Background:
Activated macrophages contribute to the pathogenesis of vascular disease. Vein graft failure is a major clinical problem with limited therapeutic options. PCSK9 (proprotein convertase subtilisin/kexin 9) increases low-density lipoprotein (LDL)-cholesterol levels via LDL receptor (LDLR) degradation. The role of PCSK9 in macrophage activation and vein graft failure is largely unknown, especially through LDLR-independent mechanisms. This study aimed to explore a novel mechanism of macrophage activation and vein graft disease induced by circulating PCSK9 in an LDLR-independent fashion.
Methods:
We used Ldlr-/- mice to examine the LDLR-independent roles of circulating PCSK9 in experimental vein grafts. Adeno-associated virus (AAV) vector encoding a gain-of-function mutant of PCSK9 (rAAV8/D377Y-mPCSK9) induced hepatic PCSK9 overproduction. To explore novel inflammatory targets of PCSK9, we used systems biology in Ldlr-/- mouse macrophages.
Results:
In Ldlr-/- mice, AAV-PCSK9 increased circulating PCSK9, but did not change serum cholesterol and triglyceride levels. AAV-PCSK9 promoted vein graft lesion development when compared with control AAV. In vivo molecular imaging revealed that AAV-PCSK9 increased macrophage accumulation and matrix metalloproteinase activity associated with decreased fibrillar collagen, a molecular determinant of atherosclerotic plaque stability. AAV-PCSK9 induced mRNA expression of the pro-inflammatory mediators IL-1β (interleukin-1 beta), TNFα (tumor necrosis factor alpha), and MCP-1 (monocyte chemoattractant protein-1) in peritoneal macrophages underpinned by an in vitro analysis of Ldlr-/- mouse macrophages stimulated with endotoxin-free recombinant PCSK9. A combination of unbiased global transcriptomics and new network-based hyperedge entanglement prediction analysis identified the NF-κB (nuclear factor-kappa B) signaling molecules, lectin-like oxidized LOX-1 (LDL receptor-1), and SDC4 (syndecan-4) as potential PCSK9 targets mediating pro-inflammatory responses in macrophages.
Conclusions:
Circulating PCSK9 induces macrophage activation and vein graft lesion development via LDLR-independent mechanisms. PCSK9 may be a potential target for pharmacologic treatment for this unmet medical need.
Keywords: graft occlusion, vascular; inflammation; macrophage activation; receptors, LDL; systems biology
Novelty and Significance.
What Is Known?
Autologous saphenous vein is a widely used surgical bypass for lower extremity peripheral artery disease and coronary artery disease.
PCSK9 (proprotein convertase subtilisin/kexin 9) targets receptors other than LDL receptor (LDLR) such as LRP1 (low-density lipoprotein receptor-related protein-1), ApoER2 (apolipoprotein E receptor 2), VLDLR (very low-density lipoprotein receptor), and CD36.
PCSK9 may induce pro-inflammatory responses in macrophages and arterial atherosclerotic lesions primarily via LDLR-dependent mechanisms.
What New Information Does This Article Contribute?
PCSK9 promotes vein graft lesion development by the mechanisms unassociated with either LDLR degradation or blood cholesterol levels.
PCSK9 induces pro-inflammatory macrophage activation: immune responses, proliferation and migration, via LDLR-independent mechanisms.
Potential PCSK9 targets mediating pro-inflammatory responses in macrophages include NF-κB (nuclear factor-kappa B) signaling molecules, lectin-like oxidized LOX-1 (LDL receptor-1), and SDC4 (syndecan-4).
The role of PCSK9 on vascular inflammation remains controversial in the clinical setting because clinical trials with PCSK9 inhibitors did not show any significant changes in high sensitivity-CRP (C-reactive protein) levels in patients with coronary artery disease. However, CRP may not solely capture changes in inflammation. Although there are a few emerging in vivo studies suggesting a direct link between PCSK9 and atherosclerosis primarily via LDLR-dependent mechanisms, the role of PCSK9 in vein graft failure, especially via LDLR-independent mechanisms, remains largely unknown. To test the hypothesis, we developed a systems approach, involving unbiased transcriptomics and network analysis. Our in vivo evidence suggests that PCSK9 promotes vein graft lesion development, macrophage proliferation, monocyte migration, foam cell formation, and collagen remodeling. We further identified NF-κB signaling molecules, LOX-1 and SDC4 as potential targets of PCSK9 mediating pro-inflammatory responses in macrophages. The evidence, including our own, has suggested that circulating PCSK9 may exert pro-inflammatory effects on vascular lesions.
In This Issue, see p 867
Meet the First Author, see p 868
Editorial, see p 890
Autologous saphenous vein grafting is a widely used surgical bypass for treatment of lower extremity peripheral artery disease and coronary artery disease. A major clinical concern associated with use of vein grafts is their high occlusion rates.1,2 Acute bypass failure within a month after surgery may result from technical failures, such as vessel wall damage and anastomotic problems, or from poor runoff due to distal disease. Aspirin and other antiplatelet drugs reduce early events resulting from thrombotic occlusion.3,4 However, later events, including progressive wall thickening and segmental neointimal or anastomotic hyperplasia, are followed (usually after approximately 2 years) by the development of changes similar to arterial atherosclerosis.5 Statins may be a therapeutic option for the later events; however, no randomized controlled trials of statins have been sufficiently powered to show their clinical impact.6,7 Despite the global burden of vein graft disease and decades of research, medical therapeutic options remain limited to prevent vein graft failure. We have recently reported that Delta-like ligand 4-Notch signaling and PPARα pathways in macrophages contribute to vein graft lesion development,8,9 suggesting that macrophage-mediated inflammation plays a key role in the pathogenesis of vein graft failure.10,11
PCSK9 (proprotein convertase subtilisin/kexin 9) is predominantly produced by the liver, and increases blood levels of low-density lipoprotein (LDL) cholesterol by escorting LDL receptors (LDLRs) on the cell plasma membrane to lysosomes for degradation in hepatocytes.12,13 PCSK9 monoclonal antibodies reduce the risk of major cardiovascular events14,15 and adverse limb events which is not possible by statin treatment alone.15,16 The Alirocumab for Stopping Atherosclerosis Progression in Saphenous Vein Grafts Pilot Trial (https://clinicaltrials.gov/ct2/show/NCT03542110), evaluating the effect of a PCSK9 inhibitor on intermediate saphenous vein graft lesion, is ongoing. The role of PCSK9 in vein graft failure, however, is largely unknown. A recent clinical study associated serum PCSK9 levels with saphenous vein graft disease after coronary artery bypass grafting (CABG), independently of LDL cholesterol levels.17 As PCSK9 is a circulating protein,18 there may be crosstalk between the liver and vein grafts via circulating PCSK9 that may have lipid-independent pro-inflammatory properties on vascular lesions. Another study demonstrated that recombinant PCSK9 induces TNFα (tumor necrosis factor alpha) mRNA in bone marrow-derived macrophages mainly, but not exclusively, in an LDLR-dependent manner.19 These studies led us to hypothesize that circulating PCSK9 induces macrophage activation and vein graft lesion development through LDLR-independent mechanisms via unknown targets associated with inflammation.
The present study used LDLR-deficient (Ldlr-/-) mice to demonstrate that circulating PCSK9 promotes macrophage activation and vein graft lesion development in an LDLR-independent fashion. We also used a systems approach involving RNA-sequencing and network-based hyperedge entanglement prediction20 to examine the global impact of PCSK9 in inflammation and identify potential novel LDLR-independent targets of PCSK9 in macrophages.
Methods
Data Availability
The online-only Supplemental Data provided detailed methods of all procedures. The data, analytic methods, and study materials will be made available to other researchers, upon request for purposes of reproducing the results or replicating the procedure.
Animal Procedures
All animal experiments were approved by the Brigham and Women’s Hospital’s Animal Welfare Assurance (protocol 2016N000219). Male Ldlr-/- mice were fed a high-fat diet (1.25% cholesterol, D12108C, Research Diets, Inc., New Brunswick, NJ). To create experimental vein grafts in Ldlr-/- mice, inferior vena cava were harvested from a donor mouse and implanted into the carotid artery of recipient mice using cuff technique, as previously described.8,21 To induce liver-specific gain-of-function mutant PCSK9, we administered adeno-associated virus (AAV) encoding a gain-of-function form of mouse PCSK9 (AAV-PCSK9, 1×1011 vg, i.v.).22 We constructed AAV encoding LacZ (AAV-LacZ) that we used as a control for AAV-PCSK9. Four weeks after implantation, grafts were evaluated. After ultrasonography and in vivo molecular imaging of vein grafts, vein graft tissues were harvested, sectioned, and used for Masson trichrome staining, picrosirius red staining, immunohistochemistry, and in situ hybridization.
Culture and Stimulation of Mouse Primary Macrophages
Mouse peritoneal macrophages cultured with RPMI 1640 containing 10% fetal bovine serum were prepared as previously described.8 Briefly, 4% Brewer thioglycolate medium (BD Diagnostic Systems) was injected into the peritoneal cavity of mice 4 days before macrophage collection. After euthanasia, 10 mL of ice-cold phosphate-buffered saline was injected into the peritoneal cavity, and cells were harvested. The cells were then washed with phosphate-buffered saline once and plated for further experiments. Overnight starvation was performed with 0.5% fetal bovine serum before each experiment. Endotoxin-free recombinant mouse PCSK9 derived from mouse myeloma cell line NS0 was used to stimulate mouse primary macrophages (R&D Systems).
Transcriptomics
RNA-sequencing—mRNAseq (polyA enriched) library prep, single-end 75bp sequencing on NextSeq of 12-16 pooled barcoded samples, and VIPER analysis23—was performed at the Molecular Biology Core Facilities at Dana-Farber Cancer Institute. The data were then analyzed using DESeq2 and Qlucore (http://www.qlucore.com/) to perform a 2-group comparison and identify differentially expressed transcripts (increase/decrease by 2-fold) between control and PCSK9 groups. STRING (version 11.5) was used to evaluate known and predicted transcript interconnectivity. The differentially expressed transcripts (increase/decrease by 2-fold, q<0.05), which were compared with the “inflammatome”24 dataset, were used for the following network analysis.
Network-Based Hyperedge Entanglement Prediction Analysis
Network-Based Hyperedge Entanglement Prediction (HEP) algorithm20 was performed to estimate the likelihood of PCSK9 directly interacting with any of the differentially expressed transcripts in the “inflammatome.” Given only a network topology and its high-order hypergraph organization, the HEP algorithm uses an ensemble of link predictors to calculate a level of statistical significance for each candidate node to hyperedge entanglement that is characterized by not having a direct connection. The link predictor is an operator that, exploiting topological information of the graph, can associate a similarity score to any disconnected node pair, suggesting the likelihood for a link between them to exist. Several link predictors perform under the evidence that 2 nodes are more likely to be linked if their common neighbors are members of a strongly inner-linked cohort, named a local community,25 and recent studies demonstrated that this growth mechanism emerges mainly considering missing links between nonadjacent nodes distanced by paths of length 2 (L2) or 3 (L3)26 following a scheme that can be well described by the Cannistraci-Hebb network automata modeling.27 The HEP analysis provides 12 different estimations of the P value that quantifies the significance of the entanglement (association) between each node and hyperedge (also called node2hyperedge entanglement) using the 12 available node-node link prediction variants.25, 27 In this study, we applied HEP algorithm according to the following specifications: the scaling of the similarity scores was not necessary, and the average operator to estimate the node2hyperedge entanglement scores was the mean.
