Abstract
Chronic graft-versus-host disease (cGvHD) is a major cause of morbidity after hematopoietic stem cell transplantation (HSCT). In large patient populations, we have shown a CD56bright natural killer (NK) population to strongly associate with a lack of cGvHD and we hypothesize that these cells function to suppress cGvHD. We aimed to isolate and define the characteristics of regulatory NK (NKreg) cells associated with suppression of cGvHD. Immunophenotypic evaluation of a large pediatric population found the CD56bright NK population associated with a lack of cGvHD to be perforin-, Granzyme B-, and CD335+. Transcriptome analysis of a small patient cohort of CD56bright compared to CD56dim NK cells found the NKreg cells to also overexpress Granzyme K, IL-7R, GPR183, RANK, GM-CSFR, TCF7, and IL23A. Further analysis of this CD56bright NKreg population found a subpopulation that overexpressed IRF1, and TNF. We also found that viable NKreg cells may be isolated by sorting on CD56+ and CD16- NK cells, and this population can suppress allogeneic CD4+ T cells, but not Treg cells or CD8+ T cells through a non-cytolytic, cell-cell contact dependent mechanism. Suppression was not reliant upon the NKp44, NKp46, or GPR183 receptors. Additionally, NKreg cells do not kill leukemic cells. Moreover, this is the first paper to clearly establish that a CD56brightCD3-CD16-perforin- NKreg population associates with a lack of cGvHD and has several unique characteristics, including the suppression of helper T-cell function in vitro. With further investigation we may decipher the mechanism of NKreg suppression and operationalize expansion of NKreg cells associated with cGvHD suppression.
Introduction
Hematopoietic stem cell transplantation (HSCT) is an important therapeutic option for patients with non-malignant diseases as well as high-risk and refractory hematopoietic malignancies.1 HSCT may be followed by chronic inflammation due to tissue self-antigen mismatches among the transplant donor and recipient. As a result, HSCT is often attributed to an increased risk of health complications, the most severe being chronic graft-versus-host disease (cGvHD).2 cGvHD is a multisystemic disorder that occurs due to foreign donor immune cells attacking the recipient’s tissues.3 cGvHD has been estimated to develop in 25% of pediatric and 60% of adult HSCT survivors, may cause chronic and often irreversible organ damage, and has a 10-25% mortality rate.4
Various immune-suppressive cell populations appear to associate with suppression of cGvHD, including Treg cells, Breg cells, M2 macrophages, dendritic cells, and CD56bright natural killer (NK) cells.5 In previous correlative studies, we observed an increased number of CD56brightCD335+CXCR3+ NK cells in adult patients who did not develop cGvHD6 and a similar lack of CD56brightCD3- NK cells in a large pediatric cohort before the onset of cGvHD.7 In a separate large adult clinical trial comparing G-CSF-mobilized marrow to G-CSF-mobilized peripheral blood, a decreased number of CD56bright NK cells in the donor product correlated with the development of cGvHD, and statistical analysis of the donor product suggested that the low number of CD56bright NK cells was the mechanism for this increased cGvHD.8
To date, strategies targeted to augment immune suppressive populations in cGvHD have focused on increasing the number and function of Treg cells. Two approaches, low dose IL-2 and extracorporeal electrophoresis (ECP), have attempted to expand Treg cells to inhibit cGvHD.9 While the association of Treg expansion and therapeutic outcome is considered the primary explanation, an alternative mechanism was identified as cGvHD inhibition being mediated by a CD56bright NK cell contact‐dependent cell cycle arrest of effector T cells through the NK receptors NKp44 and NKp46.10 Further, low dose IL-2 as treatment of systemic lupus erythematosus has shown to successfully suppress disease activity through both an increase in Treg cells and NK cells.11 Similarly, recent evidence supports that ECP therapy suppresses cGvHD primarily through increased CD56bright NK cells.12
Based on this data, we hypothesized that there is a subset of CD56bright NK cells which suppress the development of cGvHD and are functionally consistent with described regulatory NK cells (NKre g).13,14 We proposed to better characterize the CD56bright NK-cell subset associated with a lack of cGvHD. In order to meet our objective, we first identified the NKreg subpopulation most significantly associated with immune tolerance in a large HSCT patient cohort. Second, in a smaller HSCT patient cohort, we determined the genes of CD56bright NK cells as compared to CD56dim NK cells which are associated with cGvHD suppression. Third, based on the previous transcriptome results, we determined the optimal cell surface markers to isolate the NKreg cell subpopulation while also defining the phenotype of NK cells. Lastly, we investigated the cytotoxicity and suppressive capacity of NKreg cells.
Methods
Immune cell populations in hematopoietic stem cell transplantation patients associated with chronic graf-versus-host disease
Pediatric HSCT patient peripheral blood mononuclear cells (PBMC) (previously described7,15,16) (Online Supplementary Table S1) were utilized for identifying immune cell populations associated with cGvHD. Study groups included evaluable patients at the onset of cGvHD with time control samples with no cGvHD obtained at 3, 6, and 12 months after HSCT. Of the CD56brightCD3- cell population, phenotyping was performed for expression of perforin, Granzyme B, CD335(NKp46) and CD69.
Hematopoietic stem cell transplantation patient transcriptome analysis of NK cells (nanoString)
Select day 100 HSCT patient peripheral blood PBMC from three cohorts (cGvHD [n=6], late aGvHD ≥114 days after HSCT [n=6], or no late aGvHD or cGvHD [n=6]) were utilized [previously described7,1 5,16]) (Online Supplementary Table S1). Total RNA was extracted from sorted NK subpopulations using the RNeasy Mini Kit (QIAGEN, Valencia, CA, USA) for nanoString (nanoString, Seattle).
