Abstract
Microorganisms that oxidize atmospheric methane in soils were characterized by radioactive labelling with 14CH4 followed by analysis of radiolabelled phospholipid ester-linked fatty acids (14C-PLFAs). The radioactive fingerprinting technique was used to compare active methanotrophs in soil samples from Greenland, Denmark, the United States, and Brazil. The 14C-PLFA fingerprints indicated that closely related methanotrophic bacteria were responsible for the oxidation of atmospheric methane in the soils. Significant amounts of labelled PLFAs produced by the unknown soil methanotrophs coeluted with a group of fatty acids that included i17:0, a17:0, and 17:1ω8c (up to 9.0% of the total 14C-PLFAs). These PLFAs are not known to be significant constituents of methanotrophic bacteria. The major PLFAs of the soil methanotrophs (73.5 to 89.0% of the total PLFAs) coeluted with 18:1 and 18:0 fatty acids (e.g., 18:1ω9, 18:1ω7, and 18:0). The 14C-PLFAs fingerprints of the soil methanotrophs that oxidized atmospheric methane did not change after long-term methane enrichment at 170 ppm CH4. The 14C-PLFA fingerprints of the soil methanotrophs were different from the PLFA profiles of type I and type II methanotrophic bacteria described previously. Some similarity at the PLFA level was observed between the unknown soil methanotrophs and the PLFA phenotype of the type II methanotrophs. Methanotrophs in Arctic, temperate, and tropical regions assimilated between 20 and 54% of the atmospheric methane that was metabolized. The lowest relative assimilation (percent) was observed for methanotrophs in agricultural soil, whereas the highest assimilation was observed for methanotrophs in rain forest soil. The results suggest that methanotrophs with relatively high carbon conversion efficiencies and very similar PLFA compositions dominate atmospheric methane metabolism in different soils. The characteristics of the methane metabolism and the 14C-PLFA fingerprints excluded any significant role of autotrophic ammonia oxidizers in the metabolism of atmospheric methane.
Microbial oxidation of atmospheric methane in soils is a key regulator of the atmospheric concentrations of this important trace gas (8, 18). The microorganisms that oxidize atmospheric methane have not been identified conclusively, and the physiological characteristics of the process remain uncertain. Different groups of bacteria have been suggested as potential oxidizers of atmospheric methane, including conventional methanotrophic bacteria similar to the ones already in cultures as well as novel high-affinity methanotrophic bacteria (4, 8, 16, 24). It has also been suggested that autotrophic nitrifying bacteria are responsible for consumption of atmospheric methane in soils due to their ability to cooxidize methane (7, 28).
Analysis of phospholipid ester-linked fatty acids (PLFAs) has been used successfully in the characterization of methanotrophic bacteria (e.g., see references 6, 10, and 21). The phenotypic relationships predicted from analysis of the methanotrophic PLFAs compare favorably with the phylogenetic relationships predicted from analysis of 16S rRNA (10). Discrimination between methanotrophic strains on the basis of PLFAs is based on fatty acid profiles and/or the presence of signature fatty acids specific for different methanotrophs. Type I methanotrophic bacteria contain fatty acids with 14 or 16 carbon atoms as their major PLFAs, whereas the major PLFAs in type II methanotrophic bacteria contain 18 carbon atoms. In addition, some type I and type II methanotrophs produce the unusual PLFAs 16:1ω8c and 18:1ω8c, respectively. These unusual PLFAs have been used as methanotroph-specific biomarkers in environmental studies (5, 29). Conventional analysis of microbial PLFAs from environmental samples may be combined with radiolabelling of selected bacteria followed by analysis of radiolabelled PLFAs (25). This technique provides a radioactive PLFA fingerprint (14C-PLFA fingerprint) of the microorganisms that metabolize labelled organic substrate added to the sample. The method has been used previously to study microorganisms that metabolize selected natural and xenobiotic compounds in environmental samples (25, 26).
In the present study, we compared the methane metabolism and the diversity of the organisms that oxidized atmospheric methane in soil samples from Arctic, temperate, and tropical regions. Selected soil samples were incubated with low concentrations of 14CH4 to specifically label the microorganisms that metabolized atmospheric methane. Subsequent analysis of radiolabelled PLFAs provided a radioactive fingerprint of the active soil methanotrophs.
MATERIALS AND METHODS
Bacterial strains.
The methanotrophic strains Methylomonas methanica S1, Methylococcus capsulatus Bath, Methylosinus trichosporium OB3b, and Methylocystis parvus OBBP were obtained from Colin Murrell, University of Warwick, Coventry, United Kingdom. The autotrophic nitrifiers Nitrosomonas europaea ATCC 19718 and Nitrosolobus multiformis ATCC 25197 were obtained from the American Type Culture Collection, Manassas, Va.
The methanotrophic bacteria were grown in a nitrate minimal medium containing 10 mM KNO3, 3.1 mM Na2HPO4, 1.9 mM KH2PO4, 0.8 mM Na2SO4, 0.2 mM MgSO4, and 0.05 mM CaCl2. Trace elements were added after autoclaving to give the following concentrations in the medium: 0.5 μM ZnCl2, 0.25 μM Na2MoO4, 0.5 μM MnCl2, 0.5 μM NaI, 0.5 μM H3BO3, 0.25 μM CoCl2, 0.25 NiCl2, 1.0 μM CuSO4, 0.25 μM KBr, 0.25 μM Na2WO4, 0.25 μM H2SeO4, 0.25 μM VCl3, and 2.5 μM EDTA. Iron was added to autoclaved medium as filter-sterilized Fe-EDTA to give a concentration of 25 μM. Liquid cultures were incubated in sealed 500-ml Erlenmeyer flasks with an initial headspace methane concentration of 5% (vol/vol).
