Abstract
Mating type in the Gibberella fujikuroi species complex is controlled by a single locus with two alleles and is usually identified following sexual crosses with standard, female-fertile tester isolates. The mating type alleles have been arbitrarily designated “+” and “−” within each biological species, and the nomenclature is tied to the standard tester strains. We developed a pair of PCR primers that can be used to amplify a unique fragment of one of the mating type alleles (MAT-2) from at least seven of the biological species in this species complex. Based on the amplification pattern, we propose a replacement for the existing, arbitrary +/− terminology that is presently in use. The new terminology is based on DNA sequence similarities between the mating type allele fragments from the biological species of the G. fujikuroi species complex and the corresponding fragments from other filamentous ascomycetes.
Gibberella fujikuroi is a complex of at least eight different mating populations (biological species) that are reproductively isolated and are denoted by letters (e.g., G. fujikuroi mating population A). Recently, some of these mating populations have been given Gibberella species names that parallel the names of their Fusarium anamorphs, and additional anamorphs have been identified (4, 19, 22, 34). These species have a worldwide distribution and have been recovered from hosts as diverse as banana, fig, maize, mango, pine, rice, sorghum, and sugarcane (26). These species also produce a variety of secondary metabolites and mycotoxins, such as fumonisins (28, 29, 41), gibberellic acid (43), moniliformin (28), fusaric acid (3), beauvericin (30), and fusaproliferin (37), that can contaminate animal feed and human food.
Mating type in filamentous ascomycetes is usually dimictic, with one locus and two functional alleles (9), often designated MAT-1 and MAT-2 following the convention proposed by Yoder et al. (51). The alleles are idiomorphs, which means that they share no significant sequence similarity but do map to the same position on homologous chromosomes (5). Mating type genes have been cloned and sequenced from several filamentous ascomycetes, including Neurospora crassa (11), Podospora anserina (7), Cochliobolus heterostrophus (45), and Magnaporthe grisea (17). Although there is little MAT sequence similarity across broad evolutionary distances (44), there is enough sequence similarity in the HMG-box region of the MAT-2 allele to permit identification of this region by PCR methods.
Mating type has not been well-characterized at the molecular level in any of the mating populations (biological species) of the G. fujikuroi species complex. Mating type in these fungi is dimictic and is usually denoted by a base letter (A through H) that specifies the mating population and a superscript “+” or “−” that specifies the mating type. The + and − designations were arbitrarily assigned by the researchers who described the mating populations, and the + allele from one mating population may or may not be the same at the molecular level as the + allele from another.
Arie et al. (2) designed degenerate PCR primers that could be used to amplify the HMG box of the MAT-2 region of various ascomycetes, including representative species of the genera Cryphonectria, Gaeuemannomyces, Mycosphaerella, Nectria, Neurospora, Pyrenophora, and Setosphaeria and 18 species of the genus Cochliobolus. Our objectives in this study were (i) to determine if the primer sets developed by Arie et al. (2) could be used to amplify the MAT-2 HMG box from strains in the different mating populations of the G. fujikuroi species complex, (ii) to develop PCR primers that are specific for the MAT-2 allele in this species complex, and (iii) to replace the arbitrary +/− terminology presently in use with terminology that is consistent with known molecular characteristics and parallels the terminology used for other filamentous ascomycetes.
(A preliminary report of the results has been presented previously [18].)
MATERIALS AND METHODS
Strains.
Except for the progeny in the mapping population used by Xu and Leslie (48, 49) to construct a genetic map for G. fujikuroi mating population A, all of the strains which we used are listed in Table 1. Strains were routinely cultured on minimal medium (6), if they had no auxotrophic requirements, or on complete medium (6), if additional nutritional supplements were needed. All incubations were performed at 25°C under a 12 h of light-12 h of darkness diurnal cycle. Strains were preserved for long-term storage as spore suspensions in a glycerol-water mixture (15:85) frozen at −70°C.
TABLE 1.
