Abstract
Neurologic disability level at hospital discharge is an important outcome in many clinical research studies. Outside of clinical trials, neurologic outcomes must typically be extracted by labor intensive manual review of clinical notes in the electronic health record (EHR). To overcome this challenge, we set out to develop a natural language processing (NLP) approach that automatically reads clinical notes to determine neurologic outcomes, to make it possible to conduct larger scale neurologic outcomes studies. We obtained 7314 notes from 3632 patients hospitalized at two large Boston hospitals between January 2012 and June 2020, including discharge summaries (3485), occupational therapy (1472) and physical therapy (2357) notes. Fourteen clinical experts reviewed notes to assign scores on the Glasgow Outcome Scale (GOS) with 4 classes, namely ‘good recovery’, ‘moderate disability’, ‘severe disability’, and ‘death’ and on the Modified Rankin Scale (mRS), with 7 classes, namely ‘no symptoms’, ‘no significant disability’, ‘slight disability’, ‘moderate disability’, ‘moderately severe disability’, ‘severe disability’, and ‘death’. For 428 patients’ notes, 2 experts scored the cases generating interrater reliability estimates for GOS and mRS. After preprocessing and extracting features from the notes, we trained a multiclass logistic regression model using LASSO regularization and 5-fold cross validation for hyperparameter tuning. The model performed well on the test set, achieving a micro average area under the receiver operating characteristic and F-score of 0.94 (95% CI 0.93–0.95) and 0.77 (0.75–0.80) for GOS, and 0.90 (0.89–0.91) and 0.59 (0.57–0.62) for mRS, respectively. Our work demonstrates that an NLP algorithm can accurately assign neurologic outcomes based on free text clinical notes. This algorithm increases the scale of research on neurological outcomes that is possible with EHR data.
Keywords: Intensive care unit, Coronavirus, Glasgow outcome scale, Modified Rankin Scale, Natural language processing, Machine learning
1. Introduction
Neurologic disability level is an important outcome in many electronic health records (EHR)-based research studies. While extracting structured EHR data is easily automated, information about neurologic outcomes is typically obtained by manual review of semi-structured or unstructured clinical notes written by physicians, physical therapists, occupational therapists, and other healthcare workers. However, chart review is labor intensive, limiting the scope of most EHR based neurologic outcome studies.
Medical natural language processing (NLP) research aims to develop automated approaches to EHR information extraction. NLP applications in medical research have been growing rapidly (Locke et al., 2021; Sheikhalishahi et al., 2019; Yuvaraj & Ahamed, 2021). Applications to date using EHR data include detection of adverse medical events (Chu, Dong, He, Duan, & Huang, 2018), detection of adverse drug reaction (Santiso, Pérez, & Casillas, 2019), drug safety surveillance (Choi, Schuetz, Stewart, & Sun, 2017; Munkhdalai, Liu, & Yu, 2018), detection of colorectal cancer (Wang, Nguyen, Islam, Li, & Yang, 2019), information extraction from cancer pathology reports (Alawad et al., 2019; Qiu et al., 2019; Yoon et al., 2019; H. Yang, 2010; Gao et al., 2018), extraction of medical problems for disease management (Kim & Meystre, 2019), ICD-9 code assignment (Bai & Vucetic, 2019; Huang, Osorio, & Sy, 2019; M. Li et al., 2019a), early prediction of acute kidney injury in critical care setting (Sun et al., 2019), prediction of postoperative hospital stay based on operative reports in neurosurgery (Danilov et al., 2019) and early prediction of diagnostic-related groups and estimation of hospital costs (J. Liu, Capurro, Nguyen, & Verspoor, 2021).
Among some of the tasks possible with NLP and machine learning, are clinical entity recognition (Z. Liu et al., 2017; Richter-Pechanski, Amr, Katus, & Dieterich, 2019; Shi et al., 2019; J. Yang, Liu, Qian, Guan, & Yuan, 2019; Zhang, Zhang, Zhou, & Pang, 2019) and clinical entity relation extraction (Chen et al., 2018; Hu et al., 2018; Z. Li et al., 2019b; Munkhdalai et al., 2018; Shi et al., 2019), temporal relation (Choi et al., 2017), temporal matching (Lüneburg et al., 2019), semantic representation (Deng, Faulstich, & Denecke, 2017), de-identification (Lee, Filannino, & Uzuner, 2019; Obeid et al., 2019; Richter-Pechanski et al., 2019), medical question-answering (Ben Abacha & Demner-Fushman, 2019; Hu et al., 2018), and dealing with text ambiguity, such as abbreviation disambiguation (Joopudi, Dandala, & Devarakonda, 2018), prediction of ambiguous terms (Pesaranghader, Matwin, Sokolova, & Pesaranghader, 2019) and disambiguation methods (Wei, Lee, Leaman, & Lu, 2019; Weissenbacher et al., 2019).
While NLP has the advantage of being substantially faster than human chart review of medical records (Buchan et al., 2011; Nadkarni, Ohno-Machado, & Chapman, 2011; Uzuner, South, Shen, & DuVall, 2011; Wilbur, Rzhetsky, & Shatkay, 2006), extraction of neurologic outcomes from medical notes using NLP remains an unsolved problem.
Herein we describe how we developed an NLP approach to automatically extract neurological outcomes from hospital discharge summaries, physical therapy, and occupational therapy notes. Multiclass logistic regression models are developed with the one-vs-rest scheme for multi classification, using LASSO regularization for dimensionality reduction. Our models assign neurologic outcomes on two widely used scales: Glasgow Coma Scale (GOS) (Jennett & Bond, 1975) and the modified Rankin Scale (mRS) (Wilson et al., 2002). The models are developed for classification of GOS with four classes, namely good recovery, moderate disability, severe disability, and death and for mRS with seven classes, namely no symptoms, no significant disability, slight disability, moderate disability, moderately severe disability, severe disability, and death. Since the classes are imbalanced for both scales, the models are developed with a balanced class weight. We demonstrate that the model performs with acceptable accuracy, showing that our NLP algorithm is a useful tool for large-scale EHR-based research on neurologic outcomes.
2. Related work
NLP has been increasingly used in the healthcare domain to extract meaningful structured information from notes. In a previous study (Fernandes et al., 2021), we developed an NLP model to classify discharge dispositions of hospitalized patients with COVID-19 from discharge summary notes. Other studies have applied NLP to hospital discharge summaries to identify critical illness (Marafino et al., 2018; Weissman et al., 2016), detect adverse events (Murff et al., 2003), or other potential medical problems (Meystre & Haug, 2006), to extract medication information (Alfattni, Belousov, Peek, & Nenadic, 2021; H. Yang, 2010), to predict risk of rehospitalization (Kang & Hurdle, 2020), to predict risk of suicide attempts (Buckland, Hogan, & Chen, 2020) and to risk stratify patients (Lehman, Saeed, Long, Lee, & Mark, 2012). NLP has also been used to capture mobility information from physical therapy notes (Newman-Griffis & Fosler-Lussier, 2021; Thieu et al., 2021). Several data mining techniques have also been applied in the field of NLP, namely bag of words (BOW) to count individual words (or phrases) that occurred within documents (Agarwala, Anagawadi, & Reddy Guddeti, 2021; Clapp et al., 2022; Kang & Hurdle, 2020; Parvin & Hoque, 2021; Selby, Narain, Russo, Strong, & Stetson, 2018; Sterling, Patzer, Di, & Schrager, 2019; Uyeda et al., 2022), term frequency-inverse document frequency (TF-IDF) to quantify the importance of string representations (words, phrases, lemmas) in a document amongst a collection of documents (Agarwala et al., 2021; Chen et al., 2020; Gordon et al., 2022; Liu, Wan, & Su, 2019; Zhan, Humbert-Droz, Mukherjee, & Gevaert, 2021), including Word2vec (Agarwala et al., 2021; Gordon et al., 2022; Liu et al., 2019). A wide range of machine learning models have been designed to generate predictions, namely regularized regression models (Ju, Chen, Rosenberger, & Liu, 2021; Kang & Hurdle, 2020; Parvin & Hoque, 2021; Uyeda et al., 2022; Zhan et al., 2021), including LASSO regression (Clapp et al., 2022; De Silva et al., 2021), neural network regression models (Sterling et al., 2019), three-layer neural networks (Kang & Hurdle, 2020), multinomial Naïve Bayes, support vector machines (Kang & Hurdle, 2020; Liu et al., 2019; Parvin & Hoque, 2021), random forests, K-nearest neighbors (Kang & Hurdle, 2020; Parvin & Hoque, 2021), adaptive boosting (Parvin & Hoque, 2021), and extreme-gradient boosting (Gordon et al., 2022). To the best of our knowledge, the present work is the first to develop an NLP model to classify GOS and mRS based on clinical notes, thus we are not able to compare performance of our model with existing benchmarks. The code to reproduce our results will be made publicly available at the time of publication so that future researchers interested in this topic can reproduce the results and benchmark against our model.
