Significance
Three clades of mpox (monkeypox) virus are recognized: Clade I is present in the Congo Basin, causes up to 10% human mortality, and is transmitted by rodents with little human-to-human spread; clade IIa exists in West Africa, has a low mortality, and is also a zoonosis; clade IIb is currently spreading globally by human transmission. The genetic basis for differences in virulence and transmission has not been determined. A roadblock is the need for a small animal model that can be studied under required stringent safety conditions. Here, we demonstrate that the clades exhibit highly significant differences in CAST/EiJ mice in the order clade I > clade IIa > clade IIb similar to the severity of clinical disease in humans.
Keywords: human monkeypox, monkeypox virus, orthopoxvirus, castaneous mouse, virus virulence
Abstract
Human mpox (monkeypox), a disease with similarities to smallpox, is endemic in Africa where it has persisted as a zoonosis with limited human-to-human spread. Unexpectedly, the disease expanded globally in 2022 driven by human-to-human transmission outside of Africa. It is not yet known whether the latter is due solely to behavioral and environmental factors or whether the mpox virus is adapting to a new host. Genome sequencing has revealed differences between the current outbreak strains, classified as clade IIb, and the prior clade IIa and clade I viruses, but whether these differences contribute to virulence or transmission has not been determined. We demonstrate that the wild-derived inbred castaneous mouse provides an exceptional animal model for investigating clade differences in mpox virus virulence and show that the order is clade I > clade IIa > clade IIb.1. The greatly reduced replication of the clade IIb.1 major outbreak strain in mice and absence of lethality at 100 times the lethal dose of a closely related clade IIa virus, despite similar multiplication in cell culture, suggest that clade IIb is evolving diminished virulence or adapting to other species.
Although first isolated from captive Asian monkeys, mpox (monkeypox) virus (MPXV) infects rodents and is incidentally transmitted to humans and nonhuman primates in Africa (1). Human mpox, a disease resembling smallpox, was recognized upon the eradication of the latter and the cessation of vaccination (2). Historically, mpox has been a zoonosis with little human-to-human transmission and until 2022 was largely limited to regions of Africa. The genome sequence of the Congo Basin (now referred to as clade I) MPXV (3) placed it in the orthopoxvirus genus of the chordopoxvirus subfamily along with variola virus, the causative agent of smallpox and vaccinia virus used as the smallpox vaccine. A second clade of MPXV (now referred to as clade IIa) with a 95% nucleotide sequence identity to clade I was subsequently recognized (4). Clade I is associated with a mortality of up to 10%, whereas clade IIa from West Africa has a mortality of less than 1%. The current outbreak strain of MPXV has been designated clade IIb because of its sequence similarity to clade IIa (5) and like the latter has a low mortality in immunocompetent individuals. Whereas clades I and IIa are primarily zoonoses, clade IIb has exhibited extensive human-to-human spread. In laboratory experiments, respiratory transmission of MPXV occurred inefficiently (6), and direct contact with animals and close human interactions appear most important in people (7).
The number of human mpox cases has been increasing in Africa, likely due to waning population immunity from the discontinued smallpox vaccine and changing environmental and social factors (8). Human mpox due to Clade IIa has occurred in other parts of the world through importation of African rodents into the United States in 2003 and by travelers from Africa but without evident human-to-human transmission (9). However, the unprecedented increase in clade IIb cases in 2022 is due to human transmission outside of Africa. At the time of writing, more than 80,000 cases in over 100 different locations were diagnosed.
Genetic differences contributing to the greater morbidity of clade I MPXV compared to clade IIa and the increased human transmission of clade IIb are unknown. Knowledge of such differences could provide an early warning of the potential virulence of new or hybrid strains, help identify therapeutic targets, and contribute to basic knowledge of virus–host interactions. Although the genomes of clade I and II viruses are highly conserved, there are numerous sequence differences that could account for the greater virulence of the former (4, 10), whereas clades IIa and IIb viruses have fewer differences (5, 11). Animal models are crucial for investigating virus pathogenesis, and in the case of MPXV, such studies need to be carried out under stringent BSL-3 containment conditions. Moreover, clade I viruses are classified as select agents in the United States, limiting the number of laboratories working on MPXV. Parker and Buller (12) and Alakunle et al. (13) have provided extensive descriptions of natural and experimental infections with MPXV, although the primary reservoir species are unknown. Experimental animals include Asian primates, African rodents, and a variety of North American rodents including prairie dogs and squirrels. The African dormouse (Graphiurus kelleni) is of particular interest since these small squirrel-like rodents can be readily bred in captivity and were among the African rodents that tested positive for MPXV in a shipment that introduced the virus into the United States in 2003. Cotton rats (Sigmodon sp.) are another species susceptible to MPXV that can be raised in captivity. Nevertheless, African dormice and cotton rats are not inbred, and the absence of immunological reagents is a disadvantage for the study of virulence. Other small animals that can be reared in captivity such as guinea pigs, hamsters, rats, and common mouse strains do not develop severe disease upon inoculation with MPXV (12).