The autotrophic nitrifying bacteria were grown in an ammonia minimal medium that was comparable to the methanotrophic medium described above except that the KNO3 was replaced with 30 mM NH4Cl and the concentrations of Na2HPO4 and KH2PO4 were only 1.55 mM and 0.95 μM, respectively. The ammonia minimal medium was buffered with 30 mM NaHCO3 (added to the medium after autoclaving) in combination with a headspace that contained 5% CO2. Liquid cultures were incubated in 500-ml Erlenmeyer flasks sealed with rubber stoppers.
Soil sampling.
Soil was collected in Denmark and Greenland as intact soil cores in acrylic core tubes (two to three cores). Soil from Vietnam, Indonesia, Brazil, and the United States was collected as 20- to 50-g soil subsamples and stored in small plastic containers (two to three samples). The container lids were perforated to allow gas exchange during transport. Soil cores and soil subsamples were stored in the dark at 10°C and sieved just prior to use.
Methane oxidation.
Oxidation of atmospheric methane was estimated from first-order decreases in headspace methane concentrations (1.7 ppm). Sieved soil samples (10 to 20 g) were incubated in 60-ml serum bottles. Methane (0.3 ml) was sampled with a needle and syringe and analyzed on a Chrompack 438A gas chromatograph equipped with a flame ionization detector. The oven, injector, and detector temperatures were 100, 120, and 225°C, respectively. Gases were separated on a Hyasep Q column (2 m by 2 mm) with N2 as the carrier gas (20 ml min−1). The detection limit for methane was 0.1 ppm.
Radioactive labelling of methanotrophic bacteria.
Soil samples were incubated with 14CH4 (<40 ppm) to label the microorganisms that metabolized methane at low methane concentrations. The soil samples (2 to 4 g [dry weight]) were incubated in 14-ml serum vials and spiked with aliquots of 0.2 ml of 14CH4 (0.25 MBq ml−1; 2.0 GBq mmol−1; Amersham, Little Chalfont, United Kingdom). The methane concentration decreased from approximately 40 ppm to <1 ppm between the 14CH4 additions. The samples were aerated between additions to ensure oxic conditions and to remove 14CO2 produced during the oxidation of 14CH4. The labelling was generally completed within 2 to 5 days when a total of >0.2 MBq 14CH4 had been consumed. All major methanotrophic biomass components were labelled after incubation with the 14CH4 (24). The 14CH4 used in the experiments was purified prior to use in order to remove potential contaminants such as 14CO and 14CO2 (15).
Carbon conversion efficiency.
The amount of methane assimilated into microbial biomass relative to the total amount of methane consumed (carbon conversion efficiency) was determined by incubating soil samples with 14CH4 at near-atmospheric concentrations (<5 ppm methane) as described previously (24). The incubation time varied between 4 and 24 h, depending on the oxidation activity in the soil samples. The amount of 14CH4 oxidized to 14CO2 was determined by flushing the samples with air and trapping the 14CO2. The amount of 14CH4 assimilated into microbial biomass was determined after converting the soil organic matter into carbon dioxide by CrO3 oxidation (which yields CO2 + 14CO2). The recovery of labelled bacterial biomass from the soil matrix by CrO3 oxidation was 93 to 95% (24).
Extraction and analysis of PLFAs.
Total microbial PLFAs were extracted from soil samples labelled with 14CH4. The purified extract was then analyzed for the presence of radiolabelled PLFAs. Extraction of microbial lipids, separation of lipid classes, and preparation of phospholipid ester-linked fatty acid methyl esters were carried out as described previously (25). Radiolabelled PLFAs were collected as 14CO2 after gas chromatography (GC) column separation and combustion to 14CO2 in the flame ionization detector (25). The recovery of 14C-PLFAs as 14CO2 after the GC separation was >70%. Routine extraction and analysis of 14C-PLFAs were performed for duplicate samples. In general, the variation in fingerprint between parallel samples was insignificant. The maximum standard error observed for samples incubated in triplicates was below 5% (standard error <5% for each PLFA fraction).
In the initial experiments, the phospholipid ester-linked fatty acid methyl esters were separated according to a GC temperature program that also included changes in column pressure (25). This method was later successfully replaced by the following 95-min temperature program with constant pressure (150 kPa): 1 min at 60°C, increase from 60 to 170°C at a rate of 40°C/min, 0.5 min at 170°C, increase from 170 to 200°C at a rate of 0.4°C/min, increase from 200 to 300°C at a rate of 10°C/min, and finally 5.75 min at 300°C. The GC injector temperature was 270°C, the detector temperature was 300°C, and the initial column temperature was 60°C. H2 was used as a carrier (1.0 ml/min), N2 was used as makeup gas (35 ml/min), and H2 and air were used for the flame ionization detector (34 and 370 ml/min, respectively). The 14C-PLFAs were separated into 15 fractions according to their retention times and equivalent chain lengths (Table 1). Tridecanoic acid (13:0) and nonadecanoic acid (19:0) were used as internal standards (200 μM). Individual fatty acids were identified on the basis of retention times relative to authentic standard fatty acids (Nu Chek Prep Inc., Elysian, Minn.). The identities of individual fatty acids were resolved further by comparisons with parallel samples analyzed by Microbial Insights Inc. (Knoxville, Tenn.).
