Abstract
The root-colonizing bacterium Pseudomonas fluorescens CHA0 was used to construct an oxygen-responsive biosensor. An anaerobically inducible promoter of Pseudomonas aeruginosa, which depends on the FNR (fumarate and nitrate reductase regulation)-like transcriptional regulator ANR (anaerobic regulation of arginine deiminase and nitrate reductase pathways), was fused to the structural lacZ gene of Escherichia coli. By inserting the reporter fusion into the chromosomal attTn7 site of P. fluorescens CHA0 by using a mini-Tn7 transposon, the reporter strain, CHA900, was obtained. Grown in glutamate-yeast extract medium in an oxystat at defined oxygen levels, the biosensor CHA900 responded to a decrease in oxygen concentration from 210 × 102 Pa to 2 × 102 Pa of O2 by a nearly 100-fold increase in β-galactosidase activity. Half-maximal induction of the reporter occurred at about 5 × 102 Pa. This dose response closely resembles that found for E. coli promoters which are activated by the FNR protein. In a carbon-free buffer or in bulk soil, the biosensor CHA900 still responded to a decrease in oxygen concentration, although here induction was about 10 times lower and the low oxygen response was gradually lost within 3 days. Introduced into a barley-soil microcosm, the biosensor could report decreasing oxygen concentrations in the rhizosphere for a 6-day period. When the water content in the microcosm was raised from 60% to 85% of field capacity, expression of the reporter gene was elevated about twofold above a basal level after 2 days of incubation, suggesting that a water content of 85% caused mild anoxia. Increased compaction of the soil was shown to have a faster and more dramatic effect on the expression of the oxygen reporter than soil water content alone, indicating that factors other than the water-filled pore space influenced the oxygen status of the soil. These experiments illustrate the utility of the biosensor for detecting low oxygen concentrations in the rhizosphere and other soil habitats.
Adaptation of many facultative anaerobic and some obligate aerobic bacteria to oxygen limitation is mediated by a family of transcriptional regulators related to the FNR (fumarate and nitrate reductase regulation) protein of Escherichia coli (43, 48, 49). In Pseudomonas aeruginosa and other Pseudomonas species belonging to rRNA homology group I, the FNR homolog ANR (anaerobic regulation of arginine deiminase and nitrate reductase pathways) is a major regulator of genes involved in anaerobic metabolism, e.g., the genes required for anaerobic respiration (1, 55) and the arcDABC (anaerobic arginine catabolism) operon of P. aeruginosa (10, 12, 41, 54, 56). When oxygen levels decrease, the ANR protein binds to a conserved sequence, the ANR box, located about 40 bp upstream of the transcription initiation site and thereby activates transcription, as shown for the arcDABC operon (10, 12, 28a, 30).
FNR of E. coli and ANR of P. aeruginosa and Pseudomonas fluorescens share essential features in all domains that are important for sensing oxygen tension and for transcriptional activation (13, 25, 38, 53, 54, 56). The sensory domain of FNR contains four cysteine residues, Cys-20, Cys-23, Cys-29, and Cys-122, serving to bind a [4Fe-4S]2+ cluster at a ratio of one cluster per FNR monomer (20, 22, 26). This type of iron-sulfur cluster has a redox potential that can be as low as −700 mV and therefore acts as a strong reductant (39). In its reduced state, the [4Fe-4S]2+ cluster appears to mediate dimerization and thus activation of FNR (20, 47, 49). By virtue of its C-terminal helix-turn-helix motif, activated FNR recognizes specific binding sites (FNR boxes) in anaerobically inducible or repressible promoters (43, 48) and modulates transcription by interaction with RNA polymerase (38, 53). Upon exposure of the cells to oxygen, the [4Fe-4S]2+ cluster is oxidized to a [2Fe-2S]2+ cluster, which remains bound to the FNR monomers but is unable to support the dimeric structure. Prolonged oxidation may eventually lead to disassembly of the iron-sulfur cluster, leaving FNR as a monomeric apoprotein (20, 22, 26, 48). Oxygen is considered to be the direct effector of FNR by oxidation of the [4Fe-4S]2+ cluster (6). It has been calculated that the diffusion of oxygen into the cytoplasm is rapid enough to maintain ambient oxygen concentrations despite respiratory consumption, as long as the concentration of oxygen in the extracellular environment is above 102 Pa (2, 48).
Genetically engineered bacteria that can be introduced and report the physicochemical conditions in natural environments have proved very useful in recent years (28, 36). In the present study, we have developed an oxygen-sensitive biosensor based on a transcriptional fusion of a modified, ANR-dependent arcDABC promoter of P. aeruginosa PAO1 to the lacZ structural gene of E. coli. The reporter system was cloned into a mini-Tn7 transposon (3), which can be transposed into a specific and unique target site (attTn7) found in a noncoding chromosomal region of many bacteria, including P. fluorescens (4, 8, 9). Although nonspecific Tn7 insertion has been reported for some P. putida strains (44), the Tn7 transposon system generally provides a system for stable insertion with a minimal risk of affecting cell functions. We inserted the oxygen reporter construct into the obligate aerobe P. fluorescens CHA0, a strain used widely in biological control studies (21, 52). In this organism, ANR has been identified as a transcriptional activator of the hcnABC operon, encoding HCN synthase, under low-oxygen conditions (25, 56). The biosensor constructed, P. fluorescens CHA900, was used to evaluate oxygen concentrations in a plant-soil microcosm, previously developed for studying nutrient starvation responses of another root-colonizing P. fluorescens strain (24).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Bacterial cultures were pregrown in Luria-Bertani (LB) broth (40) or a defined Na-citrate (0.7%)-asparagine (0.2%) medium (CAMM; modified from reference 7) containing the appropriate antibiotic(s) (ampicillin or kanamycin). The oxygen response of the constructs was monitored in batch cultures grown in OS minimal medium (33) containing 0.1% (NH4)2SO4 and supplemented with either glucose (0.5%) or yeast extract (0.4%) and glutamate (25 mM) as carbon sources. Oxystat experiments were carried out with the yeast extract-glutamate OS medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | hsdR17 recA1 endA1 gyrA96 relA1 thi-1 supE44 ΔlacU169 (φ80lacZΔM15) | 40 |
| CC118/λpir | araD139 Δ(ara-leu)7697 ΔlacX79 phoA20 gelE thi rpsE rpoB argEamrecA1/λpir | 15 |
| P. fluorescens | ||
| CHA0 | Wild type | 52 |
| CHA21 | anr::ΩKm | 25 |
| CHA900 | Chromosomal mini-Tn7-PANR-lacZ | This study (Fig. 1) |
| CHA901 | Chromosomal mini-Tn7-PConst.-lacZ | This study (Fig. 1) |
| Plasmids | ||
| pTJ1R | pUX-BF5 derivative deleted for the SalI site upstream of the Kmr determinant, thus containing a unique SalI-HincII site | J. Wall (University of Missouri—Columbia) |
| pUX-BF5 | pUC19-based plasmid containing the mini-Tn7-Km transposon, Ap | 3 |
| pUX-BF13 | R6K replicon-based helper plasmid, providing the Tn7 transposition functions in trans, Ap. Can only replicate when the pir gene is supplied in trans. | 3 |
| pNM480 | Vector for translational ′lacZ fusions, Ap | 31 |
| pME3535 | pNM480 derivative for transcriptional lacZ fusions; lacZY genes downstream of a polylinker (EcoRI SmaI BamHI SalI PstI), Ap | This study |
| pME3781 | IncQ plasmid containing an ANR-dependent consensus promoter derived from a modified arcDABC promoter, transcriptional lacZ fusion, Ap | 54 |
| pME3771-1 | IncQ plasmid containing a noninducible promoter, transcriptional lacZ fusion, Ap | 54 |
| pME6000 | Broad-host-range vector, Tc | 29 |
| pME6502 | pTJ1R containing a transcriptional fusion between the promoter from pME3781 and lacZ, Km | This study (Fig. 1) |
| pME6504 | pTJ1R containing a transcriptional fusion between the promoter from pME3771-1 and lacZ, Km | This study (Fig. 1) |
Construction of plasmids.
