Significance
Transcriptional pausing provides a hub for gene regulation. Pausing provides a timing mechanism to coordinate regulatory interactions, cotranscriptional RNA folding and protein synthesis, and stop signals for transcriptional termination. Cellular RNA polymerases (RNAPs) are complex, with multiple mobile modules shifting positions to control its catalytic activity and pause RNAP in response to DNA-encoded pause signals. Understanding how these modules move to enable pausing is crucial for a mechanistic understanding of gene regulation. Our results clarify the picture significantly by defining multiple states among which paused RNAP partitions in response to different pause signals. This work contributes to an emerging theme wherein multiple interconverting states of the RNAP proceed through a pathway (e.g., initiation or pausing), providing multiple opportunities for regulation.
Keywords: RNA polymerase, transcriptional pausing, transcriptional regulation, cryo-EM, Escherichia coli
Abstract
Transcriptional pausing underpins the regulation of cellular RNA synthesis, but its mechanism remains incompletely understood. Sequence-specific interactions of DNA and RNA with the dynamic, multidomain RNA polymerase (RNAP) trigger reversible conformational changes at pause sites that temporarily interrupt the nucleotide addition cycle. These interactions initially rearrange the elongation complex (EC) into an elemental paused EC (ePEC). ePECs can form longer-lived PECs by further rearrangements or interactions of diffusible regulators. For both bacterial and mammalian RNAPs, a half-translocated state in which the next DNA template base fails to load into the active site appears central to the ePEC. Some RNAPs also swivel interconnected modules that may stabilize the ePEC. However, it is unclear whether swiveling and half-translocation are requisite features of a single ePEC state or if multiple ePEC states exist. Here, we use cryo-electron microscopy (cryo-EM) analysis of ePECs with different RNA–DNA sequences combined with biochemical probes of ePEC structure to define an interconverting ensemble of ePEC states. ePECs occupy either pre- or half-translocated states but do not always swivel, indicating that difficulty in forming the posttranslocated state at certain RNA–DNA sequences may be the essence of the ePEC. The existence of multiple ePEC conformations has broad implications for transcriptional regulation.
Transcriptional pausing is an evolved feature of all cellular DNA-dependent RNA polymerases (RNAPs) responsible for the regulated expression of genes. At pause sites, interactions of RNAP with certain RNA–DNA sequences temporarily halt transcription for tens to thousands of times longer than the 10 to 20 ms average nucleotide addition cycle (NAC) (for Escherichia coli RNAP). These delays allow time for folding of nascent RNA into biologically active forms, interactions with diffusible regulators, coupling with translation or splicing, termination of transcription, or other events that regulate gene expression (1, 2).
A now widely accepted model posits that RNA–DNA interactions put RNAP into an initially paused off-pathway state called the elemental pause (1, 3–5). Formation of the elemental paused elongation complex (ePEC) competes with the NAC rather than halting all RNAPs (Fig. 1A). Conformational fluctuations in the complex, multidomain RNAP, modulated by RNA–DNA sequence, govern this competition and ePEC lifetime. The ePEC can rearrange into longer-lived pauses by backtracking of the RNA and DNA chains, by interactions with nascent RNA secondary structures (pause hairpins; PHs), or by interactions with diffusible regulators (e.g., NusA or propausing NusG/Spt5) (6–9). RNA structures and regulators also can shorten or suppress elemental pausing (e.g., HK022 put RNA and λQ antiterminators or antipausing NusG/Spt5) (10–14). The elemental pause model arose from the observation of residual pausing when the E. coli his operon leader PH was deleted and from detection of nonbacktracked paused states in single-molecule experiments (3, 15, 16). The off-pathway, nonbacktracked nature of elemental pausing is confirmed by multiple studies (17–21). Genome-scale analyses reveal a consensus sequence for strong elemental pauses recognized by diverse RNAPs, including during transcription initiation and for mammalian RNAPII [5′-GGnnnnnntgYRccc, where YR corresponds to the pause RNA 3′ end and incoming nucleoside triphosphate (NTP)] (22–26).
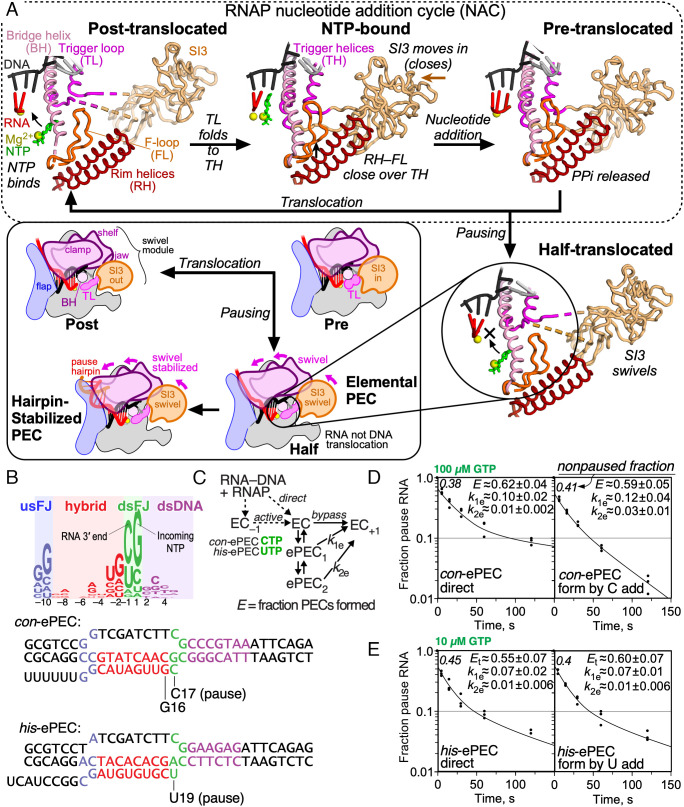
Fig. 1.
Elemental pausing and the RNAP NAC. (A) Active-site changes during the RNAP NAC. The BH, TL, SI3, RH, and FL all undergo conformational changes upon NTP binding, catalysis, pyrophosphate (PPi) release, and translocation. Pause sequences induce the formation of an elemental pause conformation in which the RNA but not DNA is translocated (half translocated). The ePECs can further rearrange by rotation of a swivel module that inhibits TL folding, t-DNA translocation, and NTP binding, and is stabilized by PH formation. The structural models are based on Protein Data Bank (PDB) coordinates 6ALF (posttranslocated), 6RH3 (NTP bound), con-ePEC-fTL (pretranslocated; this work), and his-ePEC-ufTL (half translocated; this work) (27, 28). (B) Consensus elemental pause sequence defined by nascent elongating–transcript sequencing in E. coli (22). The consensus consists of four distinct elements: the usFJ, RNA–RNA hybrid, dsFJ, and dsDNA. Two ePECs are relatively well studied: the consensus ePEC (con-ePEC) and the ePEC formed prior to PH formation in the his operon leader region (his-ePEC). Sequences shown are the scaffolds used for cryo-EM analysis. (C) Two methods to assemble ePECs were used and compared kinetically. Active ePEC formation was accomplished by reaction of cytidine 5′-triphosphate (CTP) or uridine 5′-triphosphate (UTP) with ECs assembled one nt upstream from the pause sites. Direct ePEC formation was accomplished by mixing RNAP with an RNA–t-DNA scaffold followed by binding of nt-DNA without any nucleotide addition. (D) Kinetic comparison of reaction with 100 µM guanosine 5′-triphosphate (GTP) of con-ePECs formed by the direct or active methods. The two methods yielded biphasic pause escape rates, equivalent fractions of slow and more slowly escaping ePECs, and slightly different escape rates. (E) Kinetic comparison of reaction with 10 µM GTP of his-ePEC formed by the direct or active methods. Both methods yielded biphasic pause escape rates and equivalent fractions and escape rates for the slow and more slowly escaping ePECs.
Cryo-EM of both E. coli and mammalian PECs reveals a half-translocated RNA–DNA hybrid (Fig. 1A) (6, 8, 29). In the half-translocated state, the recently added 3′ RNA nucleotide has cleared the active site to allow binding of the next NTP substrate. However, the template DNA (t-DNA) base needed to specify the next NTP remains paired to the nontemplate DNA (nt-DNA) strand in the downstream DNA channel. Completion of DNA translocation to allow NTP binding appears to limit escape from the ePEC and reentry into the NAC (19).
In the paused state, modest rotation of a swivel module of RNAP [by ~1.5° to 6° relative to an NTP-bound elongation complex (EC)] (28) appears to inhibit RNAP motions required to complete translocation, NTP binding, and catalysis (6, 8, 30). The swivel module comprises ~33% of the mass of bacterial RNAP and consists of the clamp, shelf, dock, jaw, β′C-term, and sequence insertion 3 (SI3) in E. coli RNAP (see SI Appendix, Table S1 for RNAP structural modules). Weak swiveling (~1.5° average) is evident in a low-resolution (~5.5 Å) ePEC structure and is detectable even in canonical ECs not bound to transcription factors or NTP (8, 30). Pronounced swiveling (~4.5° to 6°) is stabilized by PHs, NusA, or both. For E. coli RNAP, which contains the 188-aa SI3 insertion in the trigger loop (TL) (31), pronounced swiveling appears incompatible with SI3 movements required for TL folding and bridge helix (BH), rim helices (RH), and F-loop (FL) movements that position NTP for nucleotide addition (Fig. 1A) (6, 28, 32).
