Abstract
Calculations predict that testing of 5 000–10 000 molecules and >1 billion US dollars (£0.8 billion, £1=$1.2) are required for one single drug to come to the market. A solution to this problem is to establish more efficient protocols that reduce the high rate of re-isolation and continuous rediscovery of natural products during early stages of the drug development process. The study of ‘rare actinobacteria’ has emerged as a possible approach for increasing the discovery rate of drug leads from natural sources. Here, we define a simple genomic metric, defined as biosynthetic novelty index (BiNI), that can be used to rapidly rank strains according to the novelty of the subset of encoding biosynthetic clusters. By comparing a subset of high-quality genomes from strains of different taxonomic and ecological backgrounds, we used the BiNI score to support the notion that rare actinobacteria encode more biosynthetic gene cluster (BGC) novelty. In addition, we present the isolation and genomic characterization, focused on specialized metabolites and phenotypic screening, of two isolates belonging to genera Lentzea and Actinokineospora from a highly oligotrophic environment. Our results show that both strains harbour a unique subset of BGCs compared to other members of the genera Lentzea and Actinokineospora . These BGCs are responsible for potent antimicrobial and cytotoxic bioactivity. The experimental data and analysis presented in this study contribute to the knowledge of genome mining analysis in rare actinobacteria and, most importantly, can serve to direct sampling efforts to accelerate early stages of the drug discovery pipeline.
Keywords: Actinokineospora, BiNI, Cuatro Cienegas, genome mining, Lentzea, natural products
Data Summary
Genome sequences analysed in this study (Table 1) have been previously deposited in the National Center for Biotechnology Information (NCBI) with the accession numbers listed in Table S1 (available with the online version of this article).
Lentzea sp. CC55 and Actinokineospora sp. PR83 genome sequences produced in this study have been deposited at GenBank/ENA/DDBJ under the accession numbers JAJCXE000000000 and JAJCXD000000000, respectively.
Environmental Streptomyces 8S0 sequences analysed in this study have been deposited at GenBank/ENA/DDBJ under the accession numbers CP045031, JAJPUG000000000, JAJPUH000000000 and JAJPUI000000000.
Table 1.
Isolation and genomic information of the Actinokineospora and Lentzea strains used in this study
|
Genome name |
Source |
No. of contigs |
N50 |
Length (Mb) |
G+C (mol %) |
Reference |
|---|---|---|---|---|---|---|
|
Lentzea genomes |
||||||
|
L. waywayandensis strain DSM 44232 |
Soil, Waywayanda Lake, USA |
39 |
475 994 |
10.15 |
68.9 |
[83] |
|
L. alba strain NEAU.D13 |
Soil, Aohan Banner, Chifeng, Mongolia |
42 |
431 430 |
10.21 |
68.7 |
[84] |
|
L. albidocapillata subsp. violacea strain DSM 44796 |
Soil, gold mine cave in Kongju, Korea |
58 |
331 077 |
8.67 |
69 |
[85] |
|
L. flaviverrucosa strain CGMCC 4.578 |
Soil, Shanxi Province, China |
34 |
409 948 |
9.47 |
69.2 |
[86] |
|
L. jiangxiensis strain CGMCC 4.6609 |
Soil, Jiangxi Province, South-East China |
62 |
279 756 |
8.59 |
70.2 |
[53] |
|
L. albida strain DSM 44437 |
Soil, Jiangxi Province, South-East China |
43 |
352 714 |
9.44 |
70.2 |
[83] |
|
L. albidocapillata strain DSM 44073 |
Tissue specimen, Germany |
43 |
397 314 |
8.64 |
68.7 |
[87] |
|
L. flaviverrucosa strain DSM 44664 |
Soil, Shanxi Province, China |
38 |
462 737 |
9.47 |
69.2 |
[86] |
|
L. terrae strain NEAU-LZS 42 |
Soil, Henan Province, China |
145 |
211 542 |
10.58 |
68.6 |
[88] |
|
L. albidocapillata strain NRRL B-24057 |
Tissue specimen, Germany |
127 |
330 024 |
8.64 |
68.7 |
[87] |
|
L. pudingi strain CGMCC 4.7319 |
Limestone, South-West China |
63 |
432 100 |
9.21 |
69.1 |
[89] |
|
L. kentuckyensis strain NRRL B-24416 |
Equine placenta, USA |
317 |
92 128 |
10.21 |
68.8 |
[90] |
|
Lentzea sp. strain FXJ1.1311 |
Soil, Tibet, China |
120 |
199 708 |
9.37 |
69.5 |
[68] |
|
L. guizhouensis strain DHS C013 |
Limestone, Guizhou Province, South-West China |
1 |
9 997 872 |
9.99 |
70 |
[69] |
|
L. cavernae strain CGMCC 4.7367 |
Limestone, Guizhou Province, South-West China |
43 |
472 751 |
9.74 |
69.6 |
[26] |
|
L. aerocolonigenes NBRC 13195 |
Soil, Nagasaki City |
61 |
348 916 |
10.69 |
69 |
not found |
|
L. atacamensis DSM 45479 |
Soil, Atacama Desert, Northern Chile |
38 |
785 641 |
9.31 |
68.9 |
[91] |
|
L. nigeriaca DSM 45680 |
Soil, Abuja, Nigeria |
42 |
641 913 |
9.33 |
68.4 |
[56] |
|
L. fradiae CGMCC 4.3506 |
Soil, Wutaishan Mountain, Shanxi Province, China |
47 |
479 571 |
8.51 |
70.5 |
[92] |
|
L. deserti DSM 45480 |
Soil, Salar de Atacama, Atacama Desert, Chile |
50 |
442 485 |
9.53 |
68.8 |
[91] |
|
L. xinjiangensis CGMCC 4.3525 |
Soil, Xinjiang, North-West China |
61 |
283 838 |
8.69 |
68.9 |
[93] |
|
L. flava JCM 3296 |
Soil, Acharya Nagarjuna, India |
190 |
133 734 |
9.72 |
69 |
[94] |
|
L. aerocolonigenes NRRL B-3298 |
Soil, Nagasaki City, Japan |
204 |
163 428 |
10.65 |
69 |
[95] |
|
Lentzea sp. CC55 * |
Sediment, Cuatro Cienegas, Coahuila, México |
101 |
145 251 |
8.27 |
70.5 |
This study |
|
Actinokineospora genomes |
||||||
|
A. enzanensis strain DSM 44649 |
Forest soil, Yamanashi, Japan |
95 |
161 170 |
8.12 |
70.8 |
[65] |
|
A. xionganensis HBU206404 |
Xiongan lakeside soil, China |
70 |
325 226 |
6.81 |
69.6 |
[58] |
|
A. terrae strain DSM 44260 |
Soil, Yamanashi Prefecture, Japan |
41 |
383 933 |
7.59 |
71 |
[66] |
|
A. bangkokensis strain DSM 44EHW |
Rhizospheric soil, Bangkok, Thailand |
78 |
168 142 |
7.45 |
74 |
[96] |
|
A. mzabensis strain CECT 8578 |
Soil, Saharan desert, South Algeria |
32 |
442 119 |
7.55 |
72.8 |
[60] |
|
A. auranticolor strain YU 961–1 |
Soil, Yamanashi and Nagano Prefectures, Japan |
61 |
339 969 |
8.43 |
71.8 |
[65] |
|
A. inagensis strain DSM 44258 |
Fallen leaves, Lake Inaga, Yamanashi, Japan |
120 |
109 727 |
7.28 |
70.2 |
Not found |
|
A. spheciospongiae strain EG49 |
Marine sponge, Germany |
263 |
52 481 |
7.53 |
72.8 |
[54] |
|
A. cianjurensis strain DSM 45657 |
Leaf-litter, Indonesia |
19 |
1 225 445 |
7.62 |
70.9 |
[97] |
|
Root from Peganum harmala, China |
137 |
104 380 |
6.44 |
72.6 |
[59] |
|
|
A. fastidiosa strain JCM 3276 |
Soil, Egypt |
30 |
1 778 554 |
7.04 |
72.5 |
[98] |
|
A. alba DSM 45114 |
Soil, Xinjiang Province, China |
1 |
7 288 615 |
7.29 |
69.7 |
[62] |
|
A. baliensis DSM 45656 |
Soil and plant-litter, Indonesia |
1 |
7 681 387 |
7.68 |
70.6 |
[97] |
|
A. iranica IBRC-M 10403 |
Soil, Inche-Broun hypersaline wetland, North Iran |
44 |
291 113 |
6.58 |
70.8 |
[70] |
|
A. alba CPCC 201030 |
Soil, Xinjiang Province, China |
35 |
459 069 |
7.27 |
69.7 |
[62] |
|
A. alba IBRC-M 10655 |
Soil, Xinjiang Province, China |
34 |
443 362 |
7.27 |
69.7 |
[62] |
|
Actinokineospora sp. PR83* |
Sediment, Cuatro Cienegas, Coahuila, México |
182 |
77 654 |
7.64 |
72.7 |
This study |
Impact Statement.
We define a novel genomic metric, defined as biosynthetic novelty index (BiNI), that can be used to rapidly rank strains according to the novelty of the subset of encoding biosynthetic clusters. We used the BiNI score to prioritize a subset of potential natural product producers from different taxonomic groups and ecological contexts. We demonstrate the applicability of the BiNI score using two rare actinobacteria strains isolated from a highly oligotrophic environment. Our genomic analysis includes a deep biosynthetic gene cluster comparison for the rare actinobacteria Lentzea and Actinokineospora .
Introduction
A period of 10 to 12 years has been estimated for a natural product (NP) to become a drug available in the market. Expenses during this period of multi-stage and interdisciplinary research can exceed US$1 billion (£0.8 billion, £1=$1.2) per molecule [1]. The first step of the drug discovery process begins when entire collections of thousands of microbial isolates are subjected to different fermentation conditions to generate libraries of NPs that need to be tested against the drug target. Overall, from 5 000 to 10 000 NPs that show promising in vitro activity against a given target, 5 will enter clinical testing and only 1 will be registered for marketing and commercialization [2]. Increasing the discovery rate of microbial isolates that generate positive results against drug targets would directly impact the overall efficiency and costs of the entire drug discovery process [3, 4].
Microbes are one of the most prolific sources of bioactive NPs. Specifically, members of the phylum 'Actinobacteria' produce approximately 10 000 different bioactive molecules, which account for approximately 45 % of all microbial metabolites described to date [5]. The wide chemical diversity found in actinobacteria includes polyketides (PKs) [6], non-ribosomal peptides (NRPs) [7], hybrid PK/NRPs [8], terpenes [9], ribosomally synthesized and post-translationally modified peptides (RiPPs), among other representative chemical families [10]. While chemically diverse, NPs are produced within discrete and highly conserved genomic regions (biosynthetic gene clusters, BGCs). Recently, genome sequencing and bioinformatic tools have allowed genomic screening and predictions of the gene-structure associations of NPs [11]. However, traditional bioactive-guided approaches using microbes and plants remain hampered by low production and high rediscovery rates [12]. These limitations can be addressed by searching for less characterized microbial species in highly oligotrophic ecosystems (i.e. environments with low concentrations of nutrients, such as deep oceanic sediments, caves, glacial and polar ice, and deep subsurface soil among others) [13, 14].