A plasmid (pME3535) suitable for the construction of transcriptional lacZ fusions was constructed as follows. The 8.6-kb pUC8-based vector pNM480, which contains the lacZ and lacY genes allowing the construction of translational fusions (31), was opened with PstI and HindIII, and a linker coding for the authentic 5′ end of lacZ mRNA was inserted. This linker reconstituted the lacZ gene with its own ribosome binding site and consisted of the annealed primers 5′-G AATTGT GAGCGG ATAACA ATTTCA CACAGG AAACAGCT ATG ACC ATG ATT CA-3′ and 3′-ACGTC TTAACA CTCGCC TATTGT TAAAGT GTGTCC TTTGTCGA TAC TGG TAC TAA GTTCGA-5′. For the insertion of reporter constructs into the P. fluorescens chromosome, plasmid pTJ1R containing a mini-Tn7-Km transposon (Table 1) was used as the carrier. The arcDABC-derived FNR consensus promoter carried by a 56-bp BamHI-PstI fragment of pME3781 (54) was fused to the 3.2-kb PstI-DraI fragment of pME3535 carrying the lacZ gene. The resulting transcriptional fusion was inserted into pME6000 (29) cleaved with BamHI and PstI (with removal of the 3′ overhang by Klenow enzyme), allowing the recovery of the fusion as a 3.2-kb XbaI-XhoI fragment, which was treated with Klenow enzyme to fill in the sticky ends and ligated into the 6.3-kb plasmid pTJ1R opened at its unique HincII site. This produced plasmid pME6502 (Fig. 1). The ANR-independent, arcDABC-derived promoter of pME3771-1 (54) fused to the lacZ gene of pME3535 was recruited similarly as a BamHI-DraI fragment. After treatment with Klenow enzyme, the 3.2-kb promoter-lacZ fragment was purified and ligated into the 6.3-kb HincII fragment of pTJ1R to give plasmid pME6504 (Fig. 1). Plasmids pME6502 and pME6504 were introduced into E. coli CC118/λpir by transformation and selection on LB agar plates containing 25 μg of kanamycin per ml and 200 μg of X-Gal (5-chloro-4-bromo-3-indolyl-β-d-galactopyranoside) per ml. The carrier plasmids pME6502 and pME6504, as well as the helper plasmid pUX-BF13, were purified from E. coli by using the Qiagen midi plasmid purification kit according to the manufacturer’s instructions (Qiagen GmbH, Hilden, Germany).
FIG. 1.
Construction of reporter plasmids. pME6502 and pME6504 were constructed by insertion of PANR-lacZ and PConst.-lacZ, respectively, into a mini-Tn7-Km transposon in the carrier plasmid pTJ1R. PANR, ANR- and FNR-dependent promoter; PConst., constitutive promoter; oriColE1, origin of replication; ApR, ampicillin resistance gene; KmR, kanamycin resistance gene; mob, mobilization region; Tn7L and Tn7R, left and right Tn7 ends, respectively; ⊗, polylinker missing from pME6504.
Preparation of competent cells of P. fluorescens.
Strain CHA0 was grown in LB broth at 30°C. After incubation overnight, 0.25 ml of the cell suspension was shifted to 25 ml of fresh LB broth in a 100-ml Erlenmeyer flask and grown at 35°C (to inactivate the restriction system) until the culture reached an optical density at 600 nm (OD600) of 0.5 to 1.0. The cells were harvested by centrifugation and washed twice with 10 ml of an ice-cold solution of 15% (wt/vol) glycerol, 1 mM MOPS (3-morpholinopropanesulfonic acid). The cells were resuspended in 200 μl of the glycerol-MOPS solution, stored on ice, and used within 2 to 3 h.
Chromosomal insertion of reporter constructs by electroporation.
Competent cells (40-μl suspension) were mixed in an Eppendorf tube with ∼0.5 μg of plasmid DNA (pME6502 or pME6504) in 5 μl of TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA) and 0.5 μg of pUX-BF13 DNA in 5 μl of TE buffer. Plasmid pUX-BF13 carries the genes encoding the transposition proteins necessary for insertion of the Tn7 cassette into the genomic target site (3). The mixture was transferred to an ice-cold electroporation cuvette and treated in a Bio-Rad electroporator (25 μF, 200 Ω, 5 ms, 2.5 kV/cm). Immediately thereafter, 1 ml of prewarmed (35°C) SOC medium (2% [wt/vol] Bacto tryptone, 0.5% [wt/vol] yeast extract, 10 mM NaCl, 10 mM MgCl2, 10 mM MgSO4, 2.5 mM KCl, 20 mM glucose) was added to the cuvette. The cell suspension was transferred to an Eppendorf tube and incubated at 35°C for 3 h, followed by spread plating of the entire culture on five selective plates (LB agar with 25 μg of kanamycin per ml). The plates were incubated overnight at 35°C, transferred to 30°C, and incubated for 48 h until Kmr colonies appeared. The incubation at 35°C after electroporation was done to keep the CHA0 restriction system inactive. This procedure enhanced the recovery of Kmr transformants. Typically, about 200 transformants were obtained. Integration of mini-Tn7-Km from pME6502 and pME6504 produced strains CHA900 and CHA901, respectively.
Southern blot analysis.
