Significance
Activated microglia play a central role in the onset and progression of neurodegenerative disorders. They phagocytose cell debris or misfolded protein and contribute to neuroinflammation. However, the origin of activated microglia during neurodegeneration remains incompletely understood. Understanding their origin is essential for controlling deregulated microglial activity. Our single-cell transcriptomic analysis reveals that Cspg4high microglia represent a subset of microglia that has a distinct transcriptome from other known microglia subtypes in mice and humans. These cells undergo steady-state mitosis continuously and throughout life in the brain at rather low levels. Cspg4high microglia are expanded contributing to microgliosis during neurodegeneration. Thus, the present study identifies Cspg4high microglia as a new source of activated microglia in the diseased brain.
Keywords: Cspg4, microglia, neurodegenerative diseases, Alzheimer’s disease, Parkinson’s disease
Abstract
Microglia play a critical role in the pathogenic process of neurodegenerative diseases, such as Parkinson’s disease (PD) and Alzheimer’s disease (AD). Upon pathological stimulation, microglia are converted from a surveillant to an overactivated phenotype. However, the molecular characters of proliferating microglia and their contributions to the pathogenesis of neurodegeneration remain unclear. Here, we identify chondroitin sulfate proteoglycan 4 (Cspg4, also known as neural/glial antigen 2)-expressing microglia as a specific subset of microglia with proliferative capability during neurodegeneration. We found that the percentage of Cspg4+ microglia was increased in mouse models of PD. The transcriptomic analysis of Cspg4+ microglia revealed that the subcluster Cspg4high microglia displayed a unique transcriptomic signature, which was characterized by the enrichment of orthologous cell cycle genes and a lower expression of genes responsible for neuroinflammation and phagocytosis. Their gene signatures were also distinct from that of known disease-associated microglia. The proliferation of quiescent Cspg4high microglia was evoked by pathological α-synuclein. Following the transplantation in the adult brain with the depletion of endogenous microglia, Cspg4high microglia grafts showed higher survival rates than their Cspg4− counterparts. Consistently, Cspg4high microglia were detected in the brain of AD patients and displayed the expansion in animal models of AD. These findings suggest that Cspg4high microglia are one of the origins of microgliosis during neurodegeneration and may open up a avenue for the treatment of neurodegenerative diseases.
Neurodegenerative diseases, such as Parkinson’s disease (PD) and Alzheimer’s disease (AD), have become one of the leading causes of disability and death in the elders, bringing heavy burdens on the economy and spirit of the patients, families, and societies (1). Accumulating evidence has suggested that microglia are a key player in the onset and progression of neurodegenerative diseases (2–5). In PD, microglia directly regulate functional brain network integrity by affecting the activities of neuronal cells and other types of glial cells with soluble factors, such as inflammatory cytokines (2, 4). For instance, microglia are responsible for the astrocytic conversion from a quiescent state to a neurotoxic A1 state in a mouse model of PD (2). Meanwhile, microglia also participate in the clearance of accumulated α-synuclein (α-syn) and cell debris (6–8). In the context of AD, microglia influence core AD pathology, including Aβ accumulation, tau protein spreading, immune/inflammatory response, and neuronal survival through several signaling pathways, such as triggering receptor expressed on myeloid cells 2 (Trem2) or apolipoprotein E (5, 9). These studies suggest that activated microglia function as a key player in the AD pathology in the living human and rodent brains.
Emerging evidence has suggested that different subtypes of microglia exhibit distinct responses during neurodegeneration (10, 11). Based on the expression of known markers and the similarity of each cluster, microglia can be classified into multiple clusters, such as homeostasis, neuroinflammation, phagocytosis, and proliferation (12–14). Among them, the clusters of neuroinflammation and phagocytosis are tightly correlated with the pathology burden in PD and AD. For instance, a phagocytic microglia type designated as disease-associated microglia (DAM) was identified by single-cell sequencing in AD model, which could be further classified into two different subclusters, stage 1 DAM and stage 2 DAM, depending on disease stages (15). However, the origin of activated microglia remains elusive.
The proliferative cluster in microglia has long been ignored in previous studies of neurodegenerative diseases. As a result, this cluster is one of the most enigmatic and understudied microglial populations. Until recently, a few studies have shown the presence of proliferating microglia under certain pathological conditions, e.g., acute brain injury (16). In the adult rodent brain, resident microglia turn over in a steady state, and local proliferation maintains a stable pool of microglia (17). Such a process is mainly controlled by transforming growth factor-β and colony-stimulating factor 1 receptor (CSF1R) signaling pathways (18, 19). However, in the context of PD or AD, the two signaling pathways are dysregulated contributing to an aberrant increase in the density of microglia, especially those accumulated around the toxic protein plaques (8, 20). Further elucidation of the mechanism underlying the excessive microgliosis in the process of chronic neurodegeneration would be vital for the control of disease progression in neurodegenerative diseases.
Certain cell types in the central nervous system (CNS) with proliferation activity have been linked to chondroitin sulfate proteoglycan 4 (Cspg4) (21–23). NG2 protein encoded by Cspg4 is normally expressed by oligodendrocyte precursor cells (OPCs) and pericytes and plays an important role in cell proliferation and differentiation in both the developing and the adult brain (24–26), through several signaling pathways including integrin-dependent regulation (27, 28). Interestingly, recent studies have demonstrated that Cspg4 is expressed in microglia, some of which could be labeled by Ki67, a proliferation marker, in the transient middle cerebral artery occlusion model (16). Whether Cspg4+ microglia are correlated with microgliosis and contribute to neurodegeneration is still unclear.
In the present study, we performed high-dimensional analysis which enabled us to characterize Cspg4+ microglia and their specific subsets. We identified Cspg4high microglia as a distinct subset of microglia with high proliferative potential. These cells had a lower expression of genes associated with phagocytosis and neuroinflammation. The local expansion of Cspg4high microglia cluster contributed to microgliosis during neurodegeneration.
Results
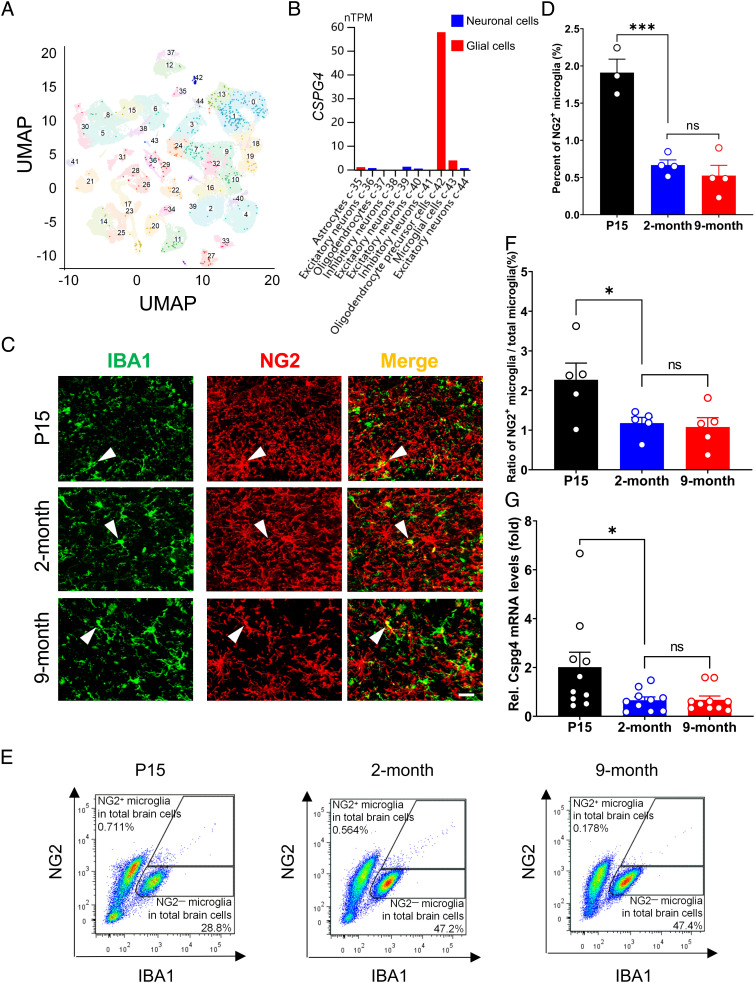
CSPG4+ Microglia Exist in the Human Brain and Show an Age-Associated Reduction in the Numbers.