TABLE 1.
Examples of fatty acids that coeluted with the radiolabelled PLFAs
| PLFA fraction | ECLa | Example(s) of fatty acids |
|---|---|---|
| 1 | <13.6 | 13:0 |
| 2 | 13.6–14.6 | 14:0, i15:1, a15:1 |
| 3 | 14.6–15.3 | i15:0, a15:0, 15:0 |
| 4 | 15.3–15.7 | i16:1, 16:1ω9, 16:1ω8 |
| 5 | 15.7–16.1 | (16:1ω8), 16:1ω7, 16:1ω5, 16:0 |
| 6 | 16.1–16.5 | i17:1, a17:1, 10Me16:0, 12Me16:0 |
| 7 | 16.5–16.8 | i17:0, a17:0, 17:1ω8, cy17:0 |
| 8 | 16.8–17.1 | (cy17:0), 17:1, 17:0 |
| 9 | 17.1–17.4 | 10Me17:0 |
| 10 | 17.4–17.6 | 18:3, 18:2, i18:1 |
| 11 | 17.6–17.9 | 18:1ω9, 18:1ω8, 18:1ω7, (18:0) |
| 12 | 17.9–18.4 | 18:0, 10Me18:0, 12Me18:0 |
| 13 | 18.4–18.8 | i19:0, a19:0, 19:1, cy19:0 |
| 14 | 18.8–19.0 | (cy19:0), 19:0 |
| 15 | >19.0 | (19:0), 20:4, 20:1, 20:0, 22ω6, 22:0 |
Equivalent chain length.
Fatty acid nomenclature.
Fatty acids are named according to the convention x:y, where x is the number of carbon atoms and y is the number of double bonds. In some cases, the position of the double bond from the methyl end (ω) is also stated. The prefix cy indicates cyclopropane fatty acids, whereas the prefixes i and a indicate iso and anteiso branching, respectively. The designation 10Me and 12Me indicate methyl groups on the 10th and 12th carbon from the carboxyl end of the fatty acid, respectively.
Liquid scintillation counting.
The radioactivity associated with trapped 14CO2 was determined by using 12 ml of Packard Instagel Plus as scintillation cocktail. Only samples exceeding twice the background radioactivity were scored as positive (>1 Bq). All radiolabelled samples were analyzed for 5 min in a Packard 1600 TR liquid scintillation counter. The measured radioactivity was corrected for quench by using external and internal standards.
RESULTS
Oxidation and assimilation of atmospheric methane.
Soil samples from Arctic, temperate, and tropical regions were assayed for oxidation and assimilation of atmospheric methane (Table 2). All soil samples were capable of oxidizing methane to levels below atmospheric concentrations (1.7 ppm). The threshold for methane oxidation was <0.5 ppm CH4. The oxidation capacity in the forest soil samples varied between 21 and 752 pmol g (dry weight)−1 h−1. The most-limited potential for oxidation of atmospheric methane was observed with agricultural soil samples (10 pmol g [dry weight]−1 h−1). The amount of carbon assimilated into microbial biomass relative to the amount of methane oxidized (carbon conversion efficiency) varied between 20% in the agricultural soil and 54% in forest soil from Vietnam. Methanotrophs in forest soils from Denmark, the United States, and Brazil showed carbon conversion efficiencies that grouped in a more narrow range (30 to 41%). No correlation was observed between the atmospheric methane oxidation capacity and the soil pHKCl (r2 < 0.3). Similarly, no correlation was observed between the atmospheric methane oxidation capacity and the soil concentration of extractable NO3− or NH4+ (r2 < 0.2).
TABLE 2.
Oxidation and assimilation of atmospheric methane in different soils
| Soil type (location) | Sampling depth (cm) | pHKCl | NO3− (μmol g−1) | NH4+ (μmol g−1) | Oxidation of atmospheric methane (pmol g−1 h−1) | Assimilation of atmospheric methane (%) |
|---|---|---|---|---|---|---|
| Heathland (Zackenberg, Greenland) | 3–6 | 4.3 | 0.06 | 0.14 | 41 | NDa |
| Pine forest (Klosterheden, Denmark) | 16–20 | 3.8 | 0.04 | 0.07 | 69 | 41 |
| Beech forest (Rold, Denmark) | 6–10 | 3.4 | 0.61 | 0.23 | 221 | 36 |
| Agricultural (Nørre Halne, Denmark) | 6–10 | 5.0 | 5.89 | 0.14 | 10 | 20 |
| Mixed hardwood forest (Connecticut) | 6–10 | 4.2 | 0.16 | 0.27 | 181 | 30 |
| Mixed hardwood forest (Maine) | 6–10 | 4.0 | 0.02 | 0.30 | 752 | 39 |
| Rain forest (Pantanal, Brazil) | 4–8 | 4.9 | 1.02 | 0.25 | 21 | 35 |
| Rain forest (Pocone, Brazil) | 4–8 | 5.2 | 1.33 | 0.48 | 33 | 38 |
| Rain forest (Sumatra, Indonesia) | 4–8 | 4.3 | 1.16 | 0.13 | 24 | 22 |
| Rain forest (Dambri Falls, Vietnam) | 4–8 | 4.5 | 1.48 | 0.25 | 52 | 54 |
ND, not determined.
14C-PLFA fingerprints of high-affinity methanotrophs.