Fusarium strains used in this study
| Straina | Mating type
|
Origin | Supplierd | Other designation(s)e | Reference(s)f | |
|---|---|---|---|---|---|---|
| Previous studiesb | This studyc | |||||
| G. fujikuroi mating population A | ||||||
| A-00015 | matA+ | MATA-2 | Laboratory cross | JEP | FGSC 6895 | 48 |
| A-00149g | matA− | MATA-1 | Sorghum, California | PTS | M-3125, FGSC 7600 | 4, 16, 47, 48, 50 |
| A-00488 | matA− | MATA-1 | Maize, Transkei, South Africa | WFOM | M-1325, MRC-826 | 16, 47, 48, 50 |
| A-00501 | matA+ | MATA-2 | Maize, Kansas | JFL | M-6461 | 16, 50 |
| A-00552 | matA− | MATA-1 | Maize, Kansas | JFL | M-6467 | 16, 50 |
| A-00606 | matA− | MATA-1 | Maize, Kansas | JFL | M-6468 | 16, 50 |
| A-00679 | matA+ | MATA-2 | Maize, Kansas | JFL | M-5124 | 16, 50 |
| A-00999g | matA+ | MATA-2 | Maize, Indiana | JFL | M-3703, FGSC 7603, ATCC 201261 | 4, 16, 48, 50 |
| A-01501 | matA− | MATA-1 | Maize, Illinois | JFL | ||
| A-01660 | matA− | MATA-1 | Maize, Beijing, People’s Republic of China | JFL | M5430 | 16, 50 |
| A-04367 | matA− | MATA-1 | Maize, Assyut, Egypt | JFL | 16, 50 | |
| A-04643 | matA− | MATA-1 | Laboratory cross | JFL | FGSC 8078 | 47, 48 |
| G. fujikuroi mating population B | ||||||
| B-00278 | matB+ | MATB-2 | Sugarcane, Taiwan | PTS | M-3127, FGSC 7608, ATCC 201262 | 16, 47, 50 |
| B-00281 | matB− | MATB-1 | Sugarcane, Taiwan | PTS | M-3128, FGSC 7609, ATCC 201263 | 16, 47, 50 |
| B-01722 | matB− | MATB-1 | Sorghum, The Philippines | JFL | M-5476 | 16, 47, 50 |
| B-03852g | matB+ | MATB-2 | Laboratory cross | JFL | M-6865, FGSC 7610, ATCC 201264 | 4, 16, 50 |
| B-03853g | matB− | MATB-1 | Laboratory cross | JFL | M-6866, FGSC 7611, ATCC 201265 | 4, 16, 50 |
| G. fujikuroi mating population C | ||||||
| C-01993g | matC+ | MATC-1 | Rice, Taiwan | EGK | M-1148 | 16, 50 |
| C-01995g | matC− | MATC-2 | Rice, Taiwan | EGK | M-1150 | 16, 50 |
| G. fujikuroi mating population D | ||||||
| D-00502 | matD+ | MATD-2 | Maize, Kansas | JFL | M-6471, FGSC 7612, ATCC 201266 | 16, 47, 50 |
| D-00526 | matD+ | MATD-2 | Maize, Kansas | JFL | M-6480 | 16, 50 |
| D-00550 | matD− | MATD-1 | Maize, Kansas | JFL | M-6482 | 16, 50 |
| D-00637 | matD+ | MATD-2 | Maize, Kansas | JFL | M-6484 | 16, 50 |
| D-00666 | matD− | MATD-1 | Maize, Kansas | JFL | M-5123 | 16, 47, 50 |
| D-00720 | matD− | MATD-1 | Sorghum, Kansas | JFL | M-6485 | 16, 47, 50 |
| D-00741 | matD+ | MATD-2 | Shattercane, Kansas | JFL | M-6492 | 16, 50 |
| D-00753 | matD+ | MATD-2 | Sorghum, Kansas | JFL | M-6495 | |
| D-00875 | matD+ | MATD-2 | Sorghum, Kansas | JFL | M-5128 | 16, 50 |
| D-02945 | matD− | MATD-1 | Sorghum, Mississippi | JFL | M-3793, FGSC 7613, ATCC 201267 | 16, 47, 50 |
| D-04853g | matD+ | MATD-2 | Laboratory cross | JFL | FGSC 7614, ATCC 201268 | 4 |
| D-04854g | matD− | MATD-1 | Laboratory cross | JFL | FGSC 7615, ATCC 201269 | 4 |
| G. fujikuroi mating population E | ||||||
| E-00434 | matE− | MATE-1 | Maize, Kansas | JFL | M-6496 | 16, 47, 50 |
| E-00505 | matE− | MATE-1 | Maize, Kansas | JFL | M-6497 | |
| E-00507 | matE− | MATE-1 | Maize, Kansas | JFL | M-5119 | 16, 47, 50 |
| Previous studiesb | This studyc | |||||
| E-00520 | matE− | MATE-1 | Maize, Kansas | JFL | M-6498 | 16, 50 |
| E-00551 | matE− | MATE-1 | Maize, Kansas | JFL | M-6501 | 16, 47, 50 |
| E-00990g | matE− | MATE-1 | Maize, Illinois | JFL | M-3696, FGSC 7616, ATCC 201270 | 4, 16, 34, 47, 50 |
| E-02192g | matE+ | MATE-2 | Maize, Illinois | JFL | M-3693, FGSC 7617, ATCC 201271 | 4, 16, 47, 50 |
| G. fujikuroi mating population F | ||||||
| F-00728 | matF+ | MATF-1 | Sorghum, Kansas | JFL | M-5598, MRC 5713 | 16, 22, 47, 50 |
| F-00779 | matF− | MATF-2 | Sorghum, Arkansas | JFL | MRC 6477 | 21, 22 |
| F-00921 | matF− | MATF-2 | Sorghum, Kansas | JFL | M-5132, MRC 5708 | 21, 22, 50 |
| F-00965 | matF+ | MATF-1 | Sorghum, Kansas | JFL | M-5134, MRC 5707 | 16, 22, 47, 50 |