The term rare actinobacteria was first defined as slow-growing actinobacteria found with lower frequency, compared to the frequency of isolation of Streptomyces using conventional methods [13, 15, 16]. Since then, rare actinobacteria (or underisolated actinobacteria) are known as a genetic repository of novel BGCs [14]. For example, Pseudonocardia sp. HS7, isolated from the cloacal aperture of sea cucumber (Holothuria moebii), can produce one of the curvularin macrolides that have shown potential cytotoxicity toward six tested cancer cell lines [17]. The endophytic actinomycete Micromonospora sp. GMKU326 isolated from the root of a leguminous plant, Maklam phueak (Abrus pulchellus Wall. Ex Thwaites subsp. pulchellus), produces maklamicin, a new PK with promising antimicrobial activity against Gram-positive pathogens [18]. Moreover, isolates from soil samples taken in a rainforest undergoing restoration contained new polycyclic antibiotics such as pradimicin-IRD, produced by Amycolatopsis sp. IRD-009, which presented antimicrobial activity against Streptococcus agalactiae , Pseudomonas aeruginosa and Staphylococcus aureus [19]. As a result of taxonomically selective isolation and genetic techniques, the number of rare genera increased to 220, with 2 500 bioactive metabolites, according to reports up to 2010 [13, 20–22].
In recent years, highly oligotrophic environments [23] have generated interest as a rich source of rare actinobacteria [24–28]. The Cuatro Cienegas Basin, located in the Chihuahuan Desert in the State of Coahuila, Mexico, has been mostly studied due to the low nutrient availability in the soil [29]. This ecosystem shows a nutrient imbalance in nitrogen-to-phosphorus stoichiometry (N:P 157 : 1), with a high calcium accumulation rising from 35 to 90 % across the microbialite matrix [30, 31]. Nutrient limitation, for example, the extremely low concentrations of phosphate and iron, generates the environmental conditions to modulate unique metabolic traits in diverse microbial communities [32–34]. Evidence of the potential for novel chemical diversity in the Cuatro Cienegas Basin was reported in two strains of Micrococcus sp. (strain CH3 and CH7) for which phylogenomic analysis and siderophore production profiles demonstrate the production of previously unreported aryl-substituted desferrioxamines [33].
Here, we present the implementation of a simple genomic metric (biosynthetic novelty index, BiNI) that allows direct classification of microbial strains according to their biosynthetic originality. We use the BiNI score to perform a strain prioritization exercise of members of different taxonomic families (and different ecological contexts), including those of rare actinobacteria. We demonstrate the potential application of the BiNI score by isolating and comparing the biosynthetic potential of two rare actinobacteria belonging to the genera Lentzea and Actinokineospora of the Cuatro Cienegas Basin. Finally, using phylogenomics and different genome mining bioinformatic tools, we report the scope of the biosynthetic potential of our isolates and all of the members from these two genera reported to date. Overall, our results highlight the importance of exploring natural extreme environments for the research of NPs and describe an efficient method for strain prioritization according to biosynthetic novelty.
Methods
Strain isolation and culture conditions
Lentzea sp. CC55 and Actinokineospora sp. PR83 were isolated from the Cuatro Cienegas Basin, as previously reported [32]. Lentzea sp. CC55 was isolated from the region of Pozas Azules (26°49'39.01" N, 102°01'26" W), while Actinokineospora sp. PR83 was isolated from Pozas Rojas (26°52'16.8" N, 102°1'11.3" W). Homogenized sediments were cultured in serial dilutions on MS solid media (per litre: mannitol, 20 g; soy flour, 20 g; agar, 20 g; in tap water) [35]. All cultures were supplemented with cycloheximide (100 µg ml−1) and nalidixic acid (30 µg ml−1). Bioactivity assays were performed for both strains using the OSMAC () approach. Lentzea sp. CC55 and Actinokineospora sp. PR83 isolates were cultured in the following media (solid and liquid media): International Streptomyces Project (ISP)-2 [36], ISP-4 [37], R5 [38], Czapek [1 l: 1 g K2HPO4, 0.01 g FeH14O11S, 0.5 g H14MgO11S, 0.5 g KCl, 3 g NaNO2, 30 g sucrose, 15 g agar], BG [1 l: 0.2 g MgSO4, 0.2 g CaCl2, 1 g KH2PO4, 1 g K2HPO4, 1 g (NH4)2SO4, 0.05 g FeCl3], BS [1 l: 0.2 g MgSO4, 0.02 g CaCl2, 1 g KH2PO4, 1 g K2HPO4, 1 g (NH4)2SO4, 0.05 g FeCl3], M1 [1 l: 10 g soluble starch, 4 g yeast extract, 2 g peptone, 20 g NaCl, 18 g agar] and oatmeal [1 l: 20 g oatmeal, 0.001 g Fe2H14O19S3, trace salts, 0.001 g Cl2H8MnO4, 0.001 g ZnSO4. 7H2O, 18 g agar]. The ISP-2, BG, BS, R5 and M1 media were prepared using glucose purchased from Biochemicals (Australia); yeast extract from Merck; peptone from Oxoid; and soluble starch, malt extract, mannitol, sucrose and agar from Sigma-Aldrich. ISP-4 and oatmeal media were purchased from DB Difco (Fisher-Scientific).
Genome sequencing and assembly
Genomic DNA was extracted from 15 ml ISP-2 liquid cultures (spore concentration of 1×106 spores ml−1). Shake flasks were inoculated with 50 µl of the spore suspension and incubated for 48 h at 28 °C, at 200 r.p.m. The biomass was transferred into a sterile porcelain dish for liquid nitrogen lysis. The crushed mycelium was mixed with 400 µl TE buffer supplemented with lysozyme (20 mg ml−1) (Sigma-Aldrich), 10 % (w/v) SDS and proteinase K (20 mg ml−1) (Fisher-Scientific), for chemical rupture. The lysate was cooled and extracted twice with equal volumes of phenol:chloroform:isoamyl alcohol (25 : 24 : 1, v/v) and phenol:chloroform (25 : 24, v/v) (Fisher-Scientific) at 10 000 r.p.m for 5 min. The aqueous phase was transferred for DNA precipitation with 70 % ethanol at −20 °C for 30 min. The pellet was formed by centrifuging at 10 000 r.p.m for 15 min. Genomic DNA was resuspended in deionized sterile water and checked for quality using 260/280 and 230/260 ratios provided by a NanoDrop 1 000 spectrophotometer (NanoDrop Technologies) and a 1 % agarose gel for electrophoresis. Whole-genome sequencing was performed using the Illumina MiSeq platform with 2×300 bp paired-end reads. The reads obtained were trimmed using Trimmomatic v0.32 [39] and assembled using Velvetg and Velveth v1.2.10 [40]. k-mers ranging from 31 to 171 (increasing 10 units per iteration) were tested, and the best assembly was selected for annotation and analysis.
A database of genomes from Lentzea (23 genomes) and Actinokineospora (16 genomes) was generated using publicly available genomes at the National Center for Biotechnology Information (NCBI) up to September 2021 (Table S1). The inclusion criteria were focused on the quality of the assembly. The mean number of contigs in each group was 81.2 and 66.3, for Lentzea and Actinokineospora , respectively. All genomes were annotated using the Rapid Annotations of the Subsystems Technology (rast) platform [41]. A core-/pan-genome analysis was performed for both datasets using the Bacterial Pan-Genome Analysis (bpga) tool [42] in the usearch algorithm, with the similarity threshold set to 0.5 orthologue identification. A core phylogenetic tree was reconstructed with the protein sequences of core genes from bpga. The process included split sequences for protein family, alignment with muscle v.3.8.31 [43] and trimming with the Gblocks package. The processed alignments were concatenated into a supermatrix with FasConCat [44] and finally the iq-tree software [45] was used for phylogeny reconstruction. The phylogenetic tree was visualized using FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). Prediction of BGCs was performed for all the genomes in both datasets using the Antibiotic and Secondary Metabolite Analysis Shell (antiSMASH), version 6.0 [46], which identifies BGCs using a signature profile hidden Markov model based on multiple sequence alignments of experimentally characterized signature proteins or protein domains. The BGC similarity networks were constructed only with complete BGCs for all Lentzea and Actinokineospora , as well as the MIBiG (Minimum Information about a Biosynthetic Gene cluster) database [47] (1802 BGCs), using the Python-based platform Biosynthetic Gene Similarity Clustering and Prospecting Engine (BiG-SCAPE) [48] with default settings. antiSMASH results were uploaded in the online repository BiG-FAM (Biosynthetic Gene Cluster Families Database) in order to search for ‘homologous’ groups of BGCs putatively encoding the production of similar specialized metabolites, through distance measurement [49].
For determination of the novelty of the biosynthetic potential encoded by a given isolate, we used a metric that mainly relates the number of clusters (n) identified by antiSMASH and the distance (d) sampling test result provided by the BiG-FAM platform (threshold value >900) with the equation . The BiNI represents an indicator of the novelty of the subset of BGCs encoded by each isolate and could be valuable in terms of obtaining insights into the contextual ecology or the biosynthetic potential of a given taxonomic gender.
Chromatography-based purification and bioactivity assays
Cultures were extracted with ethylacetate:methanol (3 : 1) (30 ml) for solid media and with ethylacetate (1.5 ml) for liquid media. The organic extracts were dried and resuspended in methanol (500 μl and 50 μl for solid and liquid, respectively). The resuspended extracts were then filtered through a 0.45 µm polytetrafluoroethylene (PTFE) membrane from Sigma-Aldrich before running in the UHPLC-DAD (ultra-high-performance liquid chromatography-diode array).
Antimicrobial activities were measured against the Gram-positive bacteria Staphylococcus aureus (ATCC 25923) and Bacillus subtilis (ATCC 6633), and the Gram-negative bacteria Escherichia coli (ATCC 25922) and P. aeruginosa (ATCC 27853), using the broth micro-dilution method [50]. The test was performed (in triplicate) in 96-well microtitre plates by serial dilution in tryptic soy broth (TSB). Crude microbial extracts (1 mg ml−1) were tested first, then an aliquot (20 µl) was transferred followed by addition of freshly prepared microbial broth (180 µl, 104–105 c.f.u. ml−1 cell density) for the dilution series. The plates were incubated at 26.5 °C for 24 h. The optical density of each well was measured spectrophotometrically at 600 nm using a POLARstar Omega plate reader (BMG LABTECH). Both media with and without microbial inoculation were tested as negative controls.