Microorganisms that metabolized atmospheric methane in soils were characterized by labelling with 14CH4 followed by analysis of radiolabelled PLFAs. Relatively comparable 14C-PLFA fingerprints were obtained with nonenriched soil samples from Arctic, temperate, and tropical regions (Table 3). The majority of the radioactivity associated with radiolabelled PLFAs from the high-affinity soil methanotrophs eluted in fraction 11 (73.5 to 88.9% of the labelled PLFAs). This PLFA fraction represents fatty acids with equivalent chain lengths between 17.6 and 17.9 (Table 1). The radiolabelled fatty acids in fraction 11 coeluted with 18:1 and, to some extent, 18:0 fatty acids. Between 2.7 and 20.5% of the radiolabelled PLFAs from the soil methanotrophs eluted in fraction 5 (Table 3). This PLFA fraction contains fatty acids with equivalent chain lengths between 15.7 and 16.1, e.g., 16:1 and 16:0 fatty acids (Table 1). In the forest and heath soils, a small but consistent amount of radiolabelled PLFAs (3.3 to 9.0%) eluted in fraction 7 (Table 3). This fraction represents fatty acids with equivalent chain lengths between 16.5 and 16.8, e.g., 17:0 and 17:1 species (Table 1). Small amounts of radioactivity (<4.2% of the total) eluted occasionally in PLFA fraction 14 or 15. However, labelling in fraction 14 or 15 was only observed for some soils, whereas radioactivity was always recovered in PLFA fractions 5, 7, and 11 (Table 3).
TABLE 3.
14C-PLFA fingerprints of methanotrophic microorganisms that metabolize atmospheric methane in different soilsa
| Soil type (location) | Recovery of 14C-PLFAs (%) in PLFA fraction:
|
||||||
|---|---|---|---|---|---|---|---|
| 5 | 7 | 9 | 11 | 12 | 14 | 15 | |
| Heathland (Zackenberg, Greenland) | 2.7 | 3.4 | 88.9 | 1.6 | 3.5 | ||
| Mixed hardwood forest (Connecticut) | 4.6 | 6.7 | 88.0 | 0.7 | |||
| Mixed hardwood forest (Maine) | 3.4 | 8.7 | 87.8 | ||||
| Rain forest (Pantanal, Brazil) | 20.5 | 3.3 | 73.5 | 2.7 | |||
| Agricultural (Nørre Halne, Denmark) | 15.1 | 1.3 | 79.5 | 4.2 | |||
| Beech forest (Rold, Denmark) | 9.3 | 9.0 | 78.4 | 3.2 | |||
| Beech forestb (Rold, Denmark) | 5.8 | 9.7 | 2.1 | 82.4 | |||
| Beech forestc (Rold, Denmark) | 30.5 | 0.9 | 66.5 | 2.1 | |||
PLFA fractions are described in Table 1. No 14C-PLFAs were recovered in fractions 1 to 4, 6, 8, 10, and 13.
Soil samples were enriched for 24 weeks at 170 ppm CH4.
Soil samples were enriched for 12 weeks at 10,000 ppm CH4.
The 14C-PLFA fingerprint of the methanotrophs that metabolized atmospheric methane was examined in more detail by using nonenriched soil samples from a Danish beech forest (Table 3). Fraction 5 (9.3% of the total 14C-PLFAs), fraction 7 (9.0% of the total 14C-PLFAs), and fraction 11 (78.4% of the total 14C-PLFAs) were subdivided into intervals to examine which fatty acids coeluted with the radiolabelled PLFAs. Fraction 14 (only 3.2% of the 14C-PLFAs) was not examined further. The analysis showed that the 14C-PLFAs from the high-affinity soil methanotrophs coeluted with the following known fatty acids: 16:1ω7c and/or 16:1ω5c in fraction 5; i17:0, a17:0, and/or 17:1ω8c in fraction 7; and/or 18:1ω9c, 18:1ω8c, and 18:1ω7c in fraction 11.
Effect of methane enrichment.
Enrichment of soil samples at a methane concentration approximately 100 times above ambient methane concentrations (170 ppm) did not result in major changes in the 14C-PLFA fingerprint of the active soil methanotrophs (Table 3). Methane-enriched soil samples were incubated for 24 weeks at 170 ppm (continuous flow) prior to radiolabelling and fingerprinting of the active soil methanotrophs. The potential for oxidation of atmospheric methane did not increase during the incubation at 170 ppm CH4. Similar to what was found with fresh soil samples, the active methanotrophs in the methane-enriched samples produced PLFAs that eluted mainly in fraction 5, 7, and 11 (Table 3).
Enrichment of forest soil methanotrophs at 10,000 ppm methane for 12 weeks resulted in a shift in labelling pattern for the active soil methanotrophic population (Table 3). Relatively more radioactivity was now recovered in fraction 5, and less was recovered in fractions 7 and 11. The methane enrichment at 10,000 ppm CH4 was accompanied by an increase in methane oxidation rates as well as an increase in apparent Km for methane from between 15 and 19 to 2,010 ppm CH4.
Two nonenriched soils with active oxidation of atmospheric methane were examined for the presence of the methanotrophic signature PLFAs 16:1ω8c (type I) and 18:1ω8c (type II). However, the signature fatty acids were not detectable (<0.1 nmol/g [dry weight]) in either of the two soils examined (Danish beech forest soil and agricultural soil). Methanotrophic signature fatty acids were only detectable after long-term methane enrichment of soil samples in the laboratory. Enrichment of the forest and agricultural soil samples for 12 weeks at 10,000 ppm CH4 resulted in the recovery of 0.7 and 5.9 nmol of 16:1ω8c per g (dry weight) and 2.2 and 2.7 nmol of 18:1ω8c per g (dry weight), respectively.