Evaluation of NKreg cell suppression
CD4+ T cells were isolated using the EasySep™ Human CD4+ T Cell Isolation Kit and CD8+ T cells were isolated using the EasySep™ Human CD8+ T Cell Isolation Kit (StemCell Technologies, Vancouver, BC, Canada). The T cells were stained using Cell Proliferation Dye eF450 (eBioscience, Mississauga, ON, Canada), and activated via the ImmunoCult™ Human CD3/CD28 T Cell Activator (StemCell Technologies, Vancouver, BC, Canada). Healthy donor PBMC samples were enriched for total NK cells using the NK Cell Enrichment Kit (StemCell Technologies, Vancouver, BC, Canada).
CD56brightCD16- and CD56dimCD16+ NK cells were sorted using sterile Beckman Coulter Astrios FACS sorting, and co-cultured with allogeneic activated CD4+ or CD8+ T cells for 96 hours (h). Treg cells were obtained from our collaborators (MKL) after expansion, as previously described (protocol F).17 In order to determine if the suppressive capacity of NKreg cells on CD4+ T cells is contact dependent the above procedures were followed using a 96-well transwell plate (Corning, Corning, NY, USA). Additionally, supernatant from a 96-h 1:1 NKreg versus CD4+ T-cell suppression assay was cultured with fresh allogeneic activated CD4+ T cells for 72 h and then analyzed.
In order to determine if the suppressive mechanism of NKreg cells is reliant on NKp44, NKp46, or GPR183 receptors the above assay preparation/analysis procedures were followed with addition of the soluble antagonists: 1 ug/mL Ultra-LEAF™ Purified anti-human NKp4610 (BioLegend, San Diego, CA, USA), 1 ug/mL Ultra-LEAF™ purified anti-human NKp4410 (BioLegend, San Diego, CA, USA), and 25 nM NIBR18918 (Sigma Aldrich, Oakville, CA, USA).
Evaluation of NKreg cell induction of apoptosis and killing
In order to investigate NK-cell induction of CD4+ T-cell apoptosis, the suppression assay co-cultured cells were harvested after 96 h, and the FITC Annexin V apoptosis detection kit (BioLegend, San Diego, CA, USA) was utilized. Further, this kit was utilized to determine the cytolytic effect of NKreg cells towards MOLT-4 (ATCC® CRL1582™), and Jurkat, Clone E61 (ATCC® TIB152™) cell lines. In order to investigate general NK-cell killing, NKreg cells and CD56dim NK cells were co-cultured with D-luciferin K562 cells (Gold Biotechnology, St Louis, MO, USA) for 24 h.
Statistical analysis
Refer to the Online Supplementary Appendix.
The study was approved by the Canadian Ethics Review
Board (H18-00022 and H16-00533).
Results
Identification of cell populations in hematopoietic stem cell transplantation patients associated with chronic graf-versus-host disease
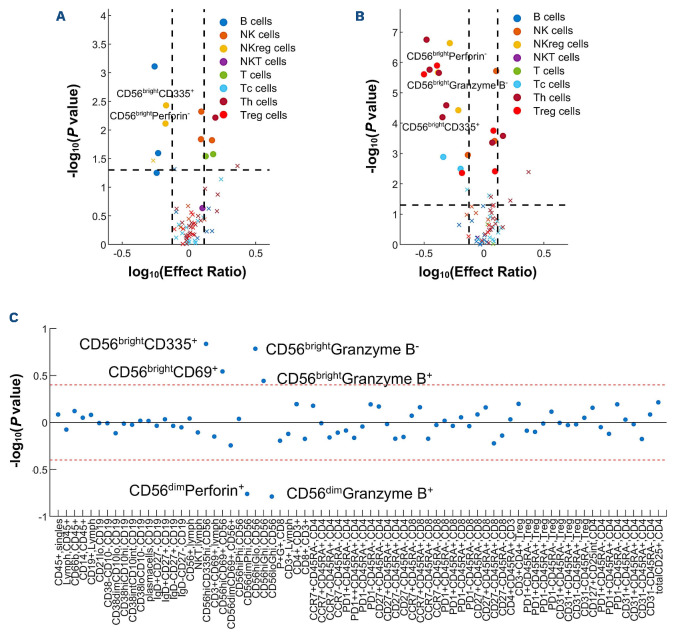
A previous analysis of a pediatric cohort (n=241) from the PBMTC 1202/ABLE1.0 study found that patients at day 100 after HSCT who later developed cGvHD had decreased CD56brightCD3- NK-cell numbers, similar to our previously described research in adult cohorts.6,8 Using the identical strict criteria of biological significance limited to significant P value, ROC AUC of ≥0.60, and effect ratio of either ≤0.75 or ≥1.3 after a Bonferroni correction, further immunophenotypic evaluation was performed on the CD56brightCD3- NK populations from the PBMTC/ABLE cohort. Of samples taken before the onset of cGvHD at day 100, we found a lack of cGvHD to be associated with CD56brightCD3- NK cells which were further characterized by being perforin-(effect ratio [ER]=0.67; P=0.007; AUC=0.63), CD335+ (ER=0.67; P=0.004; AUC=0.64), and close to meeting our criteria for Granzyme B- (ER=0.54; P=0.03; AUC=0.59) (Figure 1A). A second analysis performed at cGvHD onset compared patients with cGvHD to time-matched samples of patients that had no cGvHD (onset of cGvHD<4 months linked to non-cGvHD controls at 3 months, onset >4 months to ≥8 months to 6 month controls, and cGvHD onset >8 months to 12 month non-cGvHD controls). The samples taken at the onset of cGvHD showed that CD56brightCD3- NK cells were perforin- (ER=0.51; P=2.3x10-7 ; AUC=0.64), CD335+ (ER=0.61; P=3.7x10-5; AUC=0.64), or Granzyme B- (ER=0.41; P=2.2x10-6; AUC=0.69) (Figure 1B). Since the association of a lack of cGvHD was most significant with a subpopulation that was perforin-, a regression analysis evaluated CD56brightCD3-perforin- NK cells against all markers and found a significant correlation with CD56brightCD3-Granzyme B- NK cells (Pearson average correlation [PAC]=0.78; P<1x10-1 7 ) and inverse relationship with CD56dimperforin+ and CD56dimGranzyme B+ NK cells (PAC=-0.76; P<1x10-1 7 and PAC=-0.79; P<1x10-1 7, respectively) (Figure 1C). Based on these findings, we concluded that perforin negativity is most significantly associated with the NKreg subpopulation important in suppressing cGvHD.
