Significance
In mammals, multiple sperm acrosomal membrane proteins are indispensable for sperm–egg interaction, yet the mechanisms underlying their biosynthesis have remained unknown. This study reveals that mouse 1700029I15Rik stabilizes OST subunits important for N-glycosylation and specifically mediates the processing of acrosomal membrane proteins in the ER of early round spermatids. We propose that during spermiogenesis, the biosynthesis of proteins destined for different subcellular compartments is orchestrated in a spatiotemporal manner. Given that 1700029I15Rik is highly conserved in humans, our discoveries in mice may provide profound insights into the etiology of idiopathic male infertility and the development of a nonhormonal contraceptive approach involving molecular interventions in the biosynthesis of acrosomal membrane proteins.
Keywords: 1700029I15Rik, C11orf94/Frey, sperm–egg fusion, fertilization, male infertility
Abstract
Sperm acrosomal membrane proteins, such as Izumo sperm–egg fusion 1 (IZUMO1) and sperm acrosome-associated 6 (SPACA6), play essential roles in mammalian gamete binding or fusion. How their biosynthesis is regulated during spermiogenesis has largely remained elusive. Here, we show that 1700029I15Rik knockout male mice are severely subfertile and their spermatozoa do not fuse with eggs. 1700029I15Rik is a type-II transmembrane protein expressed in early round spermatids but not in mature spermatozoa. It interacts with proteins involved in N-linked glycosylation, disulfide isomerization, and endoplasmic reticulum (ER)–Golgi trafficking, suggesting a potential role in nascent protein processing. The ablation of 1700029I15Rik destabilizes non-catalytic subunits of the oligosaccharyltransferase (OST) complex that are pivotal for N-glycosylation. The knockout testes exhibit normal expression of sperm plasma membrane proteins, but decreased abundance of multiple acrosomal membrane proteins involved in fertilization. The knockout sperm show upregulated chaperones related to ER-associated degradation (ERAD) and elevated protein ubiquitination; strikingly, SPACA6 becomes undetectable. Our results support for a specific, 1700029I15Rik-mediated pathway underpinning the biosynthesis of acrosomal membrane proteins during spermiogenesis.
Mammalian fertilization is a complicated series of events culminating in the union of a haploid spermatozoon and a haploid egg to produce a diploid zygote. This process is a fundamental and obligatory prerequisite for successful propagation of parental genomes to the next generation. During the journey through the female reproductive tract, spermatozoa undergo the acrosome reaction to expose the acrosomal contents, including hydrolytic enzymes and acrosomal membrane proteins, to gain competence in penetrating the egg zona pellucida (ZP) and fusing with the oolemma (1, 2).
Gamete fusion is a fascinating eukaryotic cell–cell fusion event involving the merger of two morphologically distinct cells from individual organisms of opposite genders. Notwithstanding the decades-long research effort, detailed cellular and molecular mechanisms underlying this unique fusion process remain shrouded in mystery (2). To date, seven sperm proteins, Izumo sperm–egg fusion 1 (IZUMO1) (3, 4), sperm acrosome-associated protein 6 (SPACA6) (5–7), fertilization-influencing membrane protein (FIMP) (8), sperm–oocyte fusion-required 1 (SOF1) (6), transmembrane protein 95 (TMEM95) (6, 9, 10), and DC-STAMP domain-containing 1 and 2 (DCST1 and DCST2) (11, 12), have been demonstrated essential for sperm–egg binding or fusion. Mouse sperm lacking any of these molecules fail to fuse with the oolemma, despite their normal morphology, motility, and the ability to elicit the acrosome reaction. Among these proteins, IZUMO1, SPACA6, TMEM95, DCST1, and DCST2 are transmembrane glycoproteins that initially localize to the acrosomal membrane and translocate to the sperm surface during the acrosome reaction (3, 6, 12, 13).
In the early spermatids, many acrosomal protein precursors are synthesized and processed in the endoplasmic reticulum (ER), trafficked to the Golgi apparatus, and secreted as proacrosomal vesicles that subsequently coalesce to form the acrosome (14). In this study, by harnessing genetics, proteomics, and cell biology approaches, we unraveled that 1700029I15Rik, a type-II transmembrane protein, facilitates the biosynthesis of acrosomal membrane proteins involved in sperm–egg interaction.
Results
1700029I15Rik Is a Testis-Specific Type-II Transmembrane Protein Expressed during Early Spermiogenesis.
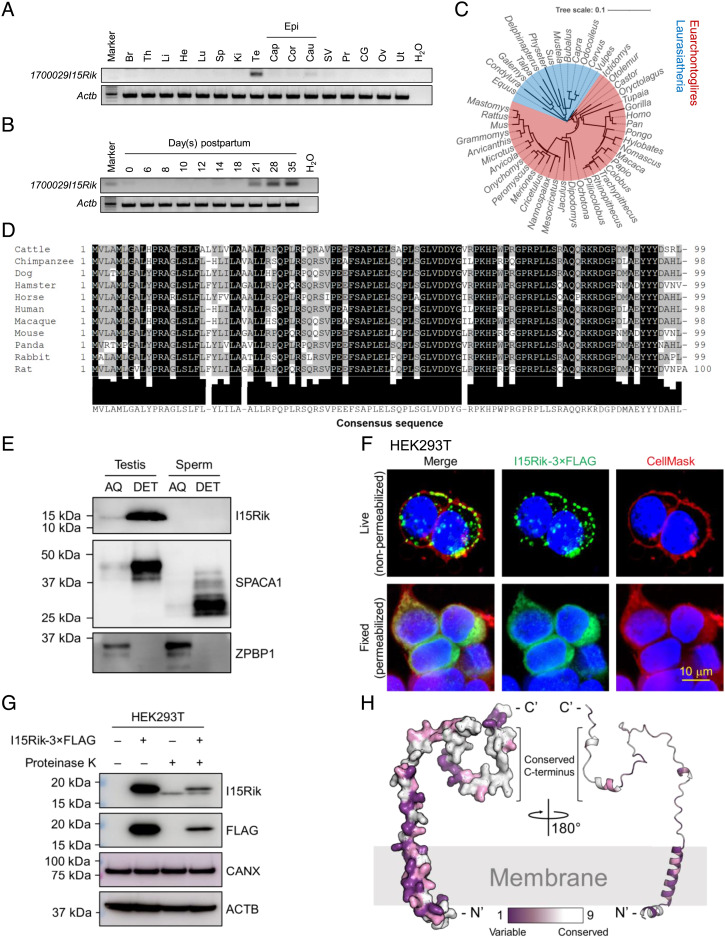
1700029I15Rik encompasses three coding exons and is localized to the forward strand of mouse chromosome 2 (SI Appendix, Fig. S1A). This gene resides on chromosome 11 in humans, thus named C11orf94. RT-PCR revealed that 1700029I15Rik exhibits a testis-biased expression, which is initiated at postnatal day 21 (Fig. 1 A and B), corresponding to the first wave of mouse spermiogenesis. Consistently, a previously published single-cell RNA-sequencing (scRNA-seq) analysis (15) indicated that 1700029I15Rik expresses predominantly in mid-round spermatids (SI Appendix, Fig. S1B). 1700029I15Rik is conserved among mammals (Fig. 1 C and D and SI Appendix, Fig. S1C). In certain vertebrates, such as black flying foxes and green sea turtles, the C terminus of peroxisomal biogenesis factor 16 (PEX16) is highly homologous to 1700029I15Rik (SI Appendix, Fig. S1 D–F). Given that Pex16 is immediately upstream of 1700029I15Rik in the genome (SI Appendix, Fig. S1A), we speculate that these species harbor a Pex16–1700029I15Rik chimeric fusion transcript.
