establishment of Two Mouse Models for CEDNIK Syndrome Reveals the Pivotal Role of SNAP29 in Epidermal Differentiation. J Invest Dermatol 136, 672–679. 10.1016/j.jid.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 41.Kugelmann D, Radeva MY, Spindler V, and Waschke J. (2019). Desmoglein 1 Deficiency Causes Lethal Skin Blistering. J Invest Dermatol 139, 1596–1599 e1592. 10.1016/j.jid.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Noguchi S, Honda S, Saitoh T, Matsumura H, Nishimura E, Akira S, and Shimizu S. (2019). Beclin 1 regulates recycling endosome and is required for skin development in mice. Commun Biol 2, 37. 10.1038/s42003-018-0279-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seaman MN, McCaffery JM, and Emr SD (1998). A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol 142, 665–681. 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasad BC, and Clark SG (2006). Wnt signaling establishes anteroposterior neuronal polarity and requires retromer in C. elegans. Development 133, 1757–1766. 10.1242/dev.02357. [DOI] [PubMed] [Google Scholar]
- 45.Wang S, and Bellen HJ (2015). The retromer complex in development and disease. Development 142, 2392–2396. 10.1242/dev.123737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, Sharma YV, Yan D, Selva EM, and Lin X. (2008). The retromer complex influences Wnt secretion by recycling wntless from endosomes to the trans-Golgi network. Dev Cell 14, 120–131. 10.1016/j.devcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Franch-Marro X, Wendler F, Guidato S, Griffith J, Baena-Lopez A, Itasaki N, Maurice MM, and Vincent JP (2008). Wingless secretion requires endosome-to-Golgi retrieval of Wntless/Evi/Sprinter by the retromer complex. Nat Cell Biol 10, 170–177. 10.1038/ncb1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Port F, Kuster M, Herr P, Furger E, Banziger C, Hausmann G, and Basler K. (2008). Wingless secretion promotes and requires retromer-dependent cycling of Wntless. Nat Cell Biol 10, 178–185. 10.1038/ncb1687. [DOI] [PubMed] [Google Scholar]
- 49.Radice G, Lee JJ, and Costantini F. (1991). H beta 58, an insertional mutation affecting early postimplantation development of the mouse embryo. Development 111, 801–811. 10.1242/dev.111.3.801. [DOI] [PubMed] [Google Scholar]
- 50.Wen L, Tang FL, Hong Y, Luo SW, Wang CL, He W, Shen C, Jung JU, Xiong F, Lee DH, et al. (2011). VPS35 haploinsufficiency increases Alzheimer’s disease neuropathology. J Cell Biol 195, 765–779. 10.1083/jcb.201105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang P, Moreno R, Lambert PF, and DiMaio D. (2020). Cell-penetrating peptide inhibits retromer-mediated human papillomavirus trafficking during virus entry. Proc Natl Acad Sci U S A 117, 6121–6128. 10.1073/pnas.1917748117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryant DM, Kerr MC, Hammond LA, Joseph SR, Mostov KE, Teasdale RD, and Stow JL (2007). EGF induces macropinocytosis and SNX1-modulated recycling of E-cadherin. J Cell Sci 120, 1818–1828. 10.1242/jcs.000653. [DOI] [PubMed] [Google Scholar]
- 53.Wassmer T., Attar N., Harterink M., van Weering JR., Traer CJ., Oakley J., Goud B., Stephens DJ., Verkade P., Korswagen HC., and Cullen PJ. (2009). The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev Cell 17, 110–122. 10.1016/j.devcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong Z, Yang Y, Zhang C, Niu Y, Li K, Zhao X, and Liu JJ (2009). The retromer component SNX6 interacts with dynactin p150(Glued) and mediates endosome-to-TGN transport. Cell Res 19, 1334–1349. 10.1038/cr.2009.130. [DOI] [PubMed] [Google Scholar]
- 55.Kottke MD, Delva E, and Kowalczyk AP (2006). The desmosome: cell science lessons from human diseases. J Cell Sci 119, 797–806. 10.1242/jcs.02888. [DOI] [PubMed] [Google Scholar]