Figure 1.
Immune cell populations in hematopoietic stem cell transplantation patients associated with chronic graf-versus-host disease. CD56bright and CD56dim NK-cell immunophenotypic expression for each group of hematopoietic stem cell transplantation (HSCT) patients (chronic graft-versus-host disease [cGvHD], acute GvHD [aGvHD], and controls) at day 100 before and at the onset of cGvHD and the differential expression between controls and GvHD subjects. (A) Regulatory NK (NKreg) cell subpopulations of HSCT patients at day 100 before the onset of cGvHD compared to HSCT patients without cGvHD. (B) NKreg cell subpopulations of HSCT patients at the onset of cGvHD compared to HSCT patients without cGvHD. (C) Correlation of CD56brightperforin- NKreg cells with other immune cell populations. For the cell subtype contrast, significance identified as P<0.05 and effect ratio >5 or <0.2. For the group contrast, significance identified as P<0.05 and effect ratio >2 or <0.5. In order to test for differential expression between CD56bright and CD56dim NK cells of each gene, a paired t test on samples from each group was performed. In order to test for group differences of each gene, a Student’s t test on samples from each cell subtype was performed. GvHD, n=44; no GvHD, n=190.
Comparison of transcriptome expression by CD56bright to CD56dim NK cells in hematopoietic stem cell transplantation patients
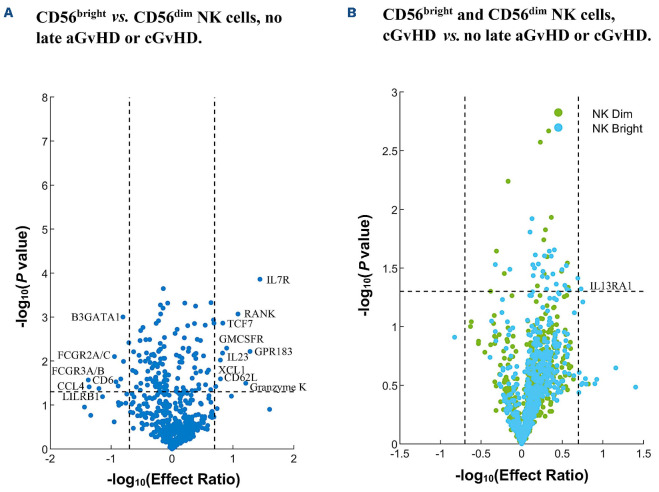
In order to further evaluate for additional markers associated with a lack of cGvHD and the CD56brightCD3- NK-cell population, we performed transcriptome analysis in a representative cohort of patients from the larger ABLE/PBMTC cohort. This was measured by nanoString on PBMC samples at day 100 after HSCT in immune-tolerant patients (n=6) that had neither late aGvHD nor cGvHD. In this patient set, we compared the expression of CD56bright to CD56dim NK cells (Online Supplementary Table S2, column 1; Figure 2A). We confirmed that the CD56bright NK cells had low expression of perforin and Granzyme B, and increased expression NKp46. Additional significant markers associated with this population were characterized by overexpression of Granzyme K (ER=16.2), IL-7R (ER=27.8), GPR183 (ER=19.1), RANK (ER=12.1), GM-CSFR (ER=7.9), TCF7 (ER=6.8), IL23A (ER=6.8), and others. We also found a significant lack of expression of select genes in CD56bright NK cells compared to CD56dim NK cells (ER≤0.2), including FcRγ (FCGR3A/B; ER=0.04), CCL4 (ER=0.04), LILRB1 (ER=0.06), and others (Online Supplementary Table S2; Figure 2A).
In order to investigate for markers that had expression most closely within induction of immune tolerance we evaluated whether the CD56bright:CD56dim NK-cell expression ratio is altered in patients with GvHD. We focused on the CD56bright population for transcriptome expressions differing in the late aGvHD (n=6) or cGvHD (n=6) patients. We found that there was a lower ratio for IL7R in the late aGvHD group (ER=25.5), and even lower in the cGvHD group (ER=20.8). Additional markers decreased in either late aGvHD or cGvHD in the CD56bright:CD56dim ratio included GPR183, Granzyme K, RANK, GM-CSFR, TCF7, IL-23A, and others. Ratios that increased in patients with either late aGvHD or cGvHD included FcRγ (FCGR3A/B), CCL4, LILRB1, and others (Online Supplementary Table S2, columns 2 and 3).
Evaluation for transcriptome expression in CD56bright NK cells associated with a lack of late acute and chronic graf-versus-host disease
In order to determine if any of the markers identified in the transcriptome were characteristic of a subpopulation of CD56bright NK cells most closely associated with a lack of cGvHD, we evaluated CD56bright NK cells in HSCT patients with no GvHD compared to patients that developed cGvHD. We found the CD56bright NK cells in the no GvHD group to overexpress interferon regulatory factor 1 (IRF1) (ER=1.4, P=0.03), and tumor necrosis factor (TNF) (ER=2.1, P=0.03) (not shown in the Online Supplementary Table S1, column 4 due to ER being less than 5) and the CD56bright NK cells in the cGvHD group compared to no late aGvHD or cGvHD group to overexpress IL13RA1 (ER=5.4, P=0.05) (Figure 2B).