Fig. 1.
1700029I15Rik Is a Testis-Enriched Type-II Transmembrane Protein Conserved in Mammals and Expressed during Spermiogenesis. (A) Analysis of 1700029I15Rik expression in various mouse tissues by RT-PCR. Br, brain; Th, thymus; Li, liver; He, heart; Lu, lung; Sp, spleen; Ki, kidney; Te, testis; Cap, caput epididymis; Cor, corpus epididymis; Cau, cauda epididymis; Epi, epididymis; SV, seminal vesicle; Pr, prostate; CG, coagulating gland; Ov, ovary; Ut, uterus. The expression of β-actin (Actb) was analyzed as a loading control. (B) Analysis of 1700029I15Rik expression in mouse testes during postnatal development. (C) Phylogenetic tree depicting the evolutionary conservation of 1700029I15Rik in mammals. The tree was visualized using the interactive Tree of Life (iTOL) (16). Red and blue highlighted species belong to Euarchontoglires and Laurasiatheria, respectively. (D) Multiple sequence alignment of 1700029I15Rik orthologous proteins in 11 mammalian species. The Lower panel indicates the consensus sequence and the extent of amino acid conservation. (E) Western blot detection of 1700029I15Rik (I15Rik) in mouse testes and sperm fractionated by Triton X-114. SPACA1 and zona pellucida binding protein 2 (ZPBP1) were analyzed as positive controls for the proteins enriched in the AQ and DET phases, respectively. (F) In vitro topological analysis of 1700029I15Rik by live cell immunostaining. HEK293T cells were transiently transfected with a plasmid encoding C-terminal 3 × FLAG-tagged 1700029I15Rik. Live or fixed HEK293T cells were probed with an anti-FLAG antibody and an Alexa Fluor™ 488-conjugated secondary antibody. Cell membranes and nuclei were visualized by CellMask™ deep red plasma membrane stain and Hoechst 33342, respectively. (G) In vitro proteinase K protection assay depicting the topology of 1700029I15Rik. Live HEK293T cells transiently expressing 1700029I15Rik-3 × FLAG were treated with proteinase K and subjected to protein extraction. The levels of 1700029I15Rik-3 × FLAG before and after the enzyme treatment were analyzed by Western blotting. CANX and ACTB were analyzed in parallel as loading controls. (H) 1700029I15Rik protein structure predicted by AlphaFold (AF-Q8CF31-F1) (17). The degree of residue conservation was determined by ConSurf (18) and plotted to the three-dimensional structures, with purple representing variable and white representing conserved.
1700029I15Rik contains a hydrophobic N-terminal region [amino acids (aa) 1 to 29], predicted as a transmembrane helix or a signal peptide by various algorithms (SI Appendix, Table S1 and Fig. S1G). For immunodetection of 1700029I15Rik, we produced a polyclonal antibody against its C terminus [aa 81 to 99 (SI Appendix, Fig. S1G)]. Triton X-114 subcellular fractionation unveiled that 1700029I15Rik is a membrane-bound protein exclusively detected in the detergent (DET) phase of testis lysates (Fig. 1E). To gain insights into the topology of 1700029I15Rik, we transiently transfected human embryonic kidney 293T (HEK293T) cells with a plasmid encoding C-terminal 3 × FLAG-tagged 1700029I15Rik, whose expression was validated by Western blotting (SI Appendix, Fig. S1H). By probing live HEK293T cells with an anti-FLAG antibody and a fluorophore-conjugated secondary antibody, we detected the fluorescence signal of 1700029I15Rik-3 × FLAG at the cell plasma membrane that was concomitantly labeled by CellMask™ plasma membrane stain (Fig. 1F). Likewise, in nonpermeabilized CV-1 in Origin Simian-7 (COS-7) monkey kidney cells, 1700029I15Rik is colocalized with the type-I transmembrane protein, IZUMO1, at the cell surface (SI Appendix, Fig. S1I). Since antibodies cannot diffuse through intact membranes, the C terminus of 1700029I15Rik is likely exposed extracellularly. To confirm this observation, we further treated live HEK293T cells overexpressing 1700029I15Rik-3 × FLAG with proteinase K, which nonspecifically cleaves cell surface proteins (19). After the enzyme treatment, the level of 1700029I15Rik was significantly reduced (Fig. 1G), indicating that the epitopes at the C terminus were proteolyzed. Taken together, these analyses demonstrate that 1700029I15Rik is a type-II transmembrane protein that is anchored to the lipid membrane via its N terminus (Fig. 1H).
1700029I15Rik Knockout Male Mice Are Infertile due to Defective Sperm–Egg Interaction.
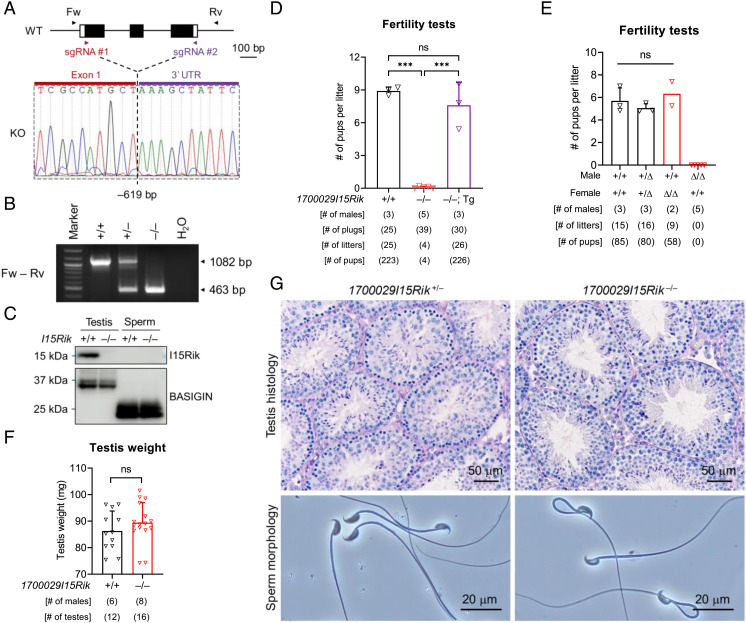
To examine the function of 1700029I15Rik in vivo, we generated a knockout mouse line (1700029I15Rik–/–) on a hybrid background of B6D2F1 by CRISPR/Cas9 (Fig. 2A and SI Appendix, SI Materials and Methods). A 619-bp deletion was introduced to the coding region of 1700029I15Rik, resulting in translation of an incorrect amino acid sequence (Fig. 2 A and B and SI Appendix, Fig. S2A). By Western blotting, we confirmed the absence of 1700029I15Rik in knockout testes (Fig. 2C). 1700029I15Rik is not detectable in wild-type sperm (Figs. 1E and 2C), implying its potential role restricted in spermatogenesis.
Fig. 2.
1700029I15Rik Knockout Males Are Infertile despite Normal Spermatogenesis and Sperm Morphology. (A) CRISPR/Cas9-mediated knockout of 1700029I15Rik in the B6D2F1 hybrid mouse strain. Single guide RNAs (sgRNAs) #1 and #2 were designed to target the first coding exon (Exon 1) and the 3′ untranslated region (UTR), respectively. Forward (Fw) and reverse (Rv) primers flanking the deletion region were employed for genotyping. (B) Genomic PCR for detecting wild-type and 1700029I15Rik knockout alleles. (C) Western blot detection of 1700029I15Rik in wild-type and knockout testes and sperm. BASIGIN was analyzed in parallel as a loading control. (D) Fertility tests of wild-type, 1700029I15Rik–/–, and 1700029I15Rik–/–; Tg male mice. ns, not significant. (E) Fertility tests of wild-type, 1700029I15Rik+/Δ, and 1700029I15RikΔ/Δ mice. The mutant mouse line was produced on an inbred genetic background of C57BL/6J. (F) Analysis of testis weight in wild-type and 1700029I15Rik–/– males. (G) Analyses of testis histology and sperm morphology in 1700029I15Rik+/– and 1700029I15Rik–/– males.
