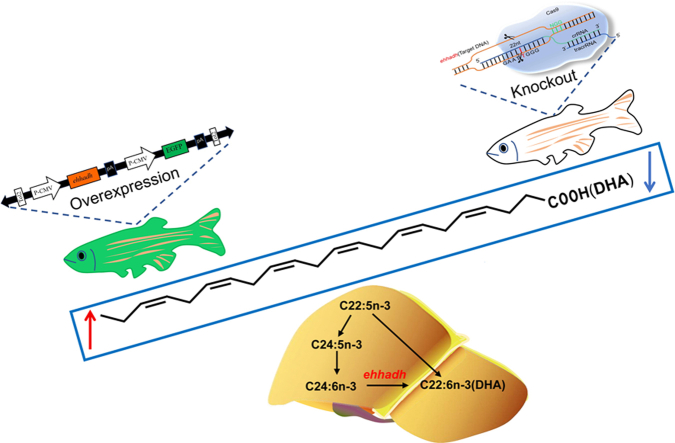
Abstract
Compared with other species, freshwater fish are more capable of synthesizing DHA via same biosynthetic pathways. Freshwater fish have a “Sprecher” pathway to biosynthesize DHA in a peroxisome-dependent manner. Enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase (Ehhadh) is involved in the hydration and dehydrogenation reactions of fatty acid β-oxidation in peroxisomes. However, the role of Ehhadh in the synthesis of DHA in freshwater fish remains largely unclear. In this study, the knockout of Ehhadh significantly inhibited DHA synthesis in zebrafish. Liver transcriptome analysis showed that Ehhadh deletion significantly inhibited SREBF and PPAR signaling pathways and decreased the expression of PUFA synthesis-related genes. Our results from the analysis of transgenic zebrafish (Tg:Ehhadh) showed that Ehhadh overexpression significantly increased the DHA content in the liver and significantly upregulated the expression of genes related to PUFA synthesis. In addition, the DHA content in the liver of Tg:Ehhadh fed with linseed oil was significantly higher than that of wildtype, but the expression of PUFA synthesis-related genes fads2 and elovl2 were significantly lower, indicating that Ehhadh had a direct effect on DHA synthesis. In conclusion, our results showed that Ehhadh was essential for DHA synthesis in the “Sprecher” pathway, and Ehhadh overexpression could promote DHA synthesis. This study provides insight into the role of Ehhadh in freshwater fish.
Supplementary key words: fatty acids, docosahexaenoic acid synthesis, “Sprecher” pathway, peroxisome β-oxidation, transgenic zebrafish, liver, PUFA synthesis, freshwater fish, SREBF signaling pathway, PPAR signaling pathway
Abbreviations: DEG, differentially expressed gene; Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase; FO, fish oil; KEGG, Kyoto Encyclopedia of Genes and Genomes; LO, linseed oil; n-3 PUFA, omega-3 PUFA; qRT-PCR, real-time quantitative PCR; WT-FO, FO-fed WT; WT-LO, LO-fed WT
Graphical abstract
The peroxisome is the main site of fatty acid β-oxidation, and it is mainly responsible for the oxidation of long-chain fatty acids, long-chain dicarboxylic acids, eicosanoids, bile acid precursors, and the side chains of certain xenobiotics (1). In addition, the peroxisome has also been reported to be the only site for β-oxidation of very long-chain fatty acids (containing ≥22 carbon atoms) (2). The enzymes encoded by genomic DNA were transported across the membrane into peroxisome, and peroxisome β-oxidation included four enzymatic steps, namely, oxidation, hydration, dehydrogenation, and thiolytic cleavage (3).
Enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase (Ehhadh) is a key enzyme involved in the β-oxidation of fatty acids by peroxisomes, and Ehhadh plays a vital role in maintaining the normal function of peroxisomes. This enzyme has two separate domains at the N-terminus and C-terminus with hydration and dehydrogenation activities, respectively. The Ehhadh is responsible for the hydration and dehydrogenation steps in fatty acid β-oxidation, and thus it is called a bifunctional enzyme (4). Ehhadh, acyl-CoA oxidase 1 (acox1), and acetyl-CoA acyltransferase 1 (acaa1) together constitute the classical peroxisome β-oxidation pathway, and this pathway is transcriptionally activated by PPARα ligand (5). The accumulation of long-chain dicarboxylic acids in the liver of Ehhadh-knockout mice results in the obstruction of medium-chain dicarboxylic acid production, and thus long-chain dicarboxylic acid is one of the substrates for Ehhadh oxidation (5, 6). In addition, a recent study has shown that 190 metabolites are significantly altered in the kidneys of Ehhadh-knockout mice, compared with those in WT, and the contents of 100 metabolites were significantly increased, including fatty acids and their conjugates, especially markers of peroxisomal dysfunction such as tetracosahexaenoic acid (C24:6n-3), pipecolate, and very long-chain acylcarnitines (7). These results indicate that the functions of Ehhadh gene remain mostly unknown, and its oxidative substrates need to be further explored. In freshwater fish, the function of Ehhadh gene has hardly been studied.
DHA (22:6n-3) is synthesized from alpha-linolenic acid (18:3n-3, ALA) through a series of biological enzymes. Many intermediate products are produced in the DHA synthesis pathway, such as EPA (20:5n-3) and docosapentaenoic acid (22:5n-3), and these fatty acids are collectively referred to as omega-3 PUFAs (n-3 PUFA) (8). n-3 PUFA plays a key role in maintaining the normal growth and development and physiological functions of humans and animals, including involvement in biofilm formation, energy supply, and disease prevention and control (9, 10). In recent years, the biosynthesis mechanism of DHA in fish has been widely explored. Some marine fish (Salmo salar and Siganus canaliculatus) and freshwater fish can convert ALA into EPA and DHA, but most marine fish exhibit limited conversion capacity (11). The difference in the biosynthesis ability of DHA between fish is related to the differences in complementarity and enzyme activity between the related synthetic genes of various species (12). However, many potential DHA synthesis pathways and related mechanisms remain to be further investigated.
In fish, DHA is mainly synthesized in the endoplasmic reticulum (13). Firstly, ALA biosynthesizes 22:5n-3 through the Δ6 pathway (Δ6 desaturation-elongation-Δ5 desaturation-elongation) or Δ8 pathway (elongation-Δ8 desaturation-Δ5 desaturation-elongation). Subsequently, some teleosts with Δ4 desaturase activity can produce DHA by desaturating 22:5n-3 at the Δ4 position, which can be known as the “Δ4 pathway” (14). In addition, another DHA synthesis pathway has been found in mice. EPA in the body first produces 24:5n-3 through two extensions under the action of fatty acyl elongases, and then product 24:5n-3 further desaturates into 24:6n-3 under the action of Δ6 desaturase. The obtained 24:6n-3 loses two carbons through fatty acid β-oxidation to produce DHA. The above serial processes take place in the peroxisome and are called the “Sprecher” pathway (15). This pathway has also been reported to exist in fish (14). However, β-oxidation to produce DHA is accomplished by a nonclassical oxidation pathway, where d-bifunctional protein (Hsd17b4) rather than Ehhadh is located (16). A recent study indicated that the deletion of Ehhadh leads to the accumulation of DHA precursor material (C24:6n-3), suggesting a possible role of Ehhadh in DHA synthesis (7). In addition, peroxisomes can not only participate in the synthesis of DHA and are the only place where DHA is oxidized into short-chain fatty acids, and thus peroxisomes play an important role in maintaining the balance between DHA synthesis and decomposition (17). However, the relevant mechanisms are currently unknown.