A mouse model for MPXV would have numerous advantages. To identify a susceptible mouse, we previously screened a panel of 38 inbred strains by intranasal (IN) inoculation of a Clade I MPXV and discovered the exceptional vulnerability of the wild-derived castaneous (CAST/EiJ) mouse and confirmed the resistance of common inbred mouse strains (14). Subsequent studies showed that the CAST mouse is also highly susceptible to other orthopoxviruses including vaccinia, cowpox, and Akhmeta viruses (15, 16). IN inoculation of CAST mice with variola virus, the causative agent of smallpox, resulted in only mild disease with some virus shedding and limited skin lesions (17) in accordance with the specificity of variola virus for humans.
CAST mice were derived from founder specimens trapped in a grain storage facility in Thailand (JAX® NOTES, Issue 456, Winter 1994) and because they are genetically distinct from common inbred laboratory mice were included in the Collaborative Cross panel (18). CAST mice are immunologically competent; they make good antibody and T cell responses upon vaccination that provide complete protection from MPXV (14). Low basal levels of natural killer cells likely contribute to their susceptibility to MPXV and other orthopoxviruses as administration of interferon gamma, IL-15, or natural killer cells expanded in vitro each provides resistance (15, 19–21). Here, we develop the CAST mouse as a model to compare MPXV clades and show that the order of virulence is clade I > IIa > IIb.1.
Results
Clade I MPXV and Clade IIa MPXV Lethally Infect African Dormice.
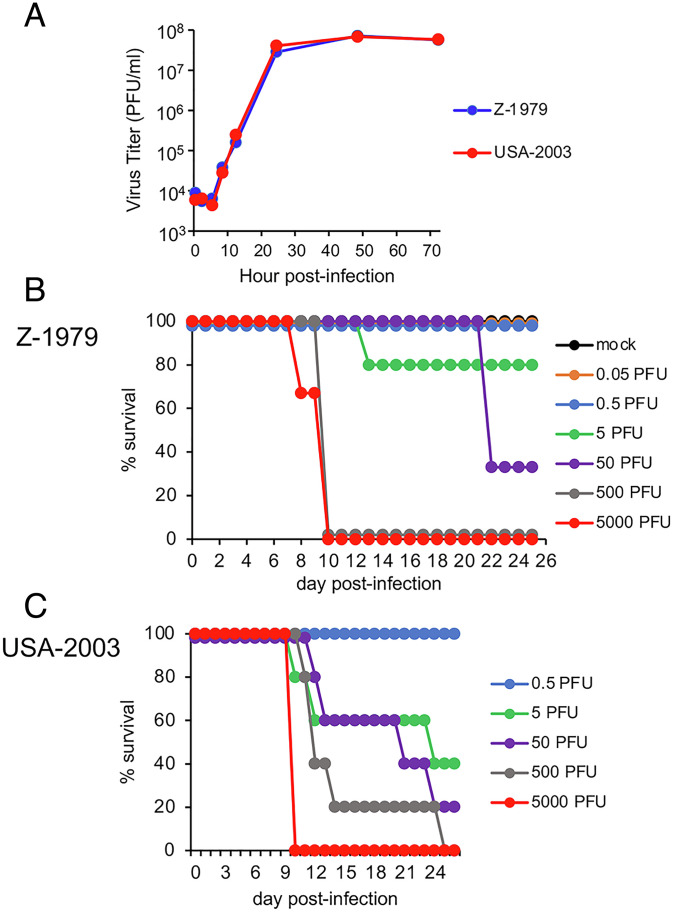
Schultz and coworkers (22) reported that African dormice are highly susceptible to MPXV by the IN route with a median 50% lethal dose (LD50) of 12 PFU for the clade I MPXV-ZAI-1979-005, and they cited a similar although undisclosed value for MPXV Copenhagen-58, a IIa clade. We confirmed and extended these studies using clonal isolates of clade I MPXV-ZAI-1979-005, abbreviated here as Z-1979, and clade IIa MPXV-USA-2003-044 (abbreviated USA-2003). No difference in replication of the two viruses was found during multistep growth in African green monkey BS-C-1 cells (Fig. 1A). Upon IN infection of African dormice with Z-1979, some deaths occurred with 5 and 50 PFU, and all died with 500 and 5,000 PFU (Fig. 1B). IN infection with USA-2003 followed a similar pattern with 100% deaths with 5,000 PFU (Fig. 1C). LD50 values of 12 and 5 PFU were calculated for Z-1979 and USA-2003, respectively. Large numbers of noninbred African dormice would be required to establish biological significance of the relatively small difference in LD50 and therefore were not considered optimal for investigating clade differences.