In parallel, we produced 1700029I15Rik mutant mice (1700029I15RikΔ/Δ) lacking 215 bp at the first coding exon on an inbred background of C57BL/6J (SI Appendix, Fig. S2 B and C). Both the knockout and mutant mice show normal development, appearance, and behavior. 1700029I15Rik–/– female mice exhibit normal fecundity, whereas 1700029I15Rik–/– males are severely subfertile despite successful coituses (SI Appendix, Fig. S2 D–F). A transgene (Tg) encoding C-terminal PA-tagged 1700029I15Rik restores the fecundity of knockout males (Fig. 2D and SI Appendix, Fig. S2 G–I), demonstrating that the male infertility is not caused by potential off-target mutations. 1700029I15RikΔ/Δ males are sterile (Fig. 2E), suggesting that the phenotype severity might correlate with the genetic background. The testis weights, spermatogenesis, sperm morphology, and sperm counts are normal in 1700029I15Rik–/– males (Fig. 2 F and G and SI Appendix, Fig. S2J). Similarly, 1700029I15RikΔ/Δ males show no abnormalities in the testis and epididymis histology, as well as the first meiotic division of spermatocytes (SI Appendix, Fig. S2 K–M). To avoid unnecessary duplication of efforts, the subsequent experiments were performed on 1700029I15Rik–/– mice.
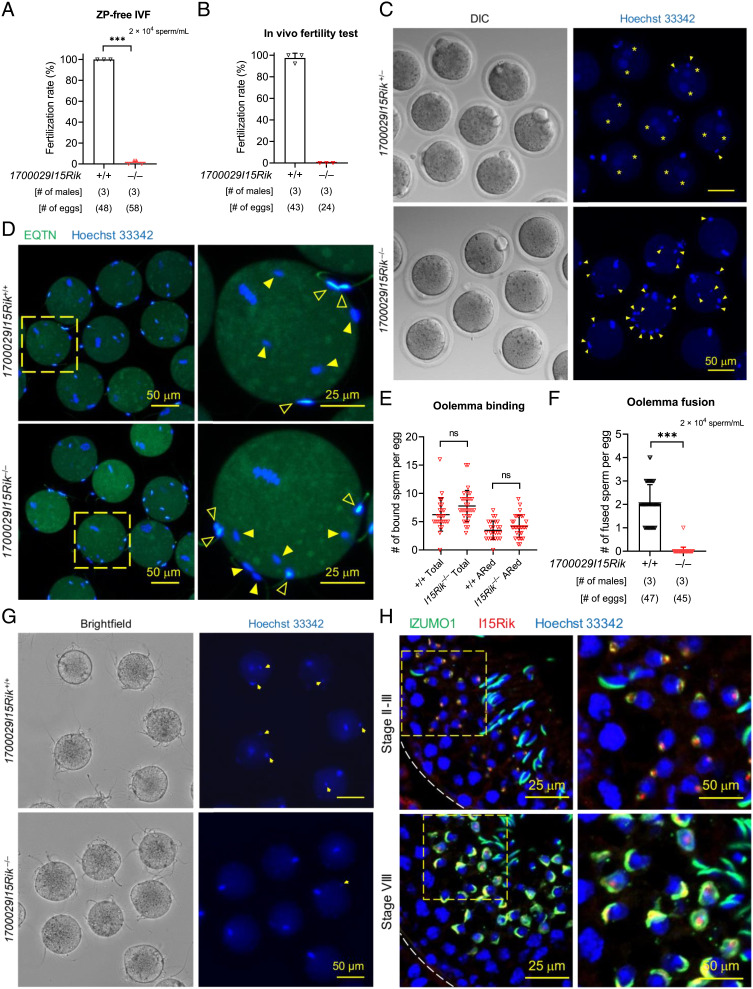
In vitro fertilization analyses revealed that 1700029I15Rik knockout sperm showed significantly reduced ability to fertilize cumulus-intact, cumulus-free, and ZP-free eggs (SI Appendix, Fig. S3 A and B and Fig. 3A). The knockout sperm were unable to fertilize eggs in vivo and accumulated in the perivitelline space, indicating that they underwent the acrosome reaction and penetrated the ZP but failed to fuse with the eggs (Fig. 3 B and C and SI Appendix, Fig. S3C). Through sperm–egg binding and fusion assays, we confirmed that 1700029I15Rik knockout sperm bind but do not fuse with the oolemma (Fig. 3 D–G and SI Appendix, SI Materials and Methods). Despite the impaired fertilizing ability, the knockout sperm exhibited normal motility and ability to bind the ZP (SI Appendix, Fig. S3 D–H).
Fig. 3.
Infertility of 1700029I15Rik Knockout Males Is Attributed to Impaired Sperm–Egg Interaction. (A) In vitro fertilization (IVF) analysis of sperm fertilizing ability using wild-type ZP-free eggs. (B and C) In vivo fertility test of wild-type and 1700029I15Rik–/– males. Eggs were harvested from superovulated B6D2F1 female mice that had copulated with wild-type or 1700029I15Rik knockout males. Sperm in the perivitelline space (yellow arrowheads) and pronuclei in the fertilized eggs (yellow asterisks) were visualized by Hoechst 33342. (D and E) In vitro analysis of sperm–egg binding. Spermatozoa pre-incubated in the Toyoda, Yokoyama, Hoshi (TYH) medium were probed with an anti-EQTN antibody and an Alexa Fluor™ 488-conjugated secondary antibody to reveal the acrosomal status. The acrosome-intact and acrosome-reacted sperm are marked by solid and hollow arrowheads, respectively. Sperm heads were stained with Hoechst 33342. (F and G) In vitro analysis of sperm–egg fusion using Hoechst 33342-preloaded ZP-free eggs. Yellow arrows indicate the fused sperm heads carrying the Hoechst dye transferred from the eggs. (H) Coimmunostaining of IZUMO1 (green) and 1700029I15Rik (red) in wild-type testis cryosections. Cell nuclei were visualized by Hoechst 33342.
Immunohistochemistry revealed that 1700029I15Rik initially appears at the center of the proacrosomal vacuole in steps 2 and 3 spermatids, distinct to IZUMO1 that localizes to the proacrosomal membrane (Fig. 3H). In step 8 spermatids, 1700029I15Rik is partially colocalized with IZUMO1 in the proacrosome. Nevertheless, 1700029I15Rik, but not IZUMO1, was detected in the acrosomal granule (Fig. 3H). From step 9 spermatids, 1700029I15Rik becomes undetectable in the proacrosome (SI Appendix, Fig. S4A). The manchette staining is nonspecific, because the same pattern of staining was observed in the knockout spermatids (SI Appendix, Fig. S4B). Together, these findings suggest that 1700029I15Rik, which is predominantly expressed in early spermatids, indirectly promotes sperm–egg interaction. In agreement with this assumption, HEK293T cells transiently expressing 1700029I15Rik could not bind or fuse with ZP-free mouse eggs (SI Appendix, Fig. S4 C and D).
1700029I15Rik Interacts with ER-Resident Proteins Implicated in Nascent Protein Processing and Stabilizes the OST Complex Subunits In Vivo.