- 56.Müller L, Hatzfeld M, and Keil R. (2021). Desmosomes as Signaling Hubs in the Regulation of Cell Behavior. Front Cell Dev Biol 9, 745670–745670. 10.3389/fcell.2021.745670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.North AJ, Chidgey MA, Clarke JP, Bardsley WG, and Garrod DR (1996). Distinct desmocollin isoforms occur in the same desmosomes and show reciprocally graded distributions in bovine nasal epidermis. Proc Natl Acad Sci U S A 93, 7701–7705. 10.1073/pnas.93.15.7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Getsios S, Simpson CL, Kojima S, Harmon R, Sheu LJ, Dusek RL, Cornwell M, and Green KJ (2009). Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J Cell Biol 185, 1243–1258. 10.1083/jcb.200809044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilleron J, Gerdes JM, and Zeigerer A. (2019). Metabolic regulation through the endosomal system. Traffic 20, 552–570. 10.1111/tra.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li JG, Chiu J, Ramanjulu M, Blass BE, and Pratico D. (2020). A pharmacological chaperone improves memory by reducing Abeta and tau neuropathology in a mouse model with plaques and tangles. Mol Neurodegener 15, 1. 10.1186/s13024-019-0350-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muzio L, Sirtori R, Gornati D, Eleuteri S, Fossaghi A, Brancaccio D, Manzoni L, Ottoboni L, Feo L, Quattrini A, et al. (2020). Retromer stabilization results in neuroprotection in a model of Amyotrophic Lateral Sclerosis. Nat Commun 11, 3848. 10.1038/s41467-020-17524-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young JE, Fong LK, Frankowski H, Petsko GA, Small SA, and Goldstein LSB (2018). Stabilizing the Retromer Complex in a Human Stem Cell Model of Alzheimer’s Disease Reduces TAU Phosphorylation Independently of Amyloid Precursor Protein. Stem Cell Reports 10, 1046–1058. 10.1016/j.stemcr.2018.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnette CR., Roth-Carter QR., Koetsier JL., Broussard JA., Burks HE., Cheng K., Amadi C., Gerami P., Johnson JL., and Green KJ. (2020). Keratinocyte cadherin desmoglein 1 controls melanocyte behavior through paracrine signaling. Pigment Cell Melanoma Res 33, 305–317. 10.1111/pcmr.12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Godsel LM, Dubash AD, Bass-Zubek AE, Amargo EV, Klessner JL, Hobbs RP, Chen X, and Green KJ (2010). Plakophilin 2 couples actomyosin remodeling to desmosomal plaque assembly via RhoA. Mol Biol Cell 21, 2844–2859. 10.1091/mbc.E10-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hegazy M, Cohen-Barak E, Koetsier JL, Najor NA, Arvanitis C, Sprecher E, Green KJ, and Godsel LM (2020). Proximity Ligation Assay for Detecting ProteinProtein Interactions and Protein Modifications in Cells and Tissues in Situ. Curr Protoc Cell Biol 89, e115. 10.1002/cpcb.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perez White BE, Ventrella R, Kaplan N, Cable CJ, Thomas PM, and Getsios S. (2017). EphA2 proteomics in human keratinocytes reveals a novel association with afadin and epidermal tight junctions. J Cell Sci 130, 111–118. 10.1242/jcs.188169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cox J, and Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 68.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. (2013). The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 10, 730–736. 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative time-lapse images of Dsg1-GFP in keratinocytes that were differentiated overnight in cells transfected with siCTL, siVPS35, or siVPS29. Yellow arrowheads illustrate Dsg1 localization in vesicle tubules and projections. Images were pseudocolored based on signal intensity using the Fire lookup table (see Figure 2A). Interval time between frames is 2.15 seconds. Movie corresponds with images in Figure 2C. Scale bars, 10 μm, 1μm.