3. Methods
3.1. Study design
This study is reported in accordance with the STrengthening the Reporting of OBservational studies in Epidemiology (STROBE) statement (Vandenbroucke et al., 2007). Data was extracted from the hospital electronic medical record under a research protocol approved by the Mass General Brigham Institutional Review Board; a waiver of informed consent was obtained. Clinical data were retrospectively analyzed for a diverse cohort of 3632 consecutive adult patients (≥18 years old) admitted to two major Boston hospitals. The cohort included 1779 patients discharged from the intensive care unit (ICU) at Massachusetts General Hospital (MGH) between January 3rd 2012 to November 3rd 2017, and 1853 patients who were discharged from MGH (n = 1273) or Brigham and Women’s Hospital (BWH, n = 580) and positive for SARS-CoV-2 infection between March 10th 2020 to June 30th 2020. Both hospitals use the EPIC EHR software system.
EHR data comprised discharge summaries, occupational therapy notes, and physical therapy notes, which consist of semi-structured free text written by physicians, physical therapists and occupational therapists. Patients included in the study had at least one of the above types of clinical notes. Patients without an assigned label due to lack of information in the notes were excluded (4 patients). To avoid double counting, patients who were admitted at different times, first in the ICU and later with COVID-19 infection were removed from the ICU cohort (6 patients). We also excluded patients assigned to the rare GOS label “persistent vegetative state” (1.4% = 52/3632 patients), for GOS classification.
3.2. Neurological outcomes
The ground truth neurological outcome for each patient was assigned by a team of fourteen physician experts who manually read each patient’s notes independently. For the cohort of patients admitted with COVID-19, the years of clinical experience of experts was two years (SIC, AAB), three and a half years (RT), six years (NV, HSA), nine years (SSM) and ten years (SAQ) and for the cohort of patients admitted to the ICU there were seven experts each with at least two years of clinical experience, where each case was independently reviewed by two experts, with any discrepancies reviewed by a third senior reviewer (Zafar et al., 2021). A second round of labels’ assignment was performed by the experts for a subset of 428 patients, to assess interrater reliability. For cases where experts were not able to generate a score (4 patients), either due to absence of notes, or severe lack of information in the patient’s record, patients were excluded from the study.
Two neurological outcome scales were utilized in this study: a modified version of the Glasgow Outcome Scale (GOS), and the Modified Rankin Scale (mRS). GOS is composed of five levels: good recovery (GOS 5), moderate disability (GOS 4), severe disability (GOS 3), persistent vegetative state (GOS 2), and death (GOS 1). We omitted GOS 2 from our analysis because this outcome was rare in our cohort. mRS is composed of seven levels: no symptoms (mRS 0), no significant disability (mRS 1), slight disability (mRS 2), moderate disability (mRS 3), moderately severe disability (mRS 4), severe disability (mRS 5), and death (mRS 6).
3.3. Interrater reliability
Pairwise interrater reliability (IRR) was assessed for a subset of patients who had a second round of label assignment. We used 100 iterations of bootstrap random sampling with replacement to calculate 95% CI for the agreement estimates between experts. The IRR was measured as percent agreement among the experts.
3.4. Data processing
All notes were extracted for the period between each patient admission up until three days after hospital discharge, to allow for cases where notes are recorded in the system only after discharge. For each patient, we selected the discharge summary and the physical therapy and occupational therapy notes with the corresponding date closest to the discharge date. These notes were then merged into one for analysis. We further address in this section the different subtypes of reports we may find for both physical and occupational therapy notes.
Discharge summaries and physical and occupational therapy notes at MGH and BWH are semi-structured, with a series of named fields containing specific types of mostly free text information. We present an example of each type of note with protected health information removed in Supplementary Table A1. The following subtypes were present in the data for both physical and occupational therapy reports: consultation, progress report, initial evaluation, re-evaluation, treatment note, service and amendments. For occupational therapy, the following additional subtypes of reports were present: discharge report, deferral note, daily treatment note, daily progress note, weekly progress update, progress update, assessment, screening assessment, screening evaluation and brief positioning evaluation. While narratives are often similar among these note subtypes, note structures vary. Thus, the methodology for preprocessing physical and occupational therapy reports differed from discharge summaries and the strategy is depicted in Fig. 1. The figure is essentially composed of two methodological parts. The upper part provides an overview of the notes preprocessing steps. The lower part of the figure shows the final stage of processing for the merged notes before modeling. The modeling steps are depicted in the lower right corner of the figure and described further in Section 3.5.
Fig. 1.
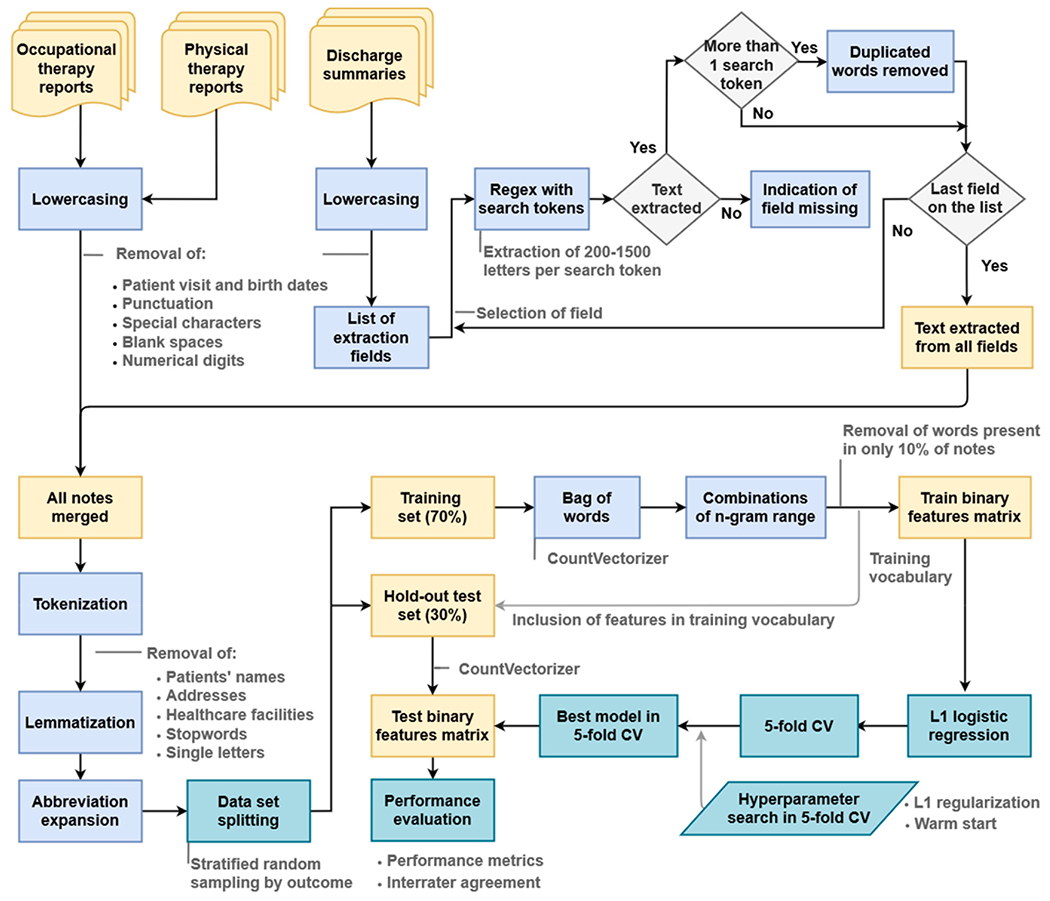
Methodology for notes preprocessing and modeling. The list of extraction fields for discharge summary processing using regular expressions (regex) is shown in Supplementary Table A.2.
Notes were subjected to lowercasing, followed by removal of visit dates, birth dates punctuation, special characters, blank spaces, and numerical digits. For discharge summaries, we generated reduced versions by applying additional preprocessing to extract the meaningful information from these long narratives, as performed in a previous study (Fernandes et al., 2021). The reduced version of the discharge summaries was then merged with the occupational and physical therapy report notes for each patient.
The merged notes were next tokenized, which enabled removal of patients’ names, addresses, healthcare facilities and hospital unit names, and single letters, leaving only words. Stopwords, which consist of frequent and less relevant words (listed in Supplementary Table A3) were removed. Notes were then lemmatized, i.e. different forms of the same word were reduced to a common root (“lemma”), using WordNetLemmatizer from the NLTK library in Python with a POS tag specified as verb. Finally, abbreviation expansion and spell correction were applied for a small list of frequently used clinical words, presented in Supplementary Table A4.