In this study, we firstly used the Ehhadh-knockout model to explore the effect of Ehhadh deletion on the content of fatty acids. We found that Ehhadh deletion reduced the content of DHA and inhibited the synthesis of n-3 PUFA. Based on transcriptomics, we explored the molecular mechanism by which Ehhadh deletion hindered the synthesis of n-3 PUFA. Finally, an Ehhadh overexpression model was constructed to verify our results. Based on the dual function of Ehhadh synthesis and oxidation, we also compared the differences in fatty acid composition between WT and transgenic zebrafish fed with linseed oil (LO) and fish oil (FO). Our results provide very useful information for us to understand the role of Ehhadh in fish.
Materials and methods
Ethics
The feeding and handling of experimental animals comply with the Management Rule of Laboratory Animals (Chinese order No. 676 of the State Council, revised 1 March, 2017). This study was approved by the Committee on the Ethics of Animal Experiments of Huazhong Agricultural University (HZAUFI-2021-0024). Fish sampling was performed under anesthesia with MS-222 (tricaine methanesulfonate, Sigma) in the concentration of 200 mg/l to minimize suffering.
Zebrafish maintenance
The WT zebrafish (AB strain) used in this study were obtained from the National Zebrafish Resource Center and reared in a dechlorinated recirculating water culture system. Specific rearing conditions were as follows: water temperature, 28 ± 0.5°C; pH, 7.6 ± 0.1; photoperiod, 14 h light/10 h dark; and fresh Artemia salina was fed three times a day (at 8:00, 13:00, and 18:00). All the experiments in this study were performed in accordance with the guidelines for the care and use of laboratory animals in the Huazhong Agricultural University.
Construction of Ehhadh−/− zebrafish model
Based on previous studies, we used CRISPR/Cas9 technology to generate Ehhadh-deficient zebrafish model (18). The Ehhadh DNA sequence was obtained from NCBI (http://www.ncbi.nlm.nih.gov/). The 20 bp target sequence was localized to the second exon and the target gene regions containing this 20 bp sequence were amplified using the primers of Ehhadh (supplemental Table S1). The in vitro transcription of Cas9 RNA and gRNA was performed by previously reported methods (19). The gRNA and CRISPR/Cas9 enzyme were mixed at a ratio of 50:150 (ng/μl), and 0.5% phenol red was added as an indicator into the mixture. The mixture was injected into zebrafish single-cell stage fertilized eggs at 2 nl per egg. After injection, fertilized eggs were incubated in constant temperature water at 28°C. The Ehhadh-knockout zebrafish were screened and validated by genomic DNA sequencing. The corresponding sequencing primers were presented in supplemental Table S1.
Construction of Ehhadh transgenic zebrafish
We established the Ehhadh overexpression zebrafish model by previously reported method using pTol2-MCS-EGFP and pCSTol2 plasmids constructed in our previous research (20). The pTol2-MCS-EGFP fusion plasmid contained two segments of P-CMV and one segment of EGFP. The Ehhadh open reading frames were ligated into the pTol2-MCS-EGFP plasmid to construct a recombinant plasmid. Specifically, first, the nucleotide sequence of the zebrafish Ehhadh gene (NM_207068.1) was obtained from the NCBI (https://www.ncbi.nlm.nih.gov/). Gene primers were designed by Primer Premier 6.0 software (supplemental Table S1). The open reading frame of the Ehhadh gene was then amplified by PCR, and the PCR product was further amplified with homology arm primers. The final products were separated through 2% agarose gel electrophoresis and purified to obtain the Ehhadh fragment with the homologous arm attached. The Ehhadh sequence was inserted into fusion plasmid pTol2-CMV-SV40 poly (A)+CMV: egfp-SV40 poly (A) using a one-step rapid cloning kit (Yeasen, China). The pCSTol2 plasmid was linearized using NotIase (New England Biolabs). DNA was recovered, and its concentration was measured using the DNA purification/ recovery kit (Yeasen, China). The mRNA was synthesized in vitro using the mMessage mMachine SP6 kit (New England Biolabs) with 1 μg linearized plasmid as the template. Approximately, 1 nl of DNA-mRNA mixture containing 25 ng/μl DNA and 25 ng/μl mRNA was injected into zebrafish-fertilized embryos at the single-cell stage. The F0 generation of Ehhadh transgenic zebrafish with green fluorescence was screened under a fluorescent stereomicroscope, and the screened F0 generation was raised to sexual maturity and crossed to produce F1 generation individuals. Afterward, the expression of Ehhadh in the F1 generation was detected.
Fatty acid composition analysis
Total lipids were extracted from WT and Ehhadh−/− by the method reported by the Bligh and Dyer (21). The extracted total lipids were then methylated at 100°C for 1 h with a methylation reagent containing a mixture of 60% boron trifluoride methanol solution and 40% dichloromethane. Fatty acid composition was determined using gas chromatography (Shimadzu emit Co., Ltd., Tokyo, Japan) according to the previously reported method (19).
C24:6n-3 assay
Methyl-tetracosahexaenoate was detected on a gas chromatography-mass spectrometry (Thermo Fisher Scientific, Waltham, MA) system using a TG-5 gas chromatography column (30 m × 0.25 mm i.d. and 0.25-μm film thickness [Thermo Fisher Scientific, Waltham, MA]). High-purity helium was used as carrier gas and passed through the gas chromatography column at 1 ml/min. The initial oven temperature was maintained at 60°C for 1.5 min, increased to 300°C at a rate of 20°C/min, and then held for 7 min. The ion source and transmission line temperatures were 280°C and 300°C, respectively. The ionization method was electron impact (EI, 70 eV). The detection range of molecular weight was 50–450.
Liver transcriptomic analysis
RNA extraction and isolation as well as lllumina Hiseq sequencing
Total RNA was extracted from the tissue samples using trizol (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. The concentration and purity of the extracted RNA were examined using Nanodrop2000 (Thermo Fisher Scientific, Waltham, MA). RNA integrity was detected by agarose gel electrophoresis, and RNA integrity number was determined by Agilent2100 (Agilent, Santa Clara, CA). A single library was constructed using 1 μg of total RNA at a concentration ≥50 ng/μl with OD260/280 range of 1.8–2.2. A-T base pairing of polyA was performed using oligo-dT–coated magnetic beads, and then mRNA was isolated from total RNA. The isolated mRNA was randomly fragmentated using fragmentation buffer. Subsequently, the broken small fragments of mRNA were used as a template to reverse-synthesize complementary DNA (cDNA). Finally, the synthesized cDNA was amplified by bridge PCR and subjected to lllumina Hiseq sequencing.