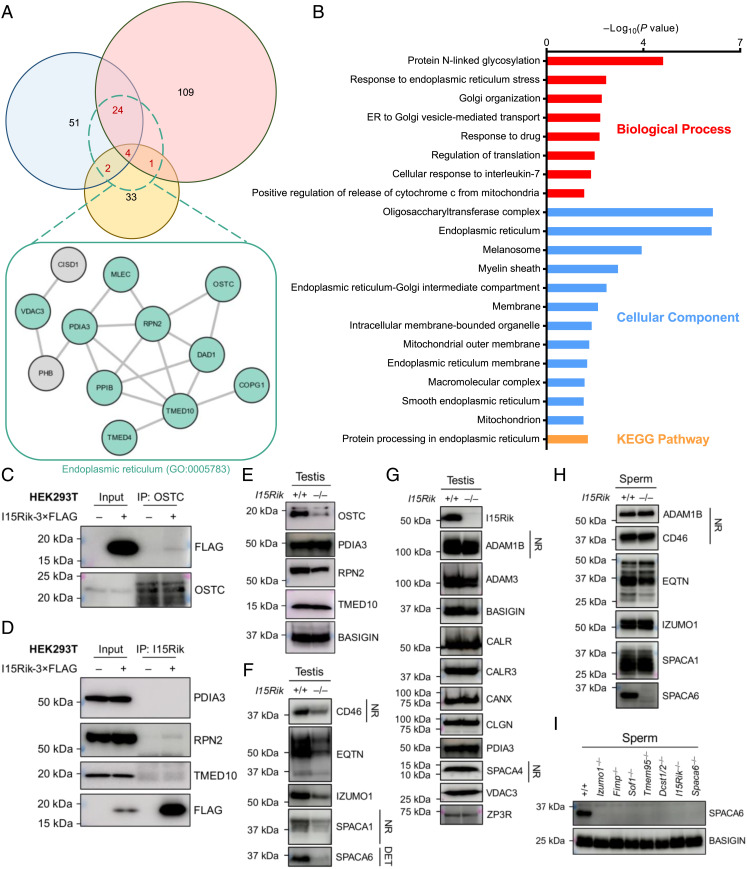
To identify the interactome of 1700029I15Rik in mouse testes, we next performed coimmunoprecipitation tandem mass spectrometry (co-IP/MS) analyses using antibody-crosslinked agarose resin (SI Appendix, Fig. S5A and SI Materials and Methods). As a result, 31 proteins, including 1700029I15Rik, were specifically detected in the wild-type samples from at least two of three biological replicates (Fig. 4 A and Dataset S1). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses revealed that the 31 hits include proteins pivotal for N-glycosylation [e.g., defender against cell death 1 (DAD1), oligosaccharyltransferase complex noncatalytic subunit (OSTC), and ribophorin II (RPN2)] and ER–Golgi vesicular trafficking [e.g., transmembrane p24 trafficking protein 4 and 10 (TMED4 and TMED10); Fig. 4 A and B].
Fig. 4.
1700029I15Rik Interacts with Proteins Involved in N-glycosylation, Disulfide Bond Formation, and Vesicular Trafficking and Facilitates the Expression of Multiple Acrosomal Membrane Proteins. (A) Venn diagram depicting shared and unique protein hits identified in three biological replicates of co-IP/MS experiments. Only the proteins specifically detected in the wild-type samples are included. The proteins detected in at least two replicates were subjected to STRING analysis (20, 21). For each replicate, 1700029I15Rik and its interacting proteins were coimmunoprecipitated from wild-type and knockout testis lysates using antibody-crosslinked agarose resin. (B) GO and KEGG analyses of 1700029I15Rik-interacting proteins. Functional annotations were conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (22). Only the 31 proteins concurrently detected in at least two replicates of the co-IP/MS experiments were analyzed. (C and D) In vitro validation of protein–protein interactions. 1700029I15Rik and its associated proteins were coimmunoprecipitated from the lysates of HEK293T cells transiently expressing 1700029I15Rik-3 × FLAG using antibody-conjugated magnetic beads. The co-IP products were subjected to SDS-PAGE and Western blot analyses to validate protein–protein interactions. (E) Western blot analyses of OSTC, PDIA3, RPN2, and TMED10 in wild-type and knockout testes. BASIGIN was analyzed in parallel as a loading control. (F and G) Western blot analyses of various proteins in wild-type and knockout testes. Unless specified otherwise, all protein samples were processed under reducing and denaturing conditions. NR, non-reducing and non-denaturing. The DET fractions of wild-type and knockout testis proteins, extracted by Triton X-114, were used for immunodetection of SPACA6. (H) Western blot analyses of various membrane proteins in wild-type and knockout sperm. (I) Western blot detection of SPACA6 in the sperm of wild-type, Izumo1–/–, Fimp–/–, Sof1–/–, Tmem95–/–, Dcst1/2–/–,1700029I15Rik–/–, and Spaca6–/– male mice. BASIGIN was analyzed as a loading control.
For determining 1700029I15Rik-interacting proteins with high confidence, another co-IP/MS analysis using non-crosslinked magnetic beads was conducted, and the relevance of protein–protein interactions was scored based on the specificity of detected spectra (SI Appendix, Fig. S5 B–D). Apart from the above-mentioned proteins, signal peptide peptidase like 2C (SPPL2C), protein disulfide isomerase family A member 3 (PDIA3), and voltage-dependent anion channel 3 (VDAC3) are also tightly associated with 1700029I15Rik (SI Appendix, Fig. S5D). SPPL2C is an intramembrane protease, homologous to signal peptide peptidases, and specific for type-II-oriented transmembrane proteins (23), but it is dispensable for male fertility (24). Interestingly, the level of 1700029I15Rik is dramatically decreased in Sppl2c knockout testes (23), suggesting that SPPL2C directly or indirectly governs the stability of 1700029I15Rik. PDIA3 is a protein disulfide isomerase that modulates protein folding, formation and remodeling of disulfide bonds, and quality control of glycoproteins (25). It might be implicated in sperm ZP binding and oolemma fusion, as revealed by previous inhibitory studies (26, 27).
We failed to verify whether OSTC, PDIA3, RPN2, or TMED10 interacts with 1700029I15Rik in testes by co-IP coupled with Western blotting (SI Appendix, Fig. S6A). This is likely owing to the low abundance of immunoprecipitated 1700029I15Rik and its interacting proteins, considering that no unique protein bands were detected in the silver-stained wild-type eluates (SI Appendix, Fig. S5 A and B). Alternatively, we overexpressed 1700029I15Rik-3 × FLAG in HEK293T cells and discovered that 1700029I15Rik interacts with the endogenous OSTC and RPN2, but not PDIA3 or TMED10 (Fig. 4 C and D). Strikingly, OSTC and RPN2, but not PDIA3 or TMED10, are significantly downregulated in the knockout testes (Fig. 4E). Nonetheless, immunohistochemistry revealed no obvious anomalies in the localization of the remaining OSTC in the knockout testes (SI Appendix, Fig. S6B). These findings indicate that 1700029I15Rik interacts with and stabilizes the OST complex subunits in the male germline.
Depletion of 1700029I15Rik Results in Reduced Expression of Multiple Acrosomal Membrane Glycoproteins.