Excel document includes gene name, spectra count, number of unique peptides, q-value, score, peptide sequence, which are output from MaxQuant/Andromeda search. Fold change of Dsg1 and E-cadherin sample spectra compared to control spectra is included. In the absence of spectra in the control sample, the number of unique peptides for the Dsg1/E-cadherin samples were listed. Proteins were considered potential protein interactors (“Hits”) if there is a 1.5 fold increase over control or there is an absence in spectra in the control with >2 unique peptides.
Excel document of Dsg1 hits (refer to Table S1 for full list of the BioID screen). Potential hits were further filtered for their % appearance in the Crapome. Hits with a Crapome % less than 40% were listed in Figure S1A.
Excel document of E-cadherin hits (refer to Table S1 for full list of the BioID screen). Potential hits were further filtered for their % appearance in the Crapome. Hits with a Crapome % less than 40% were listed in Figure S1A.
Data Availability Statement
BioID proteomics data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) partner repository: PXD036646. Accession number is also listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-VPS35 | Abcam | Cat# ab157220, RRID:AB_2636885 |
| rabbit anti-VPS35 | Life Technologies | Cat# PA5-21898, RRID:AB_11153540 |
| mouse anti-VPS35 | Santa Cruz Biotechnology | Cat# sc-374372, RRID:AB_10988942 |
| mouse anti-VPS29 | Santa Cruz Biotechnology | Cat# sc-398874, RRID:AB_2885036 |
| mouse anti-Dsg1 | PROGEN | Cat# 651111, RRID:AB_1541107 |
| mouse anti-Dsg1 | R&D Systems | Cat# MAB944, RRID:AB_2093296 |
| mouse anti-Dsg1 | Thermo Fisher Scientific | Cat# 32-6000, RRID:AB_2533088 |
| rabbit anti-Dsg1 | Abcam | Cat# ab124798, RRID:AB_10974963 |
| mouse anti- FLAG | Sigma-Aldrich | Cat# F1804, RRID:AB_262044 |
| chicken anti-plakoglobin | Aves Laboratories | 1407 and 1408 |
| mouse anti-EEA1 | BD Bioscience | Cat# 610456, RRID:AB_397829 |
| mouse anti-LAMP1 | Santa Cruz Biotechnology | Cat# sc-20011, RRID:AB_626853 |
| mouse anti-GFP | Takara | Cat# 632381, RRID: AB_2313808 |
| mouse anti-E-cadherin | M. Takeichi and O. Abe, Riken Center for Developmental Biology | HecD1 |
| mouse anti-E-cadherin | BD Biosciences | Cat# 61081, RRID:AB_397580 |
| rabbit anti-E-cadherin | R. Marsh and R. Brackenbury, University of Cincinnati | 795 |
| goat anti-E-cadherin | R&D Systems | Cat# AF748, RRID:AB_355568 |
| rabbit anti-β-catenin | Sigma | Cat# C2206, RRID:AB_476831 |
| rabbit anti-K10 | J. Segre, National Human Genome Research Institute | N/A |
| rabbit anti-M6PR (cation independent) | Abcam | Cat# ab124767, RRID:AB_10974087 |
| rabbit anti-GLUT1 | Abcam | Cat# ab115730, RRID:AB_10903230 |
| mouse anti-GLUT1 | Novus Biologicals | Cat# NBP2-75785, RRID:AB_2923123 |
| mouse anti-Dsg2 (6D8) and anti-Dsc2 (7G6) | J. Wahl III, University of Nebraska Medical Center | N/A |
| mouse anti-Dsg3 | Millipore Sigma | Cat# MABT335, RRID:AB_2923124 |
| mouse anti-Dsc1 | PROGEN | Cat# 61092, RRID:AB_1541100 |
| mouse anti-Integrin β1 | BD Biosciences | Cat# 610467, RRID:AB_2128060 |