| F-01054 | matF− | MATF-2 | Sorghum, Kansas | JFL | M-5594, MRC 5709 | 16, 22, 50 |
| F-01087 | matF− | MATF-2 | Sorghum, Kansas | JFL | M-5595, MRC 5710 | 16, 22, 50 |
| F-01106 | matF− | MATF-2 | Sorghum, Kansas | JFL | M-5589, MRC-5703 | 16, 22, 50 |
| F-01137 | matF+ | MATF-1 | Sorghum, Kansas | JFL | M-5596 | 16, 22, 50 |
| F-01183 | matF+ | MATF-1 | Sorghum, Kansas | JFL | M-5590, MRC 5704 | 16, 22, 50 |
| F-01321 | matF+ | MATF-1 | Sorghum, Kansas | JFL | M-5591, MRC 5705 | 16, 22, 50 |
| F-04093g | matF− | MATF-2 | Laboratory cross | JFL | M-6563, FGSC 7056, ATCC 200521 | 4, 21, 22, 34 |
| F-04094g | matF+ | MATF-1 | Laboratory cross | JFL | M-6564, FGSC 7057, ATCC 200522 | 21, 22 |
| G. fujikuroi mating population G | ||||||
| G-05111g | matG+ | MATG-1 | Laboratory cross | PEN | M-7492, ATCC 201272 | 16, 19 |
| G-05112g | matG− | MATG-2 | Laboratory cross | PEN | M-7491, ATCC 201273 | 16, 19 |
| G. zeae | ||||||
| Z-10588 | Homo-thallic | Andropogon gerardii, Kansas | KAZ | |||
| Z-10628 | Homo-thallic | Andropogon scoparius, Kansas | KAZ | |||
| F. oxysporum | ||||||
| O-02332 | MAT-2 | Banana, Malaysia | BS | |||
| O-02520 | MAT-1(?) | Celery, California | JEP | O-1 | 6 | |
| O-02523 | MAT-1(?) | Chrysanthemum, Florida | PEN | O-734 | 6 | |
| O-02527 | MAT-1(?) | Tomato, Florida | PEN | O-1078 | 6 | |
| O-02529 | MAT-1(?) | Watermelon, Florida | PEN | O-1087 | 6 | |
| O-02530 | MAT-2 | Tomato, Ontario, Canada | PEN | O-1090 | 6 | |
| O-02533 | MAT-2 | Cotton, Mississippi | PEN | O-1139 | 6 | |
| O-02534 | MAT-2 | Banana, Queensland, Australia | PEN | O-1222 | 6 | |
Kansas State University Strain Collection designation.
Strains that have only a MAT-1 or MAT-2 designation have not been proven to be fertile in a sexual cross.
BS, Baharuddin Salleh; JEP, John E. Puhalla; JFL, John F. Leslie; KAZ, Kurt A. Zeller; PEN, Paul E. Nelson; PTS, Philip T. Spieth; WFOM, Walter F. O. Marasas.
ATCC, American Type Culture Collection, Manassas, Va.; FGSC, Fungal Genetics Stock Center, University of Kansas Medical Center, Kansas City; M- and O-, Fusarium Research Center, Pennsylvania State University, University Park; MRC, PROMEC, Medical Research Council, Tygerberg, South Africa.
References cited by Yan et al. (50) are not included.
Standard strain for the mating type.
Sexual crossing protocol.
Crosses were made on carrot agar (20) by using standard tester strains (Table 1) as the female parents. Fertile crosses produced perithecia that exuded a cirrhus of ascospores 2 to 4 weeks after fertilization.
DNA manipulation.
DNA was isolated by a cetyltrimethylammonium bromide (CTAB) procedure modified from the procedure of Murray and Thompson (32). Briefly, a frozen sample (approximately 1.5 to 2 g [wet weight]) was ground to a fine powder under liquid nitrogen with a mortar and pestle. Then we added 8 ml of hot (65°C) 2% CTAB buffer (2% CTAB, 100 mM Tris-HCl [pH 8.0], 20 mM EDTA, 1.4 M NaCl). We incubated the sample at 65°C for 30 min, extracted the preparation once with an equal volume of chloroform-isoamyl alcohol (24:1), removed the aqueous layer, and precipitated the crude nucleic acids with an equal volume of 2-propanol. We dissolved the pellet in 1 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 7.4) and extracted it sequentially with equal volumes of phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1). The last extract (approximately 600 μl) was treated with 1.5 μl of RNase (2 mg/ml), and the remaining nucleic acids were precipitated with either 2.5 volumes of 95% ethanol or 1 volume of 2-propanol. The resulting pellet was washed twice with 70% ethanol, dried briefly, and then resuspended in 50 to 100 μl of TE buffer. We estimated final DNA concentrations by using HindIII-digested bacteriophage λ DNA and an Applied Biosystems model IS-1000 digital imaging system (Alpha Inotech Corp., San Leandro, Calif.); samples and sample dilutions were electrophoresed in 1% agarose gels.