To determine whether CSPG4+ microglia are present in the human brain, we reanalyzed the single-cell transcriptomic data of the human brain provided in the Human Protein Atlas (https://www.proteinatlas.org/ENSG00000173546CSPG4/single+cell+type/brain) (29). As expected, CSPG4 was mainly expressed in OPCs. However, CSPG4 was also found to be expressed in microglia among various cell populations that expressed relatively low levels of CSPG4, such as astrocytes and excitatory neurons. The CSPG4+ microglia accounted for 1.94% of the total number of microglia (Fig. 1 A and B). These data suggest that the human brain hosts a menagerie of CSPG4+ microglia. Consistent with this, Cspg4+ microglia were also detectable in the mouse brain. Cspg4+ microglia were distinguishable from OPCs, as Cspg4+ microglia expressed ionized calcium-binding adaptor molecule 1 (IBA1), but were not labeled by tdTomoto under the control of the platelet-derived growth factor α receptor (Pdgfra) promoter in Pdgfra-creERT2/Rosa26-tdTomoto double-transgenic mice (SI Appendix, Fig. S1), indicating that NG2+/IBA1+ microglia are an independent cell population that is distinctive from NG2+ OPCs.
Fig. 1.
CSPG4+ microglia exist in the human brain and their numbers in mice are decreased during aging. (A) UMAP showing the cell populations in the human brain based on the data extracted from the Human Protein Atlas. (B) CSPG4 expression in neuronal and glial cells of the human brain. The data are extracted from the Human Protein Atlas. (C) Representative immunofluorescence microphotographs showing colocalization of NG2 and IBA1 in WT mice at various ages, from postnatal day 15 (P15, n = 3), 2 mo (n = 4) and 9 mo (n = 4) of age. White arrow heads indicate NG2+ microglia. (Scale bar, 20 μm.) (D) Quantitative data shown in C. (E) Cytometric analysis of NG2+/IBA1+ cells in the mouse brain at P15 (n = 5), 2 mo (n = 5) and 9 mo (n = 5) of age. (F) Quantitative data shown in E. (G) Representative graph showing relative mRNA levels of Cspg4 in microglia of WT mice at P15 (n = 10), 2 mo (n = 10), and 9 mo (n = 10) of age. Data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Given that aging is the primary risk factor for most neurodegenerative diseases, we investigated the changes of Cspg4+ microglia across the age in mice. Double-label immunofluorescent staining for NG2 and IBA1 revealed that the percentages of NG2+ microglia in the total microglial population dropped sharply from 1.91% at postnatal day 15 (P15) to 0.67% at 2 mo of age. There was no further significant decrease in the percentage of this cell type at 9 mo of age as compared to 2 mo of age (Fig. 1 C and D), indicating that Cspg4+ microglia population is shrinking in young adulthood but remains relatively stable late in life. These results were corroborated by fluorescence-activated cell sorting (FACS) analysis, which showed that there was a marked reduction in the proportions of NG2+ microglia in 2-mo-old mice compared to that at P15 (Fig. 1 E and F).
Next, we assessed the change in microglial expression of Cspg4 messenger ribonucleic acid (mRNA) in mice at P15, 2 mo and 9 mo of age. Microglia were isolated from whole-brain tissues of wild-type (WT) mice using CD11b-based magnetic-activated cell sorting (MACS). The levels of Cspg4 mRNA expression in 2- or 9-mo-old adult mice dropped to approximately 30% as compared with P15 mice (Fig. 1G). These results suggest that microglial Cspg4 expression is negatively correlated with age in young healthy animals, and Cspg4+ microglia remain relatively stable in aging under physiological conditions.
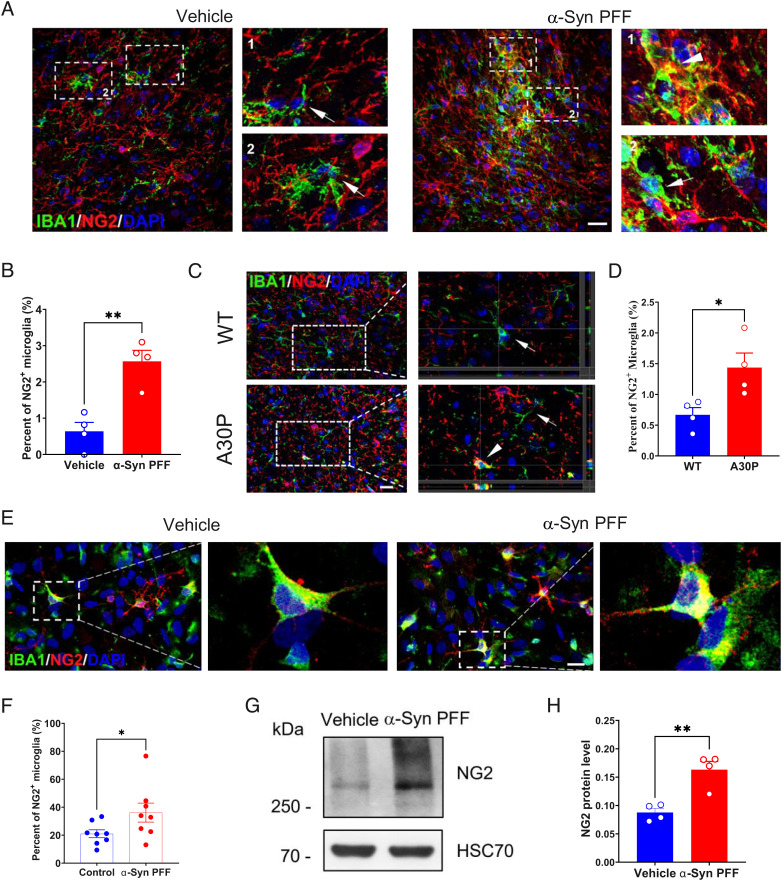
Expansion of Cspg4+ Microglia Is Induced by α-Syn.
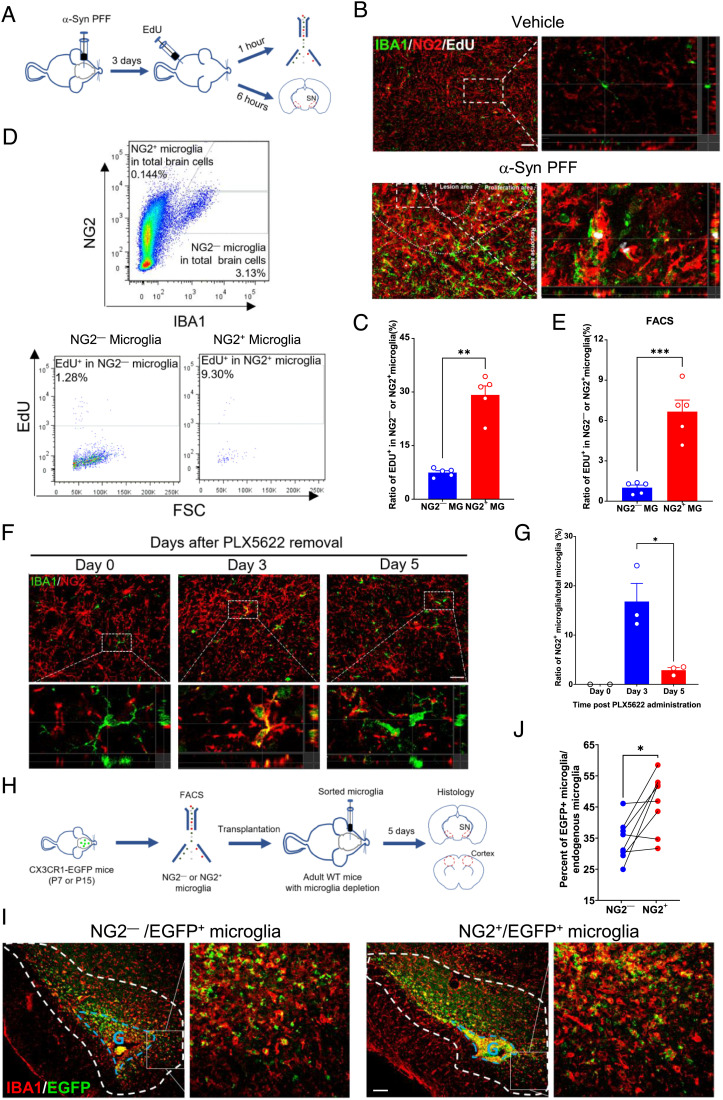
To investigate the influence of pathogenic stimuli on Cspg4+ microglia, we used an α-syn preformed fibril (PFF)-induced mouse model of PD (Fig. 2A and SI Appendix, Fig. S2A), in which α-syn PFFs were stereotaxically injected into the substantia nigra (SN) to induce glial cell activation and protein aggregation (30). Immunohistochemical analysis revealed a significant expansion of Cspg4+ microglia in the SN following α-syn PFF injection, as evidenced by a fourfold increase in the proportion of NG2+ microglia in the total microglia populations in α-syn PFF-treated animals, as compared to control (Fig. 2 A and B). Likewise, a significant increase in the number of NG2+ microglia was also found in the SN of human A30P mutant α-syn Tg mice (Fig. 2 C and D). This mouse line carries the A30P mutation in α-syn, which is known to cause familial PD (31, 32). These results indicate that Cspg4+ microglia are responsive to pathological stimuli and can be induced in mouse models of PD. This notion was further supported by the data showing that the percentage of NG2+ microglia rose from 21% in control group to 36% in α-syn PFF-treated primary microglia (Fig. 2 E and F), which was accompanied by a marked increase in NG2 protein expression levels compared with control, as revealed by western blot analysis (Fig. 2 G and H). Together, these results indicate that Cspg4+ microglia sense pathogenic stimuli, such as α-syn, in PD and proliferate in response to toxic proteins.