Confirmation of transcriptome differences by surface and intracellular expression in CD56bright NK cells
Based on the immunophenotypic characterization of the larger populations and the confirmation by nanoString, we hypothesized that a subpopulation of CD56bright NK cells that are perforin- and Granzyme B- (NKreg) may have the greatest ability to act as a cGvHD suppressor. However, isolation of this population based on cytoplasmic perforin and granzyme staining results in non-viable cells. Therefore, we aimed to determine the optimal cell surface-expression approach to isolate the NKreg population based on the candidates identified in the nanoString analysis. Our selected markers were significantly associated with a lack of GvHD development in the CD56bright NK-cell subset and expressed on the cell surface. Using these criteria, we selected the following markers: CD56, IL-7R, GPR183, GM-CSFR, CD62L, and CD16. We also investigated CXCR3 expression based on previous data which showed this marker to be highly expressed by CD56bright NK cells associated with immune tolerance.6
We verified that expression of Granzyme K, GPR183, IL-7R, CXCR3, and CD62L was present in the CD56bright NK-cell population. Additionally, we confirmed the CD25, NKp44, and NKp46 expression (Online Supplementary Figure S1), and lack of KIR expression (Online Supplementary Figure S2) among the CD56bright NK cells. In contrast, the CD56dim NK cells show RNA and protein expression of KIR, Granzyme B, and perforin. However, unlike the transcriptome findings, GM-CSF showed minimal protein expression among the CD56bright NK cells. When investigating the optimal cell-surface marker combination for sorting the NKreg cells we found that the greatest purity (>95%) for perforin- Granzyme B- NK cells (Online Supplementary Figure S3) was in a CD56bright and CD16- NK-cell population. In contrast, the CD56brightCD16+ subset is >70% perforin+ Granzyme B+. The other investigated markers (IL-7R, GPR183, GM-CSFR, and CD62L) in combination with CD56 did not result in consistent isolation of pure (>95%) perforin-, Granzyme B- NKreg cells. Therefore, we define the NKreg subpopulation as CD56brightCD16-NK cells and utilized these isolated cells for all NKreg functional evaluations.
Figure 2.
Differences between CD56bright and CD56dim NK cells that are associated with a lack of graf-versus-host disease (GvHD) and differences in gene expression between CD56bright NK cells in hematopoietic stem cell transplantation (HSCT) patients who developed GvHD versus those who were immune-tolerant (no late acute GvHD or chronic GvHD) measured on samples from day 100 post HSCT. CD56bright and CD56dim NK cell gene expression for HSCT patients (cGvHD, and no late aGvHD or cGvHD) at day 100 post HSCT and before the potential onset of cGvHD. (A) Gene expression of CD56bright NK cells as compared to CD56dim NK cells among HSCT patients that did not develop late aGvHD or cGvHD. (B) Gene expression of CD56bright and CD56dim NK cells among HSCT patients that developed cGvHD as compared to those who did not develop late aGvHD or cGvHD. For the cell subtype contrast and group contrast, significance identified as P<0.05 and effect ratio >5 or <0.2. In order to test for differential expression between CD56bright and CD56dim NK cells of each gene, a paired t test on samples from each group was performed. In order to test for group differences of each gene, a Student’s t test on samples from each cell subtype was performed. Late aGvHD, n=6; cGvHD, n=6; no GvHD, n=6.
Evaluation of regulatory CD56brightCD16- NK-cell ability to suppress T-cell proliferation
A major characteristic of Treg cells is that they suppress T-cell function,19 which is necessary to suppress the development of GvHD. The evaluation of CD56brightCD16- NKreg cell’s ability to suppress allogeneic CD4+ T cells found the NKreg cells to strongly suppress T-cell proliferation, particularly at the 1:1 (approximately 96% suppression) ratio (Figure 3A). In contrast, the CD56dimCD16+ NK cells have minimal suppressive capacity (Figure 3B). The suppressive capacity of Treg cells was comparable to that of NKreg cells (Figure 3C). Interestingly, the NKreg cells suppress CD4+ T-cell proliferation significantly stronger than CD8+ T-cell proliferation at the 1:1 ratio (7.67% suppression of CD8+ T-cell proliferation, compared to 94% of CD4+ T-cell proliferation) (Figure 4). NKreg cells also did not significantly suppress Treg cell proliferation at the 1:1 ratio compared to the Treg control (126% proliferation, SD=44.6, P=0.42) (Figure not shown).
Evaluation of the cytolytic ability of CD56brightCD16- NKreg cells
NKreg cells were found to not be cytotoxic towards K562 cells, but CD56dimCD16+ NK cells lysed the target (Online Supplementary Figure S4). Additionally, NKreg cells did not induce apoptosis of CD4+ T cells, but CD56dimCD16+ NK cells induced substantial apoptosis of CD4+ T cells (Figure 5). Further, as expected, NKreg cells did not result in significant (P>0.05) killing of leukemic cells at the 10:1 (10.5% apoptosis of Jurkat cells, SD=4.95; 6% apoptosis of MOLT-4 cells, SD=0) or 1:1 (9% apoptosis of Jurkat cells, SD=4.36; 3% apoptosis of MOLT-4 cells, SD=0) ratios (Figure not shown).