Plasmid isolation, restriction digestion, ligation, bacterial transformation, and Southern hybridization were performed by using standard procedures (38). Agarose gels containing the amplification products were capillary blotted onto a Hybond N+ membrane (Amersham International, Amersham, United Kingdom) and probed with DNA fragments labeled with [α-32P]dCTP. Stringent conditions were used for Southern hybridization; i.e., the membranes were washed first in 2× SSC–0.1% sodium dodecyl sulfate at 25°C for 20 min and then two times in 0.1× SSC–0.1% sodium dodecyl sulfate at 65°C for 20 min (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
PCR products amplified from G. fujikuroi A-00999 were extracted from an agarose gel with QIAEX II (Qiagen Inc., Chatsworth, Calif.) by following the manufacturer’s instructions. Purified DNA fragments were blunt-end ligated into the EcoRV site of plasmid pBluescript KS II+ (Stratagene, La Jolla, Calif.).
Amplification conditions.
We used degenerate oligonucleotide primers NcHMG1 and NcHMG2 (2) to amplify the putative HMG-box from the G. fujikuroi standard mating type tester strains. The PCR mixtures (50 μl) contained 1× PCR buffer (Promega, Madison, Wis.), 20 ng of fungal DNA, 2.5 mM MgCl2, each deoxynucleoside triphosphate (Promega) at a concentration of 0.25 mM, each degenerate primer at a concentration of 2 μM, and 1 U of Taq DNA polymerase (Promega). For amplification we used the following program: initial denaturation at 95°C for 3 min, followed by 30 cycles consisting of 1 min at 94°C, 30 s at 55°C, and 1 min at 72°C and a final elongation incubation step consisting of 72°C for 10 min. PCR amplification products were separated by electrophoresis in 1.5% (wt/vol) agarose gels, stained with ethidium bromide, and visualized with UV light.
We designed two Gibberella-specific primers, GfHMG1 (5′-ACCGTAAGGAGCGTCACCATT-3′) and GfHMG2 (5′-GGGGTACTGTCGGCGATGTT-3′), based on the sequence of the 262-bp putative HMG-box amplification product obtained from G. fujikuroi A-00999 (EMBL accession no. AJ 131527). These specific primers were used to confirm the results obtained from amplifications performed with the degenerate primers, to identify the mating types of previously untested field isolates, and to confirm mating type assignments made by classical genetic analysis of the progeny of laboratory crosses. When the specific primers replaced the degenerate primers, we altered the amplification protocol by increasing the annealing temperature from 55 to 60°C and by reducing the primer concentration from 2 to 0.1 μM.
Sequencing protocol.
DNA was sequenced with a Sequenase kit (version 2.0; United States Biochemicals, Cleveland, Ohio). Nucleotide sequences were compared by using the GCG software package (8).
RESULTS
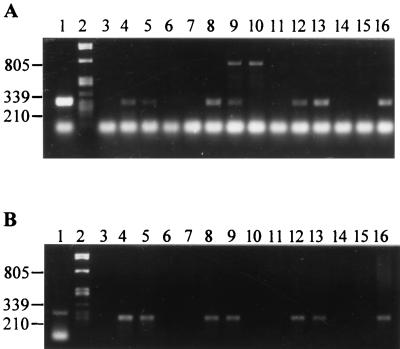
We used two degenerate oligonucleotide primers, NcHMG1 and NcHMG2 (2), to amplify the MAT-2 HMG box from the tester pairs of mating populations A through G of the G. fujikuroi species complex (Fig. 1A). All of the strains produced a bright band consisting of unincorporated primers, and the two G. fujikuroi mating population D strains each produced an additional amplification product of approximately 800 bp. As the 800-bp amplification product was found in strains of both mating types, this amplification product is not specifically associated with either mating type idiomorph. Another PCR product, which was approximately 260 bp long, was clearly amplified from one member of each of the tester pairs used for mating populations A, C, E, F, and G but not from their counterparts. A similar amplification product was obtained from the representatives of mating populations B and D, but in each case the band was less intense than the bands obtained for the other five mating populations.
FIG. 1.
(A) PCR amplification of a putative HMG-box sequence from mating type testers of the G. fujikuroi species complex with degenerate primers NcHMG1 and NcHMG2. Lane 1, N. crassa FGSC 4066; lane 2, λ phage DNA digested by PstI; lanes 3 through 16, G. fujikuroi A-00149, A-00999, B-03852, B-03853, C-01993, C-01995, D-04853, D-04854, E-00990, E-02192, F-04093, F-04094, G-05111, and G-05112, respectively. (B) PCR amplification of the HMG box with Gibberella-specific primers GfHMG1 and GfHMG2. Lanes 2 to 16 are amplification products from the same source as in panel A. Lane 1, HMG-box sequence amplified from G. fujikuroi A-00999 by PCR with the NcHMG1 and NcHMG2 primers.