Fig. 2.
The percentage of NG2+ microglia is increased in the pd models. (A) Representative immunofluorescent micrographs showing colocalization of NG2 and IBA1 in the SN of mice injected with vehicle (PBS) or α-syn PFFs. Arrows indicate NG2− microglia. Arrowheads indicate NG2+ microglia. (Scale bar, 20 μm.) (B) Quantitative data showing the percentage of NG2+ microglia in the SN of mice with vehicle (n = 4) or α-syn PFF injection (n = 4). (C) Representative immunofluorescent micrographs showing colocalization of NG2 and IBA1 in the SN of A30P Tg mice or WT mice. Arrows indicate NG2− microglia. Arrowheads indicate NG2+ microglia. (Scale bar, 20 μm.) (D) Quantitative data showing the percentage of NG2+ microglia in the SN of A30P Tg mice (n = 4) or WT mice (n = 4). (E) Representative immunofluorescent microphotographs showing colocalization of NG2 and IBA1 in primary cultured cells of rat brain exposed to α-syn PFFs. (Scale bar, 20 μm.) (F) Quantitative data shown in E, vehicle (n = 8) or α-syn PFF (n = 8). (G) Representative immunoblots showing expression of NG2 in primary cultured microglia exposed to α-syn PFFs. (H) Quantitative data shown in G, vehicle (n = 4) or α-syn PFF injection (n = 4). Data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
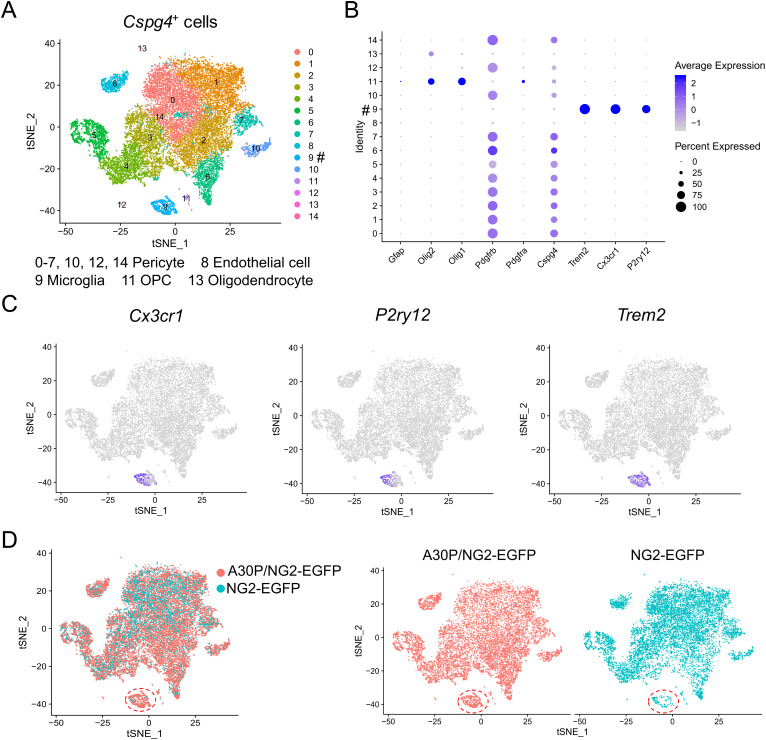
Single-Cell RNA Sequencing Unveils Unique Transcriptomic Signatures of Cspg4+ Microglia.
Cspg4+ microglia are typically characterized by simultaneous expression of NG2 and microglial markers, such as IBA1. To get insights into the molecular characteristics of Cspg4+ microglia, we crossed NG2-enhanced green fluorescent protein (EGFP) transgenic (Tg) reporter mice, expressing EGFP under the control of NG2 promoter, with A30P Tg mice, to generate A30P α-syn;NG2-EGFP double Tg mouse line. This double Tg mouse line allowed to analyze the responses of Cspg4+ cell populations labeled by EGFP, including microglia, following exposure to A30P α-syn. EGFP-labeled Cspg4+ cells were sorted by FACS and then subjected to single-cell RNA sequencing (scRNA seq). It was revealed that the whole population of Cspg4+ cells could be divided into 15 clusters (cluster 0 to 14) (Fig. 3A). Consistent with previous studies (24, 25), classical Cspg4+ cells, such as pericytes and OPCs, were detected among these clusters (Fig. 3 A and B), confirming the validity of the methods used. We found that microglia and endothelial cells could also be detected in the cell clusters (Fig. 3A). Cluster 9 of Cspg4+ cells displayed a typical gene expression profile of microglia, such as purinergic receptor P2Y, G-protein coupled 12 (P2ry12), chemokine (C-X3-C motif) receptor 1 (Cx3cr1), and triggering receptor expressed on myeloid cells 2 (Trem2) (Fig. 3 B and C), in addition to Cspg4, suggesting that cluster 9 represents Cspg4+ microglia but not macrophage. Taken together, we identify Cspg4+ microglia as a distinct cluster of microglia at the single-cell level in both mouse and human brains.
Fig. 3.
The single-cell transcriptomic analysis for egfp-labeled Cspg4+ cells isolated from A30P;NG2-EGFP double Tg mice. (A) The clusters of Cspg4+ cells. # denotes cluster 9, Cspg4+ microglia. (B) A dotplot for glial marker genes in cluster 0 to 14. (C) Expression of microglial marker genes in clusters. (D) Cluster 9 (circled) in A30P mice and control. The data represent SN tissues pooled from both NG2-EGFP Tg mice (n = 6) and A30P α-syn;NG2-EGFP double Tg mice (n = 6).
To determine the influence of A30P mutation on Cspg4+ microglia, we compared the cell numbers of different clusters between A30P;NG2-EGFP double Tg mice and NG2-EGFP Tg mice. There appeared to be an expansion of Cspg4+ microglia in mice overexpressing α-syn A30P mutant (Fig. 3D and SI Appendix, Table S1). This was comparable to the results obtained in α-syn PFF-induced PD model and A30P mice (Fig. 2). Together, these data confirm the existence of Cspg4+ microglia using two different animal models of PD. They also demonstrate that in pathological situations, such as PD, the expansion of Cspg4+ microglia can be provoked in vivo.
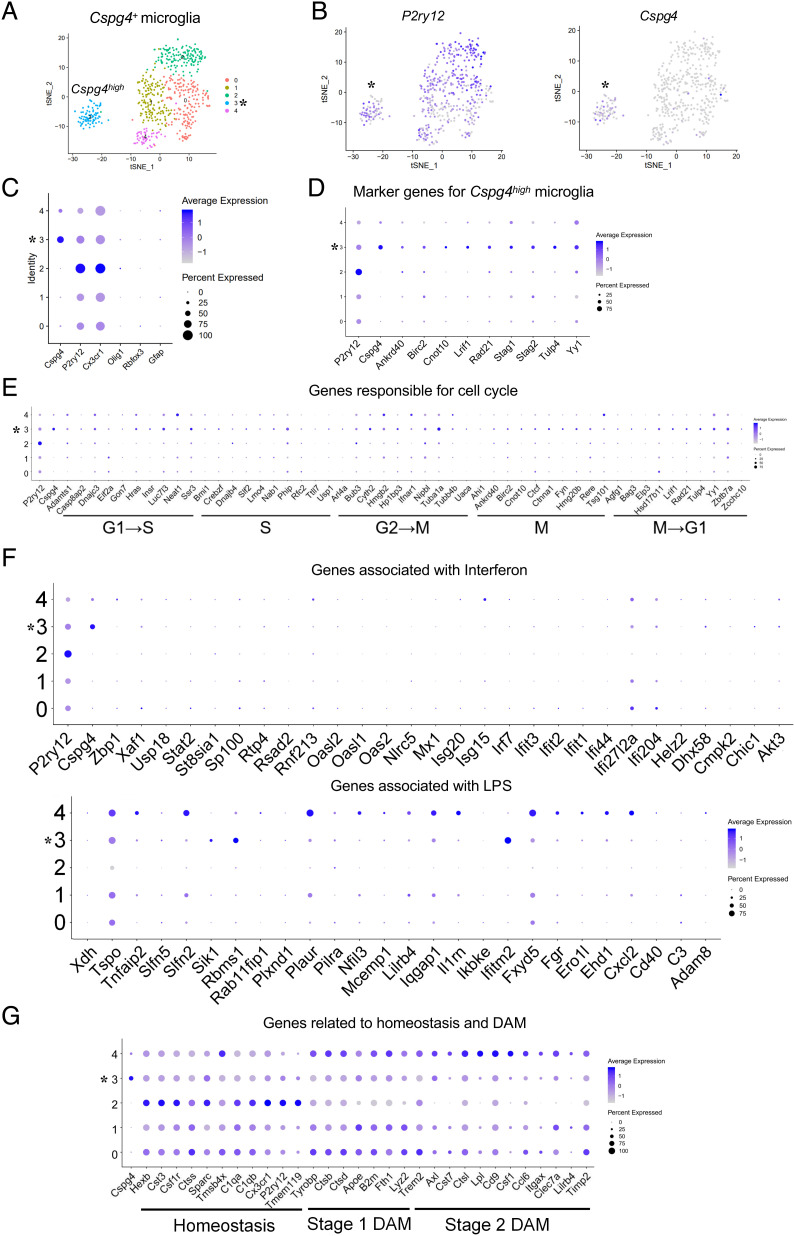
Cspg4high Microglia Are Characterized by Active Proliferation.
