Significance
Chicken is one of the most important vertebrate model organisms, yet its genome is far from complete. This study generated the complete sequence of a chicken genome, uncovering six chromosome models absent in previous genome assemblies. Ten small microchromosomes evolved distinct genomic and epigenetic features, unlike any other vertebrate chromosomes but remain stable and conserved in birds. Most chicken centromeres were found to contain higher-order repeats (HORs), resembling the centromeric organization in primates. The complete chicken chromosome models are useful to reconstruct the karyotype of the vertebrate ancestor. We reveal the evolutionary trajectory of chromosome changes from ancestral chordate to early vertebrate and Amniota through frequent fusion events before and after whole-genome duplications.
Keywords: chicken genome, dot chromosome, centromere, chromosome evolution
Abstract
Microchromosomes are prevalent in nonmammalian vertebrates [P. D. Waters et al., Proc. Natl. Acad. Sci. U.S.A. 118 (2021)], but a few of them are missing in bird genome assemblies. Here, we present a new chicken reference genome containing all autosomes, a Z and a W chromosome, with all gaps closed except for the W. We identified ten small microchromosomes (termed dot chromosomes) with distinct sequence and epigenetic features, among which six were newly assembled. Those dot chromosomes exhibit extremely high GC content and a high level of DNA methylation and are enriched for housekeeping genes. The pericentromeric heterochromatin of dot chromosomes is disproportionately large and continues to expand with the proliferation of satellite DNA and testis-expressed genes. Our analyses revealed that the 41-bp CNM repeat frequently forms higher-order repeats (HORs) at the centromeres of acrocentric chromosomes. The centromere core regions where the kinetochore attaches often encompass telomeric sequence (TTAGGG)n, and in a one of the dot chromosomes, the centromere core recruits an endogenous retrovirus (ERV). We further demonstrate that the W chromosome shares some common features with dot chromosomes, having large arrays of hypermethylated tandem repeats. Finally, using the complete chicken chromosome models, we reconstructed a fine picture of chordate karyotype evolution, revealing frequent chromosomal fusions before and after vertebrate whole-genome duplications. Our sequence and epigenetic characterization of chicken chromosomes shed insights into the understanding of vertebrate genome evolution and chromosome biology.
Chicken is one of the most important vertebrate model organisms, and its genome is widely used for study in vertebrate evolution and avian biology (1–3). The first draft chicken genome was sequenced from an inbred line UCD001 in 2004 with shotgun reads, combined with reads from bacterial artificial chromosomes (BACs) and fosmids (4) and had been subsequently improved (5). The current reference genome (GRCg6a) was created by Genome Reference Consortium (GRC) using the same DNA source but employing PacBio single-molecular long reads (6). The Vertebrate Genome Project (VGP) has also released a genome assembly (bGalGal1.mat.broiler.GRCg7b) from a female chicken using a trio-binning approach (7). Various maps have been used to improve chromosomal anchoring, including a consensus linkage map, East Lansing map, and radiation hybrid map (4). However, although karyotype analyses revealed that the chicken genome contains 2n = 78, thus far only 32 autosomes plus two sex chromosomes (Z and W) were assembled, indicating six chromosomes missing.
A typical avian genome contains 10 pairs of macrochromosomes and ~30 pairs of microchromosomes that are evolutionarily stable during avian diversification (8, 9) and is thought to resemble ancestral vertebrate karyotype (10–12). It has been recently established that Amniota microchromosomes are stable (13) and have a chordate origin (14), likely formed due to asymmetric sequence losses following vertebrate whole-genome duplications (WGD) (15, 16). The six missing chromosomes in chicken genomes are all microchromosomes, expected to be gene-dense like other microchromosomes (17). A recent chicken pan-genome construction with PacBio reads has identified more than 1,000 novel genes that are enriched on microchromosomes (18). Creating a new reference chicken genome with all chromosome models assembled will reveal a complete gene repertoire and a full picture of vertebrate karyotype evolution.
The use of Nanopore ultralong reads and PacBio HiFi reads permits telomere-to-telomere genome assembly (19, 20). The HiFi reads can resolve complex regions (21) while the ultralong reads can assist to resolve tandem duplications (22). The combination of those two sequencing technologies has successfully completed the human genome (23), which provides a promising strategy to produce finished genomes for other species (24). Here, we present a complete chicken genome with all chromosomes assembled and all gaps closed except for the W chromosome.