De novo assembly and annotation
To ensure the quality of the assembly and the accuracy of the subsequent bioinformatics analysis, the raw sequencing data were filtered by using the software SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle). Data filtering was as follows: 1) the joint sequences were removed from raw reads; 2) the bases with low quality at the end of the sequence (3′end) (quality value less than 30) were removed; 3) the reads containing more than 10% of N were removed; and 4) the sequences that were still less than 50 bp in length after adapter and quality trimming were discarded. The quality-controlled data were assessed again and compared with the reference genome to obtain mapped data (reads) for subsequent analysis. To annotate the transcriptome, we performed the Blastx with an E-value <10−5 based on the databases Gene ontology (http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/). Finally, the data were analyzed using the online platform Majorbio.
Real-time quantitative PCR
In this study, total RNAs were extracted from the liver tissues of WT, Ehhadh−/−, and Tg:Ehhadh by using RNA isoPlus (TaKaRa, Japan). Primers were designed using Primer Premier 6.0 (supplemental Table S1) for real-time quantitative PCR (qRT-PCR) amplification. The qRT-PCR was performed using a Mini Option real-time detector (Bio-Rad, Hercules, CA). The 10 μl qRT-PCR system contained 5 μl SYBR Green real-time PCR mater mix, 3.6 μl double distilled water, 0.2 μl forward primer, 0.2 μl reverse primer, and 1 μl cDNA. The PCR procedures were as follows: predenaturation at 98°C for 5 min, followed by 40 cycles of denaturation at 98°C for 30 s, quenching at 60°C for 30 s, and extension at 72°C for 30 s. The relative expression level of each gene was determined using the 2−△△Ct method.
LO diet and FO diet feeding experiment
Two experimental diets with 7% LO or 7% FO were designed and formulated in our laboratory. The formulations of the two diets are shown in supplemental Tables S2 and S3 showed the fatty acid compositions of the two diets. A total of 45 WT zebrafish with 2 months old or 45 Tg:Ehhadh zebrafish with 2 months old were selected separately for each model, and the 45 zebrafish in each model were raised in three tanks with 15 fish in each tank and fed for 4 weeks using 7% LO diet/7% FO diet. Fish were manually fed on the experimental diets three times a day at 9:00 AM, 2:00 PM, and 7:00 PM until satiation.
After the feeding experiment, fatty acid content and gene mRNA expression levels were determined in the livers of LO-fed WT (WT-LO), LO-fed transgenic zebrafish (Tg:Ehhadh-LO), FO-fed WT (WT-FO), and FO-fed transgenic zebrafish (Tg:Ehhadh-FO).
Statistical analysis
Data in this study were expressed as mean ± SEM. All data were analyzed using statistical softwares in SPSS 19.0 (SPSS 19.0, Michigan Avenue, Chicago, IL). Data from WT and Ehhadh−/− fed with LO and FO diets were analyzed by using 2-way ANOVA to test for main effects of genotypes and diets. The Tukey honestly significant difference test was used for the mean comparisons. Data from WT and Ehhadh−/− fed with A. salina diet were analyzed using Student's t-test. P < 0.05 was considered statistically significant.
Results
Generation of Ehhadh−/−
To investigate the effect of Ehhadh on DHA production, we generated Ehhadh-deletion zebrafish (Ehhadh−/−) models using CRISPR/Cas9 technology. The second exon was selected as the knockout target, and the sequence of the target site was GTGGAGAGAATGGGCGATTCTG (supplemental Fig. S1A). The Ehhadh gene was found to lack a T-base at the target site, and its amino acid translation was shortened from 718 to 69. The Ehhadh gene expression level in the liver was significantly lower in the Ehhadh−/− than in WT (supplemental Fig. S1B). As shown in supplemental Fig. S2, there were no significant differences in early survival (0–96 h) and body weight (30–90 days) between WT and Ehhadh−/−, indicating that Ehhadh deletion did not affect zebrafish survival and growth.
Ehhadh deletion reduces DHA content and impedes synthesis pathway of n-3 PUFA
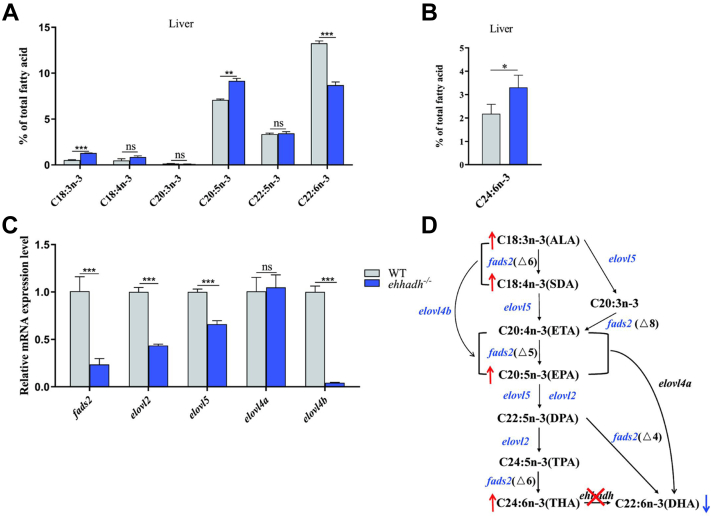
Compared with WT, Ehhadh−/− exhibited a significantly higher saturated fatty acid content but a significantly lower n-3 PUFA content in the liver, but no significant differences were observed in mono-unsaturated fatty acid and n-6 PUFA between Ehhadh−/− and WT (supplemental Fig. S3). Ehhadh−/− displayed significantly higher 18:3n-3, 20:5n-3, and 24:6n-3 contents and significantly lower 22:6n-3 content than WT (Fig. 1A, B and supplemental Fig. S4). Gene expression analysis showed that the gene expression levels of fatty acid desaturase 2 (fads2), fatty acid elongase 2 (elovl2), fatty acid elongase 5 (elovl5), and elongase 4b (elovl4b) in liver were significantly lower in Ehhadh−/− than in WT, but there was no significant difference in the expression of fatty acid elongase 4a (elovl4a) (Fig. 1C). The above results suggested that the deletion of Ehhadh inhibited n-3 PUFA synthesis (Fig. 1D).
Fig. 1.
Changes of liver fatty acid compositions and mRNA expression levels of fatty acid metabolism-related genes in Ehhadh-knockout zebrafish (Ehhadh−/−). A: liver n-3PUFA (18:3n-3; 18:4n-3; 20:3n-3; 20:5n-3; 22:5n-3; 22:6n-3) composition of WT zebrafish and Ehhadh−/−. B: liver C24:6n-3 content of WT and Ehhadh−/−. C: the mRNA expression levels of fads2, elovl2, elovl5, elovl4a, and elovl4b in livers of WT and Ehhadh−/−. D: the effect of Ehhadh knockout on DHA synthesis pathway. The blue font indicates that gene expression is significantly downregulated, the blue arrow indicates that the content of related fatty acids is significantly reduced, and the red arrow indicates that the content of related fatty acids is significantly increased. ns means no significant difference; ∗ means P < 0.05; ∗∗ means P < 0.01; ∗∗∗ means P < 0.001. Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase; elovl2, fatty acid elongase 2; elovl4a, fatty acid elongase 4a; elovl4b, fatty acid elongase 4b; elovl5, fatty acid elongase 5; fads2, fatty acid desaturase 2; MUFA, mono-unsaturated fatty acid; PUFA, poly-unsaturated fatty acid; SFA, saturated fatty acid.