Our co-IP/MS experiments suggest potential involvement of 1700029I15Rik in protein processing and trafficking. Given the impaired fertilizing ability of 1700029I15Rik knockout sperm, we next examined the expression of fertilization-related proteins in testes and sperm by Western blotting. Interestingly, the amounts of acrosomal membrane N-linked glycosylated proteins, including membrane cofactor protein (CD46), equatorin (EQTN), IZUMO1, sperm acrosome-associated 1 (SPACA1), and SPACA6 are drastically reduced in the knockout testes (Fig. 4F and SI Appendix, Fig. S6 C–E). In contrast, the levels of sperm head plasma membrane proteins [e.g., a disintegrin and metallopeptidase domain 1b and 3 (ADAM1B and ADAM3)], ER chaperones [e.g., calreticulin (CALR), calnexin (CANX), and PDIA3], sperm acrosome-associated 4 (SPACA4), a glycosylphosphatidylinositol (GPI)-anchored protein localized to the acrosomal membrane, and zona pellucida 3 receptor (ZP3R), a soluble protein expressed in the acrosomal matrix, are comparable in wild-type and knockout testes (Fig. 4G). Noticeably, the downregulated acrosomal membrane proteins are all highly expressed in mid-round spermatids, whilst the plasma membrane proteins show peak expression in spermatocytes (SI Appendix, Fig. S6F). These findings unveil that 1700029I15Rik may specifically regulate the biosynthesis of acrosomal membrane glycoproteins expressed during spermiogenesis. Co-IP and Western blot analyses indicated that 1700029I15Rik exhibits no detectable interactions with IZUMO1 or EQTN in testes (SI Appendix, Fig. S6G). Transgenic expression of 1700029I15Rik-PA, although at a trace amount, restores the amount of IZUMO1 to the wild-type level (SI Appendix, Fig. S7A). Interestingly, CD46, EQTN, IZUMO1, and SPACA1, which are downregulated in the knockout testes, exhibited normal expression levels in the knockout sperm (Fig. 4H). Moreover, these acrosomal membrane proteins show normal subcellular localization in the knockout testes and sperm (SI Appendix, Fig. S7 B and C). SPACA6, however, is absent in 1700029I15Rik knockout spermatozoa (Fig. 4H and SI Appendix, Fig. S7D), providing a plausible explanation for the defective oolemma fusion ability.
A previous study has indicated that SPACA6 is absent in Izumo1, Dcst1, and Dcst2 knockout sperm (11). Here, we found that SPACA6 is also missing in Tmem95, Fimp, and Sof1 knockout sperm (Fig. 4I). In the sperm of Izumo1–/–; Tg (Izumo1-mCherry) male mice, the amount of transgenically expressed IZUMO1 is lower than that of the wild-type protein. Intriguingly, the transgene does not fully restore SPACA6 to its endogenous level (SI Appendix, Fig. S7E). A similar phenomenon was observed in the sperm of Tmem95–/–; Tg (Tmem95-1D4) males (SI Appendix, Fig. S8A). RT-PCR revealed that the mRNA expression of Spaca6 is normal in Tmem95–/– and Tmem95–/–; Tg testes (SI Appendix, Fig. S8B). Based on these results, we propose that the presence and abundance of SPACA6 is dependent upon that of the other gamete fusion-related proteins. Further, the expression of 1700029I15Rik is normal in Izumo1 and Spaca6 knockout testes (SI Appendix, Fig. S8 C and D). These findings indicate that the loss of SPACA6 in 170029I15Rik knockout males may be directly caused by a lack of the 1700029I15Rik-mediated protein biosynthesis pathway, or indirectly induced by aberrant expression of other gamete fusion-related acrosomal membrane proteins.
1700029I15Rik Knockout Sperm Show Elevated Levels of ER Chaperones and Ubiquitination.
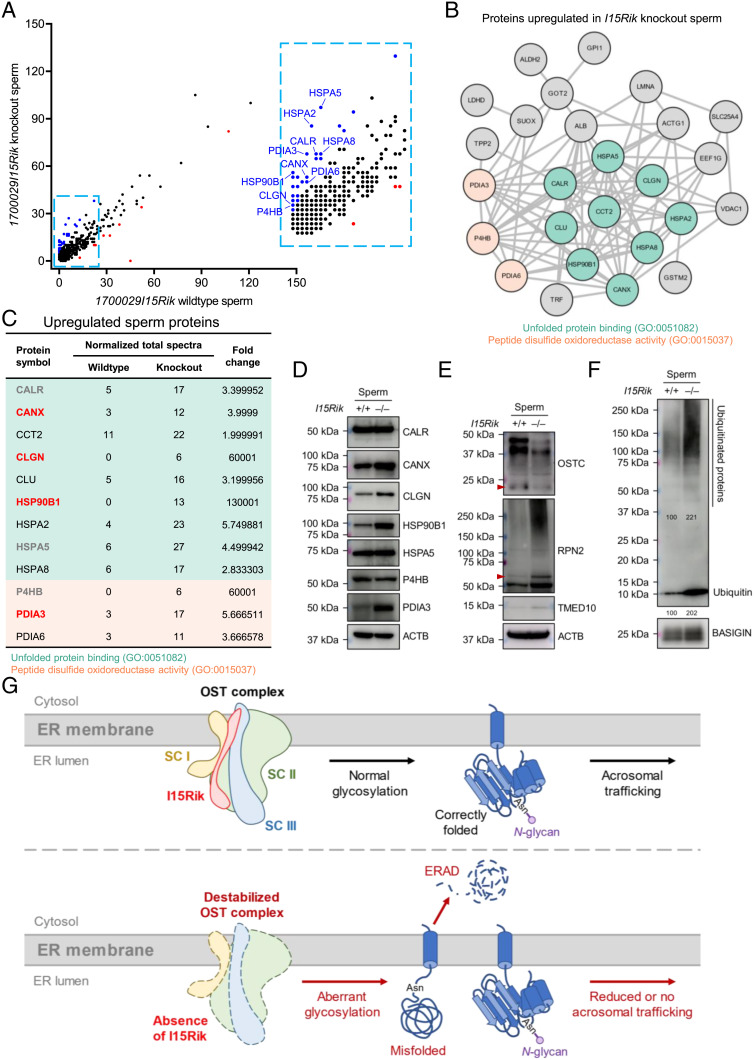
We hypothesize that apart from SPACA6, there might be other acrosomal membrane glycoproteins downregulated in response to the absence of 1700029I15Rik. Considering that whole testis lysates are extremely heterogeneous that impedes the detection of low-abundance proteins by proteomics, we performed an MS analysis to compare the proteome of wild-type and knockout spermatozoa. As a result, we detected 35 proteins significantly upregulated and nine proteins significantly downregulated in the knockout sperm (Fig. 5A). GO and STRING analyses indicated that most of the upregulated proteins are ER chaperones that bind unfolded or misfolded proteins and coordinate the ubiquitin-dependent ER-associated degradation (ERAD) pathway [GO:0051082 and GO:0030433; e.g., CANX, CALR, and heat shock protein 90 beta family member 1 (HSP90B1)], or catalyze disulfide formation or remodeling [GO:0015037; e.g., prolyl 4-hydroxylase subunit beta (P4HB), PDIA3, and protein disulfide isomerase family A member 6 (PDIA6)] (Fig. 5 B and C and SI Appendix, Fig. S8E). The nine proteins downregulated in the knockout sperm, however, do not show distinct commonalities in their molecular functions; yet, acrosin (ACR), dipeptidase 3 (DPEP3), and inactive serine protease 39 (PRSS39), are associated with the acrosomal vesicle (GO:0001669; SI Appendix, Fig. S8F). Notably, the levels of EQTN, IZUMO1, and SPACA1 are slightly but non-significantly reduced in the knockout sperm, compared with those in wild-type sperm (SI Appendix, Fig. S8G). By Western blotting, we confirmed the upregulation of CANX, CLGN, HSP90B1, and PDIA3 in 1700029I15Rik knockout spermatozoa; however, the levels of CALR, heat shock protein 5 (HSPA5), and P4HB are comparable between wild-type and knockout groups (Fig. 5D). Regarding the potential interactome of 1700029I15Rik, RPN2 and TMED10 are upregulated, whilst OSTC shows reduced abundance in the knockout sperm (Fig. 5E).