Next, we were determined to further characterize Cspg4+ microglia. To this end, the microglia cluster (cluster 9) was isolated from the total Cspg4+ cells of A30P;NG2-EGFP double Tg mice for transcriptomic analysis. It was revealed that Cspg4+ microglia could be divided into five subclusters (0 to 4) with a higher resolution (Fig. 4A). Of these subclusters, subcluster 3 displayed higher levels of Cspg4 expression as compared with the rest of the cell subclusters (Fig. 4 B and C and SI Appendix, Table S2). Thus, subcluster 3 was designated as Cspg4high microglia, while other subclusters were referred as Cspg4low microglia. Notably, Cspg4high microglia also expressed microglial markers, such as P2ry12 and Cx3cr1 with a lower abundance than subcluster 2 (Fig. 4 B and C), indicating that Cspg4high and Cspg4low microglia are two discrete microglial subpopulations.
Fig. 4.
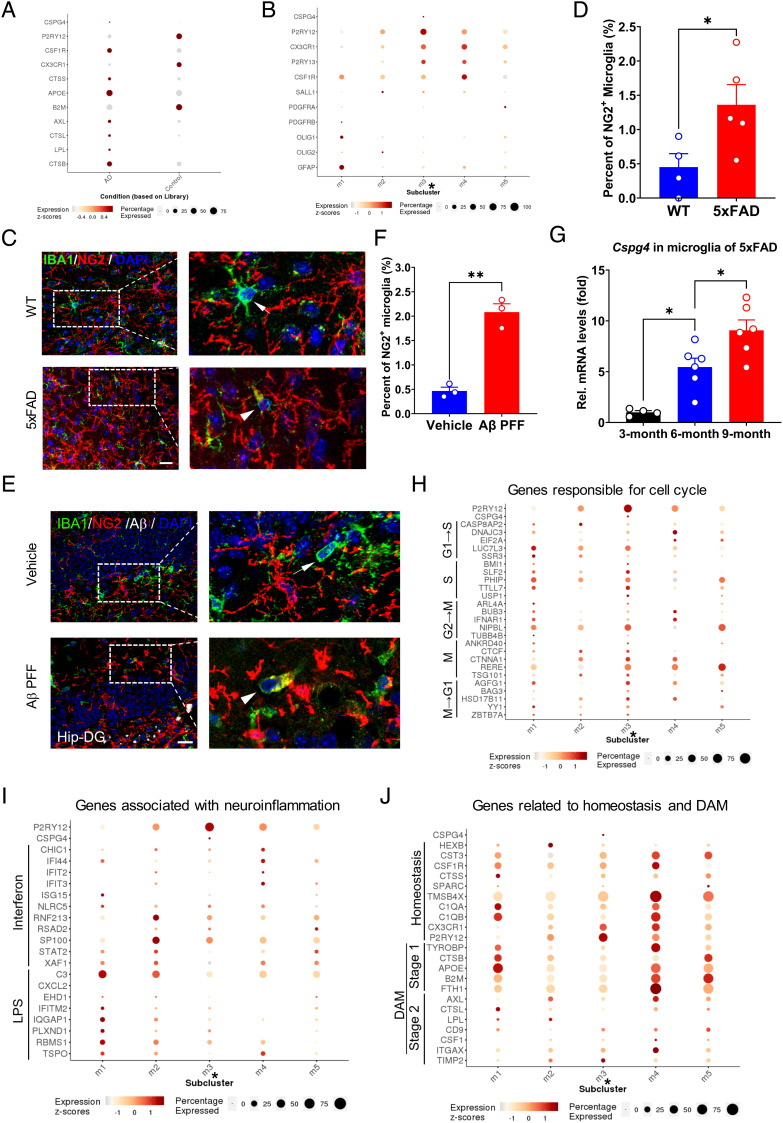
Cspg4high microglia exhibit a high expression of genes associated with cell proliferation but not disease-associated response in A30P mice. (A) Two-dimensional UMAP embedding of the scRNA-seq of cluster 9 data. Dots are colored based on the scRNA-seq clustering. Subcluster 3 represents Cspg4high microglia indicated with asterisk (*). (B) Expression of P2ry12 and Cspg4 in cluster 9. (C) A dotplot for glial marker genes in subclusters. (D) A dotplot for marker genes of Cspg4high microglia. (E) A dotplot showing the relative expression of genes responsible for cell cycle (x axis) across all subclusters (y axis). (F) A dotplot showing the relative expression of genes associated with neuroinflammation (x axis) across all subclusters (y axis). (G) A dotplot for genes related to homeostasis and DAM (x axis) across all subclusters (y axis).
To further define the Cspg4high microglia, we analyzed the differentially expressed genes (DEGs) between subcluster 3 and other subclusters. Among the DEGs, a number of genes were related to cell proliferation. Subsequently, a cluster of 10 genes was selected as the marker genes for Cspg4high microglia. They included the followings: Cspg4 (33), ankyrin repeat domain 40 (Ankrd40) (34), baculoviral IAP repeat-containing 2 (Birc2) (35), CCR4-NOT transcription complex subunit 10 (Cnot10) (36), ligand-dependent nuclear receptor-interacting factor 1 (Lrif1) (37), RAD21 cohesin complex component (Rad21) (38, 39), stromal antigen 1/2 (Stag1/2) (40, 41), tubby-like protein 4 (Tulp4) (42), and transcription factor Yin Yang 1 (Yy1) (26, 42) (Fig. 4D), given that these genes have been reported to be involved in cell proliferation. Most of them, such as Birc2, Rad21, Cnot10, and Stag1/2, are known to be key players in mitosis, indicating that Cspg4high microglia are in a state of active proliferation.
We thus evaluated the distribution of Cspg4high microglia to the different phases of microglial cell cycle, by using the gene sets of different phases including the synthesis (S), gap-2 (G2), M, and gap-1 (G1) phase, based on a previous study describing adult microglia proliferation (43). Increased abundance of Ankrd40, Birc2, and Cnot10, which are belonging to the marker gene cluster of M phase were detected in Cspg4high microglia, while higher expression of Lrif1, Tulp4, and Yy1 among the marker gene cluster of M→G1 phase were also observed. Consistent with the previous study (43), these data suggest that the genes associated with microglial cell cycle were enriched in Cspg4high microglia, particularly those related to mitosis (G2→M, M, and M→G1) (Fig. 4E and SI Appendix, Fig. S3). Collectively, these data suggest that Cspg4high microglia retain proliferative capacity.
Cspg4high Microglia Have a Distinct Transcriptomic Signature from Cspg4low Microglia or Cspg4− Microglia.
In the pathology of neurodegenerative diseases, several distinct microglial populations, such as inflammatory microglia, homeostatic microglia, and phagocytic DAM, have been described (12, 15). We asked whether Cspg4high microglia belong to a category of these microglial subgroups. The analysis of transcriptomic signatures of Cspg4high microglia showed a lower expression for genes associated with immune response to interferon/lipopolysaccharide (LPS) (Fig. 4F) and phagocytosis (Fig. 4G). In contrast, subclusters 0 and 4 shared similar gene expression patterns with stage 1 or stage 2 DAM, respectively, whereas gene expression pattern of subcluster 2 resembled that of homeostatic microglia (Fig. 4G), indicating that Cspg4low microglia are associated with the canonical functions of microglia in neurodegeneration. Together, these results support the idea that Cspg4high microglia are a unique microglial subpopulation with proliferative capacity.