We cloned and sequenced the 262-bp fragment amplified from G. fujikuroi A-00999. The nucleotide sequence of this fragment, including the intron, was 58 and 62% similar to the nucleotide sequences of the HMG boxes in Neurospora crassa mt a (39) and Podospora anserina FPR1 (7), respectively. The G. fujikuroi mating population A sequence also contains a putative intron (12) at a conserved site between bp 92 and 138.
We used the nucleotide sequence of the putative MAT-2 HMG box from strain A-00999 to design two primers to be used to amplify the homologous fragment from any MAT-2 strain of the G. fujikuroi species complex. Primer GfHMG1 corresponds to bases 29 to 49 at the 5′ end of the HMG box, and primer GfHMG2 corresponds to bases 223 to 252 at the 3′ end of the HMG box. When we used this pair of primers to amplify PCR products from genomic DNAs of the seven pairs of mating type testers, a single 213-bp PCR product was generated from only one of the mating type tester strains for each mating population (Fig. 1B). We identified PCR amplification products obtained from GfHMG1 and GfHMG2 that corresponded to the MAT-2 allele in strains representing matA+, matB+, matC−, matD+, matE+, matF−, and matG−. We probed Southern blots of these PCR amplification preparations with the 262-bp HMG fragment cloned from A-00999 and observed strong hybridization signals under high-stringency conditions (data not shown). No hybridization was observed when the PCR product amplified from N. crassa was probed under the same conditions (data not shown).
Based on amplification patterns such as those shown in Fig. 1, we assigned MAT-1 and MAT-2 allele designations to the known mating types (Table 2). Independent blind tests of the diagnostic ability of GfHMG1 and GfHMG2 were conducted in both Gödöllő, Hungary, and Manhattan, Kans. We examined 128 progeny (49 matA+ progeny and 79 matA− progeny) from the mapping population of Xu and Leslie (48, 49), the 14 standard testers representing the seven G. fujikuroi mating populations, and 48 field strains representing five of the seven mating populations, Fusarium oxysporum, and Gibberella zeae (Table 1). For these tests, the strains were coded such that we knew the mating population to which a strain belonged but did not know its mating type. The results of the PCR amplifications were identical to the results of the sexual crosses with standard strains. Thus, the PCR results were predictive of the crossing results, and the crossing results were predictive of the PCR results. Four of the eight F. oxysporum strains examined had a PCR amplification product indicative of the MAT-2 allele. No PCR amplification products were observed with either G. zeae strain.
TABLE 2.
Correspondence between previous mating type terminology and revised mating type terminology for the G. fujikuroi species complex
| Speciesa | G. fujikuroi mating population | Mating type designation
|
|
|---|---|---|---|
| Previous studies | This study | ||
| Fusarium moniliforme | A | matA+ | MATA-2 |
| (Fusarium verticillioides) (Gibberella moniliformis) | matA− | MATA-1 | |
| Fusarium subglutinans (Fusarium sacchari) | B | matB+ | MATB-2 |
| matB− | MATB-1 | ||
| Fusarium fujikuroi | C | matC+ | MATC-1 |
| matC− | MATC-2 | ||
| Fusarium proliferatum | D | matD+ | MATD-2 |
| matD− | MATD-1 | ||
| Fusarium subglutinans (Gibberella subglutinans) | E | matE+ | MATE-2 |
| matE− | MATE-1 | ||
| Fusarium thapsinum, Gibberella thapsina | F | matF+ | MATF-1 |
| matF− | MATF-2 | ||
| Fusarium nygamai, Gibberella nygamai | G | matG+ | MATG-1 |
| matG− | MATG-2 | ||
Anamorph name (Fusarium species), followed by teleomorph name (Gibberella species) if other than a mating population of G. fujikuroi. The names in parentheses are synonyms.
DISCUSSION
We sequenced and analyzed a 262-bp fragment resulting from PCR amplification of A-00999 DNA by using the NcHMG1 and NcHMG2 primers (2) and then designed two new primers, GfHMG1 and GfHMG2, based on the sequence obtained. Three lines of evidence suggest that the 262-bp fragment is part of one of the MAT alleles of G. fujikuroi. First, the fragment shares significant sequence similarity with N. crassa mt a-1 and P. anserina FPR1. Second, in the 52 field strains belonging to the seven different mating populations examined, the fragment is consistently associated with only one mating type within a mating population. Finally, the presence of the amplified fragment cosegregated with the matA+ mating type and the absence of the fragment cosegregated with the matA− mating type for 128 progeny of the mapping population previously used to construct a G. fujikuroi genetic map (48, 49). On the basis of these classical genetic mapping data, the amplified fragment maps with 95% certainty to a 2.3-map unit region that includes MAT and is unlikely to map more than 1 map unit from MAT if it is not coincident with it.
Based on the genetic mapping data and the correlations observed with mating type in the field strains, we propose that the current nomenclature (e.g., matA+ and matA−) be replaced with new terminology (e.g., MATA-1 and MATA-2) (Table 2). The new terminology includes the mating population with which an allele is associated because of the multiple biological species in the G. fujikuroi species complex and the potential for significant differences in the sequences associated with different biological species. Such differences might be exploited to design primers that are specific for both mating population and mating type. If a single species is identified by name (e.g., Gibberella moniliformis instead of G. fujikuroi mating population A), then the letter in the gene symbol for the mating population is not needed and may be deleted.