Cytotoxicity assays were carried out using the MTT [3-(4,5-dimethylthiazol-2- yl)−2,5-diphenyltetrazolium bromide; Sigma-Aldrich] method, as previously described [51], using adherent NCIH460 (human large cell lung carcinoma). Cells were harvested with trypsin (from Sigma-Aldrich) and dispensed into 96-well microtitre assay plates at 2 000 cells per well and incubated for 18 h at 37 °C with 5 % CO2 (to allow cells to attach). Crude extracts were dissolved in 5 % DMSO in PBS (v/v) and aliquots (20 µl) tested at a final concentration of 1 mg ml−1. Vinblastine was used as positive control and negative control wells were treated with 5 % aqueous DMSO. After 68 h incubation at 37 °C with 5 % CO2, an aliquot (20 µl) of MTT in PBS (4 mg ml−1) was added to each well (final concentration of 0.4 mg ml−1), and the microtitre plates incubated for a further 4 h at 37 °C with 5 % CO2. Following this final incubation, the medium was aspirated and the formazan crystals that precipitated were dissolved in DMSO (100 µl per well). The absorbance for each well was measured at 580 nm on a POLARstar Omega microtitre plate reader. IC50 values were calculated using Prism 5.0 (GraphPad Software), as the concentration of analyte required to achieve 50 % inhibition of cancer cell growth (compared to negative controls). All experiments were performed in duplicate.
Results
BiNI
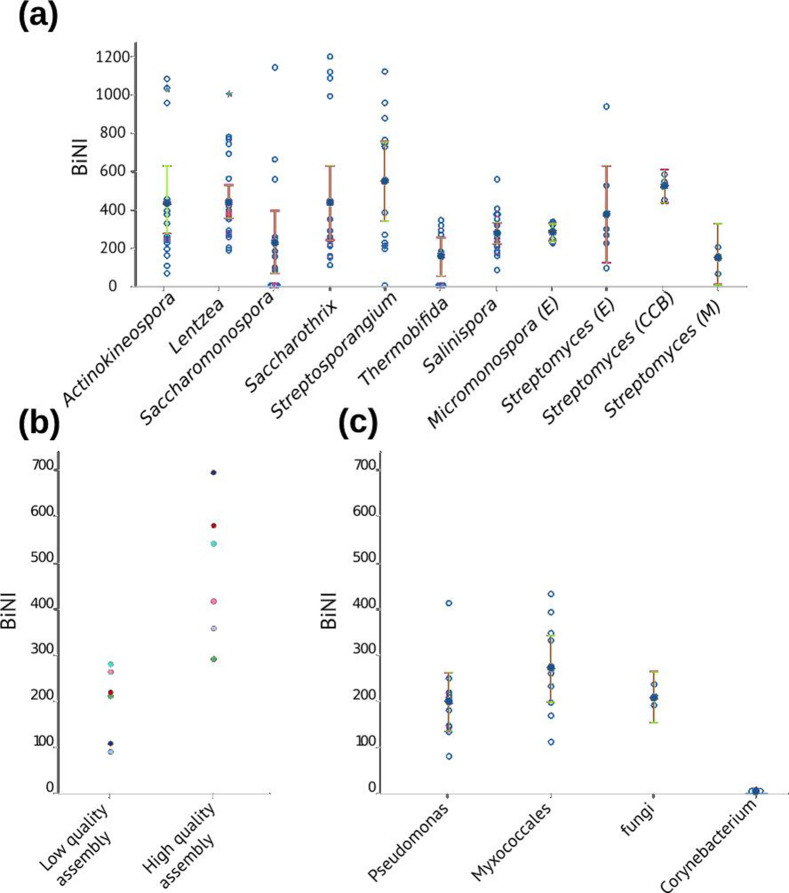
We first used a database of high-quality genomes including common and rare actinobacteria (underisolated actinobacteria) from public databases and our strain collection (Table S1). We estimated the BiNI for all the genomes and compared the taxonomic genera recognized as rare actinobacteria, including Actinokineospora (17 genomes), Lentzea (24 genomes), Saccharomonospora (14 genomes), Saccharothrix (17 genomes), Streptosporangium (14 genomes), Thermobifida (10 genomes) and Salinispora pacifica strains (18 genomes) (Fig. 1a). Taking into account the entire genera, Streptosporangium showed the highest novelty index (BiNI=548.4), followed by Actinokineospora (BiNI=451.6), Lentzea (BiNI=439.2) and Saccharothrix (BiNI=438.4). However, Saccharomonospora , Thermobifida and Salinispora showed the lowest novelty BiNIs (155.2, 231.1 and 274, respectively). While detailed information about the isolation site is missing from databases, we observed that most of highest BiNI-scored rare actinobacteria were isolated from desert soils. For example, Streptosporangium saharense strain CECT 8840 (Saharan soil, BiNI=1120), Actinokineospora fastidiosa JCM 3276 (Egyptian soil, BiNI=1 097), Actinokineospora sp. PR83 (Cuatro Cienegas, BiNI=1049), Lentzea sp. CC55 (Cuatro Cienegas, BiNI=1 008) and Saccharothrix tamanrassetensis CECT8640 (Saharan soil, BiNI=1122). We did not observe a specific environment of isolation for strains with low BiNI.
Fig. 1.
BiNI of different actinobacteria genera. (a) BiNI comparison between genomes from rare genera of actinobacteria ( Actinokineospora , Lentzea , Saccharomonospora , Saccharothrix , Streptosporangium , Thermobifida and Salinispora); endophytic actinobacteria (E) ( Micromonospora and Streptomyces ); free-living Streptomyces isolated from Cuatro Cienegas Basin (CCB); and reference Streptomyces (M) (Streptomyces xiamenensis 318, Streptomyces rapamycinicus NRRL 549 and Streptomyces coelicolor A32). Mean values (solid blue) are reported for all groups. Blue stars represent the BiNI values of Actinokineospora sp. PR83 and Lentzea sp. CC55 isolated in this study from the Cuatro Cienegas Basin. (b) BiNI comparison between low-quality and high-quality genome assemblies. Blue, Streptomyces sp. KL122B (2104 vs 18 contigs); red, Streptomyces sp. CC228A (1299 vs 26 contigs); green, Streptomyces 8s0 CC219A (1172 vs 382 contigs); purple, Streptomyces sp. KL111A (2096 vs 205 contigs); cyan, Streptomyces 8s0 CC210A (847 vs 9 contigs); pink, Streptomyces sp. CC201C (742 vs 23 contigs). (c) A comparison of the BiNI of reference strains, including genomes from Pseudomonas , Myxococcales , fungi and Corynebacterium .
In addition, we tested the robustness of the BiNI by assessing genomes of different assembly quality and length (Fig. 1b). We compared the BiNIs of highly fragmented genome assemblies of five Streptomyces isolates (sequenced with the Illumina platform) with their corresponding high-quality assemblies obtained by Oxford Nanopore sequencing: Streptomyces sp. KL122B (2104 vs 18 contigs), Streptomyces sp. CC228A (1299 vs 26 contigs), Streptomyces 8s0 CC219A (1172 vs 382 contigs), Streptomyces sp. KL111A (2096 vs 205 contigs), Streptomyces 8s0 CC210A (847 vs 9 contigs) and Streptomyces sp. CC201C (742 vs 23 contigs). We observed that the biosynthetic novelty is underestimated when it is tested with low-quality genome assemblies. Finally, we tested the BiNI of genomes from well-characterized strains, which are not expected to show a high BiNI, including Pseudomonas , Corynebacterium , Myxococcales and fungi (Fig. 1c).
Oligotrophic environments harbour strains with high BiNI
We isolated Lentzea sp. CC55 and Actinokineospora sp. PR83 from sediments of the Cuatro Cienegas Basin, Mexico. Growth on MS agar showed circular colonies with raised elevation and filiform margin, with pink mycelium for Lentzea sp. CC55 and yellow mycelium for Actinokineospora sp. PR83. Using Illumina sequencing, we determined general genome assembly features: Lentzea sp. CC55 encodes 8.27 Mb (70.5 mol% G+C, 101 contigs, N50 145251) and Actinokineospora sp. PR83 encodes 7.64 Mb (72.7 mol% G+C, 182 contigs, N50 77654) genomes (Table 1). The genome of Lentzea sp. CC55 encodes a total of 7675 proteins, 9 rRNA genes and 69 tRNA genes, while Actinokineospora sp. PR83 encodes a total of 6565 proteins, 13 rRNA genes and 50 tRNA genes. Using the Microbial Genome Atlas platform (MiGA version 1.1.2.2) [52], we determined that Lentzea sp. CC55 shares 96.90 % average nucleotide identity (ANI) with Lentzea jiangxiensis CGMCC 4.6609 [53], and Actinokineospora sp. PR83 shares 95.9 % ANI with Actinokineospora spheciospongiae EG49 [54]. Lentzea sp. CC55 and Actinokineospora sp. PR83 genome sequences produced in this study have been deposited at GenBank/ENA/DDBJ under the accession numbers JAJCXE000000000 and JAJCXD000000000, respectively.
Using the BiNI scores, we ranked according to biosynthetic novelty all the Lentze a and Actinokineospora strains reported in genomic databases. The strain Lentzea sp. CC55, isolated from sediments of an oligotrophic environment, has the highest degree of biosynthetic novelty (BiNI=1 008.6) among all the Lentzea strains in our database. Another Lentzea strain with potential of encoding novel BGCs is Lentzea flava (BiNI=777.8), isolated from soil, Acharya Nagarjuna, India [55], and Lentzea nigeriaca (BiNI=742.5) isolated from desert soil in Nigeria [56] (Table S1). Regarding the strains of Actinokineospora , based on the BiNI, we found that the strains A. fastidiosa isolated from desert soil samples in Egypt [57], 'Actinokineospora xionganensis' isolated from lakeside soil samples in China [58], and Actinokineospora sp. PR83 isolated from oligotrophic environments in Mexico had the highest BiNI values (1097.2, 1050.5 and 1049.3, respectively). In contrast, strains Actinokineospora pegani isolated from root of Peganum harmala, China [59], and Actinokineospora mzabensis isolated from Saharan desert soil [60], Southern Algeria, had the lowest BiNI scores (81.3 and 117.7, respectively) (Table S1).
To support the idea that nutrient-limiting environments harbour micro-organisms with novel biosynthetic potential, we compared the genomes of a group of free-living Streptomyces sp. 8s0 isolated in the Cuatro Cienegas Basin, seven endophytic Micromonospora (BiNI=284.3), seven endophytic Streptomyces with a group of reference Streptomyces genomes (Streptomyces_xiamenensis_strain 318, Streptomyces_rapamycinicus strain NRRL 5491 and Streptomyces_coelicolor strain A32) (Table S1). We chose the Streptomyces genomes of different length to avoid a possible bias caused by the number of BGCs. According to the BiNI score, isolates from nutrient-limited environments encode BGCs of higher novelty as the groups of Streptomyces from Cuatro Cienegas Basin and the endophytic Streptomyces showed a BiNI score of 525.8 and 376, respectively. The three reference Streptomyces genomes used showed a BiNI score of 144.5 (Fig. 1a).