Fig. 1.
MPXV Z-1979 and USA-2003: in vitro replication and African dormouse infection. (A) BS-C-1 cells were infected in triplicate with 0.05 PFU/cell of MPXV Z-1979 or USA-2003. At intervals, virus yields were determined by plaque assay on BS-C-1 cells. Bars indicate SD. (B) Each group (n = 3 to 5) of African dormice was mock infected or infected IN with 0.05 to 5,000 PFU of Z-1979 and survival plotted. (C) Same as panel B except animals were inoculated with 0.5 to 5,000 PFU of USA-2003.
Clade I MPXV Is up to 1,000 Times More Virulent Than Clade IIa MPXV in CAST Mice.
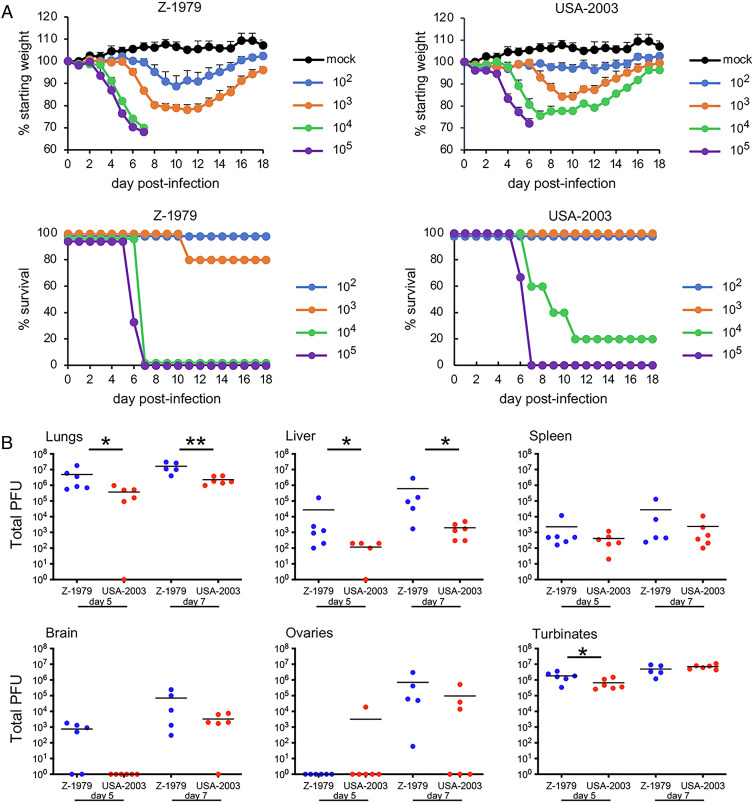
CAST mice infected IN with 103 PFU of Z-1979 lost more than 20% of their weight and one died (Fig. 2A). At 104 and 105 PFU, all mice infected with Z-1979 died or were euthanized because of 30% weight loss. Following IN infection with USA-2003, comparable weight loss occurred at slightly higher doses, and most mice died at 104 PFU and all at 105 PFU. From these data, LD50 of 2,370 and 4,200 were obtained for Z-1979 and USA-2003, respectively. In addition, there was greater recovery of virus from the nasal turbinates, lungs, brain, and abdominal organs on days 5 and 7 after IN infection of Z-1979 compared to USA-2003 (Fig. 2B).
Fig. 2.