Results
Toward the Complete Sequence of a Chicken Genome.
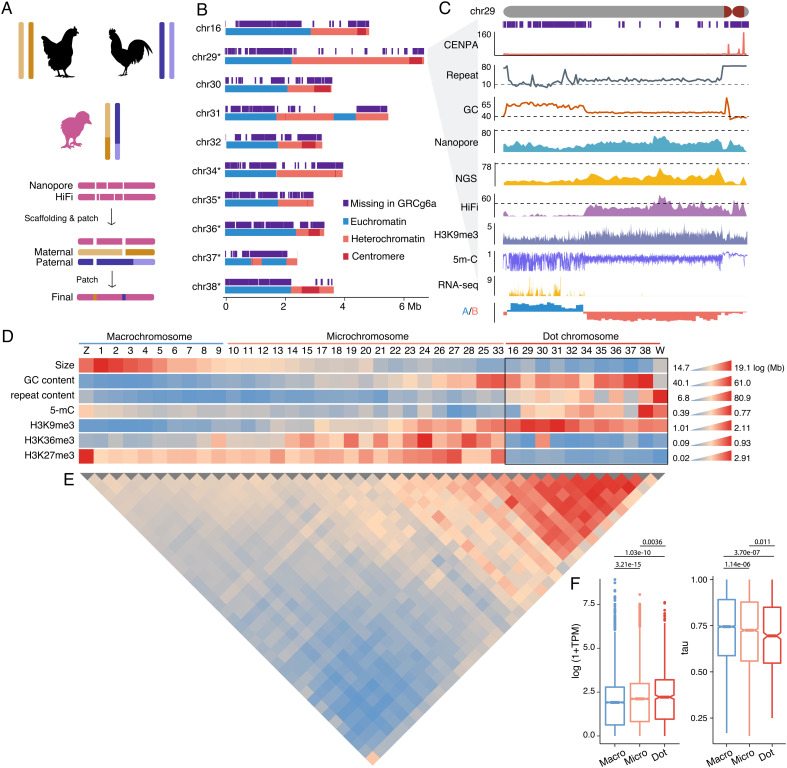
We employed a trio-sequencing approach (25) to assemble the diploid genome of a female chick (Huxu breed, Fig. 1A). We generated ~80× ultralong Nanopore and ~52× HiFi reads and both produced highly continuous genome assemblies (SI Appendix, Tables S1 and S2). We used HiFi-based contigs to replace sequences and fill gaps in the Nanopore-based backbone assembly (Fig. 1A). The integration of the two datasets resulted in contigs already at chromosome-scale, with only 26 gaps remaining. Those gaps are mostly embedded within long arrays of satellite DNA or simple repeats.
Fig. 1.
A complete chicken genome with 10 dot chromosomes. (A) A trio-based genome assembly pipeline. Rounded rectangles represent contigs. Paternal and maternal contigs were used to fill gaps in the primary contigs. (B) The dot chromosomes are in general composed of a euchromatic part and a heterochromatic part. The asterisks denote newly assembled chromosome models. (C) A zoom-in view for chr29, showing CENP-A and H3K9me3 binding, coverage of Nanopore ultralong, HiFi, NGS (BGISEQ-500, dashed lines indicate genomic average), gene expression (RNA-seq read counts in 1 kb windows), 5-mC levels, and A/B compartments. (D) The heatmap shows the chromosomal sizes (log-transformed), GC content, repeat content, chromosome-wide 5-mC levels, and ChIP/input ratios for H3K9me3, H3K36me3, and H3K27me3. (E) Interchromosomal interaction frequency measured using Hi-C data. (F) Dot chromosomes have a lower Tau value, i.e., lower level of tissue specificity but a higher expression level. P values were calculated using the Wilcoxon signed-rank tests.