| rabbit anti-Ki67 | EMD Millipore | Cat# AB9260, RRID:AB_2142366 |
| mouse anti-P-cadherin | M. Wheelock, University of Nebraska Medical Center | N/A |
| rabbit anti-HA-tag | Cell Signaling | Cat# 3724S, RRID:AB_1549585 |
| rabbit anti-GAPDH | Sigma-Aldrich | Cat# G9545, RRID: AB_796208 |
| mouse anti-tubulin | Developmental Hybridoma Studies Bank | 12G10, RRID: AB_1157911 |
| mouse anti-actin | Millipore Sigma | Cat# MAB1501, RRID:AB_2223041 |
| mIgG isotype control | Abcam | Cat# ab37355, RRID:AB_2665484 |
| rbIgG isotype control | Abcam | Cat# ab27478, RRID:AB_2616600 |
| Secondary goat anti-mouse, -rabbit, and -chicken HRP | SeraCare Life Sciences | N/A |
| Secondary goat anti-mouse conjugated with IRDye 680RD | LI-COR Biosciences | Cat# 926-68070, RRID:AB_10956588 |
| Secondary goat anti-mouse conjugated with IRDye 800CW | LI-COR Biosciences | Cat# 925-32210, RRID:AB_2687825 |
| Secondary goat anti-rabbit conjugated with IRDye 680RD | LI-COR Biosciences | Cat# 926-68071, RRID:AB_10956166 |
| Secondary goat anti-rabbit conjugated with IRDye 800CW | LI-COR Biosciences | Cat# 926-32211, RRID:AB_621843 |
| Secondary goat anti-mouse IgG1 isotype conjugated with IRDye 680LT | LI-COR Biosciences | Cat# 926-68050, RRID:AB_2783642 |
| Secondary goat anti-mouse IgG2a isotype conjugated with IRDye 800 CW | LI-COR Biosciences | Cat# 926-32351, RRID:AB_2782998 |
| Secondary goat anti-mouse IgG2a isotype conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-21131; RRID: AB_2535771 |
| Secondary goat anti-mouse IgG2a isotype conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21124; RRID: AB_2535766 |
| Secondary goat anti-mouse IgG1 isotype conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-21121; RRID: AB_2535764 |
| Secondary goat anti-mouse conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-11029; RRID: AB_2534088 |
| Secondary goat anti-mouse conjugated with Alexa Fluor-568 | Thermo Fisher Scientific | Cat# A-11004; RRID: AB_2534072 |
| Secondary goat anti-mouse conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-32728; RRID: AB_2633277 |
| Secondary goat anti-rabbit conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-11008; RRID: AB_143165 |
| Secondary goat anti-rabbit conjugated with Alexa Fluor-568 | Thermo Fisher Scientific | Cat# A-11011; RRID: AB_143157 |
| Secondary goat anti-rabbit conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21245; RRID: AB_2535813 |
| Secondary goat anti-chicken conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21449; RRID: AB_2535866 |
| Secondary donkey anti-goat conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# A-11055, RRID:AB_2534102 |
| Secondary donkey anti-goat conjugated with Alexa Fluor-647 | Thermo Fisher Scientific | Cat# A-21447, RRID:AB_2535864 |
| Secondary donkey anti-mouse conjugated with Alexa Fluor-568 | Thermo Fisher Scientific | Cat# A10037, RRID:AB_2534013 |
| Secondary goat anti-mouse AffiniPure F(ab’)2 Fragment unconjugated | Jackson Laboratory | Cat# 115-006-003, RRID:AB_2338466 |