Virulence of MPXV Z-1979 and USA-2003 upon IN infection of CAST mice. (A) CAST mice (groups of n = 4 to 5) were mock infected or infected IN with 102 to 105 PFU of MPXV Z-1979 or USA-2003 and examined and weighed daily. The percentage of starting weights and survival are indicated. Bars indicate SD. (B) CAST mice (n = 4 to 5) were infected IN with 102 PFU of MPXV Z-1979 or USA-2003. On days 5 (d5) and 7 (d7), whole organs and organ pairs were removed, homogenized, and the total PFU determined by plaque assay. Bars represent geometric means. *P < 0.05 and **P < 0.01.
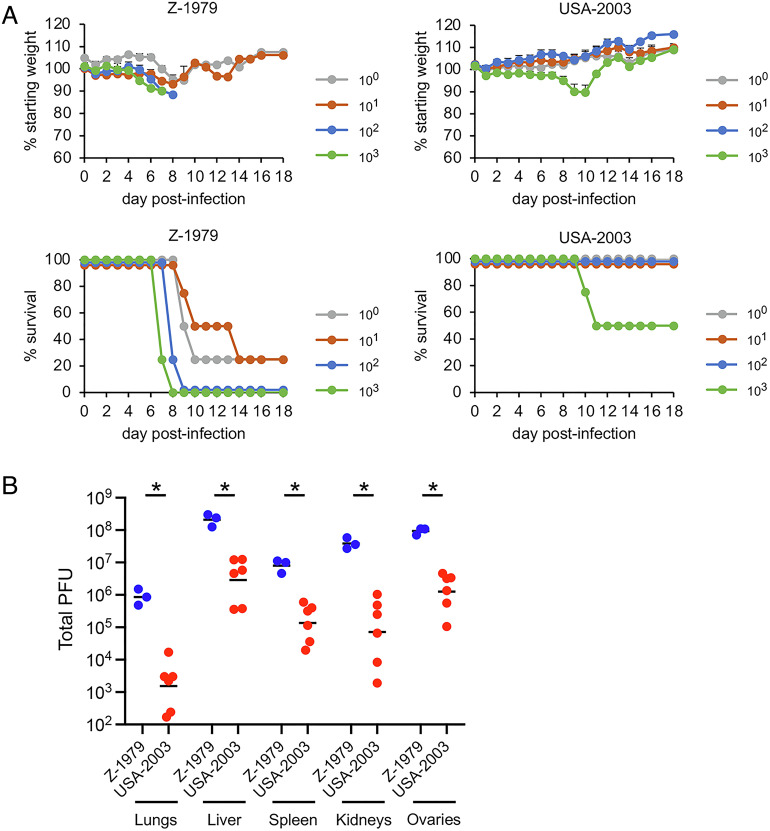
Since the upper respiratory tract is not thought to be an important route of MPXV infection and extensive spread of virus to the abdominal organs occurred by day 5 after IN infection, we investigated a systemic route of infection. Remarkably, most CAST mice infected with Z-1979 died with 1 and 10 PFU and all with 100 and 1,000 PFU administered intraperitoneally (IP), despite little weight loss (Fig. 3A). In contrast, USA-2003 caused no deaths at 100 PFU, and only 50% succumbed to 1,000 PFU. The LD50 values were <1 for Z-1979 and 1,000 for USA-2003. Furthermore, the titers of the Z-1979 were significantly higher in all organs analyzed at day 6 after infection compared to USA-2003 (Fig. 3B). Note that mortality is greater by IP infection, but weight loss is more pronounced by IN infection, possibly because the latter route reduces eating and drinking.
Fig. 3.
Virulence of MPXV Z-1979 and USA-2003 upon IP infection of CAST mice. (A) CAST mice (groups of n = 4 to 5) were mock infected or infected IP with 1 to 103 PFU of MPXV Z-1979 or USA-2003 and examined and weighed daily. The percentage of starting weights and survival are indicated. Bars indicate SD. (B) CAST mice (n = 3 to 6) were infected IP with 102 PFU of MPXV Z-1979 or USA-2003. On day 6, whole organs and organ pairs were removed, homogenized, and the total PFU determined by plaque assay. Bars represent geometric means. *P < 0.05.
MPXV Clade IIb Is More Attenuated Than Clade IIa.