Preprocessed merged notes were divided into train and test sets and notes in the training set were used to create the training vocabulary. A BoW model was used to represent each patient’s notes as a binary vector, indicating the presence of a given n-gram (single word or sequence of 2 or 3 words), disregarding grammar and word order, using the function CountVectorizer from Python.
Finally, dimensionality reduction was performed by considering only words present in at least 10% of notes in the training set, and by using multi-class logistic regression with the least absolute shrinkage and selection operator (LASSO) (Tibshirani, 1996) to sparsify the model.
The same procedure was applied to convert notes from the test set into feature vectors. Note that the feature extraction procedure was based entirely on the training data set.
3.5. Model development
A multinomial logistic regression model with the one-vs-rest scheme for multi classification was trained for each neurological outcome. A binary problem was fitted for the four classes in GOS and seven classes in mRS, with a balanced class weight. The one-vs-rest logistic regression model estimator depicted in Eq. (1) used LASSO regularization for dimensionality reduction, with the objective function indicated in Eq. (2). corresponds to the design input matrix consisting of binary values indicating the presence or absence of the features in vector , with p as the number of features, namely combinations of unigrams, bigrams and trigrams. Y corresponds to the vector of observations, in our case the neurological outcomes, where n indicates the number of patients. represents the vector of regression coefficients and ‖β‖1 corresponds to the L1 norm of this vector. The regularization parameter lambda controls the amount of shrinkage, adding a penalty on the weights, thereby preventing overfitting.
| (1) |
| (2) |
We created a training set (70%) to develop the model, and an independent hold-out set (30%) for evaluating the model, using stratified random sampling to ensure comparable distributions of neurologic outcomes. A randomized search was performed during training with 100 iterations of 5-fold cross validation (CV) for hyperparameter tuning. The model solver algorithm used in the optimization problem was set to “liblinear” and the “warm start” hyperparameter was varied between true/false, with “true” corresponding to reusing the solution of the previous call to fit as initialization, and “false” corresponding to erasing the previous solution. The inverse of the regularization strength, C, was varied among these values: 0.005, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1, 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5.
3.6. Performance measures
To select the best model configuration in the training data, the coefficient of determination R2 was used as the scoring metric in CV; higher values indicate better performance. To encourage model robustness, we applied the one standard error rule to select the regularization parameter, which favors models with fewer features over more complex models that have similar performance. The one standard error rule selects the simplest model whose R2 mean score falls within one standard deviation of the best performing model.
To evaluate the final model on the test data, we used the following metrics: area under the receiver operating characteristic curve (AUROC), area under the precision-recall curve (AUPRC), average precision (AP), accuracy, recall, specificity and F-score (Azari, Janeja, & Levin, 2015). We performed 100 iterations of bootstrap random sampling with replacement to calculate 95% confidence intervals (CI) for micro average performance metrics applied to the hold-out test set. Micro average performance is suited for problems with class imbalance and consists of calculating metrics globally by counting the total true positives, false negatives and false positives. The equations of the metrics presented in this section are depicted in Supplementary Table A5.
4. Results
4.1. Patient population
Patient selection into mRS and GOS cohorts is diagrammed in Fig. 2. To avoid double counting, 6 patients admitted at different times, first in the ICU and later with COVID-19 infection were removed from the ICU cohort. Cases of patients where experts were not able to generate a score due to lack of physical and occupational therapy notes and lack of information in the discharge summaries were not assigned a label. After removing patients admitted at different times (6 patients) and those without a label assigned (4 patients), from an initial cohort comprising 3642 patients, we were left with 3632 patients for mRS classification. We also excluded patients assigned to the rare GOS label “persistent vegetative state” (1.4% = 52/3632 patients), leaving 3581 patients for GOS classification.
Fig. 2.
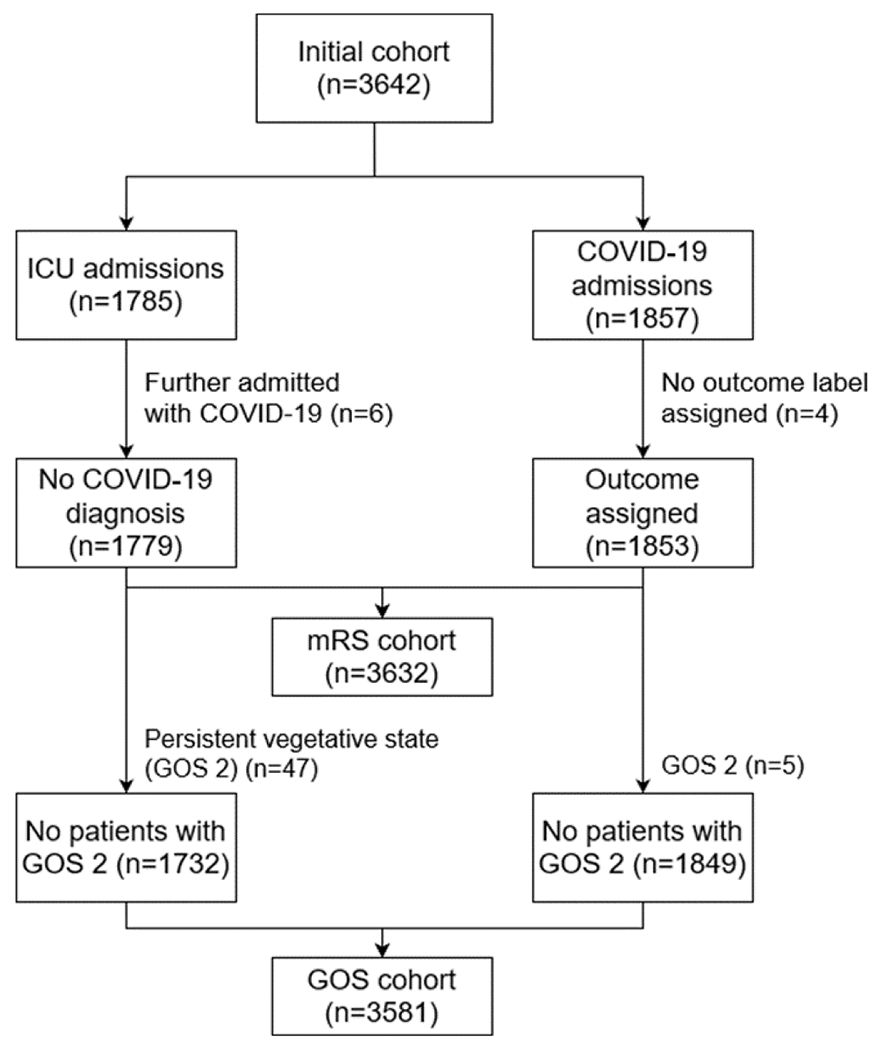
Study cohorts for classification of Glasgow Outcome Scale (GOS) and modified Rankin Scale (mRS) outcomes, with inclusion and exclusion criteria. ICU – Intensive Care Unit. Persistent vegetative state (GOS 2) was omitted from the analysis because this outcome was rare in our cohort.
Stratified random sampling by outcome was applied to split cohorts into train and test sets. Cohort baseline characteristics for train and test sets are shown in Table 1. The average age was between 59 and 61 years old in both sets. Approximately 62% of patients were White (2081/3376 in GOS and 2118/3428 in mRS) and 12% were Black or African American (407/3376 in GOS and 412/3428 in mRS). The majority of patients had GOS outcome of severe disability 1636/3581 (46%) and mRS outcome of moderately severe disability 910/3632 (25%). Among all patients with COVID-19, there were 291 (16%) non-survivors. Accounting for 382 (21%) non-survivors in the ICU dataset, there were a total of 673 deaths (19%) in our study cohort.
Table 1.
