Significance
Nearly all cells in the body have an internal biological circadian clock that is synchronized by external cues. This machinery forms a transcription–translation feedback loop that anticipates and adapts organismal physiology throughout the 24-h day–night cycle. We tested the hypothesis that timing of energetic stressors that affect glucose and energy homeostasis, namely exercise and feeding status, can differentially influence metabolism in adipose tissue. We found that acute exercise elicits a timing-specific effect on adipose tissue, which is independent of feeding status. Adipose tissue sensitivity to exercise is timing-dependent and modulated in a cell-autonomous fashion, as evidenced by enhanced transcription of metabolic genes in early active phase. Thus, exercise timing may fine-tune adipose metabolism to improve energy homeostasis in cardiometabolic disease.
Keywords: circadian rhythm, adipose tissue, metabolism, exercise, lipolysis
Abstract
The circadian clock is a cell-autonomous transcription–translation feedback mechanism that anticipates and adapts physiology and behavior to different phases of the day. A variety of factors including hormones, temperature, food-intake, and exercise can act on tissue-specific peripheral clocks to alter the expression of genes that influence metabolism, all in a time-of-day dependent manner. The aim of this study was to elucidate the effects of exercise timing on adipose tissue metabolism. We performed RNA sequencing on inguinal adipose tissue of mice immediately following maximal exercise or sham treatment at the early rest or early active phase. Only during the early active phase did exercise elicit an immediate increase in serum nonesterified fatty acids. Furthermore, early active phase exercise increased expression of markers of thermogenesis and mitochondrial proliferation in inguinal adipose tissue. In vitro, synchronized 3T3-L1 adipocytes showed a timing-dependent difference in Adrb2 expression, as well as a greater lipolytic activity. Thus, the response of adipose tissue to exercise is time-of-day sensitive and may be partly driven by the circadian clock. To determine the influence of feeding state on the time-of-day response to exercise, we replicated the experiment in 10-h-fasted early rest phase mice to mimic the early active phase metabolic status. A 10-h fast led to a similar lipolytic response as observed after active phase exercise but did not replicate the transcriptomic response, suggesting that the observed changes in gene expression are not driven by feeding status. In conclusion, acute exercise elicits timing-specific effects on adipose tissue to maintain metabolic homeostasis.
Sedentary lifestyle and obesity are prominent risk factors for type 2 diabetes, while exercise and caloric restriction are effective risk-reducing therapeutics (1). Exercise training enhances insulin sensitivity and glucose metabolism in skeletal muscle, even in individuals with obesity or type 2 diabetes (2). The effects of exercise on metabolism are not limited to skeletal muscle, with widespread alterations in metabolomic profiles of multiple organs in response to acute exercise at different times of the day (3). Long-term exercise is associated with reduced body fat, indicating that repeated elevations in energy expenditure associated with each bout of training directly influences adipocyte metabolism (4). In rodents, exercise training remodels subcutaneous fat tissue by reducing cell size and increasing insulin-stimulated glucose uptake and metabolism in isolated adipocytes (5). Exercise training also increases adipocyte mitochondrial activity and reduces tissue inflammatory markers that interfere with insulin sensitivity (6–8). Exercise-mediated adaptations in adipocytes are also tissue-autonomous, as isoproterenol-stimulated lipolysis is greater in trained versus sedentary mice (9). Thus, habitual exercise not only affects working skeletal muscle, but also adipose tissue.
The sleep-wake cycle profoundly influences organismal physiology and behavior (10). This circadian cycle relies on intrinsic molecular timekeepers, and an interaction between central and peripheral clocks with external Zeitgebers (i.e., timekeepers) such as light and feeding (11). Within the cell, this machinery operates in a transcription–translation feedback loop, consisting of the positive elements CLOCK (clock circadian regulator) and BMAL (basic helix-loop-helix ARNT like 1), which heterodimerize to initiate transcription of negative regulators CRY (cryptochrome circadian regulator 1) and PER (period), which in turn are translated to repress CLOCK-BMAL activity (12). The molecular circadian clock rhythmicity is influenced by Zeitgebers, which can serve to ensure or disrupt homeostasis. For example, shift work in humans or altered light exposure in mouse models disrupts circadian rhythms and leads to weight gain and impaired glucose tolerance (13, 14). Conversely, metabolism can be realigned by time-restricted feeding paradigms, which protect against obesity and metabolic impairments (15–17). Thus, timed exercise or dietary interventions may fine-tune metabolism to influence energy homeostasis.
Body weight is affected by circadian misalignment, suggesting a time-of-day-specific response of energy homeostasis (18). In human adipocytes, genes implicated in fatty acid metabolism show transcriptomic rhythmicity, indicating lipid homeostasis is under circadian control (19). Several lines of evidence suggest that metabolic genes are directly under control of the clock machinery (20). For example, clock mutant mouse models lack diurnal oscillations in lipolytic gene expression and fatty acid metabolism, and develop obesity, highlighting the importance of circadian control of adipose function (21, 22). Thus, the response of adipose tissue to changes in systemic energy demands may be influenced by time of day. Exercise timing specifies phase differences in inter- and intratissue metabolism with tissue-specific metabolic responses (3). Given the importance of adipose tissue in chronic disease and the circadian dynamics of metabolic stimuli and function, we hypothesize that time of day influences the metabolic response to exercise in adipose tissue.
Results
Exercise Elicits Time-Dependent Effects on Lipid Metabolism without Disturbing the Adipose Molecular Clock.
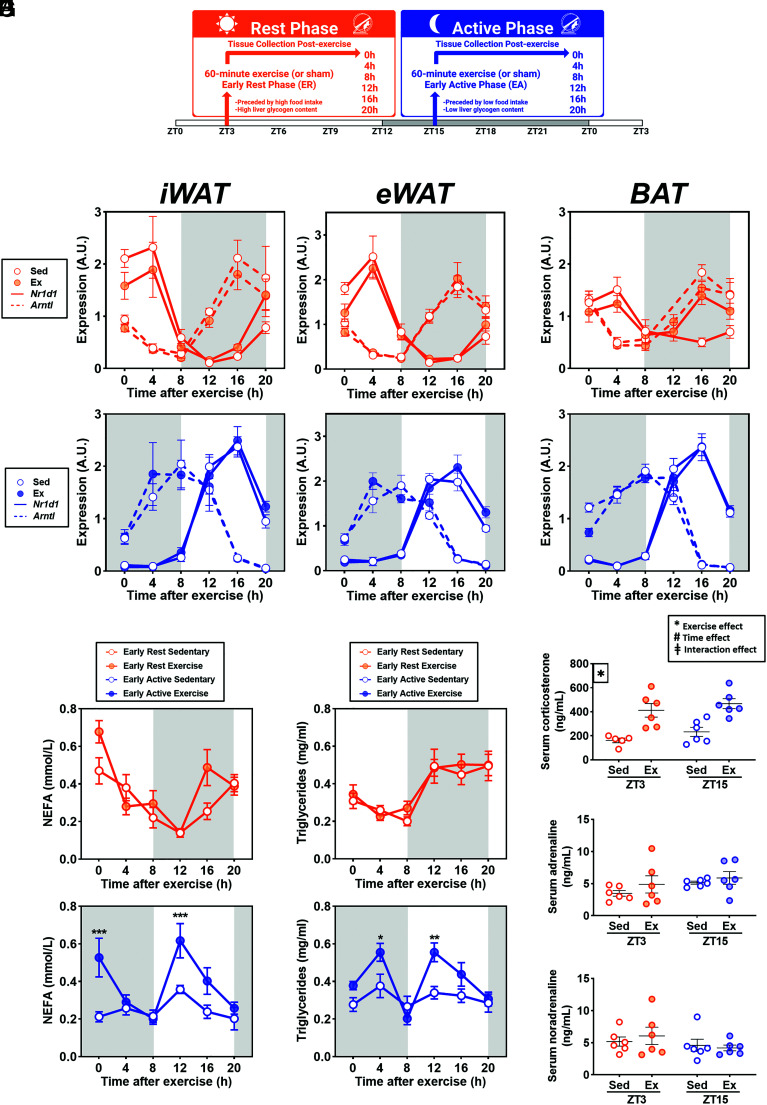
To assess the effect of acute exercise at the onset of the rest and active phases, mice were subjected to an acute exercise bout with adipose tissue collected every 4 h over 20-h period [Fig. 1A; (23)]. First, we determined the effects of time and exercise on adipose tissue peripheral clocks by measuring the expression of core clock genes Arntl and antiphase counterpart Nr1d1 in inguinal (iWAT) and epidydimal (eWAT) white adipose tissues, as well as in interscapular brown adipose tissue (BAT). Clock gene transcripts in inguinal and epididymal white adipose tissue displayed rhythmic expression in all adipose tissue depots from 0 to 20 h postexercise, with amplitudes and periods remaining unperturbed by exercise at either phase [Fig. 1 B and C; RAIN, P < 0.05]. Rhythmicity of Arntl and Nr1d1 was not significant in BAT of either sedentary or exercised mice (Fig. 1D). However, in the BAT, expression of Nr1d1 was increased 16 h postexercise in early rest phase mice (P = 0.0008; Fig. 1D). This indicates a BAT-specific effect of exercise in rest phase mice only, which may alter the clock-metabolic axis in the subsequent behavioral phase. Thus, molecular clock rhythmicity was not disrupted by exercise.