Comparison of PLFA fingerprints.
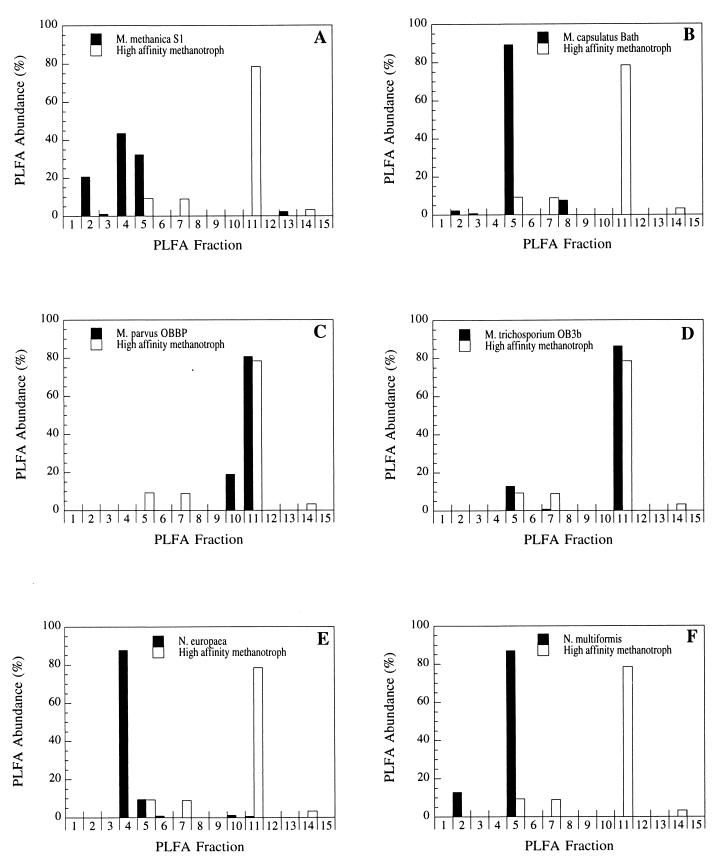
The 14C-PLFA fingerprint of the high-affinity soil methanotrophs was compared with the PLFA fingerprints of autotrophic nitrifying bacteria and representative type I and II methanotrophic bacteria (Fig. 1). A representative 14C-PLFA fingerprint of active methanotrophs from a Danish beech forest soil (Table 3) was used in the comparison. At the PLFA level, little similarity was observed between the fingerprint of the soil methanotrophs and the fingerprints of the type I methanotrophic bacteria M. methanica S1 and M. capsulatus Bath (Fig. 1A and B). Likewise, no similarity was observed between the soil methanotrophs and the autotrophic nitrifying bacteria N. europaea and N. multiformis (Fig. 1E and F). In contrast, the 14C-PLFA fingerprint of the unknown soil methanotrophs showed some similarity with the PLFA fingerprints of the type II methanotrophic bacteria and in particular M. trichosporium OB3b (Fig. 1C and D). The high-affinity soil methanotrophs were different from M. trichosporium OB3b in that they produced significant amounts of PLFAs (>1%) that eluted in fractions 7 and 14. This was not observed with M. trichosporium OB3b or any other reference culture tested.
FIG. 1.
PLFA fingerprints of the type I methanotrophic bacteria M. methanica S1 (A) and M. capsulatus (B), the type II methanotrophic bacteria M. parvus OBBP (C) and M. trichosporium OB3b (D), and the autotrophic nitrifying bacteria N. europaea (E) and N. multiformis (F). An example of a 14C-PLFA fingerprint of the soil methanotrophs that oxidized atmospheric methane has been included for comparison (fingerprint of active methanotrophs from the Danish beech forest soil in Table 3).
DISCUSSION
Consumption of atmospheric methane in soils is carried out by unknown aerobic microorganisms with a high affinity for methane. Different physiological mechanisms in these unknown methane oxidizers may account for the oxidation process (2, 8, 13, 19, 24). For example, atmospheric methane may be oxidized fortuitously or the microorganisms involved may be specialized for use of atmospheric methane as a primary substrate. Different groups of bacteria have been suggested as potential oxidizers of atmospheric methane in soils, including conventional methanotrophic bacteria, conventional autotrophic nitrifying bacteria, and novel high-affinity methanotrophic bacteria (8, 16). It has also been suggested that physiologically different microorganisms (autotrophic nitrifiers and methanotrophs) are responsible for atmospheric methane consumption in different soil types (7, 11, 13, 28).
Known obligate methanotrophic bacteria are unique in their ability to use methane as the sole source of carbon and energy. The majority of the methanotrophs in cultures have been isolated with inorganic minimal media and relatively high methane concentrations (e.g., 1 to 50% CH4 in the headspace). The known methanotrophs can be divided into two main groups (types I and II) based on their morphology, physiology, and phylogeny (14, 20). For example, type I methanotrophic bacteria produce 16:0 and 16:1 fatty acids as their major PLFAs, and the unusual fatty acid 16:1ω8c appears indicative of certain type I methanotrophs. Type II methanotrophs produce fatty acids with 18 carbon atoms as their major PLFAs, and the unusual fatty acid 18:1ω8c appears indicative of some type II organisms. Detailed analysis of two soils in the present study showed that the methanotrophic signature PLFAs 16:1ω8c and 18:1ω8c were not detectable in fresh samples from an agricultural soil (Nørre Halne, Denmark) and a beech forest soil (Rold forest, Denmark). However, methanotrophic microorganisms in both soils oxidized and assimilated atmospheric methane (Table 2). The methanotrophic signature fatty acids 16:1ω8c and 18:1ω8c were only detectable after long-term methane enrichment of soil samples in the laboratory (12 weeks at 10,000 ppm CH4). This indicates that methane-oxidizing bacteria similar to some of the known type I and II methanotrophs were present in fresh soil but in a very low abundance. Hence, it is questionable whether methanotrophs containing 16:1ω8c and 18:1ω8c played any quantitative role in the oxidation of atmospheric methane in these soils.