Results to date suggest that half-translocation and swiveling are characteristic of the paused state, but it remains unclear whether all PEC states are half-translocated and swiveled, whether multiple PEC states interconvert, and whether different pause signals may generate different pause states. To gain greater insight into elemental pause state(s), we used cryo-EM and biochemical probes of RNAP conformation and translocation to examine ePECs formed on a strong, consensus elemental pause signal (con-ePEC) and a weaker signal that forms ePECs prior to stabilization by a PH in the his operon leader region (his-ePEC). We find that pause sequences lead to both pre- and half-translocated states, the common feature being inhibition of achieving the posttranslocated state. In addition, multiple ePECs that differ in global RNAP conformation are observed on each pause sequence. Kinetic modeling suggests how these ePEC states might interconvert upon RNAP entry into a pause.
Results
Multiple Interconverting Pause States Form In Vitro.
To provide a kinetic framework for structural analysis of elemental pause states, we first compared formation and escape kinetics of the long-lived con-ePEC (19, 22) and the shorter-lived his-ePEC lacking the PH (6, 8, 15) (Fig. 1B; see SI Appendix, Table S2 for oligonucleotides and plasmids used in this study). We examined both ePECs using two different ways to make them (Fig. 1C). One method, often used for cryo-EM, was to form ePECs by direct reconstitution from RNAP mixed with RNA and DNA strands (Fig. 1C; direct). The other method was to form the same ePECs by nucleotide addition from the ECs reconstituted at the −1 position by incubation with CTP (con-ePEC) or UTP (his-PEC) (Fig. 1C; active). Consistent with prior analyses (15, 19), both con-ePEC and his-ePEC exhibited biphasic apparent rates of pause escape and a nonpaused subpopulation for both types of complexes (Fig. 1 D and E). Biphasic escape rates reflect the formation of multiple ePEC states. The nonpaused subpopulations reflect fractions of ECs that do not pause and confirm that ePECs form as off-pathway states. Also consistent with prior analyses, con-ePEC paused states were roughly ten times longer lived than his-ePEC states. This conclusion derives from their similar pause lifetimes at 10-fold different GTP concentrations (Fig. 1 D and E) well below the mM KNTPs for ePECs (22) where rate vs. [NTP] becomes pseudolinear, and is confirmed by much slower, monophasic con-ePEC escape at 10 µM GTP (19). Slight differences with prior analyses and between the two ways to generate con-ePECs reflect the known kinetic heterogeneity of ePECs. Different RNAP preps, scaffold lengths, and GTP concentrations readily shift the distribution of ePECs among paused (and nonpaused) states (19). The structural difference between fast and slow escaping ePECs has been unclear to date but is unlikely to involve backtracking. A 1-base pair (bp) backtrack is possible on the con-ePEC scaffold but it does not contribute to slow escape rates (19). Even 1-bp backtracking is strongly disfavored on the his-ePEC scaffold by an RNA–DNA base mismatch at −11 (Fig. 1B). We concluded that ePECs formed either by direct reconstitution or by one round of nucleotide addition were kinetically similar and were both suitable for structural analyses by cryo-EM.
The con-ePEC Occupies Pretranslocated States That Differ in TL Folding and SI3 Location.
Understanding of PEC structure to date has relied on the comparison of directly reconstituted PECs to nonpaused ECs formed on unrelated DNA–RNA sequences. To compare the con-ePEC to a mechanistically related EC, we formed con-ePECs by single-round nucleotide addition at 23 °C with 200 µM CTP from 10 µM con-ePEC−1 (i.e., actively formed con-ePECs from a reconstituted EC poised 1 bp preceding the pause). con-ePEC formed rapidly upon CTP addition to con-ePEC−1. Its pause escape behavior upon subsequent addition of GTP exhibited indistinguishable biphasic escape kinetics with ~8% nonpaused ECs (14 s to 2 min between CTP and GTP additions; SI Appendix, Fig. S1). Thus, at least two kinetically distinguishable con-ePEC states formed quickly remained unchanged in proportion for at least 2 min.
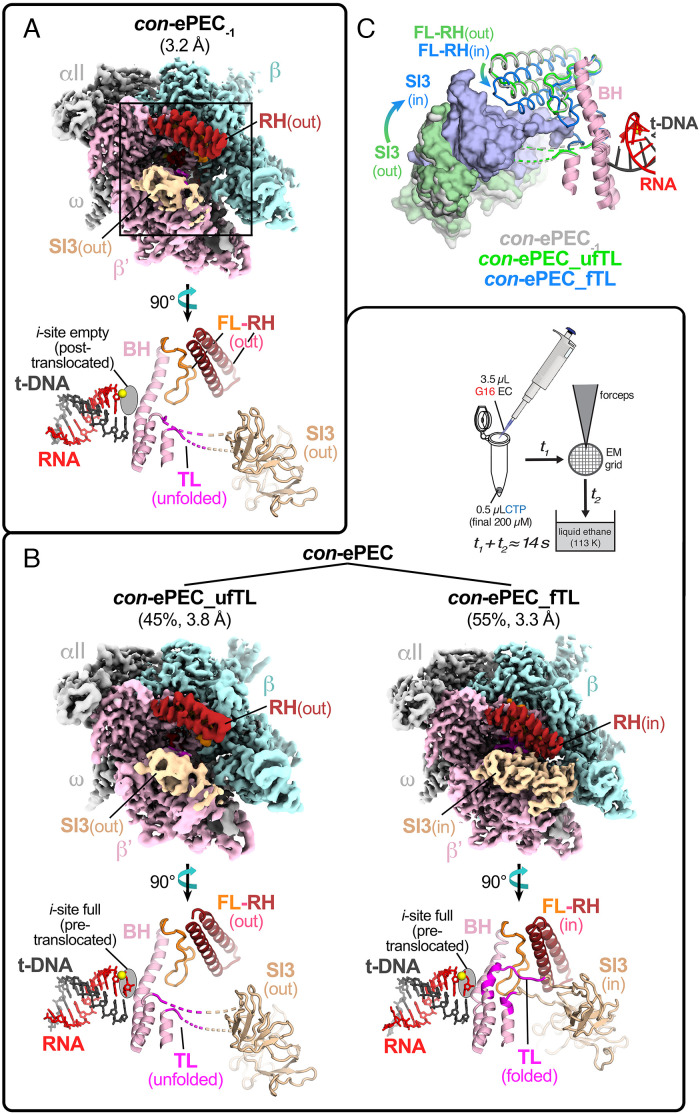
Guided by these results, we determined cryo-EM structures for both con-ePEC−1 and con-ePEC using >200K polished particles for each. con-ePEC was formed by adding 200 μM CTP to 10 μM con-ePEC−1 and plunge freezing in liquid ethane after 14 s at 23 °C (Fig. 2). In each case, we used three-dimensional classification to identify complexes containing intact scaffold (SI Appendix, Fig. S2 and Table S3). As expected, con-ePEC−1 was posttranslocated with an unfolded TL (3.2 Å resolution; Fig. 2A). It generally resembled the conformation of a previously determined posttranslocated EC structure containing an RNA 3′ A in the i site and t-DNA G in the open i + 1 site (27) (vs. 3′ rG and t-DNA C for con-ePEC−1).
Fig. 2.
Cryo-EM analysis of actively formed con-ePEC. (A) The overall cryo-EM structure and active-site conformation of con-ePEC−1. The cryo-EM map is colored by RNAP subunit or feature with β′ in light pink, β in pale cyan, α and ω in light gray, SI3 in wheat, and the RH in dark red. An enlarged and rotated region of con-ePEC−1 around the active site is shown using secondary structure cartoons below the cryo-EM map with additional features of FL in orange, BH in light pink, TL in light pink or magenta (flexible region), RNA in red, t-DNA in gray, and active-site Mg2+ in yellow sphere. (B) Inset (Upper Right of panel): con-ePECs were formed by ~14 s reaction at ~23 °C of directly reconstituted con-ePEC−1 with 200 µM CTP that included time on cryo-EM grid before plunge freezing in liquid ethane. The overall cryo-EM structures and active-site conformations of con-ePEC_ufTL and con-ePEC_fTL are shown, with cryo-EM maps colored by RNAP subunits and features as described in A. The percentages refer to the relative amounts of the two structures observed by cryo-EM (SI Appendix, Fig. S3). Enlarged and rotated regions around the active site are shown for each con-ePEC structure below the cryo-EM maps as described for con-ePEC−1 in B. (C) Comparison of the locations of SI3, RH, and FL in con-ePEC−1 (light gray), con-ePEC_ufTL (green), and con_ePEC-fTL (light blue). The BH and TL-helices (light pink) are shown for reference.
con-ePEC was exclusively pretranslocated, in contrast to the half-translocated his-ePEC observed previously (6, 8). The pretranslocated con-ePECs sorted into two distinct classes (SI Appendix, Tables S3 and S4). One subpopulation (con-ePEC_ufTL, ~45% of particles; 3.8 Å resolution) contained an unfolded TL and SI3 in the open position (Fig. 2B). A second subpopulation (con-ePEC_fTL, ~55% of particles; 3.2 Å resolution) contained a folded TL with SI3 in the closed position (similar to a previously described pretranslocated initiation complex except lacking σ70) (32, 33). In con-ePEC_fTL, the RH–FL module was rotated 11.9° toward the top of the folded TL (Fig. 2C). Thus, con-ePEC_fTL resembled most closely an NTP-bound EC in which the TL was folded and the RH–FL module was rotated (PDB 6RH3) (28). However, con-ePEC_fTL and con-ePEC_ufTL differed from each other at the upstream fork junction (usFJ). In con-ePEC_fTL, the RNA–DNA hybrid was 10 bp; the −11 RNA and partner t-DNA nucleotides were separated. In con-ePEC_ufTL, the hybrid was nearly 11 bp because the −11 RNA and t-DNA nucleotides rotated back toward the hybrid on the main cleft side of the lid, approaching bp distance (~6 Å) (SI Appendix, Fig. S3). Assuming con-ePEC_fTL is the first paused state formed after nucleotide addition, formation of a partial −11 bp in con-ePEC_ufTL may reflect failure to translocate upon TL unfolding and a conformational shift that, instead, allows the usFJ to rearrange. Formation of this −11 near-bp may stabilize the pretranslocated con-ePEC and explain the conservation of the strong −11 rG–dC bp in the consensus ePEC pause signal (22, 24).
