Abstract
We tested coatis (Nasua nasua) living in an urban park near a densely populated area of Brazil and found natural SARS-CoV-2 Zeta variant infections by using quantitative reverse transcription PCR, genomic sequencing, and serologic surveillance. We recommend a One Health strategy to improve surveillance of and response to COVID-19.
Keywords: COVID-19, respiratory infections, severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, SARS, coronavirus disease, zoonoses, viruses, coronavirus, spillback, natural infections, coatis, wildlife, urban parks, Brazil
By November 2022, the COVID-19 pandemic had resulted in >630 million cases of disease worldwide (1). During the outbreak, natural occurrence of SARS-CoV-2 infections in animals was a hallmark; infections have been reported mainly in companion, domestic, captive, and farmed animals but also in wildlife (2,3). As of September 2022, the World Organisation for Animal Health had recorded 26 animal species infected with SARS-CoV-2 in 36 countries (2), indicating that the virus is able to cross the species barrier, thereby increasing risk of new transmission cycles and animal reservoirs (2,3).
Coatis (Nasua nasua) from South America are small diurnal mammals (family Procyonidae) that are omnivorous, terrestrial, synanthropic, and opportunistic. Coatis interact easily with humans and are often seen foraging for human food, especially from trash (4,5). We investigated the transmission of SARS-CoV-2 to a coati population living in an urban park near a large anthropized area of Brazil.
We collected serum samples and anal and oral swab samples during February–August 2021 from 40 free-living coatis inhabiting Mangabeiras Municipal Park in Belo Horizonte, Brazil (Appendix Table, Figure 1). Trained professionals captured coatis during 4 periods (February, June, July, and August), using appropriate personal protective equipment (laboratory coats, gloves, N95 face masks, and face shields) in accordance with all biosafety guidelines. Ethics approval was obtained for this study (Appendix).
Coatis were captured in Tomahawk Live Traps (https://www.livetrap.com/index.php) (70 cm × 35 cm × 40 cm) baited with banana pieces. Animals were anesthetized with Zoletil 100 (Virbac, https://vet-uk.virbac.com) by intramuscular injection (7–10 mg/kg body weight), clinically evaluated, identified, and marked with polypropylene earrings and microchips. After anesthesia recovery, each coati was released at their capture site.
We stored anal and oral swab specimens by using RNAlater (ThermoFisher Scientific, https://www.thermofisher.com) and extracted RNA by using QIAmp Viral RNA Mini Kits (QIAGEN, https://www/qiagen.com). We performed quantitative reverse transcription PCR targeting the nucleocapsid N1 and N2 regions (6) and sequenced PCR positive samples by using nanopore technology. We performed phylogenetic analysis by using IQ‐TREE2 (7) and maximum-likelihood reconstruction.
We detected SARS-CoV-2 RNA in 2 (5%) female coatis that had no clinical signs of infection (Table). We obtained a complete genomic sequence from the anal swab specimen from coati 535 (99% average coverage). The genomic sequence of SARS-CoV-2 obtained from the anal swab specimen from coati 535 indicated this variant belonged to the Zeta lineage (B.1.1.28.2, P.2) (Figure). The P.2 variant was initially detected in the state of Rio de Janeiro, Brazil, in July 2020 and was considered a variant of interest (9).
Table. Specimens from 2 SARS-CoV-2 RNA–positive coatis in study of SARS-CoV-2 spillback to wild coatis in sylvatic–urban hotspot, Brazil*.
| Coati ID | Collection date | Sex | Sample | SARS-CoV-2† | N1 Count‡ | N2 Count‡ |
|---|---|---|---|---|---|---|
| C341 |
2021 Feb 17 |
F |
Oral swab | Positive | 33 | 37 |
| Anal swab | Negative | NA | NA | |||
| Serum |
Negative |
NA |
NA |
|||
| C535 | 2021 Feb 18 | F | Oral swab | Positive | 20 | 24 |
| Anal swab | Positive | 30 | 33 | |||
| Serum | Negative | NA | NA |
*Oral and anal swab and serum samples were collected from 40 wild coatis inhabiting Mangabeiras Municipal Park in Belo Horizonte, Brazil. We performed quantitative reverse transcription PCR targeting the nucleocapsid N1 and N2 regions of SARS-CoV-2 RNA for each sample. ID, identification, NA, not applicable. †Specimens positive or negative for SARS-CoV-2 RNA by PCR. ‡PCR cycle threshold count.
Figure.