Phylogenomic analysis of Lentzea sp. CC55 and Actinokineospora sp. PR83 isolates
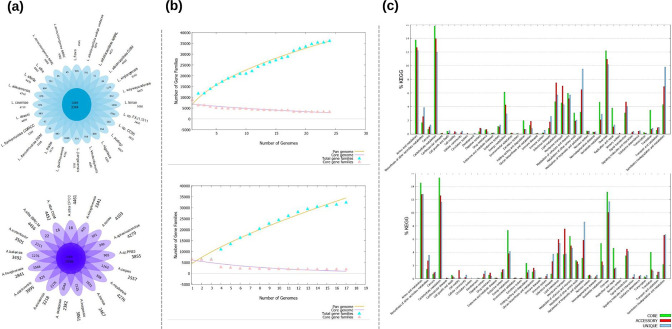
To provide insights into the genetic diversity that accounts for adaptation capabilities and population structure, we analysed Lentzea and Actinokineospora pan-genomes using the bpga tool. Our analysis includes all of the genomes reported for both Lentzea and Actinokineospora up to September 2021 (see Table 1). The pan-genome of Lentzea is estimated at 33 321 genes, including a core genome of 3344 genes, a mean accessory genome of 4513 genes and a total of 17 800 unique genes (Fig. 2a). Lentzea aerocolonigenes (strains NBRC 131195 and NRRL B-3298), Lentzea albidocapillata (strains DSM 44073 and NRRL B-24057) and Lentzea flaviverrucosa (strains CGMCC 4.578 and DSM 44664) encode the smallest number (range 45–85) of unique genes within the pan-genome. In contrast, Lentzea terrae DSM 44260 (1 016 genes), Lentzea xinjiangensis CGMCC 4.3525 (1 176 genes), Lentzea guizhouensis DHS C013 (1 668 genes) and Lentzea sp. FXJ1.1311 (2561 genes) encode an almost 20 times larger number of unique genes (Table S2). Our strain, Lentzea sp. CC55, encodes 568 unique genes. The pan-genome of Actinokineospora is estimated at 32 010 genes, including a core genome of 1956 genes, a mean accessory genome of 3573 genes and a total of 17 821 unique genes (Fig. 2a). Actinokineospora alba strains DSM 45114 (16 genes), CPCC 201030 (18 genes) and IBRC-M 10655 (22 genes) present a small number of unique genes compared to A. fastidiosa JCM 3276, Actinokineospora auranticolor YU 961–1 and Actinokineospora enzanensis DSM 44649, which encode much larger numbers of unique genes (2044, 2111 and 2176 genes, respectively) (Table S2). Our strain, Actinokineospora sp. PR83, encodes 965 unique genes.
Fig. 2.
Pan-genome analysis for Lentzea and Actinokineospora genomes. (a) Flower diagrams representing the core, unique and accessory genes for each genus. The numbers underneath the names represent the number of accessory genes. (b) Core-/pan-genome plot over 500 iterations using the bpga analysis tool. The pan-genome is open. (c) KEGG functional analysis of the pan-genome. The graph shows the predicted function of proteins encoded by core (green), accessory (red) and unique (blue) genes of the pan-genome using KEGG identification.
Inspection of the core–pan plots indicated that both pan-genomes are open. This highlights the strong requirement to increase efforts for isolation in these genera (Fig. 2b). Functional annotation using the KEGG (Kyoto Encyclopedia of Genes and Genomes) database for the pan-genomes indicates that most of the core, accessory and unique genes are related to central metabolic pathways (Fig. S1). According to the KEGG functional classification, 13–15 % of the pan-genome for Lentzea (core 20, unique 31, accessory 21) and Actinokineospora (core 28, unique 32, accessory 29) belong to amino acid metabolism and carbohydrate metabolism and, overall, the genes involved in these functions would represent the major contributors to the genetic diversity observed in each genus. As expected, for both genera, most of the unique genes pertain to the specialized metabolism of terpenoids and PKs, as well as the biodegradation of xenobiotics metabolism (Fig. 2c).
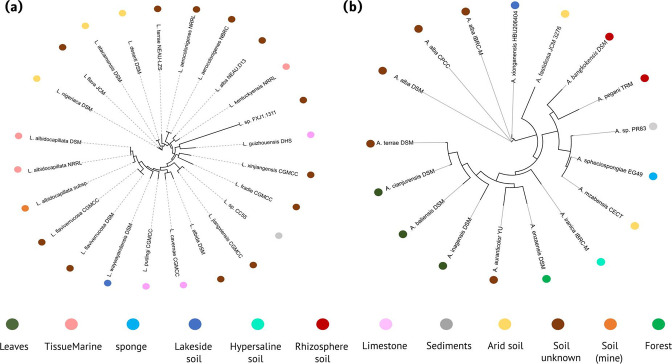
To infer the phylogenetic relationships among members of both Lentzea and Actinokineospora , we reconstructed a phylogenetic tree using the core genome derived from the pan-genome analysis. For the Lentzea phylogenetic tree (Fig. 3a), 24 strains were grouped into two clades, with the exception of L. flava (soil, Acharya Nagarjuna, India) and L. nigeriaca (arid soil from Nigeria). One clade contained two Lentzea strains isolated from arid soils (Atacama Desert of Northern Chile) [61], and the second clade contained 20 strains isolated from different soil sources (Table 1). The strains Lentzea deserti and Lentzea atacamensis (isolated from arid soils); L. albidocapillata DSM and NRRL (isolated from samples of tissue); L. aerocolonigenes NRRL and NRBC (isolated from soil, Japan); L. xinjiangensis and Lentzea fradiae (soil, China); and L. flaviverrucosa DSM and CGMCC (soil, Shanxi Province, China) were subgrouped according to the place of isolation. Our strain, Lentzea sp. CC55, was closely related to L. jiangxiensis strain CGMCC 4.6609 (soil isolate from Jiangxi Province, South-East China) [50].
Fig. 3.
Phylogeny trees based on the pan-matrix from bpga analysis for the Lentzea (a) and Actinokineospora (b) strains. The alignment was performed with muscle v.3.8.31. The characteristics of isolation for each genome are visualized by colour.
For the phylogenetic analysis of Actinokineospora (Fig. 3b), 17 strains were grouped into two clades, with the exception of the A. alba strains (soil, Xinjiang Province, China) [62], 'A. xionganensis' (lakeside soil Xiongan, China) [58] and A. fastidiosa (arid soil, Egypt) [57]. In the first clade, the strains Actinokineospora bangkokensis DSM [63] and A. pegani TRM (both from rhizosphere soils) [59] are grouped together, while our strain, Actinokineospora sp. PR83, A. spheciospongiae (marine sponge) [54] and A. mzabensis CECT (desert soil) [60] form another subclade. A. enzanensis DSM (soil, Yamanashi Japan) [64] and A. auranticolor YU [65] form a monophyletic group with three strains isolated from tree leaves as well as Actinokineospora terrae (soil, Japan) [66]. Only the strains of A. alba isolated from soil, China; A. bangkokensis and A. pegani isolated from rhizospheric soil; and Actinokineospora cianjurensis , Actinokineospora baliensis and Actinokineospora inagensis isolated from leaves were subgrouped according to the ecological context.
Lentzea sp. CC55 presents a higher number of BGCs grouped as singletons compared to other Lentzea strains
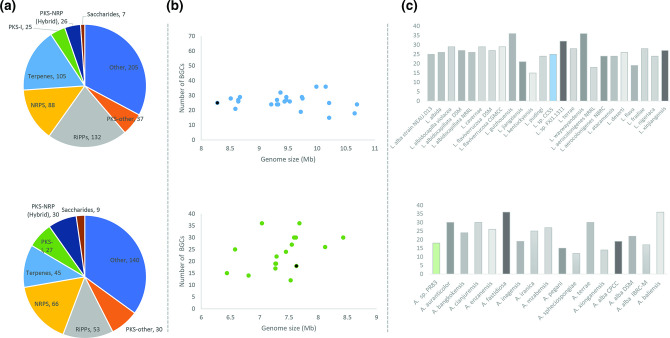
Using antiSMASH (version 6.0) [46], we identified 32 BGCs (25 complete) for Lentzea sp. CC55, including 6 NRPSs, 7 PKSs, 4 terpenes, 4 RiPPs and 4 classified as ‘other’. A total of seven BGCs showed medium-to-high (50–100 %) homology with previously identified clusters (two geosmins, 2-methylisoborneol, coelichelin, Ery-9 and methoxyhydroquinones). Fourteen clusters were identified with low homology (1–36 % identity) and four BGCs presented no match in the database (see Table S3). We analysed the biosynthetic potential (number of complete BGCs) for all the genomes of Lentzea publicly available, taking into account genome size and quality of the assembly (number of contigs). We identified a total of 625 BGCs in all of the genomes (mean=26 BGCs per genome), where 14 % were classified as non-ribosomal peptides, 17 % as terpenes, 21 % as RiPPs, 10 % as PK synthases, 4 % as hybrid PKS-NRP and 2 % as saccharides. A total of 205 BGCs either belonged to other chemical families or had no defined classification according to available databases (Fig. 4a). We observed an even number of BGCs (mean=25±10) (Fig. 4b). The highest numbers of BGCs in Lentzea were found in Lentzea waywayandensis (36 BGCs, 10.15 Mb, 39 contigs), L. guizhouensis (36 BGCs, 9.99 Mb, 1 contig) and Lentzea sp. FXJ1.1311 (32 BGCs, 9.37 Mb, 120 contigs), while Lentzea sp. CC55 genome encodes 25 BGCs in a genome of 8.28 Mb assembled in 101 contigs. The strain with the lowest number of BGCs was Lentzea kentuckyensis (15 BGCs, 10.21 Mb, 317 contigs) (Fig. 4c).
Fig. 4.
Distribution of BGCs according to antiSMASH identity. (a) Circular representation of BGC family distribution for Lentzea (circle at the top) and Actinokineospora (circle at the bottom). (b) Genome length versus number of clusters for Lentzea (blue; graph at the top) and Actinokineospora (green; graph at the bottom). Black circles represent Lentzea sp. CC55 and Actinokineospora sp. PR83, respectively. (c) Histogram for number of clusters by strain for Lentzea (blue; graph at the top) and Actinokineospora (green; graph at the bottom).
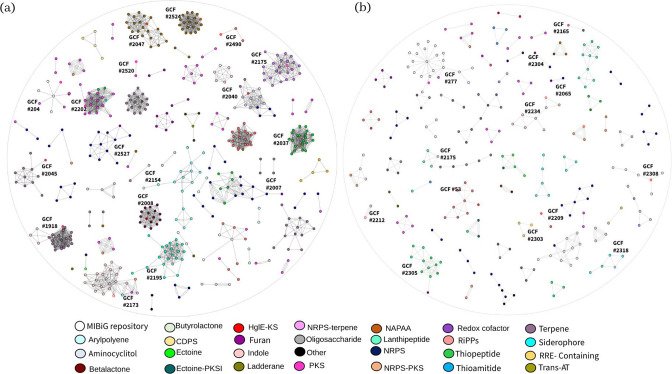
In order to study the repertoire of specialized metabolism-related clusters of all of the Lentzea strains, we constructed BGC sequence similarity networks including the 1802 BGCs reported in the MIBiG database (September 2021). We used a raw distance cut-off of 0.3 to observe the BGC relationships among strains, highlighting the principal gene cluster families (GCFs) related to Lentzea sp. CC55 (Fig. 5a, Table S3). Most of the GCFs conserved by the Lentzea strains were related to the category other, including siderophores, redox cofactors and indole, followed by RiPPs such as non-alpha poly-amino acids like e-Polylysin (NAPPA), lanthipeptides and thiopeptides; terpenes; NRPS and PKS. Based on this analysis, we observed that the BGCs from L. flaviverrucosa DSM and L. flaviverrucosa CGMCC (isolated from soils from Shanxi Province, China) [67] are conserved among all members of the genus Lentzea . In contrast, Lentzea sp. FXJ1.1311 isolated from soil in Tibet, China [68], and L. guizhouensis isolated from limestone [69], Guizhou Province in South-Western China, were the strains with the highest number of unique BGCs (25/32 and 20/36, respectively). On comparison of the BGCs to the MIBiG database, we found a match for only 5/280 GCFs (GCF 2362, GCF 79, GCF 204, GCF 1278 and GCF 1951). We assigned 17 out of 25 BGCs from Lentzea sp. CC55 to a known GCF, while 8 BGCs were identified as singletons (Table S3).