We scored MAT-1 as the absence of the MAT-2 amplification product. The accuracy of scoring could be increased if primers were available to amplify a specific fragment from MAT-1 as well, since at the moment, anything that results in no amplification automatically leads to a diagnosis of MAT-1. Development of a system in which positive results are used to identify both MAT alleles (40) is probably essential for studying the molecular basis of homothallism in G. zeae (33) and G. fujikuroi mating population B (4).
The lack of amplification in the G. zeae strains may have several causes. First, the HMG box-containing MAT allele may be absent, as it is in some homothallic Neurospora species (10, 11). Alternatively, the sequence may be present but divergent enough from the sequences of strains in the G. fujikuroi species complex that amplification does not occur. Finally, it is possible that some G. zeae strains have MAT-1 and some have MAT-2 and that the two strains which we examined had only MAT-1. Based on preliminary reports (52), it appears that some G. zeae strains have coding sequences from both MAT-1 and MAT-2. Presumably, the region corresponding to either GfHMG1 or GfHMG2 was altered in the fusion process, which resulted in no amplification in a PCR when this primer pair was used.
We examined both of the strains (B-03852 and B-03853) that Britz et al. (4) identified as occasionally homothallic. One of these strains (B-03852) clearly yielded a MAT-2 PCR amplification product, while the other strain did not. Thus, the homothallism in these strains cannot be due to mating type switching, as has been observed in some yeasts and filamentous fungi (13–15, 31, 36, 42). Molecular scoring of mating types should reduce the amount of effort required to screen field populations for sexual fertility and should increase the efficiency of the process through which new mating populations are identified by 50 to 75%, since only strains known to differ in mating type need to be crossed.
The availability of molecular diagnostic tools for mating type also may enable workers to analyze purportedly asexual fungi, including Fusarium oxysporum and 12 of the 13 new Fusarium taxa recently described by Nirenberg and O’Donnell (34) and Nirenberg et al. (35). There is circumstantial evidence that sexual reproduction occurs in F. oxysporum, and this evidence is in the form of high levels of diversity with respect to the multilocus vegetative compatibility trait (1, 23, 25, 46), especially in populations of putatively nonpathogenic strains. As Leslie and Klein (27) have shown, sexual reproduction need not be frequent to play an important role in the maintenance and generation of genotypic diversity in field populations of fungi. The availability of mating type data should make it easier to identify potentially cross-fertile strains that can be used to test some of these hypotheses.
In conclusion, our work shows that one of the MAT idiomorphs of G. fujikuroi contains an HMG fragment that is similar to a mating type idiomorph found in other model and economically important filamentous ascomycetes. We anticipate that the similarities which have been observed mean that we will be able to apply much of the information obtained from studies of other filamentous ascomycetes, such as Neurospora, Podospora, and Cochliobolus, to members of the G. fujikuroi species complex without any major changes. By standardizing the nomenclature from the present A+/A− form to the MAT-1/MAT-2 form, we hope to improve communication between researchers who study filamentous fungi and to establish a terminology in which all of the mating types are named based on molecular structure rather than the arbitrary choices of numerous researchers.
ACKNOWLEDGMENTS
This research was supported in part by Hungarian State Research Grant OTKA T 019315, by the Kansas Agricultural Experiment Station, and by the International Sorghum and Millet Collaborative Research Support Program (INTSORMIL) USAID.
Footnotes
Contribution no. 99-376-J from the Kansas Agricultural Experiment Station, Manhattan.