Statistics
Statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software). For experiments with a small sample size (n<6), P values were determined by nonparametric analysis except for RNA-sequencing analysis. Other data were examined for normality before analysis by Shapiro-Wilk test. For all normally distributed data, an unpaired 2-tailed Student t test was used for comparisons between 2 groups, 1-way ANOVA followed by Bonferroni or Dunnett post-hoc test was used to compare multiple groups. When there were 2 experimental factors, 2-way ANOVA for comparisons between multiple groups was used. If the data were not normally distributed, nonparametric unpaired 2-tailed Mann-Whitney U test was used to compare 2 groups, and Kruskal-Wallis test with Dunn post-hoc test was used to compare multiple groups. Wald Test in DESeq2 was used for bulk RNA-sequencing analysis. Data are expressed as mean ± SEM for continuous variables. P values of <0.05 were considered statistically significant.
Results
AAV-PCSK9 Promotes Vein Graft Lesion Development in an LDLR-Independent Manner
In vivo studies have suggested a direct link between PCSK9 and atherosclerosis28,29; however, no evidence has yet implicated PCSK9 in vein graft lesion development. Furthermore, these in vivo studies indicate that the underlying mechanism of atherosclerotic lesion development by PCSK9 may depend mainly on LDLR. Whether PCSK9 can exert pro-inflammatory effects via LDLR-independent mechanisms is unclear. To test the hypothesis that circulating PCSK9 promotes vein graft lesion development in vivo, we used experimental vein grafts in mice.21 A single intravenous injection of AAV-PCSK9 induced liver production of PCSK9. To exclude potential systemic toxicity of AAV, AAV-LacZ was developed and used as a control (Figure 1A; Online Supplemental Methods). All experiments used Ldlr-/- mice to explore LDLR-independent mechanisms unless otherwise noted.
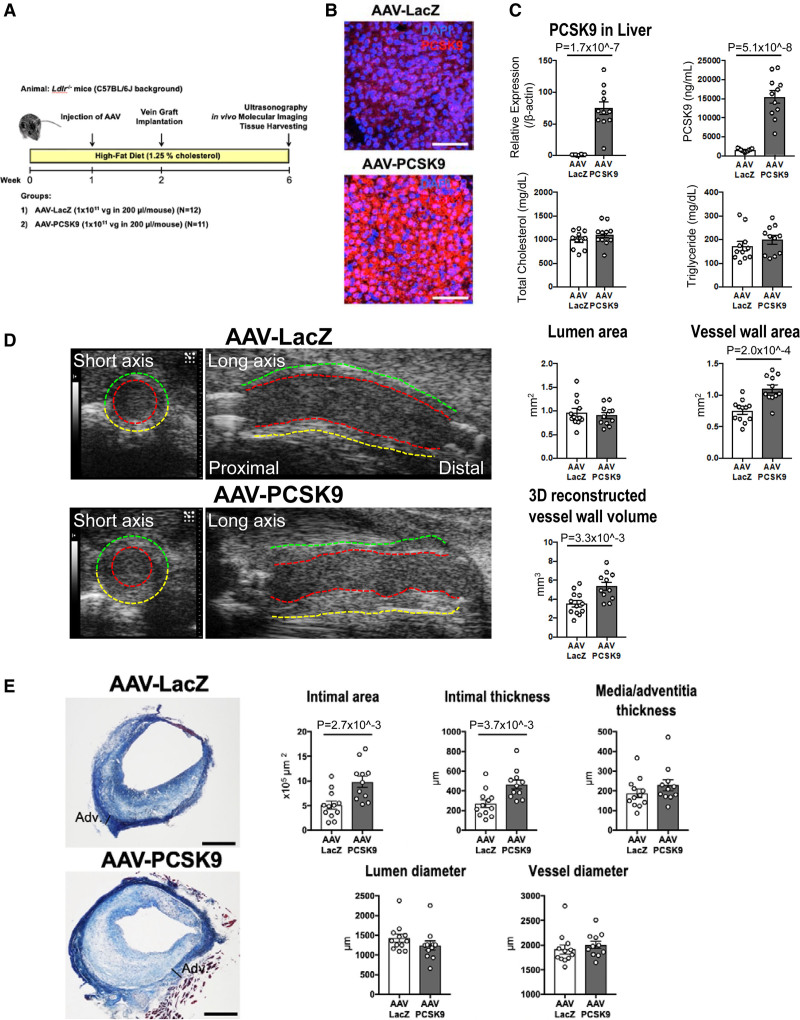
Figure 1.
The effects of Adeno-associated virus (AAV)-PCSK9 (proprotein convertase subtilisin/kexin 9) on vein graft lesion development in Ldlr-/- mice. A, An experimental protocol for the treatments in Ldlr-/- mice. B, In situ hybridization of PCSK9 (red) in the liver of Ldlr-/- mice 1 week after intravenous injection with AAV-LacZ or AAV-PCSK9. Nuclei are visualized with DAPI. The data represent 4 mice per group. Scale bars indicate 100 μm. C, PCSK9 mRNA level in the liver of AAV treated Ldlr-/- mice (n=12 and 11 for AAV-LacZ and AAV-PCSK9 groups, respectively), and serum levels of PCSK9, total cholesterol, and triglyceride in AAV treated Ldlr-/- mice (n=11 per group). D, Left, ultrasonography images of vein grafts from Ldlr-/- mice treated with AAV-LacZ or AAV-PCSK9 28 days after implantation. The red, green, and yellow dotted lines indicate lumen, near wall, and far wall, respectively. Right, The quantitative analysis of lumen and vessel wall area in short axis view and 3D reconstructed vessel wall volume of vein grafts by ultrasonography (n=12 and 11 for AAV-LacZ and AAV-PCSK9 groups, respectively). E, Left, Masson’s trichrome staining of vein grafts from Ldlr-/- mice treated with AAV-LacZ or AAV-PCSK9 28 days after implantation. Scale bars indicate 500 μm. Right, The quantitative analysis of intimal area, intimal and media/adventitia thickness, and lumen and vessel diameter of vein grafts (n=12 and 11 for AAV-LacZ and AAV-PCSK9 group, respectively). P value was calculated by Mann-Whitney U-test (triglyceride in C, media/adventitia thickness, lumen diameter, and vessel diameter in E) and unpaired Student t test (A, B, and D). Data are reported as mean ± SEM.
Body weight and blood pressure did not differ significantly between the AAV-LacZ and AAV-PCSK9 groups 4 weeks after vein graft implantation (Supplemental Table 1). In situ hybridization showed that AAV-PCSK9 markedly increased PCSK9 mRNA levels in the liver (Figure 1B). AAV-PCSK9 increased PCSK9 mRNA expression in the liver and serum levels of PCSK9 (Figure 1C; Supplemental Table 1). Serum levels of total cholesterol and triglycerides did not differ between the AAV-LacZ and AAV-PCSK9 groups (Figure 1C; Supplemental Table 1). These findings indicate that the effects of AAV-PCSK9 on vein graft lesions described below did not depend on blood cholesterol and triglyceride concentrations (Supplemental Table 1).
Noninvasive ultrasonography visualized that AAV-PCSK9 injection caused increased vessel wall area, but not lumen area, in the short axis view and 3-dimensional reconstructed vessel wall volume of vein grafts (Figure 1D). Consistent with the ultrasonography, histological analysis by Masson trichrome staining showed that AAV-PCSK9 increased intimal area and thickness of vein grafts. Lumen diameter, vessel diameter, and media/adventitia thickness did not differ significantly, suggesting compensatory outward remodeling of vein grafts (Figure 1E).
AAV-PCSK9 Increases Thin Collagen Fibers and Macrophage Accumulation in Vein Graft Lesions
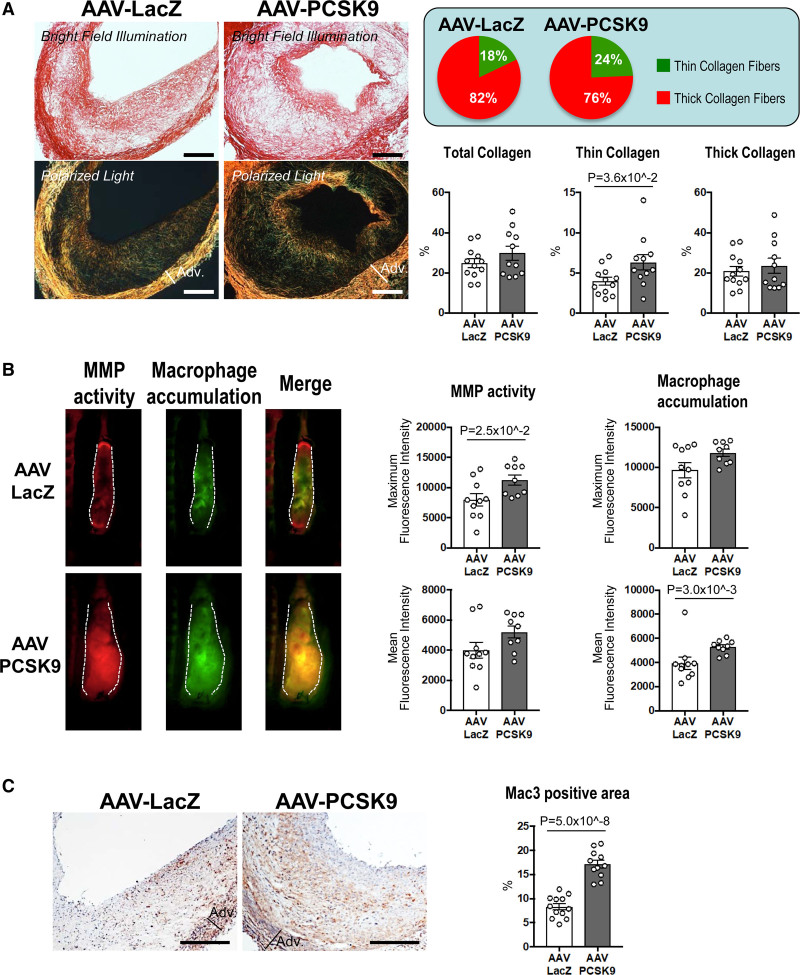
Evidence suggests that vein grafts in humans can develop lesions similar to those of advanced arterial atherosclerosis, and plaque rupture can occur in inflamed vein grafts.30,31 Therefore, exploring vein graft lesion composition in depth is critical to understanding what contributes to this occurrence. We then examined the collagen content of vein grafts by picrosirius red staining. AAV-PCSK9 increased the percentage of thin collagen fibers as a ratio of the total collagen fibers in the vein grafts, suggesting that circulating PCSK9 induced pathological features similar to those in thin-capped atherosclerotic plaques (Figure 2A). In vivo molecular imaging further provided insight into the effects of circulating PCSK9 on matrix metalloproteinase activity and macrophage accumulation in vein grafts. We intravenously co-injected 2 spectrally different imaging agents that elaborate near-infrared signals for visualization of matrix metalloproteinase activity (MMPsense, 680 nm) and macrophage accumulation (AminoSPARK, 750 nm). Following the evidence that activated macrophages express MMPs that degrade interstitial collagen as previously reported by our group,32,33 in vivo molecular imaging demonstrated that increased matrix metalloproteinase activity co-localized with accumulated macrophages (Figure 2B, left). In addition, AAV-PCSK9 treatment promoted matrix metalloproteinase activity and macrophage accumulation in vein grafts (Figure 2B), suggesting a potential mechanism of collagen remodeling as determined by Picrosirius red staining. Immunohistochemical analysis further demonstrated increased macrophage accumulation in vein grafts by AAV-PCSK9 (Mac3; Figure 2C). Macrophage proliferation and migration may contribute to the mechanisms of macrophage accumulation in vascular diseases.34 Supporting this mechanism, AAV-PCSK9 increased the percentage of macrophages positive for Ki-67, an indicator of proliferating cells (Supplemental Figure 1A, B). Consistent with this in vivo finding, recombinant PCSK9 augmented M-CSF-induced cell proliferation in mouse bone marrow-derived macrophages (BMDMs) (Supplemental Figure 2A, B). Monocyte migration assay using the human monocytic cell line THP-1 showed that recombinant PCSK9 tended to promote MCP-1 (monocyte chemoattractant protein-1)-induced chemotaxis, suggesting lipid-independent effects of PCSK9 on monocyte migration (Supplemental Figure 2C). On the other hand, AAV-PCSK9 did not increase apoptosis determined by TUNEL staining in vein grafts of Ldlr-/- mice (Supplemental Figure 3A). Accordingly, AAV-PCSK9 did not change necrotic core area in the vein grafts (Supplemental Figure 3B).