Differences in the PLFA profiles of known type I and II methanotrophic bacteria make it possible to distinguish between these populations in environmental samples after labelling with 14CH4 followed by analysis of radiolabelled PLFAs (25). In the present study, this technique was used to study the PLFA profiles of the methanotrophs that metabolize atmospheric methane in soils. This was possible because the unknown methanotrophs assimilated significant amounts of methane into microbial biomass (Table 2). Although methanotrophs may discriminate against 14CH4 (at the per mille level), all major macromolecules eventually become labelled in experiments with 14CH4 as long as all the available methane (12CH4 + 13CH4 + 14CH4) is consumed completely (24). This was the case in our labelling experiments, where the total concentration of methane was depleted before analysis of the methanotrophic biomarkers. Control experiments have confirmed that methanotrophic bacteria that were fed a mixture of 12CH4 and 14CH4 had a 14C-PLFA profile similar to the 12C-PLFA profile of methanotrophs fed only 12CH4 (25).
Analysis of the radiolabelled phospholipids from the unknown soil methanotrophs resulted in recovery of >95% of the radioactivity in PLFA fractions 5, 7, and 11 (Table 3). The labelling pattern was comparable for selected soil samples labelled in short-term (48 h) and long-term assays (168 h) (data not shown). The 14C-PLFA fingerprints were also relatively similar despite differences in soil characteristics and origin of the samples. Differences in transport and storage conditions may have affected the absolute methane oxidation capacity and perhaps also the carbon conversion efficiency of the soil methanotrophs, but the 14C-PLFA fingerprints appeared surprisingly comparable (Table 3). The similarity among the 14C-PLFA fingerprints does not indicate a labelling pattern that was due to carbon assimilation by nonmethanotrophic microorganisms. Uptake by soil heterotrophs of labelled metabolites excreted by the methane oxidizers would likely have resulted in 14C-PLFA fingerprints that were much more diverse than the profiles shown in Table 3. Similarly, significant autotrophic assimilation of 14CO2 produced during the oxidation of 14CH4 would likely have resulted in more-diverse fingerprints in the different soils. In addition, 14CO2 produced from 14CH4 was diluted substantially by the large pool of unlabelled CO2 in the soils, resulting in low specific activities and limited autotrophic 14CO2 assimilation. This conclusion is also supported by the soil 14C-PLFA fingerprints, which were very different from the PLFA profiles of known soil autotrophs such as ammonia oxidizers (see below). On the basis of these findings, we find it highly probable that the labelled PLFAs in fractions 5, 7, and 11 represent fatty acids strictly associated with methanotrophic microorganisms. The source of the radioactivity in fractions 14 and 15 (≤4% of the labelled PLFAs) was less clear, as these fractions were only labelled in some soils.
It is noteworthy that methane enrichment of soil samples at 170 ppm did not result in major changes in the labelling pattern in fractions 5, 7, and 11 (Table 3). The methane concentration was increased to evaluate whether some of the PLFAs were produced as a response to methane limitations (e.g., the unusual PLFAs in fraction 7). A methane concentration of 170 ppm was well above the apparent Km of 15 to 19 ppm determined for the high-affinity methanotrophic activity in the forest soil (22). This methane affinity (apparent Km) is comparable to that reported for other soils with active consumption of atmospheric methane (e.g., see references 2, 3, 12, and 24). In contrast, long-term methane enrichment at 10,000 ppm resulted in a methanotrophic population with a somewhat different PLFA labelling pattern (Table 3). This change was accompanied by a >100-times increase in apparent Km for methane and by the appearance in the soil of the methanotrophic signature PLFAs 16:1ω8c and 18:1ω8c. Thus, methane enrichment at 10,000 ppm CH4 was likely associated with activation and growth of conventional type I and type II methanotrophs in the soil. However, it was not possible to determine whether the change in the labelling pattern after enrichment at 10,000 ppm CH4 was also associated with physiological changes in the methanotrophs active at low methane concentrations.