Fig. 5.
Ablation of 1700029I15Rik Results in Upregulated ER Chaperones and Ubiquitinated Proteins in the Sperm. (A) MS analysis of wild-type and 1700029I15Rik knockout sperm proteome. Blue and red dots indicate proteins upregulated and downregulated in the knockout spermatozoa, respectively. (B) STRING (20, 21) and GO (22) analyses of proteins upregulated in 1700029I15Rik knockout spermatozoa. The proteins related to unfolded protein binding (GO:0051082) or exhibited peptide disulfide oxidoreductase activity (GO:0015037) are highlighted in teal and light orange, respectively. (C) Upregulated ER chaperones in 1700029I15Rik knockout spermatozoa. The upregulation of CANX, CLGN, HSP90B1, and PDIA3 (highlighted in bold font and red color), but not CALR, HSPA5, and P4HB (in bold font and gray color), have been verified by Western blot analyses shown in Fig. 5D. (D) Western blot analyses of multiple ER chaperones in wild-type and knockout spermatozoa. ACTB was analyzed as a loading control. (E) Western blot analyses of OSTC, RPN2, TMED10 in wild-type and knockout spermatozoa. ACTB was analyzed as a loading control. (F) Western blot detection of ubiquitin and ubiquitinated proteins in wild-type and knockout spermatozoa. BASIGIN was analyzed in parallel as a loading control. The band intensities relative to BASIGIN were measured by ImageJ. The numbers represent the relative intensities of ubiquitinated proteins (Upper) or ubiquitin (Lower). (G) A diagram depicting 1700029I15Rik-mediated processing of acrosomal membrane proteins. OST complex is composed of subcomplex (SC) I, II, and III. Briefly, 1700029I15Rik interacts with and stabilizes OSTC and RPN2, which localize to SC II and SC III, respectively (28). In the absence of 1700029I15Rik, the destabilized OST complex causes aberrant glycosylation of acrosomal membrane proteins, resulting in increased protein misfolding. The misfolded proteins are eliminated by ubiquitin-dependent ERAD. Thus, the acrosomal trafficking of mature proteins is reduced or impaired.
Concerning that low-abundance proteins might not be readily detected by MS in whole sperm lysates, we performed individual analyses on Triton X-114-extracted soluble and membrane fractions of wild-type and knockout sperm. Interestingly, multiple protein bands were specifically detected in the aqueous (AQ) phase, but not the DET phase of knockout sperm (SI Appendix, Fig. S9A). The MS analyses collectively uncovered 50 significantly upregulated and 34 significantly downregulated proteins in 1700029I15Rik knockout sperm (Dataset S1). In agreement to the whole sperm proteomic analysis, multiple ERAD-associated chaperones are increased in the knockout sperm (SI Appendix, Fig. S9B). Apart from these chaperones, the OST complex subunits, ribophorin I (RPN1) and RPN2, and the ER–Golgi intermediate compartment (ERGIC) proteins, TMED4 and TMED10, are also upregulated in the knockout sperm (SI Appendix, Fig. S9B). GO and KEGG analyses revealed that the downregulated hits include proteins exhibited oxidoreductase activity (GO:0016491; 4 proteins), involved in the biosynthesis of amino acids (KEGG: mmu01230; 4 proteins), or associated with the acrosomal vesicle (GO:0001669; 4 proteins) or mitochondria (GO:0005739; 13 proteins). Notably, renalase (RNLS) and succinate-Coenzyme A ligase, ADP-forming, beta subunit (SUCLA2) are downregulated in both trials of sperm proteomic analyses (SI Appendix, Figs. S8F and S9B). However, whether these proteins play important roles in protein biosynthesis or fertilization remains unknown.
The increased ER chaperones involved in the unfolded protein response and ubiquitin-dependent ERAD pathway may reflect accumulation of misfolded proteins in the male germline. In corroboration with this assumption, we detected prominent elevation of ubiquitin and ubiquitinated proteins in the knockout spermatozoa (Fig. 5F). Collectively, our findings suggest that 1700029I15Rik, in collaboration with the OST complex subunits and folding enzymes, ensures proper biosynthesis of sperm acrosomal membrane glycoproteins in early spermatids (Fig. 5G). This working hypothesis is further consolidated by our in vitro observations that 1700029I15Rik promotes the expression of TMEM95 and prevents the formation of a nonnative, SDS-resistant dimer of TMEM95 in vitro (SI Appendix, SI Results and Discussion and Fig. S10 A–G).
Discussion
Nascent polypeptides translocated across the ER membrane usually undergo folding, modifications, and assembly to acquire their native conformations. To maintain protein homeostasis, unfolded or misfolded proteins are eliminated by sophisticated quality control systems (29). In somatic cells, lectin-like ER chaperones CANX and CALR target misfolded proteins for degradation via the ubiquitin-dependent ERAD pathway (29, 30). In the male germline, calmegin (CLGN) and calreticulin-3 (CALR3) are the testis-specific paralogs of CANX and CALR, respectively, and regulate the biosynthesis of sperm plasma membrane proteins (e.g., ADAM1B and ADAM3) required for sperm penetration through the uterotubal junction and ZP binding (31–33). Ablation of CLGN or CALR3 does not impair sperm–egg fusion (33, 34), suggesting that the biosynthesis of gamete fusion-related proteins is regulated by an independent pathway.
Mammalian cells express two distinct forms of OST complexes, centered around catalytic subunits staurosporine and temperature sensitive 3A and 3B (STT3A and STT3B), respectively. STT3A–OST is directly associated with the translocon and cotranslationally regulates N-glycosylation of nascent polypeptides, whereas STT3B–OST post-translationally glycosylates fully synthesized proteins (35). 1700029I15Rik interacts with non-catalytic OST subunits DAD1, OSTC, and RPN2; 1700029I15Rik knockout testes exhibit reduced levels of OSTC and RPN2 (Fig. 4 A and E). DAD1 and RPN2 are shared accessory subunits of STT3A and STT3B complexes, whereas OSTC exclusively exists in STT3A–OST (28). Previous in vitro studies have demonstrated that depletion of DAD1 destabilizes the OST subunits RPN1, RPN2, and dolichyl-di-phosphooligosaccharide-protein glycotransferase (DDOST) and causes hypoglycosylation (36, 37), whereas ablation of OSTC impairs glycosylation of STT3A-dependent substrates (38). Thus, it is tempting to speculate that loss of 1700029I15Rik might disrupt the structural integrity of the OST complex and potentially hamper the STT3A-mediated cotranslational glycosylation. 1700029I15Rik also interacts with folding chaperones PDIA3 and peptidyl-prolyl cis–trans isomerase B (PPIB), and malectin (MLEC; Fig. 4A), a carbohydrate-binding protein involved in the quality control of glycoproteins (39), raising a possibility that 1700029I15Rik has multiple functions in protein processing. In 1700029I15Rik knockout testes, CD46, EQTN, IZUMO1, and SPACA1 are significantly reduced but remain glycosylated and trafficked to the acrosomal membrane (Fig. 4F and SI Appendix, Fig. S7C). Thus, we hypothesize that deletion of 1700029I15Rik partially impairs protein glycosylation and disulfide bond formation, resulting in increased protein misfolding; an accumulation of misfolded proteins presumably induces ER stress that transcriptionally upregulates the ERAD machinery. In support of this assumption, we observed elevated ERAD chaperones and ubiquitinated proteins in 1700029I15Rik knockout sperm (Fig. 5 C and F and SI Appendix, Fig. S9B).