These con-ePEC structures represent paused conformations in which the EC remains pretranslocated with multiple TL conformations. Occupancy of predominantly pretranslocated rather than half-translocated states by the con-ePEC is consistent with prior findings that it is sensitive to pyrophosphorolysis and that the incoming t-DNA base is still paired to the nt-strand (19, 22).
The his-ePEC Is Mostly Half-Translocated and Swiveled but Includes Unique Pretranslocated States.
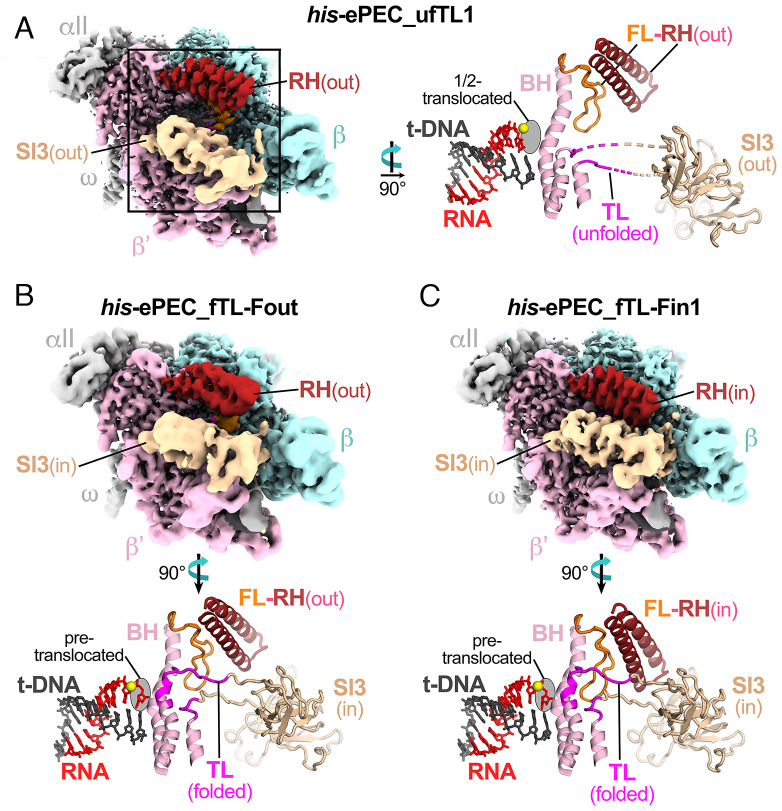
Although the differences between the con-ePEC and previously determined his-ePEC are striking, the his-ePEC structure was determined at relatively low resolution (5.5 Å) from a limited number of particles (6). Thus, we sought a more complete, higher-resolution structure of his-ePEC for comparison to con-ePEC. Since formation by direct reconstitution gave pause kinetics indistinguishable from actively formed his-ePEC (Fig. 1E), we reconstituted the his-ePEC on the same scaffold used previously for the hairpin-stabilized his-PEC but using an RNA lacking the PH (Fig. 1B). The resulting his-ePECs determined from ~900K polished particles yielded five distinct conformations (SI Appendix, Figs. S4 and S8 and Tables S3 and S4). The dominant his-ePEC states (73% of the his-ePEC particles) were half translocated with an unfolded TL but differed from the previously determined his-ePEC structure and from each other by varying degrees of swiveling. The most swiveled his-ePEC, his-ePEC_ufTL1, was the most populated, representing 49% of the total his-ePEC particles (Fig. 3A). The less-swiveled state, his-ePEC_ufTL2, represented 24% of the his-ePEC particles (Fig. 3B). The his-ePEC_ufTL2 state is likely populated with many intermediate-swiveled states, giving rise to the poor resolution of the reconstruction (5.5 Å nominal resolution despite being populated with ~211K particles; SI Appendix, Table S3). Thus, we propose that the his-ePEC_ufTL states comprise a favored, relatively homogeneous swiveled state (his-ePEC_ufTL1) and a population of particles sampling less-swiveled states.
Fig. 3.
Cryo-EM analysis of directly reconstituted his-ePEC. (A) The overall cryo-EM structure and active-site conformation of his-ePEC_ufTL1. RNAP subunits and features in the cryo-EM map and rotated active site view are depicted as described for con-ePEC−1 in Fig. 2B. The half-translocated state of his-ePEC_ufTL1 is evident from translocation of the RNA 3′ nucleotide but not its t-DNA partner. The swiveled conformation of his-ePEC-ufTL1 is depicted in Fig. 4. (B) The overall cryo-EM structure and active-site conformation of his-ePEC_fTL-Fout. Graphic details are as described for con-ePEC−1 in Fig. 2B. (C) The overall cryo-EM structure and active-site conformation of his-ePEC_fTL-Fin1. Graphic details are as described for con-ePEC−1 in Fig. 2B.
The remaining ~27% of the his-ePEC particles were pretranslocated with the folded TL and SI3 in the closed conformation similar to con-ePEC_fTL (Fig. 2C). Three distinct conformations of the RH–FL module could be resolved among these particles. In his-ePEC fTL-Fout (6.6% of particles; Fig. 3C), the RH–FL module position was similar to that observed in ECs with an unfolded TL (such as con-ePEC−1; Fig. 2A) as well as many other E. coli RNAP structures. In his-ePEC_fTL-Fin1 (17% of his-ePEC particles; Fig. 3C), the RH–FL module rotates 12.3° onto the TL (Fig. 3B). In his-ePEC_fTL-Fin2 (3.4% of the his-ePEC particles), the RH–FL module rotates an additional 1.6° toward the TL (SI Appendix, Fig. S8). The movement of the RH–FL module in his-ePEC_fTL-Fin structures resembles that in a previously reported CTP-bound EC in which the RH–FL module was rotated down over the folded TL (28).
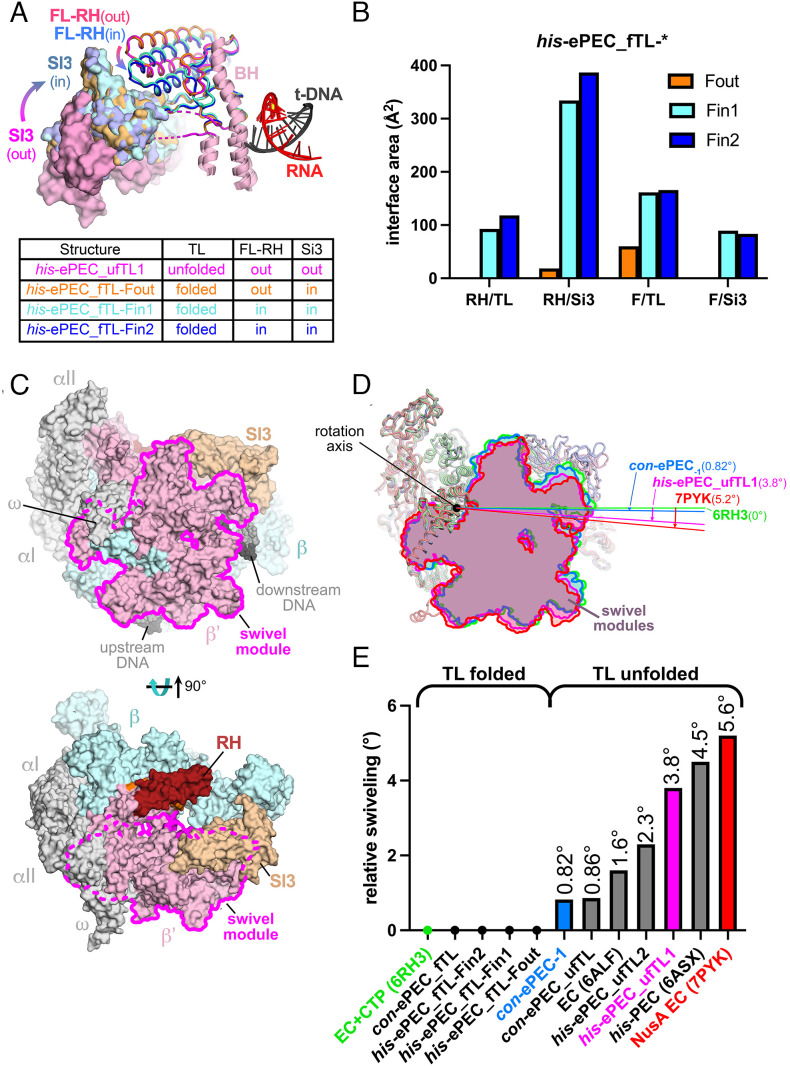
Other than the RH–FL module changes, the folded-TL his-ePEC structures were nearly identical (maximum rmsd of 0.577 Å over 3,104 α-carbon positions), giving us an opportunity to assess the effects of the RH–FL conformational changes on contacts between important RNAP structural modules. Rotation of the RH–FL module onto the TL in the his-ePEC_fTL-Fin structures generated substantial interface areas between the RH and TL (RH–TL interface area ~105 Å2) and the FL–SI3 (~86 Å2), whereas there were no contacts (0 Å2 interface area) in his-ePEC_fTL-Fout (Fig. 4 A and B). The RH–SI3 and FL–TL contacts also increased substantially upon RH–FL closing onto the folded TL (Fig. 4B).