REFERENCES
- 1.Appel D J, Gordon T J. Local and regional variation in populations of Fusarium oxysporum from agricultural field soils. Phytopathology. 1994;84:786–791. [Google Scholar]
- 2.Arie T, Christiansen S K, Yoder O C, Turgeon B G. Efficient cloning of ascomycete mating type genes by PCR amplification of the conserved MAT HMG box. Fungal Genet Biol. 1997;21:118–130. [PubMed] [Google Scholar]
- 3.Bacon C W, Porter J K, Norred W P, Leslie J F. Production of fusaric acid by Fusarium species. Appl Environ Microbiol. 1996;62:4039–4043. doi: 10.1128/aem.62.11.4039-4043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britz H, Coutinho T A, Wingfield M J, Marasas W F O, Gordon T J, Leslie J F. Fusarium subglutinans f. sp. pini represents a distinct mating population in the Gibberella fujikuroi species complex. Appl Environ Microbiol. 1999;65:1198–1201. doi: 10.1128/aem.65.3.1198-1201.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coppin E, Debuchy R, Arnaise S, Picard M. Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev. 1997;61:411–428. doi: 10.1128/mmbr.61.4.411-428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correll J C, Klittich C J R, Leslie J F. Nitrate nonutilizing mutants and their use in vegetative compatibility tests. Phytopathology. 1987;77:1640–1646. [Google Scholar]
- 7.Debuchy R, Coppin E. The mating types of Podospora anserina: functional analysis and sequence of the fertilization domains. Mol Gen Genet. 1992;233:113–121. doi: 10.1007/BF00587568. [DOI] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fincham J R S, Day P R, Radford A. Fungal genetics. 4th ed. Berkeley: University of California Press; 1979. [Google Scholar]
- 10.Glass N L, Metzenberg R L, Raju N B. Homothallic Sordariaceae from nature: the absence of strains containing only the a mating type sequence. Exp Mycol. 1990;14:274–289. [Google Scholar]
- 11.Glass N L, Vollmer S J, Staben C, Grotelueschen J, Metzenberg R L, Yanofsky C. DNAs of the two mating-type alleles of Neurospora crassa are highly dissimilar. Science. 1988;241:570–573. doi: 10.1126/science.2840740. [DOI] [PubMed] [Google Scholar]
- 12.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, United Kingdom: IRL Press; 1987. pp. 93–139. [Google Scholar]
- 13.Gutz H, Schmidt H. The genetic basis of homothallism and heterothallism in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Semin Dev Biol. 1990;1:169–176. [Google Scholar]
- 14.Haber J E. A locus control region regulates yeast recombination. Trends Genet. 1998;14:317–321. doi: 10.1016/s0168-9525(98)01501-7. [DOI] [PubMed] [Google Scholar]
- 15.Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huss M J, Campbell C L, Jennings D B, Leslie J F. Isozyme variation among biological species in the Gibberella fujikuroi species complex (Fusarium section Liseola) Appl Environ Microbiol. 1996;62:3750–3756. doi: 10.1128/aem.62.10.3750-3756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang S, Chumley F G, Valent B. Isolation of the mating-type genes of the phytopathogenic fungus Magnaporthe grisea using genomic subtraction. Genetics. 1994;138:289–296. doi: 10.1093/genetics/138.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerényi Z, Leslie J F, Hornok L. Proceedings of the Annual Meeting of the Hungarian Society for Microbiology. 1998. A PCR-based method for the detection of isolates belonging to the mat+ mating type in Gibberella fujikuroi; p. 65. [Google Scholar]
- 19.Klaasen J A, Nelson P E. Identification of a mating population, Gibberella nygamai sp. nov., within the Fusarium nygamai anamorph. Mycologia. 1996;88:965–969. [Google Scholar]
- 20.Klittich C J R, Leslie J F. Nitrate reduction mutants of Fusarium moniliforme (Gibberella fujikuroi) Genetics. 1988;118:417–423. doi: 10.1093/genetics/118.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klittich C J R, Leslie J F. Identification of a second mating population within the Fusarium moniliforme anamorph of Gibberella fujikuroi. Mycologia. 1992;84:541–547. [Google Scholar]
- 22.Klittich C J R, Leslie J F, Nelson P E, Marasas W F O. Fusarium thapsinum (Gibberella thapsina): a new species in section Liseola from sorghum. Mycologia. 1997;89:643–652. [Google Scholar]
- 23.Kondo N, Kodama F, Ogoshi A. Vegetative compatibility groups of Fusarium oxysporum f. sp. adzukicola and nonpathogenic Fusarium oxysporum on adzuki bean isolated from adzuki bean fields in Hokkaido. Ann Phytopathol Soc Jpn. 1997;63:8–12. [Google Scholar]
- 24.Leslie J F. Mating populations in Gibberella fujikuroi (Fusarium section Liseola) Phytopathology. 1991;81:1058–1060. [Google Scholar]
- 25.Leslie J F. Fungal vegetative compatibility. Annu Rev Phytopathol. 1993;31:127–150. doi: 10.1146/annurev.py.31.090193.001015. [DOI] [PubMed] [Google Scholar]
- 26.Leslie J F. Gibberella fujikuroi: available populations and variable traits. Can J Bot. 1995;73:S282–S291. [Google Scholar]