Fig. 5.
BGC network showing chemical diversity forLentzea (a) and Actinokineospora (b) strains. Each colour node corresponds to a BGC associated with a chemical family. BGCs with red circular borders are those of . Lentzea sp. CC55 and Actinokineospora sp. PR83. Singletons were not included in the sequence similarity networks due to having a distance lower than 0.3. HglE-KS, Heterocyst glycolipid synthase-like; NAPAA, non-alpha poly-amino acids (such as e-polylysine).
Actinokineospora sp. PR83 encodes a unique subset of BGCs
Actinokineospora sp. PR83 encodes 18 complete BGCs, including 6 PKSs, 3 NRPSs, 1 terpene, 2 RiPPs and 6 classified as other. Amongst all BGCs, only two showed medium-to-high homology with previously characterized NPs, including PKs similar to streptovaricin, and JBIR-76/JBIR-77. Twelve BGCs that showed 1–44 % identity included hybrids (such as kosinostatin, kalimantacin A and oxalomycin B), PKs (saquayamycin A, chlorothricin, macrotetrolide and dynemicin), non-ribosomal peptides similar to atratumycin and gobichelin A, the saccharides apramycin, methylenomycin A, and cetoniacytone A, classified as other. A total of four BGCs presented no match in the database (Table S4). When we consider the entire genus Actinokineospora , we identified a total of 400 BGCs (23.5 BGCs per genome), of which 17 % were classified as non-ribosomal peptides, 14 % were PK synthases, 13 % were RiPPs, 11 % were terpenes, 8 % hybrid PKS-NRPS, 2 % were saccharides and 35 % were not classified into a principal chemical family (Fig. 4a). In contrast to the genus Lentzea , the number of clusters does not present a linear relationship to genome size (Fig. 4b). The genomes with the highest number of BGCs were A. fastidiosa (36 BGCs, 7.04 Mb, 30 contigs), A. baliensis (36 BGCs, 7.68 Mb, 1 contig), A. auranticolor (30 BGCs, 8.43 Mb, 61 contigs) and A. terrae (30 BGCs, 7.59 Mb, 41 contigs), while the genome with the lowest number of BGCs was A. spheciospongiae (12 BGCs, 7.53 Mb, 263 contigs) (Fig. 4c).
When we compared the biosynthetic space of all of the Actinokineospora strains using a homology network, we found that most of the GCFs were related to NRPS, followed by the category other, including siderophores, ectoines, indole, terpenes and RiPPs. The BGCs from Actinokineospora iranica (isolated from soil, Inche-Broun hypersaline wetland, North Iran) [70] and A. alba IBRC-M (isolated from soil, Xinjiang Province, China) [62] are part of the ‘biosynthetic core genome’ for the genus Actinokineospora . In contrast, A. fastidiosa and A. auranticolor were the strains with the highest number of singletons. When comparing the BGCs to the MIBiG database, we found a match for only nine GCFs (Fig. 5b, Table S4). We assigned 14 out of 18 BGCs from Actinokineospora sp. PR83 to a known GCF, including BGCs similar to oxalomycin B, NRPS, saquayamycin A, indole, atratumycin, dynemicin A, RiPP-like, apramycin, gobichelin A, macrotetrolide and streptovaricin.
Lentzea sp. CC55 and Actinokineospora sp. PR83 show antimicrobial activity against Gram-positive pathogens and cytotoxicity
After studying the in silico biosynthetic potential (number of BGCs) of Lentzea sp. CC55 and Actinokineospora sp. PR83, we used the OSMAC strategy to screen BGCs with antimicrobial or anticancer activity. In order to cover different N and C sources, we analysed extracts from eight different media (ISP-2, ISP-4, R5, Czapek, oatmeal, BS, BG and M1), from both liquid and solid cultures. We correlated the results obtained from the antimicrobial (against B. subtilis , Staphylococcus aureus , E. coli and P. aeruginosa ) and anticancer (NCIH460 cell line) activity assays to their corresponding UHPLC (ultra-HPLC) chromatograms. This approach allowed us to identify potential signals in the chromatograms for use as leads for the purification of molecules responsible for the bioactivity.
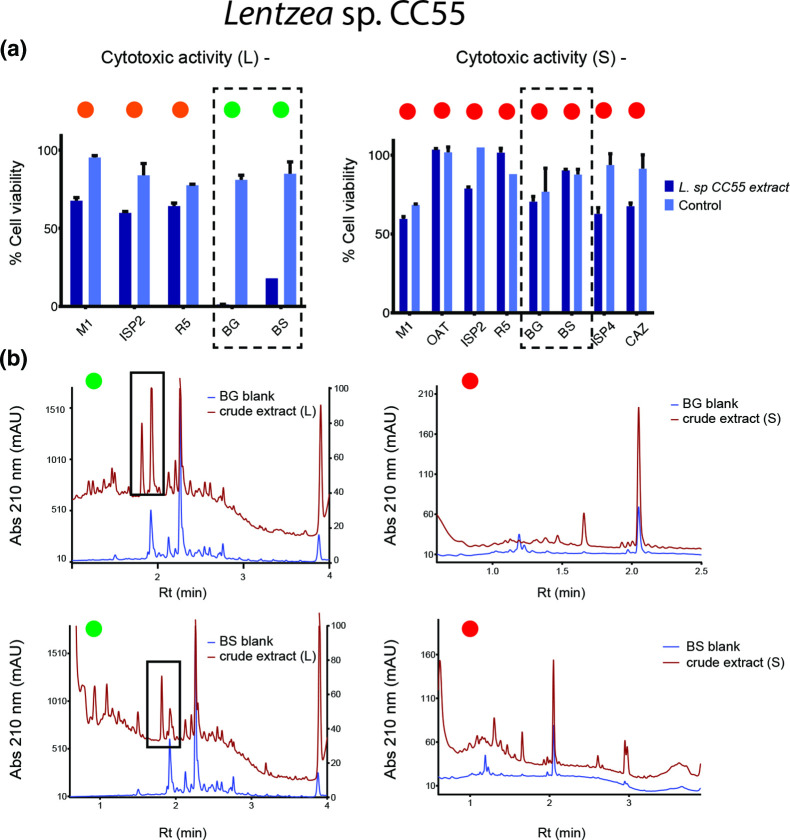
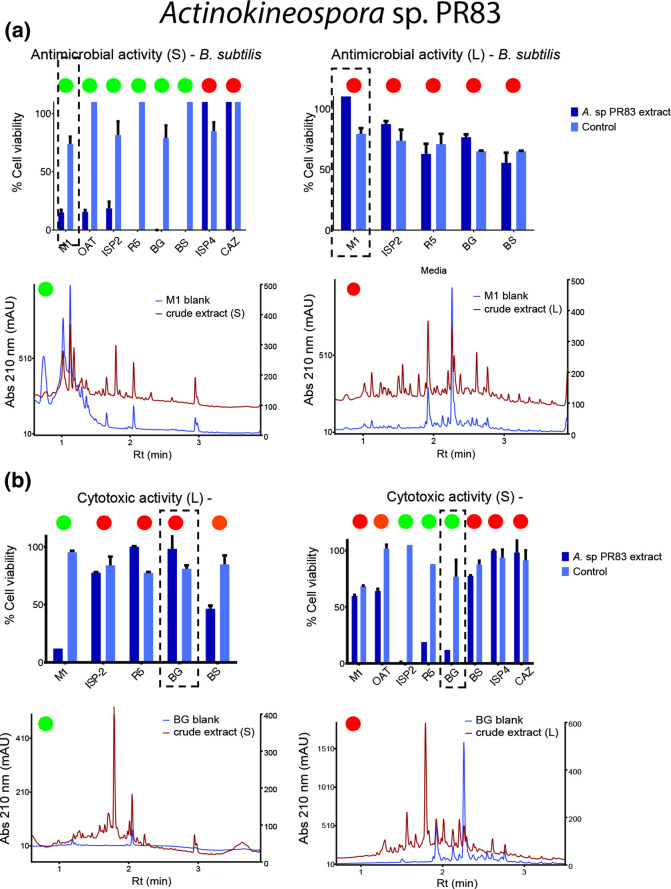
Lentzea sp. CC55 culture extracts from BG and BS media showed cytotoxic activity only in liquid cultures (Fig. 6a). We used the chromatograms obtained from BG and BS to identify a peak (1.8 min retention time) that might be responsible for the observed activity, since such signals are not observed in BG and BS solid media chromatograms (Fig. 6b). Regarding Actinokineospora sp. PR83 extracts, we observed antimicrobial activity against Gram-positive pathogens such as B. subtilis for all the solid media tested (<10 % cell viability). In contrast, the extracts of liquid media did not present a significant antimicrobial effect on pathogen cell viability. Positive chromatograms were correlated with the signals found in negative extracts for peak identification. For example, the bioactivity of extracts from solid M1 medium was related to one peak closest to 0.5 min retention time, while this signal is absent in liquid M1 extracts (Fig. 7a). However, Actinokineospora sp. PR83 extracts also showed a cytotoxic effect on the carcinoma cell line in M1 and BS liquid extracts (M1, <10 % cell viability; BS, <50 % cell viability) and in ISP-2, R5 and BG solid extracts (ISP-2, R5, BG, <10 % cell viability). Given that we observed two signals with more than one significant bioactivity, it is difficult to differentiate chromatography signals for each bioactivity (Fig. 7b).
Fig. 6.
Cytotoxicity assays and chromatography results from liquid and solid culture media of Lentzea sp. CC55. (a) Dashed line boxes highlight the highest cytotoxic effect on NCIH460 cells related to a reduction in cell viability. Null, moderate and high cytotoxic effect can be observed as red, orange and green circles at the top of the bioactivities. (b) Comparison between chromatograms from extracts from liquid BG and BS media that presented cytotoxicity and their counterparts from solid media with no bioactivity. Presence (green) or absence (red) of peaks in chromatograms related to bioactivities are shown at the top with green and red circles, respectively. Rt, retention time. mAU, milli-absorbance unit
Fig. 7.
Bioactivity assays and chromatography results from liquid and solid culture media of Actinokineospora sp. PR83. (a) Antimicrobial activity against B. subtilis . (b) Cytotoxic effect on NCIH460 cells. For a and b, dashed line boxes highlight the highest antimicrobial or cytotoxic effect related to a reduction in cell viability. Null, moderate and high antimicrobial or cytotoxic effect can be observed as red, orange and green circles at the top of the bioactivities, respectively. Presence (green) or absence (red) of peaks in chromatograms related to bioactivities are shown at the top with green and red circles, respectively. Rt, retention time. mAU, milli-absorbance unit.