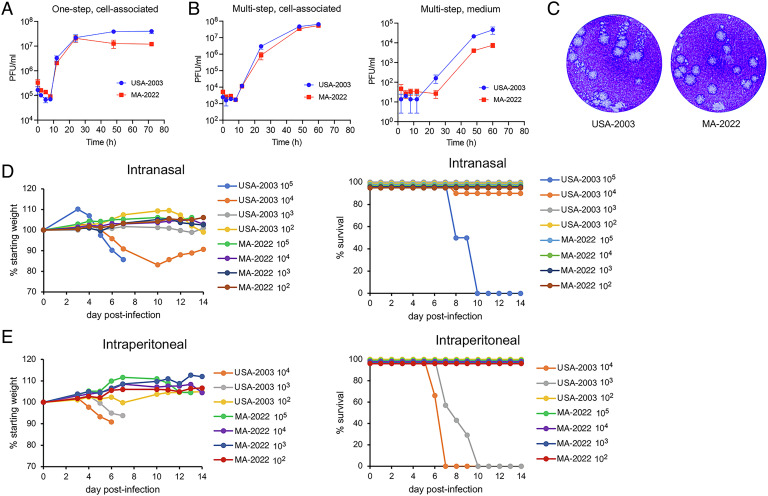
The above experiments were carried out with MPXV isolated decades before the global 2022 outbreak due to MPXV clade IIb. We received a low-passage MPXV isolated in 2022 from an individual in Massachusetts that is designated MPXV-USA-2022-MA001 (abbreviated MA-2022) and is a prototype for the major outbreak variant called b.1. Replication of the USA-2003 and MA-2022 at low (0.05 PFU/cell) and high (3 PFU/cell) multiplicities in African green monkey BS-C-1 cells was not significantly different, although there was a trend for greater release of USA-2003 into the medium compared to MA-2022 (Fig. 4 A and B). Greater release of USA-2003 was corroborated by a “comet assay”; in the absence of a methylcellulose overlay, small satellite plaques (comets) radiating out from larger round plaques were apparent in the stained monolayers infected with USA-2003 but were infrequent in cells infected with MA-2022 (Fig. 4C).
Fig. 4.
In vitro replication and virulence of USA-2003 and MA-2022. (A) One-step replication. BS-C-1 cells were infected in triplicate with 3 PFU/cell of USA-2003 or MA-2022 and harvested at the indicated times. Infectious virus in cell lysates was determined by plaque assay on BS-C-1 cells. Bars represent SDs. (B) Multistep replication. BS-C-1 cells were infected in triplicate with 0.05 PFU of USA-2003 or MA-2022. At the indicated times, the medium was removed, centrifuged, and supernatant collected. The pelleted cells were combined with the cells that adhered to the dish and lysed. Infectious virus in the lysates and supernatant was determined by plaque assay on BS-C-1 cells using a methylcellulose overlay. Bars indicate SDs. (C) Satellite (comet) plaque assay. BS-C-1 cell monolayers were infected with ~25 PFU of USA-2003 or MA-2022 for 1 h at 37 °C. Unadsorbed virus was removed, and the incubation was continued for 48 h without a methylcellulose overlay and then stained with crystal violet. (D) IN infection of CAST mice. CAST mice were inoculated with 102 PFU (n = 3), 103 PFU (n = 4), 104 PFU (n = 6 to 7), and 105 PFU (n = 2) of USA-2003 and MA-2022. Daily weights and survival are plotted. (E) IP infection of CAST mice. CAST mice were inoculated with 102 PFU (n = 3), 103 PFU (n = 4 to 7), and 104 PFU (n = 6) of USA-2003 and MA-2022. Daily weights and survival are plotted.
Upon IN infection of CAST mice, USA-2003 caused severe disease with weight loss and deaths at doses of 104 and 105 PFU (Fig. 4D) similar to that obtained for USA-2003 in the prior experiment shown in Fig. 2. In contrast, neither weight loss nor disease occurred with MA-2022. Furthermore, following IP infection, USA-2003 caused weight loss and 100% lethality at 103 and 104 PFU, whereas neither weight loss nor lethality occurred at doses up to 105 PFU of MA-2022 (Fig. 4E).