Figure 2.
The effects of Adeno-associated virus (AAV)-PCSK9 (proprotein convertase subtilisin/kexin 9) on collagen thinning and macrophage accumulation in vein graft lesions of Ldlr-/- mice. A, Picrosirius red staining of vein grafts without (top) or with (bottom) polarized light (n=12 and 11 for AAV-LacZ and AAV-PCSK9 group, respectively). Scale bars indicate 500 μm. Circle graphs show the percentage of thin (green) and thick (red) collagen fibers compared with total fibers. B, Intravital microscopy images of MMPSense 680 (red) and AminoSPARK750 (green) in vein grafts for visualization of MMP activity and macrophage accumulation, respectively (n=10 and 9 for AAV-LacZ and AAV-PCSK9 group, respectively). C, Mac3 (macrophages) staining in vein grafts. Scale bars indicate 500 μm. P value was calculated by Mann-Whitney U-test (thick collagen in A, macrophage accumulation mean fluorescence intensity in B) and unpaired Student t test (C). Data are reported as mean ± SEM.
AAV-PCSK9 Induces Macrophage Activation via an LDLR-Independent Route In Vivo
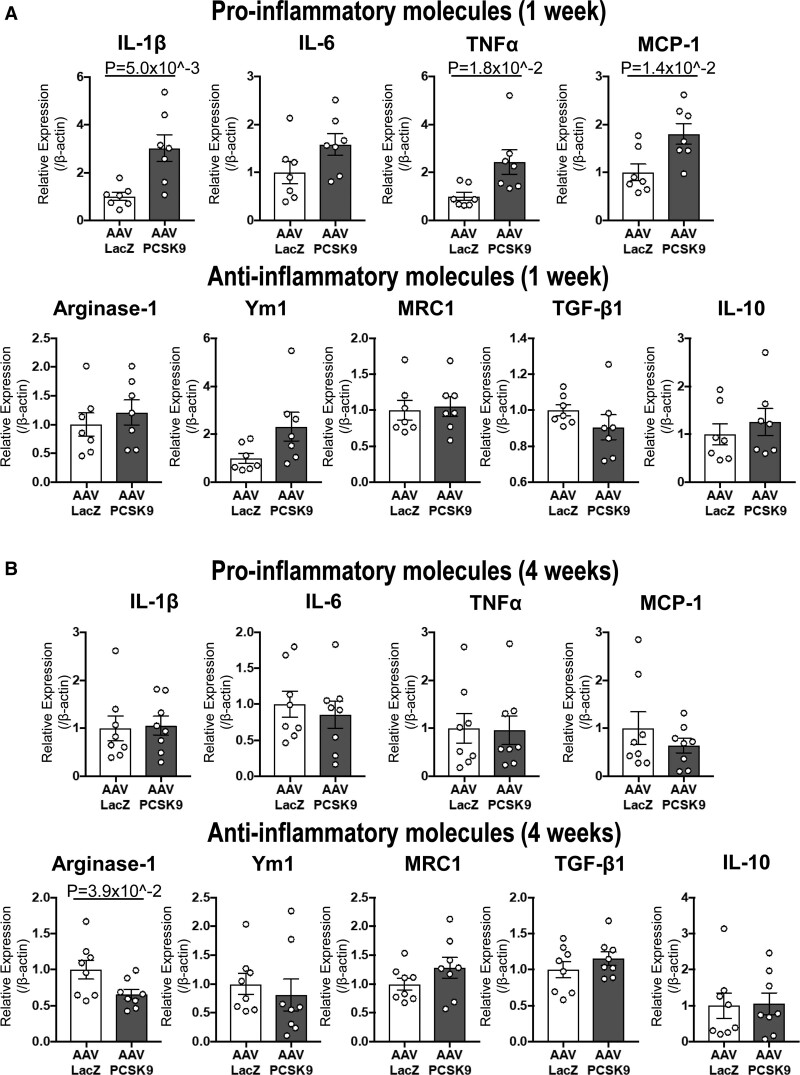
We previously reported that macrophage activation plays a key role in vein graft lesion development.8 To explore the effects of circulating PCSK9 on macrophage activation, we examined mRNA levels of pro-inflammatory (IL-1β [interleukin-1 beta], IL-6, TNFα, and MCP-1) and antiinflammatory molecules (Arginase-1, Ym1, MRC1, TGF-β1 [transforming growth factor-beta 1], and IL-10) in the peritoneal F4/80+ macrophages of Ldlr-/- mice 1 or 4 weeks after intravenous injection of AAVs. AAV-PCSK9 promoted pro-inflammatory IL-1β, IL-6, TNFα, and MCP-1 mRNA levels at 1 week (Figure 3A). AAV-PCSK9 also suppressed Arginase-1 mRNA levels at 4 weeks when the increase in the mRNA levels of pro-inflammatory molecules at 1 week had subsided (Figure 3B). We have recently reported that predominant pathways of the vein grafts change over time during lesion development. In the same study, pathways with immune responses represent the early responsive proteins in proteomics analysis.9 These results indicate that PCSK9 can induce pro-inflammatory responses in macrophages initially and later impair resolution of inflammation in vivo.
Figure 3.
Adeno-associated virus (AAV)-PCSK9 (proprotein convertase subtilisin/kexin 9) induced macrophage activation in murine peritoneal macrophages via LDL receptor (LDLR)-independent mechanisms in vivo. mRNA levels of pro-inflammatory molecules (IL-1β, IL-6, TNFα, MCP-1) and antiinflammatory molecules (Arginase-1, Ym1, MRC1, TGF-β1, IL-10) were measured in murine peritoneal macrophages of Ldlr-/- mice 1 week (A) or 4 weeks (B) after intravenous injection with AAV-LacZ or AAV-PCSK9 (n=7 to 8 per group). P value was calculated by Mann-Whitney U-test (TNFa in A, IL-1b, TNFa, MCP-1, Arginase-1, Ym1, and IL-10 in B) and unpaired Student t test. Data are reported as mean ± SEM.
Endotoxin-Free Recombinant PCSK9 Induced Macrophage Activation in Peritoneal Macrophages of Ldlr -/- Mice In Vitro
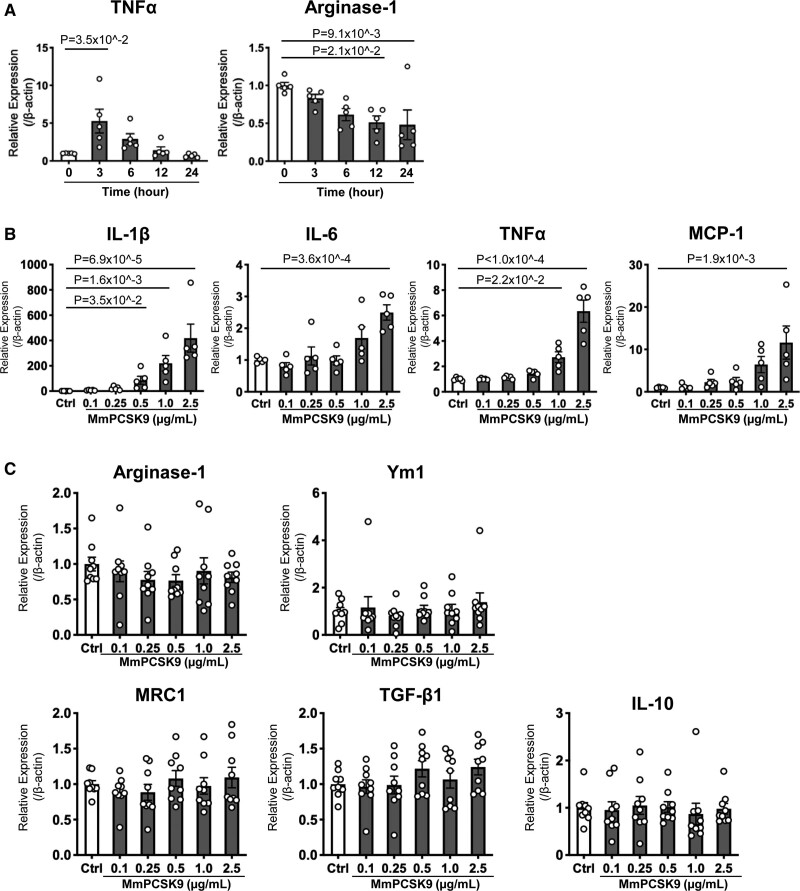
To support our in vivo evidence, we examined in vitro the effects of physiologically relevant levels of recombinant mouse PCSK9 (0.1–2.5 μg/mL)35 on the mRNA levels of the same pro-inflammatory and antiinflammatory molecules examined in Ldlr-/- mouse macrophages. First, we conducted a time-course study of recombinant PCSK9-induced TNFα and Arginase-1 mRNA expression levels to determine an optimal time point for harvesting Ldlr-/- mouse macrophages. The increase of TNFα mRNA levels peaked 3 hours after stimulation, whereas the decrease of Arginase-1 mRNA levels plateaued 12 hours after stimulation with recombinant PCSK9 (Figure 4A). For all experiments, Ldlr-/- mouse macrophages were therefore harvested 3 and 12 hours after stimulation to measure pro- and antiinflammatory molecules, respectively. Peritoneal macrophages treated with recombinant PCSK9 exhibited increased mRNA levels of IL-1β, IL-6, TNFα, and MCP-1 in a concentration-dependent manner (Figure 4B). In contrast, recombinant PCSK9 did not decrease the mRNA levels of antiinflammatory molecules (Figure 4C). These results support the in vivo evidence of macrophage activation by circulating PCSK9 in an LDLR-independent fashion.