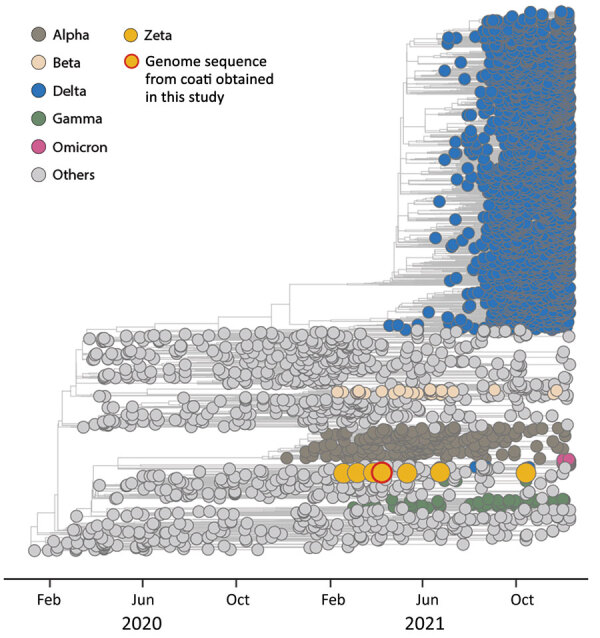
Time-scaled phylogenetic analysis of SARS-CoV-2 sequences, by variant type, in study of SARS-CoV-2 spillback to wild coatis in sylvatic–urban hotspot, Brazil. Maximum-likelihood method was used to compare the complete genomic sequence of SARS-CoV-2 obtained from coati (Nasua nasua) 535 (red-outlined yellow circle) and 3,441 SARS-CoV-2 reference genomic sequences (GISAID, https://www.gisaid.org) from around the world collected through October 2021. Colors represent clades corresponding to different SARS-CoV-2 variants of concern described by the World Health Organization; yellow indicates Zeta variant sequences. The SARS-CoV-2 sequence generated in this study was deposited in the GISAID database (accession no. EPI_ISL_8800460) and SisGen (Sistema Nacional de Gestão do Patrimônio Genético, https://www.sisgen.gov.br; no. A627307).
We performed plaque reduction neutralization tests (PRNT) on serum samples from all captured coatis to detect SARS-CoV-2 neutralizing antibodies (8). We serially diluted serum samples to obtain 1:20, 1:40, and 1:80 dilutions and measured 50% and 90% neutralizing activity against SARS-CoV-2. Twenty (50%) coatis had SARS-CoV-2 neutralizing antibodies in >1 dilution at the 50% level; at the 90% level, 13 (32.5%) coatis had detectable neutralizing antibodies in >1 dilutions and 7 (17.5%) coatis had SARS-CoV-2 neutralizing antibodies in all 3 dilutions. We observed neutralizing antibodies in all 3 serum dilutions for coati 535 (Appendix Figure 2).
We were unable to confirm the mode of SARS-CoV-2 transmission to the coati population. However, we found evidence for human-to-animal transmission; the P.2 genomic sequence from coati 535 was the same variant circulating in humans within the area during the study period. Furthermore, 50% of the coati population had antibodies against SARS-CoV-2, suggesting a cluster of natural exposure and infections within this population. Our results support indirect contact of coatis with contaminated human trash and food scraps in dumpsters and in the bordering urban areas of the park or potential direct close contact with infected human visitors (Appendix Figure 1).
Our findings agree with results from a zoo in Illinois, USA, that also confirmed SARS-CoV-2 in a coati by using molecular methods (2,10). Those results reinforce the susceptibility of coatis to SARS-CoV-2 infection and suggest possible virus shedding and transmission capacity of coatis. Viral RNA detection in both oral and anal swab specimens from coati 535 (Table) and presence of neutralizing antibodies indicate that viral replication occurred in this host. Therefore, our findings highlight possible SARS-CoV-2 enzootic maintenance in nature, including in fragmented green areas close to urban settings. Because of the potential for SARS-CoV-2 interspecies transmission, we recommend establishing a One Health strategy to improve surveillance and ability to respond to COVID-19 emergency health events.
Additional information for SARS-CoV-2 spillback to wild coatis in sylvatic–urban hotspot, Brazil.
Acknowledgments
We thank colleagues from the Laboratório de Vírus Instituto de Ciências Biológicas for their excellent technical support; Andrea Garcia de Oliveira, Marcelo Camargos, Kelly Nascimento, and Laboratório Federal de Defesa Agropecuária, Ministry of Agriculture Livestock and Food Supply (MAPA), for supporting our activities in the Biosafety Level Office International des Epizooties laboratory; Fundação de Parques Municipais e Zoobotânica of Belo Horizonte and Sérgio Augusto Domingues for allowing the field work; Augusto Gomes for coati photographs; Brazilian Post and Telegraph Company (Correios) for transporting our samples; and Jônatas Abrahão for excellent scientific discussions.