Fig. 1.
Response to exercise or sham intervention in mice at the early rest or early active phase. (A) Experimental schematic. Mice were exposed to sham- or maximal-exercise intervention for 60 min at ZT3 (Orange, early rest) or ZT15 (Blue, early active), and samples were collected every 4 h up to 20 h postintervention (n = 6 per group). (B–D) Expression of core clock genes Arntl and Nr1d1 following exercise intervention at early active or early rest phase in iWAT (B), eWAT (C), BAT (D). Serum NEFA (E) and TG (F) following exercise at ZT3 or ZT15. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (G–I) Serum corticosterone (G), adrenaline (H), and noradrenaline (I) from immediate postexercise or sham intervention at the early rest (Orange) and early active phase (Blue). *Exercise effect P < 0.05. #Time effect P < 0.05. ‡Interaction effect P < 0.05.
We measured serum nonesterified fatty acids (NEFA) as a read-out of lipolysis (24) at all postexercise time points. Serum NEFA displayed rhythmic variation from 0 to 20 h postintervention, peaking at the onset of the rest phase and declining at the beginning of the active phase (Fig. 1E). Serum NEFA levels were increased immediately after exercise during the early active phase (Fig. 1E; P < 0.05), a time at which serum NEFA was at a nadir in sedentary mice, and again 12 h later; this effect of exercise was absent in the early rest phase mice. The expression level of lipolytic enzymes such as ATGL (adipocyte triglyceride lipase) and HSL (hormone sensitive lipase) was not altered by exercise (SI Appendix, Fig. S1 A and B). Serum triglycerides (TG) showed an opposite oscillation pattern to NEFA by peaking at the early active phase and declining at the early rest phase (Fig. 1F). Serum levels of TG were altered at 4- and 12 h post exercise at the early active phase (Fig. 1F), suggesting delayed compensatory feeding in response to early active phase exercise, an effect not observed following early rest phase exercise.
To assess the influence of circulating hormones on adipose tissue lipolytic activity, levels of corticosterone, adrenaline, and noradrenaline were measured immediately postexercise. Corticosterone was elevated in exercised mice, with similar responses at the early rest and early active phases (Fig. 1G). Neither adrenaline nor noradrenaline were influenced by exercise or time-of-day (Fig. 1 H and I). Collectively, these data indicate that early active phase exercise acutely increased NEFA release without an increase in canonical lipolytic factors.
Adipose Tissue Transcriptomic Profile Is Altered by Active Phase Exercise.
The effect of exercise timing on iWAT gene expression was assessed by performing RNA sequencing. The transcriptomic profile showed timing-divergent expression of transcripts in the sedentary (192) and postexercise (744) state, while 121 transcripts were altered with early active phase exercise and 0 with early rest phase exercise (Fig. 2A and SI Appendix, Fig. S1C; False Discovery Rate (FDR) < 0.05). We identified 113 genes that showed a time-of-day effect, regardless of exercise status (visualized by blue dots, Fig. 2B). Conversely, 75 genes were altered by the time of day in sedentary mice specifically (visualized by green dots, Fig. 2B), while 592 genes were specifically altered within the exercise condition between early active and early rest phases (visualized by red dots, Fig. 2B). A total of 479 genes showed a time-of-day effect in exercised, but not sedentary, mice while the time-of-day effect in sedentary mice was modest (75 genes), suggesting that there is cumulative time-specific and exercise effect. In response to early active phase exercise, 121 transcripts were altered compared with sedentary mice, most of which were up-regulated (visualized by pink dots, Fig. 2C); conversely, no transcripts were recognized as differentially expressed between exercised and sedentary mice at the early rest phase, suggesting a timing-sensitive transcriptomic effect of exercise in the iWAT (Fig. 2 A–C).
Fig. 2.
RNA sequencing results from iWAT collected immediately postexercise or sham intervention during the early rest or early active phase. (A) Upset plot of altered transcripts at 0 h post exercise at the early rest and early active phases. (B) Differential expression of logFC between early active and early rest phase sedentary (y; green dots) and exercised (x; red dots) groups. Common transcripts between groups are shown as blue dots. (C) Differential expression of logFC between early rest and early active (x; pink dots) phase exercise effects. (D) Gene ontology of transcripts differentially altered within the early active exercise group (versus Sed). (E) Target gene enrichment analysis comparing early rest phase exercise, early active phase exercise, time of day effect exercise, and time of day effect sedentary with public RNAseq data sets from dexamethasone or iso-stimulated 3T3 mouse adipocytes and (CL316) adrenergic-agonist-treated mice. (F) Heatmap of stress-responsive and thermogenesis-associated transcripts from iWAT (FDR < 0.05).
Gene ontology analysis of altered transcripts from early active phase exercise showed an enrichment of pathways involved in DNA-binding transcription activator activity, fibronectin binding, steroid hormone receptor activity, nuclear receptor activity, ligand-activated transcription factor activity, and glucocorticoid receptor binding (Fig. 2D). Due to the association of transcripts from early active phase exercise with hormone signaling-associated processes, a target gene enrichment analysis was performed using publicly available data sets of 3T3-L1 mouse adipocytes exposed to β-adrenergic (isoproterenol) or glucocorticoid-receptor (dexamethasone) agonists, as well as white adipose tissue from mice exposed to exogenous β3-adrenergic stimuli (CL316,243). The postexercise iWAT transcriptome of early active phase mice showed a similar transcriptional response to dexamethasone- and isoproterenol-treated 3T3-L1 cells, whereas this effect was not observed with early rest phase exercise (Fig. 2E). To further characterize the early active phase exercise response, the regulation of genes showing previous connections to stress- and adrenergic-responsive processes was investigated. Accordingly, genes responsive to stress or glucocorticoid stimuli (such as Nr4a1, Nr4a3, Vegfα, and Adrb2) (25–27), as well as genes associated with adipose tissue browning and thermogenesis (such as Cpeb2, Cyp2b10, Pdk4, Slc25a25, Cpeb2, Vegfα, Pgc1α, Ucp1, Dio2, Gadd45g, Irf4, Zfp516, and Crem) (28–37), were uniquely up-regulated by exercise at the early active phase (Fig. 2F). Thus, there is a unique, timing-specific transcriptomic response to exercise in iWAT, with early active phase exercise conferring a greater stimulative potential on metabolically relevant genes.
Gene Expression and Lipolytic Response to Isoproterenol Are Clock Associated in 3T3-L1 Adipocytes.