| Secondary goat anti-mouse AffiniPure F(ab’)2 Fragment conjugated with Alexa Fluor-488 | Jackson Laboratory | Cat# 715-546-150, RRID:AB_2340849 |
| Bacterial and virus strains | ||
| pLZRS-GFP | Getsios et al.58 | N/A |
| pLZRS-WT-Dsg1-FLAG | Getsios et al.58 | N/A |
| pLZRS-shDsg1 | Getsios et al.58 | N/A |
| pLZRS-SAM Dsg1-FLAG | Cohen-Barak et al.35 | N/A |
| pLZRS-shNT (Non-targeting) | Arnette et al.63 | N/A |
| pLZRS-E-cadherin-mCherry | Godsel et al.64 | N/A |
| pLZRS-Dsg1-BirA* | This manuscript | N/A |
| pLZRS-E-cadherin-BirA* | This manuscript | N/A |
| pLZRS-WT-Dsg1-FLAG/GFP (miRNA Resistant) | This manuscript | N/A |
| pLZRS-SAM Dsg1-FLAG/GFP (miRNA Resistant) | This manuscript | N/A |
| Biological samples | ||
| Neonatal human foreskin | NU SBDRC | N/A |
| Control adult human skin biopsies | NU SBDRC | N/A |
| SAM syndrome patient c.49-1 G>A skin biopsies | Samuelov et al.15 and Cohen-Barak et al.35 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 4’,6-Diamido-2-Phenylindole (DAPI) | Sigma-Aldrich | Cat# D9542 |
| Dispase II | Roche | Cat# 04942078001 |
| Goat serum | Sigma-Aldrich | Cat# G9023 |
| LysoBrite NIR | AAT Bioquest | Cat# 22641 |
| R55 | Sigma-Aldrich | Cat# 531084 |
| R55 | MedKoo Biosciences | Cat # 504210 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat# D8418 |
| Primaquine bisphosphate | Sigma-Aldrich | Cat # 160393 |
| Ingenio Electroporation Solution | Mirus Bio | Cat# MIR 50117 |
| Polybrene | Sigma-Aldrich | Cat# H9268 |
| Albumin from bovine serum (BSA) | Sigma-Aldrich | Cat# A7906 |
| 16% Formaldehyde (w/v), Methanol-free | Thermo Fisher Scientific | Cat# PI28908 |
| Urea | Thermo Fisher Scientific | Cat# BP169-212 |
| Sodium Dodecyl Sulfate (SDS) | Thermo Fisher Scientific | Cat# BP166-500 |
| Tris Hydrochloride | Thermo Fisher Scientific | Cat# BP153-500 |
| Pyronin-Y | Sigma-Aldrich | Cat# P7017 |
| β-Mercaptoethanol | Sigma-Aldrich | Cat# M6250 |
| Ethylenediaminetetraacetic acid (EDTA) | Thermo Fisher Scientific | Cat# BP120-1 |
| Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) | Sigma-Aldrich | Cat# E4378 |
| Acrylamide/Bis-acrylamide, 30% solution | Sigma-Aldrich | Cat# A3574 |
| Formalin solution, neutral buffered, 10% | Sigma-Aldrich | Cat# HT501320 |
| Xylene | Thermo Fisher Scientific | Cat# X3P |
| Citric acid | Sigma-Aldrich | Cat# C0759 |
| Glycine | Sigma-Aldrich | Cat# G8790 |
| Methanol | Sigma-Aldrich | Cat# 179337 |
| Glycerol | Thermo Fisher Scientific | Cat# BP229 |
| Ethyl Alcohol, 200 proof | Thermo Fisher Scientific | Cat# A4094 |
| Triton X-100 | Sigma-Aldrich | Cat# X100 |
| Tween20 | Sigma-Aldrich | Cat# P1379 |
| ProLong Gold antifade reagent | Thermo Fisher Scientific | Cat# P36930 |
| PBS | Thermo Fisher Scientific | Cat# 21040CV |
| Opti-MEM I Reduced Serum Medium | Thermo Fisher Scientific | Cat# 11058021 |
| Intercept (PBS) Blocking Buffer | LI-COR Biosciences | Cat# 927-70001 |
| Sodium Chloride (NaCl) | Sigma-Aldrich | Cat# S9625 |
| Empigen | Sigma-Aldrich | Cat# 30326 |
| HEPES | Sigma-Aldrich | Cat# H4034 |
| Medium 154CF, Kit (M154): With gentamicin/amphotericin B solution and CaCl2 | Thermo Fisher Scientific | Cat# M154CF500 |