Liver transcriptomic analysis
Differentially expressed gene screening and KEGG pathway enrichment analysis
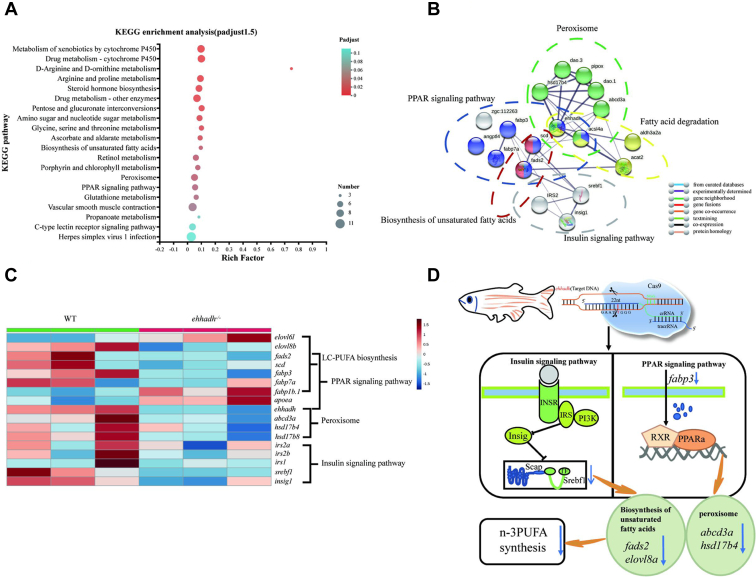
Transcriptome sequencing data results were shown in supplemental Tables S4 and S5. Differentially expressed genes (DEGs) were identified according to the thresholds of P < 0.05 and |log2 (fold change)| ≥1.5. A total of 368 DEGs were identified from WT and Ehhadh−/−, of which 150 DEGs were upregulated and 218 DEGs were downregulated (supplemental Fig. S5A, B). To characterize the functions of DEGs, we performed a KEGG pathway enrichment analysis (Fig. 2A). The results showed that the top five pathways in which DEGs were enriched were the metabolism of xenobiotics by cytochrome P450, drug metabolism-cytochrome P450, D-arginine and D-ornithine metabolism, arginine and proline metabolism, and steroid hormone biosynthesis pathways. In addition, DEGs were also significantly enriched in 3 PUFA metabolism-related signaling pathways including biosynthesis of unsaturated fatty acids, peroxisome, and PPAR signaling pathway.
Fig. 2.
Liver transcriptome analyses of WT zebrafish and Ehhadh-knockout zebrafish (Ehhadh−/−). A: KEGG pathway enrichment analysis of DEG from WT and Ehhadh−/−. B: Network interaction analysis between DEG corresponding proteins. C: DEG heat map analysis related to fatty acid metabolism. D: Flow chart of Ehhadh regulating DHA synthesis through the PPARα and insulin signaling pathways. The blue arrow indicates a significant decrease in expression. DEG, differentially expressed gene; Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase.
Protein-protein interaction network and DEG heat map analysis
For the proteins corresponding to the DEGs, protein-protein interactions were analyzed based on the STRING database (Fig. 2B). The 18 proteins corresponding to DEGs showed correlation with each other. Ehhadh, as the central node of the network, showed strong correlation with other proteins, indicating that alteration of Ehhadh protein activity would significantly affect the associated proteins. The KEGG pathway enrichment analysis of differentially expressed proteins showed that these 18 proteins fell into five functional categories, namely, biosynthesis of unsaturated fatty acids, peroxisome, PPAR signaling pathway, fatty acid degradation, and insulin signaling pathway, which was partially consistent with our results of KEGG analyses of DEGs. Based on transcriptome data, we selected fatty acid biometabolism-related DEGs and above-mentioned five functional category-related DEGs for heat map analysis (Fig. 2C). A total of 17 DEGs were screened, of which fatty acid elongase 6 (elovl6), fatty acid binding protein 1b, tandem duplicate 1 (fabp1b.1), apolipoprotein Ea (apoea) were upregulated in Ehhadh−/−; and elovl fatty acid elongase 8b (elovl8b), fatty acid desaturase 2 (fads2), stearoyl-CoA desaturase (scd), fatty acid binding protein 3 (fabp3), fatty acid binding protein7a (fabp7a), Ehhadh, ATP-binding cassette, sub-family D, member 3a (abcd3a), hydroxysteroid 17-beta dehydrogenase 4 (hsd17b4), hydroxysteroid 17-beta dehydrogenase 8 (hsd17b8), insulin receptor substrate 2a (irs2a), insulin receptor substrate 2b (irs2b), insulin receptor substrate 1 (irs1), sterol regulatory element binding transcription factor 1 (srebf1), and insulin-induced gene 1 (insig1) were downregulated in Ehhadh−/−.
Further, 12 DEGs were randomly selected for qPCR validation. The qPCR validation results were consistent with the transcriptome analysis results of gene expression changes, indicating that our transcriptome data were reliable (supplemental Fig. S5C).
Ehhadh overexpression increases DHA content and promotes synthesis of n-3 unsaturated fatty acids
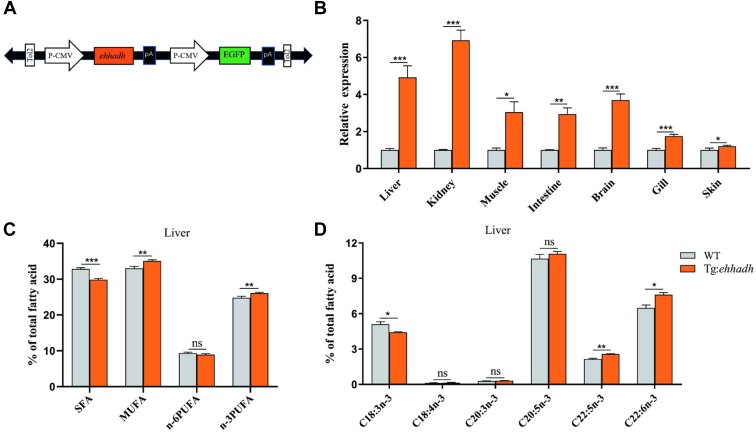
To confirm the above findings, an Ehhadh transgenic zebrafish model (Tg:Ehhadh) was constructed in this study (Fig. 3A). Compared to that in WT, Ehhadh expression in Tg:Ehhadh was significantly increased in liver, kidney, intestine, muscle, eye, brain, skin, and gill (Fig. 3B). Further, we determined the liver fatty acid composition. Compared to WT, Tg:Ehhadh zebrafish liver exhibited a significantly lower saturated fatty acid content but significantly higher n-3 PUFA and mono-unsaturated fatty acid contents (Fig. 3C). In addition, 22:5n-3 and 22:6n-3 contents were significantly higher, but 18:3n-3 content was significantly lower in Tg:Ehhadh than in WT (Fig. 3D). The determination results of Tg:Ehhadh zebrafish liver fatty acid were opposite to those of Ehhadh−/− zebrafish liver fatty acid results, implying that Ehhadh overexpression promoted n-3 PUFA synthesis.