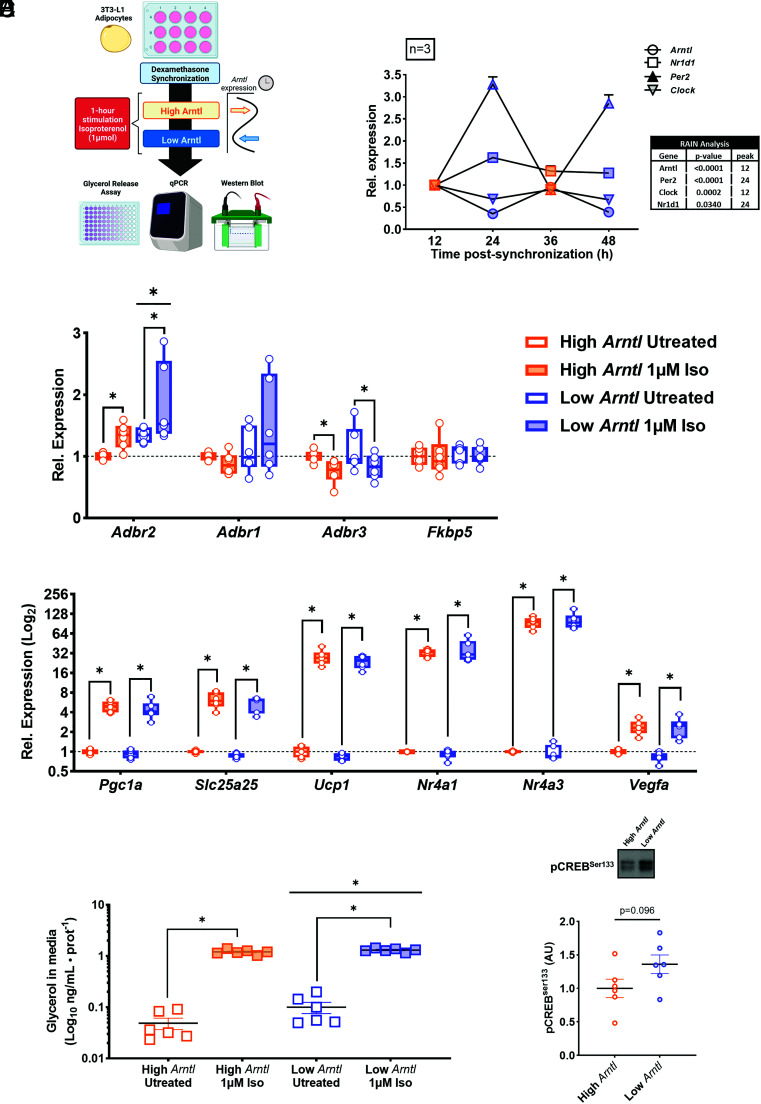
To address the influence of cell-autonomous circadian clock oscillation on adipocyte metabolism and function, 3T3-L1 adipocytes were synchronized and the metabolic profile was studied at 12 h, 24 h, 36 h, and 48 h (Fig. 3 A and B). Expression of core clock genes Arntl, Per2, Clock, and Nr1d1 displayed rhythmic oscillation, with Per2 and Nr1d1 peaking at 24 h, while Arntl and Clock showed an opposing pattern (RAIN analysis, P < 0.05, Fig. 3B).
Fig. 3.
Metabolic response in synchronized 3T3-L1 mouse adipocytes in response to time-point-specific exposure to 1 µM isoproterenol (n = 3). (A) Experimental schematic of synchronization (dexamethasone 200 nM) and subsequent isoproterenol (1 µM) treatment. (B) Relative mRNA of clock genes over 48 h with 4 selected time points previously selected to indicate greatest difference in Arntl expression. (C) Relative mRNA level of genes encoding for adrenergic receptor subunits and dexamethasone-responsive marker FK506-binding protein 5 (Fkbp5) in untreated and 1 µM isoproterenol-treated adipocytes. (D) Relative mRNA of previously identified exercise-associated genes in high-Arntl (orange dots) and low-Arntl (blue dots) states. *P < 0.05. (E) Glycerol release assay from untreated and 1 µM isoproterenol-treated 3T3-L1 adipocytes at high-Arntl and low-Arntl states.*P < 0.05. (F) Western blot showing relative abundance of pCREBSer133 in untreated synchronized 3T3-L1 adipocytes in high-Arntl (orange dots) and low-Arntl (blue dots) states.
To replicate the clock state observed in vivo, we selected two time points separated by 12 h, coinciding with time differences between phases in mice that displayed a peak in Arntl expression (corresponding to early rest phase) and nadir in Arntl expression (corresponding to early active phase). Expression of the adrenergic receptor subunits β1, β2, and β3 may affect the response to adrenergic stimulus in vitro. Expression of Adrb2 was increased at the Low-Arntl state, as well as in response to isoproterenol treatment (Fig. 3C; Two-way ANOVA, P < 0.05). Expression of Adrb1 and Adrb3 were unaffected by Arntl status; however, Adrb3 expression was decreased in response to isoproterenol treatment (Fig. 3C; Treatment effect, P < 0.05). The level of FK506-binding protein 5 (Fkbp5) mRNA expression, a marker of dexamethasone exposure (38), was unaffected by time point, indicating that observed effects relate to clock function rather than proximity to dexamethasone exposure (Fig. 3C). Expression of genes displaying a timing-selective exercise response (Fig. 2F), including Pgc1α, Slc25a25, Ucp1, Nr4a1, Nr4a3, and Vegfα, were up-regulated in response to isoproterenol, yet no interaction effect with Arntl status was observed (Fig. 3D).
To assess whether adrenergic-mediated lipolytic activity is associated with clock function, basal and stimulated lipolysis were measured between high and low Arntl states. Basal glycerol release was greater in the Low-Arntl state (Two-way ANOVA, P = 0.034; Fig. 3E). Isoproterenol treatment increased glycerol release (P < 0.0001) and showed an interaction effect between clock state and treatment (P = 0.045), suggesting that the stimulative potential of lipolysis depends on clock status (Fig. 3E). Gene expression and glycerol release were assessed for rhythmicity over all four time points using R package RAIN (SI Appendix, Fig. S2 A–F); rhythmicity was identified in the expression of several genes including Adrb2 (untreated and treated), Vegfα (untreated), Nr4a3 (untreated), and glycerol release (untreated) (RAIN: P < 0.05; SI Appendix, Fig. S2 A–C). Because untreated cells showed clock-associated differences in lipolysis, potential signaling mechanisms were evaluated. Relative abundance of pCREBSer133 (phospho-cAMP response element-binding protein) signaling showed a trending increase (Unpaired t test; P = 0.096) at the low-Arntl state (Fig. 3F), suggesting a clock-controlled intracellular energy state. Levels of pAMPKThr172 and pACCSer79 were not significantly affected by Arntl status (SI Appendix, Fig. S2 E and F). Thus, differentiated 3T3-L1 adipocytes show timing-associated differences in adrenergic receptor expression and lipolytic function, which may be influenced by clock control of the intracellular energy state.
Adipose Tissue Response to Exercise Is Partially Independent of Nutritional Status.
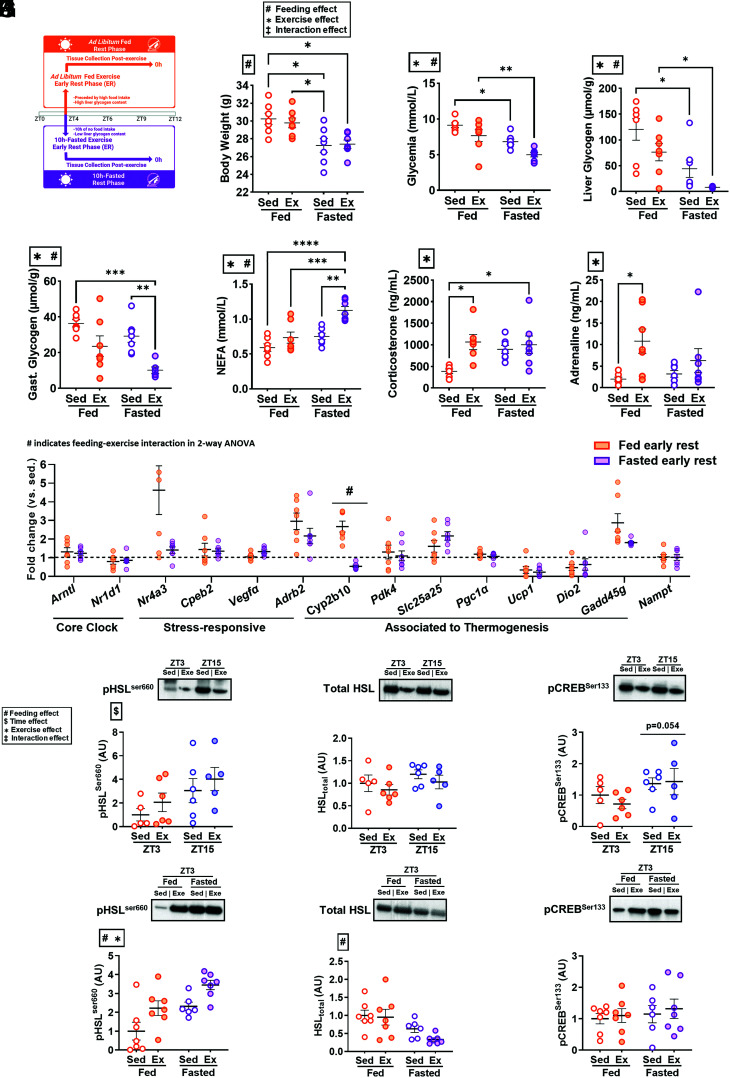
Food intake, liver glycogen, and serum lipids regulate exercise metabolism and display circadian regulation with differences between activity phases (3, 23). Because serum NEFA was increased only following exercise at the early active phase (Fig. 1E), we speculated that substrate utilization and/or feeding state may account for the distinct postexercise transcriptome in the iWAT of early active phase mice. To decipher the influence of substrate availability on the time-of-day response to exercise, the substrate availability profile of early active phase mice was replicated during the early rest phase by removing food 10 h prior to the exercise bout (fasted early rest) (Fig. 4A).