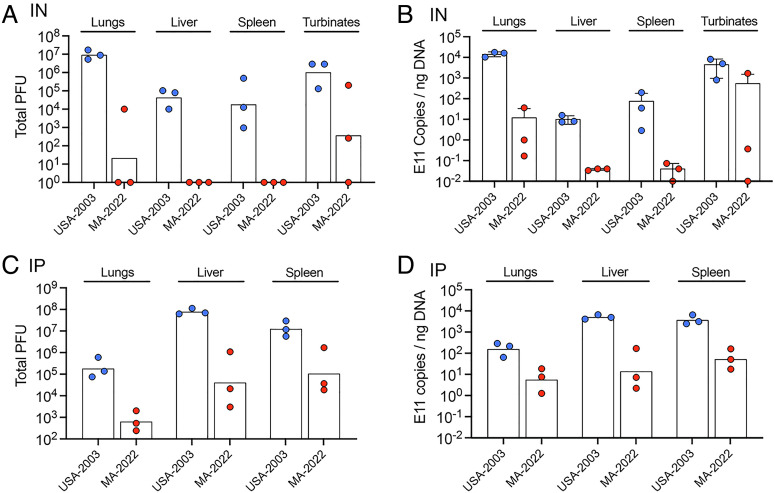
Next, we determined virus titers and genome copies in the organs of mice infected with 104 PFU of USA-2003 and MA-2022. At 7 d after IN infection, the titers of USA-2003 were >106 PFU and >105 PFU in the lungs and nasal turbinates, respectively, and about 104 PFU in the liver and spleen (Fig. 5A). In contrast, only 1 of 3 mice infected with MA-2022 had detectable virus in the lungs; none had detectable virus in the liver or spleen, and the mean titer in the nasal turbinates was three logs lower than that of USA-2003. Those results were consistent with an analysis of genomic DNA in the same samples by digital droplet (dd) PCR (Fig. 5B). We also determined virus titers and genomic DNA following IP infection with USA-2003 and MA-2022. The titers of USA-2003 were more than 100-fold higher than that of MA-2022 (Fig. 5C), and again, this was corroborated by analysis of genomic DNA by ddPCR. Thus, the lower virulence of MA-2022 in CAST mice correlated with lower virus replication.
Fig. 5.
Infectious virus and genome copies of USA-2003 and MA-2022 in organs of CAST mice. (A and B) CAST mice (groups of n = 3) were infected IN as in Fig. 5C, and infectious virus and MPXV genome copies determined. (C and D) CAST mice (groups of n = 3) were infected IP as in Fig. 5D, and infectious virus and MPXV genome copies determined.
Discussion
Numerous reports have suggested that observed genome sequence variations might account for differences in virulence and transmission of MPXV clades. However, few such predictions have been tested experimentally. Reasons for this include the need for a suitable animal model and the stringent biosafety conditions that must be met, particularly for clade I MPXV. Using objective criteria, we showed that clade I, clade IIa, and clade IIb.1 MPXVs exhibit large differences in morbidity and virus replication in CAST mice. By the IP route of infection, MPXV clades I and IIa had LD50 values of 1 PFU and 103 PFU, respectively, whereas all mice survived even 105 PFU of the clade IIb.1 virus. The difference in virulence between clades I and IIa was less by the IN route but here too mice survived 105 PFU of the clade 2b.1 virus. For both routes, replication in the lungs and abdominal organs also followed the order of clade I > IIa > IIb.1. Although we have not yet compared clades using other routes for infection of CAST mice, we previously reported that skin scarification of the clade I virus causes local lesions but no other signs of disease (14), whereas footpad inoculation caused swelling of the lower leg, and one of four mice died on day 14 with virus in the lungs, liver, and spleen (14). Similar studies with clade IIa and IIb viruses are planned. The increase in human transmission of clade IIb and the striking reduction in virulence for CAST mice raise the possibility that MPXV is adapting to humans with a concomitant loss of fitness for certain other species.
Determination of which genes are responsible for the virulence differences of MPXV clades remains to be determined. MPXV like other members of the orthopoxvirus genus encodes about 100 accessory genes that are largely dispensable for replication in vitro but affect host range and virulence. Interestingly, recent gene loss has been a major factor in the evolution of orthopoxviruses that can account for differences in their host range (23). For example, cowpox virus with the most accessory genes has a wide host range, whereas variola virus with the fewest is specific for humans. Of interest, human-transmitted variola virus (17) and MPXV clade 2b.1 cause only mild disease in CAST mice. While differences in accessory genes frequently affect host range, alterations in essential genes should not be overlooked. In this respect, the significance of our finding that USA-2003 made more extracellular virus than MA-2022 in vitro needs to be assessed. We have started to investigate the genetic determinants responsible for virulence differences of clade I and IIa viruses and plan to extend this to clade IIb pending institutional approval. It has been suggested that orthopoxvirus immune modulators might reduce an overreaction of the host immune response, and loss of such genes might increase virulence (24). Thus, both gain and loss of gene function should be considered in assessing virulence.
Methods
Ethics Statement.