Chromosomal DNA was purified from mini-Tn7-Km-transformed P. fluorescens strains by phenol extraction (11). DNA samples (∼2 μg) were digested with SmaI or HindIII, separated electrophoretically on a 0.7% agarose gel, and transferred to a Hybond N membrane (Amersham) according to the instructions of the supplier. The 1.37-kb PstI fragment of pTJ1R constituting the Kmr gene was labeled with digoxigenin-11-dUTP (DIG) (Boehringer Mannheim) and used as a probe. After hybridization at 60°C for 12 h and being washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (40) with 0.2% sodium dodecyl sulfate (SDS) at room temperature, the DIG-labeled fragments were detected by reaction with anti-DIG antibodies coupled to alkaline phosphatase, according to a protocol supplied by the manufacturer (Boehringer Mannheim). Nylon membranes were exposed to an X-ray film for 3 min.
TSO test.
The genetic instability of strain CHA0 can be monitored by using tryptophan side chain oxidase (TSO) as a marker (52). Kmr transformants were streaked on King’s B agar (23) and incubated at 30°C for 3 days. The colonies were then overlaid with 20 ml of TSO agar and incubated at room temperature for 4 h, after which time, TSO-positive and TSO-negative colonies could be observed as black and white colonies, respectively. TSO agar consisted of 1.2 g of agar melted in 54 ml of water and mixed at 50°C with 6 ml of 10% (wt/vol) SDS and 60 ml of 20 mM l-tryptophan in 1 M glycine-HCl buffer (pH 3.0) (45).
Growth measurements.
P. fluorescens CHA0 (wild type), CHA900 (oxygen reporter), and CHA901 (constitutive control) grown in LB broth were used to inoculate CAMM (100 ml) in 500-ml baffled Erlenmeyer flasks. Cultures were incubated on a rotary shaker (300 rpm [IKA KS250 Basic; Janke and Kunkel, Staufen, Germany]) at 30°C, and the OD600 was monitored for 15 h.
Determination of β-galactosidase activities.
For pure cultures, β-galactosidase activity was measured in toluenized cells with o-nitrophenol-β-d-galactoside (ONPG) as a substrate and expressed in Miller kilounits (40). For soil experiments, samples of 0.5 g of soil were suspended in 5 ml of phosphate-buffered saline (PBS) (40), and 2 ml of this was then centrifuged (14,000 rpm, 3 min, model 5415C; Eppendorf-Netheler-Hinz GmbH, Hamburg, Germany) and resuspended in 200 μl of PBS. Toluene (10 μl) was added, and β-galactosidase activities were determined by addition of ONPG, as described above. Soil particles were removed by centrifugation (14,000 rpm, 3 min). Enzyme activities (A420 units) were related to the number of P. fluorescens CHA900 or CHA901 cells in the soil slurries determined as CFU on LB agar containing kanamycin (25 μg per ml) by the drop plate method (16). An OD600 value of 1 equals ∼2 × 109 cells per ml.
Oxygen response measurements of reporter constructs.
In preliminary experiments, E. coli CC118/λpir carrying pME6502 or pME6504 and P. fluorescens CHA900 and CHA901 were grown in LB broth overnight. These cultures were used to inoculate 60 ml of OS minimal medium (in 125-ml serum bottles) or 100 ml of OS minimal medium (in 500-ml Erlenmeyer flasks). The serum bottles were sealed with Teflon-coated rubber stoppers. Bacterial cultures in these bottles consumed the oxygen initially present. P. fluorescens CHA0 cells, being strictly aerobic, stopped growth at ca. 5 × 108 cells per ml. Bottles and flasks were incubated on a rotary shaker (200 rpm) at 30°C. Cells from both the oxygen-limited and the well-aerated cultures thus obtained were harvested at an OD600 of ∼0.5 for determination of β-galactosidase activity.
The oxygen response of carbon-starved P. fluorescens CHA900 was determined by growing the cells to early exponential phase (OD600 of 0.25 to 0.50) in CAMM and shifting them to 25 ml of OS buffer (OS medium without addition of carbon source or electron donor) in 100-ml serum bottles. The bottles were flushed with nitrogen gas, and the oxygen concentration was adjusted to 210 × 102, 70 × 102, or 7 × 102 Pa, respectively of O2. At intervals, samples were taken from the cell suspension for determination of β-galactosidase activity. The oxygen concentration was monitored by analyzing 200-μl headspace samples on a gas chromatograph equipped with a thermal conductivity detector (HP 6890 series; Hewlett-Packard).
Oxystat experiments were carried out by growing P. fluorescens at defined oxygen tensions (partial O2 pressure [pO2]) in a 3.5-liter fermentor (Bioengineering AG, Wald, Switzerland). Oxygen levels were measured with an electrochemical electrode inserted into the medium. The oxygen level in the fermentor was controlled by supplying (i) N2 (0 to 1 liter per min) through a manually controlled valve and (ii) compressed air through an automatic valve connected to the pO2 electrode via a programmable control unit (pO2 controller). OS medium (final volume, 2 liters) was allowed to stabilize at 30°C and stirred with a magnetic rotary stirrer at 600 rpm. The oxystat was inoculated with 1% of an overnight culture, and the medium was flushed with a constant flow of air (1 liter per min). To decrease the oxygen level, the N2 valve was opened (1 liter per min), and the pO2 controller was turned on. In this way, the air supply was arrested until the O2 level approached the set point, after which the air valve was turned on and off by the pO2 controller to maintain the oxygen level in the medium. The oxygen concentration fluctuated by ±2.5% to 5% around the set value. The pH typically increased from 6.7 to 7.0 during growth.
Soil and plant experiments.
A sandy loam soil (coarse sand, 26.9%; fine sand, 39.9%; silt, 15.4%; clay, 15.3%; humic particles, 2.5%) was sampled from an agricultural field (Højbakkegaard, Taastrup, Denmark), passed through a 2-mm-pore-diameter sieve, and stored at 4°C until use. Barley seeds (Hordeum vulgare, var. Lamba) were allowed to germinate for 48 h on wetted filter paper, resulting in ∼1-cm-long roots. Precultures of P. fluorescens CHA900 and CHA901 were grown in LB broth, shifted to 100 ml of CAMM (OD600 of ∼0.005) in baffled 500-ml Erlenmeyer flasks, and incubated on a rotary shaker (300 rpm) at 30°C. The cells were harvested from early-exponential-phase cells (OD600 of between 0.25 and 0.50), washed once, and resuspended in PBS to a final cell density of ∼2 × 1010 cells per ml. The barley seedlings were placed in the cell suspension for 30 min. Bulk soil was inoculated by spraying the cell suspension onto the soil, reaching a final concentration of ∼1 × 108 to 5 × 108 cells per g of soil and a water content of 15% (wt/wt), i.e., 60% of the field capacity of this soil. The soil was packed loosely in polyvinyl chloride (PVC) tubes (2.8 × 11.5 cm) to a final density of ∼1.1 g of soil per cm3. Two bacterium-coated barley seedlings were planted in each tube. The tubes were placed in sealed transparent plastic bags containing wetted soil to avoid desiccation and incubated at 20°C with 12 h of light and 12 h of dark. Throughout the experiments, the water content of the soil was monitored by weighing of the tubes and was adjusted when needed.