Fig. 4.
Response to exercise or sham intervention in mice ad libitum-fed or 10-h fasted prior to the early rest phase. (A) Experimental schematic. Mice were exposed to sham- or maximal-exercise intervention for 60 min at ZT3 in the ad libitum-fed and 10-h-fasted state. Samples were collected immediately following the exercise bout (n = 28). (B–H) Body weight (B), glycemia (C), liver glycogen (D), gastrocnemius glycogen (E), NEFA (F), corticosterone (G), and adrenaline (H) measurements in ad libitum-fed (orange dots) and 10-h fasted (purple dots) early rest phase mice. Main effect: #Feeding effect P < 0.05; *Exercise effect P < 0.05; ‡Interaction effect P < 0.05. Individual comparisons: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (I) mRNA fold change in expression (relative to sedentary) of clock and previously selected genes identified via transcriptomics in iWAT. Fold change was calculated as relative expression to sedentary state within feeding state. Differential fold change was represented as the interaction between fasting and exercise main effects in a two-way ANOVA of relative mRNA within each gene. #P < 0.05. Comparison in pHSLser660(J), HSLtotal (K), pCREBser133(L) between early rest and early active phase-exercised mice, as well as between fed and fasted early rest phase mice. #Feeding effect P < 0.05; $Time effect P < 0.05; *Exercise effect P < 0.05; ‡Interaction effect P < 0.05.
Liver glycogen is expected to be low at the onset of the active phase (23). After a 10-h fast, body weight and glycemia, as well as hepatic and muscular glycogen content was lower, with further reductions in glycemia, hepatic, and muscular glycogen after exercise (Fig. 4 B–E). Furthermore, serum NEFA was increased by exercise in the fasted early rest phase, suggesting exercise-induced lipolysis is selective to the fasted state (Fig. 4F) and replicating the metabolic state observed following early active phase exercise (Fig. 1E). Serum TG and insulin were unaffected by fasting or exercise (SI Appendix, Fig. S1 D and E). Levels of corticosterone and adrenaline were increased due to exercise, but not fasting (Fig. 4 G and H).
The expression of core clock genes, as well as exercise-responsive genes identified in the previous RNA sequencing analysis (Fig. 2F), was assessed in iWAT of fed and fasted mice during the early rest phase. Core clock genes Arntl and Nr1d1 were not altered by feeding status or exercise in the fed or fasted state during the early rest phase (Fig. 4I), consistent with the earlier experiment (Fig. 1 B–D). The fold change expression of Cpeb2, Vegfα, Pdk4, Slc25a25, Pgc1α, Ucp1, Dio2, Nampt, Nr4a3, Adrb2, and Gadd45g in response to exercise was not different between fed or fasted mice during the early rest phase (Fig. 4I), suggesting that the upregulation of these genes following early active phase exercise occurs independently of feeding state. Conversely, the fold change of Cyp2b10, a brown adipose-associated mitochondrial gene (39), was lower in fasted early rest phase versus fed early rest phase mice (Fig. 4I). These data suggest that the differential time-of-day transcriptomic response to exercise in iWAT is not governed by feeding status.
Because exercise during the active phase or the fasted rest phase elicits a lipolytic response, we next characterized the canonical signaling events involved in the regulation of adipose tissue lipolysis. Phosphorylation of HSLser660, a central lipolytic enzyme (40), was affected by both feeding status and time of day (Fig. 4J). Total HSL protein was diminished in fasted mice during the early rest phase (versus fed mice studied at the early rest phase) but not in mice studied in the early active versus early rest phase (Fig. 4K). These results highlight that adipose tissue enzymatic status at the early active phase is distinct from the fasted state. Nonspecific downstream PKA (protein kinase A) signaling showed no influence of time of day, feeding, or exercise (SI Appendix, Fig. S1F); however, phosphorylation of CREBSer133, which is induced by the adrenergic-PKA signaling axis, showed a trend for increased phosphorylation (P = 0.054) in the early active phase (versus early rest), which was absent in fasted early rest phase mice (Fig. 4L). These results suggest a distinct metabolic state of the iWAT between early active and early rest phase that is partially independent of feeding state.
Discussion
The molecular clock operates via a transcription–translation feedback loop that responds to external Zeitgebers including exercise and diet to fine-tune metabolism throughout the day. Thus, timing of these energetic stressors over the day with the molecular circadian clock may maximize the health promoting benefits to improve metabolism. Recently, active phase food consumption has been shown to increase energy expenditure via clock-controlled energy dissipation in adipose tissue (17). In this context, further evaluation of the phase-specific effects of energetic stressors, on adipose tissue metabolism is warranted. Here, we found that the oscillation of adipose clock gene expression was unaffected by exercise, consistent with previous reports in skeletal muscle (23). Conversely, exercise timing differentially regulated the transcriptomic and metabolic response in adipose tissue. Early active phase exercise increased serum NEFA, which did not occur in response to early rest phase exercise. The elevated serum NEFA level indicates an increase in subcutaneous adipose tissue lipolysis (41, 42), suggesting that the metabolic response to exercise is time-of-day dependent, and that during the early rest phase, mice may lack metabolic flexibility in exercise-responsiveness. Of note, an effect of intervention was observed in early active phase mice, wherein the sedentary group (but not the exercised group) show diminished variance in light-dark oscillations of NEFA and TG levels (Fig. 1 E and F). These data suggest a timing-specific sensitivity to behavioral disruption (sham intervention) during the early active phase, which may elicit metabolic effects over time; however, the metabolic stress of exercise appears to supersede the acute stress of handling.
The molecular clock modulates the expression of lipolytic enzymes (such as Hsl and Atgl) and diurnal rhythms in serum fatty acids (21). Phosphorylation of hormone-sensitive lipase was greater in iWAT of sedentary mice during the early active phase, further indicating that regulation of adipose tissue lipolysis is timing-sensitive. Further, the increase in NEFA observed 12 h following exercise only at the early active phase indicates a sustained effect of acute exercise on adipose tissue lipolysis that is timing-specific. Thus, timing influences adipose tissue metabolism, with heightened and sustained effects observed when exercise is performed during the early active phase.