Evaluation of cell-cell contact dependence for CD56brightCD16- NKreg cell suppression
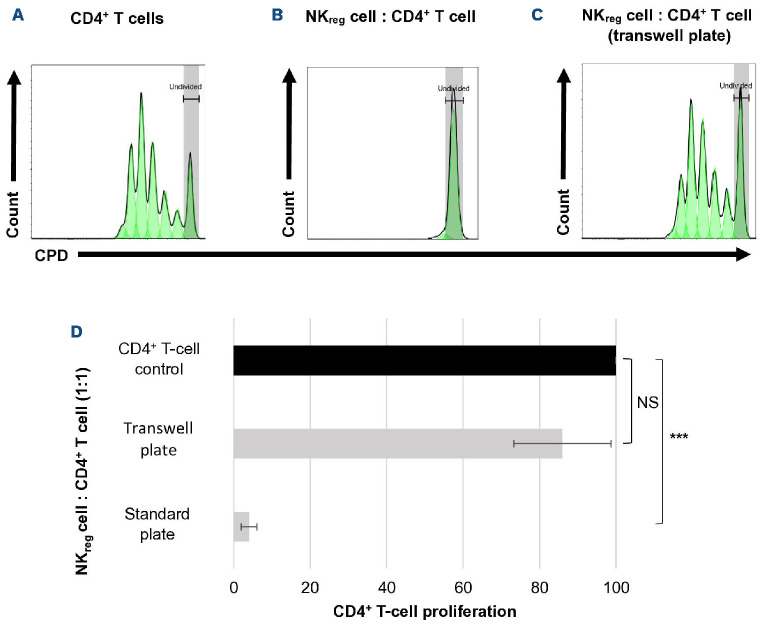
In order to evaluate whether NKreg cells suppress through cell-cell contact, NKreg cells were co-cultured with allogeneic CD4+ T cells in a transwell plate to prevent direct contact between the two cell types, but still allow transfer of soluble factors through the permeable membrane. In this environment, there was found to be a significant decrease in the suppressive effect of the NKregs (91% of CD4+ T cells continued to proliferate at the 1:1 ratio) (Figure 6). In confirming that suppression was not through a soluble factor secreted by NKreg cells, we found no suppression of CD4+ T-cell proliferation when co-culturing with the NKreg suppression assay supernatant (Online Supplementary Figure S5).
Figure 3.
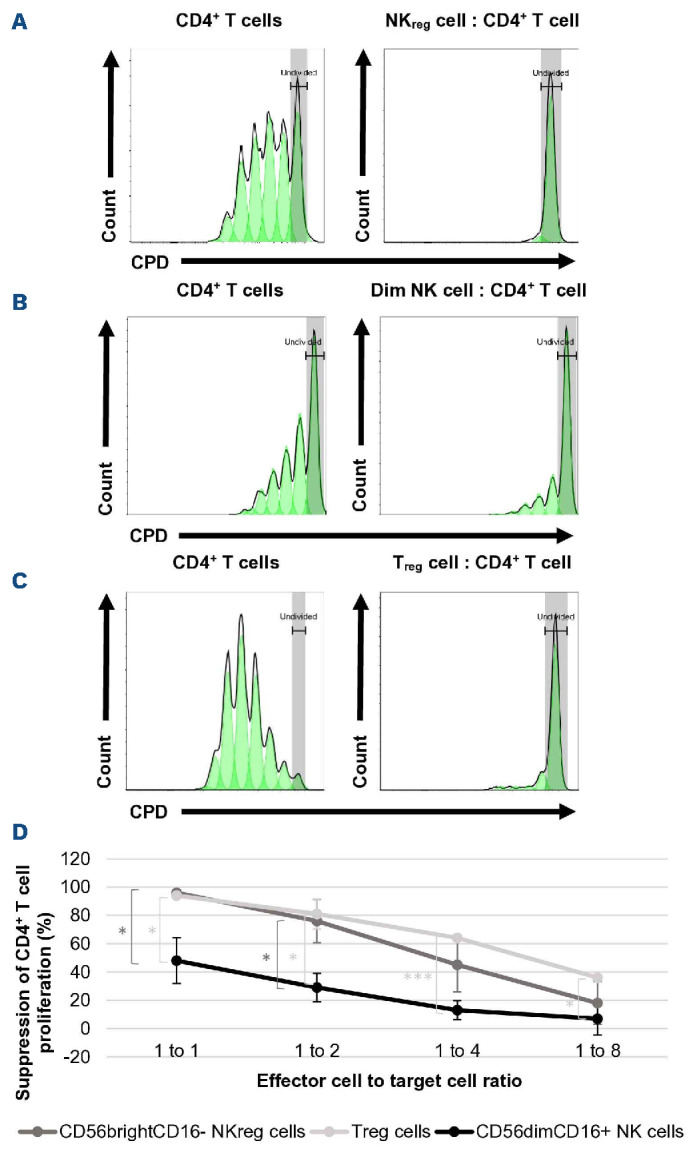
Evaluation of CD56brightCD16- NKreg cell ability to suppress CD4+ T-cell proliferation. Suppressive effect of (A) CD56brightCD16- regulatory NK (NKreg) cells, (B) CD56dimCD16+ NK cells, and (C) Treg cells towards allogeneic CD4+ T cells. On the histograms, the x-axis displays the cell proliferation dye (CPD) eF450 and the y-axis displays the cell count. (D) Suppressive capacity of CD56brightCD16- NKreg cells, CD56dimCD16+ NK cells, and Treg cells towards allogeneic CD4+ T cells at the 1:1, 1:2, 1:4, and 1:8 ratios as compared to the activated CD4+ T-cell control (mean +/- standard deviation). *P<0.05, ***P<0.0005. Statistical analyses for suppression experiments were performed using Microsoft Excel version 2110 and a two-tailed t test – two-sample assuming unequal variance. The data is from a single experiment representative of 5 experiments for the NK-cell vs. CD4+ T-cell assays, and 2 experiments for the Treg cell vs. CD4+ T-cell assay.