All procedures with infectious MPXV were performed in registered select agent BSL-3 and ABSL-3 laboratories by trained and smallpox vaccinated investigators using protocols approved by the NIH Institutional Biosafety Committee and judged not to have the potential for dual-use research of concern. Experiments and procedures using mice were approved by the National Institute of Allergy and Infectious Diseases (NIAID) Animal Care and Use Committee according to standards set forth in the NIH guidelines, Animal Welfare Act and US Federal Law. Euthanasia was carried out when weight loss of mice reached 30% using carbon dioxide inhalation in accordance with the American Veterinary Medical Association (AVMA) guidelines (2013 Report of the AVMA panel on euthanasia).
Cells.
African green monkey BS-C-1 cells (ATCC CCL-26) were maintained at 37 °C and 5% CO2 in modified Eagle minimal essential medium (Quality Biological, Inc.) supplemented with 8% fetal bovine serum, 2 mM L-glutamine, 10 U penicillin/mL, and 10 µg streptomycin/mL.
Viruses.
Low-passage stocks of MPXV-ZAI-1979-005, MPXV-USA-2003-044, and MPXV-USA-2022-MA1 were obtained from the Centers for Disease Control and Prevention, and procedures for isolation of clones of the 1979 and 2003 viruses have been reported (14).
Analysis of Virus Replication.
BS-C-1 cells in 12-well plates were infected with MPXV. After 1 h, monolayers were washed three times and overlayed with fresh medium. At various times, cells from triplicate wells were harvested and stored at −80 °C. Cells were lysed by three rounds of freeze-thawing, and samples were sonicated, and virus yields determined by plaque assay on BS-C-1 cells (25). For determination of extracellular virus, the medium was collected before harvesting cells, centrifuged, and the supernatant analyzed.
Comet Assay.
BS-C-1 monolayers in 12-well dishes were incubated with diluted samples of MPXV for 1 h at 37 °C. The medium was aspirated, and the monolayers washed with fresh medium to remove free virus. The plates were further incubated at 37 °C for 48 h and then fixed and stained with crystal violet.
Mice.
Five- to 7-wk-old female CAST/EiJ mice were obtained from Jackson Laboratories and were maintained in small ventilated microisolator cages. IN and IP inoculations were performed as described previously (14). Titers of each virus inoculum were confirmed by plaque assay.
African Dormice (Graphiurus kelleni).
Animals were originally obtained from R. M. Buller (deceased) and bred and maintained in large ventilated microisolater cages at the NIH, and 5- to 8-mo mixed females and males were used for infection.
Titration of Virus from Organs.
Whole organs and organ pairs were removed aseptically from animals, placed in 2 mL of balanced salt solution supplemented with 0.1% bovine serum albumin, and stored at −80 °C until used. Homogenization and titration were performed as described previously (14).
Quantitation of Viral DNA.
Viral DNA from infected organs was determined by ddPCR as previously described (26). Briefly, DNA was isolated from homogenized MPXV-infected tissues using the DNeasy Blood and Tissue Kit (Qiagen). Several 10-fold dilutions of each purified DNA were prepared in nuclease-free H2O and added to a reaction mixture containing ddPCR EvaGreen Supermix (Bio-Rad). Forward (5′-GAATACATTCACATTGACCAATCAGAA-3′) and reverse (5′GGTTCGTCAAAGACATAAAACTCATT-3′) primers were specific for the conserved VACV WR E11L open reading frame that encodes an essential core protein (27). Reaction oil droplets were synthesized using an Automated Droplet Generator (Bio-Rad) prior to PCR thermocycling. Droplets were subsequently acquired using a QX200 Droplet Reader (Bio-Rad), and the number of genome copies was determined using QuantaLife software (Bio-Rad).
Statistics.
Significance was calculated by the Mann–Whitney using Prism.
Acknowledgments
We thank Catherine Cotter for assistance with some experiments and discussions. Excellent animal care was provided by the technical staff of the National Institute of Allergy and Infectious Diseases Comparative Medical Branch. The work was funded by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases.
Author contributions
B.M. designed research; J.L.A. and P.L.E. performed research; J.L.A. and P.L.E. analyzed data; and B.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: M.C., University of Oxford; and S.N.I., University of Pennsylvania.
Data, Materials, and Software Availability
All study data are included in the main text and are freely available.