We further partitioned ultralong and HiFi reads derived from parental and maternal haploids and produced two haploid assemblies using the same approach described above (Fig. 1A). The paternal and maternal contigs are expected to resolve complex regions that often lead to gaps in haploid (primary) genome assembly. Indeed, the remaining gaps, except for those on the W chromosome, are all resolved by one of the haploid assemblies or both (Fig. 1A). For instance, a ~3 Mb subtelomeric sequence at the 5′ end of chromosome 1 (chr1) was assembled into four contigs in the primary assembly but a single contig in both paternal and maternal haploid assembly (SI Appendix, Fig. S1).
The final assembly (GGswu1) contains 38 autosomes, a Z and a W chromosome (SI Appendix, Fig. S2), consistent with the known female karyotype (Fig. 2A) (26). Compared with the reference genome GRCg6a, the total length increases by 50.4 Mb to 1.1 Gb, mainly attributed to the addition of satellite DNA or segments with GC content larger than 55% (GC plateaus, SI Appendix, Table S3). Satellite DNA occupied 5.0% of the new assembly compared to 3.0% in GRCg6a. Importantly, our new assembly contains six chromosomal models (chr29, chr34 to 38) that are absent in GRCg6a. The homologous sequences of newly assembled chromosomes are scattered in the unanchored scaffolds or incorrectly anchored to chr31 and chr33 in GRCg6a (SI Appendix, Fig. S3). The GRCg7b assembly has proposed seven new chromosome models homologous to the six new ones in GGswu1, but their total size is only 13.4 Mb compared with 40.4 Mb in GGswu1 (SI Appendix, Table S4). We mapped more than 1,000 BAC clones against GGswu1 and found that 99.9% of the BAC sequences were aligned, and the one-to-one alignments have an average identity of 99.2% (SI Appendix, Table S5).
Fig. 2.
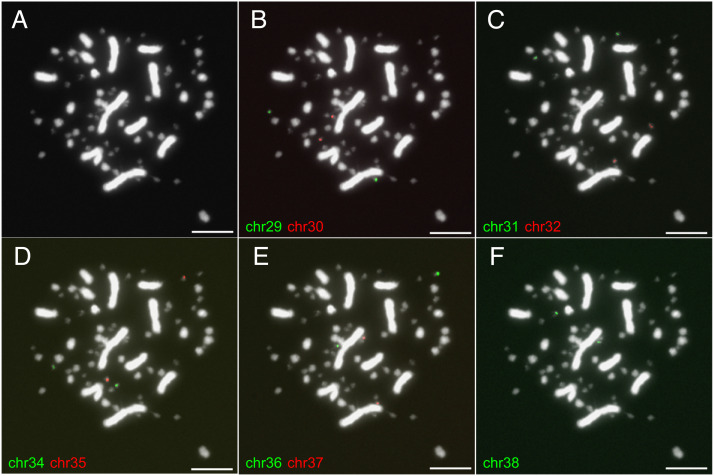
FISH verification for dot chromosomes. (A) The karyotype of a chicken cell. (B–F) The probes of nine dot chromosomes bind to different chromosomes in the same cell. The probes are composed of unique oligonucleotides (Dataset S2) and chromosome-specific amplicons (Fig. 5F). Chromosome 16 has been verified by previous studies (27). The scale for the white bar: 10 μm.
Approximately 44.6% of heterochromatic parts of the newly uncovered chromosomes are absent in GRCg6a, but unexpectedly 77.6% of the euchromatic parts carrying coding genes were absent (Fig. 1B and SI Appendix, Fig. S4 and Table S6). This is likely because the euchromatin has a much higher GC content than heterochromatin (58.7% vs. 51.4%), despite a lower repeat content (29.5% vs. 61.9%, SI Appendix, Fig. S5). The extremely high GC content makes the euchromatin inaccessible for HiFi or next-generation sequencing (NGS) but fortunately less so for Nanopore sequencing (Fig. 1C and SI Appendix, Fig. S4).
The Chicken Genome Contains 10 Dot Chromosomes.