Plants were harvested by gently removing the soil cores from the PVC tubes. The roots were loosened from the soil, and excess soil was removed by gentle shaking. The soil adhering to the roots was defined as rhizosphere soil. Root pieces of ∼2 cm were cut from the root base immediately below the seed, resulting in four to six 0.5-g samples per plant-soil microcosm. In some experiments, samples were taken at the root tip usually 5 to 7 cm below the seed. The samples were placed in 10-ml test tubes, and 5 ml of cold PBS was added. The tubes were shaken for 1 min on a rotary shaker and sonicated for 0.5 min to extract the bacteria from soil particles and roots. The root pieces were then removed, and the β-galactosidase activity determined as described above.
The effect of soil compaction was studied in soil, inoculated with P. fluorescens CHA900, and amended with 2% (wt/wt) ground wheat straw, packed in 10-ml test tubes at five different bulk densities (1.0, 1.1, 1.3, 1.6, or 1.8 g of soil per cm3, respectively). The water content was adjusted to 60% of field capacity in half of the tubes and 85% in the other half. The tubes were incubated at 20°C for 24 h. The soil cores (1.5 × 5 cm) were then removed from the tubes, and 0.5-g samples were taken from the center of the cores (2.5 cm from the top) for determination of β-galactosidase activity.
For determination of the half-maximal oxygen response of the reporter bacteria in soil, a set of microcosms were placed in an anaerobic jar. The jar was sealed and flushed with nitrogen, and the oxygen concentration was adjusted to ∼5 × 102 Pa of O2 by addition of atmospheric air (controlled by gas chromatography analysis of gas samples). The jar was then incubated at 20°C for 12 h before determination of β-galactosidase activity.
RESULTS
Construction of reporter strains.
An artificial promoter (PANR), which was derived from the arcD promoter of P. aeruginosa and contains the consensus FNR box (TTGAT…ATCAA) as well as an optimal −10 hexamer (TATAAT) for ς70 RNA polymerase (Fig. 1), was recognized equally well by FNR in E. coli and ANR in P. aeruginosa during oxygen limitation (54). This promoter was used to construct a transcriptional lacZ fusion, which was inserted into a mini-Tn7 transposon carried by the suicide plasmid pME6502 (Fig. 1). Chromosomal integration of mini-Tn7-PANR-lacZ from pME6502 was achieved in P. fluorescens CHA0 by coelectroporation with a nonreplicating helper plasmid expressing transiently the Tn7 transposition functions (see Materials and Methods). The reporter strain obtained was designated CHA900. In parallel, a constitutive promoter (PConst.) previously characterized in E. coli and P. aeruginosa (54) was used for the construction of a control strain, CHA901, via the suicide plasmid pME6504 (Fig. 1).
Insertion of mini-Tn7-PANR-lacZ and mini-Tn7-PConst.-lacZ into the unique Tn7 attachment site of P. fluorescens was checked by Southern blotting. Chromosomal DNA was isolated from strains CHA900 and CHA901, digested with SmaI or HindIII, and probed with the 1.3-kb Kmr gene of pME6502 (Fig. 1). The band patterns obtained were identical for the three independent isolates of both strains (data not shown). Digestion with SmaI, for which there is one site in the Kmr gene and none in the lacZ gene (Fig. 1), resulted in bands of 5.7 and 2.6 kb. Digestion with HindIII, cutting once in the Kmr gene and once at the 5′ end of the lacZ gene (Fig. 1), resulted in bands of >20 kb and 0.75 kb, i.e., the 3′ end of the Kmr gene (data not shown). These results indicate that P. fluorescens CHA0 has a unique Tn7 attachment site into which a single copy of the Tn7 cassette can be inserted in one specific orientation.
Grown in CAMM at 30°C, the generation times of P. fluorescens CHA0, CHA900, and CHA901 were 1.12 ± 0.03, 1.16 ± 0.02, and 1.14 ± 0.01 h, respectively (mean of three replicates ± standard error). Thus, the Tn7 cassette insertions in CHA900 and CHA901 did not influence the growth rates, in agreement with data previously reported for another strain of P. fluorescens (9). Furthermore, strains CHA900 and CHA901 were TSO positive, as was the wild-type CHA0, indicating that a frequent class of pleiotropic mutation entailing loss of TSO expression in P. fluorescens (52) had not occurred in the reporter constructs.
Oxygen response of reporter constructs in batch experiments.
The PANR-lacZ reporter was strongly induced in E. coli CC118/λpir carrying pME6502 and in P. fluorescens CHA900 during oxygen limitation, compared to the basal expression in well-aerated cultures (Table 2), indicating that the construct could indeed be used as an oxygen reporter. The control strains E. coli CC118/λpir carrying pME6504 and P. fluorescens CHA901 showed little if any response to oxygen limitation (Table 2). The response of the PANR-lacZ reporter to oxygen limitation was enhanced in E. coli and P. fluorescens when glucose was replaced by yeast extract and glutamate in the growth medium (Table 2), probably reflecting faster growth of the bacteria with the latter two substrates. In a control experiment designed to demonstrate regulation by ANR, the PANR-lacZ cassette was cloned into the high-copy-number shuttle vector pME6000. The recombinant plasmid obtained gave anaerobically inducible β-galactosidase activities in the wild-type CHA0: i.e., oxygen limitation caused a shift from 8.7 to 118.8 Miller kilounits in glucose medium. In the anr-negative mutant CHA21, anaerobic induction was essentially prevented; i.e., expression amounted to 11.6 Miller kilounits under oxygen limitation.
TABLE 2.
Induction of the PANR-lacZ reporter by oxygen limitation in E. coli and P. fluorescens
| Strain | β-Galactosidase activitya (Miller kilounits) after growthb in:
|
|||
|---|---|---|---|---|
| OS + 0.5% glucose
|
OS + 0.4% yeast extract + 25 mM glutamate
|
|||
| +O2 | −O2 | +O2 | −O2 | |
| E. coli CC118/λpir | ||||
| pME6502 (PANR-lacZ) | 0.54 ± 0.05 | 8.54 ± 0.92 | 0.45 ± 0.02 | 51.63 ± 7.21 |
| pME6504 (PConst.-lacZ) | 0.60 ± 0.03 | 0.79 ± 0.03 | NDc | ND |
| P. fluorescens | ||||
| CHA900 (PANR-lacZ) | 0.18 ± 0.01 | 2.12 ± 0.20 | 0.20 ± 0.04 | 5.16 ± 0.58 |
| CHA901 (PConst.-lacZ) | 0.10 ± 0.02 | 0.10 ± 0.01 | ND | ND |
Values represent the mean of three replicates (± standard error). All samples were taken at an OD600 of between 0.3 and 0.5.
Good aeration (+O2) was achieved by incubation in Erlenmeyer flasks, with oxygen limitation (−O2) by incubation in sealed serum bottles. OS refers to the basal salts medium described in Materials and Methods.
ND, not determined.