Discussion
The first milestone of the microbial-based drug discovery process is the establishment of the correlation strain–metabolite–bioactivity. During this process, re-discovery of known metabolites is inevitable, since isolation methods using conventional culture media have been over utilized. A plausible solution to this problem is the development of protocols for the systematic prioritization of strains based on the biosynthetic novelty. For instance, the isolation of uncommon/underisolated actinobacteria, also called rare actinobacteria, has created new opportunities for the discovery of new metabolites [13–15, 17–19]. In this study, we defined the BiNI to provide a quantitative framework for classification of strains according to their biosynthetic novelty. We demonstrated that the BiNI score can be used to compare biosynthetic novelty within a subset of genomes and to draw comparisons between ecological niches and taxonomic groups.
At the level of taxonomic genera, Streptosporangium , Actinokineospora and Saccharothrix showed the highest novelty index, while Thermobifida and Saccharomonospora showed the lowest BiNIs. When comparing strains of the same genus, our results suggest that bacteria inhabiting highly oligotrophic environments (e.g. desert soils) encode a novel subset of BGCs. We observed that for the Actinokineospora and Lentzea genomes tested, the strains with highest BiNI values were isolated from the Cuatro Cienegas Basin and desert soil samples from Egypt ( A. fastidiosa ). In addition, when we compared Streptomyces strains from different environments, including endophytic strains, we observed that environments with low levels of nutrients harbour strains with higher biosynthetic novelty.
Lentzea sp. CC55 and Actinokineospora sp. PR83 were isolated from the Cuatro Cienegas Basin in Mexico. The soils of the Cuatro Cienegas Basin are characterized by a extremely unbalanced N:P (157 : 1) soil stoichiometry, and rich sulfur, magnesium and calcium sulfate concentrations [71]. It has previously been shown that these conditions conserve a great diversity of endemic microbes with novel biosynthetic potential [32, 72]. For instance, our research group has previously described new diversity in desferrioxamine siderophores (propionyl-ferrioxamine, m/z=628.17; aryl-ferrioxamine, m/z=690.26) in species of Microbacterium , Micrococcus and Streptomyces [33].
We performed a pan-genomic analysis using publicly available genomes and found a high number of unique genes for both genera (~17 800 per each genus for Lentzea and Actinokineospora ) compared to other rare actinobacteria isolated from extreme polar environments [73]. We found that novel genes are mostly distributed in specialized metabolite-related genes. The rarefaction curves obtained indicate that more genomes are required in order to cover the entire genetic space within these taxonomic genera. The phylogenetic analysis derived from the pan-genomes found no correlation between the ecological contexts of the Lentzea and Actinokineospora strains.
Genomic analysis using GCFs is useful to classify BGCs according to their functional families and biosynthetic novelty [74, 75]. Initial inspection of the GCFs from our strains showed a high number of singletons, which represent potential novel BGCs. While a direct correlation between genome size and number of BGC is expected [76], we observed a correlation between the taxonomic genus and number of clusters. Expression of the biosynthetic potential of environmental isolates in the laboratory is limited by the capacity to artificially reproduce culture conditions that resemble those of the isolation site. In our study, we evaluated the capacity of our strains to express BGCs with antimicrobial and cytotoxic activity using the OSMAC strategy. We correlated the chromatograms obtained from different media settings (liquid or solid cultures) and media formulations (different C and N sources). The bioactivity test allowed us to identify the most suitable culture conditions for directing purification of the NPs with relevant bioactivity. As expected, we found that solid media favours a higher expression of BGCs responsible for antimicrobial and cytotoxicity activity compared to liquid cultures [77–81]. Solid substrates maintain the membrane skeleton, allowing the hydrophobicity for aerial mycelium growth and, consequently, metabolite production [82]. However, at large-scale, solid media cultures are not feasible for metabolite characterization (chemical composition determination) since the production requires high-yield processes performed in a bioreactor. The OSMAC approach combined with bioactivity assays helped us to correlate chromatography signals to bioactivity. We consider that this method can be used as the primary experimental approach for the chemical characterization of novel compounds. However, it is important to note that this method can only be used with extracts that show a single positive bioactive chromatography signal.
Overall, our study provides a full genomic and phenotypic characterization of two rare actinobacteria from the genera Lentzea and Actinokineospora . Our genomic analysis also provides a metric for quickly prioritizing strains according to the novelty of their BGCs. While the biosynthetic novelty of the strains we analysed from the Cuatro Cienegas Basin was not as high as that of other genera of rare actinobacteria, our strains show high novelty when comparing isolates from other ecological niches. This highlights the potential of exploring highly oligotrophic environments in the search for novel NPs. More importantly, we consider that our study contributes to the development of strategies for the rapid prioritization of strains isolated from different ecological contexts.
Supplementary Data
Funding information
L.A.G.-S. and L.R.-O. received a scholarship from the Mexican National Council of Science and Technology (CONACYT) and a scholarship from Tecnológico de Monterrey, grant no. 20240I-14 (Tec de Monterrey), and UCMEXUS, no. CN-18-10, to C.L.-C. The laboratory of F.B.-G. was supported by CONACYT, Mexico (FINNOVA no. 214716), Laboratorios Liomont and the UK Royal Society via a Newton Advanced Fellowship (NAF\R2\180631). StrainBiotech provided financial support to the Industrial Genomics Laboratory to perform experimental work.
Acknowledgements
We wish to thank the LANGEBIO Sequencing Facility and TECBASE laboratory for providing all of the sequencing services. We thank Dr Pablo Cruz Morales from the Novo Nordisk Center for Biosustainability, Technical University of Denmark, and Dr Susana de Torre Zavala from the Universidad Autónoma de Nuevo León, Mexico, for productive scientific discussions. Finally, we would also like to thank two anonymous reviewers for improving the manuscript through their comments.
Author contributions
L.A.G.-S., F.B.-G. and C.L.-C. designed and performed all of the genomic analysis. M.Q. and R.R.C. performed all the media extract analysis and bioactivity assays. F.B.-G., H.R.-A., V.S.-S. and L.R.-O. performed genomic sequencing and microbial isolations. F.B.-G. and C.L.-C. designed, financed and performed genome sequencing, and discussed the results. L.A.G.-S. and C.L.-C. wrote the paper. All authors have read and critically reviewed the manuscript, and consent to its publication.
Conflicts of interest
The authors declare that there are no conflicts of interest
Footnotes
Abbreviations: BGC, biosynthetic gene cluster; BiNI, biosynthetic novelty index; GCF, gene cluster family; KEGG, Kyoto Encyclopedia of Genes and Genomes; MIBiG, Minimum Information about a Biosynthetic Gene cluster; NP, natural product; NRP, non-ribosomal peptide; OSMAC, one strain many compounds; PK, polyketide; RiPP, ribosomally synthesized and post-translationally modified peptide.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure and four supplementary tables are available with the online version of this article.
References
- 1.Mullard A. $1.3 billion per drug? Nat Rev Drug Discov. 2020;19:226. doi: 10.1038/d41573-020-00043-x. [DOI] [PubMed] [Google Scholar]
- 2.Deore AB, Dhumane JR, Wagh R, Sonawane R. The stages of drug discovery and development process. Asian J Pharm Res Dev. 2019;7:62–67. doi: 10.22270/ajprd.v7i6.616. [DOI] [Google Scholar]
- 3.Marmann A, Aly AH, Lin W, Wang B, Proksch P. Co-cultivation--a powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar Drugs. 2014;12:1043–1065. doi: 10.3390/md12021043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu M, El-Hossary EM, Oelschlaeger TA, Donia MS, Quinn RJ, et al. Potential of marine natural products against drug-resistant bacterial infections. Lancet Infect Dis. 2019;19:e237–e245. doi: 10.1016/S1473-3099(18)30711-4. [DOI] [PubMed] [Google Scholar]
- 5.Ranjani A, Dhanasekaran D, Gopinath PM. In: Actinobacteria – Basics and Biotechnological Applications. Dhanasekaran D, Jiang Y, editors. London: InTech; 2016. An introduction to actinobacteria. [Google Scholar]
- 6.Metsä-Ketelä M, Salo V, Halo L, Hautala A, Hakala J, et al. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol Lett. 1999;180:1–6. doi: 10.1111/j.1574-6968.1999.tb08770.x. [DOI] [PubMed] [Google Scholar]
- 7.Miao V, Brost R, Chapple J, She K, Coëffet-Le Gal M-F, et al. The lipopeptide antibiotic A54145 biosynthetic gene cluster from Streptomyces fradiae . J Ind Microbiol Biotechnol. 2006;33:129–140. doi: 10.1007/s10295-005-0028-5. [DOI] [PubMed] [Google Scholar]
- 8.Sottorff I, Wiese J, Lipfert M, Preußke N, Sönnichsen FD, et al. Different secondary metabolite profiles of phylogenetically almost identical Streptomyces griseus strains originating from geographically remote locations. Microorganisms. 2019;7:166. doi: 10.3390/microorganisms7060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manivasagan P, Kang K-H, Sivakumar K, Li-Chan ECY, Oh H-M, et al. Marine actinobacteria: an important source of bioactive natural products. Environ Toxicol Pharmacol. 2014;38:172–188. doi: 10.1016/j.etap.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Poorinmohammad N, Bagheban-Shemirani R, Hamedi J. Genome mining for ribosomally synthesised and post-translationally modified peptides (RiPPs) reveals undiscovered bioactive potentials of actinobacteria. Antonie Van Leeuwenhoek. 2019;112:1477–1499. doi: 10.1007/s10482-019-01276-6. [DOI] [PubMed] [Google Scholar]
- 11.Ren H, Shi C, Zhao H. Computational tools for discovering and engineering natural product biosynthetic pathways. iScience. 2020;23:100795. doi: 10.1016/j.isci.2019.100795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubert J, Nuzillard J-M, Renault J-H. Dereplication strategies in natural product research: how many tools and methodologies behind the same concept? Phytochem Rev. 2017;16:55–95. doi: 10.1007/s11101-015-9448-7. [DOI] [Google Scholar]