Figure 4.
Recombinant PCSK9 (proprotein convertase subtilisin/kexin 9) induced macrophage activation in LDL receptor (LDLR)-independent mechanisms in vitro. A, Time course of TNFα and Arginase-1 mRNA levels in peritoneal macrophages of wild-type mice after stimulation with recombinant mouse PCSK9 (MmPCSK9; 2.5 μg/mL) (n=5 per group). P value was calculated by Kruskal-Wallis test, followed by Dunn multiple comparison test. B, mRNA levels of pro-inflammatory molecules (IL-1β, IL-6, TNFα, MCP-1) and (C) antiinflammatory molecules (Arginase-1, Ym1, MRC1, TGF-β1, IL-10) were measured in murine peritoneal macrophages from Ldlr-/- mice 3 and 12 hours after stimulation with recombinant mouse PCSK9 for pro-inflammatory and antiinflammatory molecules, respectively (n=9 per group). P value was calculated by 1-way ANOVA, followed by Dunnett multiple comparison test (IL-6, TNFa, MCP-1‚ MRC1) or Kruskal-Wallis test, followed by Dunn multiple comparison test. Data are reported as mean ± SEM.
Endotoxin contamination in recombinant proteins can induce pro-inflammatory responses. Therefore, we chose a recombinant protein derived from a mammalian system, mouse myeloma cell line (NS0). The endotoxin level of recombinant mouse PCSK9 used in this study was 0.00441 EU/µg or less, well below the threshold of 0.1 EU/µg required for accurate cell-based assays.19 To exclude further endotoxin contamination in the recombinant PCSK9 protein affecting any measurements, we performed the chromogenic Limulus amebocyte lysate endotoxin assay. The final endotoxin level of the maximum dose of the recombinant protein (2.5 µg/mL) was 0.005 EU/mL or less, which coincides with the endotoxin levels of the commercially available endotoxin-free medium. In addition, mass spectrometry conducted on the solvent derived from reconstituted recombinant mouse PCSK9 protein solution did not produce signals, suggesting that there were no small molecule impurities present (Supplemental Figure 4A). Furthermore, stimulation with heat-inactivated recombinant mouse PCSK9 protein (2.5 μg/mL) caused no changes in the mRNA levels of IL-1β, TNF-α, and MCP-1, demonstrating that endotoxin contamination did not drive the effects observed (Supplemental Figure 4B).
A combination of Unbiased Global Transcriptomics and Network-Based Hyperedge Entanglement Prediction Analysis Revealed Potential Targets of PCSK9 in Macrophages
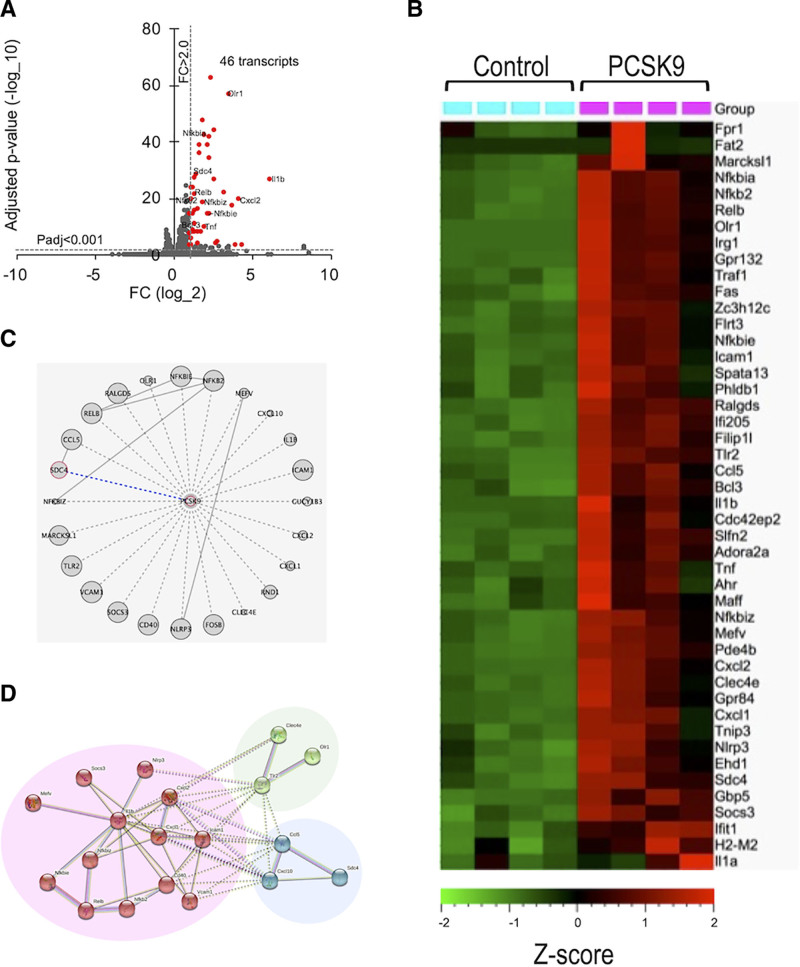
We explored further LDLR-independent pro-inflammatory signaling pathways in an unbiased manner, using RNA-sequencing of Ldlr-/- mouse macrophages stimulated with recombinant PCSK9. When analyzing the differentially expressed transcripts (FC>2.0, q<0.001), we observed 46 transcripts significantly increased by PCSK9 stimulation in Ldlr-/- macrophages compared with controls (Figure 5A). A hierarchical heat map of Ldlr-/- mouse macrophages filtered by differentially expressed transcripts showed a clear separation between control (no stimulus) and PCSK9 group (Figure 5B). We provided statistically significantly overrepresented transcripts ranked according to their fold change abundance (Supplemental Table 3A) to further examine these differentially expressed transcripts. PCSK9 treatment increased many transcripts implicated in pro-inflammatory responses in Ldlr-/- macrophages (eg, IL-1b, TNF, CXCL2, NF-κB [nuclear factor-kappa B]-related genes).19
Figure 5.
Transcriptomics, network analysis, and hyperedge entanglement prediction (HEP) of mouse macrophages stimulated with recombinant PCSK9 (proprotein convertase subtilisin/kexin 9). A, Volcano plot for Ldlr-/- mouse macrophages stimulated with recombinant PCSK9. Red markers indicate significantly expressed transcripts with a fold change cutoff of 2.0 and adjusted P value cutoff of 0.001. The enriched transcripts that were previously reported were denoted with their names. B, Heat map of the transcriptomics of Ldlr-/- mouse macrophages (FC>2, q<0.001-filtered data for the difference between macrophages stimulated with and without recombinant PCSK9, n=4 per group). C, The schematic network visualization by HEP prediction. The significantly predicted association of PCSK9 to the 24 mapped inflammation-related transcripts derived from the list of transcripts (FC>2.0, q<0.05) indicates every unknown pairs of interaction (dashed line) that have to be proven yet experimentally. One of them was validated by silencing Sdc4 (syndecan-4) mRNA (blue dashed line). All other links represent known physical interactions (solid gray lines). The node size illustrates the degree of the proteins in the protein-protein interaction network. D, Network analysis of the 24 overrepresented transcripts in Ldlr-/- mouse macrophages using STRING database. Nodes are colored according to k-means clustering (number of clusters=3). Only connected nodes are shown here.
To provide additional mechanistic links, we attempted to identify PCSK9-regulated proteins other than LDLR that mediate the aforementioned pro-inflammatory downstream responses. Many cascading effects in the transcriptional regulatory machinery could control the differentially expressed mRNAs. This set of differentially expressed transcripts were identified as the result of a genome-wide transcriptional profiling method, which might not necessarily interact directly with PCSK9. To estimate the likelihood of PCSK9 directly interacting with those differentially expressed transcripts, we tested this hypothesis verification through network computational analysis. In brief, PCSK9 can be seen as a node in a protein-protein interaction network,36 and the set of differentially expressed transcripts can be seen as a hyperedge projected on the protein-protein interaction network (a group of nodes that are associated by a common feature of biological functions). Muscoloni et al recently introduced a novel algorithm for HEP that exploits an ensemble of link predictors and that provides a level of statistical significance for the entanglement between each node and hyperedge that does not have a direct interaction in the network.20 We then performed the HEP analysis to quantify the significance of the entanglement between PCSK9 and the hyperedge of 24 differentially expressed transcripts (FC>2.0, q<0.05) in the dataset (Supplemental Figure 5A and 5B, Supplemental Table 3B). The resulting P values of 7/12 node-node link prediction methods (RA, CH2-L2, CH3-L2, and all 4 L3-based methods) established a significant association between PCSK9 and the hyperedge of the 24 differentially expressed transcripts (Figure 5C; Supplemental Table 2). These data suggest that any of the 24 differentially expressed transcripts have a high likelihood of directly interacting with PCSK9, enabling us to validate a direct interaction between PCSK9 and one of these 24 differentially expressed transcripts. To understand the biological relevance of these transcripts, we explored the network connectivity of these transcripts identified by HEP analysis using the STRING database (confidence interaction score ≥0.7).37 The network analysis showed a highly clustered network (average local clustering coefficient: 0.547) containing 24 nodes with 52 edges (expected number of edges=3), providing significantly more interactions than expected for a random set of genes of similar size (P value ≤1.0e−16). The k-means clustering (number of clusters=3) showed that one of these 3 clusters mainly consisted of the NF-κB signaling pathway based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (IL-1b, CXCL1 [chemokine (C-X-C motif) ligand 1], CXCL2 [chemokine (C-X-C motif) ligand 2], ICAM1, VCAM1 [vascular cell adhesion molecule 1], RELB [RELB Proto-Oncogene, NF-kB subunit], NFKB2 [nuclear factor kappa B subunit 2]), and the other 2 clusters included OLR1 (LOX-1) and SDC4, respectively (Figure 5D).
It is already reported that NF-κB signaling pathway is involved in PCSK9-induced atherosclerotic inflammation.38 In the present study, AAV-PCSK9 promoted NF-κB p65 mRNA level in the peritoneal F4/80+ macrophages of Ldlr-/- mice (Supplemental Figure 6A). Inhibition of NF-κB signaling pathway with IκB kinase inhibitor, 2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide (TPCA1), decreased pro-inflammatory responses to recombinant PCSK9 in Ldlr-/- mouse macrophages (Supplemental Figure 6B). These data indicate that NF-κB may mediate PCSK9-induced pro-inflammatory responses in vein graft lesions.
PCSK9 stimulates the expression of LOX-1, which in turn takes up ox-LDL in macrophages.39 AAV-PCSK9 increased foam cell formation determined by Oil red O positive area in vein grafts (Supplemental Figure 7A). We further found that recombinant PCSK9 increased ox-LDL uptake in bone marrow-derived macrophages from Ldlr-/- mice (Supplemental Figure 7B). These results indicate that PCSK9 also induces foam cell formation by increased ox-LDL uptake, contributing not only to atherosclerotic but also to vein graft lesion development.