Oxygen response of reporter constructs in oxystat experiments.
P. fluorescens CHA900 and E. coli CC118/λpir(pME6502) were grown at defined oxygen tensions in an oxystat. A fully aerated growth medium (OS medium supplemented with yeast extract and glutamate) was inoculated at 1% with the reporter strain, and exponential growth was allowed to be established before the oxygen tension was decreased to a defined constant level. At intervals, β-galactosidase activities in samples were determined. In the representative experiment shown (Fig. 2), the oxygen tension was reduced from 210 × 102 Pa to 50 × 102 Pa (102 Pa corresponds to ∼1 μM dissolved O2). The reporter strain CHA900, initially adapted to optimal aeration (210 × 102 Pa) by a decrease in β-galactosidase activity, reflecting a dilution of the β-galactosidase present in the stationary-phase, partially-oxygen-limited inoculum. Strain CHA900 then responded rapidly to the decrease in the oxygen tension to 50 × 102 Pa by an increase in β-galactosidase activity. The activity reached a steady-state level within a couple of hours while the culture was still growing exponentially (Fig. 2). Similar results were obtained with E. coli harboring pME6502 (data not shown).
FIG. 2.
Oxygen response of P. fluorescens CHA900 in an oxystat experiment. OS medium supplemented with yeast extract and glutamate was inoculated with 1% of an overnight culture. The oxygen concentration (solid line) was initially kept close to 210 mbar (210 × 102 Pa) by flushing the medium with atmospheric air. After 4 h of growth, the oxygen concentration was decreased to 50 mbar (50 × 102 Pa) by flushing with a mixture of nitrogen gas and atmospheric air. The β-galactosidase activity (○) then increased to a steady-state level in the growing culture. Growth was measured as OD600 units (●).
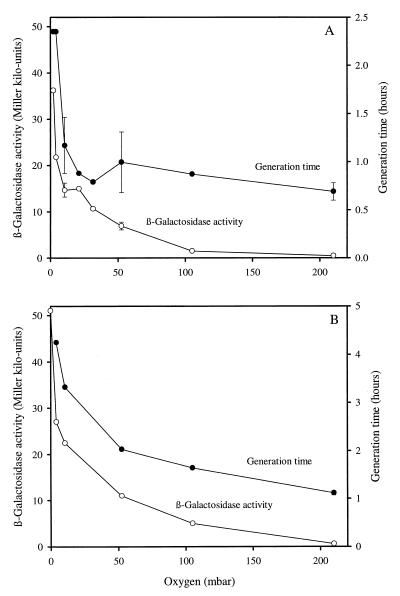
In these oxystat experiments, cultures were shifted to different oxygen tensions, and the steady-state activities obtained were plotted against the respective oxygen levels. For P. fluorescens CHA900, experiments were done in triplicate (Fig. 3A). Similar experiments were also run once with E. coli CC118/λpir(pME6502) as a control (Fig. 3B). The response curves were similar for the two strains, showing a hyperbolic-like increase in β-galactosidase activity as the oxygen tension decreased. Half-maximal induction occurred at ca. 5 × 102 Pa of O2, and the induction factor was close to 100 for both P. fluorescens and E. coli when the β-galactosidase levels at 2 × 102 Pa of O2 were compared to those measured at 210 × 102 Pa of O2 (Fig. 3). The obligate aerobe P. fluorescens CHA900 grew quite well under microaerobic conditions (with a continuous supply of 2 × 102 Pa of O2 the generation time was 2.4 h), but below 2 × 102 Pa of O2, P. fluorescens, unlike E. coli, was unable to maintain growth (Fig. 3). The results obtained for the reporter construct in E. coli are in good agreement with previous data published by Becker et al. (6). The main conclusion is that ANR-mediated induction of PANR-lacZ in P. fluorescens was practically the same as FNR-dependent induction of the reporter in E. coli, suggesting that ANR and FNR sense intracellular O2 concentrations similarly.
FIG. 3.
Steady-state levels of β-galactosidase activities (○) and generation times (●) of P. fluorescens CHA900 (A) and E. coli CC118/λpir(pME6502) (B) grown in the oxystat at different oxygen concentrations. Triplicate experiments were run for some of the oxygen concentrations (data points with standard error bars). The other points represent single oxystat experiments. In panel A, the highest β-galactosidase values were obtained at 2 mbar (2 × 102 Pa) of O2; in panel B, the highest values correspond to 0 mbar (0 × 102 Pa) of O2.
Expression of the reporter construct in carbon-free medium.
The oxygen response of P. fluorescens CHA900 was examined after shifting exponentially growing, well-aerated cells to OS medium without a carbon source under three different oxygen tensions (7 × 102, 70 × 102, and 210 × 102 Pa of O2). This shift resulted in cessation of growth at an OD600 of ∼0.13 in the three cultures. Nevertheless, the expression of the reporter increased with decreasing oxygen tension (Fig. 4), although a steady-state level was reached only after 10 to 12 h of incubation. Incubation at 7 × 102 Pa of oxygen resulted in a β-galactosidase activity of 0.9 to 1.0 Miller kilounits, a value which is considerably lower than that (15 to 20 Miller kilounits) obtained in an exponentially growing culture at the same oxygen level in the oxystat experiment (Fig. 3A). Under conditions of carbon starvation, the anaerobic induction factor was below 10 (Fig. 4), i.e., much smaller than the corresponding value determined during growth in the oxystat (Fig. 3A).
FIG. 4.
Oxygen response of P. fluorescens CHA900 shifted from an exponentially growing culture to 25 ml of carbon-free OS medium in sealed 100-ml serum bottles. At time zero, the headspace was flushed with nitrogen gas and the oxygen concentration was adjusted to 7 (●), 70 (▴), or 210 (■) mbar (7 × 102, 70 × 102, or 210 × 102 Pa, respectively) of O2 by injection of atmospheric air. All results are given as means ± standard error (n = 3).
Induction factor in plant-soil experiments.
P. fluorescens CHA900 (oxygen reporter) and CHA901 (constitutive control strain) were introduced into soil microcosms with and without barley. At intervals, expression of β-galactosidase was analyzed in harvested microcosms. The ability of the reporter bacteria to respond to decreased oxygen tensions was tested by incubating some of the microcosms in an anaerobic jar at ∼5 × 102 Pa of O2 for 12 h; the other microcosms remained exposed to atmospheric air.