Fig. 4.
Conformational changes among his-ePEC states. (A) Comparison of the locations of SI3, RH, and FL in four his-ePEC states. his-ePEC states are colored as indicated in the figure. The table indicates the status of the TL, the FL–RH modules, and SI3 in each state. (B) The area of the solvent-inaccessible interface between RNAP modules in three his-ePEC folded-TL states. The interface areas (34) in Å2 between two modules separated by a forward slash are plotted for his-ePEC_Fout (orange), his-ePEC_Fin1 (cyan), and his-ePEC_Fin2 (blue). (C) The boundary of the swivel module (magenta) comprised the RNAP shelf, clamp, jaw, β′ C-terminal region, and SI3 when viewed from the ω side (Top, corresponding to the axis of swivel module rotation) or secondary-channel side (Bottom) of RNAP. RNAP subunits are colored as described for con-ePEC−1 in Fig. 2B. (D) Rotation of the swivel module around the swivel axis for representative ECs and PECs colored as depicted in the figure. (E) The angle of swivel module rotation for ECs and PECs relative to an NTP-bound EC (PDB 6RH3) (30). The differences in rotational angle among the ECs and PECs plotted at 0° are not reliably distinguishable.
The his-ePEC_fTL-Fout and his-ePEC_fTL-Fin structures all have a folded TL, revealing that FL movement and TL unfolding are not tightly coupled. Retention of the folded TL in the his-ePEC_fTL-Fout conformation indicates that RNA–DNA sequence–determined interactions in the ePEC may inhibit TL unfolding. These interactions may disfavor forward translocation and favor retention of folded TL–nucleic acid interactions in the ePEC as one way for pause sequences to prolong pausing.
TL Unfolding Permits RNA–DNA Sequence-Dependent RNAP Swiveling.
The ePEC structures described here represent a large range of potential swivel angles (Fig. 4C shows the swivel module in the context of the con-ePEC−1 structure). To compare swivel angles, we performed structural superpositions based on the RNAP core module (SI Appendix, Table S1). As posited by Zhu et al. (30), a structure bound to the incoming CTP substrate with folded TL poised for catalysis (PDB 6RH3) is likely most representative of the catalytically active state and was used as a reference to compare other structures (0° swiveling). Swivel angles computed to be <0.5° were set to 0° (such small rotation angles are not meaningful because the rotation axis cannot be determined reliably). By contrast, an E. coli EC bound to NusA (generally pause promoting; 7PYK) (30) was found by our analysis to be swiveled 5.6° (Fig. 4 D and E). The seven ePEC structures reported here, along with the reference (6RH3) and some other relevant structures (6ALF, E. coli EC; 6ASX, his-PEC; 7PYK, E. coli NusA-EC) (6, 30), exhibit a range of swivel angles from ~0° to 5.6° (Fig. 4 D and E).
Previously, we found that swiveling promoted by the his-PEC PH (6ASX) allosterically inhibited TL folding by inhibiting SI3 closure (6). Our current results indicate that TL folding in turn inhibits swiveling; all of the compared structures containing a folded TL were unswiveled (~0° swiveling; Fig. 4E). Unfolding of the TL allows swiveling to occur, but to varying degrees (Fig. 4 D and E). All of the swiveled his-ePEC structures are substantially more swiveled (2.3° to 3.8°) than the swiveled con-ePEC structure (~0.8°), suggesting that swiveling is RNA–DNA sequence dependent.
Cys-Triplet Reporter (CTR) Verifies Large Differences in con-ePEC and his-ePEC States.
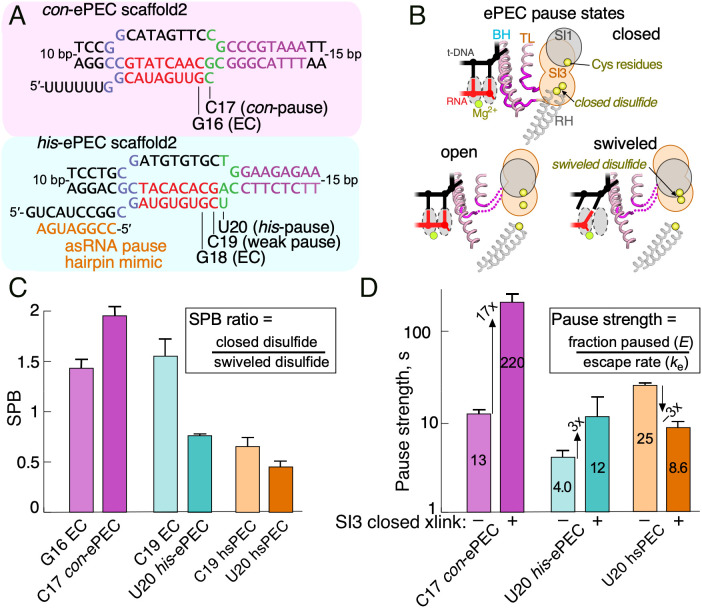
The strong pretranslocation bias of the con-ePEC differs from the half-translocated, swiveled states that dominated the directly reconstituted his-ePEC. We next asked whether these differences were also evident under conditions of active transcription in solution, using longer, fully complementary scaffolds that lack perturbing influences of an artificial transcription bubble and short downstream DNA present in cryo-EM complexes (Fig. 5A and SI Appendix, Fig. S5). We first used a CTR assay to distinguish closed and swiveled positions of SI3 in con-ePEC and his-ePEC (Fig. 5B and SI Appendix, Fig. S5B) (35). In the CTR assay, the closed SI3 forms a disulfide between engineered β′1051C in the SI3 hairpin loop and β′671C at the tip of the RH. The swiveled SI3 instead forms a β′1051C disulfide with β267C in SI1 and the two disulfides are distinguishable by nonreducing sodium dodecyl sulfate–polyacryamide gel electrophoresis (SI Appendix, Fig. S5C). The ratio of the closed to swiveled disulfide, denoted SI3 positional bias (SPB), is a relative measure of influence of transcription complex conformation on SI3 position (35).
Fig. 5.
Measurement of SI3 location by CTR assay and pause kinetics for con-ePEC and his-ePEC. (A) The scaffold sequences used for CTR and pausing assays of con-ePEC and his-ePEC. These scaffolds are fully complementary and contain sufficient duplex downstream DNA to avoid perturbing effects on translocation register (see SI Appendix, Fig. S5A for complete sequences). (B) Diagrammatic representation of Cys residue locations near the active site of RNAP for CTR assay in SI3 closed, open, and swiveled states (35). (C) SPB as calculated from the CTR assay for ECs and PECs. A higher SPB indicates greater occupancy of the SI3 closed conformation. Note that SPB reflects changes in conformational ratios and not their absolute values (35). The increase in SPB for con-ePEC vs. con-ePEC−1 and decrease in SPB for his-ePEC vs. his-ePEC−1 indicate that con-ePEC favors the closed SI3, presumably pretranslocated conformation, whereas his-ePEC favors the swiveled SI3, presumably half-translocated conformation. (D) Pause strengths of paused ECs with and without a disulfide cross-link that restrains SI3 in the closed position (SI3 closed xlink). The his-ePEC results depicted here are from experiments reported previously (35).
To ask whether SPB differed in con-ePEC and his-ePEC, we reconstituted the two ePECs by active elongation from an EC positioned one nucleotide upstream of the pause site (G16 for con-ePEC or C19 for his-ePEC). his-ePEC−1 is known to also pause weakly (36). When compared to the −1 ECs, SPB increased for con-ePEC and decreased for his-ePEC (Fig. 5C). Consistent with previous results, formation of a PH mimic using an antisense RNA further decreased SPB for his-ePEC. These results indicate that SI3 became more biased toward the closed position in con-ePEC, consistent with the pretranslocated con-ePECs observed by cryo-EM. Conversely, SI3 became more swiveled in the his-ePEC, also consistent with predominant occupancy of swiveled states observed by cryo-EM. Using an assay orthogonal to cryo-EM and scaffolds devoid of potentially perturbing influences, these results confirm that active con-ePEC and his-ePEC formed on scaffolds occupy predominantly pretranslocated and predominantly swiveled conformations, respectively.
The disulfides used in the CTR assay also can be used individually to bias ECs toward the SI3-closed or swiveled conformations (35). To ask whether biasing SI3 toward the closed conformation would affect the pause strength of con-ePEC and his-ePEC similarly or differently, we measured the effect of the closed disulfide on pause kinetics (Fig. 5D and SI Appendix, Fig. S5 D and E). Trapping the closed SI3 with a disulfide dramatically enhanced the pause strength and lifetime of the con-ePEC (Fig. 5D and SI Appendix, Fig. S5D). In contrast, the same disulfide modestly increased the strength of the his-ePEC and decreased the strength of the PH-stabilized hisPEC (Fig. 5D and SI Appendix, Fig. S5E). These contrasting effects of the closed SI3 disulfide are consistent with bias toward the pretranslocated state in the con-ePEC that is greatly strengthened by the disulfide and a bias toward the swiveled conformation in the his-ePEC that is countered by the disulfide, resulting in a modest effect on pause lifetime.