- 13.Subramani R, Aalbersberg W. Culturable rare Actinomycetes: diversity, isolation and marine natural product discovery. Appl Microbiol Biotechnol. 2013;97:9291–9321. doi: 10.1007/s00253-013-5229-7. [DOI] [PubMed] [Google Scholar]
- 14.Ding T, Yang L-J, Zhang W-D, Shen Y-H. The secondary metabolites of rare actinomycetes: chemistry and bioactivity. RSC Adv. 2019;9:21964–21988. doi: 10.1039/c9ra03579f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhakal D, Pokhrel AR, Shrestha B, Sohng JK. Marine rare actinobacteria: isolation, characterization, and strategies for harnessing bioactive compounds. Front Microbiol. 2017;8:1106. doi: 10.3389/fmicb.2017.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lechevalier HA, Lechevalier MP. Biology of actinomycetes. Annu Rev Microbiol. 1967;21:71–100. doi: 10.1146/annurev.mi.21.100167.000443. [DOI] [PubMed] [Google Scholar]
- 17.Ye X, Anjum K, Song T, Wang W, Yu S, et al. A new curvularin glycoside and its cytotoxic and antibacterial analogues from marine actinomycete Pseudonocardia sp. HS7. Nat Prod Res. 2016;30:1156–1161. doi: 10.1080/14786419.2015.1047775. [DOI] [PubMed] [Google Scholar]
- 18.Igarashi Y, Ogura H, Furihata K, Oku N, Indananda C, et al. Maklamicin, an antibacterial polyketide from an endophytic Micromonospora sp. J Nat Prod. 2011;74:670–674. doi: 10.1021/np100727h. [DOI] [PubMed] [Google Scholar]
- 19.Bauermeister A, Calil FA, das C L Pinto F, Medeiros TCT, Almeida LC, et al. Pradimicin-IRD from Amycolatopsis sp. IRD-009 and its antimicrobial and cytotoxic activities. Nat Prod Res. 2019;33:1713–1720. doi: 10.1080/14786419.2018.1434639. [DOI] [PubMed] [Google Scholar]
- 20.Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 21.Tiwari K, Gupta RK. Diversity and isolation of rare actinomycetes: an overview. Crit Rev Microbiol. 2013;39:256–294. doi: 10.3109/1040841X.2012.709819. [DOI] [PubMed] [Google Scholar]
- 22.Subramani R, Sipkema D. Marine rare actinomycetes: a promising source of structurally diverse and unique novel natural products. Mar Drugs. 2019;17:249. doi: 10.3390/md17050249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasol JM. Bacteria inoligotrophic environments: starvation-survival lifestyle – Richard Y. Morita. Int Microbiol. 1998;1:241–242. [Google Scholar]
- 24.Zhang B, Wu X, Tai X, Sun L, Wu M, et al. Variation in actinobacterial community composition and potential function in different soil ecosystems belonging to the arid Heihe River Basin of Northwest China. Front Microbiol. 2019;10:2209. doi: 10.3389/fmicb.2019.02209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adam D, Maciejewska M, Naômé A, Martinet L, Coppieters W, et al. Isolation, characterization, and antibacterial activity of hard-to-culture actinobacteria from cave moonmilk deposits. Antibiotics. 2018;7:28. doi: 10.3390/antibiotics7020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang B-Z, Salam N, Han M-X, Jiao J-Y, Cheng J, et al. Insights on the effects of heat pretreatment, pH, and calcium salts on isolation of rare Actinobacteria from Karstic Caves. Front Microbiol. 2017;8:1535. doi: 10.3389/fmicb.2017.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arocha-Garza HF, Canales-Del Castillo R, Eguiarte LE, Souza V, De la Torre-Zavala S. High diversity and suggested endemicity of culturable Actinobacteria in an extremely oligotrophic desert oasis. PeerJ. 2017;5:e3247. doi: 10.7717/peerj.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebollar EA, Avitia M, Eguiarte LE, González-González A, Mora L, et al. Water-sediment niche differentiation in ancient marine lineages of Exiguobacterium endemic to the Cuatro Cienegas Basin. Environ Microbiol. 2012;14:2323–2333. doi: 10.1111/j.1462-2920.2012.02784.x. [DOI] [PubMed] [Google Scholar]
- 29.Souza V, Siefert JL, Escalante AE, Elser JJ, Eguiarte LE. The Cuatro Ciénegas Basin in Coahuila, Mexico: an astrobiological Precambrian Park. Astrobiology. 2012;12:641–647. doi: 10.1089/ast.2011.0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elser JJ, Schampel JH, Garcia-Pichel F, Wade BD, Souza V, et al. Effects of phosphorus enrichment and grazing snails on modern stromatolitic microbial communities. Freshwater Biol. 2005;50:1808–1825. doi: 10.1111/j.1365-2427.2005.01451.x. [DOI] [Google Scholar]
- 31.Nitti A, Daniels CA, Siefert J, Souza V, Hollander D, et al. Spatially resolved genomic, stable isotopic, and lipid analyses of a modern freshwater microbialite from Cuatro Ciénegas, Mexico. Astrobiology. 2012;12:685–698. doi: 10.1089/ast.2011.0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallegos-Lopez S, Mejia-Ponce PM, Gonzalez-Salazar LA, Rodriguez-Orduña L, Souza-Saldivar V, et al. Draft genome sequence of Streptomyces sp. strain C8S0, isolated from a highly oligotrophic sediment. Microbiol Resour Announc. 2020;9:e01441-19. doi: 10.1128/MRA.01441-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cruz-Morales P, Ramos-Aboites HE, Licona-Cassani C, Selem-Mójica N, Mejía-Ponce PM, et al. Actinobacteria phylogenomics, selective isolation from an iron oligotrophic environment and siderophore functional characterization, unveil new desferrioxamine traits. FEMS Microbiol Ecol. 2017;93:fix086. doi: 10.1093/femsec/fix086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos-Aboites H, Yáñez-Olvera A, Barona-Gómez F. In: Ecosystem Ecology and Geochemistry of Cuatro Cienegas. García-Oliva F, Elser J, Souza V, editors. Cham: Springer; 2018. Bacterial siderophore-mediated iron acquisition in Cuatro Cienegas Basin: a complex community interplay made simpler in the light of evolutionary genomics. [Google Scholar]
- 35.Hobbs G, Frazer CM, Gardner DCJ, Cullum JA, Oliver SG. Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol. 1989;31:272–277. doi: 10.1007/BF00258408. [DOI] [Google Scholar]
- 36.International Streptomyces Project-2 Medium (ISP-2), ActinoBase. http://actinobase.org/index.php/ISP2 n.d.
- 37.ISP4 Medium, ActinoBase. [ October 31; 2021 ]. http://actinobase.org/index.php/ISP4 n.d. accessed.
- 38.R5, ActinoBase. [ November 1; 2021 ]. http://actinobase.org/index.php/R5 n.d. accessed.
- 39.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaudhari NM, Gupta VK, Dutta C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci Rep. 2016;6:24373. doi: 10.1038/srep24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kück P, Meusemann K. FASconCAT: convenient handling of data matrices. Mol Phylogenet Evol. 2010;56:1115–1118. doi: 10.1016/j.ympev.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49:W29–W35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020;48:D454–D458. doi: 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarro-Muñoz JC, Selem-Mojica N, Mullowney MW, Kautsar SA, Tryon JH, et al. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. 2020;16:60–68. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kautsar SA, Blin K, Shaw S, Weber T, Medema MH. BiG-FAM: the biosynthetic gene cluster families database. Nucleic Acids Res. 2021;49:D490–D497. doi: 10.1093/nar/gkaa812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jorgensen JH, Ferraro MJ. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis. 2009;49:1749–1755. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 51.Kumar P, Nagarajan A, Uchil PD. Analysis of cell viability by the MTT assay. Cold Spring Harb Protoc. 2018;2018:prot095505. doi: 10.1101/pdb.prot095505. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez-R LM, Gunturu S, Harvey WT, Rosselló-Mora R, Tiedje JM, et al. The microbial genomes atlas (MiGA) webserver: taxonomic and gene diversity analysis of archaea and bacteria at the whole genome level. Nucleic Acids Res. 2018;46:W282–W288. doi: 10.1093/nar/gky467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Zhang L, Ding Y, Gao Y, Ruan J, et al. Lentzea jiangxiensis sp. nov., isolated from acidic soil. Int J Syst Evol Microbiol. 2012;62:2342–2346. doi: 10.1099/ijs.0.033795-0. [DOI] [PubMed] [Google Scholar]
- 54.Kämpfer P, Glaeser SP, Busse H-J, Abdelmohsen UR, Ahmed S, et al. Actinokineospora spheciospongiae sp. nov., isolated from the marine sponge Spheciospongia vagabunda . Int J Syst Evol Microbiol. 2015;65:879–884. doi: 10.1099/ijs.0.000031. [DOI] [PubMed] [Google Scholar]
- 55.Nouioui I, Carro L, García-López M, Meier-Kolthoff JP, Woyke T, et al. Genome-based taxonomic classification of the phylum Actinobacteria . Front Microbiol. 2018;9:02007. doi: 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Camas M, Veyisoglu A, Tatar D, Saygin H, Cetin D, et al. Lechevalieria nigeriaca sp. nov., isolated from arid soil. Int J Syst Evol Microbiol. 2013;63:3750–3754. doi: 10.1099/ijs.0.052266-0. [DOI] [PubMed] [Google Scholar]
- 57.Labeda DP, Price NP, Tan GYA, Goodfellow M, Klenk HP. Emended description of the genus Actinokineospora Hasegawa 1988 and transfer of Amycolatopsis fastidiosa Henssen et al. 1987 as Actinokineospora fastidiosa comb. nov. Int J Syst Evol Microbiol. 2010;60:1444–1449. doi: 10.1099/ijs.0.016568-0. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Shi J, Liu T, Zhang Y, Zhang L, et al. Actinokineospora xionganensis sp. nov., a filamentous actinomycete isolated from the lakeside soil of Baiyangdian. Antonie van Leeuwenhoek. 2021;114:487–496. doi: 10.1007/s10482-021-01532-8. [DOI] [PubMed] [Google Scholar]
- 59.Lei Y-J, Xia Z-F, Luo X-X, Zhang L-L. Actinokineospora pegani sp. nov., an endophytic actinomycete isolated from the surface-sterilized root of Peganum harmala L. Int J Syst Evol Microbiol. 2020;70:4358–4363. doi: 10.1099/ijsem.0.004299. [DOI] [PubMed] [Google Scholar]
- 60.Aouiche A, Bouras N, Mokrane S, Zitouni A, Schumann P, et al. Actinokineospora mzabensis sp. nov., a novel actinomycete isolated from Saharan soil. Antonie van Leeuwenhoek. 2015;107:291–296. doi: 10.1007/s10482-014-0328-8. [DOI] [PubMed] [Google Scholar]
- 61.Ping M, Yun-Lin Z, Jun L, Jian G, Zheng-Gang X. Proposal of Lentzea deserti (Okoro et al. 2010) Nouioui et al. 2018 as a later heterotypic synonym of Lentzea atacamensis (Okoro et al. 2010) Nouioui et al. 2018 and an emended description of Lentzea atacamensis . PLoS One. 2021;16:e0246533. doi: 10.1371/journal.pone.0246533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan L-J, Zhang Y-Q, Yu L-Y, Liu H-Y, Guan Y, et al. Alloactinosynnema album gen. nov., sp. nov., a member of the family actinosynnemataceae isolated from soil. Int J Syst Evol Microbiol. 2010;60:39–43. doi: 10.1099/ijs.0.010744-0. [DOI] [PubMed] [Google Scholar]
- 63.Intra B, Greule A, Bechthold A, Euanorasetr J, Paululat T, et al. Thailandins A and B, new polyene macrolactone compounds isolated from Actinokineospora bangkokensis strain 44EHW(T), possessing antifungal activity against anthracnose fungi and pathogenic yeasts. J Agric Food Chem. 2016;64:5171–5179. doi: 10.1021/acs.jafc.6b01119. [DOI] [PubMed] [Google Scholar]
- 64. Actinokineospora enzanensis, NamesforLife. [ April 3; 2022 ]. https://www.namesforlife.com/10.1601/nm.6783 n.d. accessed.