Throughout these experiments, the P. fluorescens CHA901 control strain showed a relatively constant expression of β-galactosidase at ∼25 A420 units per 108 CFU in both bulk and rhizosphere soils under normal or reduced oxygen concentrations (data not shown). The reporter strain CHA900 showed a relatively constant, low level of β-galactosidase in bulk soil microcosms incubated under atmospheric air (Fig. 5). In bulk soil microcosms exposed to low-oxygen conditions, strain CHA900 responded to the oxygen decrease with a 5- to 10-fold increase in β-galactosidase activity immediately after introduction into the soil. A similar induction factor was found in the rhizosphere samples (Fig. 5). However in bulk soil, the response to low oxygen tensions decreased within 2 days, and only an insignificant response could be observed after 3 to 4 days of incubation (Fig. 5). This behavior probably reflects starvation conditions. In contrast, in the nutrient-rich rhizosphere, strain CHA900 responded to decreasing O2 levels during a period of up to 6 days (Fig. 5). A similar difference in metabolic status between bacteria in bulk and rhizosphere soil has previously been reported for an introduced strain of P. fluorescens (24, 51) as well as for the indigenous population (32). A bacterium less capable of utilizing root exudates, however, may show higher metabolic activity in the bulk soil than in the rhizosphere (14).
FIG. 5.
Oxygen response of P. fluorescens CHA900 (oxygen reporter) retrieved from bulk soil (A) and the rhizosphere (B) of plant-soil microcosms. For each sampling time, samples were taken from parallel sets of microcosms, half of them previously incubated at 5 mbar (5 × 102 Pa) of O2 for 12 h (○) and half of them incubated at atmospheric air (210 mbar [210 × 102 Pa] of O2) (●). β-Galactosidase activities were calculated as 1,000 × A420 × (Δt)−1 × (108 CFU)−1, where Δt is the assay time (in minutes). All results are given as means ± standard error (n = 6, two microcosms with 3 samples from each).
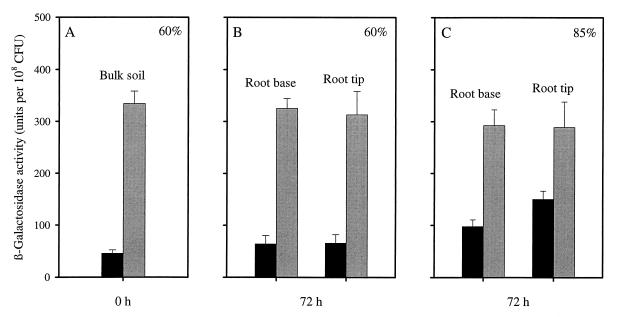
A series of plant-soil microcosms were established with P. fluorescens CHA900 introduced into the soil. The water content of the soil was initially adjusted to 60% of field capacity; after 24 h, the water content was increased to 85% of field capacity in half of the microcosms. After a further 48 h of incubation, soil samples were taken from the root base (0 to 2 cm below the seed) and from the root tip (5 to 7 cm below the seed). P. fluorescens CHA900 expressed a basal level of β-galactosidase activity of ∼50 units per 108 CFU when incubated at 60% of field capacity under atmospheric air and a level of ∼300 units per 108 CFU when exposed to low-oxygen conditions, whether sampled from the root base or the root tip (Fig. 6B). However, when incubated under atmospheric air at 85% of field capacity, the β-galactosidase activity of P. fluorescens CHA900 at the root tip was ∼150 units per 108 CFU and was significantly higher than the basal level (P < 0.001; t test of means) (Fig. 6C). The β-galactosidase activity observed at the root base was not significantly higher than the basal level (P > 0.05, t test of means) (Fig. 6C).
FIG. 6.
Oxygen response of P. fluorescens CHA900 retrieved from bulk soil (A) and rhizosphere soil (B and C) of plant-soil microcosms. The water content of the soil was initially adjusted to 60% of field capacity. After 24 h, the water content was increased to 85% of field capacity in half of the plant microcosms. For each sampling time, half of the microcosms were incubated at 5 mbar (5 × 102 Pa) of O2 for 12 h (gray bars) and the other half were kept at atmospheric air (210 mbar [210 × 102 Pa] of O2) (black bars). All results represent mean ± standard error (n = 6, three microcosms with two samples from each).
The effect of soil compaction was studied by introducing P. fluorescens CHA900 into a series of soil microcosms. Soil compaction was adjusted to five different bulk density levels, and for each level of compaction, soil water content was adjusted to either 60 or 85% of field capacity. After 24 h of incubation, the degree of compaction was seen to have a clear effect on the expression of the oxygen reporter. The β-galactosidase activity was ∼10 times higher at a bulk density of 1.8 g per cm3 than that determined at 1.0 g per cm3 (Fig. 7) and corresponded to the level of enzyme expression obtained by incubating the soil at 2 × 102 Pa of oxygen for 12 h. The difference in water content, on the other hand, had no significant effect on the response pattern of the oxygen reporter in this experiment.
FIG. 7.
Oxygen response of P. fluorescens CHA900 retrieved from bulk soil packed at five different densities. The water content was adjusted to 60% (●) or 85% (○) of field capacity, and the soil was incubated at 20°C for 24 h. Two sets of samples were previously incubated at 2 mbar (2 × 102 Pa) (■) and 5 mbar (5 × 102 Pa) (□) of O2, respectively, for 12 h. All results are given as means ± standard error (n = 3).
DISCUSSION
It may seem surprising, at first sight, that an obligate aerobe such as P. fluorescens CHA0 should have an anaerobic regulator, ANR, which is very similar (53% identical) to the anaerobic regulator FNR of E. coli, a facultative anaerobe. However, as we have found here, strain CHA0 grows quite well with as little as 2 × 102 Pa of O2 (Fig. 3). This level of O2 would not stop growth of, e.g., Clostridium haemolyticum, an organism usually considered to be an oxygen-sensitive, strict anaerobe (27). Thus, P. fluorescens CHA0 is well adapted to ecological niches containing little O2, and it is only in environments containing less than 2 × 102 Pa of O2 that growth of strain CHA0 is handicapped. The oxygen biosensor construct PANR-lacZ was regulated by FNR in E. coli and by ANR in P. fluorescens in virtually the same way (Fig. 3A). This implies that FNR and ANR are also very similar in terms of mechanisms. All essential features of FNR (i.e., the cysteine residues involved in [4Fe-4S]2+ binding, the RNA polymerase α subunit interacting sites, the dimerization domain, the contacts with the ς70 factor, and the helix-turn-helix motif) are highly conserved in ANR (25, 38).
Electrochemical microsensors have been widely used to study distribution of oxygen and oxygen consumption in biofilms and sediments (37). Such microsensors have also been used in artificial soil systems like waterlogged aggregates (18, 42) and gel-stabilized soil systems (19, 34). For optimal utilization of these microsensors, however, it is crucial to determine the diffusion characteristics, e.g., porosity and diffusion coefficients. However, this is difficult in natural soil environments because of heterogeneity in texture and distribution of water (18). Here the use of oxygen reporter bacteria is an improvement, because this technology meets the criteria of in situ detection and high sensitivity (28). Recently, a technique has been developed allowing quantification of β-galactosidase in single cells (35). Combination of this technique with the biosensor strain CHA900 may further extend the usefulness of the biosensor.