Baseline characteristics of the patient population stratified by train and test sets, for the classification of Glasgow Outcome Scale (GOS) and modified Rankin Scale (mRS) outcomes.
| Characteristic | GOS classification |
mRS classification |
||
|---|---|---|---|---|
| Train set (n = 2506) | Test set (n = 1075) | Train set (n = 2542) | Test set (n = 1090) | |
| Age (years) mean (SD) | 60.55 (18.0) | 59.44 (17.7) | 60.12 (18.0) | 60.50 (17.8) |
| Gender Male, n (%) | 1320 (52.7) | 581 (54.0) | 1330 (52.3) | 590 (54.1) |
| Race, n (%) | ||||
| White | 1551 (61.9) | 631 (58.7) | 1562 (61.4) | 657 (60.3) |
| Hispanic or Latino | 59 (2.4) | 37 (3.4) | 66 (2.6) | 30 (2.8) |
| Black or African American | 297 (11.9) | 129 (12.0) | 287 (11.3) | 143 (13.1) |
| Asian | 95 (3.8) | 38 (3.5) | 95 (3.7) | 43 (3.9) |
| Othera | 504 (20.0) | 240 (22.4) | 532 (21.0) | 217 (19.9) |
| Institution MGH, n (%) | 2114 (84.4) | 889 (82.7) | 2121 (83.4) | 932 (85.5) |
| COVID-19 positive, n (%) | 1286 (51.3) | 563 (52.4) | 1308 (51.5) | 545 (50.0) |
| Neurologic outcome, n (%) | ||||
| Good recovery (GOS 5) | 595 (23.7) | 255 (23.7) | – | – |
| No symptoms (mRS 0) | – | – | 420 (16.5) | 180 (16.5) |
| No significant disability (mRS 1) | – | – | 166 (6.5) | 71 (6.5) |
| Slight disability (mRS 2) | – | – | 188 (7.4) | 81 (7.4) |
| Moderate disability (mRS 3) | – | – | 304 (12.0) | 130 (11.9) |
| Moderate disability (GOS 4) | 307 (12.3) | 132 (12.3) | – | – |
| Moderately severe disability (mRS 4) | – | – | 637 (25.1) | 273 (25.0) |
| Severe disability (mRS 5) | – | – | 369 (14.5) | 158 (14.5) |
| Severe disability (GOS 3) | 1145 (45.7) | 491 (45.7) | – | – |
| Death (GOS 1, mRS 6) | 459 (18.3) | 197 (18.3) | 458 (18.0) | 197 (18.1) |
The number of patients is represented by n.
Other includes ‘unknown’, ‘declined’, ‘unavailable’ and race with a number less than 10, to preserve patient privacy. MGH – Massachusetts General Hospital.
Patients’ notes were preprocessed as described in Section 3.4. We obtained 7314 notes from the cohort of 3632 patients, including 3485 discharge summaries, 1472 occupational therapy and 2357 physical therapy notes. We indicate here the number of tokens (and respective outcome for the classification task) present in the training vocabulary, throughout the dimensionality reduction steps after data splitting. Before modeling, notes were composed of 37,693 (GOS) and 37,968 (mRS) tokens. After including only words present in at least 10% of the notes, there were 1258 (GOS) and 1267 (mRS) tokens. The tokens removed, such as “aaaom”, “aai”, “aala”, are not present in the English language, and may consist of typing errors; other words appeared only in a few rare notes. Thus, removal of these tokens was important for model generalizability and applicability. From these token sets, 2671 (GOS) and 2687 (mRS) combinations of n-grams were generated. Thus, the candidate features in the training vocabulary consisted of 1238 (GOS) and 1248 (mRS) unigrams, 864 (GOS) and 879 (mRS) bigrams, 569 (GOS) and 560 (mRS) trigrams.
4.2. Interrater reliability for extracting neurologic outcomes from charts
Four thousand and twenty-eight cases were reviewed by two experts and determined the IRR for mRS and GOS scores from medical charts. For mRS levels 0–6, IRR (95% CI) was 0.83 (0.65–1.00), 0.67 (0.36–0.94), 0.53 (0.30–0.72), 0.43 (0.25–0.61), 0.50 (0.29–0.64), 0.33 (0.10–0.65), 1 (1.00–1.00), respectively. For GOS levels 1 and 3–5, IRR (95% CI) was 1 (1.00–1.00) and 0.73 (0.59–0.86), 0.60 (0.42–0.76), 0.91 (0.78–1.00), respectively. Overall IRR across all levels was 72% for mRS, and 81% for GOS. We specified a priori that an 80% cut off for GOS would be acceptable, while for mRS we specified a 70% cut off, due to a higher number of labels and thus higher chance of disagreement. Cases with discrepancies were reviewed, and for cases where experts disagreed, it was observed that there was room for disagreement. Thus, we considered these levels of IRR adequate, and for the remaining 3204 cases only one expert reviewed each chart to extract mRS and GOS scores.
4.3. Modeling performance
The one-vs-rest logistic regression model with the best configuration parameters was evaluated in the hold-out test set. Performance results for each neurological outcome are shown in Table 2. Performance metrics for the GOS model were generally more robust than for the mRS model, potentially due to the greater number of choices on the mRS scale (Gupta, Bengio, & Weston, 2014). For mRS, class labels in the extremes, namely “no symptoms” (mRS 0) and “death” (mRS 6), were classified more accurately than intermediate outcomes (mRS 1 – mRS 5), as shown in Fig. 3. In the GOS model, “moderate disability” was less accurately classified compared to other outcome labels, with a recall (0.48 95% CI 0.42–0.57) and F1 (0.45 95% CI 0.38–0.52). A possible explanation is the lower number of patients with this outcome label (12%) used to train the model. This is reflected in the confusion the model makes with the nearest labels “severe disability” and “good recovery”, as shown in Fig. 4(a). “Severe disability”, when misclassified, also tends to be confused with “moderate disability”. Nevertheless, the majority of patients are correctly classified (recall 0.78 95% CI 0.74–0.81, and F1 0.81 95% CI 0.78–0.84).
Table 2.
Model performance in the hold-out test set and configuration parameters.
| AUROC | ACC | Recall | Spec | F1 | AP | No. Features (1, 2, 3 grams) | |
|---|---|---|---|---|---|---|---|
| GOS | 0.94 [0.93–0.95] | 0.77 [0.75–0.80] | 0.77 [0.75–0.80] | 0.92 [0.92–0.93] | 0.77 [0.75–0.80] | 0.65 [0.62–0.69] | 243 (166, 65, 12) |
| mRS | 0.90 [0.89–0.91] | 0.59 [0.567–0.62] | 0.59 [0.57–0.62] | 0.93 [0.93–0.94] | 0.59 [0.57–0.62] | 0.41 [0.38–0.44] | 536 (397, 116, 23) |
The bootstrapping results in 95% confidence intervals are in parenthesis. AUROC – Area under the receiver operating characteristic curve, ACC – accuracy, Spec – specificity, AP – average precision, GOS – Glasgow Outcome Scale, mRS – modified Rankin Scale, C – inverse of regularization strength. For both outcome models, the configuration parameters selected for C and warm start were 0.05 and “True”, respectively.
Fig. 3.
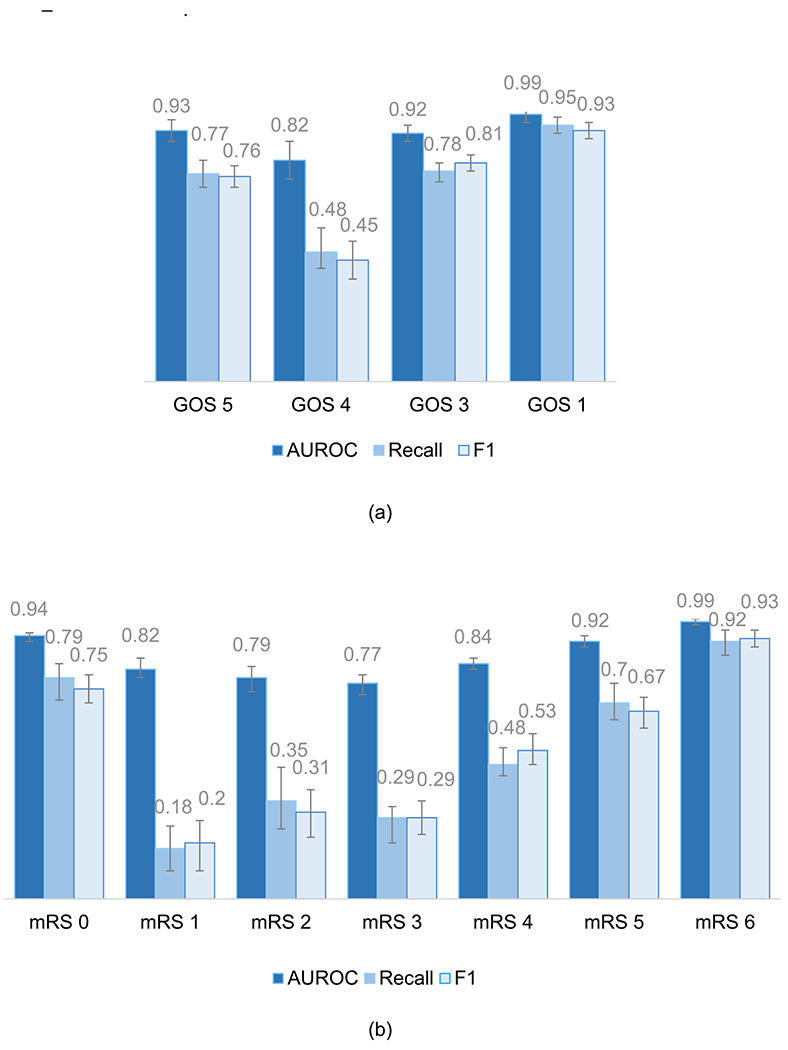
Models’ performance on the hold-out test set by class label, for (a) Glasgow Outcome Scale and (b) modified Rankin Scale. Labels: GOS 1, mRS 6 – death; GOS 3, mRS 5 – severe disability; mRS 4 – moderately severe disability; GOS 4, mRS 3 – moderate disability; mRS 2 – slight disability; mRS 1 – no significant disability; mRS 0 – no symptoms; GOS 5 – good recovery.