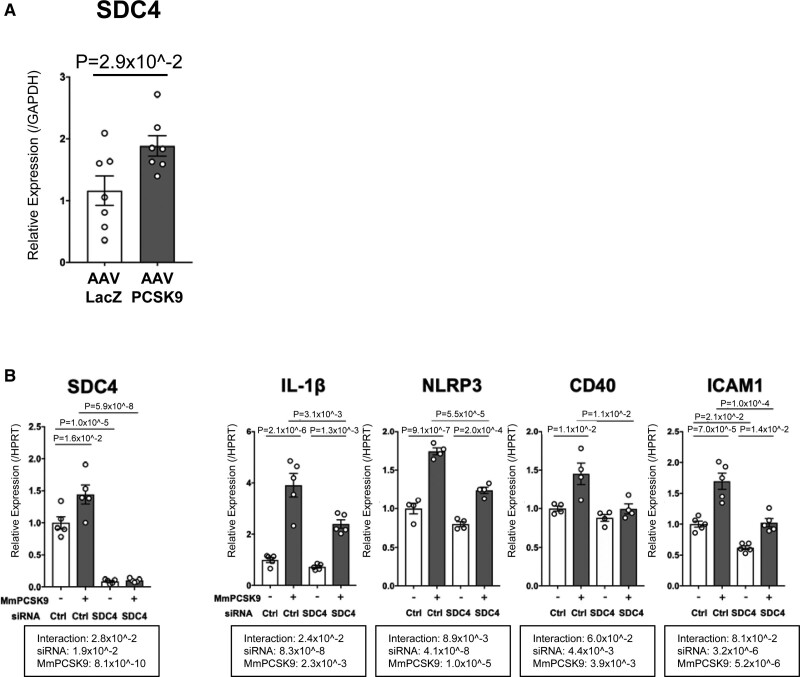
Among the members of this differentially expressed transcript hyperedge, we found that SDC4 included in the last cluster could be a novel target of PCSK9 (Figure 5D). While SDC4 was not a high-ranking molecule in the transcript ranking lists (30th in Supplemental Table 3A and 67th in Supplemental Table 3B), its fold change after stimulation with recombinant PCSK9 in Ldlr-/- mice macrophages was statistically significant (2.65, log2FC>1). AAV-PCSK9 also increased SDC4 mRNA level in the peritoneal F4/80+ macrophages of Ldlr-/- mice (Figure 6A). SDC4 is a heparan sulfate proteoglycan40 expressed on the surface of human macrophages.41 In addition, SDC4 mRNA is increased in M(LPS) but not M(IL-4/IL-13) or M(IL-10) in bone marrow-derived macrophages.42 A recent study reported that heparan sulfate proteoglycans physically bind to PCSK9 on the hepatocyte surface43 However, the investigators did not address the interaction between PCSK9 and any specific individual syndecans. Our in silico protein-protein docking analysis using HPEPDOCK, a web server for protein-protein docking based on a hierarchical algorithm, predicted binding between PCSK9 and SDC4 (Supplemental Figure 8A).44 We entered the pdb files of PCSK9 (PDB ID: 2PMW), LDLR (PDB ID: 1N7D), and SDC4 (PDB ID: 1EJP) into HPEPDOCK server, which in turn presented the top 100 docking models based on docking energy minimized scores. Protein-protein docking analysis showed that the binding efficiency of SDC4 and PCSK9 (−212.23 to −235.6 kJ/mol) was comparable to that of LDLR and PCSK9 (−259.85 to −303.21 kJ/mol) based on the docking score of the top 5 binding predictions (Supplemental Figure 8A). Besides, the ligand root mean square deviation, often used to evaluate the correctness of the docking geometry and optimal superimposition of the receptor-ligand binding, was lower in the binding of SDC4 and PCSK9 (39.12–85.16) compared with that of LDLR and PCSK9 (43.76–156.25) for the top 5 binding predictions, suggesting better binding between SDC4 and PCSK9 than in LDLR and PCSK9. Furthermore, co-immunoprecipitation analysis of mouse liver tissue lysates validated the occurrence of the predicted PCSK9-SDC4 binding in vivo (Supplemental Figure 8B).
Figure 6.
RNA silencing validated SDC4 (syndecan-4) as a potential target of PCSK9 (proprotein convertase subtilisin/kexin 9)-induced pro-inflammatory response in macrophages, identified via hyperedge entanglement prediction (HEP) prediction. A, SDC4 mRNA level was measured in murine peritoneal macrophages of Ldlr-/- mice 1 week after intravenous injection with AAV-LacZ or AAV-PCSK9 (n=7 per group). B, mRNA levels of SDC4, IL-1b, NLRP3, CD40, and ICAM1 were measured by RT-qPCR in mouse Ldlr-/- peritoneal macrophages after pretreatment with siSDC4 or siControl (Ctrl) for 48 hours and stimulation with 2.5 μg/mL recombinant mouse PCSK9 (MmPCSK9) for 3 hours (n=4–5). P value was calculated by 1-way ANOVA, followed by Bonferroni multiple comparison test. Error bars indicate ± SEM. Data are reported as mean ± SEM.
SDC4 Silencing Suppresses PCSK9-Induced Pro-Inflammatory Responses in Macrophages
We further validated our prediction that SDC4 is a potential target of PCSK9 using siRNA silencing of mouse Sdc4 in mouse peritoneal macrophages. Silencing Sdc4 with siSDC4 treatment achieved 93% reduction of SDC4 mRNA expression (Figure 6B). siSDC4 was then used to determine whether it could suppress PCSK9-induced pro-inflammatory molecules, including those predicted to be linked with PCSK9 by the HEP prediction (Figure 5C). siSDC4 suppressed mRNAs of the pro-inflammatory molecules IL-1β, NLRP3, CD40, and ICAM1, but not TNFα, MCP-1, TLR2, CXCL1, CXCL2, or CXCL10 after stimulation with recombinant PCSK9 (Figure 6B, Supplemental Figure 9). These data suggest that SDC4 partially regulates a PCSK9-induced pro-inflammatory response. Furthermore, these mRNA silencing results substantiate the validity of the prediction, yielding the integration of PCSK9 in our hyperedge of 24 disconnected differentially expressed transcripts.
Local Production of PCSK9 in the Liver Was Greater Than in Vein Graft Lesions
Our study demonstrated that circulating PCSK9 produced various changes in Ldlr-/- mice. We, however, attempted to examine the potential impact of local PCSK9 production in vein grafts. Immunohistochemical analysis using an anti-PCSK9 antibody in previous studies showed that vascular smooth muscle cells in human atherosclerotic plaque and collar-induced neointima of murine carotid artery express PCSK9.45,46 Based on these results, we examined whether locally produced PCSK9 in vein grafts may also contribute to vein graft lesion development. The local expression of PCSK9, as determined by in situ hybridization in the intima of vein grafts, was minimal compared with that in the liver (Figure 1B; Supplemental Figure 10A). RT-qPCR analysis further confirmed numerically significantly less PCSK9 mRNA expression in vein grafts than in the liver, the primary source of circulating PCSK9 (Supplemental Figure 10B). Moreover, we examined which cells within vein grafts can produce PCSK9 in humans by comparing PCSK9 mRNA levels in human saphenous vein endothelial cells, smooth muscle cells, and human primary macrophages derived from peripheral blood mononuclear cells. While we detected substantial PCSK9 expression in human saphenous vein smooth muscle cells, the level was significantly lower than that in HepG2 (liver) cells (Supplemental Figure 10C). Human primary macrophages and human saphenous vein endothelial cells contained minimal PCSK9 mRNA (Supplemental Figure 10C). Recent studies reported a modest increase of PCSK9 mRNA and protein levels in macrophages exposed to pro-inflammatory stimuli.39,47 We, therefore, examined PCSK9 mRNA expression in human primary macrophages treated with pro-inflammatory stimuli. Although LPS and IFNγ induced a pro-inflammatory response in the macrophages, as determined by TNFα mRNA expression (Supplemental Figures 10D and 10E, right graphs), PCSK9 mRNA expression did not increase after LPS or IFNγ (Supplemental Figures 10D and 10E, left graphs). These results indicate that smooth muscle cells (SMC), but not macrophages, may primarily produce PCSK9 in vein grafts, but to a much lower extent than the liver.
Macrophages Rather Than Endothelial Cells and SMCs May Mediate the Effects of PCSK9 on Vein Graft Lesion Development
Although our study focuses on macrophages, we examined the role of PCSK9 in the activation of endothelial cells and SMCs because other investigators reported potential interactions between PCSK9 and these cell types.46,48 Endothelial cell activation precedes vein graft lesion development.49 Due to the scarcity of mRNA in CD31-positive endothelial cells in vein grafts, we sorted them from the liver and lungs of Ldlr-/- mice injected with AAV-PCSK9 instead to examine the effects of PCSK9 on endothelial cell activation in an LDLR-independent fashion. AAV-PCSK9 did not increase mRNA levels of adhesion molecules, such as VCAM-1, ICAM-1, and E-selectin, in EC from either organ when compared with AAV-LacZ (Supplemental Figure 11A). We further found in vitro that recombinant PCSK9 did not increase mRNA levels of these adhesion molecules in human saphenous vein endothelial cells (Supplemental Figure 11B). These results indirectly indicate that PCSK9 may not induce endothelial activation in vein grafts.
SMC modulation, migration, and proliferation contribute to the development of vascular disorders, including atherosclerosis50 and vein graft.51 A recent study showed that PCSK9 may sustain SMC dedifferentiation, migration, and proliferation in neointimal hyperplasia in response to vascular injury,46 suggesting the involvement of SMCs in PCSK9-induced vein graft lesion development. To explore this possibility, we examined the effects of recombinant PCSK9 on SMC dedifferentiation, migration, and proliferation in human saphenous vein smooth muscle cells. Recombinant PCSK9 slightly decreased SM22α mRNA levels (0.74 and 0.68-fold for 1.0 and 5.0 µg/mL, respectively), but did not affect differentiation molecules, such as αSMA, calponin, SM1, or SM2 (Supplemental Figure 12A). In addition, recombinant PCSK9 did not affect PDGF-induced migration activity of human saphenous vein smooth muscle cells (Supplemental Figure 12B). A BrdU assay using human saphenous vein smooth muscle cells showed that recombinant PCSK9 did not enhance 10% fetal bovine serum-induced proliferation (Supplemental Figure 12C). AAV-PCSK9 did not affect the SMC content in vein grafts (Supplemental Figure 12D). Collectively, in our experimental settings, our data did not provide the mechanistic evidence in vitro and in vivo that activation of endothelial cells or SMCs contribute to PCSK9-induced vein graft lesion formation. These results indicate that circulating PCSK9 promotes vein graft lesion development predominantly through macrophage activation.
Discussion
This study investigated LDLR-independent mechanisms by which PCSK9 induces pro-inflammatory activation of macrophages and accelerates vein graft lesion development. The key findings include: (1) AAV-PCSK9 did not change lipid profile but promoted vein graft lesion development in Ldlr-/- mice, suggesting LDLR-independent mechanisms; (2) AAV-PCSK9 increased macrophage accumulation and decreased fibrillar collagen in vein graft lesions; (3) PCSK9 induced macrophage activation in Ldlr-/- mice; (4) unbiased global transcriptomics of Ldlr-/- mouse macrophages stimulated with recombinant PCSK9 revealed activation of several pro-inflammatory pathways; (5) a combination of the global transcriptomics and novel network-based hyperedge entanglement prediction analysis showed NF-κB signaling pathway and OLR1(LOX-1) could play a key role in vein graft lesion development; (6) SDC4, a heparan sulfate proteoglycan, could be a novel target of PCSK9 that mediates pro-inflammatory responses in macrophages.