Evaluation of GPR183R, NKp46, and NKp44 dependence for CD56brightCD16- NKreg cell suppression
Based on the transcriptome study results and previous studies,10 we investigated the role of the GPR183, NKp44, and NKp46 receptors in the NKreg suppressive mechanism. We blocked GPR183R using the antagonist NIBR189 and NKp44 and NKp46 using blocking monoclonal antibodies. We found no significant difference in NKreg cell suppressive capacity when GPR183R, NKp44, or NKp46 were blocked in solution (Online Supplementary Figure S6).
Discussion
Previous large patient population correlative studies have found CD56bright NKreg cells to be the consistent cell population associated with lack of cGvHD and development of immune tolerance. In this study, we further characterized the NKreg population through immunophenotypic, transcriptomic, and functional analysis as a NKreg subpopulation, identifying CD56brightperforin- cells to have the most significant association both before and at the onset of cGvHD, with Granzyme B- cells strongly correlating with the perforin- group. Within a smaller cohort of HSCT patient samples, transcriptome analysis identified additional markers that may be characteristic of this distinct CD56bright NK subpopulation associated with the suppression of GvHD. Based on these findings, we determined that we could sort CD56+ and CD16- NK cells to obtain a high purity of the NKreg subpopulation for manipulation in vitro. The identified NKreg population is similar to Treg cells in its strong cell-cell contact-dependent suppression of CD4+ T cells. However, NKreg cells do not appear to suppress Treg cells or CD8+ T cells, suggesting that NKreg cells selectively regulate T-cell subsets, differentiating their function from Treg cells which strongly suppress both CD4+ and CD8+ T cells.20 This finding may correlate to NK cells contribution to the graft-versus-leukemia affect in HSCT patients, which is strongly mediated by CD8+ T cells.21 As expected, the NKreg cells did not result in significant killing of leukemic cells. Further, the NKreg cells exhibited a lack of cytotoxicity towards CD4+ T cells and K562 cells, differentiating their function from CD56dimCD16+ NK cells, which significantly lysed both target cell populations.
Contrary to others, our studies show there to be no significant difference in NKreg cell suppressive capacity when the NKp44 or NKp46 receptors were blocked in solution. However, these studies utilized CD56bright NK cells expanded with low-dose IL-2, which may activate and mature the cells towards the cytolytic CD56bright population.22 Our ana lyses were performed on freshly isolated, unexpanded NKreg cells to be more reflective of their in vivo function. Additionally, we found no significant difference in the NKreg cell suppressive capacity when the candidate receptor GPR183R was blocked in solution. Further, our nanoString study revealed no significant differences in PD-1 and LAG3 expression of NK cells, thus we also ruled out these immune checkpoints as being important in the NKreg suppressive mechanism. Therefore, like Treg cells,23 the contact-dependent mechanism of NKreg suppression is unknown and requires further study. Other studies have proposed soluble mediators of suppression by cytokine induced NKreg cells, though these cells are both phenotypically and functionally different from the freshly isolated NKreg cells we describe.22,24
Figure 4.
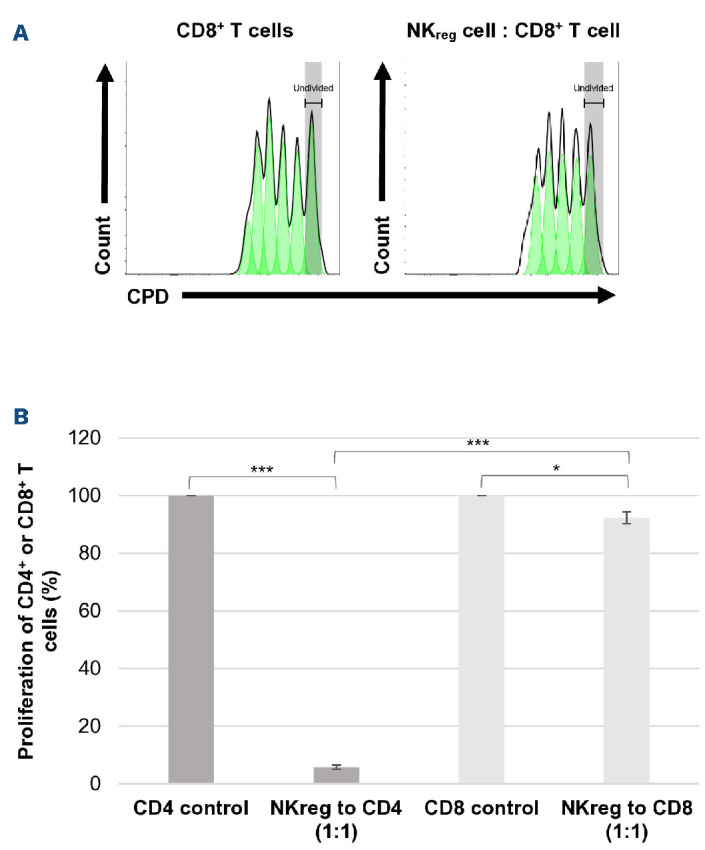
Evaluation of CD56brightCD16- NKreg cell ability to suppress CD8+ T-cell proliferation. Suppressive effect of (A) CD56brightCD16- regulatory NK (NKreg) cells towards allogeneic CD8+ T cells compared to the CD8+ T-cell control. On the histograms, the x-axis displays the cell proliferation dye (CPD) eF450 and the y-axis displays the cell count. (B) Proliferation of CD8+ or CD4+ T cells after 96-hour co-culture with allogeneic CD56brightCD16- NKreg cells at the 1:1 ratio as compared to the activated CD8+ or CD4+ T-cell controls (mean +/- standard deviation). *P<0.05, ***P<0.0005. Statistical analyses for suppression experiments were performed using Microsoft Excel version 2110 and a two-tailed t test – two-sample assuming unequal variance. The data is from a single experiment representative of 3 experiments.