Fig. 3.
The effect of Ehhadh overexpression on fatty acid composition of zebrafish. A: Schematic depicting of the recombinant plasmid containing the Ehhadh transgene. The Ehhadh gene was driven by the CMV promoter and conjugated to the DNA sequences of Tol2 and GFP fluorescent protein. B: Quantitative expression of Ehhadh in different tissues of WT and Ehhadh transgenic zebrafish (Tg:Ehhadh). C: Liver ∑SFA, ∑MUFA, ∑n-6PUFA, and ∑n-3PUFA composition of WT and Tg:Ehhadh. D: Liver n-3PUFA (18:3n-3, 18:4n-3, 20:3n-3, 20:5n-3, 22:5n-3, and 22:6n-3) composition of WT and Tg:Ehhadh. ns means no significant difference; ∗ means P < 0.05; ∗∗ means P < 0.01; ∗∗∗ means P < 0.001. Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase; MUFA, mono-unsaturated fatty acid; PUFA, poly-unsaturated fatty acid; SFA, saturated fatty acid.
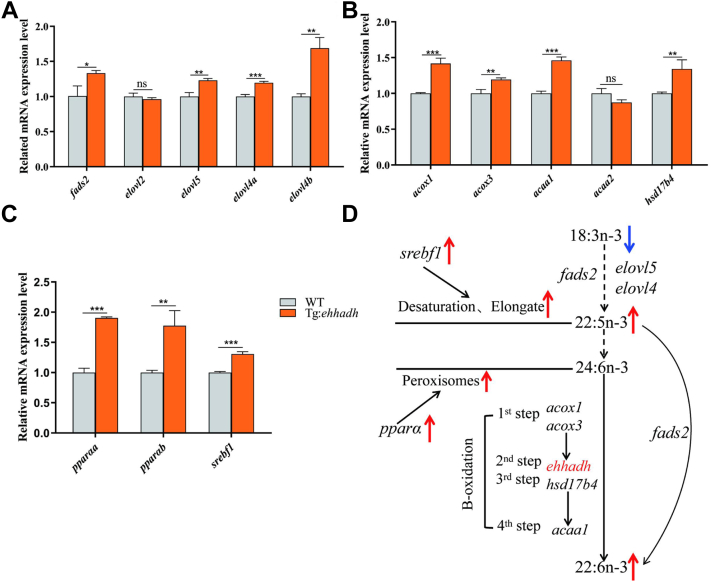
Based on the above transcriptome results, we selected related genes from Tg:Ehhadh and WT for gene expression analysis (Fig. 4). The expression levels of n-3 PUFA synthesis-related genes such as fads2, elovl5, elovl4a, and elovl4b in liver of Tg:Ehhadh zebrafish were significantly upregulated, compared with those of WT (Fig. 4A). Gene expression analysis results revealed that acetyl-CoA acyltransferase 1 (acaa1), acyl-CoA oxidase 1 (acox1), and acyl-CoA oxidase 3 (acox3), and hsd17b4 in peroxisome were significantly upregulated in Tg:Ehhadh (Fig. 4B). The expressions of both sterol regulatory element binding transcription factor 1 (srebf1) and peroxisome proliferator activated receptor α (pparα) were significantly elevated in Tg:Ehhadh zebrafish liver (Fig. 4C). The above results suggested that Ehhadh overexpression may promote zebrafish liver n-3 PUFA fatty acid synthesis by affecting the expressions of pparα and srebf1 signaling axis-related genes (Fig. 4D).
Fig. 4.
The mRNA expression levels of fatty acid metabolism-related genes in liver of WT and Ehhadh transgenic zebrafish (Tg:Ehhadh). A: Effects of Ehhadh overexpression on mRNA expression of PUFA synthesis-related genes in liver. B: Effects of Ehhadh overexpression on mRNA expression of Peroxisome β-oxidation related genes in liver. C: Effects of Ehhadh overexpression on mRNA expression of pparα and srebf1 in liver. D: The effect of Ehhadh overexpression on DHA synthesis pathway. The blue arrow indicates the downward adjustment, and the red arrow indicates the upward adjustment. ns means no significant difference; ∗ means P < 0.05; ∗∗ means P < 0.01; ∗∗∗ means P < 0.001. acaa1, acetyl-CoA acyltransferase 1; acaa2, acetyl-CoA acyltransferase 2; acox1, acyl-CoA oxidase 1; acox3, acyl-CoA oxidase 1; Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase; elovl2, fatty acid elongase 2; elovl4a, fatty acid elongase 4a; elovl4b, fatty acid elongase 4b; elovl5, fatty acid elongase 5; fads2, fatty acid desaturase 2; hsd17b4, hydroxysteroid 17-beta dehydrogenase 4; pparαa, peroxisome proliferator activated receptor alpha a; pparαb, peroxisome proliferator activated receptor alpha b; srebf1, sterol regulatory element binding transcription factor 1.
Effects of LO diet and FO diet feeding on PUFA synthesis in WT and Tg:Ehhadh
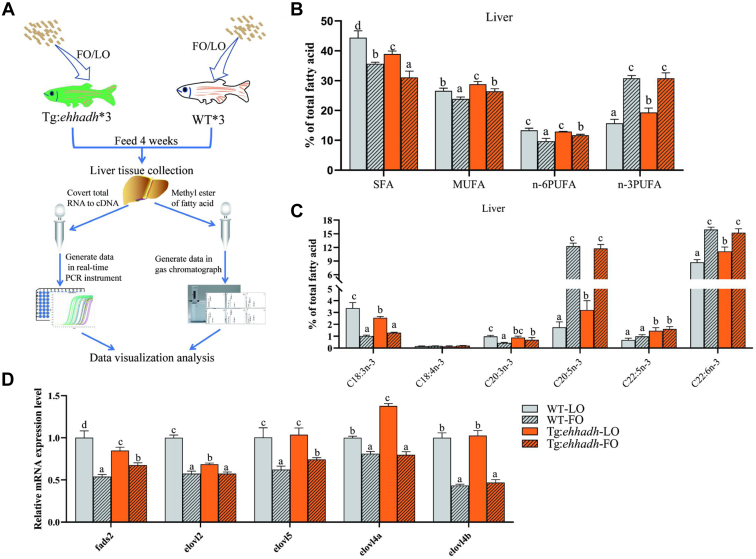
Next, we conducted a 4-week feeding experiment with LO and FO as dietary fat sources (Fig. 5A), and we divided WT and Tg:Ehhadh into four groups (WT-LO, Tg:Ehhadh-LO, WT-FO, and Tg:Ehhadh-FO). The results showed that the n-3 PUFA content in zebrafish liver was significantly higher in the Tg:Ehhadh-LO group than WT-LO, but it was significantly lower in the two FO groups (Fig. 5B). In addition, the n-3 PUFA content in zebrafish liver was not significantly different between the two FO groups (Fig. 5B). The contents of C20:5n-3 and C22:6n-3 in two FO groups were significantly higher than in two LO groups, whereas the contents of C18:3n-3 and C20:3n-3 were significantly lower in two FO groups than in two LO groups (Fig. 5C). In the LO group, the contents of C20:5n-3, C22:5n-3, and C22:6n-3 in the liver of Tg:Ehhadh zebrafish were significantly higher than those of WT (Fig. 5C). In the FO group, the contents of C20:5n-3 and C22:6n-3 in Tg:Ehhadh zebrafish liver were not significantly different from those in WT (Fig. 5C).