Fig. 4.
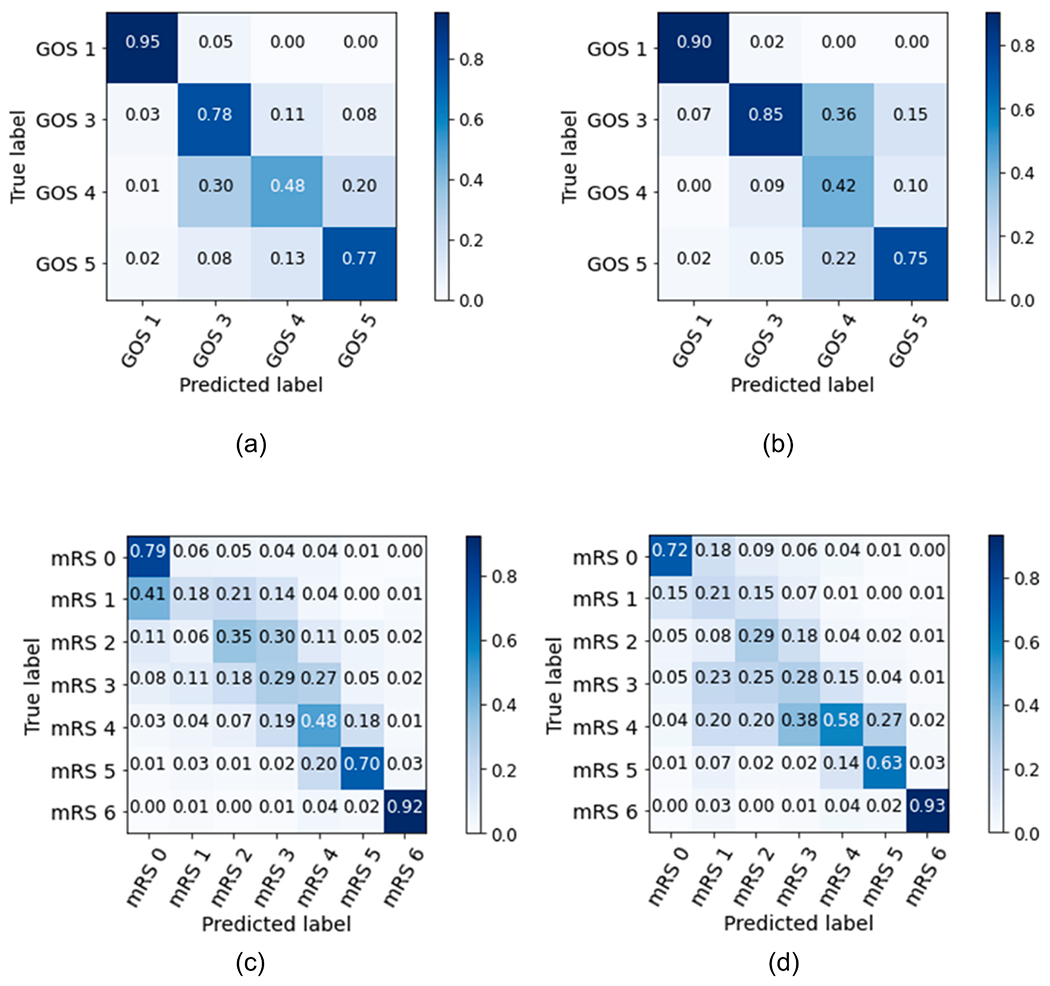
Confusion matrices normalized by (a) recall and (b) precision, for the GOS model, and normalized by (c) recall and (d) precision, for the mRS model, evaluated in the hold-out test sets. Labels: GOS 1, mRS 6 – death; GOS 3, mRS 5 – severe disability; mRS 4 – moderately severe disability; GOS 4, mRS 3 – moderate disability; mRS 2 – slight disability; mRS 1 – no significant disability; mRS 0 – no symptoms; GOS 5 – good recovery.
Models’ performance on the hold-out test set for each outcome label is presented for all metrics in Supplementary Table A6. AUROC and AUPRC curves are presented in Supplementary Fig. A1.
We also created models for mRS grouped into 3 or 4 levels: no symptoms to slight disability (mRS 0–2); moderate to severe disability (mRS 3–5); and death (mRS 6); or: no significant disability (mRS 0); slight to moderately severe disability (mRS 1–4); severe disability (mRS 5); and death (mRS 6). For these groupings, the model achieved an AUROC of 0.96 (95% CI 0.96–0.97) and 0.95 (95% CI 0.94–0.96), and F1 of 0.87 (95% CI 0.85–0.89) and 0.82 (95% CI 0.79–0.84), for the 3 and 4 level mRS groupings, respectively. Performance results are shown in Supplementary Table A7 and confusion matrices in Supplementary Fig. A2.
To assess potential bias related to under-representation of minorities in datasets, we assessed the GOS and mRS models performance by race, as presented in Table A8. For the patients identified as White in the EHR (60% of test data), the confidence intervals range was narrower across metrics compared with those of other races, indicating higher confidence in the classification. This is likely explained by the fact that the models were developed with a higher amount of data from White patients, therefore are more fitted to classify the neurological scores for this race. For patients identified as White, the models GOS and mRS achieved an AUROC of 0.94 (95% CI 0.92–0.95) and 0.91 (95% CI 0.89–0.92), and a recall of 0.77 (95% CI 0.73–0.80) and 0.58 (95% CI 0.54–0.61), respectively. However, when assessing the models’ performance in other race categories, GOS and mRS performed the best for Asian patients (4% of test data), achieving an AUROC of 0.96 (95% CI 0.93–0.99) and 0.92 (95% CI 0.87–0.96), and a recall of 0.86 (95% CI 0.81–0.98) and 0.67 (95% CI 0.56–0.81), respectively. For Black or African American patients (13% of test data), GOS and mRS achieved an AUROC of 0.94 (95% CI 0.90–0.96) and 0.90 (95% CI 0.86–0.92), and a recall of 0.78 (95% CI 0.69–0.86) and 0.59 (95% CI 0.49–0.67), respectively. Among patients identifying as Hispanic or Latino (3% of test data), GOS and mRS achieved an AUROC of 0.91 (95% CI 0.83–0.97) and 0.90 (95% CI 0.84–0.96), and a recall of 0.71 (95% CI 0.50–0.86) and 0.67 (95% CI 0.53–0.85), respectively.
4.4. Feature importance
The GOS and mRS feature selection steps reduced the initial training feature sets by approximately 91% (243/2671) and 80% (536/2687). These numbers were obtained with regularization constant values C of 0.05 for both models. Training performance curves as a function of C are presented in Supplementary Fig. A3.
We plot the importance of the top 15 features selected by LASSO regularization in Supplementary Figs. A4 and A5. Blue bars correspond to features with positive coefficients values and red bars to features with negative coefficients.
For both GOS and mRS, ‘decease’ was considered most important for classifying death (GOS 1, mRS 6). For the remaining outcome labels, this feature was assigned high importance with a negative coefficient. Features related with discharge disposition, such as home, inpatient rehab or skilled nursing facility (snf), were also assigned high importance to determine level of disability. For severe disability (GOS 3, mRS 5), ‘discharged home’ was assigned a negative coefficient, while ‘inpatient rehab’, ‘skilled nurse’ and ‘snf’ for GOS 3 and ‘peg’ (percutaneous endoscopic gastrostomy), ‘tube feed’ and ‘dnr’ (do-not-resuscitate order) for mRS 5, were assigned positive coefficients. On the contrary, for no symptoms (mRS 0) and good recovery (GOS 5), ‘inpatient rehab’ and ‘rehab’ were assigned negative coefficients while ‘discharge home’ a positive one for GOS 5. From mild to good recovery or no symptoms (mRS 0–3, GOS 4–5), features such as ‘home care’, ‘home support’, ‘ambulation’, ‘home pt’ (home care physical therapy), ‘iadl’ (instrumental activities of daily living) were all assigned positive coefficients, while ‘mechanical’, ‘dnr’, ‘feed’, ‘fibrillation’ and ‘htn’ (hypertension) were assigned negative ones.