The PLFA fingerprint of the unknown soil methanotrophs was very different from that of representative autotrophic nitrifying bacteria (Fig. 1E and F). Nitrifying bacteria such as N. europaea, N. multiformis, and Nitrosococcus oceanus all produce fatty acids with 16 carbon atoms as their major PLFAs (up to 97 to 99% of the total PLFAs) (23). PLFAs with 16 carbon atoms elute mainly in fractions 4 and 5 (Table 1), whereas the majority of the radiolabelled PLFAs from the soil methanotrophs eluted in fraction 11 (73.5 to 89.0%). Thus, the 14C-PLFA fingerprints of the high-affinity methanotrophs exclude any quantitative role of known autotrophic nitrifying bacteria in the metabolism of atmospheric methane in our soils. This conclusion is supported by the high carbon conversion efficiency for atmospheric methane, which does not suggest a fortuitous oxidation-assimilation mechanism (Table 2). Analysis of the 14C-PLFAs from methanotrophs from nonenriched soils showed that the radiolabelled fatty acids coeluted with groups of PLFAs that included the following known species: 16:1ω7c and 16:1ω5c (fraction 5); i17:0, a17:0, and 17:1ω8c (fraction 7); and 18:1ω9c, 18:1ω8c, and 18:1ω7c (fraction 11). Radiolabelled fatty acids that eluted in fraction 7 made up 3.3 to 9.0% of the PLFAs from methanotrophs from heathland and forest soils. PLFAs with 17 carbon atoms (e.g., i17:0, a17:0, and 17:1ω8c) have not been reported as significant constituents (>1%) of known autotrophic nitrifying bacteria or methanotrophic bacteria (6, 10, 23). Guckert et al. (10) reported the presence of i17:0 and a17:0 in M. trichosporium OB3b but at a relative abundance of only 0.06 and 0.1%, respectively. Bowman et al. (6) did not measure any i17:0 or a17:0 in M. trichosporium OB3b but reported trace amounts (0.1 to 0.6%) of these fatty acids in Methylosinus sporium. We have subsequently analyzed M. trichosporium OB3b to evaluate whether altered incubation conditions would produce a PLFA profile similar to the 14C-PLFA fingerprint of the unknown soil methanotrophs. M. trichosporium OB3b was selected because this organism showed some similarity with the high-affinity soil methane oxidizers (Fig. 1D). M. trichosporium OB3b was incubated at a low methane concentration (1.7 and 1,700 ppm CH4 in the headspace) for 7 to 10 days to examine whether methane starvation would initiate synthesis of PLFAs not found in cells grown at higher methane concentrations. However, it was not possible to obtain fingerprints comparable to the 14C-PLFA fingerprints measured in the soil samples. The PLFA composition of M. trichosporium OB3b cells grown on CH3OH rather than CH4 was also examined. The composition of CH3OH-grown methanotrophs was evaluated because this substrate has been suggested recently as a potential cosubstrate for methanotrophs that oxidize atmospheric methane (4, 17, 24). However, the fingerprints of the methanol-grown cells were comparable to those of methane-grown cells and did not match the fingerprints of the unknown soil methanotrophs.
The above findings do not completely exclude the possibility that known type II methanotrophs produce a PLFA fingerprint comparable to the fingerprints of the active soil methanotrophs. However, we find it more likely that the methanotrophs responsible for oxidation of atmospheric methane in soils represent a novel group of closely related methanotrophic bacteria. At the PLFA level, these organisms are likely related to the type II methanotrophs Methylosinus and Methylocystis. The presence of novel methanotrophic bacteria in soils with active oxidation of atmospheric methane is supported by several observations, including the unusual kinetic characteristics of the process, the unusual starvation responses, the unusual responses to water stress, and the unusual pH optimum of the organisms (1, 8, 19, 24, 27). The presence of novel methanotrophs in soils that consume atmospheric methane is also supported by direct sequence analysis of monooxygenase gene libraries (16). On the basis of these analyses, novel groups of methane-oxidizing bacteria related to the type II methanotrophs have been detected in three of the soils examined in the present study (16). The likely contribution of novel methanotrophs is also supported by a recent study in which a novel high-affinity methane-oxidizing bacterium was characterized (9).
In conclusion, the above findings suggest that novel methane-oxidizing microorganisms related to the type II methanotrophs are responsible for oxidation of atmospheric methane in soils. 14C-PLFA analysis of soil samples from Arctic, temperate, and tropical regions suggests that these methanotrophs are closely related. No evidence that indicated any quantitative role of autotrophic ammonia oxidizers in the oxidation of atmospheric methane in the soils was found. The novel methane oxidizers produced significant amounts of unusual methanotrophic PLFAs. The unusual methanotrophic PLFAs produced by the novel soil methanotrophs may have potential as a biomarker(s) in comparisons with known methanotrophic bacteria.
ACKNOWLEDGMENTS
We thank Kirsten Maagaard for excellent technical assistance and Søren O. Petersen, Kaj Henriksen, and Nanna Høegh for valuable input. We also thank the following people for collecting soil samples: Niels Kildemark, Søren Roslev Kristensen, Torben Røjle Christensen, and the Brute Boar Tour of 1996.
This work was supported by European Commission grant BIO4-CT96-0419 and Danish Technical Research Council grants 9502651 and 9701310.