CD46, EQTN, IZUMO1, and SPACA1 are downregulated in 1700029I15Rik knockout testes, but show normal protein levels in the knockout sperm (Fig. 4F). The expression of these acrosomal membrane proteins initiates in early round spermatids (SI Appendix, Fig. S6F). Western blot analyses of acrosomal membrane proteins in whole testis lysates mainly reflect their levels in cytoplasm-rich spermatids, because spermatozoa devoid of cytoplasm only account for a small population in testes. It might be possible that in early spermiogenesis, acrosomal membrane proteins are synthesized at an excess amount in the ER; only a fraction of the proteins is subsequently incorporated into the acrosomal membrane and eventually retained in mature spermatozoa, while the remainder is abandoned with the residual bodies. Therefore, in 1700029I15Rik knockout males, although the acrosomal membrane proteins show decreased abundance in testes relative to the wild-type, the amounts are sufficient to secure endogenous protein levels in mature spermatozoa. This hypothesis might explain the discrepancies in the protein levels observed in the immunoblots of testis and sperm samples but requires experimental verification.
Proteins destined for sperm plasma membrane and acrosomal membrane exhibit striking differences in the timing of mRNA expression. For example, sperm plasma membrane proteins, ADAM1B and ADAM3, show peak expression in late spermatocytes, whereas acrosomal membrane proteins, IZUMO1 and SPACA1, exhibit biased expression in mid-round spermatids (SI Appendix, Fig. S6F). Consistent to the expression timing of their putative substrates, CLGN and 1700029I15Rik are highly expressed in late spermatocytes and early spermatids, respectively (SI Appendix, Figs. S1B and S6F). Thus, we propose that the biosynthesis pathways of sperm plasma membrane and acrosomal membrane proteins are independently regulated in a spatiotemporal manner (SI Appendix, Fig. S11). However, further investigation is warranted to determine whether the substrates of 1700029I15Rik are restricted to acrosomal membrane glycoproteins.
1700029I15Rik, which supposedly functions in the ER, is initially detected in the acrosomal granule of round spermatids and becomes absent in elongating spermatids (SI Appendix, Fig. S4A). Early studies have reported that the ER chaperone CALR also localizes to the acrosomal granule of mouse and rat spermatids (40, 41). Similarly, the ER-resident protein OSTC first appears as a granule-like structure in spermatocytes, subsequently relocates to the acrosomal granule in round spermatids, and is eventually discarded via shedding of residual bodies (SI Appendix, Fig. S6B). This disposal mechanism of ER-resident proteins may explain how 1700029I15Rik becomes absent in elongating spermatids. Future studies are necessary to explore whether 1700029I15Rik has a function in the acrosomal granule.
Recently, two independent studies on 1700029I15Rik were published (42, 43). Despite that the knockout mouse lines exhibit the same phenotype, the works by three groups show great discrepancies in terms of the proposed molecular functions of 1700029I15Rik. Contreras et al. (42) discovered that 1700029I15Rik retains IZUMO1 in the ER of HeLa cells, contradicting with our observations that IZUMO1 colocalizes with 1700029I15Rik at the surface of COS-7 cells (SI Appendix, Fig. S1I) and that coexpression with 1700029I15Rik does not prominently alter the subcellular expression pattern of IZUMO1 in HEK293T cells (SI Appendix, Fig. S10H). We reason that the use of different cell lines may account for the different localization of ectopically expressed 1700029I15Rik. By immunohistochemistry, Contreras et al. observed that IZUMO1 shows reduced colocalization with BIP (also known as HSPA5) in the acrosomal granule of knockout spermatids as compared to the wild-type (42). We argue that additional evidence is required to support the conclusion that 1700029I15Rik mediates ER retention of IZUMO1 in vivo. More importantly, IZUMO1 is an acrosomal membrane protein that does not localize to the acrosomal granule of wild-type spermatids. We reason that the fluorescence signals detected in the proacrosome might be affected by the angle of sectional plane during imaging (e.g., Lower Right panel of Fig. 3H). Based on a previous discovery of an IZUMO1-containing multiprotein complex (44), the authors proposed a working model that 1700029I15Rik transiently retains IZUMO1 in the ER, thereby allowing IZUMO1 to coassemble with other fertilization-related proteins (42). However, it remains open questions whether this high-molecular-weight protein complex is assembled in the ER, and whether the aberrant assembly of this complex would negatively affect sperm–egg binding or fusion.
Distinct to the observations made by Contreras et al. (42) and our study (SI Appendix, Fig. S2J), Hao et al. (43) reported that 1700029I15Rik knockout male mice show significantly reduced sperm counts. The authors performed proteomic analyses on wild-type and knockout whole testis and sperm and identified a multitude of downregulated proteins in the knockout groups (43). We are concerned that the decreased sperm counts in that mouse line may reflect changes in the germ cell populations (i.e., reduced percentages of round spermatids or testicular sperm in knockout testes), which potentially complicates the interpretation of the MS outcomes. Different from our Western blot outcomes (Fig. 4H), the proteomic analysis by Hao et al. revealed that IZUMO1 and EQTN are downregulated in the knockout sperm (43).
It is worth noting that gamete fusion-related acrosomal membrane proteins, such as TMEM95 and SPACA6, had not been detected in any of the testis or sperm proteomic analyses in the present study (Dataset S1). This is likely due to their extremely low levels of expression, which also hinder us from visualizing their subcellular localization by immunohistochemistry. These proteins might be effectively enriched in round spermatids isolated from testes; however, the velocity sedimentation (STA-PUT) or fluorescence-activated cell sorting (FACS)-based germ cell purification involves protease treatments and prolonged exposure to an in vitro environment, thus potentially introducing unexpected alterations in the proteome. To comprehensively elucidate the characteristics of such low-abundance proteins, we envision that immunostaining with signal amplification by exchange reaction (Immuno-SABER) and proximity-dependent biotinylation identification (BioID) could be employed in future investigations (45, 46).
In summary, this study suggests that 1700029I15Rik specifically regulates the biosynthesis of acrosomal membrane proteins by ensuring precise and timely coordination of multiple protein modification events. 1700029I15Rik interacts with proteins involved in N-glycosylation, disulfide isomerization, and vesicular trafficking. Ablation of 1700029I15Rik destabilizes the OST subunits and likely disrupts N-glycosylation of acrosomal membrane proteins. The aberrantly glycosylated or improperly disulfide-bonded proteins are misfolded and eliminated by the ubiquitin-dependent ERAD pathway, thereby resulting in the reduced abundance or absence of mature proteins (Fig. 5G). This pathway acts independently to the CLGN-mediated processing of sperm plasma membrane proteins (SI Appendix, Fig. S11). Given that 1700029I15Rik is highly conserved in humans, this work may open avenues for the development of a nonhormonal contraceptive targeting the biosynthesis of gamete fusion-required acrosomal proteins during spermiogenesis.