Subcutaneous adipose tissue is the primary source of circulating NEFAs (41, 42), thus we prioritized this tissue for further transcriptomic analysis. We determined a time-specific response to exercise, with a distinct early active phase transcriptomic response characterized by activation of pathways such as DNA-binding transcription activator activity, fibronectin binding, steroid hormone receptor activity, and glucocorticoid receptor binding. Specifically, the steroid hormone receptor and glucocorticoid receptor binding annotations were driven by the upregulation of nuclear receptor subfamily A transcripts, which have been empirically characterized as exercise-responsive in skeletal muscle (43). The Nr4a subfamily conducts signaling from fat-soluble hormones, which include exercise-responsive steroids such as corticosterone (44). In contrast to previous reports (45), the Nr4a subfamily was not rhythmic, however an exercise-mediated increase in Nr4a1 and Nr4a3 was noted only after early active phase exercise. To assess whether the timing-specific exercise effects can be attributed to systemic factors, we measured serum glucocorticoids and catecholamines. Additionally, our comparative transcriptomic analysis with publicly available data sets from dexamethasone- or isoproterenol-stimulated 3T3-L1 adipocytes, as well as from mice treated with a β3-adrenergic agonist revealed that early active phase exercise induced a similar signature to dexamethasone- or isoproterenol-treated adipocytes. Corticosterone, the primary glucocorticoid in rodents (46), was increased in serum by exercise, irrespective of time, consistent with our previous report of iWAT corticosterone levels (3). The persistence of a timing-specific transcriptomic response, despite similar levels of circulating factors including corticosterone and catecholamines, suggests that intrinsic tissue-specific factors may regulate gene expression. Of note, we cannot exclude that local modulation of sympathetic activation could contribute to the time-of-day-specific adipose tissue response to exercise (47). Additionally, as adipose tissue is a highly heterogenous organ (48), cell-specific-selectivity for transcriptomic analyses may be relevant to consider. For instance, macrophages show circadian rhythmicity in cytokine expression (49); thus, paracrine effects of cell populations within heterogenous tissues such as adipose may become relevant when considering time-of-day effects. Nevertheless, the fold-induction of transcripts related to steroid and glucocorticoid receptor activity due to exercise at the early active phase suggests a heightened sensitivity to these stimuli, which is perhaps driven by rhythmic factors such as the Nr4a subfamily. Conversely, no genes were significantly altered by exercise at the early rest phase, concomitant with a blunted NEFA release following exercise at this time point. Thus, the early rest phase is characterized by a lack of transcriptomic and lipolytic plasticity in response to exercise, whereas the early active phase shows a heightened exercise responsiveness in each of these domains.
Adrenergic signaling, as well as glucocorticoid signaling, regulate the expression of genes associated with browning and thermogenic activity in adipose tissue (50, 51). Accordingly, beiging-associated markers (39, 52) were prominent among the 121 genes up-regulated following early active phase exercise. The phenotypic shift of white adipose toward a brown-adipose-like phenotype (beiging) is associated with increased energy expenditure and metabolic flexibility of adipose tissue (53). Each exercise bout induces a series of transcriptional events that ultimately modulate the abundance of proteins that enhance metabolism and beiging of white adipose (54–56). The time-of-day-specific effect of exercise is consistent with evidence linking beiging-associated gene expression to the adipose tissue clock (57). BAT mass and thermoregulatory capacity are increased in whole-body Arntl knockout mice due to an interference with TGF‐β and BMP signaling (57). Moreover, thermogenic genes such as Ucp1, Cidea, and Dio2 are up-regulated in Per2-null mice (52). Alternatively, metabolites of lipolysis (metabokines) increase expression of beiging markers such as Ucp1, Pgc1α, and Cidea (58). As serum NEFA was increased only with early active phase exercise, phase-specific lipolysis may play a role in the beiging-associated transcriptional response during the early active phase. Indeed, we have reported the global metabolomic profile of iWAT from these early active phase mice (3), and found a phase-specific increase of beiging-associated metabokines such as 3-hydroxybutyrate (BHBA) (59) and 13-hydroxyoctadecadienoic acid (13-HODE) and 9-HODE (60). Thus, our data suggest that the molecular clock interacts with exercise cues to amplify the degree of adipose tissue beiging in a time-of-day-specific manner and may play a role in long-term adipose tissue adaptation to exercise.
We further probed the interaction between catecholamines, clock gene expression levels and metabolism in a cell-autonomous manner using synchronized 3T3-L1 adipocytes. Adrenergic receptors β-1 and β-2 (Adrb1 and Adrb2) regulate catecholamine responsiveness in adipose tissue (61). Phase-specific variation in expression of these genes may reveal a link between the circadian clock and adrenergic sensitivity. Indeed, expression of Adrb2 is clock associated in vitro (Fig. 3C). Likewise, glycerol release was higher in the low-Arntl state, with an interaction between Arntl expression and isoproterenol—suggesting a link between clock status and lipolytic sensitivity. Stable time effects have been observed in human adipose tissue, where explant adipocytes retain clock-oriented lipolytic function (62). Similarly, explant adipocytes from rats exhibited diurnal sensitivity to adrenergic stimuli (63). Thus, our data provide evidence to suggest that adrenergic-mediated lipolysis is rhythmic in synchronized mouse adipocytes, which may result from an interaction between the intracellular clock and lipolytic signaling. While we do not replicate the time-of-day change in gene regulation in vitro, the rhythmic expression of adrenergic receptors may provide a molecular explanation for timing-sensitive exercise response in mouse adipose tissue.
Clock proteins shape exercise capacity in a daytime-dependent manner through changes in liver glycogen levels and feeding behavior (64). A synergistic effect exists between nutritional state and the adaptive response to exercise, which may be sensitive to time of day. Feeding behavior of mice during the rest phase is reduced, resulting in a greater depletion of hepatic glycogen content to maintain glucose homeostasis (23). Hepatic glycogen is a primary reservoir for glycemic regulation, and its level can alter the metabolic response to exercise in mice and humans (65–67). Skeletal muscle metabolism during exercise is dependent on time of day, with a higher reliance on lipids in the active phase (23), consistent with an activation of lipolysis in adipose tissue. This switch in substrate utilization during exercise may be driven by feeding status, as exercise in a fasted state leads to higher expression of glucose and fatty acid metabolic genes in adipose tissue (68). We found that fasting and early active phase exercise elicit a similar induction of NEFA, suggesting that the increased lipolysis is related to the feeding status. However, fasting did not recapitulate the transcriptomic response to early active phase exercise. The lack of exercise-mediated transcriptional similarity between fasted early rest and the early active phase exercise signifies that feeding status is secondary to time of day as a regulator of exercise-responsive transcriptional activity in adipose tissue.
While the molecular mechanisms involved remain to be deciphered, we identified that CREB phosphorylation, a regulator of thermogenesis-associated genes (69), trended to be higher in the active phase only. CREB is phosphorylated at Ser133 in response to an increase in intracellular cAMP (cyclic adenosine monophosphate) levels, which have shown to be rhythmic and regulated by the molecular clock (70, 71). Interestingly, pCREB in adipose tissue was unaltered by exercise in this study; nonetheless, a phase-specific difference in pCREB suggests a differential energetic state between the phases. In concordance with greater pCREB, active phase exercise increased expression of cAMP response element modulator (Crem), which is a member of the CREB transcription factor family (72). This timing-specific difference was also observed in vitro, with low-Arntl 3T3-L1 adipocytes showing increased pCREB levels, coinciding with increased lipolytic function. Therefore, a diurnal contrast in cAMP-responsive element activations may play a role in the transcriptomic response to exercise, independent of feeding status.
In conclusion, exercise elicits a time-of-day-specific transcriptomic and metabolic response in mouse adipose tissue, which is most pronounced during the early active phase. This time-of-day-specific effect is partly independent of feeding status and circulating levels of glucocorticoids and catecholamines. Adipose tissue timing-dependent sensitivity to exercise may be modulated in a cell-autonomous fashion, as evidenced by enhanced transcriptomic induction of metabolic genes in early active phase. Additionally, adipocytes show clock-associated adrenergic receptor expression and lipolysis in a cell-autonomous fashion. Collectively, our results provide evidence that time-of-day drives specific adaptations to acute exercise in the adipose tissue of healthy mice. Whether timing of exercise can fine-tune adipose metabolism to improve energy homeostasis in obesity or type 2 diabetes warrants further investigation.
Materials and Methods
Animal Experiments.