Next, we sought to determine the transcriptional difference between Cspg4high microglia and Cspg4− microglia. We carried out cluster analysis for Cspg4− microglia and Cspg4+ cells, which gave rise to 16 distinct clusters. Cspg4− microglia consisted of seven clusters (0, 1, 3, 5, 6, 11, and 12), as expected, all of which were devoid of Cspg4, and Cspg4+ cells included nine clusters (2, 4, 7-10, 13-15) (SI Appendix, Fig. S4 A and B). Cspg4− microglia showed fairly low expression of cell cycle genes, especially those related to M phase as compared with Cspg4high microglia (Fig. 4E and SI Appendix, Fig. S4C), indicating their cell division activities are suppressed.
Moreover, among multiple NG2− microglia clusters, cluster 12 had a high expression of genes responding to neuroinflammation, particularly the interferon-related pathway, which were down-regulated in Cspg4high microglia (Fig. 4F and SI Appendix, Fig. S4 D and E). Furthermore, Cspg4 − microglia displayed distinct expression patterns of representative genes corresponding to homeostasis and DAM from Cspg4high microglia (Fig. 4G and SI Appendix, Fig. S4F). Together, Cspg4high microglia exhibit a characteristic transcriptome that is distinguishable from Cspg4low microglia and Cspg4 − microglia.
Cspg4high Microglia Undergo Active Proliferation In Vitro and In Vivo.
To functionally assess the proliferative activity of Cspg4high microglia, WT adult mice received stereotaxic injection of α-syn PFFs into the SN followed by 5-ethynyl-2′-deoxyuridine (EdU) administration, a nucleoside analog of thymidine incorporated into DNA during active DNA synthesis (44) (Fig. 5A). Immunohistochemical analysis showed that the proportion of EdU+ cells in NG2+ microglia was four times higher than that in NG2− microglia in α-syn PFF-treated mice following single pulse labeling with EdU for 6 h (Fig. 5 B and C). In accordance with this, FACS analysis showed that there was a sevenfold increase in the percentage of EdU+/NG2+ microglia than EdU+/NG2− microglia in the brain administered with α-syn PFF following single pulse labeling with EdU for 1 h (Fig. 5 D and E). This was confirmed by immunohistochemical analysis using an in vitro α-syn PFF model, which also showed a higher percentage of EdU+ cells in NG2+ microglia in primary microglia (SI Appendix, Fig. S5 A and B). Comparatively, under normal conditions, the percentage of proliferating NG2+ microglia was still higher than NG2− microglia, although they showed a sharp decline with advancing age in vivo (SI Appendix, Fig. S5 C and D). Indeed, NG2+ microglia showed an apparently higher cell growth rate than NG2− microglia in vitro (SI Appendix, Fig. S5 E and F). These results suggest that Cspg4high microglia are in a state of active proliferation in the context of neurodegeneration.
Fig. 5.
NG2+ microglia are in a state of active proliferation and contribute to microgliosis during microglial repopulation. (A) A diagram showing timeline for EdU administration in mice with injection of α-syn PFFs. (B) Immunofluorescent histochemical staining for NG2, IBA1, and EdU in α-syn PFF-induced PD model. (Scale bar, 50 μm.) (C) Quantitative data showing the percentage of EDU+ cells in NG2− microglia (n = 5) or NG2+ microglia (n = 5) in α-syn PFF-treated animals. (D) Cytometric analysis of EdU+/NG2+/IBA1+ cells in α-syn PFF-induced PD model. (E) Quantitative data shown in D, NG2− microglia (n = 5) and NG2+ microglia (n = 5). (F) Representative immunofluorescent staining for NG2 and IBA1 in WT mice during microglial repopulation post PLX5622 treatment. (Scale bar, 30 μm.) (G) Quantitative data shown in F, Day 0 post PLX5622 (n = 2), Day 3 post-PLX5622 (n = 3), Day 5 post-PLX5622 (n = 3). (H) A diagram showing timeline for microglia transplantation. (I) Immunofluorescent histochemical staining for IBA1 in the SN of mice received transplantation of NG2+ or NG2− CX3CR1-EGFP microglia (P7). G indicates the graft. (Scale bar, 100 μm.) (J) Quantitative data showing the percentage of transplanted microglia derived from NG2+ microglia (n = 8) or NG2− microglia (n = 8) in the region surrounding the graft (circled with dashed white lines). Data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
To investigate whether Cspg4high microglia are able to expand and contribute to microgliosis in the adult brain, we established a microglial depletion and repopulation model using PLX5622, a selective inhibitor of CSF1R that eliminates microglia (45, 46). After 7-d administration, PLX5622 was removed and microglial repopulation started. A burst of proliferative activity occurred on day 3 post PLX5622 administration, and the proportion of NG2+ microglia was upregulated to 16.8% from 0% on day 0 post PLX5622 treatment (Fig. 5 F and G). These data indicate that Cspg4high microglia could not only respond to the depletion of microglia but also contribute to microgliosis during microglial repopulation.
To extend the characterization of the contribution of Cspg4high microglia to microgliosis in vivo, we took one step further to examine their behavior in adult brain by using microglial cell transplantation. NG2+ and NG2− microglia were isolated from CX3CR1-EGFP knock-in mice at P7 or P15 of age using FACS (SI Appendix, Fig. S5G), and followed by transplantation into the SN or cerebrocortex of adult mouse brain. Prior to microglial cell transplantation, profound depletion of endogenous microglia was achieved through administration with PLX5622. This strategy allowed correct identification of exogenous EGFP-labeled NG2+ or NG2− microglia in recipient animals. Immunohistochemical analysis revealed that in the brain regions surrounding the grafts (P7) that were implanted in the SN, the percentage of NG2+/EGFP+ microglia was significantly higher than that derived from NG2− microglia grafts (Fig. 5 H–J) Consistent with this, there was also more EGFP-labeled NG2+ microglia migrating from the P15 grafts in the cerebrocortex to the host brain following transplantation (SI Appendix, Fig. S5 H and I), confirming that NG2+ microglia are an active player in the microglial repopulation.
In the PD brain, microglia play a key role in the clearance of misfolded protein α-syn. We asked whether Cspg4high microglia displayed phagocytic capacity upon α-syn PFF stimulation. Immunohistochemistry for phosphorylated α-syn showed that NG2+ microglia with α-syn aggregates accounted for 6.9% of the total NG2+ microglia in the brain with injection of α-syn PFFs (SI Appendix, Fig. S5 J and K). In contrast, approximately 29.9% of NG2− microglia showed phagocytosis of α-syn aggregates (SI Appendix, Fig. S5 J and K), indicating a poor phagocytic capability of Cspg4high microglia. Taken together, these data suggest that in the context of neurodegeneration, Cspg4high microglia differ from other microglial subsets in that they retain a high capacity of proliferation and low capability for phagocytosis of misfolded proteins.
Cspg4high Microglia Are Detected in the AD Brain.
To examine whether the finding that pathogenic stimuli evoke Cspg4high microglia can be extended to other neurodegenerative diseases, such as AD, we re-analyzed the single-cell transcriptomic atlas of the entorhinal cortex in the patients with AD from Polo laboratory (47). It was revealed that in agreement with the observations in animal models of PD (Figs. 3 and 4), in the AD brain, the expression of CSPG4 mRNA in microglia was found to be elevated as compared to healthy control subjects (Fig. 6A). Microglial cells in the AD brain could be divided into five subclusters (m1 to m5) (47). Only subcluster m3, but not other clusters, was found to express CSPG4 (Fig. 6B). These data suggest that Cspg4high microglia in the human brain are also responsive to pathological stimuli in AD progression.
Fig. 6.
Presence of CSPG4high microglia in the brain of AD patients and the expansion of Cspg4+ microglia in animal models of AD. (A) Single-cell analysis for microglial CSPG4 level in the entorhinal cortex of patients with AD. (B) Microglial clusters (m1 to m5) in the entorhinal cortex of subjects with AD. Cluster m3 represents Cspg4high microglia indicated with asterisk (*). (C) Representative immunofluorescent staining for NG2 and IBA1 in WT and 5×FAD mice. Arrows indicate NG2− microglia. Arrowheads indicate NG2+ microglia. (Scale bar, 15 μm.) (D) Quantitative data shown in C, WT (n = 4), 5×FAD mice (n = 5). (E) Representative immunofluorescent staining for NG2 and IBA1 in mice received injection of Aβ PFF or vehicle. Arrows indicate NG2− microglia. Arrowheads indicate NG2+ microglia. (Scale bar, 20 μm.) Hip-DG, hippocampal dentate gyrus. (F) Quantitative data shown in E, vehicle (n = 3), Aβ PFF (n = 3). (G) Representative graph showing relative mRNA levels of Cspg4 in microglia of 5×FAD mice at 3 mo (n = 4), 6 mo (n = 6), and 9 mo (n = 6) of age. (H) A dotplot showing the relative expression of genes responsible for cell cycle (x axis) across all subclusters (y axis). (I) A dotplot showing the relative expression of genes associated with neuroinflammation (x axis) across all subclusters (y axis). LPS, lipopolysaccharide. (J) A dotplot for genes related to homeostasis and DAM (x axis) across all subclusters (y axis). Data are presented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
In support of these data, an increase in the percentage of NG2+ microglia was prominently detected in 5×FAD mice, a transgenic model of AD overexpressing human APP and PSEN1 transgenes with a total of five AD-linked mutations (48), compared to WT littermate control, as evaluated by immunohistochemistry (Fig. 6 C and D). To investigate the causal link between Aβ deposition and induction of Cspg4high microglia, Aβ PFFs were stereotaxically injected into the hippocampus and their impact on Cspg4high microglia was evaluated (SI Appendix, Fig. S2B). We found that there was a 4.4-fold increase in the ratio of NG2+ microglia over IBA1+ microglia in Aβ PFF-treated animals as compared with control (Fig. 6 E and F), which was consistent with the data obtained from 5×FAD mice (Fig. 6 C and D). Given that AD is a progressive disease, we speculated that elevated Cspg4high microglia response is correlated with AD progression. To test this, by using MACS, CD11b+ microglia were isolated from the brain of 5×FAD mice at 3, 6, and 9 mo of age, during which the mice rapidly develop severe amyloid pathology and microgliosis (48). We found that expression levels of Cspg4 mRNA in microglia were steadily increased with disease progression (Fig. 6G), indicating that the pathological environment of AD facilitates the expansion of Cspg4high microglia contributing to microgliosis.