The six newly assembled chromosomes all have a small size (on average 3.9 Mb), much smaller than other microchromosomes (~10.2 Mb), but exhibit a much higher GC content and repeat content, a higher level of chromosome-wide DNA methylation (5 mC), and H3K9me3 histone modifications but depletion of H3K36me3 and H3K27me3 (Fig. 1D and SI Appendix, Table S7). Four previously partially assembled chromosomes (chr16, 30 to 32) share similar features (Fig. 1D and SI Appendix, Table S7). Collectively, we termed those 10 chromosomes (chr16, 29, 30 to 32, 34 to 38) as dot chromosomes based on their morphology and heterochromatic nature. This term does not necessarily imply common features with the Drosophila dot chromosome (28). Other authors have used the term “D group” to refer to those dot-like chromosomes (26). The average size of the dot chromosome is 4.0 Mb, in line with the estimated size by a pulse electrophoresis method for the smallest chicken microchromosomes (3.4 to 4.8 Mb) (29). The ten dot chromosomes collectively account for only 3.7% of the chicken genome. We synthesized unique oligonucleotides across the euchromatin and amplified chromosome-specific duplicated genes (see the section ‘Testis-Expressed Amplicons in pericentromeric heterochromatin (PCH)’) as probes to verify the dot chromosomes. As a result, each chromosome-specific probe binds to one pair of chromosomes (Fig. 2).
The dot chromosomes typically have a highly compartmentalized chromatin organization, with the compact euchromatin occupying a part of the long-arm, and centromeric and PCH on the other part of the chromosomes (Fig. 1 B and C and SI Appendix, Fig. S4). In line with the abovementioned disparity in GC and repeat content, the heterochromatin and euchromatin parts of the dot chromosomes display distinct epigenetic features, including an extremely high methylation level for the heterochromatin (Fig. 1C and SI Appendix, Fig. S4). Similar to microchromosomes (30, 31), the dot chromosomes show intensive interchromosomal interactions between themselves (Fig. 1E), consistent with their physical clustering in the nuclear center (32) and are mainly driven by interactions between euchromatin (SI Appendix, Fig. S6). Despite intensive interchromosomal interactions, the dot chromosomes clearly show their own chromosomal territories (SI Appendix, Fig. S2B), supporting isolated assembly of chromosome models.
There is no sharp borderline between macrochromosomes and microchromosomes or between microchromosomes and dot chromosomes (Fig. 1D and SI Appendix, Fig. S7). For instance, chr25 and chr33 share some features with dot chromosomes, including small sizes (~4.1 Mb) and high GC contents (Fig. 1D). Their repeat content, epigenetic features (5-mC, H3K36me3 and H3K27me3), and the intensity of interchromosomal interactions, on the other hand, make them distanced from dot chromosomes, though the H3K9me3 landscape appears to be similar (Fig. 1D and SI Appendix, Table S7).
The gene density of dot chromosomes is much higher than that of macrochromosomes, but similar to microchromosomes, with an average gene number of 89. The newly assembled or completed dot chromosomes supplied additional 307 genes previously missed. Collectively, dot-chromosome genes have higher expression than microchromosomes or macrochromosomes and are more widely expressed (Fig. 1F and SI Appendix, Fig. S8), thus more likely housekeeping genes.
Reconstruction of Vertebrate Ancestral Karyotype.