Male C57BL6/JBomTac mice (n = 96) were purchased from Taconic Biosciences and housed in the University of Copenhagen animal facility in Denmark. Animal experiments complied with the European directive 2010/63/EU of the European Parliament and were approved by the Danish Animal Experiments Inspectorate (2012-15-2934-26 and 2015-15-0201-796). Housing conditions for mice included 12-h/light/dark cycles with ad libitum access to standard rodent chow (#1310, Altromin, Germany). For the exercise intervention, 10- to 11-wk-old mice were separated into sham- or exercise- treatment at the early rest (ZT3) or early active (ZT15) phase (Fig. 1A). Exercised mice were exposed to a 1-h exercise bout (ZT3 with lights on; ZT15 in the dark with the use of a red-light lamp) while sham-exercise counterparts were placed on an artificial treadmill for 1 h. Following the exercise bout, mice were sacrificed under isoflurane anesthesia and samples of inguinal white adipose tissue, epididymal white adipose tissue, intrascapular BAT, and serum were collected at 0 h, 4 h, 8 h, 12 h, 16 h, and 20 h (n = 6 per group) (23). Prior to the exercise bout, all mice were acclimated to the rodent treadmill using a previously established 4-d acclimation protocol (23). Exercise began at a speed of 6 m min−1 and increased by 2 m min−1 every 2 min until a speed of 16 m min−1 was reached. This experimental protocol was repeated in a subset of mice (n = 24), with samples collected only at 0 h postintervention to obtain additional samples.
To evaluate the influence of fasting on the diurnal exercise transcriptome, we subjected mice (n = 28; C57BL/6NTac; Taconic, Ejby, DK) to 10 h of fasting (fasted early rest) or ad libitum feeding (fed early rest) prior to an exercise bout at ZT3. Fasting was achieved by removing food from the cage 10 h prior to exercise. Following the exercise bout, samples were collected from each mouse as mentioned previously.
Cell Culture.
3T3-L1 fibroblasts were maintained in growth medium (Dulbecco's Modified Eagle Medium (DMEM) 4.5 g/L glucose + 10% Calf Serum). At full confluence, cells were then switched to differentiation media supplemented with insulin (1 mM), Rosiglitazone (1 µM), 3-isobutyl-1-methylxanthine (IBMX; 500 µM), Dexamethasone (500 nM), and T3 (50 nM) for 2 d. On day 2, cells were switched to differentiation media with insulin (1 mM), Rosiglitazone (1 µM), 3-isobutyl-1-methylxanthine (IBMX; 500 µM), and T3 (50 nM) for 2 d. On day 4, cells were switched to differentiation media only. At day 8, cells were synchronized with 200 nM of Dexamethasone for 30 min, washed once with differentiation media, and then maintained in differentiation media (73, 74). The end of the 30-min dexamethasone treatment was considered at Cell-Time (CT) 0. For samples collected at 36- and 48 h postsynchronization, the medium was changed at CT24. Cells were incubated with 1 uM of isoproterenol in differentiation medium for 2 h at 12-, 24-, 36-, and 48 h postsynchronization. Samples of RNA and protein were collected for qPCR and western blot analysis. Additionally, cells were treated with 1 uM isoproterenol in low-glucose DMEM (1 g/L) with 2% bovine serum albumin (BSA) for 1 h for lipolysis measurement. Following treatment, the medium was collected from the wells and assessed for glycerol content using a Free Glycerol Kit (Sigma Aldrich).
Gene Expression Analysis.
RNA was isolated using a trizol-chloroform extraction procedure (Fisher Scientific) (75). Synthesis of cDNA was performed using a High-Capacity cDNA Reverse Transcription Kit (ThermoFisher). qPCR was performed using a ViiA 7 Real-Time PCR System (Applied Biosciences) using both SYBR Green Master Mix (Bio-Rad) and TaqMan Fast Universal PCR Master Mix (ThermoFisher). Quantitative PCR was performed for 40 cycles at 95 °C for 1 s followed by 60 °C for 20 s. Quantstudio Real-time PCR software (ThermoFisher; version 1.3) set to ΔΔCt was used to evaluate Cycle Threshold. All results were normalized to expression of TATA box binding protein (Tbp).
Biochemistry Analysis.
Blood was collected via tail vein immediately following the exercise bout. Blood glucose concentration was measured via a hand-held glucometer (Contour XT, Bayer). Serum non-esterified fatty acids (NEFA) were assessed using the NEFA-HR Assay (FujiFilm). Serum Catecholamines (Adrenaline and Noradrenaline) were assessed using a 2-CAT Research ELISA (LDN). Serum Corticosterone was assessed using a Corticosterone ELISA kit (ab108821) (Abcam). Serum triglycerides were determined using a Serum Triglyceride Determination Kit (Sigma Aldrich). Serum insulin was determined using an Ultra-Sensitive Mouse Insulin ELISA Kit (Crystal Chem). Liver and gastrocnemius glycogen content were assessed using a Glycogen Assay Kit II (Abcam).
Library Preparation and RNA Sequencing Library.
RNA concentration and purity were assessed by absorbance at 260 and 280 nm using NanoDrop One (Thermo Fisher Scientific). RNA was checked for quality using the Agilent RNA 600 nano kit and Bioanalyzer instrument (Agilent Technologies). Aliquots of RNA (1,000 ng) were analyzed using the Illumina TruSeq Stranded Total RNA with Ribo-Zero Gold protocol (Illumina) as described (76). Samples were cleaned and validated for DNA concentration using the Qubit dsDNA HS assay kit (Invitrogen) and for base pair size and purity using the Agilent High Sensitivity DNA chip and Bioanalyzer instrument. The libraries were subjected to 38-bp paired-end sequencing on a NextSeq500 (Illumina).
Bioinformatic Analysis.
Reads were mapped to Ensembl mm10 release 92 using STAR (2.5.3a), and transcripts counted with FeatureCounts (1.6.0). Sequencing depth ranged from 54.2 to 78.6 million with a mean of 66.3 million reads. Transcriptomic data are deposited under accession number GSE199429. Reads were filtered using filterbyExpression, with a minimum count of 10 and design matrix modeling group. Differential gene expression analysis was performed using edgeR, quasi-likelihood negative binomial generalized log-linear model, and the design ~0 + group. Gene ontology (GO) enrichment analysis was performed utilizing clusterProfiler and molecular function GO terms, with all detected genes serving as background. Enrichment signatures of genes affected by isoprenaline (GSE43658) (77), dexamethasone (only 24-h treatments; GSE62635) (78), and CL316243 beta adrenergic stimulation (GSE55934) (79) was performed using fry. All P-values were adjusted using false discovery rate correction, and alpha was set to 0.05. Rhythmic oscillations in gene expression and glycerol release were analyzed with R package RAIN (Rhythmicity Analysis Incorporating Non-parametric Methods, release 3.15) (80), with period set to 24 h, method independent, and period delta set to 12 h. Outcomes with a P < 0.05 were considered as rhythmic.
Protein Isolation and Western Blot Analysis.
Inguinal adipose tissue samples were mechanically homogenized on dry ice using a mortar and pestle. Crushed tissue or 3T3-L1 adipocytes were homogenised in ice-cold homogenization buffer [20 mM Tris, pH 7.8, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 0.5 mM Na3VO4, 1% Triton X-100, 10% glycerol, 10 mM NaF, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM ethylenediaminetetraacetic acid (EDTA), 5 mM Na4P2O7, and 1% (vol/vol) Protease Inhibitor Cocktail (Calbiochem)]. Protein content was quantified using a bicinchoninic acid protein assay kit (Pierce). Protein lysates were diluted into Laemmli buffer, then heated at 56 °C for 20 min. Proteins were resolved with SDS-PAGE (4 to 12% polyacrylamide gels) and transferred to the PVDF (polyvinylidene fluoride) membrane via wet electrotransfer. Following this, membranes were stained with Ponceau S [0.1% (w/v) in 5% (v/v) acetic acid] to evaluate standardization of sample loading and transfer. Membranes were then blocked with 7.5% (w/v) dry milk in Tris-buffered saline with Tween 20 [TBST: 20 mM Tris, 150 mM NaCl, 0.02% (v/v) Tween 20, pH 7.5] for 1 h at room temperature. After blocking, membranes were incubated with a primary antibody in the primary antibody buffer [20 mM Tris, 150 mM NaCl, pH 7.5, 0.1% (w/v) BSA and 0.1% (w/v) sodium azide] overnight at 4 °C and then with the secondary antibody–horseradish peroxidase conjugate in TBST with 5% (w/v) dry milk for 1 h at room temperature. Finally, membranes were incubated with ECL reagent and then immunolabeled proteins were visualized on the X-ray films. Films were scanned with GS-800 Densitometer (Bio-Rad) and analyzed with Quantity One 1-D Analysis Software (Bio-Rad). Intensities of individual bands were expressed in arbitrary units relative to the total intensity of all the bands. The primary antibodies used were anti-pHSLSer660 (cat. no. 4126; Cell Signaling Technology), anti-HSL (cat. no. 4107; Cell Signaling Technology), anti-CREB Ser133 (cat. no. 9198; Cell Signaling Technology), anti-pAMPKαThr172 (cat. no. 2535; Cell Signaling Technology) and anti-pAcetyl-CoA CarboxylaseSer79 (cat. no. 3661; Cell Signaling Technology). Quantified values are normalized to ponceau staining, then normalized to the average of sedentary ZT3 animals.