Reversed autologous saphenous veins are widely used for surgical bypass for treatment of lower extremity peripheral artery disease and coronary artery disease. Peripheral artery disease affects approximately 8.5 million people in the United States aged ≥40 years52 and a total of 202 million worldwide, as of 2010.53 The incidence of peripheral artery disease has been rising by 23.5% globally from 2000 to 2010 largely due to aging populations.53 Of note, 40% of vein grafts for peripheral artery disease are failing or fail within a year.1 The volume of CABG procedures for severe coronary artery disease has declined since 1998; however, 371 000 bypass procedures were still performed in the United States in 2014,52 and approximately 30% of these costly procedures are failing or fail 12 to 18 months after surgery.2
Evidence suggests that pro-inflammatory responses to PCSK9 in macrophages and arterial atherosclerotic lesions may primarily depend on LDLR. Recombinant PCSK9 induced TNFα mRNA in bone marrow-derived macrophages mainly in an LDLR-dependent fashion.19 Furthermore, overexpression of human PCSK9 in macrophages promoted atherosclerotic lesions.28 Deletion of the PCSK9 gene in the liver reduced atherosclerotic lesions, primarily via mechanisms dependent on LDLR.29 To the best of our knowledge, no in vivo studies have demonstrated LDLR-independent pro-inflammatory effects of PCSK9 on vascular lesion development. The present study demonstrated that circulating PCSK9 induces macrophage activation and vein graft lesion development in LDLR-independent mechanisms. The relative contributions of the LDLR-dependent and independent mechanisms, however, remain unknown.
Clinical reports demonstrated that cholesterol-lowering with PCSK9 inhibitors reverses the pro-inflammatory profile of monocytes in patients with hypercholesterolemia,54 indicating the lipid-dependent pro-inflammatory effects of PCSK9 in humans. Clinical trials also demonstrated that PCSK9 inhibitors lower not only serum levels of LDL cholesterol but also lipoprotein (a) (Lp(a)).55 Mice lack apolipoprotein (a) or Lp(a),56 which enabled us to rule out the role of Lp(a) in the effects of PCSK9 in our in vivo study. Although genetic evidence suggests LDL-dependent effects of PCSK9 in gain-of-function and loss-of-function carriers57 and the clinical benefits of antiPCSK9 strategies derive largely from LDL lowering, nonLDL-dependent effects might also contribute.58 As serum total cholesterol and triglyceride levels did not differ in AAV-LacZ and AAV-PCSK9 groups, our findings in Ldlr-/- mice do not seem to depend on PCSK9’s effects on the profile of circulating lipids.
High sensitivity C-reactive protein is the most established marker of inflammation to predict cardiovascular events independently in primary and secondary prevention.59 Clinical trials with PCSK9 inhibitors did not show any significant changes in high sensitivity C-reactive protein levels in patients with coronary artery disease.60,61 Antiinflammatory changes in monocyte phenotype with PCSK9 antibodies were not accompanied by a CRP (C-reactive protein) reduction in patients with familial hypercholesterolemia, suggesting that CRP may not solely capture changes in local inflammation produced by PCSK9 inhibition.54 In addition, the ATHEROREMO-IVUS study (The European Collaborative Project on Inflammation and Vascular Wall Remodeling in Atherosclerosis - Intravascular Ultrasound) demonstrated a linear relationship between serum levels of PCSK9 and the fraction or amount of the necrotic core in the nonculprit lesions of patients with acute coronary syndrome, independently of serum levels of LDL cholesterol. Our study provides support for the notion that circulating PCSK9 may exert pro-inflammatory effects on vascular lesions. Further studies are needed to explore whether PCSK9 inhibition ameliorates local inflammation in vascular lesions in patients.
In general, multi-omics technologies, such as proteomics and transcriptomics, identify many differentially expressed molecules. It is challenging to assess direct interactions between these molecules or their causal roles in a biological or disease context by examining each molecule or interaction among many. This challenge has driven efforts in computational network biomedicine to develop unbiased prediction methods. By exploiting the mere topology of a protein-protein interaction network, one can quantify the likelihood of direct interactions between a molecule of interest (eg, PCSK9 in the present study) and a hyperedge of differentially expressed molecules (which represents a network functional module). We used a novel node2hyperedge entanglement method20 and showed any of the 24 differentially expressed transcripts, including NF-κB signaling molecules, OLR1(LOX-1), and SDC4, as a potential target of PCSK9 associated with inflammation. In the present study, we identified SDC4 as a potential novel target of PCSK9. Although Sdc4 was not a high-ranking differential expressed transcript, its fold change after stimulation with recombinant PCSK9 in the RNA-sequencing was statistically significant. We further performed in vitro experiments involving siRNA silencing in primary macrophages to substantiate our computational prediction platform and demonstrated that SDC4 indeed binds to PCSK9 in vivo and mediates PCSK9-induced pro-inflammatory responses. The contribution of macrophage SDC4 to PCSK9-induced pro-inflammatory responses in vivo remains to be elucidated. This novel prediction method may offer a powerful and necessary tool for the completion of the human interactome and for generating new hypotheses.
SDC4 regulates several robust pro-inflammatory molecules, such as IL-1β, NLRP3 (NLR family pyrin domain containing 3), CD40 (cluster of differentiation 40) and ICAM1. Various endogenous signals abundantly present in atherosclerotic lesions activates NLRP3, a key mediator of IL-1β maturation.62,63 T cell-mediated macrophage activation involves CD40 to produce interstitial collagens and tissue-factor pro-coagulant.64 The role of macrophage ICAM1 in atherosclerosis remains obscure, but a recent study reported that macrophage ICAM1 suppresses “M2-like” macrophages during tumor progression. SDC4 silencing inhibited these pro-inflammatory molecules. The magnitude of inhibition, however, was modest, suggesting that while we propose SDC4 is a novel target of PCSK9, but PCSK9 may also target other molecules (eg, NF-κB signaling molecules). The mechanisms mediated via SDC4 we report here appears to be one of multiple pathways affected by PCSK9. Heparan sulfate proteoglycans modulate macrophage activation by maintaining cells in a quiescent state through the capture and sequestration of IFNβ.65,66 We speculate that SDC4 serves as a mediator of PCSK9-induced pro-inflammatory responses through the capture of PCSK9 on the surface of macrophages as seen in the hepatocyte.43 Our data demonstrated that NF-κB may mediate pro-inflammatory responses downstream of the PCSK9 and SDC4 binding. Understanding more detailed mechanisms by which SDC4 mediates the pro-inflammatory responses and their downstream signaling induced by PCSK9 requires future investigations.
Our study does not demonstrate definitively that local PCSK9 plays an important role in vein graft lesion development. In situ hybridization and qPCR data suggest that SMCs may be the primary source of PCSK9 in these lesions. The expression level of PCSK9 in SMCs was much lower than in the liver, although the relative gene expression level may not be a valid determinant of its biological role or importance. Whether and how circulating PCSK9 affects lesional macrophages through the contact with endothelial cells (which in turn affects macrophage phenotype or the entry into the subendothelial space, which results in direct contact with macrophages) remains to be elucidated.67 Future work needs to define detailed molecular mechanisms.
Our data do not demonstrate that human primary macrophages can express PCSK9, even when stimulated with pro-inflammatory stimuli, which was consistent with previous studies.45,68 Evidence suggests, however, the participation of endogenous PCSK9 in macrophages during pro-inflammatory responses. In vitro experiments showed that overexpression of PCSK9 in macrophage-like THP-1 cells enhanced the pro-inflammatory response induced by oxidized LDL via NF-κB activation.38 In contrast, PCSK9 gene silencing inhibited the response.38,47 In vivo evidence also showed that overexpression of human PCSK9 in macrophages increased atherosclerotic lesion formation in Apoe-/- mice.28 These results suggest that not only circulating PCSK9 produced from the liver but also local PCSK9 produced from SMCs and macrophages may induce a pro-inflammatory response in macrophages and contribute to vascular lesion development. What cell types other than SMCs contribute to monocyte/macrophage infiltration, and activation remains to be elucidated.
The present study demonstrates several lines of novel evidence that circulating PCSK9 promotes macrophage activation and vein graft lesion development via mechanisms independent of blood cholesterol and LDLR degradation. Despite current therapeutics and research advancements, effective medical solutions are limited to prevent vein graft failure. Our study provides molecular bases that circulating PCSK9 deserves investigations as a potential therapeutic target for vein graft failure.
Article Information
Acknowledgments
We thank Ryo Kawakami, Yuki Tsukano, Dayanna C. Romero, Whitney S. Golden, Keishi Nihira, Andrew K. Mlynarchik, Edward Guzman, and Daniel G. Anderson for their technical assistance. The graphical figure was created with Biorender.com. Additional Information: Coauthor Amitabh Sharma died on November 24, 2019.
Sources of Funding
This work was supported by the National Institute of Health grants R01HL126901 and R01HL149302, and Pfizer ASPIRE Award to MA; R01HL136431, R01HL147095, and R01HL141917 to EA; and the MSD Scholarships for Overseas Study, and Japan Society for the Promotion of Science Fellowships for Overseas Researchers to SK.
Disclosures
Pfizer was not involved in the study other than funding. MA also received research grants from Kowa Company and Sanofi.
Supplemental Materials
Online Supplemental Materials and Methods
Supplemental Figures 1–14
Supplemental Tables 1–4
Supplementary Material
Nonstandard Abbreviations and Acronyms
- AAV
- adeno-associated virus
- HEP
- hyperedge entanglement prediction
- IL-1β
- interleukin-1 beta
- LDL
- low-density lipoprotein
- LDLR
- LDL receptor
- MCP-1
- monocyte chemoattractant protein-1
- NF-κB
- nuclear factor-kappa B
- PCSK9
- proprotein convertase subtilisin/kexin 9
- SDC4
- syndecan-4
- SMC
- smooth muscle cell
- TNFα
- tumor necrosis factor alpha
S. Katsuki and P.K. Jha contributed equally.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCRESAHA.121.320056.
For Sources of Funding and Disclosures, see page 887.