Fig. 5.
Fatty acid composition of liver and mRNA expression levels of fatty acid metabolism-related genes of zebrafish (WT) and Ehhadh transgenic zebrafish (Tg:Ehhadh) fed with linseed oil (LO) or fish oil (FO) diet for 4 weeks. A: The flow chart of feeding experiment. B: Liver ∑SFA, ∑MUFA, ∑n-6PUFA, and ∑n-3PUFA composition of WT and Tg:Ehhadh. C: Liver fatty acid n-3PUFA compositions of WT and Tg:Ehhadh. D: The mRNA expression levels of fads2, elovl2, elovl5, elovl4a, and elovl4b in livers of WT and Tg:Ehhadh. The different letters above the bars indicated significant difference (P < 0.05). Ehhadh, enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase; elovl2, fatty acid elongase 2; elovl4a, fatty acid elongase 4a; elovl4b, fatty acid elongase 4b; elovl5, fatty acid elongase 5; fads2, fatty acid desaturase 2; MUFA, mono-unsaturated fatty acid; PUFA, poly-unsaturated fatty acid; SFA, saturated fatty acid.
In order to explore the effect of different feeding treatments on PUFA synthesis, we detected the expression of genes related to PUFA synthesis (Fig. 5D). The results showed that the mRNA expression levels of fads2, elovl2, elovl5, elovl4a, and elovl4b were significantly higher in LO group than in FO group. In addition, the mRNA expression levels of fads2 and elovl2 in liver were significantly lower in Tg:Ehhadh-LO than in WT-LO, whereas the mRNA expression level of elovl4a was significantly higher in Tg:Ehhadh-LO than in WT-LO. The mRNA expression levels of fads2 and elovl5 in liver were significantly higher in Tg:Ehhadh-FO than in WT-FO.
Discussion
In this study, we used gene mutation analysis to clarify the role of Ehhadh in DHA synthesis. The results showed that Ehhadh deletion/overexpression resulted in a significant decrease/increase in DHA content, indicating that Ehhadh played an important role in DHA synthesis in zebrafish.
Freshwater fish have a stronger DHA synthesis capacity than mammals and marine fish (22). In general, DHA can be synthesized from ALA directly by Elovls and Fads in freshwater fish (14, 23). In addition, DHA can be synthesized in the “Sprecher” pathway, which is a peroxisome-dependent process (15). In the peroxisome, C24:6n-3 is oxidized into DHA in the presence of straight-chain acyl-CoA oxidase (Scox), Hsd17b4, 3-ketoacyl-CoA thiolase (Kat), and sterol carrier protein X (Scpx) through a series of enzymatic reactions (24). According to reports, Hsd17b4 is the only bifunctional enzyme involved in C24:6n-3 hydration and dehydrogenation during C24:6n-3 β-oxidation process, and very long-chain fatty acid accumulation was observed in the liver of Hsd17b4 knockout mice, while no phenotypic abnormality was found in Ehhadh-knockout mice (25, 26). In this study, the Ehhadh-deletion zebrafish showed C24:6n-3 accumulation and a decrease in DHA content, while Ehhadh-overexpressing zebrafish exhibited increased DHA content, suggesting that Ehhadh might be involved in the “Sprecher” pathway. This conclusion is also supported by related studies. The in vitro studies showed that Hsd17b4 deletion in mice did not completely prevent β-oxidation of [3-14C]24:6n-3, since radio-labeled C22:6n-3 could still be detected (16). The most potential explanation lies in that Ehhadh can act on substrate [3-14C]24:6n-3. A recent study has demonstrated that Ehhadh may play a role in DHA synthesis. C24:6n-3 has been reported to be significantly accumulated in the kidneys of Ehhadh-knockout mice (7). In this study, the downregulation or upregulation of fads2 and elovl gene expression respectively caused by Ehhadh deletion or overexpression might be an important reason for the significant decrease or increase of DHA content. Moreover, LO diet-feeding experiment further demonstrated that Ehhadh played a direct role in DHA synthesis. Compared with that of WT, the DHA content in liver of Tg:Ehhadh was significantly increased, while the mRNA level of fads2 and elovl2 genes was significantly decreased. Therefore, it could be concluded that Ehhadh was an essential for DHA synthesis in the “Sprecher” pathway.
In addition to direct participation of Ehhadh in the “Sprecher” pathway to synthesize DHA, Ehhadh deficiency inhibited the synthesis pathway of n-3 PUFA, thus leading to significant accumulation of C18:3n-3 and C20:5n-3 (abundant fatty acids in A. salina, supplemental Fig. S6 and Fig. 1D). In order to explore the possible influence of Ehhadh on n-3 PUFA synthesis, we analyzed WT and Ehhadh−/− liver transcriptional profiles. Our protein-protein interaction results showed that Ehhadh was strongly correlated with the 18 DEGs. KEGG functional enrichment analysis showed that the 18 DEGs fell into five functional categories namely, biosynthesis of unsaturated fatty acids, peroxisome, PPAR signaling pathway, fatty acid degradation, and insulin signaling pathway. Our further investigation revealed that in PPAR signaling pathway and insulin signaling pathway, there were two key regulators, sterol regulatory element binding transcription factor 1 (srebf1) and peroxisome proliferator activated receptor a (pparα), and srebf1 and pparα have been reported to regulate lipid metabolism by mediating the expression of downstream target genes, and thus they play an important role in maintaining cell homeostasis (27). Of these two regulators, srebf1 promotes fat synthesis by regulating the genes related to fatty acid, triglyceride, cholesterol synthesis (28). The other regulator ppara regulates target genes to promote fatty acid oxidation and thermogenesis for energy supply (29). This study showed that the expression levels of genes regulated by srebf1 and pparα were significantly lower in Ehhadh−/− relative to WT. The expressions of numerous genes involved in fatty acid transport, fatty acid synthesis, and fatty acid catabolism were significantly decreased, including abcd3, fads2, elovl8b, fabp3, and hsd17b4, four of which (abcd3a, fads2, elovl8b, and hsd17b4) have been reported to be involved in the synthesis of n-3 PUFA, and the downregulation of these four genes will significantly reduce the content of n-3 PUFA (30, 31, 32, 33). In addition, Ehhadh overexpression significantly upregulated the expressions of pparα, srebf1, and their downstream genes (Fig. 4D), thus promoting the synthesis of n-3 PUFA. Based on these findings, we speculated that Ehhadh deletion might interfere with n-3 PUFA synthesis by affecting the expression of transcriptional regulators and genes related to n-3 PUFA synthesis. However, the mechanism by which Ehhadh affects n-3 PUFA synthesis via PPAR and SREBF signaling pathways remains to be further explored.