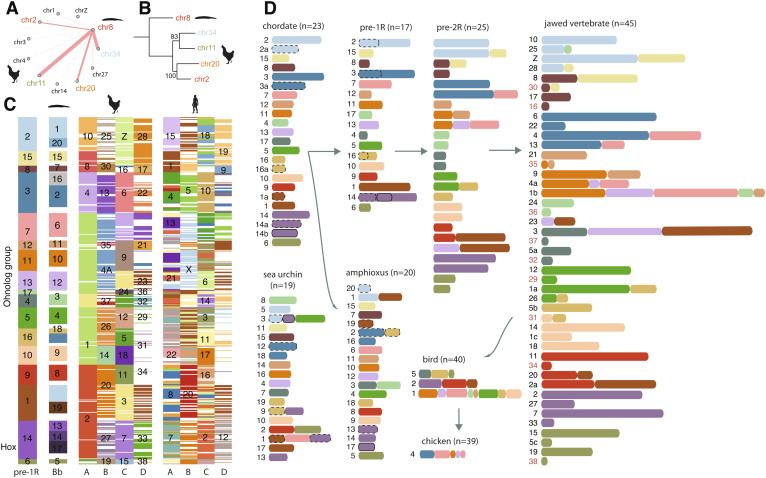
We used the complete chromosomal assembly of the chicken genome to reconstruct the full picture of vertebrate karyotype evolution. Our recent efforts have demonstrated that most amphioxus chromosomes are homologous with four different chicken chromosomes (1:4 relationships) due to two-round (2R) vertebrate WGD, except for six chromosomes with 1:3 relationships (15). Here, we rediscovered the 1:4 relationships for those six 1:3 cases by finishing the six new dot-chromosomes (Fig. 1A and SI Appendix, Fig. S9). For example, genes on amphioxus chr8 are homologous with those on chicken chr2, chr11, chr20, and chr34 (Fig. 3 A and B), among which chr34 is a newly uncovered dot-chromosome. Assigning homologous chicken chromosomes in turn led to the identification of 478 homologous gene (ohnolog) groups that have at least three ohnolog members in chicken (Fig. 3C and Dataset S1). Those ohnolog groups then led to the identification of 197 new ohnologs in the human genome based on chicken-human orthologous relationships (Fig. 3C and Dataset S1) and are helpful to corroborate the origin of microchromosomes and dot-chromosomes due to asymmetric sequence losses on one of the duplicated chromosomes following WGDs (15, 16).
Fig. 3.
Chordate karyotype evolution. (A) Genes from amphioxus chromosome 8 are primarily homologous with those from four chicken chromosomes. The thickness of connecting lines indicates the relative abundance of homologous genes for chicken chromosomes. (B) Maximum-likelihood phylogeny of concatenated homologous genes from the homologous chromosomes in (A). Bootstrapping values are shown at the nodes. (C) 478 ohnolog groups that have at least three ohnologs in chicken. Each row represents one ohnolog group. The numbers indicate the chromosomes or ancestral linkage groups. White colors imply the absence of ohnologs. Chicken and human have up to four homologous chromosomes (A–D) for each amphioxus (Bb) chromosome. (D) The reconstructed evolutionary history of vertebrate chromosomes. The IDs of dot chromosomes are colored in red in the jawed vertebrate panel.
While we reconstructed 17 ancestral chromosomes prior to the vertebrate first WGD (1R), in agreement with other studies (33, 34), we found that the chordate ancestral karyotype likely had 23 chromosomes, and they experienced six fusions in the branch leading to proto-vertebrate and three independent fusions in the branch to Cephalochordata (Fig. 3D). We further revealed drastic chromosomal changes following WGD, including nine post-1R fusions and five post-2R fusions (Fig. 3D), largely agreeing with a recent study (16). The common ancestor of jawed vertebrates likely had 45 pairs of chromosomes which experienced only five fusion events over ~400 My of evolution leading to the bird common ancestor inferred to have 40 chromosome pairs. Those five fusions led to the formation of bird chr1, chr2, and chr5 which are all macrochromosomes.
Origin and Diversity of Chicken Centromeres.
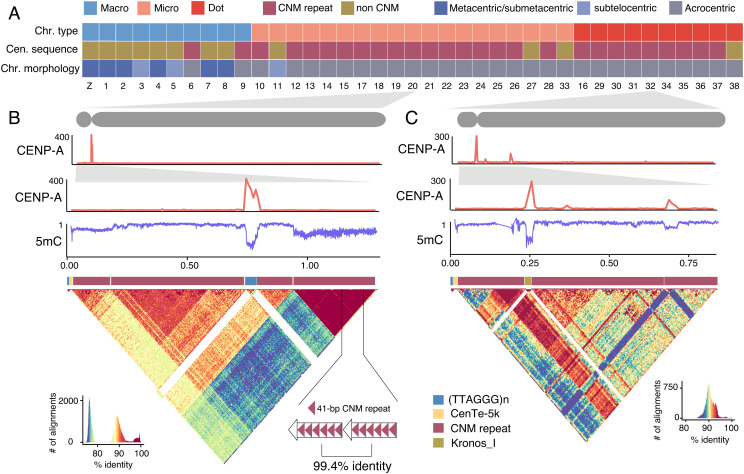
Chicken macrochromosomes tend to be metacentric or submetacentric (8 out of 10) whose unique centromeric repeats are largely known (35), in contrast to microchromosomes or dot-chromosomes that are mostly (28 out of 29) acrocentric (Fig. 4A), with their centromeric sequences remaining elusive (36). This is in part due to the difficulty in resolving the short-arm sequences of acrocentric chromosomes (23). With the new chicken assembly, we show that the acrocentric centromeres are 0.2 to 0.8 Mb long and are almost always associated with tandem arrays of a 41-bp repeat (Fig. 4A and SI Appendix, Figs. S10–S12) known as the CNM repeat (SI Appendix, Fig. S13) (35, 37–39). The CNM repeat-associated centromeres are also present in two unusually acrocentric macrochromosomes (chr6 and chr9, SI Appendix, Fig. S10) (38), suggesting that the morphology (centromere position), rather than the size of chromosomes, determines the composition of centromeric sequences.