The rhizosphere is a habitat in which living roots release a range of low-molecular-weight substrates, including sugars, oligosaccharides, and amino acids. These compounds allow microbial maintenance or growth (5). Oxygen consumption by microorganisms or by the root cells themselves can lead to low oxygen levels in the rhizosphere (17, 19). However, the oxygen status in the rhizosphere depends on the texture, structure, and water content of the soil (46). In the present study, this tenet has been verified experimentally, either by an increase of soil water or by soil compaction (Fig. 6 and 7). The prevalence of microaerobic conditions in the lower part of wheat roots has been previously observed by the use of a nifH-gusA reporter construct in Azospirillum brasilense (50). In further agreement, it is known that ANR-dependent HCN production by P. fluorescens CHA0 occurs predominantly in poorly aerated, wet soils, as judged from a strong suppressive effect on Thielaviopsis basicola-induced black root rot of tobacco, whereas in more aerated soil, the cyanogenic capacity of strain CHA0 does not appear to be expressed (25). In conclusion, the oxygen-sensing reporter strain presented here seems a promising tool for in situ studies of spatial and temporal variations in bioavailability of oxygen in natural habitats.
ACKNOWLEDGMENTS
We thank Elisabeth Koluda and Ulla Rasmussen for technical assistance with the plant-soil experiments, Gary P. Roberts and J. Wall for supplying plasmids, and Christoph Keel and Geneviéve Défago for discussion.
This study was supported by grants from the OECD Cooperative Research Program: Biological Resource Management for Sustainable Agricultural Systems, the Danish Agricultural and Veterinary Research Council (grant 9313839), the Swiss National Science Foundation (grant 31-50522.97), and the Swiss Priority Programme Biotechnology (project 5002-04502311).
REFERENCES
- 1.Arai H, Kodama T, Igarashi Y. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol Microbiol. 1997;25:1141–1148. doi: 10.1046/j.1365-2958.1997.5431906.x. [DOI] [PubMed] [Google Scholar]
- 2.Arras T, Schirawski J, Unden G. Availability of O2 as a substrate in the cytoplasm of bacteria under aerobic and microaerobic conditions. J Bacteriol. 1998;180:2133–2136. doi: 10.1128/jb.180.8.2133-2136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao Y, Lies D P, Fu H, Roberts G P. An improved Tn7 system for the single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene. 1991;109:167–168. doi: 10.1016/0378-1119(91)90604-a. [DOI] [PubMed] [Google Scholar]
- 4.Barry G F. Permanent insertion of foreign genes into the chromosomes of soil bacteria. Bio/Technology. 1986;4:446–449. [Google Scholar]
- 5.Bazin M J, Markham P, Scott E M, Lynch J M. Population dynamics and rhizosphere interactions. In: Lynch J M, editor. The rhizosphere. Chichester, England: Wiley and Sons; 1990. pp. 99–127. [Google Scholar]
- 6.Becker S, Holighaus G, Gabrielczyk T, Unden G. O2 as the regulatory signal for FNR-dependent gene regulation in Escherichia coli. J Bacteriol. 1996;178:4515–4521. doi: 10.1128/jb.178.15.4515-4521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binnerup S J, Sørensen J. Nitrate and nitrite microgradients in barley rhizosphere as detected by a highly sensitive denitrification bioassay. Appl Environ Microbiol. 1992;58:2375–2380. doi: 10.1128/aem.58.8.2375-2380.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig N L. Transposon Tn7. Curr Top Microbiol Immunol. 1996;204:27–48. doi: 10.1007/978-3-642-79795-8_2. [DOI] [PubMed] [Google Scholar]
- 9.Drahos D J, Hemming B C, McPherson S. Tracking recombinant organisms in the environment: β-galactosidase as a selectable non-antibiotic marker for fluorescent pseudomonads. Bio/Technology. 1986;4:439–443. [Google Scholar]
- 10.Galimand M, Gamper M, Zimmermann A, Haas D. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol. 1991;173:1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamper M, Ganter B, Polito M R, Haas D. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J Mol Biol. 1992;173:4742–4750. doi: 10.1016/0022-2836(92)91044-p. [DOI] [PubMed] [Google Scholar]
- 12.Gamper M, Zimmermann A, Haas D. Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J Bacteriol. 1991;173:4742–4750. doi: 10.1128/jb.173.15.4742-4750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas D, Gamper M, Zimmermann A. Anaerobic control in Pseudomonas aeruginosa. In: Silver S, Galli E, Witholt B, editors. Pseudomonas: molecular biology and biotechnology. Washington, D.C: American Society for Microbiology; 1992. pp. 177–187. [Google Scholar]
- 14.Heijnen C E, Page S, Van Elsas J D. Metabolic activity of Flavobacterium strain P25 during starvation and after introduction to bulk soil and the rhizosphere of wheat. FEMS Microbiol Ecol. 1995;18:129–138. [Google Scholar]
- 15.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertions of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoben H J, Somasegaran P. Comparison of the pour, spread, and drop plate methods for enumeration of Rhizobium spp. in inoculants made from presterilized peat. Appl Environ Microbiol. 1982;44:1246–1247. doi: 10.1128/aem.44.5.1246-1247.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Højberg O, Binnerup S J, Sørensen J. Potential rates of ammonium oxidation, nitrite oxidation, nitrate reduction and denitrification in the young barley rhizosphere. Soil Biol Biochem. 1996;28:47–54. [Google Scholar]
- 18.Højberg O, Revsbech N P, Tiedje J M. Denitrification in soil aggregates analyzed with microsensors for nitrous oxide and oxygen. Soil Sci Soc Am J. 1994;58:1691–1698. [Google Scholar]
- 19.Højberg O, Sørensen J. Microgradients of microbial oxygen consumption in a barley rhizosphere model system. Appl Environ Microbiol. 1993;59:431–437. doi: 10.1128/aem.59.2.431-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan P A, Thomson A J, Ralph E T, Guest J R, Green J. FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett. 1997;416:349–352. doi: 10.1016/s0014-5793(97)01219-2. [DOI] [PubMed] [Google Scholar]
- 21.Keel C, Défago G. Interactions between beneficial soil bacteria and root pathogens: mechanisms and ecological impact. In: Gange A C, Brown V K, editors. Multitrophic interactions in terrestrial systems. Oxford, England: Blackwell Science; 1997. pp. 27–46. [Google Scholar]