5. Discussion
5.1. Principal findings
In this study we developed a machine-learning-based NLP pipeline to extract neurologic outcomes GOS and mRS from hospital discharge summaries, occupational therapy and physical therapy notes of hospitalized adult patients. The analysis included a diverse cohort of patients admitted to the ICU and patients admitted with COVID-19 infection. Performance was excellent for extreme outcomes, including no symptoms (mRS 0), good recovery (GOS 5), and death (GOS 1, mRS 6). For intermediate mRS and GOS outcome labels performance was overall lower which corresponds to lower IRR by experts for intermediate scores and remains a challenge in fields utilizing mRS and GOS. However, when NLP misclassifications occurred, they were largely to neighboring outcome levels, and combining mRS levels into 3 or 4 meaningful groups resulted in higher performance. Our method is able to process discharge summaries, physical and occupational therapy reports in an automated fashion at scale, extending the scope of feasible research beyond what is possible by manual chart review. Using real-world EHR data has been having increased recognition for designing outcomes studies in neurology (Biggin, Emsley, & Knight, 2020).
6. Limitations
The analysis included two academic medical centers located in the same geographic region (Boston, United States), both of which use the EPIC EHR and may not be representative of other US and non-US populations limiting the generalizability of the model across populations and hospital settings. An example of this limitation is that the model was developed with data from a patient population where White race was the majority, and the confidence in the classification for this race was overall higher than for other races. While we were encouraged that for other races or for patients who identified as Hispanic or Latino in the EHR, classification was not diminished, it is necessary to adapt these models in patient populations that reflect different communities. Thus, future studies will utilize this algorithm across different hospitals and EHR systems in the United States. Even though the analysis is based on data from highly specialized tertiary centers, the specific terms found in ICU notes might be also found for example in emergency departments notes, suggesting this NLP approach could perform a classification task in different medical settings. Although the model was developed with textual information, we did not consider the addition of other clinical features, such as age, gender or vital signs, medications and comorbidities. While clinicians do not use these factors in generating mRS or GOS scores, unconscious biases may occur when determining between levels of disability (mild, moderate or severe) in people who are older or have multimorbidity and maybe useful information when generating NLP algorithms. We did not consider using bidirectional encoder representations from transformers approaches, the current state-of-the-art in many NLP tasks, since their application presents limitations on classification of long clinical texts (Gao et al., 2021) and they are more complex and time costly. However, we propose to use this method as future work to compare the results with the method developed in this study. Another limitation in this work was the difficulty in classifying intermediate disability scores, due to the lack of training data, where there was a misrepresentation of these classes. Even though the models were trained with a balanced class weight, the reduced number of these classes decreased the model learning capability. Thus, future work entails acquiring more data, especially from patients assigned intermediate neurologic disability scores. Furthermore, we were not able to generate scores for four patients, due to lack of physical and occupational therapy notes and lack of information in the discharge summaries.
6.1. Conclusions
We developed a machine-learning-based NLP model to automatically and accurately extract neurological outcomes in a diverse cohort of hospitalized patients and showed good performance overall. The scale of research can be accelerated by using the methodological approach and model developed in this study with EHR data.
Supplementary Material
Acknowledgements
M. Brandon Westover (MBW) was supported by the Glenn Foundation for Medical Research and American Federation for Aging Research (Breakthroughs in Gerontology Grant); American Academy of Sleep Medicine (AASM Foundation Strategic Research Award); Football Players Health Study (FPHS) at Harvard University; Department of Defense through a subcontract from Moberg ICU Solutions, Inc; by the National Institutes of Health (NIH) ([R01NS102190], [R01NS102574], [R01NS107291], [RF1AG064312], [R01AG062989]), and National Science Foundation (NSF) [2014431]. MBW is a co-founder of Beacon Biosignals. Sahar F. Zafar (SFZ) is supported by the NIH [K23NS114201]. Shibani S. Mukerji (SSM) is supported by the NIH [K23MH115812, R01MH131194], James S. McDonnell Foundation, and Rappaport Fellowship.
Footnotes
CRediT authorship contribution statement
Marta B. Fernandes: Conceptualization, Methodology, Data curation, Visualization, Formal analysis, Validation, Software, Writing – original draft. Navid Valizadeh: Conceptualization, Methodology, Data curation, Investigation, Writing – review & editing. Haitham S. Alabsi: Conceptualization, Methodology, Data curation, Investigation. Syed A. Quadri: Data curation, Investigation. Ryan A. Tesh: Data curation, Investigation. Abigail A. Bucklin: Data curation, Investigation. Haoqi Sun: Conceptualization, Writing – review & editing. Aayushee Jain: Data curation, Software. Laura N. Brenner: Conceptualization, Methodology, Investigation. Elissa Ye: Data curation, Software. Wendong Ge: Conceptualization, Data curation, Software. Sarah I. Collens: Data curation. Stacie Lin: Data curation. Sudeshna Das: Conceptualization, Methodology, Writing – review & editing. Gregory K. Robbins: Methodology, Investigation, Writing – review & editing. Sahar F. Zafar: Supervision, Funding acquisition, Investigation, Writing – review & editing. Shibani S. Mukerji: Supervision, Funding acquisition, Investigation, Conceptualization, Methodology, Writing – review & editing. M. Brandon Westover: Supervision, Funding acquisition, Investigation, Conceptualization, Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eswa.2022.119171.
Data availability
The code to reproduce our results will be made publicly available at the time of publication so that future researchers can reproduce the results and benchmark against our model.
References
- Agarwala S, Anagawadi A, & Reddy Guddeti RM (2021). Detecting Semantic Similarity Of Documents Using Natural Language Processing. Procedia Computer Science, 189, 128–135. 10.1016/j.procs.2021.05.076 [DOI] [Google Scholar]
- Alawad M, Gao S, Qiu J, Schaefferkoetter N, Hinkle JD, Yoon H-J, Christian JB, Wu X-C, Durbin EB, Jeong JC, Hands I, Rust D, & Tourassi G (2019). Deep Transfer Learning Across Cancer Registries for Information Extraction from Pathology Reports. IEEE EMBS International Conference on Biomedical Health Informatics (BHI), 2019, 1–4. 10.1109/BHI.2019.8834586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfattni G, Belousov M, Peek N, & Nenadic G (2021). Extracting Drug Names and Associated Attributes From Discharge Summaries: Text Mining Study. JMIR Medical Informatics, 9(5), e24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azari A, Janeja VP, & Levin S (2015). Imbalanced learning to predict long stay Emergency Department patients. IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2015, 807–814. [Google Scholar]
- Bai T, & Vucetic S (2019). Improving Medical Code Prediction from Clinical Text via Incorporating Online Knowledge Sources. The World Wide Web Conference, 72–82. 10.1145/3308558.3313485 [DOI] [Google Scholar]
- Ben Abacha A, & Demner-Fushman D (2019). A question-entailment approach to question answering. BMC Bioinformatics, 20(1), 511. 10.1186/s12859-019-3119-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin F, Emsley HCA, & Knight J (2020). Routinely collected patient data in neurology research: A systematic mapping review. BMC Neurology, 20(1), 431. 10.1186/s12883-020-01993-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan NS, Rajpal DK, Webster Y, Alatorre C, Gudivada RC, Zheng C, Sanseau P, & Koehler J (2011). The role of translational bioinformatics in drug discovery. Drug Discovery Today, 16(9–10), 426–434. [DOI] [PubMed] [Google Scholar]
- Buckland RS, Hogan JW, & Chen ES (2020). Selection of Clinical Text Features for Classifying Suicide Attempts. AMIA Annual Symposium Proceedings. AMIA Symposium, 2020, 273–282. [PMC free article] [PubMed] [Google Scholar]