In general, there are two main DHA metabolic pathways. One is that DHA acts as a precursor substance to form the compound required by the body, and the other metabolic pathway is that DHA is oxidized into other active substances (34). One previous study has shown that β-oxidation of DHA occurs only in peroxisomes, since only peroxisomes contain very long-chain acyl-CoA synthetases to activate DHA (22). In this study, we found that the elevated expression of peroxisomal fatty acid β-oxidation genes in the liver of Tg:Ehhadh zebrafish did not lead to a decrease in DHA content. The possible reason for this might be that DHA β-oxidation is inefficient in vivo, and thus DHA tends to be deposited in various tissues (35), which was confirmed by our subsequent feeding experiments on LO diet (rich in ALA) and FO diet (rich in EPA and DHA). In our experiments, when LO diet was fed, the DHA content in the liver of Tg:Ehhadh was increased significantly, compared with that of WT. In the FO diet group, there was no significant difference between WT and Tg:Ehhadh in liver. The DHA content in fish has been reported to be affected by dietary fatty acids. When dietary DHA lacks, the fish can synthesize DHA from the substrate fatty acids through a series of biological enzymes (36). This study found that the overexpression of Ehhadh further promoted the synthesis of DHA in LO diet feeding group. When the dietary DHA is sufficient, the fish′s endogenous synthesis pathway is blocked. Dietary DHA is absorbed by fish, most of which was synthesized into triglycerides, rather than being oxidized and decomposed as the body's main energy source (37). In this study, the DHA content in Tg:Ehhadh fed with FO diet was not significantly different from that in WT, indicating that the oxidation of DHA in vivo remained inefficient even at high DHA content and β-oxidation capacity. Therefore, this study showed that Ehhadh had a greater effect on DHA synthesis than on β-oxidative decomposition in zebrafish, which might be attributed to the fact that at low DHA diet, overexpression of Ehhadh can improve DHA endogenous synthesis, whereas at high DHA diet, overexpression of Ehhadh does not affect DHA deposition in vivo.
In conclusion, this study first revealed the role of Ehhadh in zebrafish by gene knockout and overexpression techniques. Our results indicate that 1) Ehhadh is essential for DHA synthesis in the “Sprecher” pathway, and 2) Ehhadh contributes more to DHA synthesis than to DHA oxidative decomposition in fish. This study not only provides new insights into the functions of Ehhadh in fish but also offers some references for increasing DHA content in fish.
Data availability
The transcriptome data have been uploaded into NCBI (SRA accession number: PRJNA896107). Further inquiries can be directed to the corresponding author.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
G. Y. and J. G. conceptualization; G. Y., S. S., J. H., and Y. W. investigation; G. Y., S. S., J. H., and Y. W. visualization; T. R. and H. H. formal analysis; G. Y. writing–original draft; J. G. funding acquisition; J. G. project administration; J. G. supervision; J. G. writing–review and editing.
Funding and additional information
This study was supported by the National Natural Science Foundation of China (31872579).
Supplemental data
References
- 1.Veldhoven P.V. Biochemistry and genetics of inherited disorders of peroxisomal fatty acid metabolism. J. Lipid Res. 2010;51:2863–2895. doi: 10.1194/jlr.R005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi R.L., Jiang L.L. Recent advances in peroxisomal fatty acid β-oxidation. Chin. J. Biochem. Mol. Biol. 2009;25:12–16. [Google Scholar]
- 3.Mayerhofer, Peter U. Targeting and insertion of peroxisomal membrane proteins: ER trafficking versus direct delivery to peroxisomes. Biochim. Biophys. Acta. 2016;1863:870–880. doi: 10.1016/j.bbamcr.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 4.Ranea-Robles P., Violante S., Argmann C., Dodatko T., Bhattacharya D., Chen H.J., et al. Murine deficiency of peroxisomal L-bifunctional protein (EHHADH) causes medium-chain 3-hydroxydicarboxylic aciduria and perturbs hepatic cholesterol homeostasis. Cell. Mol. Life Sci. 2021;78:5631–5646. doi: 10.1007/s00018-021-03869-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houten S.M., Denis S., Argmann C.A., Jia Y., Ferdinandusse S., Reddy J.K., et al. Peroxisomal L-bifunctional enzyme (Ehhadh) is essential for the production of medium-chain dicarboxylic acids. J. Lipid Res. 2012;53:1296–1303. doi: 10.1194/jlr.M024463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding J., Loizides-Mangold U., Rando G., Zoete V., Michielin O., Reddy J., et al. The peroxisomal enzyme L-PBE is required to prevent the dietary toxicity of medium-chain fatty acids. Cell Rep. 2013;5:248–258. doi: 10.1016/j.celrep.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 7.Ranea-Robles P., Portman K., Bender A., Lee K., He J.C., Mulholland D.J., et al. Peroxisomal L-bifunctional protein (EHHADH) deficiency causes male-specific kidney hypertrophy and proximal tubular injury in mice. Kidney. 2021;360:1441–1454. doi: 10.34067/KID.0003772021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metherel A.H., Bazinet P.R. Updates to the n-3 polyunsaturated fatty acid biosynthesis pathway: DHA synthesis rates, tetracosahexaenoic acid and (minimal) retroconversion. Prog. Lipid Res. 2019;76:101008. doi: 10.1016/j.plipres.2019.101008. [DOI] [PubMed] [Google Scholar]
- 9.Harayama T., Shimizu T. Roles of polyunsaturated fatty acids, from mediators to membranes. J. Lipid Res. 2020;61:1150–1160. doi: 10.1194/jlr.R120000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B., Tontonoz P. Phospholipid remodeling in physiology and disease. Annu. Rev. Physiol. 2019;10:165–188. doi: 10.1146/annurev-physiol-020518-114444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie D.Z., Chen C.Y., Dong Y.W., You C.H., Wang S.Q., Monroig Ó., et al. Regulation of long-chain polyunsaturated fatty acid biosynthesis in teleost fish. Prog. Lipid Res. 2021;82:101095. doi: 10.1016/j.plipres.2021.101095. [DOI] [PubMed] [Google Scholar]
- 12.Fonseca-Madrigal J., Navarro J.C., Hontoria F., Tocher D.R., Martínez-Palacios C.A., Monroig Ó. Diversification of substrate specificities in teleostei fads2: characterization of Δ4 and Δ6Δ5 desaturases of Chirostoma estor. J. Lipid Res. 2014;55:1408–1419. doi: 10.1194/jlr.M049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meesapyodsuk D., Xiao Q. The front-end desaturase: structure, function, evolution and biotechnological use. Lipids. 2012;47:227–237. doi: 10.1007/s11745-011-3617-2. [DOI] [PubMed] [Google Scholar]