- 27.Leslie J F, Klein K K. Female fertility and mating type effects on effective population size and evolution in filamentous fungi. Genetics. 1996;144:557–567. doi: 10.1093/genetics/144.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leslie J F, Marasas W F O, Shephard G S, Sydenham E W, Stockenström S, Thiel P G. Duckling toxicity and the production of fumonisin and moniliformin by isolates in the A and F mating populations of Gibberella fujikuroi (Fusarium moniliforme) Appl Environ Microbiol. 1996;62:1182–1187. doi: 10.1128/aem.62.4.1182-1187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leslie J F, Plattner R D, Desjardins A E, Klittich C J R. Fumonisin B1 production by strains from different mating populations of Gibberella fujikuroi (Fusarium moniliforme) Phytopathology. 1992;82:341–345. [Google Scholar]
- 30.Logrieco A, Moretti A, Castella G, Kostecki M, Golinski P, Ritieni A, Chelkowski J. Beauvericin production by Fusarium species. Appl Environ Microbiol. 1998;64:3084–3088. doi: 10.1128/aem.64.8.3084-3088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milgroom M G, Lipari S E, Powell W A. DNA fingerprinting and analysis of population structure in the chestnut blight fungus Cryphonectria parasitica. Genetics. 1992;131:297–306. doi: 10.1093/genetics/131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray M G, Thompson W F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson P E, Toussoun T A, Marasas W F O. Fusarium species: an illustrated manual for identification. University Park: Pennsylvania State University Press; 1983. [Google Scholar]
- 34.Nirenberg H I, O’Donnell K. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia. 1998;90:434–458. [Google Scholar]
- 35.Nirenberg H I, O’Donnell K, Kroschel J, Andrianaivo A P, Frank J M, Mubatanhema W. Two new species of Fusarium: Fusarium brevicatenuulatum from the noxious weed Striga asiatica in Madagascar and Fusarium pseudoanthophilum from Zea mays in Zimbabwe. Mycologia. 1998;90:459–464. [Google Scholar]
- 36.Perkins D D. Mating-type switching in filamentous ascomycetes. Genetics. 1987;115:215–216. doi: 10.1093/genetics/115.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritieni A, Fogliano V, Randozzo G, Scarallo A, Logrieco A, Moretti A, Mannina L, Bottalico A. Isolation and characterization of fusaproliferin, a new toxic metabolite from Fusarium proliferatum. Nat Toxins. 1995;3:17–20. doi: 10.1002/nt.2620030105. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Staben C, Yanofsky C. Neurospora crassa a mating-type region. Proc Natl Acad Sci USA. 1990;87:4917–4921. doi: 10.1073/pnas.87.13.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steenkamp E T, Wingfield B D, Coutinho T A, Wingfield M J, Marasas W F O, Leslie J F. PCR-based differentiation of MAT-1 and MAT-2 from Gibberella fujikuroi mating population H. Phytopathology. 1999;89:S75. [Google Scholar]
- 41.Thiel P G, Marasas W F O, Sydenham E W, Shephard G S, Gelderblom W C A, Nieuwenhuis J J. Survey of fumonisin production by Fusarium species. Appl Environ Microbiol. 1991;57:1089–1093. doi: 10.1128/aem.57.4.1089-1093.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thon G, Friis T. Epigenetic inheritance of transcriptional silencing and switching competence in fission yeast. Genetics. 1997;145:685–696. doi: 10.1093/genetics/145.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tudzynski B, Hölter K. Gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet Biol. 1998;25:157–170. doi: 10.1006/fgbi.1998.1095. [DOI] [PubMed] [Google Scholar]
- 44.Turgeon B G. Application of mating-type gene technology to problems in fungal biology. Annu Rev Phytopathol. 1998;36:115–137. doi: 10.1146/annurev.phyto.36.1.115. [DOI] [PubMed] [Google Scholar]
- 45.Turgeon B G, Bohlmann H, Ciuffetti L M, Christiansen S K, Yang G, Schafer W, Yoder O C. Cloning and analysis of the mating-type genes from Cochliobolus heterostrophus. Mol Gen Genet. 1993;238:270–284. doi: 10.1007/BF00279556. [DOI] [PubMed] [Google Scholar]
- 46.Woudt L P, Neuvel A, Sikkema A, van Grinsven M Q J M, de Milliano W A J, Campbell C L, Leslie J F. Genetic variation in Fusarium oxysporum from cyclamen. Phytopathology. 1995;85:1348–1355. [Google Scholar]
- 47.Xu J-R, Yan K, Dickman M B, Leslie J F. Electrophoretic karyotypes distinguish the biological species of Gibberella fujikuroi (Fusarium section Liseola) Mol Plant-Microbe Interact. 1995;8:74–84. [Google Scholar]
- 48.Xu J-R, Leslie J F. A genetic map of Fusarium moniliforme (Gibberella fujikuroi mating population A) Genetics. 1996;143:175–189. doi: 10.1017/s0016672300034066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu J-R, Leslie J F. Strain genotypes of Gibberella fujikuroi mating population A (Fusarium moniliforme) mapping population. Fungal Genet Newsl. 1996;43:61–65. doi: 10.1017/s0016672300034066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan K, Dickman M B, Xu J-R, Leslie J F. Sensitivity of field strains of Gibberella fujikuroi (Fusarium section Liseola) to benomyl and hygromycin B. Mycologia. 1993;85:206–213. [Google Scholar]
- 51.Yoder O C, Valent B, Chumley F. Genetic nomenclature and practice for plant pathogenic fungi. Phytopathology. 1986;76:383–385. [Google Scholar]
- 52.Yun S-H, Arie T, Klein M, Lee S-H, Yoder O C, Turgeon B G. Molecular analyses of mating type loci of Gibberella/Fusarium spp. and Hypocrea spinulosa (Chromocrea spinulosa) Fungal Genet Newsl. 1999;46(Suppl.):93. [Google Scholar]