Fig. 4.
The sequences and epigenetics of chicken centromeres. (A) The type of centromeric repeat and the type and morphology of chromosomes. Non-CNM repeats include chromosome-specific tandem or non-tandem repeat, simple repeats, and TEs. (B and C) The ChIP/input ratio for CENP-A ChIP-seq, and 5 mC levels (0 to 1) estimated with Nanopore reads on the centromeres of chr20 and chr32. The CENP-A peaks have the lowest 5-mC levels. The heatmaps show the pair-wise sequence identity (%) between 4-kb sequences. CNM repeats are divergent from each other but often form HORs.
Given that chordate ancestral chromosomes were likely all acrocentric (15), we hypothesize that CNM repeat-associated acrocentric centromeres are ancestral and stable during avian evolution. On the contrary, sub/metacentric macrochromosomes usually contain chromosome-specific tandem repeats in the centromeres (35) that are likely derived and were formed upon chromosomal fusions (40) following WGDs (Fig. 3D). Supporting this, the centromere of chr1 locates near a fusion point and is only 2.2 Mb away from a large interstitial telomeric repeat (SI Appendix, Fig. S14) (38), suggesting its likely formation following chromosomal fusions and the loss of CNM repeats. The disparity in centromere sequence composition and property between macrochromosomes and microchromosomes or dot chromosomes may play a role in driving their spatial segregation in the interphase nucleus (41).
In 19 acrocentric microchromosomes and dot chromosomes, the short arm encompasses merely a 5-kb conserved sequence (CenTe_5k) apart from the telomeric repeats (TTAGGG)n and CNM clusters (Fig. 4 B and C and SI Appendix, Table S8). The binding sites of CENP-A that define kinetochores and centromere cores are usually not on the body of CNM clusters, but at or close to their boundaries. In seven chromosomes, the centromere core sequences consist of (TTAGGG)n embedded within CNM tandem arrays, and in eight chromosomes, other simple repeats, such as (GCCTT)n, were recruited at centromere cores (SI Appendix, Figs. S11 and S12). Curiously, on the dot chromosome 32, the centromere core sequence consists of three tandemly arrayed full-length Kronos_I, an endogenous retrovirus (ERV) element (Fig. 4C). In spite of the differences in sequence composition or origin, the centromere cores show a deficient level of DNA methylation (Fig. 4 B and C and SI Appendix, Figs. S10–S12), though in some chromosomes the patterns are obscure, possibly due to centromere shifts (42).
The human centromeres are characteristic of higher-order repeats (HORs) (43) that evolved through “layered expansions” (44). Similarly, we discovered that the CNM monomer frequently forms HORs in acrocentric chromosomes in spite of their large intra- and inter-chromosomal divergence (Fig. 4B). The CNM HOR units contain 26 to 98 copies of the CNM repeat, and each HOR cluster contains 5 to 162 HOR units. In seven acrocentric chromosomes (chr9–10, 12–14, 17, 20), the active HORs sit at the distal end of centromeres, suggesting the expansion of HORs in the direction away from short-arm telomeres (Fig. 4B and SI Appendix, Figs. S10–S12), while in others (e.g., chr32) the active HORs flank the centromere cores (Fig. 4C). These two modes (outward and inward) of tandem-repeat expansion also apply to macrochromosome centromeres, with younger expansions at the centromere edge in, for example, chr1, but at the centromere centers in chr2 to 4 (SI Appendix, Fig. S10).