Currently, the focus on regulatory mechanisms in cGvHD have primarily evaluated the role of Tregs25 and to a lesser extent, Bregs.26 Thus, strategies to induce immune tolerance through augmentation of regulatory mechanisms have also focused on increasing the function of Tregs, either by strategies that do not suppress Tregs, such as ruxolitinib,27 or expansion of Tregs by low dose IL-2 or ECP.9 Our studies suggest that NKreg cells may have an early innate-mediated tolerogenic stimulus on the development of cGvHD with a lack of NKreg cells both before the onset of cGvHD (Figure 1A) and at the onset of cGvHD (Figure 1B). This may be similar to physiological settings, such as in placental tolerance where CD56bright NK cells are present in the first trimester, while only in the second and third trimester are Treg cells present.28 Interestingly, the successful cGvHD therapies of low dose IL-2 and ECP, utilised to augment Treg cells, also simultaneously augment CD56bright NK cells, and some studies have suggested that the CD56bright NK population most closely correlates with suppression of cGvHD.12 Moreover, NKreg cells have been found to be important in the suppression of other autoimmune diseases, including solid organ transplantation, and rheumatoid arthritis, and are part of the suppressive tumor immune microenvironment.29 Our findings support that in addition to Treg cells and Breg cells, NKreg cells should also be considered important as part of the regulatory mechanisms that suppress cGvHD.
Figure 5.
CD56brightCD16- NKreg cell induction of CD4+ T-cell apoptosis. Apoptotic CD4+ T cells after the 96-hour suppression assay co-culture were identified with use of Annexin V and 7-AAD flow cytometric staining. Conditions tested included (A) activated CD4+ T cells in the absence of suppressor cells (control), (B) CD56brightCD16- regulatory NK (NKreg) cells, and (C) CD56dimCD16+ NK cells co-cultured with allogeneic CD4+ T cells at the 1:1 ratio. With use of FSC-A and SSC-A, lymphocytes were gated and using CD4 and 7-AAD staining live and dead CD4+ T cells were gated (plots not shown). After, Annexin V and 7-AAD were plotted. For the histogram, Annexin V-negative and 7-AAD-negative (live) CD4+ T cells are shown in blue, and Annexin V-positive, 7-AAD-negative (early apoptotic) and annexin V-positive, 7-AAD-positive (late apoptotic) CD4+ T cells are shown in purple. (D) The percentage of apoptotic CD4+ T cells in the CD4+ T-cell control as compared to the NKreg and NKdim conditions are shown (mean +/- standard deviation). NS: not significant. Statistical analyses were performed using Microsoft Excel version 2110 and a two-tailed t test – two-sample assuming unequal variance. The data is from a single experiment representative of 3 experiments.
Figure 6.
Evaluation of cell-cell contact dependence for CD56brightCD16- NKreg cell suppression. Suppressive effect of CD56brightCD16-relulatory NK (NKreg) cells towards allogeneic CD4+ T cells when cells are co-cultured for 96 hours at the ratio 1:1 in a standard round-bottom 96-well plate or 96-well transwell plate. (A) Proliferation of CD4+ T cells (control), (B) proliferation of CD4+ T cells co-cultured with NKreg cells in a standard plate, (C) proliferation of CD4+ T cells co-cultured with NKreg cells in a transwell plate. On the histograms, the x-axis displays the cell proliferation dye (CPD) eF450 and the y-axis displays the cell count. (D) Proliferation of CD4+ T-cell control compared to CD4+ T cells co-cultured with NKreg cells in a standard or transwell plate (mean +/- standard deviation). ***P<0.0005, NS: not significant. Statistical analyses were performed using Microsoft Excel version 2110 and a two-tailed t test – two-sample assuming unequal variance. The data is from a single experiment representative of 5 experiments.
As a result of our studies, we have determined an optimized cell sorting approach to isolate NKreg cells, verified their in vitro suppressive function which may correlate with cGvHD suppression, and outlined additional phenotypic and functional characteristics of NKreg cells. With further investigation we may decipher the mechanism of NKreg suppression and move forward in optimizing and operationalizing the expansion of NKreg cells associated with cGvHD suppression.
Supplementary Material
Acknowledgments
We would also like to acknowledge the Reid lab members from BCCHR, including Mohammad Reza Rahavi and Ali Farokhi for creating Luciferin-transfected K562 cells and imaging the plates. We further acknowledge the Lim lab members from BCCHR, including Pascal Leclair for contributing Jurkat and MOLT-4 leukemic cell lines. Additionally, we acknowledge the Center for Heart Lung Innovation at St. Paul’s Hospital (Vancouver, BC, Canada) for performing the nanoString assays.
Funding Statement
Funding: The authors would like to acknowledge the funding which supported this study, including a CIHR Foundation grant (to KRS), CIHR Team grant (to KRS and GDC), the Bjorknas Foundation (to KRS), Michael Cuccione Fellowship (to MPL), and CIHR CGS-M and Vanier scholarships (to MPL). Additional resources for the project were supported by collaborators at BCCHR (Megan Levings), BCCHR FlowCore, Canadian Blood Services, and healthy blood donor volunteers.