Next, we characterized molecular feature of Cspg4high microglia (subcluster m3) in the AD brain (47). These cells were under a unique proliferative state, manifested by high expression of genes related to cell cycle progression, as compared to other subclusters (Fig. 6H). As expected, the expression of genes associated with neuroinflammation, homeostasis, and DAM was downregulated in subcluster m3 (Fig. 6 I and J), indicating that this specific cell cluster has lower canonical activities of microglia, such as phagocytosis and inflammation. These characteristics of Cspg4high microglia in the AD brain were highly similar to those of the PD brain, implying that the activation of Cspg4high microglia is one of common cellular events shared among different neurodegenerative diseases.
Discussion
Despite the key role of activated microglia in the onset and progression in neurodegenerative diseases, little is known about the origin of microgliosis in the aged and neurodegenerative brain and how microgliosis is regulated during the processes. In the present study, we found that Cspg4high microglia represent a distinct subset of microglia in both human and mouse brain at the single-cell level. This subset of microglia is characterized by high proliferative capability and lower gene expression associated with phagocytosis and inflammation. In the context of PD and AD, these cells were responsive to pathogenic stimuli resulting in a marked expansion with advancing disease stage. These data suggest that Cspg4high microglia may contribute significantly to microgliosis and chronic neurodegeneration in the AD or PD brain.
AD and PD are characterized by progressive degeneration in the CNS. A growing body of evidence has suggested that microglia play irreplaceable roles in the maintenance of CNS homeostasis and immune surveillance. The dysregulation of microglia functionality contributes to the imbalance of CNS homeostasis and immune responses. While an attempt to rebalance microglial function and improve microglia fitness is important, it is equally important to understand the origin of activated microglia under pathological conditions. We found Cspg4high microglia to be a distinct subset of microglia that has major characteristics of proliferating cells. We considered Cspg4high microglia are proliferative microglia based on the following evidence shown in the present study. Genes associated with cell cycle were abundantly expressed in this group of cells. It has been shown that proliferation of certain types of cells in the CNS is linked to Cspg4 expression. NG2 protein encoded by Cspg4 is often found in cells with high proliferative potential, such as OPCs, during brain development (26, 49), indicating that Cspg4 plays a vital role in cell division. Indeed, there is evidence that myeloid-specific ablation of Cspg4 in mice could lead to a great reduction in myeloid cell abundance (27, 50). And the numbers of IBA1+ cells were profoundly reduced in Cspg4-deficient (NG2 knockout) mice compared to WT control under basal conditions and during experimental autoimmune encephalomyelitis, a well-accepted animal model of multiple sclerosis (51). These data suggest that Cspg4 is a common requirement for the maintenance of cell division across different types of proliferative cells, including microglia. Importantly, Cspg4+ microglia were labeled with EdU and their labeling ratio was much higher than Cspg4− microglia. In microglial depletion model, Cspg4+ microglia showed accelerated repopulation upon removal of PLX5622. Moreover, those genes related to microglial responses, such as phagocytosis and inflammation, showed negative correlation with Cspg4high microglia activity. Thus, Cspg4high microglia represent a unique subset of proliferating microglia and may be one of sources of activated microglia during neurodegeneration.
We found that Cspg4+ (NG2+) microglia harboring Cspg4high and Cspg4low subclusters were heterogenous in the brain of animal model of PD. The discordance in NG2 protein and Cspg4 mRNA may be due to the fact that the turnover of NG2 protein is relatively slow, whereas Cspg4 mRNA expression is highly dynamic in Cspg4high microglia. This notion is supported by previous findings that the efficiency of LysM-Cre-mediated Cspg4 ablation in myeloid cells is barely determined due to its quite transient expression in these cells (50). It is likely that Cspg4+ microglia are mixed microglial populations harboring multiple subclusters, each of which represents the different states of the activities, thus highlighting the complexity and heterogeneity of Cspg4+ microglia and the microglia population as a whole.
Among various subtypes of microglia identified so far, DAM represent a unique microglial subtype in the AD brain that correlates with the disease progression (15). Our data suggest that Cspg4high microglia differ from DAM in several ways. First, the two subtypes exert different biological functions. Cspg4high microglia serve as a cell source of activated microglia, whereas DAM are responsible for phagocytosis, as reflected by their distinct transcriptomic profiles. Second, percentages of each subtype in the AD brain vary greatly. For example, the ratio of Cspg4+ microglia vs. total microglial cells was ~2%, and Cspg4high microglia account for 13.5% of Cspg4+ microglia, whereas DAM accounts for more than 20% of entire microglial population in the AD brain (15). Despite these differences, the two subtypes share a few similarities. For instance, they were responsive to accumulated Aβ which stimulates the expansion of Cspg4high microglia and the phenotype switch of DAM, respectively, implying that environmental factors in the CNS milieu differentially influence the behavior and function of microglial subpopulations during neurodegenerative process. These changes on Cspg4high microglia and DAM are enforced and become more evident along with disease progression, suggesting a potential link between each other. We speculate that Cspg4high microglia may be an ancestor of DAM during the disease progression, as accumulated Aβ PFFs stimulate the proliferation of Cspg4high microglia, which are quiescent in aging brain but retain a remarkable capacity to undergo mitosis. These cells may be subsequently induced to differentiate into DAM in the AD brain. Further studies are required to test this hypothesis.
In summary, we found that Cspg4high microglia are a distinct subset of microglia with the proliferation potential but rather low phagocytic capacity and neuroinflammatory response. In the context of PD and AD, Cspg4high microglia are expanded in response to neurodegenerative signals, such as α-syn and Aβ. Activated Cspg4high microglia may give rise to multiple subtypes of microglia contributing to a variety of pathological processes. Our data indicate that Cspg4high microglia may be a potential target for intervention of neurodegenerative diseases.
Materials and Methods
Animals.
Neonatal or adult C57BL/6 mice were from the Shanghai Laboratory Animal Center, Chinese Academy of Sciences. NG2-EGFP Tg reporter mice [Tg(Cspg4-EGFP*)HDbe/J] (52), CX3CR1-EGFP knock-in mice (Cx3cr1tm1Litt/J) (53), Pdgfra-CreERT2 mice [Tg(Pdgfra-cre/ERT)467Dbe/J] (54), ROSA26-loxP-stop-loxP-tdTomato reporter mice [Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J] (55), A30P mutant human α-syn transgenic mice [Tg(THY1-SNCA*A30P)TS2Sud] (56), and the 5×FAD transgenic mice [Tg(APPSwFlLon,PSEN1* M146L*L286V)6799Vas/Mmjax] (48) were purchased from the Jackson Laboratory. All mice were maintained on a 12-h light/dark cycle at 23 °C with food and water available ad libitum. Animal maintenance and handling were approved by the Institutional Animal Care and Use Committee (IACUC No. NA-004-2016) and were in accordance with the US NIH Guide for the Care and Use of Laboratory Animals.
Western Blot Analysis.
Western blotting was performed as described previously (57). The membranes were incubated with either a) rabbit anti-NG2 pAb (1:500; Millipore, ab5320) or b) mouse anti-HSC70 mAb (1:2,000; Santa Cruz, sc-7298). The membranes were washed and incubated for 1 h at room temperature with the corresponding secondary antibodies: a) HRP-conjugated goat antirabbit IgG (1:10,000; Jackson ImmunoResearch Laboratories); b) HRP-conjugated goat antimouse IgG (1:10,000; Jackson ImmunoResearch Laboratories). Peroxidase activity was detected with SuperSignal WestPico chemiluminescent substrate (Pierce Biotechnology) and visualized and digitized with ImageQuant (LAS-4000, Fujifilm, Japan). Optical densities of bands were analyzed by ImageJ (NIH, USA).