Materials and Methods
For immunodetection of 1700029I15Rik, a polyclonal antibody against its C terminus was raised in rabbits. The mutant mouse lines lacking 1700029I15Rik were generated by CRISPR/Cas9. Fertility, histological, sperm motility, and in vitro fertilization analyses were carried out to investigate the phenotype of knockout males. Co-IP/MS analyses were conducted to identify the interacting proteins of 1700029I15Rik in mouse testes. Proteomic changes in 1700029I15Rik knockout testes and sperm were analyzed by MS and Western blotting. In vitro analyses were performed using HEK293T and COS-7 cells to elucidate the protein features and molecular functions of 1700029I15Rik. Detailed experimental procedures are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We thank Natsuki Furuta and Saki Nishioka for technical assistance, Akinori Ninomiya and the Core Instrumentation Facility at the Research Institute for Microbial Diseases, Osaka University for assistance in MS analyses, Dr. Julio M. Castañeda for critical reading of this manuscript, and Dr. Peter S. Kim and members of Department of Experimental Genome Research for discussions on this study. This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI grants (JP18K16735 and JP22K15103 to Y.L., JP18K14612 and JP20H03172 to T.N., and JP19H05750 and JP21H05033 to M.I.), the Takeda Science Foundation grants to T.N. and M.I., the Japan Science and Technology Agency (JST) PRESTO grant JPMJPR2148 to T.N., the Nakajima Foundation grant to T.N., the Eunice Kennedy Shriver National Institute of Child Health and Human Development grants K99HD104924 to S.T., and P01HD087157 and R01HD088412 to M.I., the Swedish Research Council grant 2018-03426 to H.S., and the Bill & Melinda Gates Foundation grant INV-001902 to M.I. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author contributions
Y.L., K.S., S.T., J.Z., H.S., and M.I. designed research; Y.L., K.S., S.T., J.Z., Y.O., T.N., and H.S. performed research; Y.L., S.T., H.S., and M.I. contributed new reagents/analytic tools; Y.L., K.S., S.T., J.Z., H.S., and M.I. analyzed data; and Y.L., S.T., and M.I. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Mass spectrometry raw data have been deposited in ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) with the dataset identifier (47). All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Ikawa M., Inoue N., Benham A. M., Okabe M., Fertilization: A sperm’s journey to and interaction with the oocyte. J. Clin. Investig. 120, 984–994 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Y., Ikawa M., Eukaryotic fertilization and gamete fusion at a glance. J. Cell Sci. 135, jcs260296 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoue N., Ikawa M., Isotani A., Okabe M., The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434, 234–238 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Matsumura T., et al. , Sperm IZUMO1 is required for binding preceding fusion with oolemma in mice and rats. Front. Cell Dev. Biol. 9, 810118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorenzetti D., et al. , A transgenic insertion on mouse chromosome 17 inactivates a novel immunoglobulin superfamily gene potentially involved in sperm–egg fusion. Mamm. Genome 25, 141–148 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Noda T., et al. , Sperm proteins SOF1, TMEM95, and SPACA6 are required for sperm–oocyte fusion in mice. Proc. Natl. Acad. Sci. U.S.A. 117, 11493–11502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbaux S., et al. , Sperm SPACA6 protein is required for mammalian sperm-egg adhesion/fusion. Sci. Rep. 10, 5335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujihara Y., et al. , Spermatozoa lacking fertilization influencing membrane protein (FIMP) fail to fuse with oocytes in mice. Proc. Natl. Acad. Sci. U.S.A. 117, 9393–9400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamas-Toranzo I., et al. , TMEM95 is a sperm membrane protein essential for mammalian fertilization. Elife 9, e53913 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang S., et al. , Human sperm TMEM95 binds eggs and facilitates membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 119, e2207805119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue N., Hagihara Y., Wada I., Evolutionarily conserved sperm factors, DCST1 and DCST2, are required for gamete fusion. Elife 10, e66313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noda T., et al. , Sperm membrane proteins DCST1 and DCST2 are required for sperm-egg interaction in mice and fish. Commun. Biol. 5, 332 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satouh Y., Inoue N., Ikawa M., Okabe M., Visualization of the moment of mouse sperm–egg fusion and dynamic localization of IZUMO1. J. Cell Sci. 125, 4985–4990 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Khawar M. B., Gao H., Li W., Mechanism of acrosome biogenesis in mammals. Front. Cell Dev. Biol. 7, 195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann B. P., et al. , The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 25, 1650–1667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashkenazy H., et al. , ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H., Kim H., Membrane topology of transmembrane proteins: Determinants and experimental tools. Biochem. Biophys. Res. Commun. 453, 268–276 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Szklarczyk D., et al. , The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D., et al. , STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherman B. T., et al. , DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niemeyer J., et al. , The intramembrane protease SPPL2c promotes male germ cell development by cleaving phospholamban. EMBO Rep. 20, e46449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park S., et al. , CRISPR/Cas9-mediated genome-edited mice reveal 10 testis-enriched genes are dispensable for male fecundity. Biol. Reprod. 103, 195–204 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turano C., Gaucci E., Grillo C., Chichiarelli S., ERp57/GRP58: A protein with multiple functions. Cell. Mol. Biol. Lett. 16, 539–563 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellerman D. A., Myles D. G., Primakoff P., A role for sperm surface protein disulfide isomerase activity in gamete fusion: Evidence for the participation of ERp57. Dev. Cell 10, 831–837 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Wong C.-W., et al. , The roles of protein disulphide isomerase family A, member 3 (ERp57) and surface thiol/disulphide exchange in human spermatozoa–zona pellucida binding. Hum. Reprod. 32, 733–742 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Braunger K., et al. , Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 360, 215–219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M., Kaufman R. J., Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Hoseki J., Ushioda R., Nagata K., Mechanism and components of endoplasmic reticulum-associated degradation. J. Biochem. 147, 19–25 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Ikawa M., et al. , The putative chaperone calmegin is required for sperm fertility. Nature 387, 607–611 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Ikawa M., et al. , Calmegin is required for fertilin α/β heterodimerization and sperm fertility. Dev. Biol. 240, 254–261 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Tokuhiro K., Ikawa M., Benham A. M., Okabe M., Protein disulfide isomerase homolog PDILT is required for quality control of sperm membrane protein ADAM3 and male fertility. Proc. Natl. Acad. Sci. U.S.A. 109, 3850–3855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamagata K., et al. , Sperm from the calmegin-deficient mouse have normal abilities for binding and fusion to the egg plasma membrane. Dev. Biol. 250, 348–357 (2002). [PubMed] [Google Scholar]
- 35.Harada Y., Ohkawa Y., Kizuka Y., Taniguchi N., Oligosaccharyltransferase: A gatekeeper of health and tumor progression. Int. J. Mol. Sci. 20, 6074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanjay A., Fu J., Kreibich G., DAD1 Is required for the function and the structural integrity of the oligosaccharyltransferase complex. J. Biol. Chem. 273, 26094–26099 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Roboti P., High S., The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation. J. Cell Sci. 125, 3474–3484 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Shrimal S., Cherepanova N.A., Gilmore R., DC2 and KCP2 mediate the interaction between the oligosaccharyltransferase and the ER translocon. J. Cell Biol. 216, 3625–3638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin S.-Y., et al. , Malectin forms a complex with ribophorin I for enhanced association with misfolded glycoproteins. J. Biol. Chem. 287, 38080–38089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura M., Oshio S., Tamura A., Okinaga S., Arai K., Antisera to calreticulin inhibits sperm motility in mice. Biochem. Biophys. Res. Commun. 186, 984–990 (1992). [DOI] [PubMed] [Google Scholar]
- 41.Nakamura M., et al. , An endoplasmic reticulum protein, calreticulin, is transported into the acrosome of rat sperm. Exp. Cell Res. 205, 101–110 (1993). [DOI] [PubMed] [Google Scholar]
- 42.Contreras W., et al. , C11orf94/Frey is a key regulator for male fertility by controlling Izumo1 complex assembly. Sci. Adv. 8, eabo6049 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hao H., et al. , The vertebrate- and testis- specific transmembrane protein C11ORF94 plays a critical role in sperm-oocyte membrane binding. Mol. Biomed. 3, 27 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellerman D. A., et al. , Izumo is part of a multiprotein family whose members form large complexes on mammalian sperm. Mol. Reprod. Dev. 76, 1188–1199 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saka S. K., et al. , Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37, 1080–1090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D. I., et al. , An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 27, 1188–1196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research 47, D442–D450 (2018), 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
Mass spectrometry raw data have been deposited in ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) with the dataset identifier (47). All study data are included in the article and/or SI Appendix.