- 14.Oboh A., Kabeya N., Carmona-Antoñanzas G., Castro L.F.C., Castro R.D., James D.R., et al. Two alternative pathways for docosahexaenoic acid (DHA, 22:6n-3) biosynthesis are widespread among teleost fish. Sci. Rep. 2017;7:3889. doi: 10.1038/s41598-017-04288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprecher H. Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim. Biophys. Acta. 2000;1486:219–231. doi: 10.1016/s1388-1981(00)00077-9. [DOI] [PubMed] [Google Scholar]
- 16.Ferdinandusse S., Denis S., Mooijer P.A.W., Zhang Z.Y., Reddy J.K., Spector A.A., et al. Identification of the peroxisomal β-oxidation enzymes involved in the biosynthesis of docosahexaenoic acid. J. Lipid Res. 2001;42:1987–1995. [PubMed] [Google Scholar]
- 17.Beek M.C.V.D., Dijkstra I.M.E., Kemp S. Method for measurement of peroxisomal very long-chain fatty acid beta-oxidation and de novo C26:0 synthesis activity in living cells using stable-isotope labeled docosanoic acid. Methods Mol. Biol. 2017;1595:45–54. doi: 10.1007/978-1-4939-6937-1_5. [DOI] [PubMed] [Google Scholar]
- 18.Li J.X., Yang C., Huang L.F., Zeng K.W., Cao X.J., Gao J. Inefficient ATP synthesis by inhibiting mitochondrial respiration causes lipids to decrease in MSTN-lacking muscles of loach Misgurnus anguillicaudatus. Funct. Integr. Genomics. 2019;19:889–900. doi: 10.1007/s10142-019-00688-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y., Cao X.J., Fu L.L., Gao J. n-3 PUFA reduction caused by fabp2 deletion interferes with triacylglycerol metabolism and cholesterolhomeostasis in fish. Appl. Microbiol. Biotechnol. 2020;104:2149–2161. doi: 10.1007/s00253-020-10366-9. [DOI] [PubMed] [Google Scholar]
- 20.Sun S.X., Castro F., Monroig Ó., Cao X.J., Gao J. fat-1 transgenic zebrafish are protected from abnormal lipid deposition induced by high-vegetable oil feeding. Appl. Microbiol. Biotechnol. 2020;104:7355–7365. doi: 10.1007/s00253-020-10774-x. [DOI] [PubMed] [Google Scholar]
- 21.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 22.Tocher D.R. Metabolism and functions of lipids and fatty acids in teleost fish. Rev. Fish. Sci. 2003;11:107–184. [Google Scholar]
- 23.Sun S.X., Ren T.Y., Li X., Cao X.J., Gao J. Polyunsaturated fatty acids synthesized by freshwater fish: a new insight to the roles of elovl2 and elovl5 in vivo. Biochem. Biophys. Res. Commun. 2020;532:414–419. doi: 10.1016/j.bbrc.2020.08.074. [DOI] [PubMed] [Google Scholar]
- 24.Moczulski D., Majak I., Mamczur D. An overview of beta-oxidation disorders. Postepy Hig. Med. Dosw. (Online) 2009;63:266–277. [PubMed] [Google Scholar]
- 25.Baes M. Inactivation of the peroxisomal multifunctional protein-2 in mice impedes the degradation of not only 2-methyl-branched fatty acids and bile acid intermediates but also of very long chain fatty acids. J. Biol. Chem. 2000;275:16329–16336. doi: 10.1074/jbc.M001994200. [DOI] [PubMed] [Google Scholar]
- 26.Ferdinandusse S., Denis S., Overmars H., Eeckhoudt L.V., Veldhoven P.P.V., Duran M., et al. Developmental changes of bile acid composition and conjugation in L- and D-bifunctional protein single and double knockout mice. J. Biol. Chem. 2005;280:18658–18666. doi: 10.1074/jbc.M414311200. [DOI] [PubMed] [Google Scholar]
- 27.Abeyrathna P., Su Y.C. The critical role of Akt in cardiovascular function. Vascul. Pharmacol. 2015;74:38–48. doi: 10.1016/j.vph.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krycer J.R., Sharpe L.J., Luu W., Brown A.J. The Akt-SREBP nexus: cell signaling meets lipid metabolism. Trends Endocrinol. Metab. 2010;21:268–276. doi: 10.1016/j.tem.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Lemberger T. Peroxisome proliferator-activated receptors: a nuclear receptor signaling pathway in lipid physiology. Annu. Rev. Cell Dev. Biol. 1996;12:335–363. doi: 10.1146/annurev.cellbio.12.1.335. [DOI] [PubMed] [Google Scholar]
- 30.Benedetto R.D., Denti M.A., Salvati S., Sanchez M., Attorri L., David G., et al. RNAi-mediated silencing of ABCD3 gene expression in rat C6 glial cells: a model system to study PMP70 function. Neurochem. Int. 2008;52:1106–1113. doi: 10.1016/j.neuint.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Stoffel W., Holz B., Jenke B., Binczek E., Günter R.H., Kiss C., et al. Delta6-desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 2008;27:2281–2292. doi: 10.1038/emboj.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Wen Z.Y., You C.H., Xie Z.Y., Tocher D.R., Zhang Y.L., et al. Genome wide identification and functional characterization of two LC-PUFA biosynthesis elongase (elovl8) genes in rabbitfish (Siganus canaliculatus) Aquaculture. 2020;522:735127. [Google Scholar]
- 33.Verheijden S., Bottelbergs A., Krysko O., Krysko D.V., Beckers L., Munter S.D., et al. Peroxisomal multifunctional protein-2 deficiency causes neuroinflammation and degeneration of purkinje cells independent of very long chain fatty acid accumulation. Neurobiol. Dis. 2013;58:258–269. doi: 10.1016/j.nbd.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Madsen L., Frøyland L., Dyrøy E., Helland K., Berge R.K. Docosahexaenoic and eicosapentaenoic acids are differently metabolized in rat liver during mitochondria and peroxisome proliferation. J. Lipid Res. 1998;39:583–593. [PubMed] [Google Scholar]
- 35.Demar J.C., Ma K., Bell J.M., Rapoport S.I. Half-lives of docosahexaenoic acid in rat brain phospholipids are prolonged by 15 weeks of nutritional deprivation of n-3 polyunsaturated fatty acids. J. Neurochem. 2010;91:1125–1137. doi: 10.1111/j.1471-4159.2004.02789.x. [DOI] [PubMed] [Google Scholar]
- 36.Li B.S., Wang J.Y., Huang Y., Hao T.T., Wang S.X., Huang B.S., et al. Effects of replacing fish oil with wheat germ oil on growth, fat deposition, serum biochemical indices and lipid metabolic enzyme of juvenile hybrid grouper (Epinephelus fuscoguttatus♀ × Epinephelus lanceolatus♂) Aquaculture. 2019;505:54–62. [Google Scholar]
- 37.Kitson A.P., Metherel A.H., Chen C.T., Domenichiello A.F., Trépanier M.O., Berger A., et al. Effect of dietary docosahexaenoic acid (DHA) in phospholipids or triglycerides on brain DHA uptake and accretion. J. Nutr. Biochem. 2016;33:91–102. doi: 10.1016/j.jnutbio.2016.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome data have been uploaded into NCBI (SRA accession number: PRJNA896107). Further inquiries can be directed to the corresponding author.