REFERENCES
- 1.Amaral J A, Ren T, Knowles R. Atmospheric methane consumption by forest soils and extracted bacteria at different pH values. Appl Environ Microbiol. 1998;64:2397–2402. doi: 10.1128/aem.64.7.2397-2402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender M, Conrad R. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol Ecol. 1992;101:261–270. [Google Scholar]
- 3.Benstead J, King G M. Response of methanotrophic activity in forest soil to methane availability. FEMS Microbiol Ecol. 1997;23:333–340. [Google Scholar]
- 4.Benstead J, King G M, Williams H G. Methanol promotes atmospheric methane oxidation by methanotrophic cultures and soils. Appl Environ Microbiol. 1998;64:1091–1098. doi: 10.1128/aem.64.3.1091-1098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borjeson G, Sundh I, Tunlid A, Svenson B H. Methane oxidation in landfill cover soils as revealed by potential oxidation measurements and phospholipid fatty acid analyses. Soil Biol Biochem. 1998;30:1423–1433. [Google Scholar]
- 6.Bowman J P, Skerratt J H, Nichols P D, Sly L I. Phospholipid fatty acid and lipopolysaccharide fatty acid signature lipids in methane-utilizing bacteria. FEMS Microbiol Ecol. 1991;85:15–22. [Google Scholar]
- 7.Castro M S, Peterjohn W T, Melillo J M, Steudler P A, Gholz H L, Lewis D. Effect of nitrogen fertilization on the fluxes of N2O, CH4, and CO2 from soils in a Florida slash pine plantation. Can J For Res. 1994;24:9–13. [Google Scholar]
- 8.Conrad R. Soil microbial processes involved in production and consumption of atmospheric trace gases. In: Gwynfryn Jones J, editor. Advances in microbial ecology. New York, N.Y: Plenum Press; 1995. pp. 207–250. [Google Scholar]
- 9.Dunfield P F, Liesack W, Henckel T, Knowles R, Conrad R. High-affinity methane oxidation by a soil enrichment culture containing a type II methanotroph. Appl Environ Microbiol. 1999;65:1009–1014. doi: 10.1128/aem.65.3.1009-1014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guckert J B, Ringelberg D B, White D C, Hanson R S, Bratina B J. Membrane fatty acids as phenotypic markers in the polyphasic taxonomy of methylotrophs within the proteobacteria. J Gen Microbiol. 1991;137:2631–2641. doi: 10.1099/00221287-137-11-2631. [DOI] [PubMed] [Google Scholar]
- 11.Gulledge J, Doyle A P, Schimel J P. Different NH4+ inhibition patterns of soil CH4 consumption: a result of distinct CH4 oxidizer populations across sites? Soil Biol Biochem. 1997;29:13–21. [Google Scholar]
- 12.Gulledge J, Schimel J P. Low-concentration kinetics of atmospheric CH4 oxidation in soil and mechanism of NH4+ inhibition. Appl Environ Microbiol. 1998;64:4291–4298. doi: 10.1128/aem.64.11.4291-4298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulledge J, Steudler P A, Schimel J P. Effect of CH4-starvation on atmospheric CH4 oxidizers in taiga and temperate forest soils. Soil Biol Biochem. 1998;30:1463–1467. [Google Scholar]
- 14.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harder J. Anaerobic methane oxidation by bacteria employing 14C-methane uncontaminated with 14C-carbon monoxide. Mar Geol. 1997;137:13–23. [Google Scholar]
- 16.Holmes A J, Roslev P, McDonald I R, Iversen N, Henriksen K, Murrell J C. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ, Microbiol. 1999;65:3312–3318. doi: 10.1128/aem.65.8.3312-3318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen S, Priemé A, Bakken L. Methanol improves methane uptake in starved methanotrophic microorganisms. Appl Environ Microbiol. 1998;64:1143–1146. doi: 10.1128/aem.64.3.1143-1146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King G M. Ecological aspects of methane oxidation, a key determinant of global methane dynamics. Adv Microb Ecol. 1992;12:431–469. [Google Scholar]
- 19.King G M. Ecophysiological characteristics of obligate methanotrophic bacteria and methane oxidation in situ. In: Murrell J C, Kelly D P, editors. Microbial growth on C1 compounds. Andover, United Kingdom: Intercept; 1993. pp. 303–313. [Google Scholar]
- 20.Murrell C, McDonald I R, Bourne D G. Molecular methods for the study of methanotroph ecology. FEMS Microbiol Ecol. 1998;27:103–114. [Google Scholar]
- 21.Nichols P D, Smith G A, Antworth C P, Hanson R S, White D C. Phospholipid and lipopolysaccharide normal and hydroxy fatty acids as potential signatures for methane-oxidizing bacteria. FEMS Microbiol Ecol. 1985;31:327–335. [Google Scholar]
- 22.Rasmussen, L. H., N. Iversen, and P. Roslev. Characterization of two methane oxidation activities in soil. Unpublished data.
- 23.Ratledge C, Wilkinson S G, editors. Microbial lipids. Vol. 1. London, United Kingdom: Academic Press; 1988. [Google Scholar]
- 24.Roslev P, Iversen N, Henriksen K. Oxidation and assimilation of atmospheric methane by soil methane oxidizers. Appl Environ Microbiol. 1997;63:874–880. doi: 10.1128/aem.63.3.874-880.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roslev P, Iversen N, Henriksen K. Direct fingerprinting of metabolically active bacteria in environmental samples by substrate specific radiolabelling and lipid analysis. J Microbiol Methods. 1998;31:99–111. [Google Scholar]
- 26.Roslev P, Madsen P L, Thyme J B, Henriksen K. Degradation of phthalate and di-(2-ethylhexyl)phthalate by indigenous and inoculated microorganisms in sludge-amended soil. Appl Environ Microbiol. 1998;64:4711–4719. doi: 10.1128/aem.64.12.4711-4719.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schnell S, King G M. Response of methanotrophic activity in soils and cultures to water stress. Appl Environ Microbiol. 1996;62:3203–3209. doi: 10.1128/aem.62.9.3203-3209.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steudler P A, Jones R D, Castro M S, Melillo J M, Lewis D. Microbial controls of methane oxidation in temperate forest and agricultural soils. In: Murrell J C, Kelly D P, editors. The microbiology of atmospheric trace gases. Berlin, Germany: Springer-Verlag; 1996. pp. 69–84. [Google Scholar]
- 29.Sundh I, Borgå P, Nilsson M, Svensson B H. Estimation of cell numbers of methanotrophic bacteria in boreal peatlands based on analysis of specific phospholipid fatty acids. FEMS Microbiol Ecol. 1995;18:103–112. [Google Scholar]