- 65.Otoguro M, Hayakawa M, Yamazaki T, Tamura T, Hatano K, et al. Numerical phenetic and phylogenetic analyses of Actinokineospora isolates, with a description of Actinokineospora auranticolor sp. nov. and Actinokineospora enzanensis sp. nov. Actinomycetologica. 2001;15:30–39. doi: 10.3209/saj.15_30. [DOI] [Google Scholar]
- 66.Tamura T, Hayakawa M, Nonomura H, Yokota A, Hatano K. Four new species of the genus Actinokineospora: Actinokineospora inagensis sp. nov., Actinokineospora globicatena sp. nov., Actinokineospora terrae sp. nov., and Actinokineospora diospyrosa sp. nov. Int J Syst Bacteriol. 1995;45:371–378. doi: 10.1099/00207713-45-2-371. [DOI] [Google Scholar]
- 67.Xie Q, Wang Y, Huang Y, Wu Y, Ba F, et al. Description of Lentzea flaviverrucosa sp. nov. and transfer of the type strain of Saccharothrix aerocolonigenes subsp. staurosporea to Lentzea albida . Int J Syst Evol Microbiol. 2002;52:1815–1820. doi: 10.1099/00207713-52-5-1815. [DOI] [PubMed] [Google Scholar]
- 68.Huang J, Huang Y. Lentzea tibetensis sp. nov., a novel Actinobacterium with antimicrobial activity isolated from soil of the Qinghai-Tibet Plateau. Int J Syst Evol Microbiol. 2021;71:004976. doi: 10.1099/ijsem.0.004976. [DOI] [PubMed] [Google Scholar]
- 69.Cao C-L, Zhou X-Q, Qin S, Tao F-X, Jiang J-H, et al. Lentzea guizhouensis sp. nov., a novel lithophilous actinobacterium isolated from limestone from the Karst area, Guizhou, China. Antonie Van Leeuwenhoek. 2015;108:1365–1372. doi: 10.1007/s10482-015-0589-x. [DOI] [PubMed] [Google Scholar]
- 70.Nikou MM, Ramezani M, Amoozegar MA, Fazeli SAS, Schumann P, et al. Alloactinosynnema iranicum sp. nov., a rare actinomycete isolated from a hypersaline wetland, and emended description of the genus Alloactinosynnema . Int J Syst Evol Microbiol. 2014;64:1173–1179. doi: 10.1099/ijs.0.049189-0. [DOI] [PubMed] [Google Scholar]
- 71.Souza V, Moreno-Letelier A, Travisano M, Alcaraz LD, Olmedo G, et al. The lost world of Cuatro Ciénegas Basin, a relictual bacterial niche in a desert oasis. Elife. 2018;7:e38278. doi: 10.7554/eLife.38278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elser JJ, Watts J, Schampel JH, Farmer J. Early Cambrian food webs on a trophic knife-edge? A hypothesis and preliminary data from a modern stromatolite-based ecosystem. Ecol Lett. 2006;9:295–303. doi: 10.1111/j.1461-0248.2005.00873.x. [DOI] [PubMed] [Google Scholar]
- 73.Dai D, Lu H, Xing P, Wu Q. Comparative genomic analyses of the genus Nesterenkonia unravels the genomic adaptation to polar extreme environments. Microorganisms. 2022;10:233. doi: 10.3390/microorganisms10020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kautsar SA, van der Hooft JJJ, de Ridder D, Medema MH. BiG-SLiCE: a highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters. Gigascience. 2021;10:giaa154. doi: 10.1093/gigascience/giaa154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gavriilidou A, Kautsar SA, Zaburannyi N, Krug D, Müller R, et al. Compendium of specialized metabolite biosynthetic diversity encoded in bacterial genomes. Nat Microbiol. 2022;7:726–735. doi: 10.1038/s41564-022-01110-2. [DOI] [PubMed] [Google Scholar]
- 76.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Webb C, Manan MA. Design aspects of solid state fermentation as applied to microbial bioprocessing. J Appl Biotechnol Bioeng. 2017;4:511–532. doi: 10.15406/jabb.2017.04.00094. [DOI] [Google Scholar]
- 78.Rajaram SK, Ahmad P, Sujani Sathya Keerthana S, Jeya Cressida P, Ganesh Moorthy I, et al. Extraction and purification of an antimicrobial bioactive element from lichen associated Streptomyces olivaceus LEP7 against wound inhabiting microbial pathogens. J King Saud Univ Sci. 2020;32:2009–2015. doi: 10.1016/j.jksus.2020.01.039. [DOI] [Google Scholar]
- 79.Wichner D, Idris H, Houssen WE, McEwan AR, Bull AT, et al. Isolation and anti-HIV-1 integrase activity of lentzeosides A-F from extremotolerant Lentzea sp. H45, a strain isolated from a high-altitude Atacama Desert soil. J Antibiot. 2017;70:448–453. doi: 10.1038/ja.2016.78. [DOI] [PubMed] [Google Scholar]
- 80.Hussain A, Rather MA, Dar MS, Aga MA, Ahmad N, et al. Novel bioactive molecules from Lentzea violacea strain AS 08 using one strain-many compounds (OSMAC) approach. Bioorg Med Chem Lett. 2017;27:2579–2582. doi: 10.1016/j.bmcl.2017.03.075. [DOI] [PubMed] [Google Scholar]
- 81.Tawfike A, Attia EZ, Desoukey SY, Hajjar D, Makki AA, et al. New bioactive metabolites from the elicited marine sponge-derived bacterium Actinokineospora spheciospongiae sp. nov. AMB Express. 2019;9:12. doi: 10.1186/s13568-018-0730-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Niladevi KN, Sukumaran RK, Prema P. Utilization of rice straw for laccase production by Streptomyces psammoticus in solid-state fermentation. J Ind Microbiol Biotechnol. 2007;34:665–674. doi: 10.1007/s10295-007-0239-z. [DOI] [PubMed] [Google Scholar]
- 83.Labeda DP, Hatano K, Kroppenstedt RM, Tamura T. Revival of the genus Lentzea and proposal for Lechevalieria gen. nov. Int J Syst Evol Microbiol. 2001;51:1045–1050. doi: 10.1099/00207713-51-3-1045. [DOI] [PubMed] [Google Scholar]
- 84.Sun X, Zhao J, Luo X, Hou W, Xiang W, et al. Lentzea alba sp. nov., a novel actinobacterium isolated from soil. Int J Syst Evol Microbiol. 2021;71 doi: 10.1099/ijsem.0.004661. [DOI] [PubMed] [Google Scholar]
- 85.Lee SD, Kim ES, Roe JH, Kim J, Kang SO, et al. Saccharothrix violacea sp. nov., isolated from a gold mine cave, and Saccharothrix albidocapillata comb. nov. Int J Syst Evol Microbiol. 2000;50 Pt 3:1315–1323. doi: 10.1099/00207713-50-3-1315. [DOI] [PubMed] [Google Scholar]
- 86.Yan XC, Deng YX. Streptomyces flavoverrucosus sp. nov., a novel actinomycete from soil. Acta Microbiol Sin. 1966;12:207–216. [Google Scholar]
- 87.Yassin AF, Rainey FA, Brzezinka H, Jahnke KD, Weissbrodt H, et al. Lentzea gen. nov., a new genus of the order Actinomycetales . Int J Syst Bacteriol. 1995;45:357–363. doi: 10.1099/00207713-45-2-357. [DOI] [PubMed] [Google Scholar]
- 88.Li D, Jiang H, Han L, Li Y, Zhao J, et al. Lentzea terrae sp. nov., isolated from soil and an emended description of Lentzea soli . Int J Syst Evol Microbiol. 2018;68:3528–3533. doi: 10.1099/ijsem.0.003024. [DOI] [PubMed] [Google Scholar]
- 89.Cao C, Yuan B, Qin S, Jiang J, Tao F, et al. Lentzea pudingi sp. nov., isolated from a weathered limestone sample in a karst area. Int J Syst Evol Microbiol. 2017;67:4873–4878. doi: 10.1099/ijsem.0.002400. [DOI] [PubMed] [Google Scholar]
- 90.Labeda DP, Donahue JM, Sells SF, Kroppenstedt RM. Lentzea kentuckyensis sp. nov., of equine origin. Int J Syst Evol Microbiol. 2007;57:1780–1783. doi: 10.1099/ijs.0.64245-0. [DOI] [PubMed] [Google Scholar]
- 91.Okoro CK, Bull AT, Mutreja A, Rong X, Huang Y, et al. Lechevalieria atacamensis sp. nov., Lechevalieria deserti sp. nov. and Lechevalieria roselyniae sp. nov., isolated from hyperarid soils. Int J Syst Evol Microbiol. 2010;60:296–300. doi: 10.1099/ijs.0.009985-0. [DOI] [PubMed] [Google Scholar]
- 92.Zhang J, Xie Q, Liu Z, Goodfellow M. Lechevalieria fradiae sp. nov., a novel actinomycete isolated from soil in China. Int J Syst Evol Microbiol. 2007;57:832–836. doi: 10.1099/ijs.0.64777-0. [DOI] [PubMed] [Google Scholar]
- 93.Wang W, Zhang Z, Tang Q, Mao J, Wei D, et al. Lechevalieria xinjiangensis sp. nov., a novel actinomycete isolated from radiation-polluted soil in China. Int J Syst Evol Microbiol. 2007;57:2819–2822. doi: 10.1099/ijs.0.65134-0. [DOI] [PubMed] [Google Scholar]
- 94.Narayana KJP, Rao V, Vijayalakshmi M. Studies on bioactive metabolites produced by lechevalieria flava. Res J Microbiol. 2007;2:871–875. [Google Scholar]
- 95.Labeda DP. Transfer of “Nocardia aerocolonigenes” (Shinobu and Kawato 1960) Pridham 1970 into the Genus Saccharothrix Labeda, Testa, Lechevalier, and Lechevalier 1984 as Saccharothrix aerocolonigenes sp. nov. Int J Syst Bacteriol. 1986;36:109–110. doi: 10.1099/00207713-36-1-109. [DOI] [Google Scholar]
- 96.Intra B, Matsumoto A, Inahashi Y, Ōmura S, Takahashi Y, et al. Actinokineospora bangkokensis sp. nov., isolated from rhizospheric soil. Int J Syst Evol Microbiol. 2013;63:2655–2660. doi: 10.1099/ijs.0.047928-0. [DOI] [PubMed] [Google Scholar]
- 97.Lisdiyanti P, Otoguro M, Ratnakomala S, Lestari Y, Hastuti RD, et al. Actinokineospora baliensis sp. nov., Actinokineospora cibodasensis sp. nov. and Actinokineospora cianjurensis sp. nov., isolated from soil and plant litter. Int J Syst Evol Microbiol. 2010;60:2331–2335. doi: 10.1099/ijs.0.013276-0. [DOI] [PubMed] [Google Scholar]
- 98.Henssen A, Kothe HW, Kroppenstedt RM. Transfer of Pseudonocardia azurea and “Pseudonocardia fastidiosa” to the genus Amycolatopsis, with emended species description. Int J Syst Bacteriol. 1987;37:292–295. doi: 10.1099/00207713-37-3-292. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.