- Chen C-H, Hsieh J-G, Cheng S-L, Lin Y-L, Lin P-H, & Jeng J-H (2020). Early short-term prediction of emergency department length of stay using natural language processing for low-acuity outpatients. The American Journal of Emergency Medicine, 38(11), 2368–2373. 10.1016/j.ajem.2020.03.019 [DOI] [PubMed] [Google Scholar]
- Chen L, Li Y, Chen W, Liu X, Yu Z, & Zhang S (2018). Utilizing soft constraints to enhance medical relation extraction from the history of present illness in electronic medical records. Journal of Biomedical Informatics, 87, 108–117. 10.1016/j.jbi.2018.09.013 [DOI] [PubMed] [Google Scholar]
- Choi E, Schuetz A, Stewart WF, & Sun J (2017). Using recurrent neural network models for early detection of heart failure onset. Journal of the American Medical Informatics Association : JAMIA, 24(2), 361–370. 10.1093/jamia/ocw112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Dong W, He K, Duan H, & Huang Z (2018). Using neural attention networks to detect adverse medical events from electronic health records. Journal of Biomedical Informatics, 87, 118–130. 10.1016/j.jbi.2018.10.002 [DOI] [PubMed] [Google Scholar]
- Clapp MA, Kim E, James KE, Perlis RH, Kaimal AJ, & Mccoy TH (2022). Natural Language Processing of Admission Notes to Predict Severe Maternal Morbidity during the Delivery Encounter. American Journal of Obstetrics and Gynecology. 10.1016/j.ajog.2022.04.008 [DOI] [PubMed] [Google Scholar]
- Danilov G, Kotik K, Shifrin M, Strunina U, Pronkina T, & Potapov A (2019). Prediction of Postoperative Hospital Stay with Deep Learning Based on 101 654 Operative Reports in Neurosurgery. ICT for Health Science Research, 125–129. 10.3233/978-1-61499-959-1-125 [DOI] [PubMed] [Google Scholar]
- De Silva K, Mathews N, Teede H, Forbes A, Jönsson D, Demmer RT, & Enticott J (2021). Clinical notes as prognostic markers of mortality associated with diabetes mellitus following critical care: A retrospective cohort analysis using machine learning and unstructured big data. Computers in Biology and Medicine, 132, Article 104305. 10.1016/j.compbiomed.2021.104305 [DOI] [PubMed] [Google Scholar]
- Deng Y, Faulstich L, & Denecke K (2017). Concept Embedding for Relevance Detection of Search Queries Regarding CHOP. Studies in Health Technology and Informatics, 245, 1260. [PubMed] [Google Scholar]
- Fernandes M, Sun H, Jain A, Alabsi HS, Brenner LN, Ye E, Ge W, Collens SI, Leone MJ, Das S, Robbins GK, Mukerji SS, & Westover MB (2021). Classification of the Disposition of Patients Hospitalized with COVID-19: Reading Discharge Summaries Using Natural Language Processing. JMIR Medical Informatics, 9(2), e25457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Alawad M, Young MT, Gounley J, Schaefferkoetter N, Yoon H-J, Wu X-C, Durbin EB, Doherty J, Stroup A, Coyle L, & Tourassi GD (2021). Limitations of Transformers on Clinical Text Classification. IEEE Journal of Biomedical and Health Informatics. 10.1109/JBHI.2021.3062322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Young MT, Qiu JX, Yoon H-J, Christian JB, Fearn PA, Tourassi GD, & Ramanthan A (2018). Hierarchical attention networks for information extraction from cancer pathology reports. Journal of the American Medical Informatics Association: JAMIA, 25(3), 321–330. 10.1093/jamia/ocx131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AJ, Banerjee I, Block J, Winstead-Derlega C, Wilson JG, Mitarai T, Jarrett M, Sanyal J, Rubin DL, Wintermark M, & Kohn MA (2022). Natural language processing of head CT reports to identify intracranial mass effect: CTIME algorithm. The American Journal of Emergency Medicine, 51, 388–392. 10.1016/j.ajem.2021.11.001 [DOI] [PubMed] [Google Scholar]
- Gupta M, Bengio S, & Weston J (2014). Training Highly Multiclass Classifiers. Journal of Machine Learning Research, 15, 1461–1492. [Google Scholar]
- Hu Y, Wen G, Ma J, Li D, Wang C, Li H, & Huan E (2018). Label-indicator morpheme growth on LSTM for Chinese healthcare question department classification. Journal of Biomedical Informatics, 82, 154–168. 10.1016/j.jbi.2018.04.011 [DOI] [PubMed] [Google Scholar]
- Huang J, Osorio C, & Sy LW (2019). An empirical evaluation of deep learning for ICD-9 code assignment using MIMIC-III clinical notes. Computer Methods and Programs in Biomedicine, 177, 141–153. 10.1016/j.cmpb.2019.05.024 [DOI] [PubMed] [Google Scholar]
- Jennett B, & Bond M (1975). Assessment of Outcome After Severe Brain Damage: A Practical Scale. The Lancet, 305(7905), 480–484. 10.1016/S0140-6736(75)92830-5 [DOI] [PubMed] [Google Scholar]
- Joopudi V, Dandala B, & Devarakonda M (2018). A convolutional route to abbreviation disambiguation in clinical text. Journal of Biomedical Informatics, 86, 71–78. 10.1016/j.jbi.2018.07.025 [DOI] [PubMed] [Google Scholar]
- Ju X, Chen VCP, Rosenberger JM, & Liu F (2021). Fast knot optimization for multivariate adaptive regression splines using hill climbing methods. Expert Systems with Applications, 171, Article 114565. 10.1016/j.eswa.2021.114565 [DOI] [Google Scholar]
- Kang Y, & Hurdle J (2020). Predictive Model for Risk of 30-Day Rehospitalization Using a Natural Language Processing/Machine Learning Approach Among Medicare Patients with Heart Failure. Journal of Cardiac Failure, 26(10), S5. [Google Scholar]
- Kim Y, & Meystre SM (2019). A Study of Medical Problem Extraction for Better Disease Management. Studies in Health Technology and Informatics, 264, 193–197. 10.3233/SHTI190210 [DOI] [PubMed] [Google Scholar]
- Lee K, Filannino M, & Uzuner, Ö. (2019). An Empirical Test of GRUs and Deep Contextualized Word Representations on De-Identification. Studies in Health Technology and Informatics, 264, 218–222. 10.3233/SHTI190215 [DOI] [PubMed] [Google Scholar]
- Lehman L, Saeed M, Long W, Lee J, & Mark R (2012). Risk stratification of ICU patients using topic models inferred from unstructured progress notes. AMIA Annual Symposium Proceedings, 2012, 505. [PMC free article] [PubMed] [Google Scholar]
- Li M, Fei Z, Zeng M, Wu F-X, Li Y, Pan Y, & Wang J (2019a). Automated ICD-9 Coding via A Deep Learning Approach. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 16(4), 1193–1202. 10.1109/TCBB.2018.2817488 [DOI] [PubMed] [Google Scholar]
- Li Z, Yang Z, Shen C, Xu J, Zhang Y, & Xu H (2019b). Integrating shortest dependency path and sentence sequence into a deep learning framework for relation extraction in clinical text. BMC Medical Informatics and Decision Making, 19(1), 22. 10.1186/s12911-019-0736-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Capurro D, Nguyen A, & Verspoor K (2021). Early prediction of diagnostic-related groups and estimation of hospital cost by processing clinical notes. NPJ Digital Medicine, 4(1), 103. 10.1038/s41746-021-00474-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wan Y, & Su X (2019). Identifying individual expectations in service recovery through natural language processing and machine learning. Expert Systems with Applications, 131, 288–298. 10.1016/j.eswa.2019.04.063 [DOI] [Google Scholar]
- Liu Z, Yang M, Wang X, Chen Q, Tang B, Wang Z, & Xu H (2017). Entity recognition from clinical texts via recurrent neural network. BMC Medical Informatics and Decision Making, 17(2), 67. 10.1186/s12911-017-0468-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke S, Bashall A, Al-Adely S, Moore J, Wilson A, & Kitchen GB (2021). Natural language processing in medicine: A review. Trends in Anaesthesia and Critical Care, 38, 4–9. 10.1016/j.tacc.2021.02.007 [DOI] [Google Scholar]
- Lüneburg N, Reiss N, Feldmann C, van der Meulen P, van de Steeg M, Schmidt T, Wendl R, & Jansen S (2019). Photographic LVAD Driveline Wound Infection Recognition Using Deep Learning. Studies in Health Technology and Informatics, 260, 192–199. [PubMed] [Google Scholar]
- Marafino BJ, Park M, Davies JM, Thombley R, Luft HS, Sing DC, Kazi DS, DeJong C, Boscardin WJ, & Dean ML (2018). Validation of prediction models for critical care outcomes using natural language processing of electronic health record data. JAMA Network Open, 1(8), e185097–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meystre S, & Haug P (2006). Improving the sensitivity of the problem list in an intensive care unit by using natural language processing. AMIA Annual Symposium Proceedings, 2006, 554. [PMC free article] [PubMed] [Google Scholar]