A Translocation Register Assay Confirms Different con-ePEC and his-ePEC States.
As a further test of differences in translocation bias of the con-ePEC and his-ePEC, we next examined reconstituted ePECs using an assay that traps ECs or PECs in their current translocation register with the phage HK022 Nun protein (27, 37). This assay reports the posttranslocated state based on its capacity for nucleotide addition and the pretranslocated state based on its sensitivity to pyrophosphorolysis (Fig. 6A and SI Appendix, Fig. S6). For this assay, we reconstituted con-ePEC and his-PEC on scaffolds similar to those used for cryo-EM but fully complementary to eliminate possible perturbation of translocation register caused by noncomplementarity (SI Appendix, Fig. S6A). It was unclear how the half-translocated PEC would be affected by Nun. We formed ePEC−1 by reconstitution, bound to Ni2+-nitrilotriacetic acid beads via an RNAP His10 tag, extended to ePECs by incubation with CTP (for con-ePEC) or UTP (for his-ePEC), treated with Nun, washed away NTPs, and then incubated with 1 mM GTP or 2.5 mM PPi to assay translocation register (SI Appendix, Fig. S6B). Nun-treated con-ePEC was more sensitive to pyrophosphorolysis and incorporated less GTP, whereas the opposite was true for his-ePEC and to an even greater extent for the hairpin-stabilized hisPEC (Fig. 6B and SI Appendix, Fig. S6 D–G). Since the hisPEC is known to be half translocated, we infer that the Nun-bound, half-translocated EC can react with NTP substrate. Taken together, these data are thus fully consistent with the cryo-EM structures, suggesting that con-ePEC is principally pretranslocated and his-ePEC is principally half translocated.
Fig. 6.
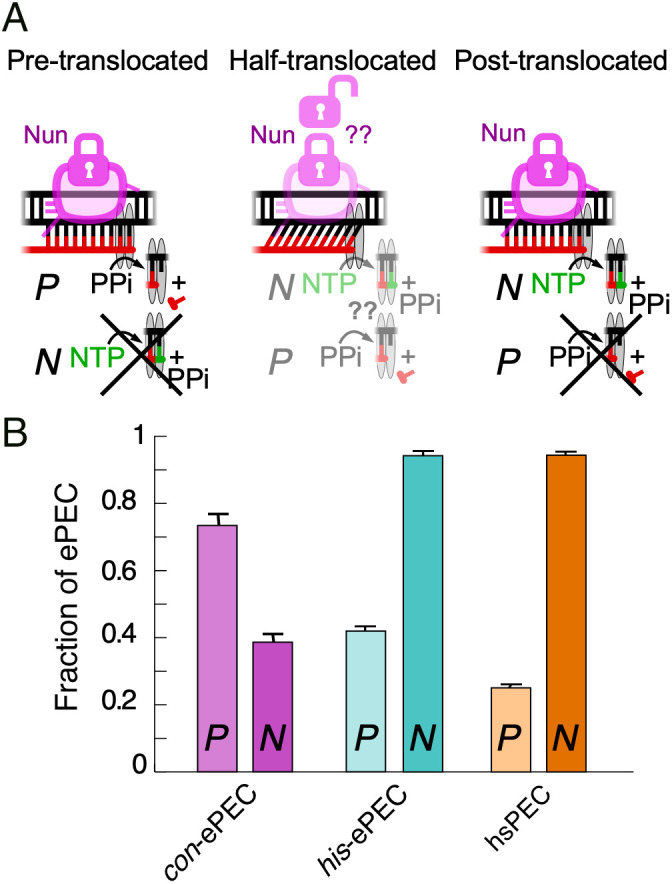
Translocation registers of con-ePEC and his-ePEC determined by Nun-locked assay (37). (A) Diagrammatic representation of Nun-locked translocation assay for pre-, half-, and post-translocated states. Nun locks translocation register via interactions with the usFJ and dsDNA. Pretranslocated ECs are susceptible to pyrophosphorolysis but not nucleotide addition. Posttranslocated ECs are susceptible to nucleotide addition but not pyrophosphorolysis. Whether Nun locks both the half-translocated state and its reactivity is uncertain, but these results are consistent with Nun-induced conversion to a posttranslocated state reactive with NTP but not PPi. (B) Fractions of PECs reactive with PPi (P) or NTP (N) after incubation with Nun. Greater reactivity with PPi and lesser reactivity with NTP of con-ePEC than his-ePEC or hairpin stabilized (hs) his-hsPEC are consistent with greater occupancy of pretranslocated register for con-ePEC and greater occupancy of half-translocated registers by his-ePEC and his-hsPEC as also seen by cryo-EM. Data are average and SD of three independent replicates from gels shown in SI Appendix, Fig. S6 C–E.
Multiple ePEC States Can Explain Biphasic Pause Kinetics and RNA–DNA Sequence Effects on Elemental Pausing.
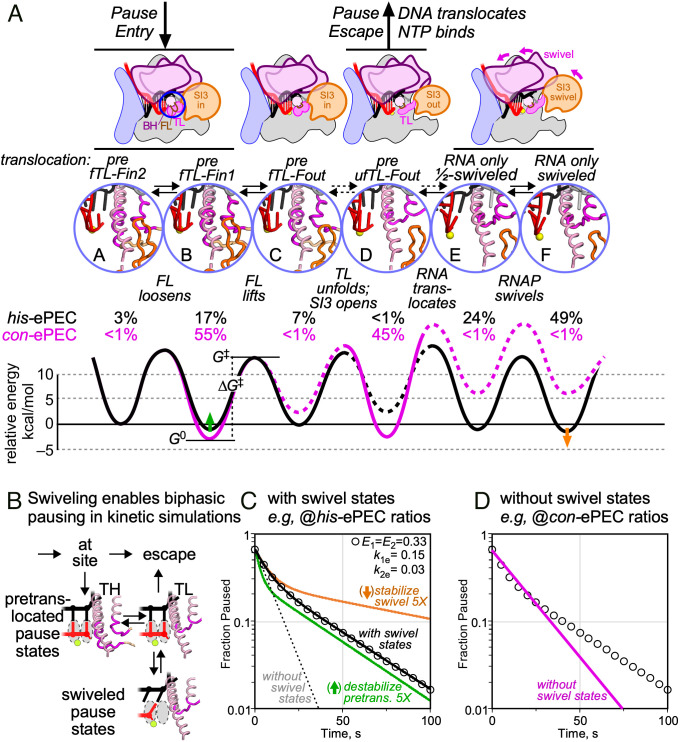
The identification of multiple coexisting ePEC states provides a potential explanation for multiphasic ePEC pause kinetics that were found previously not to involve backtracking (19). Sequence-dependent differences in the preferred states also may help explain how multipartite RNA–DNA sequences modulate pausing. To explore these explanations, we considered how the different ePEC states may be connected structurally and energetically (Fig. 7A and SI Appendix, Figs. S7–S9). The pretranslocated-folded TL (closed SI3) state should be the first to form when the EC arrives at a pause site because it is directly generated by the nucleotide-addition reaction. We thus assigned this state as the first paused state, either fTL–Fin2 state or fTL–Fin1 (states A or B, Fig. 7A).
Fig. 7.
The multiple intermediate model of elemental transcriptional pausing. (A) Distinct ePEC intermediates revealed by con-ePEC (intermediates B and D) and his-ePEC (intermediates A, B, C, E, and F) cryo-EM and their deduced energetic relationships (see SI Appendix, Table S4 for comparison to con-ePEC and his-ePEC intermediate names). All complexes are assigned to paused states in this analysis although it is possible that small amounts (e.g., portions of states A and C) could represent online ECs. The relative stabilities of the intermediates were calculated from cryo-EM particle distributions (percentages shown in black or magenta), assuming a constant rapid forward rate (100 s−1, left to right) and fitting reverse rates to observed intermediate occupancies (SI Appendix, Expanded Materials and Methods). The relative energy levels (G0) but not the interconversion rates (determined by ∆G‡) are constrained by the distributions of ePEC states (scale is relative not absolute). (B) The swivel model of pausing accounts for biphasic pause escape kinetics. (C) Fitting of the swivel model to a typical set of biphasic pause escape kinetics. Fraction of paused RNA remaining as a function of time was calculated using the parameters listed and a two-exponential rate of decay (open circles). A kinetic model that included off-pathway swiveled states E and F and fixed observed ratios of states A to F for his-ePEC fitted these data successfully (black line), whereas a kinetic model lacking off-pathway swiveled states was unable to fit the data (dotted line) (SI Appendix, Expanded Materials and Methods). Fivefold changes in the stabilities of pretranslocated state B (green line, corresponding to green arrow in A) or swiveled state F (orange line, corresponding to orange arrow in A) altered either the slow or slower components of the biphasic pause escape rates. Kinetic constants used to generate these data are shown in SI Appendix, Fig. S9. (D) Fitting of a kinetic model lacking off-pathway swiveled states using fixed ratios of states B and D as observed for con-ePEC was unable to give biphasic kinetics (magenta line; predicted data are same as for C; pause entry was to state B in this case).