Statistical Analysis.
Data are presented as Mean ± SEM. All analyses were performed using a two-way ANOVA in GraphPad Prism version 8.0.0 (GraphPad Software). All data for two-way ANOVAs were tested for normality using the Shapiro–Wilk test and tested for equal variance using Levene's test. If data failed these tests, it was log-transformed prior to the performance of the two-way ANOVA.
Ethical Review.
Specific Pathogen Free male C57BL6/JBomTac and C57BL/6NTac mice were purchased from Taconic Biosciences and maintained at the animal facilities of the University of Copenhagen, Denmark. Animal experiments complied with the European directive 2010/63/EU of the European Parliament and were approved by the Danish Animal Experiments Inspectorate (2012-15-2934-26, 2015-15-0201-796, 2017-15-0201-01276, and 2020-15-0201-00450). Mice were maintained on a 12-h light/dark cycle.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This study was supported by the Novo Nordisk Foundation (NNF14OC0011493), Swedish Diabetes Foundation (DIA2021-645), Swedish Research Council for Sport Science (P2022-0013) and the Swedish Research Council (2015-00165). L.A.P. was supported by KID funding from Karolinska Institutet (2019). L.D. was supported by a Novo Nordisk postdoctoral fellowship run in partnership with Karolinska Institutet and by a Novo Nordisk Foundation postdoctoral fellowship for research abroad (NNF20OC0060969), as well as by EFSD/Novo Nordisk and EFSD/Lilly. The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research center at the University of Copenhagen, partially funded by an unrestricted donation from the Novo Nordisk Foundation.
Author contributions
L.A.P., L.S.L., A.M.E., S.P.A., A.L.B., A.K., J.T.T., L.D., and J.R.Z. designed research; L.A.P., L.S.L., A.M.E., S.P.A., M.S., A.L.B., and L.D. performed research; A.K., J.T.T., and J.R.Z. contributed new reagents/analytic tools; L.A.P., L.S.L., A.M.E., S.P.A., M.S., A.L.B., J.T.T., and L.D. analyzed data; and L.A.P., L.S.L., A.K., L.D., and J.R.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Lucile Dollet, Email: lucile.dollet@sund.ku.dk.
Juleen R. Zierath, Email: Juleen.Zierath@ki.se.
Data, Materials, and Software Availability
The RNA-Seq dataset reported in this paper has been deposited in the Gene Expression Omnibus database (ID: GSE199429). All scripts used to analyze data are available on http://www.github.com/leonidaslundell/GSE199429. Previously published data were used for this work and are available in Gene Expression Omnibus with accession numbers: GSE43658 (77); GSE62635 (78); GSE55934 (79).
Supporting Information
References
- 1.Kanaley J. A., et al. , Exercise/physical activity in individuals with type 2 diabetes: A consensus statement from the American college of sports medicine. Med. Sci. Sports Exerc. 54, 353–368 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabriel B. M., Zierath J. R., The limits of exercise physiology: From performance to health. Cell Metab. 25, 1000–1011 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Sato S., et al. , Atlas of exercise metabolism reveals time-dependent signatures of metabolic homeostasis. Cell Metab. 34, 329–345.e328 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Armstrong A., et al. , Effect of aerobic exercise on waist circumference in adults with overweight or obesity: A systematic review and meta-analysis. Obes. Rev. 23 e13446 (2022), 10.1111/obr.13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig B. W., Hammons G. T., Garthwaite S. M., Jarett L., Holloszy J. O., Adaptation of fat cells to exercise: Response of glucose uptake and oxidation to insulin. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 51, 1500–1506 (1981). [DOI] [PubMed] [Google Scholar]
- 6.Mendham A. E., et al. , Exercise training results in depot-specific adaptations to adipose tissue mitochondrial function. Sci. Rep. 10, 3785 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutherland L. N., Bomhof M. R., Capozzi L. C., Basaraba S. A., Wright D. C., Exercise and adrenaline increase PGC-1{alpha} mRNA expression in rat adipose tissue. J. Physiol. 587, 1607–1617 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawanishi N., Yano H., Yokogawa Y., Suzuki K., Exercise training inhibits inflammation in adipose tissue via both suppression of macrophage infiltration and acceleration of phenotypic switching from M1 to M2 macrophages in high-fat-diet-induced obese mice. Exerc. Immunol. Rev. 16, 105–118 (2010). [PubMed] [Google Scholar]
- 9.Higa T. S., Spinola A. V., Fonseca-Alaniz M. H., Evangelista F. S., Remodeling of white adipose tissue metabolism by physical training prevents insulin resistance. Life Sci. 103, 41–48 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Hastings M. H., Maywood E. S., Brancaccio M., Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Heyde I., Oster H., Differentiating external zeitgeber impact on peripheral circadian clock resetting. Sci. Rep. 9, 20114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wefers J., et al. , Circadian misalignment induces fatty acid metabolism gene profiles and compromises insulin sensitivity in human skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 115, 7789–7794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonken L. K., et al. , Light at night increases body mass by shifting the time of food intake. Proc. Natl. Acad. Sci. U.S.A. 107, 18664–18669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaix A., Lin T., Le H. D., Chang M. W., Panda S., Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 29, 303–319.e304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manoogian E. N. C., Chow L. S., Taub P. R., Laferrere B., Panda S., Time-restricted eating for the prevention and management of metabolic diseases. Endocr. Rev. 43, 405–436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hepler C., et al. , Time-restricted feeding mitigates obesity through adipocyte thermogenesis. Science 378, 276–284 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reutrakul S., Van Cauter E., Sleep influences on obesity, insulin resistance, and risk of type 2 diabetes. Metabolism 84, 56–66 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Christou S., et al. , Circadian regulation in human white adipose tissue revealed by transcriptome and metabolic network analysis. Sci. Rep. 9, 2641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox K. H., Takahashi J. S., Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 63, R93–R102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shostak A., Meyer-Kovac J., Oster H., Circadian regulation of lipid mobilization in white adipose tissues. Diabetes 62, 2195–2203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paschos G. K., et al. , Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 18, 1768–1777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato S., et al. , Time of exercise specifies the impact on muscle metabolic pathways and systemic energy homeostasis. Cell Metab. 30, 92–110.e114 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Horowitz J. F., Fatty acid mobilization from adipose tissue during exercise. Trends Endocrinol. Metab. 14, 386–392 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Fu M., et al. , A nuclear receptor atlas: 3T3-L1 adipogenesis. Mol. Endocrinol. 19, 2437–2450 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Xue Y., et al. , Hypoxia-independent angiogenesis in adipose tissues during cold acclimation. Cell Metabol. 9, 99–109 (2009). [DOI] [PubMed] [Google Scholar]
- 27.de Fatima Silva F., et al. , Dexamethasone-induced adipose tissue redistribution and metabolic changes: Is gene expression the main factor? An animal model of chronic hypercortisolism. Biomedicines 10, 2328 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H. F., Hsu C. M., Huang Y. S., CPEB2-dependent translation of long 3’-UTR Ucp1 mRNA promotes thermogenesis in brown adipose tissue. Embo J. 37, e99071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y., Kang B. E., Ryu D., Oh S. W., Oh C. M., Comparative transcriptome profiling of young and old brown adipose tissue thermogenesis. Int. J. Mol. Sci. 22, 13143 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao Q., et al. , Transcriptome profiling of brown adipose tissue during cold exposure reveals extensive regulation of glucose metabolism. Am. J. Physiol. Endocrinol. Metab. 308, E380–E392 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Anunciado-Koza R. P., et al. , Inactivation of the mitochondrial carrier SLC25A25 (ATP-Mg2 + /Pi transporter) reduces physical endurance and metabolic efficiency in mice. J. Biol. Chem. 286, 11659–11671 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puigserver P., et al. , A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92, 829–839 (1998). [DOI] [PubMed] [Google Scholar]