PLP Fixation and Immunohistochemistry.
In brief, mice were anesthetized and perfused with 4% paraformaldehyde containing 0.1 M L-lysine and 0.01 M sodium metaperiodate (PLP) as described previously (58). Immunohistochemistry was carried out according to previously published protocols (57). The following primary antibodies were used: a) rabbit anti-NG2 pAb (1:1,000; Millipore, ab5320); b) goat anti-IBA1 pAb (1:200; Abcam, ab5076); c) mouse anti-β-amyloid (1:5,000; Biolegend, 800712); d) mouse anti-p-α-syn (1:10,000 to 25,000; WAKO, 014-20281). The following secondary antibodies were used: a) Alexa Fluor 647 donkey antimouse IgG (Invitrogen A-31571); b) Alexa Fluor 647 goat antirabbit IgG (Invitrogen, A27040); c) Alexa Fluor 555 donkey antirabbit IgG (Beyotime, A0453); d) Alexa Fluor 555 donkey antigoat IgG (Invitrogen, A-31570); e) FITC-conjugated donkey antirabbit IgG (Sangon Biotech, D110051); f) Alexa Fluor 488 donkey antigoat IgG (Invitrogen, A-11055); g) DAPI (Sigma-Aldrich, D9542). Brain sections were imaged using a laser confocal microscope (Nikon TiE-A1 plus). Data were obtained and processed using ImageJ.
EdU Injection and Detection.
Mice were intraperitoneally injected with EdU (Beyotime, ST067) (100 μg/mice). The duration of EdU labeling was 2 h for cell experiments in vitro, 1 h for FACS, and 6 h for immunohistochemistry, respectively. Detection was performed using the BeyoClick™ EdU-647 kit (Beyotime, C0081S) according to the manufacturer’s instructions.
Primary Microglial Culture.
Neonatal mice were used for microglia isolation, as described previously (57). Briefly, the neonatal brain was dissociated and cells were plated at a density of 5 × 107 cells/75 cm2 poly-L-lysine-coated flask (Corning) with Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum, penicillin, and streptomycin at 37 °C in humidified 5% CO2/95% air. On day 10 in vitro, cells were shaken at 250 rpm for 90 min in a 37 °C incubator. Cells in DMEM were seeded into a 24-well plate and the medium was changed after 30 min. Cultured microglia were treated with α-syn PFF (1 μg/mL, 2 h).
Isolation of RNA and RNA-Sequencing Analysis.
Isolation of RNA and RNA-sequencing analysis were performed as described previously (59). Cells were homogenized in TRIzol reagent. Total RNA was purified and cDNA was sequenced. Sequencing libraries were generated using NEB Next® Ultra RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample.
Stereotaxic Injection of α-Syn PFF or Aβ PFF.
Mice were anesthetized and modeled by stereotaxic injection with α-syn PFF (5 μg) or Aβ PFF (2 μg). α-Syn PFFs were injected into the SN at the coordinates of AP −3.0 mm, ML ±1.3 mm, DV −4.5 mm and Aβ PFFs were injected in the hippocampus (AP −2 mm, ML ±1 mm, DV −2 mm). Injections were performed at a rate of 0.1 μL/min with the needle left in place for 10 min.
Magnetic Isolation of Microglia.
Microglia were isolated using MACS according to the manufacturer’s instructions. Briefly, the brain tissues were cut into small pieces and dissociated by enzymatic digestion using the Neural Tissue Dissociation Kit P (Miltenyi Biotec, 130-092-628) to obtain the cell suspensions. Following digestion with enzymes, cells were labeled with magnetic anti-CD11b microbeads. The labeled suspensions were loaded onto the MACS column and sorted. The remaining microglia were collected and stored at −80 °C.
FACS.
Mice were anesthetized and perfused with phosphate-buffered saline (PBS), and the brain tissues were cut into small pieces and dissociated by enzymatic digestion using the Neural Tissue Dissociation Kit P to obtain the cell suspensions. After the digestion was completed, cells were fixed with 2% PFA for 10 min. Samples were blocked by CD16/CD32 monoclonal antibody for 10 min in FACS buffer (eBioscience, 14016182) followed by incubation with the following primary antibodies for 30 min: a) goat anti-IBA1 pAb (1:50; Abcam, ab5076); b) rabbit anti-NG2 pAb (1:50; Millipore, ab5320). The following secondary antibodies were used: a) Alexa Fluor 488 donkey antigoat IgG (1:200; Invitrogen, A-11055); b) Alexa Fluor 647 goat antirabbit IgG (1:200; Invitrogen, A27040); c) Hoechst (1:1,000; Sigma-Aldrich, 382065). The detection of EdU was performed using BeyoClick™ EdU-647 kit (Beyotime, C0081S) according to the manufacturer’s instructions.
Microglial Cell Transplantation.
Before transplantation, the PLX5622-formulated chow diet was prepared, which contained 5 g PLX5622 per kilogram. To delete brain microglia, recipient mice were fed with PLX5622 diet for consecutive 7 d. The brain tissue of CX3CR1-EGFP mice (donor) at P7 or P15 was cut into small pieces and dissociated by enzymatic digestion using the Neural Tissue Dissociation Kit P to obtain the cell suspensions. During the process, the concentration of DNase used was three times higher than standard level to prevent cell adhesion caused by excessive DNA. Following centrifugation at 800×g for 3 min, cell pellets were resuspend with D-PBS to the total volume of 1,700 μL, and mixed with 300 μL Debris Removal Solution (Miltenyi Biotec, 130-109-398). Overlay very gently with 2,000 μL D-PBS. Centrifugation was carried out at 4 °C and 3,000×g for 10 min. The bottommost cells were collected, followed by filling up with D-PBS, and centrifugation at 800×g for 3 min. Samples were blocked for 10 min in FACS buffer followed by incubation with rabbit anti-NG2 pAb for 30 min, and finally were incubated with Alexa Fluor 555 donkey antirabbit IgG (1:200). The same number (≥4,000) of NG2+ and NG2− microglia were collected by FACS. Up to 2,000 NG2+ or NG2− microglia were stereotaxically microinjected into the SN (AP −3.0 mm, ML ±1.3 mm, DV −4.5 mm) or cerebrocortex (AP 1.42 mm, ML ±1.25 mm, DV −1.75 mm). Five to six days postsurgery, mice were anesthetized and perfused with PLP.
Single-Cell Sequencing and Analysis for NG2-EGFP Mice.
The SN tissues were dissociated and digested with enzymes followed by incubation with cold Debris Removal Solution (Miltenyi Biotec, 130-109-398). After removal of cell debris, the cell suspensions were sorted by FACS. NG2-EGFP cells and EGFP-negative cells were collected and checked for their quality and cell identifies. Cells were loaded into the wells of a 10× Chromium chip (10× Genomics, USA). Then single-cell cDNA was obtained and sequenced. Single-cell transcriptional information was analyzed using Seurat package (https://satijalab.org/seurat/) (60). And the analysis was run in the R environment.
Statistical Analysis.
Statistical analysis was performed using GraphPad software. Data presented as mean ± SEM. were submitted to student t test or one-way ANOVA followed by the Student–Newman–Keul’s test (as a post hoc test). P < 0.05 was considered significant in statistics.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Ms. W. Su for excellent technical assistance in animal genotyping; Ms. L. Han, Ms. Q. Liu and their team at the small animal house for support in mouse breeding; Dr. Q. Hu and his colleagues at the Optical Imaging Center of CEBSIT for technical support in confocal microscopy. This work was supported by grants from the Ministry of Science and Technology of China (2020YFC2002800, 2021ZD0200900), the Natural Science Foundation of China (32230049, U1801681), Strategic Priority Research Program of Chinese Academy of Science (XDB32020100), Shanghai Municipal Science and Technology Major Projects (2018SHZDZX05, AD project), Key Realm R&D Program of Guangdong Province (2018B030337001), Innovative Research Team of High-Level Local Universities in Shanghai.
Author contributions
Y.-j.L., G.H., and J.-w.Z. designed research; Y.-j.L., Y.D., Y.-q.Y., and H.X. performed research; Y.-j.L. contributed new reagents/analytic tools; Y.-j.L. analyzed data; Y.-q.Y. animal management; and Y.-j.L. and J.-w.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. All the sequencing data will be shared on the Sequence Read Archive (SRA), the primary NIH-funded archive for high throughput datasets. This data received the accession code BioProject ID PRJNA929920, and can be accessed at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA929920/.