The active site would then relax by movement of the FL and RH away from the folded TL (state C). This movement of the FL and RH would allow subsequent unfolding of the TL and shifting of SI3 to the open position to yield a pretranslocated complex ready for translocation (state D). We postulate that full translocation of both RNA and DNA from state D would create a posttranslocated EC ready for nucleotide addition and would constitute pause escape. However, translocation of only the RNA and not the DNA would form a half-translocated paused state associated with swiveling (state E). It is unclear whether swiveling leads to half translocation, or vice versa, or whether swiveling and half translocation are mutually reinforcing. Further swiveling would yield the swiveled ePEC observed in the his-ePEC that can be stabilized by PH formation (state F).
Because we knew the equilibrium distributions of the different states for the con-ePEC and his-ePEC from the cryo-EM particle distributions, we could estimate the relative energetics of their interconversion. In this simple model, con-ePEC principally occupies states B and D (magenta energy diagram; dotted lines indicate unoccupied states for which only limits on the energy diagram can be ascertained). The height of energy barriers between states dictates their rates of interconversion, whereas their relative occupancies at equilibrium depend on the relative positions of the troughs. The his-ePEC, in contrast, occupies states A, B, C, E, and F (black energy diagram).
This analysis provides a parsimonious hypothesis that can explain biphasic pause escape kinetics by formation of swiveled states (Fig. 7B). To illustrate this point, we fit a kinetic model with fixed ratios of the paused states corresponding to the his-ePEC cryo-EM states to a calculated two-exponential decay dataset that resembles the biphasic experimental data observed for his-ePECs (black circles, Fig. 7C; 0.33 pause bypass fraction and two 0.33 pause fractions escaping at 0.15 s−1 and 0.03 s−1). Fitting was accomplished using predictions of numerically integrated rate equations and least-squares minimization (Materials and Methods and SI Appendix, Fig. S9). These fixed his-ePEC state ratios readily fit the biphasic pause dataset when escape back from the swiveled states was slow (black line, Fig. 7C and SI Appendix, Fig. S9). However, a his-ePEC kinetic model lacking swiveled states was unable to generate biphasic pausing behavior (dotted line, Fig. 7C). Biphasic kinetics are possible with the swiveled states because they provide additional off-pathway states that ePECs can enter and escape from more slowly.
Small changes (by factors of 5) in the stabilities of the pause states predicted obvious changes in distinct phases of pause escape (green and orange lines, Fig. 7C). These changes illustrate how small changes caused by different RNA–DNA sequences or interactions with transcription factors may modulate pause kinetics.
However, swiveled states were not observed for con-ePEC even though it also exhibits biphasic pause escape kinetics. At least two considerations might explain this discrepancy. First, the method, scaffold, and conditions used for cryo-EM of con-ePECs may minimize biphasic kinetics. The actively formed con-ePECs exhibited less pronounced biphasic kinetics compared to direct reconstitution (Fig. 1D and SI Appendix, Fig. S1) and to con-ePECs formed on scaffolds with longer downstream DNA (19). Swivel states may arise and explain con-ePEC biphasic kinetics in these other cases.
Second, the contribution of swiveled states per se to biphasic kinetics is a hypothesis. It is attractive because it readily explains what has been otherwise mysterious: how biphasic pause escape kinetics arise without involvement of backtracking. However, any off-line pause state that is not an obligate intermediate to pause escape can explain multiphasic pause kinetics. Such states may appear similar or even indistinguishable from other states by cryo-EM even when they differ significantly in thermodynamic stability (G 0) and thus rates of change (see Discussion). For example, con-ePEC pre-fFLin (states A + B) could separate into two currently indistinguishable states—one that isomerizes to pre-ufTLout (state C) and one in which the TL remains folded and gives rise to the slower phase of con-ePEC escape. The observation of two similar fTL-Fin states in his-ePEC (states A and B) reinforces this point.
We favor the swivel hypothesis because it is parsimonious. Swiveling is only observed when the TL is unfolded (Fig. 4E). If nucleotide addition cannot be completed once RNAP swivels, then swiveling conveniently enables modulation of pause strength by interactions that stabilize the swiveled state(s) (e.g., of PHs or NusA) (6, 8, 30).
Discussion
RNA–DNA sequence-dependent pausing underpins transcriptional regulation. RNAP responds to pause signals by initially entering an offline, elemental paused state that is neither backtracked nor stabilized by nascent RNA structures or diffusible regulators. Half translocation (RNA but not DNA) has been associated with elemental pausing and swiveling with pause stabilization (6, 8, 29, 30), but a definitive structural description of the ePEC has remained elusive. By comparing two ePECs formed on different nucleic acid sequences, we found that the ePEC actually comprises a family of distinct paused states. Different ePEC conformations dominate for different pause signals. A strong consensus pause signal was predominantly pretranslocated, whereas a weaker signal found in the his biosynthetic operon leader region was predominantly swiveled even in the absence of the swivel-stabilizing, nascent PH. However, for each ePEC, multiple distinct conformations were resolved by cryo-EM. This view of the ePEC as an ensemble of interconverting states, rather than a single species, resolves some mysteries about elemental pausing, has implications for both transcriptional regulation and the general mechanism of nucleotide addition by RNAP, and raises questions for future study.
Multiple ePEC States Increase Regulatory Options.
The ability of ePECs to occupy multiple conformations creates opportunities for differential regulation of transcriptional pausing. Both RNA–DNA sequences and diffusible regulators may modulate the dwell time of RNAP at a pause as well as the fraction of RNAP molecules that isomerize into the ePEC vs. continuing rapid transcription. Together, the pause fraction and pause dwell time determine pause strength, which is the mathematical product of the two parameters (38, 39). When a single pause state exists, pause fraction and dwell time may be interdependent. This relationship has been observed for short-lived pauses by single-molecule methods at 23 °C (40). The existence of multiple interconverting pause states, on the contrary, could allow unlinked modulation of pause fraction and pause dwell time, creating increased regulatory flexibility.
For example, transcriptional regulators may modulate the dwell time of pauses by stabilizing or destabilizing swiveling without changing the fraction of RNAP molecules that pause (Figs. 7C and SI Appendix, Fig. S9B). PHs are thought to operate this way. Formation of the ePEC allows time for nascent PH formation. PH–RNAP interactions may then stabilize the swiveled state, which increases pause dwell time to allow time for regulator interactions. In E. coli, for example, ribosomes translating the nascent RNA may destabilize the swiveled state, trigger pause escape, and thus link RNAP movement to translational status (41).
Other cellular components that interact with nascent RNA may substitute for the ribosome in this type of indirect regulation of pausing (i.e., via RNA structure). For example, small molecules can bind the nascent RNA during riboswitch attenuation (42). The unlinking of pause dwell times from pause fraction that is possible due to the existence of multiple ePEC states allows these mechanisms to operate with high efficiency by ensuring that most RNAPs enter pauses that can be modulated by different interactions. For example, modulation of pausing by the universal transcription factor NusG and its paralogs like RfaH may operate via enhancement or suppression of swiveling (13).
Conversely, other interactions with RNAP may favor nonswiveled ePECs with different regulatory impacts. Nonswiveled pause states may facilitate backtracking, for example. Strong RNAP–nucleic acid interactions may stabilize the pretranslocated paused state so much that pause bypass becomes hard to detect (4, 19, 43). Nonswiveled pauses also might be especially important for halting antiterminated ECs. Diverse antitermination regulators modify ECs to promote transcript elongation, including by inhibiting swiveling (44–47). Antiterminated ECs appeared to be dissociated by specialized intrinsic terminators. Pausing in the pretranslocated register may be important for these specialized terminators where swiveling would remain suppressed by the antitermination modifications.
Regulation of Multiple Pause States May Be Analogous to Regulation of Multistep Transcription Initiation.
Decades of work have elucidated how RNAP initiates transcription at multipartite promoters [i.e., containing upstream (UP), −35, −10 elements, etc.] by a multistep process involving sequential intermediate states from promoter recognition to promoter escape (48, 49). Each step is subject to differential regulation by transcription factors and promoter sequences that affect different steps in the multistep initiation process (48, 50). For example, some factors or sequences may increase initial promoter melting, whereas others may stabilize the open complex.
The multiple paused–state model (Fig. 7) suggests regulatory opportunities that are strikingly parallel to the regulation of multistep transcription initiation. Like the multipartite promoter, pause sequences also are multipartite [e.g., PH, usFJ, hybrid, downstream fork junction (dsFJ), and downstream duplex DNA (dsDNA); Fig. 1B] (19, 51). Different regulators affect pausing through different interactions with these sequences (e.g., NusA with RNA structures, NusG with the usFJ and nt-DNA) (7, 8, 52). The multiple ePEC states defined here thus suggest regulatory opportunities for pausing that parallel the regulation of multistep transcriptional initiation. However, the roles of different pause signal components in controlling steps in the pause mechanism remain unknown (e.g., it is unclear which sequence elements control swiveling). Elucidating these connections will be an important focus for future research.
ePEC States: Steps in the NAC or Off-Pathway Paused States?
Whether swiveling and half translocation could be substeps that occur in every NAC or are uniquely off-pathway paused states requires consideration. Some ePEC states clearly resemble active NAC intermediates that occur at least transiently in every catalytic cycle (e.g., pretranslocated states before and after TL unfolding). However, ePEC states must differ in some way from active NAC states because a fraction of ECs rapidly bypasses the same template position at which ePECs form (Fig. 1 and SI Appendix, Fig. S1). This kinetic difference must reflect energetic differences in ePEC and active NAC states located at the same DNA position. The ground state free energy(s) (G0) of the ePEC state(s) may be lower than the corresponding active NAC state(s); the transition state free energy(s) (G‡) connecting one or more ePEC states to the active NAC may be higher than the analogous G‡ for the active NAC state(s); or both. Both effects increase the barrier (∆G‡) to ePEC escape back to the active NAC. Hence, both decrease the rate of pause escape.