Testis-Expressed Amplicons in PCH.
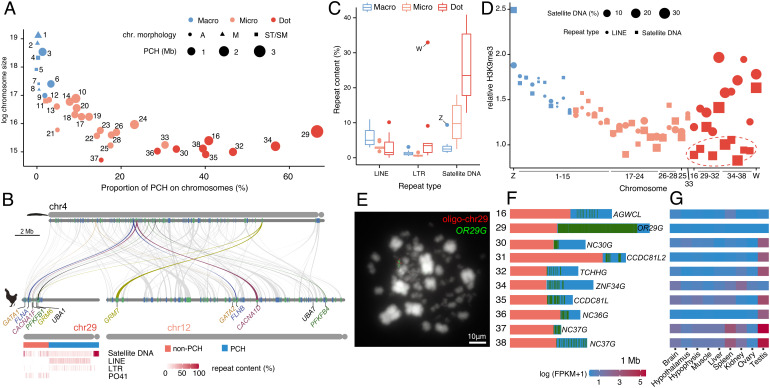
The morphology of chromosomes also determines the extent of PCH expansion (20). In particular, PCH is smaller in non-acrocentric chromosomes but expands to a larger size in acrocentric chromosomes (Fig. 5A). As a consequence, PCH occupies ~40.8% of dot chromosomes, compared to ~12.3% in microchromosomes and ~1.0% in macrochromosomes (Fig. 5A). In one extreme case, the proportion of PCH reaches 66.6% on chr29, leaving coding genes crowding in a small euchromatic region depleted of transposable elements (TEs) (Fig. 5B). Satellite DNA is the major component of dot-chromosome repeat sequences (Fig. 5C) and shows a lower level of H3K9me3 modifications compared to LINEs and LTRs (Fig. 5D). We propose that the less repressed satellite DNA is likely a major contributor to PCH expansion in dot chromosomes.
Fig. 5.
The expansion of PCH and amplicons. (A) The dot-chromosomes have larger PCH and the proportion of PCH is higher relative to microchromosomes and macrochromosomes. (B) Chicken chr12 and chr29 are homologous with amphioxus chr4. Six ohnolog pairs between chr29 and chr12 are highlighted. The PCH region of chr29 is rich in TEs while the non-PCH region is depleted of retrotransposons but is enriched for the PO41 repeats (37). (C) Satellite DNA is the most abundant repeat sequence on dot-chromosomes. (D) In macrochromosome and microchromosomes the relative H3K9me3 levels are similar for LINEs and satellite DNA while in dot-chromosomes the satellite DNA tends to have a lower relative H3K9me3 level (red dashed ellipse). (E) The PCH of dot chromosomes contains a large number of multicopy genes. Each green vertical bar represents one copy. (F) OR29G was detected in the PCH of chr29 while the synthesized oligonucleotides (oligo-chr29) were detected in the other part of chr29. (G) The expression of multicopy genes is testis-biased for seven out of the 10 dot chromosomes.
The repetitive PCH regions in dot-chromosomes, however, surprisingly contain a large number of duplicated protein-coding genes or lncRNAs (Fig. 5F). For instance, on chr29, an olfactory receptor gene (named OR29G) has 386 intact copies (Fig. 5 E and F). Interestingly, in seven out of 10 dot-chromosomes, the amplified genes are strongly testis-biased or testis-specific (Fig. 5G). Those testis-expressed amplicons tend to be closer to PCH boundaries (Fig. 5 F and G) and likely are still increasing their copy numbers and invading the euchromatin.
The W Chromosome as a Putative Dot Chromosome.