References
- 1.Conte MS, Bandyk DF, Clowes AW, Moneta GL, Seely L, Lorenz TJ, Namini H, Hamdan AD, Roddy SP, Belkin M, et al. Results of PREVENT III: a multicenter, randomized trial of edifoligide for the prevention of vein graft failure in lower extremity bypass surgery. J Vasc Surg. 2006;43:742–751; discussion 751. doi: 10.1016/j.jvs.2005.12.058. [DOI] [PubMed] [Google Scholar]
- 2.Alexander JH, Hafley G, Harrington RA, Peterson ED, FergusonLorenz TBTJ, Goyal A, Gibson M, Mack MJ, Gennevois D, Califf RM, et al. Efficacy and safety of edifoligide, an E2F transcription factor decoy, for prevention of vein graft failure following coronary artery bypass graft surgery: PREVENT IV: a randomized controlled trial. JAMA. 2005;294:2446–2454. doi: 10.1001/jama.294.19.2446. [DOI] [PubMed] [Google Scholar]
- 3.Goldman S, Copeland J, Moritz T, Henderson W, Zadina K, Ovitt T, Doherty J, Read R, Chesler E, Sako Y, et al. Improvement in early saphenous vein graft patency after coronary artery bypass surgery with antiplatelet therapy: results of a Veterans Administration Cooperative Study. Circulation. 1988;77:1324–1332. doi: 10.1161/01.cir.77.6.1324 [DOI] [PubMed] [Google Scholar]
- 4.Solo K, Lavi S, Kabali C, Levine GN, Kulik A, John-Baptiste AA, Fremes SE, Martin J, Eikelboom JW, Ruel M, et al. Antithrombotic treatment after coronary artery bypass graft surgery: systematic review and network meta-analysis. BMJ. 2019;367:l5476. doi: 10.1136/bmj.l5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yahagi K, Kolodgie FD, Otsuka F, Finn AV, Davis HR, Joner M, Virmani R. Pathophysiology of native coronary, vein graft, and in-stent atherosclerosis. Nat Rev Cardiol. 2016;13:79–98. doi: 10.1038/nrcardio.2015.164 [DOI] [PubMed] [Google Scholar]
- 6.Post Coronary Artery Bypass Graft Trial I. The effect of aggressive lowering of low-density lipoprotein cholesterol levels and low-dose anticoagulation on obstructive changes in saphenous-vein coronary-artery bypass grafts. N Engl J Med. 1997;336:153–162. doi: 10.1056/NEJM199701163360301 [DOI] [PubMed] [Google Scholar]
- 7.Kulik A, Abreu AM, Boronat V, Ruel M. Intensive versus moderate statin therapy and early graft occlusion after coronary bypass surgery: the aggressive cholesterol therapy to inhibit vein graft events randomized clinical trial. J Thorac Cardiovasc Surg. 2019;157:151–161.e1. doi: 10.1016/j.jtcvs.2018.05.123 [DOI] [PubMed] [Google Scholar]
- 8.Koga JI, Nakano T, Dahlman JE, Figueiredo JL, Zhang H, Decano J, Khan OF, Niida T, Iwata H, Aster JC, et al. Macrophage notch ligand delta-like 4 promotes vein graft lesion development: implications for the treatment of vein graft failure. Arterioscler Thromb Vasc Biol. 2015;35:2343–2353. doi: 10.1161/ATVBAHA.115.305516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decano JL, Singh SA, Bueno CG, Lee LH, Halu A, Chelvanambi S, Matamalas JT, Zhang H, Mlynarchik AK, Qiao J, et al. Systems approach to discovery of therapeutic targets for vein graft disease pparalpha pivotally regulates metabolism, activation, and heterogeneity of macrophages and lesion development. Circulation. 2021;143:2454–2470. doi: 10.1161/CIRCULATIONAHA.119.043724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vries MR, Simons KH, Jukema JW, Braun J, Quax PH. Vein graft failure: from pathophysiology to clinical outcomes. Nat Rev Cardiol. 2016;13:451–470. doi: 10.1038/nrcardio.2016.76 [DOI] [PubMed] [Google Scholar]
- 11.de Vries MR, Quax PHA. Inflammation in vein graft disease. Front Cardiovasc Med. 2018;5:3. doi: 10.3389/fcvm.2018.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161 [DOI] [PubMed] [Google Scholar]
- 13.Horton JD, Cohen JC, Hobbs HH. PCSK9: a convertase that coordinates LDL catabolism. J Lipid Res. 2009;50:S172–S177. doi: 10.1194/jlr.R800091-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridker PM, Revkin J, Amarenco P, Brunell R, Curto M, Civeira F, Flather M, Glynn RJ, Gregoire J, Jukema JW, et al. Cardiovascular efficacy and safety of bococizumab in high-risk patients. N Engl J Med. 2017;376:1527–1539. doi: 10.1056/NEJMoa1701488 [DOI] [PubMed] [Google Scholar]
- 15.Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664 [DOI] [PubMed] [Google Scholar]
- 16.Bonaca MP, Nault P, Giugliano RP, Keech AC, Pineda AL, Kanevsky E, Kuder J, Murphy SA, Jukema JW, Lewis BS, et al. Low-density lipoprotein cholesterol lowering with evolocumab and outcomes in patients with peripheral artery disease: insights from the FOURIER Trial (Further Cardiovascular Outcomes Research With PCSK9 Inhibition in Subjects With Elevated Risk). Circulation. 2018;137:338–350. doi: 10.1161/CIRCULATIONAHA.117.032235 [DOI] [PubMed] [Google Scholar]
- 17.Gao J, Wang HB, Xiao JY, Ren M, Reilly KH, Li YM, Liu Y. Association between proprotein convertase subtilisin/kexin type 9 and late saphenous vein graft disease after coronary artery bypass grafting: a cross-sectional study. Bmj Open. 2018;8:e021951. doi: 10.1136/bmjopen-2018-021951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagace TA, Curtis DE, Garuti R, McNutt MC, Park SW, Prather HB, Anderson NN, Ho YK, Hammer RE, Horton JD. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricci C, Ruscica M, Camera M, Rossetti L, Macchi C, Colciago A, Zanotti I, Lupo MG, Adorni MP, Cicero AFG, et al. PCSK9 induces a pro-inflammatory response in macrophages. Sci Rep. 2018;8:2267. doi: 10.1038/s41598-018-20425-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muscoloni A, Abdelhamid I, Decano JL, Souza E, Maiorino E, Aikawa M, Silverman EK, Sharma A, Cannistraci CV. Hyperedge entanglement in high-order multilayer networks. Preprints. Preprint posted online December 21, 2020. doi: 10.20944/preprints202012.200500.v202011 [Google Scholar]
- 21.Yu P, Nguyen BT, Tao M, Campagna C, Ozaki CK. Rationale and practical techniques for mouse models of early vein graft adaptations. J Vasc Surg. 2010;52:444–452. doi: 10.1016/j.jvs.2010.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjorklund MM, Hollensen AK, Hagensen MK, Dagnaes-Hansen F, Christoffersen C, Mikkelsen JG, Bentzon JF. Induction of atherosclerosis in mice and hamsters without germline genetic engineering. Circ Res. 2014;114:1684–1689. doi: 10.1161/CIRCRESAHA.114.302937 [DOI] [PubMed] [Google Scholar]
- 23.Cornwell M, Vangala M, Taing L, Herbert Z, Koster J, Li B, Sun H, Li T, Zhang J, Qiu X, et al. VIPER: visualization pipeline for RNA-seq, a snakemake workflow for efficient and complete RNA-seq analysis. BMC Bioinformatics. 2018;19:135. doi: 10.1186/s12859-018-2139-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang IM, Zhang B, Yang X, Zhu J, Stepaniants S, Zhang C, Meng Q, Peters M, He Y, Ni C, et al. Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol Syst Biol. 2012;8:594. doi: 10.1038/msb.2012.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cannistraci CV, Alanis-Lobato G, Ravasi T. From link-prediction in brain connectomes and protein interactomes to the local-community-paradigm in complex networks. Sci Rep. 2013;3:1613. doi: 10.1038/srep01613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muscoloni A, Abdelhamid I, Cannistraci CV. Local-community network automata modelling based on length-three-paths for prediction of complex network structures in protein interactomes, food webs and more. bioRxiv. Preprint posted online June 18, 2018. doi: 10.1101/346916 [Google Scholar]
- 27.Muscoloni A, Michieli U, Cannistraci C. Adaptive network automata modelling of complex networks. Preprints. Preprint posted online May 11, 2022. doi: 10.20944/preprints202012.0808.v1 [Google Scholar]
- 28.Giunzioni I, Tavori H, Covarrubias R, Major AS, Ding L, Zhang Y, DeVay RM, Hong L, Fan D, Predazzi IM, et al. Local effects of human PCSK9 on the atherosclerotic lesion. J Pathol. 2016;238:52–62. doi: 10.1002/path.4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denis M, Marcinkiewicz J, Zaid A, Gauthier D, Poirier S, Lazure C, Seidah NG, Prat A. Gene inactivation of proprotein convertase subtilisin/kexin type 9 reduces atherosclerosis in mice. Circulation. 2012;125:894–901. doi: 10.1161/CIRCULATIONAHA.111.057406 [DOI] [PubMed] [Google Scholar]
- 30.Yazdani SK, Farb A, Nakano M, Vorpahl M, Ladich E, Finn AV, Kolodgie FD, Virmani R. Pathology of drug-eluting versus bare-metal stents in saphenous vein bypass graft lesions. JACC Cardiovasc Interv. 2012;5:666–674. doi: 10.1016/j.jcin.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao JH, Walts AE, Fishbein MC. The severity of atherosclerosis at sites of plaque rupture with occlusive thrombosis in saphenous-vein coronary-artery bypass grafts. Am Heart J. 1991;122:955–958. doi: 10.1016/0002-8703(91)90457-S [DOI] [PubMed] [Google Scholar]
- 32.Aikawa M, Rabkin E, Okada Y, Voglic SJ, Clinton SK, Brinckerhoff CE, Sukhova GK, Libby P. Lipid lowering by diet reduces matrix metalloproteinase activity and increases collagen content of rabbit atheroma: a potential mechanism of lesion stabilization. Circulation. 1998;97:2433–2444. doi: 10.1161/01.cir.97.24.2433 [DOI] [PubMed] [Google Scholar]
- 33.Deguchi JO, Aikawa E, Libby P, Vachon JR, Inada M, Krane SM, Whittaker P, Aikawa M. Matrix metalloproteinase-13/collagenase-3 deletion promotes collagen accumulation and organization in mouse atherosclerotic plaques. Circulation. 2005;112:2708–2715. doi: 10.1161/CIRCULATIONAHA.105.562041 [DOI] [PubMed] [Google Scholar]
- 34.Aikawa M, Rabkin E, Sugiyama S, Voglic SJ, Fukumoto Y, Furukawa Y, Shiomi M, Schoen FJ, Libby P. An HMG-CoA reductase inhibitor, cerivastatin, suppresses growth of macrophages expressing matrix metalloproteinases and tissue factor in vivo and in vitro. Circulation. 2001;103:276–283. doi: 10.1161/01.cir.103.2.276 [DOI] [PubMed] [Google Scholar]