However, the distinguishing features of similar ePEC and NAC states may not be obvious from cryo-EM structures alone. Strong vs. weak atomic contacts and thus contributions to G0 or G‡ may appear the same at currently achievable cryo-EM resolutions; key contacts may even remain unresolved. Thus, similar motions of the TL, FL, and RH may occur in the active NAC and in ePECs (Movie S1). This distinction between kinetic states and cryo-EM structural states may also explain why we did not resolve ~8% active EC distinct from con-ePEC states (Fig. 2 and SI Appendix, Figs. S1 and S2B). Further study will be required to understand the energetic contributions of NAC-resembling ePEC states to pausing.
Additionally, whether or not the swiveled and half-translocated paused states have analogous intermediates in the active NAC is uncertain. Swiveling can occur in halted ECs (30) and could in principle mediate RNA-first translocation in the NAC (53, 54). However, to be mechanistically important, swiveling, half translocation, or both must be energetically linked to RNA–DNA movements. ECs and ePECs exhibit large thermal motions on a microsecond time scale (55) and may transiently occupy many conformations. These transient conformations could include swiveled or half-translocated states during the active NAC. They become mechanistically significant only if the energy landscape of translocation required tight linkage among them. In other words, if translocation during the active NAC almost always proceeds with RNA before DNA or is almost always accompanied by swiveling, then half translocation and swiveling become mechanistically important. If not, then they may still occur as transient conformational excursions without mechanistic significance.
In contrast, available evidence indicates that both swiveling and half translocation are mechanistically important for pausing. Both states are catalytically inactive, represent the majority of some ePEC populations, and can be stabilized by PHs or pause-enhancing regulators (6, 8, 9). Swiveling kinks the BH toward the active site where it occludes DNA translocation and NTP binding and inhibits TL folding (6, 9). t-DNA translocation in the half-translocated ePEC appears to be the final energy barrier to pause escape (19). Thus, a parsimonious interpretation is that swiveling occurs in offline ePECs and is neither a necessary aid to translocation nor compatible with rapid catalysis during the NAC (30, 56).
Finally, we note that manual time-resolved detection of paused states did not yield clear evidence of intermediates that precede the apparently equilibrated states formed by direct reconstitution. For example, we did not detect complexes retaining pyrophosphate. Direct reconstitution of the his-ePEC revealed states similar to those formed by freeze capture of con-ePECs after nucleotide addition. Nonetheless, the approach of capturing intermediates shortly after nucleotide addition holds promise and may provide further insight into ePEC formation using methods able to achieve subsecond time resolution (57).
Materials and Methods
Further details are provided in SI Appendix, Expanded Materials and Methods.
Reagents and Materials.
Plasmids and oligonucleotides used in this study are listed in SI Appendix, Table S2. RNAPs were purified as described previously (35, 58) using expression plasmids pEcrpoA(-X234–241H)BCZ + pACYCDuet-1rpoZ (58, 59), pRM756 (60), or derived from pRM756 or pRM843 (61) and described in detail in SI Appendix, Expanded Materials and Methods.
Cryo-EM.
Transcription complex preparation and cryo-EM data collection and analyses were conducted as described previously and in detail in SI Appendix, Expanded Materials and Methods.
In Vitro Transcription Assays and Biochemical Assays.
In vitro transcription assays and biochemical analyses of disulfide cross-links and Nun-locked translocation were performed as described previously (19, 35) and in detail in SI Appendix, Expanded Materials and Methods.
Thermodynamic and Kinetic Modeling.
Thermodynamic and kinetic modeling was performed using Kintek Explorer (62) as described in SI Appendix, Expanded Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
The movie shows the movements of active-site modules (BH, TL, RH, FL) and nucleic acids that connect the different elemental PEC states observed for con-ePEC and his-ePEC in two side-by-side views of the active site of RNAP. Movements of the BH, RH, and FL that accompany TL folding and unfolding are also likely to occur during the active NAC.
Acknowledgments
We thank members of the Darst-Campbell and Landick Laboratories for experimental advice and helpful discussions and M. Ebrahim, J. Sotiris, and H. Ng at The Rockefeller University Evelyn Gruss Lipper Cryo-electron Microscopy Resource Center for help with cryo-EM data collection. This work was supported by Burroughs Wellcome Fund CASI award 1016945 to T.V.M. and NIH grants R35 GM118130 to S.A.D and R01 GM38330 to R.L.
Author contributions
J.Y.K., T.V.M., Y.B., J.C., S.A.D., and R.L. designed research; J.Y.K., T.V.M., Y.B., E.L., J.L., and R.L. performed research; J.Y.K., T.V.M., Y.B., J.C., J.L., S.A.D., and R.L. analyzed data; and J.Y.K., T.V.M., Y.B., J.C., S.A.D., and R.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Seth A. Darst, Email: darst@rockefeller.edu.
Robert Landick, Email: rlandick@wisc.edu.
Data, Materials, and Software Availability
Cryo-EM data have been deposited in the RCSB Protein Data Bank (www.pdb.org) and in the Electron Microscopy Data Bank (www.emdatabank.org). The PDB accession codes for the coordinates of con-ePEC−1, con-ePEC_fTL, con-ePEC_ufTL, his-ePEC_fTL-Fin1, his-ePEC-fTL-Fin2, his-ePEC_fTL-Fout, his-ePEC_ufTL1, and his-ePEC_ufTL2 are 8EG7 (63), 8EG8 (64), 8EGB (65), 8EH8 (66), 8EH9 (67), 8EHA (68), 8EHF (69), and 8EHI (70), respectively, and the accession codes for the cryo-EM maps are EMD-28109 (71), EMD-28110 (72), EMD-28113 (73), EMD-28143 (74), EMD-28144 (75), EMD-28145 (76), EMD-28146 (77), and EMD-28148 (78), respectively. All study data are included in the article, SI Appendix, or both. Previously published data were used for this work (DOI: 10.1073/pnas.2101805118).
Supporting Information
References
- 1.Landick R., Transcriptional pausing as a mediator of bacterial gene regulation. Annu. Rev. Microbiol. 75, 291–314 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Mayer A., Landry H. M., Churchman L. S., Pause & go: From the discovery of RNA polymerase pausing to its functional implications. Curr. Opin. Cell Biol. 46, 72–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landick R., The regulatory roles and mechanism of transcriptional pausing. Biochem. Soc. Trans. 34, 1062–1066 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Qian J., Dunlap D., Finzi L., Basic mechanisms and kinetics of pause-interspersed transcript elongation. Nucleic Acids Res. 49, 15–24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang J. Y., Mishanina T. V., Landick R., Darst S. A., Mechanisms of transcriptional pausing in bacteria. J. Mol. Biol. 431, 4007–4029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang J. Y., et al. , RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol. Cell 69, 802–815.e805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yakhnin A. V., Kashlev M., Babitzke P., NusG-dependent RNA polymerase pausing is a frequent function of this universally conserved transcription elongation factor. Crit. Rev. Biochem. Mol. Biol. 55, 716–728 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo X., et al. , Structural basis for NusA stabilized transcriptional pausing. Mol. Cell 69, 816–827.e814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delbeau M. Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis. bioRxiv [Preprint] (2022). 2022.2010.2021.513233 (Accessed 3 December 2022). [DOI] [PMC free article] [PubMed]
- 10.Hwang S., et al. , Structural basis of transcriptional regulation by a nascent RNA element, HK022 putRNA. Nat. Commun. 13, 4668 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin Z., Kaelber J. T., Ebright R. H., Structural basis of Q-dependent antitermination. Proc. Natl. Acad. Sci. U.S.A. 116, 18384–18390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grayhack E. J., Yang X., Lau L. F., Roberts J. W., Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell 42, 259–269 (1985). [DOI] [PubMed] [Google Scholar]
- 13.Kang J. Y., et al. , Structural basis for transcript elongation control by NusG/RfaH universal regulators. Cell 173, 1650–1662.e1614 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dey S., et al. , Structural insights into RNA-mediated transcription regulation in bacteria. Mol. Cell 82, 3885-3900.e3810 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Kyzer S., Ha K. S., Landick R., Palangat M., Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J. Biol. Chem. 282, 19020–19028 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Herbert K.M., et al. , Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell 125, 1083–1094 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabizon R., Lee A., Vahedian-Movahed H., Ebright R. H., Bustamante C. J., Pause sequences facilitate entry into long-lived paused states by reducing RNA polymerase transcription rates. Nat. Commun. 9, 2930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janissen R., Eslami-Mossallam B., Artsimovitch I., Depken M., Dekker N. H., High-throughput single-molecule experiments reveal heterogeneity, state switching, and three interconnected pause states in transcription. Cell Rep. 39, 110749 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Saba J., et al. , The elemental mechanism of transcriptional pausing. eLife 8, e40981 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strobel E. J., Roberts J. W., Two transcription pause elements underlie a sigma70-dependent pause cycle. Proc. Natl. Acad. Sci. U.S.A. 112, e4374–e4380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toulokhonov I., Zhang J., Palangat M., Landick R., A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell 27, 406–419 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Larson M. H., et al. , A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344, 1042–1047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vvedenskaya I. O., et al. , Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science 344, 1285–1289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imashimizu M., et al. , Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome. Biol. 16, 98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winkelman J. T., et al. , XACT-seq comprehensively defines the promoter-position and promoter-sequence determinants for initial-transcription pausing. Mol. Cell 79, 797–811.e798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gajos M., et al. , Conserved DNA sequence features underlie pervasive RNA polymerase pausing. Nucleic Acids Res. 49, 4402–4420 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang J. Y., et al. , Structural basis of transcription arrest by coliphage HK022 nun in an Escherichia coli RNA polymerase elongation complex. Elife 6, e25478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdelkareem M., et al. , Structural basis of transcription: RNA polymerase backtracking and its reactivation. Mol. Cell 75, 298–309.e294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vos S. M., Farnung L., Urlaub H., Cramer P., Structure of paused transcription complex Pol II-DSIF-NELF. Nature 560, 601–606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu C., et al. , Transcription factors modulate RNA polymerase conformational equilibrium. Nat. Commun. 