References
- 1.Park B, Yoo KH, Kim C. Hematopoietic stem cell expansion and generation: the ways to make a breakthrough. Blood Res. 2015;50(4):194-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogonek J, Kralj Juric M, Ghimire S, et al. Immune reconstitution after allogeneic hematopoietic stem cell transplantation. Front Immunol. 2016;7:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeiser R, Blazar BR. Pathophysiology of chronic graft-versus-host disease and therapeutic targets. N Engl J Med. 2017;377(26):2565-2579. [DOI] [PubMed] [Google Scholar]
- 4.Boyiadzis M, Arora M, Klein JP, et al. Impact of chronic graft-versus-host disease on late relapse and survival on 7,489 patients after myeloablative allogeneic hematopoietic cell transplantation for leukemia. Clin Cancer Res. 2015;21(9):2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filipovich AH, Weisdorf D, Pavletic S, et al. National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. Diagnosis and staging working group report. Biol Blood Marrow Transplant. 2005;11(12):945-956. [DOI] [PubMed] [Google Scholar]
- 6.Kariminia A, Holtan SG, Ivison S, et al. Heterogeneity of chronic graft-versus-host disease biomarkers: association with CXCL10 and CXCR3+ NK cells. Blood. 2016;127(24):3082-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultz KR, Kariminia A, Ng B, et al. Immune profile differences between chronic GVHD and late acute GVHD: results of the ABLE/PBMTC 1202 studies. Blood. 2020;135(15):1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kariminia A, Ivison S, Ng B, et al. CD56bright natural killer regulatory cells in filgrastim primed donor blood or marrow products regulate chronic graft-versus-host disease: the Canadian Blood and Marrow Transplant Group randomized 0601 study results. Haematologica. 2017;102(11):1936-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koreth J, Kim HT, Belizaire R, et al. Extra-corporeal photopheresis plus low-dose interleukin-2 for steroid-refractory chronic graft-vs.-host disease: efficacy, safety and immune correlates. Blood. 2017;130(Suppl 1):S515. [Google Scholar]
- 10.McQuaid SL, Loughran ST, Power PA, Maguire P, Szczygiel A, Johnson PA. Low-dose IL-2 induces CD56bright NK regulation of T cells via NKp44 and NKp46. Clin Exp Immunol. 2020;200(3):228-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, Zhang R, Shao M, et al. Efficacy and safety of low-dose IL-2 in the treatment of systemic lupus erythematosus: a randomised, double-blind, placebo-controlled trial. Ann Rheum Dis. 2020;79(1):141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iniesta P, Revilla N, Chen-Liang TH, et al. An early increase of CD56bright natural killer subset as dominant effect and predictor of response to extracorporeal photopheresis for graft-versus-host disease. Transfusion. 2018;58(12):2924-2932. [DOI] [PubMed] [Google Scholar]
- 13.Fu B, Tian Z, Wei H. Subsets of human natural killer cells and their regulatory effects. Immunology. 2014;141(4):483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kucuksezer UC, Aktas Cetin E, Esen F, et al. The role of natural killer cells in autoimmune diseases. Front Immunol. 2021;12:622306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuvelier GDE, Kariminia A, Nemecek ER, et al. Naïve helper T-cell and regulatory T- and NK-cell subsets are associated with pediatric chronic graft-versus-host disease: results of the ABLE/PBMTC 1202 study. Blood. 2020;136(Suppl 1):S11-12. [Google Scholar]
- 16.Cuvelier GDE, Nemecek ER, Wahlstrom JT, et al. Benefits and challenges with diagnosing chronic and late acute GVHD in children using the NIH consensus criteria. Blood. 2019;134(3):304-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dijke E, Ellis T, Larsen I, et al. Human leukocyte antigen (HLA) class II expression on regulatory T cells (Tregs) isolated from discarded human thymus is induced by in vitro expansion conditions. J Heart Lung Transplant. 2020;39:S178-S179. [Google Scholar]
- 18.Clottu AS, Mathias A, Sailer AW, et al. EBI2 expression and function: robust in memory lymphocytes and increased by Natalizumab in multiple sclerosis. Cell Rep. 2017;18(1):213-224. [DOI] [PubMed] [Google Scholar]
- 19.Sakaguchi S, Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T. Regulatory T cells: how do they suppress immune responses? Int Immunol. 2009;21(10):1105-1111. [DOI] [PubMed] [Google Scholar]
- 20.McMurchy AN, Levings MK. Suppression assays with human T regulatory cells: a technical guide. Eur J Immunol. 2012;42(1):27-34. [DOI] [PubMed] [Google Scholar]
- 21.Locatelli F, Pende D, Falco M, Della Chiesa M, Moretta A, Moretta L. NK cells mediate a crucial graft-versus-leukemia effect in haploidentical-HSCT to cure high-risk acute leukemia. Trends Immunol. 2018;39(7):577-590. [DOI] [PubMed] [Google Scholar]
- 22.Konjević GM, Vuletić AM, Mirjačić Martinović KM, Larsen AK, Jurišić VB. The role of cytokines in the regulation of NK cells in the tumor environment. Cytokine. 2019;117:30-40. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt A, Oberle N, Krammer PH. Molecular mechanisms of treg-mediated T cell suppression. Front Immunol. 2012;3:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laroni A, Gandhi R, Beynon V, Weiner HL. IL-27 imparts immunoregulatory function to human NK cell subsets. PLoS One. 2011;6(10):e26173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whangbo JS, Antin JH, Koreth J. The role of regulatory T cells in graft-versus-host disease management. Expert Rev Hematol. 2020;13(2):141-154. [DOI] [PubMed] [Google Scholar]
- 26.Nakasone H, Sahaf B, Miklos DB. Therapeutic benefits targeting B-cells in chronic graft-versus-host disease. Int J Hematol. 2015;101(5):438-451. [DOI] [PubMed] [Google Scholar]
- 27.Spoerl S, Mathew NR, Bscheider M, et al. Activity of therapeutic JAK 1/2 blockade in graft-versus-host disease. Blood. 2014;123(24):3832-3842. [DOI] [PubMed] [Google Scholar]
- 28.Vacca P, Vitale C, Munari E, Cassatella MA, Mingari MC, Moretta L. Human innate lymphoid cells: their functional and cellular interactions in Decidua. Front Immunol. 2018;9:1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian Z, Gershwin ME, Zhang C. Regulatory NK cells in autoimmune disease. J Autoimmun. 2012;39(3):206-215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.