| Human Keratinocyte Growth Supplement (HKGS) Kit: | Thermo Fisher Scientific | Cat# S001K |
| FluoroBrite DMEM | Thermo Fisher | Cat# A1896701 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher | Cat# 11668019 |
| Streptavidin conjugated with Alexa Fluor-488 | Thermo Fisher Scientific | Cat# S11223 |
| Streptavidin conjugated with HRP | Cell Signaling Technology | Cat# 3999 |
| Pierce Streptavidin Magnetic Beads | Thermo Fisher | Cat# 88816 |
| Dynabeads Protein G | Thermo Fisher | Cat# 10004D |
| Critical commercial assays | ||
| Duolink® In Situ PLA® Probe Anti-Rabbit PLUS | Sigma-Aldrich | Cat# DUO92002 |
| Duolink® In Situ PLA® Probe Anti-Mouse MINUS | Sigma-Aldrich | Cat# DUO92004 |
| Duolink® In Situ Detection Reagents Red | Sigma-Aldrich | Cat# DUO92008 |
| Duolink® In Situ Detection Reagents Far Red | Sigma-Aldrich | Cat# DUO92013 |
| Deposited data | ||
| Dsg1 and E-cadherin BioID | This manuscript | PXD036646 |
| Experimental models: Cell lines | ||
| Neonatal human epidermal keratinocytes (NHEKs) | N/A | Isolated from neonatal human foreskin |
| Phoenix cells | G. Nolan, Stanford University | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: wild-type C57BL/6 | The Jackson Laboratory | Strain #:000664 |
| Mouse: Dsg1−/− C57BL/6 | Godsel et al. 25 | N/A |
| Oligonucleotides | ||
| siGENOME non-targeting siRNA pool #2 | Dharmacon | cat# D-001206-14 |
| Silencer Select VPS35 siRNA (siVPS35 #1): 5’- CCATGGATTTTGTACTGCTTT-3’ | Sigma | Product # s31375 |
| VPS35 siRNA (siVPS35 #2): 5’- GAGATATTCAATAAGCTCAACCTTG-3’ | Integrated DNA Technologies | N/A |
| VPS29 siRNA: 5’- ATGATGTGAAAGTAGAACGAATCGA-3’ | Integrated DNA Technologies | N/A |
| Dsg1 siRNA: 5′-CCATTAGAGAGTGGCAATAGGATGA-3′ | Integrated DNA Technologies | N/A |
| Dsg2 siRNA: 5′- CCTGGAAGCAGAGACAGTGTGGTCCTT −3′ | Integrated DNA Technologies | N/A |
| siGENOME SMARTpool Dsg3 siRNA | Dharmacon | cat# M-011646-01-0005 |
| siGENOME SMARTpool Dsc1 siRNA | Dharmacon | cat# M-011995-01-0005 |
| Dsc2 siRNA: 5′-AGAGAGACACTATAAACAAGTACAC-3′ | Integrated DNA Technologies | N/A |
| Software and algorithms | ||
| LI-COR Image Studio (Version 5.2) | LI-COR Biosciences | RRID:SCR_015795 |
| ZEN 2.3 software | Carl Zeiss | RRID: SCR_013672 |
| NIS-Elements | Nikon | RRID: SCR_014329 |
| Leica Application Suite | Leica | RRID:SCR_016555 |
| ImageJ | NIH | RRID: SCR_003070 |
| Prism | GraphPad | RRID: SCR_002798 |
| Microsoft Excel | Microsoft | RRID: SCR_016137 |
| Photoshop | Adobe | RRID: SCR_014199 |
| Illustrator | Adobe | RRID: SCR_010279 |
| MaxQuant (Version 2.1.3.0) | Max Planck Institute of Biochemistry | RRID:SCR_014485 |
| Other | ||
| Amersham™ Protran® Western blotting membranes, nitrocellulose (0.2 μm pore size) | Cytiva | Cat# GE10600001 |
| Amersham™ Protran® Western blotting membranes, nitrocellulose (0.45 μm pore size) | Cytiva | Cat# GE10600002 |
| 4-Chamber 35mm glass bottom dish with 20 mm microwell, #1.5 cover glass | CellVis | Cat# D35C4-20-1.5-N |
| Nucleofector 2b Device | Lonza | Cat# AAB-1001 |
| Amicon Ultra-15 Centrifugal Filter Unit | Sigma-Aldrich | Cat# UFC9030 |