References
- 1.Doty J. B., et al. , Assessing monkeypox virus prevalence in small mammals at the human-animal interface in the Democratic Republic of the Congo. Viruses-Basel 9, 283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simpson K., et al. , Human monkeypox - After 40 years, an unintended consequence of smallpox eradication. Vaccine 38, 5077–5081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shchelkunov S. N., et al. , Analysis of the monkeypox virus genome. Virology 297, 172–194 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Likos A. M., et al. , A tale of two clades: Monkeypox viruses. J. Gen. Virol. 86, 2661–2672 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Gigante C. M., et al. , Multiple lineages of monkeypox virus detected in the United States, 2021–2022. Science eadd4153 (2022), 10.1126/science.add4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutson C. L., et al. , Transmissibility of the monkeypox virus clades via respiratory transmission: Investigation using the prairie dog-monkeypox virus challenge system. Plos One 8, e55488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleischauer A. T., et al. , Evaluation of human-to-human transmission of monkeypox from infected patients to health care workers. Clin. Infect. Dis. 40, 689–694 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Nguyen P. Y., Ajisegiri W. S., Costantino V., Chughtai A. A., MacIntyre C. R., Reemergence of human monkeypox and declining population immunity in the context of urbanization, Nigeria, 2017–2020. Emerging Infect. Dis. 27, 1007–1014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mauldin M. R., et al. , Exportation of monkeypox virus from the African continent. J. Infect. Dis. 225, 1367–1376 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N. H., et al. , Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology 340, 46–63 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isidro J., et al. , Multi-country outbreak of monkeypox virus. Nat. Med. 28, 2220–2221 (2022), 10.1038/s41591-022-01907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker S., Buller R. M., A review of experimental and natural infections of animals with monkeypox virus between 1958 and 2012. Future Virol. 8, 129–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alakunle E., Moens U., Nchinda G., Okeke M. I., Monkeypox virus in Nigeria: Infection biology, epidemiology, and evolution. Viruses 12, 1257 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Americo J. L., Moss B., Earl P. L., Identification of wild-derived inbred mouse strains highly susceptible to monkeypox virus infection for use as small animal models. J. Virol. 84, 8172–8180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Americo J. L., et al. , Susceptibility of the wild-derived inbred CAST/Ei mouse to infection by orthopoxviruses analyzed by live bioluminescence imaging. Virology 449, 120–132 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan C. N., et al. , Laboratory infection of novel Akhmeta virus in CAST/EiJ mice. Viruses-Basel 12, 1416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallardo-Romero N. F., et al. , Use of live Variola virus to determine whether CAST/EiJ mice are a suitable surrogate animal model for human smallpox. Virus Res. 275, 197772 (2019), 10.1016/j.virusres.2019.197772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Threadgill D. W., Churchill G. A., Ten years of the collaborative cross. Genetics 190, 291–294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earl P. L., Americo J. L., Moss B., Lethal monkeypox virus infection of CAST/EiJ mice is associated with a deficient interferon-gamma response. J. Virol. 86, 9105–9112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earl P. L., Americo J. L., Moss B., Insufficient innate immunity contributes to the susceptibility of the castaneous mouse to orthopoxvirus infection. J. Virol. 91, e01042-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earl P. L., Americo J. L., Moss B., Natural killer cells expanded in vivo or ex vivo with IL-15 overcomes the inherent susceptibility of CAST mice to lethal infection with orthopoxviruses. PLoS Pathog. 16, e1008505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultz D. A., Sagartz J. E., Huso D. L., Buller R. M. L., Experimental infection of an African dormouse (Graphiurus kelleni) with monkeypox virus. Virology 383, 86–92 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senkevich T. G., Yutin N., Wolf Y. I., Koonin E. V., Moss B., Ancient gene capture and recent gene loss shape the evolution of orthopoxvirus-host interaction genes. mBio 12, e0149521 (2021), 10.1128/mBio.01495-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alcami A., Smith G. L., A mechanism for inhibition of fever by a virus. Proc. Natl. Acad. Sci. U.S.A. 93, 11029–11034 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cotter C. A., Earl P. L., Wyatt L. S., Moss B., Preparation of cell cultures and vaccinia virus stocks. Curr. Protoc. Mol. Biol. 117, 16 16 11-16 16 18 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Americo J. L., Earl P. L., Moss B., Droplet digital PCR for rapid enumeration of viral genomes and particles from cells and animals infected with orthopoxviruses. Virology 511, 19–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S. P., Shuman S., A temperature sensitive mutation of the vaccinia virus E11 gene encoding a 15-kDa component. Virology 216, 252–257 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the main text and are freely available.