- 22.Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley P J. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 24.Kragelund L, Hosbond C, Nybroe O. Distribution of metabolic activity and phosphate starvation response of lux-tagged Pseudomonas fluorescens reporter bacteria in the barley rhizosphere. Appl Environ Microbiol. 1997;63:4920–4928. doi: 10.1128/aem.63.12.4920-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laville J, Blumer C, von Schroetter C, Gaia V, Défago G, Keel C, Haas D. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. J Bacteriol. 1998;180:3187–3196. doi: 10.1128/jb.180.12.3187-3196.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazazzera B A, Beinert H, Khoroshilova N, Kennedy M C, Kiley P J. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 27.Loesche W J. Oxygen sensitivity of various anaerobic bacteria. Appl Microbiol. 1969;18:723–727. doi: 10.1128/am.18.5.723-727.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loper J E, Lindow S E. Reporter gene systems useful in evaluating in situ gene expression by soil- and plant-associated bacteria. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C: ASM Press; 1997. pp. 482–492. [Google Scholar]
- 28a.Lu C-D, Winteler H, Abdelal A, Haas D. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J Bacteriol. 1999;181:2459–2464. doi: 10.1128/jb.181.8.2459-2464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurhofer M, Reimmann C, Schmidli-Sacherer P, Heeb S, Haas D, Défago G. Salicylic acid biosynthesis genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology. 1998;88:678–684. doi: 10.1094/PHYTO.1998.88.7.678. [DOI] [PubMed] [Google Scholar]
- 30.Mercenier A, Simon J-P, Vander Wauven C, Haas D, Stalon V. Regulation of enzyme synthesis in the arginine deiminase pathway of Pseudomonas aeruginosa. J Bacteriol. 1980;144:159–163. doi: 10.1128/jb.144.1.159-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minton N. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene. 1984;31:269–273. doi: 10.1016/0378-1119(84)90220-8. [DOI] [PubMed] [Google Scholar]
- 32.Norton J M, Firestone M K. Metabolic status of bacteria and fungi in the rhizosphere of ponderosa pine seedlings. Appl Environ Microbiol. 1991;57:1161–1167. doi: 10.1128/aem.57.4.1161-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ornston L N, Stanier R Y. The conversion of catechol and protocatechuate to β-ketoadipate by Pseudomonas putida. J Biol Chem. 1966;241:3776–3786. [PubMed] [Google Scholar]
- 34.Petersen S O, Nielsen T H, Henriksen K. Direct measurements of oxygen microprofiles and distribution of phospholipid-P in a two-phase soil-manure system. Geoderma. 1991;56:549–559. [Google Scholar]
- 35.Poulsen L K, Dalton H M, Angles M L, Marshall K C, Molin S, Goodman A E. Simultaneous determination of gene expression and bacterial identity in single cells in defined mixtures of pure cultures. Appl Environ Microbiol. 1997;63:3698–3702. doi: 10.1128/aem.63.9.3698-3702.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prosser J I. Molecular marker systems for detection of genetically engineered micro-organisms in the environment. Microbiology. 1994;140:5–17. doi: 10.1099/13500872-140-1-5. [DOI] [PubMed] [Google Scholar]
- 37.Revsbech N P, Jørgensen B B. Microelectrodes: their use in microbial ecology. Adv Microbiol Ecol. 1986;9:293–352. [Google Scholar]
- 38.Rhodius V A, Busby S J W. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. doi: 10.1016/s1369-5274(98)80005-2. [DOI] [PubMed] [Google Scholar]
- 39.Rouault T A, Klausner R D. Iron-sulfur clusters as biosensors of oxidants and iron. Trends Biol Sci. 1996;21:174–177. [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Sawers R G. Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol Microbiol. 1991;5:1469–1481. doi: 10.1111/j.1365-2958.1991.tb00793.x. [DOI] [PubMed] [Google Scholar]
- 42.Sexstone A J, Revsbech N P, Parkin T B, Tiedje J M. Direct measurement of oxygen profiles and denitrification rates in soil aggregates. Soil Sci Soc Am J. 1985;49:645–651. [Google Scholar]
- 43.Spiro S. The FNR family of transcriptional regulators. Antonie Leeuwenhoek. 1994;66:23–36. doi: 10.1007/BF00871630. [DOI] [PubMed] [Google Scholar]
- 44.Staley T E, Lawrence E G, Drahos D J. Variable specificity of Tn7::lacZY insertion into the chromosome of root-colonizing Pseudomonas putida strains. Mol Ecol. 1997;6:85–87. [Google Scholar]
- 45.Takai K, Hayaishi O. Purification and properties of tryptophan side chain oxidase types I and II from Pseudomonas. Methods Enzymol. 1987;142:195–217. doi: 10.1016/s0076-6879(87)42029-6. [DOI] [PubMed] [Google Scholar]
- 46.Torbert H A, Wood C W. Effects of soil compaction and water-filled pore space on soil microbial activity and N losses. Commun Soil Sci Plant Anal. 1992;23:1321–1331. [Google Scholar]
- 47.Unden G. Transcriptional regulation and energetics of alternative respiratory pathways in facultatively anaerobic bacteria. Biochim Biophys Acta. 1998;1365:220–224. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 48.Unden G, Becker S, Bongaerts J, Holighaus G, Schirawski J, Six S. O2-sensing and O2-dependent gene regulation in facultatively anaerobic bacteria. Arch Microbiol. 1995;164:81–90. [PubMed] [Google Scholar]
- 49.Unden G, Schirawski J. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signals and reactions. Mol Microbiol. 1997;25:205–210. doi: 10.1046/j.1365-2958.1997.4731841.x. [DOI] [PubMed] [Google Scholar]
- 50.Vande Broek A, Michiels J, Van Gool A, Vanderleyden J. Spatial-temporal colonization patterns of Azospirillum brasilense on the wheat root surface and expression of bacterial nifH gene during association. Mol Plant-Microbe Interact. 1993;6:592–600. [Google Scholar]
- 51.Van Overbeek L S, van Elsas J D, van Veen J A. Pseudomonas fluorescens Tn5-B20 mutant RA92 responds to carbon limitation in soil. FEMS Microbiol Ecol. 1997;24:57–71. [Google Scholar]
- 52.Voisard C, Bull C T, Keel C, Laville J, Maurhofer M, Schnider U, Défago G, Haas D. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches. In: O’Gara F, Dowling D N, Boesten B, editors. Molecular ecology of rhizosphere microorganisms. Weinheim, Germany: VCH Publishers; 1994. pp. 67–89. [Google Scholar]
- 53.Williams S M, Savery N J, Busby S J W, Wing H J. Transcription activation at class I FNR-dependent promoters: identification of the activating surface of FNR and the corresponding contact site in the C-terminal domain of the RNA polymerase alfa subunit. Nucleic Acids Res. 1997;25:4028–4034. doi: 10.1093/nar/25.20.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winteler H V, Haas D. The homologous regulators ANR of Pseudomonas aeruginosa and FNR of Escherichia coli have overlapping but distinct specificities for anaerobically inducible promoters. Microbiology. 1996;142:685–693. doi: 10.1099/13500872-142-3-685. [DOI] [PubMed] [Google Scholar]
- 55.Ye R W, Haas D, Ka J-O, Krishnapillai V, Zimmermann A, Baird C, Tiedje J M. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J Bacteriol. 1995;177:3606–3609. doi: 10.1128/jb.177.12.3606-3609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimmermann A, Reimmann C, Galimand M, Haas D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol Microbiol. 1991;5:1483–1490. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]