13, 1546 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lane W. J., Darst S. A., Molecular evolution of multisubunit RNA polymerases: Sequence analysis. J. Mol. Biol. 395, 671–685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuo Y., Steitz T. A., Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol. Cell 58, 534–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B., Zuo Y., Steitz T. A., Structures of E. coli sigmaS-transcription initiation complexes provide new insights into polymerase mechanism. Proc. Natl. Acad. Sci. U.S.A. 113, 4051–4056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Bao Y., Landick R., Obligate movements of an active site-linked surface domain control RNA polymerase elongation and pausing via a Phe pocket anchor. Proc. Natl. Acad. Sci. U.S.A. 118, e2101805118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan C., Wang D., Landick R., Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3’ end spacing distinguishes pause and termination pathways. J. Mol. Biol. 268, 54–68 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Vitiello C. L., Kireeva M. L., Lubkowska L., Kashlev M., Gottesman M., Coliphage HK022 Nun protein inhibits RNA polymerase translocation. Proc. Natl. Acad. Sci. U.S.A. 111, E2368–E2375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landick R., Wang D., Chan C., Quantitative analysis of transcriptional pausing by RNA polymerase: The his leader pause site as a paradigm. Meth. Enzymol. 274, 334–352 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Theissen G., Pardon B., Wagner R., A quantitative assessment for transcriptional pausing of DNA-dependent RNA polymerases in vitro. Anal. Biochem. 189, 254–261 (1990). [DOI] [PubMed] [Google Scholar]
- 40.Zhou J., Ha K. S., La Porta A., Landick R., Block S. M., Applied force provides insight into transcriptional pausing and its modulation by transcription factor NusA. Mol. Cell 44, 635–646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landick R., Turnbough C. L. J., Yanofsky C., “Transcription attenuation” in Escherichia coli and Salmonella: Cellular and Molecular Biology, Neidhardt F. C., et al., Eds. (ASM, Washington, DC, 1996), ed. 2, pp. 1263–1286. [Google Scholar]
- 42.Scull C. E., Dandpat S. S., Romero R. A., Walter N. G., Transcriptional riboswitches integrate timescales for bacterial gene expression control. Front. Mol. Biosci. 7, 607158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bochkareva A., Yuzenkova Y., Tadigotla V. R., Zenkin N., Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. EMBO J. 31, 630–639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodson J. R., Winkler W. C., Processive antitermination. Microbiol. Spectr. 6, RWR0031-2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin Z., Bird J. G., Kaelber J. T., Nickels B. E., Ebright R. H.. Structural basis of transcription antitermination by Qλ: NusA induces refolding of Qλ to form a nozzle that extends the RNA polymerase RNA-exit channel. Proc. Natl. Acad. Sci. U.S.A. 119, e2205278119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krupp F., et al. , Structural basis for the action of an all-purpose transcription anti-termination factor. Mol. Cell 74, 143–157 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Huang Y. H., et al. , Structure-based mechanisms of a molecular RNA polymerase/chaperone machine required for ribosome biosynthesis. Mol. Cell 79, 1024–1036.e1025 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Jensen D., Galburt E. A., The context-dependent influence of promoter sequence motifs on transcription initiation kinetics and regulation. J. Bacteriol. 203, e00512-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruff E. F., et al. , E. coli RNA polymerase determinants of open complex lifetime and structure. J. Mol. Biol. 427, 2435–2450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J., Boyaci H., Campbell E. A., Diverse and unified mechanisms of transcription initiation in bacteria. Nat. Rev. Microbiol. 19, 95–109 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan C. L., Landick R., Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J. Mol. Biol. 233, 25–42 (1993). [DOI] [PubMed] [Google Scholar]
- 52.Turtola M., Belogurov G. A., NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. eLife 5, e18096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tagami S., et al. , Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 468, 978–982 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Shu B., Gong P., Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl. Acad. Sci. U.S.A. 113, E4005–E4014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silva D. A., et al. , Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc. Natl. Acad. Sci. U.S.A. 111, 7665–7670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekine S., Murayama Y., Svetlov V., Nudler E., Yokoyama S., The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol. Cell 57, 408–421 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Dandey V. P., et al. , Time-resolved cryo-EM using Spotiton. Nat. Methods 17, 897–900 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Twist K. A., et al. , A novel method for the production of in vivo-assembled, recombinant Escherichia coli RNA polymerase lacking the alpha C-terminal domain. Protein Sci. 20, 986–995 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bae B., et al. , Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma70 domain 1.1. Proc. Natl. Acad. Sci. U.S.A. 110, 19772–19777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Windgassen T., et al. , Trigger-helix folding pathway and SI3 mediate catalysis and hairpin-stabilized pausing by Escherichia coli RNA polymerase. Nucleic Acids Res. 42, 12707–12721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hein P. P., et al. , RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat. Struct. Mol. Biol. 21, 794–802 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marcoline F. V., Furth J., Nayak S., Grabe M., Macey R. I., Berkeley Madonna Version 10-A simulation package for solving mathematical models. CPT Pharmacometrics Syst. Pharmacol. 11, 290–301 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of pre-consensus elemental paused elongation complex. Protein Data Bank. https://www.rcsb.org/structure/8EG7. Deposited 11 September 2022. [Google Scholar]
- 64.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of consensus elemental paused elongation complex with a folded TL. Protein Data Bank. https://www.rcsb.org/structure/8EG8. Deposited 12 September 2022. [Google Scholar]
- 65.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of consensus elemental paused elongation complex with an unfolded TL. Protein Data Bank. https://www.rcsb.org/structure/8EGB. Deposited 12 September 2022. [Google Scholar]
- 66.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (1). Protein Data Bank. https://www.rcsb.org/structure/8EH8. Deposited 13 September 2022. [Google Scholar]
- 67.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (2). Protein Data Bank. https://www.rcsb.org/structure/8EH9. Deposited 13 September 2022. [Google Scholar]
- 68.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (out). Protein Data Bank. https://www.rcsb.org/structure/8EHA. Deposited 14 September 2022. [Google Scholar]
- 69.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of his-elemental paused elongation complex with an unfolded TL (1). Protein Data Bank. https://www.rcsb.org/structure/8EHF. Deposited 14 September 2022. [Google Scholar]
- 70.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of his-elemental paused elongation complex with an unfolded TL (2). Protein Data Bank. https://www.rcsb.org/structure/8EHI. Deposited 14 September 2022. [Google Scholar]
- 71.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of pre-consensus elemental paused elongation complex. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-28109. Deposited 11 September 2022. [Google Scholar]
- 72.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of consensus elemental paused elongation complex with a folded TL. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-28110. Deposited 12 September 2022. [Google Scholar]
- 73.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of consensus elemental paused elongation complex with an unfolded TL. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-28113. Deposited 12 September 2022. [Google Scholar]
- 74.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (1). Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-28143. Deposited 13 September 2022. [Google Scholar]
- 75.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (2). Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-28144. Deposited 13 September 2022. [Google Scholar]
- 76.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (out). Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-28145. Deposited 14 September 2022. [Google Scholar]
- 77.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of his-elemental paused elongation complex with an unfolded TL (1). Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-28146. Deposited 14 September 2022. [Google Scholar]
- 78.Kang J.Y., Chen J., Llwellyn E., Landick R., Darst S.A., Cryo-EM structure of his-elemental paused elongation complex with an unfolded TL (2). Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-28148. Deposited 14 September 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
The movie shows the movements of active-site modules (BH, TL, RH, FL) and nucleic acids that connect the different elemental PEC states observed for con-ePEC and his-ePEC in two side-by-side views of the active site of RNAP. Movements of the BH, RH, and FL that accompany TL folding and unfolding are also likely to occur during the active NAC.
Data Availability Statement
Cryo-EM data have been deposited in the RCSB Protein Data Bank (www.pdb.org) and in the Electron Microscopy Data Bank (www.emdatabank.org). The PDB accession codes for the coordinates of con-ePEC−1, con-ePEC_fTL, con-ePEC_ufTL, his-ePEC_fTL-Fin1, his-ePEC-fTL-Fin2, his-ePEC_fTL-Fout, his-ePEC_ufTL1, and his-ePEC_ufTL2 are 8EG7 (63), 8EG8 (64), 8EGB (65), 8EH8 (66), 8EH9 (67), 8EHA (68), 8EHF (69), and 8EHI (70), respectively, and the accession codes for the cryo-EM maps are EMD-28109 (71), EMD-28110 (72), EMD-28113 (73), EMD-28143 (74), EMD-28144 (75), EMD-28145 (76), EMD-28146 (77), and EMD-28148 (78), respectively. All study data are included in the article, SI Appendix, or both. Previously published data were used for this work (DOI: 10.1073/pnas.2101805118).