This study was funded by National Council for Scientific and Technological Development (CNPq) (grant nos. 440593/2016-6 and 403761/2020-4); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), process no. 88882.348380/2010-1; DECIT (Departamento de Ciência e Tecnologia)/MS (Brazilian Ministry of Health) no. 14/2016, Prevenção e Combate ao vírus Zika, FAPEMIG (Fundação de Amparo à Pesquisa do estado de Minas Gerais); PRPq-UFMG (Pró Reitoria de Pesquisa) and Finep/RTR/PRPq/Rede (Financiadora de Estudos e Projetos) COVID-19 (no. 0494/20-0120002600). This work was partially funded by the Ministry of Science, Technology, and Innovation (MCTI-Brazil); US National Institutes of Health (grant no. U01 AI151698) for the United World Antiviral Research Network; CRP- ICGEB RESEARCH GRANT 2020 (project no. CRP/BRA20-03, contract no. CRP/20/03); Oswaldo Cruz Foundation (no. VPGDI-027-FIO-20-2-2-30); and Brazilian Ministry of Health (no. SCON2021-00180). L.C.D.O. and M.M.T. were supported by a grant from MCTI/CNPQ/CAPES and FAPEMIG (project no. 465425/2014-3, Dengue and host-microbial interactions), M.G. and L.C.J.A. were supported by Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ), and M.G. was funded by PON “Ricerca e Innovazione” 2014–2020. Findings reported herein are the sole deduction, view, and responsibility of the researchers and do not reflect the official positions and sentiments of the funders.
G.S.T., B.P.D., M.G., M.M.T., H.L.F., C.W.A., and L.C.J.A are CNPq researchers; M.M.T., C.W.A., and E.L.D. are members of the Redevirus-MCTI; and H.L.F., Z.I.P.L., B.P.D., C.W.A., and E.L.D. are members of the Previr-MCTI.
Biography
Ms. Stoffella-Dutra is a PhD student in microbiology at the Federal University of Minas Gerais. Her primary research interests focus on virology, viral zoonoses, epidemiology, and wildlife disease ecology.
Footnotes
Suggested citation for this article: Stoffella-Dutra AG, de Campos BH, e Silva PHB, Dias KL, Domingos IJS, Hemetrio NS, et al. SARS-CoV-2 spillback to wild coatis in sylvatic–urban hotspot, Brazil. Emerg Infect Dis. 2023 Mar [date cited]. https://doi.org/10.3201/eid2903.221339
References
- 1.World Health Organization. WHO coronavirus (COVID-19) dashboard, 2022. [cited 2022 Nov 1]. https://covid19.who.int
- 2.World Organisation for Animal Health. SARS-CoV-2 in animals—situation report 17, 2022. [cited 2022 Nov 1]. https://www.woah.org/app/uploads/2022/10/sars-cov-2-situation-report-17.pdf
- 3.Delahay RJ, de la Fuente J, Smith GC, Sharun K, Snary EL, Flores Girón L, et al. Assessing the risks of SARS-CoV-2 in wildlife. One Health Outlook. 2021;3:7. 10.1186/s42522-021-00039-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues DH, Calixto E, Cesario CS, Repoles RB, de Paula Lopes W, Oliveira VS, et al. Feeding ecology of wild brown-nosed coatis and garbage exploration: a study in two ecological parks. Animals (Basel). 2021;11:2412. 10.3390/ani11082412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Projeto Quatis. Parque das Mangabeiras [cited 2022 Jun 6]. https://sites.google.com/site/projetoquatis/parque-das-mangabeiras
- 6.Centers for Disease Control and Prevention. CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel [cited 2022 Jun 8]. https://www.fda.gov/media/134922/download
- 7.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaves DG, de Oliveira LC, da Silva Malta MCF, de Oliveira IR, Barbosa-Stancioli EF, Teixeira MM, et al. Pro-inflammatory immune profile mediated by TNF and IFN-γ and regulated by IL-10 is associated to IgG anti-SARS-CoV-2 in asymptomatic blood donors. Cytokine. 2022;154:155874. 10.1016/j.cyto.2022.155874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voloch CM, da Silva Francisco R Jr, de Almeida LGP, Cardoso CC, Brustolini OJ, Gerber AL, et al. ; Covid19-UFRJ Workgroup, LNCC Workgroup, Adriana Cony Cavalcanti. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol. 2021;95:e00119–21. 10.1128/JVI.00119-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.US Department of Agriculture, Animal and Plant Health Inspection Service. Confirmation of COVID-19 in a coatimundi at an Illinois zoo, 2021. [cited 2022 Jun 6]. https://www.aphis.usda.gov/aphis/newsroom/stakeholder-info/sa_by_date/sa-2021/sa-10/covid-coatimundi
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information for SARS-CoV-2 spillback to wild coatis in sylvatic–urban hotspot, Brazil.