Supporting Information
References
- 1.Feigin V. L., et al. , Global, regional, and national burden of neurological disorders, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 18, 459–480 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yun S. P., et al. , Block of A1 astrocyte conversion by microglia is neuroprotective in models of Parkinson’s disease. Nat. Med. 24, 931–938 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baror R., et al. , Transforming growth factor-beta renders ageing microglia inhibitory to oligodendrocyte generation by CNS progenitors. Glia 67, 1374–1384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abe N., Nishihara T., Yorozuya T., Tanaka J., Microglia and macrophages in the pathological central and peripheral nervous systems. Cells 9, 2132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman B. A., et al. , Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Rep. 22, 832–847 (2018). [DOI] [PubMed] [Google Scholar]
- 6.George S., et al. , Microglia affect alpha-synuclein cell-to-cell transfer in a mouse model of Parkinson’s disease. Mol. Neurodegener. 14, 34 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheiblich H., et al. , Microglia jointly degrade fibrillar alpha-synuclein cargo by distribution through tunneling nanotubes. Cell 184, 5089–5106.e5021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song N., Chen L., Xie J., Alpha-synuclein handling by microglia: Activating, combating, and worsening. Neurosci. Bull. 37, 751–753 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascoal T. A., et al. , Microglial activation and tau propagate jointly across Braak stages. Nat. Med. 27, 1592–1599 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Kettenmann H., Hanisch U. K., Noda M., Verkhratsky A., Physiology of microglia. Physiol. Rev. 91, 461–553 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Hickman S., Izzy S., Sen P., Morsett L., El Khoury J., Microglia in neurodegeneration. Nat. Neurosci. 21, 1359–1369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young A. M. H., et al. , A map of transcriptional heterogeneity and regulatory variation in human microglia. Nat. Genet. 53, 861–868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond T. R., et al. , Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavin Y., et al. , Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keren-Shaul H., et al. , A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e1217 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Huang W., Bai X., Meyer E., Scheller A., Acute brain injuries trigger microglia as an additional source of the proteoglycan NG2. Acta Neuropathol. Commun. 8, 146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd A. F., Davies C. L., Miron V. E., Microglia: Origins, homeostasis, and roles in myelin repair. Curr. Opin. Neurobiol. 47, 113–120 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Elmore M. R., et al. , Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Utz S. G., et al. , Early fate defines microglia and non-parenchymal brain macrophage development. Cell 181, 557–573.e518 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Sosna J., et al. , Early long-term administration of the CSF1R inhibitor PLX3397 ablates microglia and reduces accumulation of intraneuronal amyloid, neuritic plaque deposition and pre-fibrillar oligomers in 5XFAD mouse model of Alzheimer’s disease. Mol. Neurodegener. 13, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stallcup W. B., The NG2 antigen, a putative lineage marker: Immunofluorescent localization in primary cultures of rat brain. Dev. Biol. 83, 154–165 (1981). [DOI] [PubMed] [Google Scholar]
- 22.Nishiyama A., Dahlin K. J., Prince J. T., Johnstone S. R., Stallcup W. B., The primary structure of NG2, a novel membrane-spanning proteoglycan. J. Cell Biol. 114, 359–371 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y., Li Z., Shi X., Ding J., Wang X., Roles of NG2 glia in cerebral small vessel disease. Neurosci Bull. Nov 18. doi: 10.1007/s12264-022-00976-w. Online ahead of print (2022). [DOI] [PMC free article] [PubMed]
- 24.Nishiyama A., Chang A., Trapp B. D., NG2+ glial cells: A novel glial cell population in the adult brain. J. Neuropathol. Exp. Neurol. 58, 1113–1124 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Ozerdem U., Grako K. A., Dahlin-Huppe K., Monosov E., Stallcup W. B., NG2 proteoglycan is expressed exclusively by mural cells during vascular morphogenesis. Dev. Dyn. 222, 218–227 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Hsieh S. Y., Zhuang F. H., Wu Y. T., Chen J. K., Lee Y. L., Profiling the proteome dynamics during the cell cycle of human hepatoma cells. Proteomics 8, 2872–2884 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Yotsumoto F., et al. , NG2 proteoglycan-dependent recruitment of tumor macrophages promotes pericyte-endothelial cell interactions required for brain tumor vascularization. Oncoimmunology 4, e1001204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makagiansar I. T., Williams S., Mustelin T., Stallcup W. B., Differential phosphorylation of NG2 proteoglycan by ERK and PKCalpha helps balance cell proliferation and migration. J. Cell Biol. 178, 155–165 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sjostedt E., et al. , An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367, eaay5947 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Thakur P., et al. , Modeling Parkinson’s disease pathology by combination of fibril seeds and alpha-synuclein overexpression in the rat brain. Proc. Natl. Acad. Sci. U.S.A. 114, E8284–E8293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruger R., et al. , Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat. Genet. 18, 106–108 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Taylor T. N., et al. , Region-specific deficits in dopamine, but not norepinephrine, signaling in a novel A30P alpha-synuclein BAC transgenic mouse. Neurobiol. Dis. 62, 193–207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Mayhani M. T., et al. , NG2 expression in glioblastoma identifies an actively proliferating population with an aggressive molecular signature. Neuro. Oncol. 13, 830–845 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosavi L. K., Cammett T. J., Desrosiers D. C., Peng Z. Y., The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13, 1435–1448 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samuel T., et al. , cIAP1 Localizes to the nuclear compartment and modulates the cell cycle. Cancer Res. 65, 210–218 (2005). [PubMed] [Google Scholar]
- 36.Rambout X., et al. , The transcription factor ERG recruits CCR4-NOT to control mRNA decay and mitotic progression. Nat. Struct. Mol. Biol. 23, 663–672 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Akram S., et al. , LRIF1 interacts with HP1alpha to coordinate accurate chromosome segregation during mitosis. J. Mol. Cell Biol. 10, 527–538 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deardorff M. A., et al. , RAD21 mutations cause a human cohesinopathy. Am. J. Hum. Genet. 90, 1014–1027 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonoda E., et al. , Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell 1, 759–770 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Viny A. D., et al. , Cohesin members Stag1 and Stag2 display distinct roles in chromatin accessibility and topological control of HSC self-renewal and differentiation. Cell Stem Cell 25, 682–696.e688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prieto I., et al. , STAG2 and Rad21 mammalian mitotic cohesins are implicated in meiosis. EMBO Rep. 3, 543–550 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerek E. M., et al. , A conserved acetylation switch enables pharmacological control of tubby-like protein stability. J. Biol. Chem. 296, 100073 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan W., et al. , Distinct phases of adult microglia proliferation: A Myc-mediated early phase and a Tnfaip3-mediated late phase. Cell Discov. 8, 34 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salic A., Mitchison T. J., A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 2415–2420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosin J. M., Vora S. R., Kurrasch D. M., Depletion of embryonic microglia using the CSF1R inhibitor PLX5622 has adverse sex-specific effects on mice, including accelerated weight gain, hyperactivity and anxiolytic-like behaviour. Brain Behav. Immun. 73, 682–697 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Dagher N. N., et al. , Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice. J. Neuroinflammation 12, 139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grubman A., et al. , A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22, 2087–2097 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Oakley H., et al. , Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raff M. C., Miller R. H., Noble M., A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature 303, 390–396 (1983). [DOI] [PubMed] [Google Scholar]
- 50.Kucharova K., Stallcup W. B., Dissecting the multifactorial nature of demyelinating disease. Neural. Regen Res. 13, 628–632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferrara G., et al. , NG2, a common denominator for neuroinflammation, blood-brain barrier alteration, and oligodendrocyte precursor response in EAE, plays a role in dendritic cell activation. Acta Neuropathol. 132, 23–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hughes E. G., Kang S. H., Fukaya M., Bergles D. E., Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat. Neurosci. 16, 668–676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung S., et al. , Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol. 20, 4106–4114 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miwa H., Era T., Generation and characterization of PDGFRalpha-GFPCreERT2 knock-In mouse line. Genesis 53, 329–336 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Madisen L., et al. , Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chandra S., Gallardo G., Fernandez-Chacon R., Schluter O. M., Sudhof T. C., Alpha-synuclein cooperates with CSPalpha in preventing neurodegeneration. Cell 123, 383–396 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Shao W., et al. , Suppression of neuroinflammation by astrocytic dopamine D2 receptors via alphaB-crystallin. Nature 494, 90–94 (2013). [DOI] [PubMed] [Google Scholar]
- 58.McLean I. W., Nakane P. K., Periodate-lysine-paraformaldehyde fixative., A new fixation for immunoelectron microscopy. J. Histochem. Cytochem. 22, 1077–1083 (1974). [DOI] [PubMed] [Google Scholar]
- 59.Zhang S. Z., et al. , NG2 glia regulate brain innate immunity via TGF-beta2/TGFBR2 axis. BMC Med. 17, 204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix. All the sequencing data will be shared on the Sequence Read Archive (SRA), the primary NIH-funded archive for high throughput datasets. This data received the accession code BioProject ID PRJNA929920, and can be accessed at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA929920/.