- 35.Lakoski SG, Lagace TA, Cohen JC, Horton JD, Hobbs HH. Genetic and metabolic determinants of plasma PCSK9 levels. J Clin Endocrinol Metab. 2009;94:2537–2543. doi: 10.1210/jc.2009-0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng F, Kovacs IA, Barabasi AL. Network-based prediction of drug combinations. Nat Commun. 2019;10:1197. doi: 10.1038/s41467-019-09186-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang ZH, Peng J, Ren Z, Yang J, Li TT, Li TH, Wang Z, Wei DH, Liu LS, Zheng XL, et al. New role of PCSK9 in atherosclerotic inflammation promotion involving the TLR4/NF-kappaB pathway. Atherosclerosis. 2017;262:113–122. doi: 10.1016/j.atherosclerosis.2017.04.023 [DOI] [PubMed] [Google Scholar]
- 39.Ding Z, Liu S, Wang X, Theus S, Deng X, Fan Y, Zhou S, Mehta JL. PCSK9 regulates expression of scavenger receptors and ox-LDL uptake in macrophages. Cardiovasc Res. 2018;114:1145–1153. doi: 10.1093/cvr/cvy079 [DOI] [PubMed] [Google Scholar]
- 40.Elfenbein A, Simons M. Syndecan-4 signaling at a glance. J Cell Sci. 2013;126:3799–3804. doi: 10.1242/jcs.124636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamon M, Mbemba E, Charnaux N, Slimani H, Brule S, Saffar L, Vassy R, Prost C, Lievre N, Starzec A, et al. A syndecan-4/CXCR4 complex expressed on human primary lymphocytes and macrophages and HeLa cell line binds the CXC chemokine stromal cell-derived factor-1 (SDF-1). Glycobiology. 2004;14:311–323. doi: 10.1093/glycob/cwh038 [DOI] [PubMed] [Google Scholar]
- 42.Tanino Y, Chang MY, Wang X, Gill SE, Skerrett S, McGuire JK, Sato S, Nikaido T, Kojima T, Munakata M, et al. Syndecan-4 regulates early neutrophil migration and pulmonary inflammation in response to lipopolysaccharide. Am J Respir Cell Mol Biol. 2012;47:196–202. doi: 10.1165/rcmb.2011-0294OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gustafsen C, Olsen D, Vilstrup J, Lund S, Reinhardt A, Wellner N, Larsen T, Andersen CBF, Weyer K, Li JP, et al. Heparan sulfate proteoglycans present PCSK9 to the LDL receptor. Nat Commun. 2017;8:503. doi: 10.1038/s41467-017-00568-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou P, Jin B, Li H, Huang SY. HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res. 2018;46:W443–W450. doi: 10.1093/nar/gky357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferri N, Tibolla G, Pirillo A, Cipollone F, Mezzetti A, Pacia S, Corsini A, Catapano AL. Proprotein convertase subtilisin kexin type 9 (PCSK9) secreted by cultured smooth muscle cells reduces macrophages LDLR levels. Atherosclerosis. 2012;220:381–386. doi: 10.1016/j.atherosclerosis.2011.11.026 [DOI] [PubMed] [Google Scholar]
- 46.Ferri N, Marchiano S, Tibolla G, Baetta R, Dhyani A, Ruscica M, Uboldi P, Catapano AL, Corsini A. PCSK9 knock-out mice are protected from neointimal formation in response to perivascular carotid collar placement. Atherosclerosis. 2016;253:214–224. doi: 10.1016/j.atherosclerosis.2016.07.910 [DOI] [PubMed] [Google Scholar]
- 47.Tang ZH, Jiang L, Peng J, Ren Z, Wei DH, Wu CY, Pan LH, Jiang ZS, Liu LS. PCSK9 siRNA suppresses the inflammatory response induced by oxLDL through inhibition of NF-kappa B activation in THP-1-derived macrophages. Int J Mol Med. 2012;30:931–938. doi: 10.3892/ijmm.2012.1072 [DOI] [PubMed] [Google Scholar]
- 48.Wu CY, Tang ZH, Jiang L, Li XF, Jiang ZS, Liu LS. PCSK9 siRNA inhibits HUVEC apoptosis induced by ox-LDL via Bcl/Bax-caspase9-caspase3 pathway. Mol Cell Biochem. 2012;359:347–358. doi: 10.1007/s11010-011-1028-6 [DOI] [PubMed] [Google Scholar]
- 49.Ward AO, Caputo M, Angelini GD, George SJ, Zakkar M. Activation and inflammation of the venous endothelium in vein graft disease. Atherosclerosis. 2017;265:266–274. doi: 10.1016/j.atherosclerosis.2017.08.023 [DOI] [PubMed] [Google Scholar]
- 50.Aikawa M, Sivam PN, Kuro-o M, Kimura K, Nakahara K, Takewaki S, Ueda M, Yamaguchi H, Yazaki Y, Periasamy M, et al. Human smooth muscle myosin heavy chain isoforms as molecular markers for vascular development and atherosclerosis. Circ Res. 1993;73:1000–1012. doi: 10.1161/01.res.73.6.1000 [DOI] [PubMed] [Google Scholar]
- 51.Wadey K, Lopes J, Bendeck M, George S. Role of smooth muscle cells in coronary artery bypass grafting failure. Cardiovasc Res. 2018;114:601–610. doi: 10.1093/cvr/cvy021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Cheng S, Delling FN, et al. Heart disease and stroke statistics-2021 update: a report from the American Heart Association. Circulation. 2021;143:e254–e743. doi: 10.1161/CIR.0000000000000950 [DOI] [PubMed] [Google Scholar]
- 53.Fowkes FG, Rudan D, Rudan I, Aboyans V, Denenberg JO, McDermott MM, Norman PE, Sampson UK, Williams LJ, Mensah GA, et al. Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: a systematic review and analysis. Lancet. 2013;382:1329–1340. doi: 10.1016/S0140-6736(13)61249-0 [DOI] [PubMed] [Google Scholar]
- 54.Bernelot Moens SJ, Neele AE, Kroon J, van der Valk FM, Van den Bossche J, Hoeksema MA, Hoogeveen RM, Schnitzler JG, Baccara-Dinet MT, Manvelian G, et al. PCSK9 monoclonal antibodies reverse the pro-inflammatory profile of monocytes in familial hypercholesterolaemia. Eur Heart J. 2017;38:1584–1593. doi: 10.1093/eurheartj/ehx002 [DOI] [PubMed] [Google Scholar]
- 55.Stein EA, Mellis S, Yancopoulos GD, Stahl N, Logan D, Smith WB, Lisbon E, Gutierrez M, Webb C, Wu R, et al. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N Engl J Med. 2012;366:1108–1118. doi: 10.1056/NEJMoa1105803 [DOI] [PubMed] [Google Scholar]
- 56.Lawn RM, Wade DP, Hammer RE, Chiesa G, Verstuyft JG, Rubin EM. Atherogenesis in transgenic mice expressing human apolipoprotein(a). Nature. 1992;360:670–672. doi: 10.1038/360670a0 [DOI] [PubMed] [Google Scholar]
- 57.Guo Q, Feng X, Zhou Y. PCSK9 Variants in familial hypercholesterolemia: a comprehensive synopsis. Front Genet. 2020;11:1020. doi: 10.3389/fgene.2020.01020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karagiannis AD, Liu M, Toth PP, Zhao S, Agrawal DK, Libby P, Chatzizisis YS. Pleiotropic anti-atherosclerotic effects of PCSK9 inhibitorsfrom molecular biology to clinical translation. Curr Atheroscler Rep. 2018;20:20. doi: 10.1007/s11883-018-0718-x [DOI] [PubMed] [Google Scholar]
- 59.Ridker PM. High-sensitivity C-reactive protein and cardiovascular risk: rationale for screening and primary prevention. Am J Cardiol. 2003;92:17K–22K. doi: 10.1016/s0002-9149(03)00774-4 [DOI] [PubMed] [Google Scholar]
- 60.Bohula EA, Giugliano RP, Leiter LA, Verma S, Park JG, Sever PS, Lira Pineda A, Honarpour N, Wang H, Murphy SA, et al. Inflammatory and cholesterol risk in the FOURIER trial. Circulation. 2018;138:131–140. doi: 10.1161/CIRCULATIONAHA.118.034032 [DOI] [PubMed] [Google Scholar]
- 61.Pradhan AD, Aday AW, Rose LM, Ridker PM. Residual inflammatory risk on treatment with PCSK9 inhibition and statin therapy. Circulation. 2018;138:141–149. doi: 10.1161/CIRCULATIONAHA.118.034645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Libby P. Interleukin-1 beta as a target for atherosclerosis therapy: biological basis of CANTOS and beyond. J Am Coll Cardiol. 2017;70:2278–2289. doi: 10.1016/j.jacc.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grebe A, Hoss F, Latz E. NLRP3 Inflammasome and the IL-1 Pathway in atherosclerosis. Circ Res. 2018;122:1722–1740. doi: 10.1161/CIRCRESAHA.118.311362 [DOI] [PubMed] [Google Scholar]
- 64.Libby P. Mechanisms of acute coronary syndromes and their implications for therapy. N Engl J Med. 2013;368:2004–2013. doi: 10.1056/NEJMra1216063 [DOI] [PubMed] [Google Scholar]
- 65.Gordts P, Foley EM, Lawrence R, Sinha R, Lameda-Diaz C, Deng L, Nock R, Glass CK, Erbilgin A, Lusis AJ, et al. Reducing macrophage proteoglycan sulfation increases atherosclerosis and obesity through enhanced type I interferon signaling. Cell Metab. 2014;20:813–826. doi: 10.1016/j.cmet.2014.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gordts PLSM. Esko JD. Heparan sulfate proteoglycans fine-tune macrophage inflammation via IFN-beta. Cytokine. 2015;72:118–119. doi: 10.1016/j.cyto.2014.12.013 [DOI] [PubMed] [Google Scholar]
- 67.Shapiro MD, Tavori H, Fazio S. PCSK9: from basic science discoveries to clinical trials. Circ Res. 2018;122:1420–1438. doi: 10.1161/CIRCRESAHA.118.311227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grune J, Meyborg H, Bezhaeva T, Kappert K, Hillmeister P, Kintscher U, Pieske B, Stawowy P. PCSK9 regulates the chemokine receptor CCR2 on monocytes. Biochem Biophys Res Commun. 2017;485:312–318. doi: 10.1016/j.bbrc.2017.02.085 [DOI] [PubMed] [Google Scholar]
- 69.Nan J, Du Y, Chen X, Bai Q, Wang Y, Zhang X, Zhu N, Zhang J, Hou J, Wang Q, et al. TPCA-1 is a direct dual inhibitor of STAT3 and NF-kappaB and regresses mutant EGFR-associated human non-small cell lung cancers. Mol Cancer Ther. 2014;13:617–629. doi: 10.1158/1535-7163.MCT-13-0464 [DOI] [PubMed] [Google Scholar]
- 70.Beltrami-Moreira M, Vromman A, Sukhova GK, Folco EJ, Libby P. Redundancy of IL-1 isoform signaling and its implications for arterial remodeling. PLoS One. 2016;11:e0152474. doi: 10.1371/journal.pone.0152474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cannistraci CV, Alanis-Lobato G, Ravasi T. Minimum curvilinearity to enhance topological prediction of protein interactions by network embedding. Bioinformatics. 2013;29:i199–i209. doi: 10.1093/bioinformatics/btt208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu M, Pan Q, Muscoloni A, Xia H, Cannistraci CV. Modular gateway-ness connectivity and structural core organization in maritime network science. Nat Commun. 2020;11:2849. doi: 10.1038/s41467-020-16619-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun X, Lin J, Zhang Y, Kang S, Belkin N, Wara AK, Icli B, Hamburg NM, Li D, Feinberg MW. MicroRNA-181b improves glucose homeostasis and insulin sensitivity by regulating endothelial function in white adipose tissue. Circ Res. 2016;118:810–821. doi: 10.1161/CIRCRESAHA.115.308166 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The online-only Supplemental Data provided detailed methods of all procedures. The data, analytic methods, and study materials will be made available to other researchers, upon request for purposes of reproducing the results or replicating the procedure.