- Munkhdalai T, Liu F, & Yu H (2018). Clinical Relation Extraction Toward Drug Safety Surveillance Using Electronic Health Record Narratives: Classical Learning Versus Deep Learning. JMIR Public Health and Surveillance, 4(2), e9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murff HJ, Forster AJ, Peterson JF, Fiskio JM, Heiman HL, & Bates DW (2003). Electronically screening discharge summaries for adverse medical events. Journal of the American Medical Informatics Association, 10(4), 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni PM, Ohno-Machado L, & Chapman WW (2011). Natural language processing: An introduction. Journal of the American Medical Informatics Association, 18(5), 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman-Griffis D, & Fosler-Lussier E (2021). Automated Coding of Under-Studied Medical Concept Domains: Linking Physical Activity Reports to the International Classification of Functioning, Disability, and Health. Frontiers in Digital Health, 3, Article 620828. 10.3389/fdgth.2021.620828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeid JS, Heider PM, Weeda ER, Matuskowitz AJ, Carr CM, Gagnon K, Crawford T, & Meystre SM (2019). Impact of De-Identification on Clinical Text Classification Using Traditional and Deep Learning Classifiers. Studies in Health Technology and Informatics, 264, 283–287. 10.3233/SHTI190228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin T, & Hoque MM (2021). An Ensemble Technique to Classify Multi-Class Textual Emotion. Procedia Computer Science, 193, 72–81. 10.1016/j.procs.2021.10.008 [DOI] [Google Scholar]
- Pesaranghader A, Matwin S, Sokolova M, & Pesaranghader A (2019). deepBioWSD: Effective deep neural word sense disambiguation of biomedical text data. Journal of the American Medical Informatics Association, 26(5), 438–446. 10.1093/jamia/ocy189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JX, Gao S, Alawad M, Schaefferkoetter N, Alamudun F, Yoon H-J, Wu X-C, & Tourassi G (2019). Semi-Supervised Information Extraction for Cancer Pathology Reports. IEEE EMBS International Conference on Biomedical Health Informatics (BHI), 2019, 1–4. 10.1109/BHI.2019.8834470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Pechanski P, Amr A, Katus HA, & Dieterich C (2019). Deep Learning Approaches Outperform Conventional Strategies in De-Identification of German Medical Reports. Studies in Health Technology and Informatics, 267, 101–109. 10.3233/SHTI190813 [DOI] [PubMed] [Google Scholar]
- Santiso S, Pérez A, & Casillas A (2019). Exploring Joint AB-LSTM With Embedded Lemmas for Adverse Drug Reaction Discovery. IEEE Journal of Biomedical and Health Informatics, 23(5), 2148–2155. 10.1109/JBHI.2018.2879744 [DOI] [PubMed] [Google Scholar]
- Selby LV, Narain WR, Russo A, Strong VE, & Stetson P (2018). Autonomous detection, grading, and reporting of postoperative complications using natural language processing. Surgery, 164(6), 1300–1305. 10.1016/j.surg.2018.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Rinaldi F, & Osmani V (2019). Natural Language Processing of Clinical Notes on Chronic Diseases: Systematic Review. JMIR Medical Informatics, 7(2), e12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Yi Y, Xiong Y, Tang B, Chen Q, Wang X, Ji Z, Zhang Y, & Xu H (2019). Extracting entities with attributes in clinical text via joint deep learning. Journal of the American Medical Informatics Association, 26(12), 1584–1591. 10.1093/jamia/ocz158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling NW, Patzer RE, Di M, & Schrager JD (2019). Prediction of emergency department patient disposition based on natural language processing of triage notes. International Journal of Medical Informatics, 129, 184–188. 10.1016/j.ijmedinf.2019.06.008 [DOI] [PubMed] [Google Scholar]
- Sun M, Baron J, Dighe A, Szolovits P, Wunderink RG, Isakova T, & Luo Y (2019). Early Prediction of Acute Kidney Injury in Critical Care Setting Using Clinical Notes and Structured Multivariate Physiological Measurements. Studies in Health Technology and Informatics, 264, 368–372. 10.3233/SHTI190245 [DOI] [PubMed] [Google Scholar]
- Thieu T, Maldonado JC, Ho P-S, Ding M, Marr A, Brandt D, Newman-Griffis D, Zirikly A, Chan L, & Rasch E (2021). A comprehensive study of mobility functioning information in clinical notes: Entity hierarchy, corpus annotation, and sequence labeling. International Journal of Medical Informatics, 147, Article 104351. 10.1016/j.ijmedinf.2020.104351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R (1996). Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society: Series B (Methodological), 58(1), 267–288. [Google Scholar]
- Uyeda AM, Curtis JR, Engelberg RA, Brumback LC, Guo Y, Sibley J, Lober WB, Cohen T, Torrence J, Heywood J, Paul SR, Kross EK, & Lee RY (2022). Mixed-methods evaluation of three natural language processing modeling approaches for measuring documented goals-of-care discussions in the electronic health record. Journal of Pain and Symptom Management. 10.1016/j.jpainsymman.2022.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Ö, South BR, Shen S, & DuVall SL (2011). 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association, 18(5), 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbroucke JP, Elm E. von, Altman DG, Gøtzsche PC, Mulrow CD, Pocock SJ, Poole C, Schlesselman JJ, Egger M, & Initiative, for the S. (2007). Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): Explanation and Elaboration. PLOS Medicine, 4(10), e297. 10.1371/journal.pmed.0040297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-H, Nguyen P-A, Islam MM, Li Y-C, & Yang H-C (2019). Development of Deep Learning Algorithm for Detection of Colorectal Cancer in EHR Data. Studies in Health Technology and Informatics, 264, 438–441. 10.3233/SHTI190259 [DOI] [PubMed] [Google Scholar]
- Wei C-H, Lee K, Leaman R, & Lu Z (2019). Biomedical Mention Disambiguation using a Deep Learning Approach. Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, 307–313. 10.1145/3307339.3342162. [DOI] [Google Scholar]
- Weissenbacher D, Sarker A, Klein A, O’Connor K, Magge A, & Gonzalez-Hernandez G (2019). Deep neural networks ensemble for detecting medication mentions in tweets. Journal of the American Medical Informatics Association, 26(12), 1618–1626. 10.1093/jamia/ocz156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman GE, Harhay MO, Lugo RM, Fuchs BD, Halpern SD, & Mikkelsen ME (2016). Natural language processing to assess documentation of features of critical illness in discharge documents of acute respiratory distress syndrome survivors. Annals of the American Thoracic Society, 13(9), 1538–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur WJ, Rzhetsky A, & Shatkay H (2006). New directions in biomedical text annotation: Definitions, guidelines and corpus construction. BMC Bioinformatics, 7(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JTL, Hareendran A, Grant M, Baird T, Schulz UGR, Muir KW, & Bone I (2002). Improving the Assessment of Outcomes in Stroke. Stroke, 33(9), 2243–2246. 10.1161/01.STR.0000027437.22450.BD [DOI] [PubMed] [Google Scholar]
- Yang H (2010). Automatic extraction of medication information from medical discharge summaries. Journal of the American Medical Informatics Association, 17(5), 545–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu Y, Qian M, Guan C, & Yuan X (2019). Information Extraction from Electronic Medical Records Using Multitask Recurrent Neural Network with Contextual Word Embedding. Applied Sciences, 9(18), 3658. 10.3390/app9183658 [DOI] [Google Scholar]
- Yoon H-J, Gounley J, Gao S, Alawad M, Ramanathan A, & Tourassi G (2019). Model-based Hyperparameter Optimization of Convolutional Neural Networks for Information Extraction from Cancer Pathology Reports on HPC. IEEE EMBS International Conference on Biomedical Health Informatics (BHI), 2019, 1–4. 10.1109/BHI.2019.8834674 [DOI] [Google Scholar]
- Yuvaraj D, Mohamed Uvaze Ahamed A, & Sivaram M (2021). A study on the role of natural language processing in the healthcare sector. Materials Today: Proceedings. 10.1016/j.matpr.2021.02.080. [DOI] [Google Scholar]
- Zafar SF, Rosenthal ES, Jing J, Ge W, Tabaeizadeh M, Aboul Nour H, Shoukat M, Sun H, Javed F, Kassa S, Edhi M, Bordbar E, Gallagher J, Moura V, Ghanta M, Shao Y-P, An S, Sun J, Cole AJ, & Westover MB (2021). Automated Annotation of Epileptiform Burden and Its Association with Outcomes. Annals of Neurology, 90(2), 300–311. 10.1002/ana.26161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Humbert-Droz M, Mukherjee P, & Gevaert O (2021). Structuring clinical text with AI: Old versus new natural language processing techniques evaluated on eight common cardiovascular diseases. Patterns, 2(7), Article 100289. 10.1016/j.patter.2021.100289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZC, Zhang Y, Zhou T, & Pang YL (2019). Medical assertion classification in Chinese EMRs using attention enhanced neural network. Mathematical Biosciences and Engineering: MBE, 16(4), 1966–1977. 10.3934/mbe.2019096 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code to reproduce our results will be made publicly available at the time of publication so that future researchers can reproduce the results and benchmark against our model.