- 33.de Jesus L. A., et al. , The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J. Clin. Invest. 108, 1379–1385 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gantner M. L., Hazen B. C., Conkright J., Kralli A., GADD45γ regulates the thermogenic capacity of brown adipose tissue. Proc. Natl. Acad. Sci. U.S.A. 111, 11870–11875 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong X., et al. , IRF4 is a key thermogenic transcriptional partner of PGC-1α. Cell 158, 69–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dempersmier J., et al. , Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol. Cell 57, 235–246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thonberg H., Lindgren E. M., Nedergaard J., Cannon B., As the proliferation promoter noradrenaline induces expression of ICER (induced cAMP early repressor) in proliferative brown adipocytes, ICER may not be a universal tumour suppressor. Biochem. J. 354, 169–177 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira M. J., et al. , FKBP5 expression in human adipose tissue increases following dexamethasone exposure and is associated with insulin resistance. Metabolism 63, 1198–1208 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi S., et al. , Adipose tissue NAD( + ) biosynthesis is required for regulating adaptive thermogenesis and whole-body energy homeostasis in mice. Proc. Natl. Acad. Sci. U.S.A. 116, 23822–23828 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anthonsen M. W., Rönnstrand L., Wernstedt C., Degerman E., Holm C., Identification of novel phosphorylation sites in hormone-sensitive lipase that are phosphorylated in response to isoproterenol and govern activation properties in vitro. J. Biol. Chem. 273, 215–221 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Nielsen S., Guo Z., Johnson C. M., Hensrud D. D., Jensen M. D., Splanchnic lipolysis in human obesity. J. Clin. Invest. 113, 1582–1588 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo Z., Hensrud D. D., Johnson C. M., Jensen M. D., Regional postprandial fatty acid metabolism in different obesity phenotypes. Diabetes 48, 1586–1592 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Pillon N. J., et al. , Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat. Commun. 11, 470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chawla A., Repa J. J., Evans R. M., Mangelsdorf D. J., Nuclear receptors and lipid physiology: Opening the X-files. Science 294, 1866–1870 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Yang X., et al. , Nuclear receptor expression links the circadian clock to metabolism. Cell 126, 801–810 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Boucher E., Provost P. R., Tremblay Y., Ontogeny of adrenal-like glucocorticoid synthesis pathway and of 20alpha-hydroxysteroid dehydrogenase in the mouse lung. BMC Res. Notes 7, 119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartness T. J., Liu Y., Shrestha Y. B., Ryu V., Neural innervation of white adipose tissue and the control of lipolysis. Front. Neuroendocrinol. 35, 473–493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eto H., et al. , Characterization of structure and cellular components of aspirated and excised adipose tissue. Plast Reconstr. Surg. 124, 1087–1097 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Keller M., et al. , A circadian clock in macrophages controls inflammatory immune responses. Proc. Natl. Acad. Sci. U.S.A. 106, 21407–21412 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luijten I. H. N., Cannon B., Nedergaard J., Glucocorticoids and brown adipose tissue: Do glucocorticoids really inhibit thermogenesis? Mol. Aspects Med. 68, 42–59 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Ramseyer V. D., Granneman J. G., Adrenergic regulation of cellular plasticity in brown, beige/brite and white adipose tissues. Adipocyte 5, 119–129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimaldi B., et al. , PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 12, 509–520 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giralt M., Villarroya F., White, brown, beige/brite: Different adipose cells for different functions? Endocrinology 154, 2992–3000 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Stanford K. I., et al. , A novel role for subcutaneous adipose tissue in exercise-induced improvements in glucose homeostasis. Diabetes 64, 2002–2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stallknecht B., Vinten J., Ploug T., Galbo H., Increased activities of mitochondrial enzymes in white adipose tissue in trained rats. Am. J. Physiol. 261, E410–E414 (1991). [DOI] [PubMed] [Google Scholar]
- 56.Vidal P., Stanford K. I., Exercise-induced adaptations to adipose tissue thermogenesis. Front. Endocrinol. (Lausanne) 11, 270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nam D., et al. , The adipocyte clock controls brown adipogenesis through the TGF-beta and BMP signaling pathways. J. Cell Sci. 128, 1835–1847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehead A., et al. , Brown and beige adipose tissue regulate systemic metabolism through a metabolite interorgan signaling axis. Nat. Commun. 12, 1905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carrière A., et al. , Browning of white adipose cells by intermediate metabolites: An adaptive mechanism to alleviate redox pressure. Diabetes 63, 3253–3265 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Dieckmann S., et al. , Fatty acid metabolite profiling reveals oxylipins as markers of brown but not brite adipose tissue. Front. Endocrinol. (Lausanne) 11, 73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evans B. A., Merlin J., Bengtsson T., Hutchinson D. S., Adrenoceptors in white, brown, and brite adipocytes. Br. J. Pharmacol. 176, 2416–2432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arredondo-Amador M., et al. , Circadian rhythms in hormone-sensitive lipase in human adipose tissue: Relationship to meal timing and fasting duration. J. Clin. Endocrinol. Metab. 105, e4407–e4416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki M., Shimomura Y., Satoh Y., Diurnal changes in lipolytic activity of isolated fat cells and their increased responsiveness to epinephrine and theophylline with meal feeding in rats. J. Nutr. Sci. Vitaminol. (Tokyo) 29, 399–411 (1983). [DOI] [PubMed] [Google Scholar]
- 64.Adamovich Y., et al. , Clock proteins and training modify exercise capacity in a daytime-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 118, e2101115118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jamart C., Naslain D., Gilson H., Francaux M., Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. Am. J. Physiol. Endocrinol. Metab. 305, E964–E974 (2013). [DOI] [PubMed] [Google Scholar]
- 66.De Bock K., et al. , Effect of training in the fasted state on metabolic responses during exercise with carbohydrate intake. J. Appl. Physiol. 1985, 1045–1055 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Izumida Y., et al. , Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization. Nat. Commun. 4, 2316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y. C., et al. , Feeding influences adipose tissue responses to exercise in overweight men. Am. J. Physiol. Endocrinol. Metab. 313, E84–E93 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Cao W., et al. , p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell Biol. 24, 3057–3067 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H., Xu J., Lazarovici P., Quirion R., Zheng W., cAMP response element-binding protein (CREB): A possible signaling molecule link in the pathophysiology of schizophrenia. Front. Mol. Neurosci. 11, 255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Neill J. S., Maywood E. S., Chesham J. E., Takahashi J. S., Hastings M. H., cAMP-dependent signaling as a core component of the Mammalian Circadian pacemaker. Science 320, 949–953 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brindle P. K., Montminy M. R., The CREB family of transcription activators. Curr. Opin. Genet. Dev. 2, 199–204 (1992). [DOI] [PubMed] [Google Scholar]
- 73.Balsalobre A., et al. , Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289, 2344–2347 (2000). [DOI] [PubMed] [Google Scholar]
- 74.Nagoshi E., et al. , Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Rio D. C., Ares M. Jr., Hannon G. J., Nilsen T. W., Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc., pdb prot5439 (2010). [DOI] [PubMed]
- 76.Gabriel B. M., et al. , Disrupted circadian oscillations in type 2 diabetes are linked to altered rhythmic mitochondrial metabolism in skeletal muscle. Sci. Adv. 7, eabi9654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kang S., et al. , Identification of nuclear hormone receptor pathways causing insulin resistance by transcriptional and epigenomic analysis. Nat. Cell Biol. 17, 44–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rohm M., et al. , Transcriptional cofactor TBLR1 controls lipid mobilization in white adipose tissue. Cell Metab. 17, 575–585 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Fischer C., et al. , A miR-327-FGF10-FGFR2-mediated autocrine signaling mechanism controls white fat browning. Nat. Commun. 8, 2079 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thaben P. F., Westermark P. O., Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29, 391–400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The RNA-Seq dataset reported in this paper has been deposited in the Gene Expression Omnibus database (ID: GSE199429). All scripts used to analyze data are available on http://www.github.com/leonidaslundell/GSE199429. Previously published data were used for this work and are available in Gene Expression Omnibus with accession numbers: GSE43658 (77); GSE62635 (78); GSE55934 (79).