The Z and W chromosomes of birds evolved from an ordinary autosomal pair more than 100 Mya (45); while the Z chromosome remains the fifth largest chromosome, the W chromosome has experienced massive sequence loss, repeat accumulation, and extensive heterochromatinization (46). The length of the new W chromosome assembly reaches 14.2 Mb (SI Appendix, Fig. S15), double the size of the one of GRCg6a, and 55% larger than that of GRCg7b. The centromere has also been assembled with a strong CENP-A signal at ~7 Mb, closely linked to CNM and (TTAGGG)n repeats (SI Appendix, Fig. S16). Repetitive sequences occupy 87% of the W chromosome, including 4.9 Mb satellite DNA which is the most abundant repeat class. Interestingly, all sequence gaps on the W chromosomes are flanked by large arrays of satellite DNA (SI Appendix, Fig. S16), suggesting that some satellite DNA is still missing in the assembly. The most abundant satellite DNA is a 21-bp element (TTTTCnnnnnGAAAAnnnnnn) named sate-21, accounting for 19.7% of the W chromosome sequence. The sate-21 sequences contain a conserved dyad symmetry, similar to that in CNM (39), and they form divergent HORs (SI Appendix, Fig. S16).
Similar to some dot-chromosome amplicons, HINT1W thought to be the only multicopy gene on the chicken W chromosome was amplified through the expansion of satellite DNA. We identified a 5,648-bp sequence containing the complete sequence of HINT1W that has been tandemly duplicated 52 times (SI Appendix, Fig. S16). This satellite DNA forms two distinct clusters, suggesting two waves of HINT1W amplification (SI Appendix, Fig. S16). This region constantly shows a high GC content (53.6%), possibly driven by GC-biased gene conversion between HINT1W copies. In addition to HINT1W, we discovered another two multicopy genes: two copies of RPL17 and three copies of TXNL1 (SI Appendix, Fig. S16).
Apart from differences in gene and sequence composition between the Z and W chromosomes, the two also displayed apparent epigenetic disparity. For instance, H3K27me3 modifications are sparse on the W chromosome, but are significantly enriched on the Z chromosome (Fig. 1D). On the contrary, the W chromosome has the highest level of DNA methylation; the Z chromosome has a relatively lower methylation level though higher than other macrochromosomes and contains a ~9-Mb hypermethylated region (SI Appendix, Fig. S17) encompassing testis-expressed amplicons (47).
Discussion
A complete chicken genome (except for the W) is achieved thanks to the Nanopore ultralong and PacBio HiFi sequencing technology and a trio-binning strategy (48). The capacity of Nanopore sequencers in sequencing through high GC content regions (49, 50) allows the assembly of six dot chromosomes missing in older versions of chicken genomes. The dot chromosomes are typically acrocentric, largely compartmentalized into two parts: a compact and gene-rich euchromatin region and a hypermethylated PCH region. This compartmentalized chromatin organization, in particular a large portion of PCH, together with extremely small size (~4 Mb), makes dot chromosomes distinct from microchromosomes. Like microchromosomes, dot chromosomes evolved from ancestral vertebrate chromosomes, only losing much more genes and sequences. It has been inferred that saurischian dinosaur already had a small genome (51) and large chromosome number (52), therefore likely possessing microchromosomes, but it is unclear whether dot chromosomes had evolved. It is possible that dot chromosomes evolved as early as in Archelosauria but were fused with macrochromosomes or microchromosomes in turtles and crocodiles (12) while persisted in modern birds and possibly dinosaurs. Studying the evolutionary history of homology of bird dot chromosomes in turtles and crocodiles can potentially reveal the origin of and evolutionary fate of dot chromosomes.
Materials and Methods
Genome Sequencing.
We collected a chicken (Huxu breed) trio from a local breeder in Huizhou, Guangdong. The F1 female chick was killed for DNA extraction and long-read sequencing. For each ultralong Nanopore library, approximately 8 to 10 μg gDNA was size-selected (>50 kb) with SageHLS HMW library system (Sage Science) and processed using the Ligation sequencing 1D kit (SQK-LSK109, Oxford Nanopore Technologies). About 800 ng DNA libraries were constructed and sequenced on the Promethion platform at Grandomics Biosciences (Wuhan, China). For HiFi (CCS) sequencing, SMRTbell target size libraries were constructed according to PacBio’s standard protocol (Pacific Biosciences) using the 15-kb preparation solutions. A total amount of 15 μg DNA per muscle sample was used for the DNA library preparations. Sequencing was performed on a PacBio Sequel II instrument with Sequencing Primer V2 and Sequel II Binding Kit 2.0 at Grandomics. To prepare the Hi-C library, fixed muscle tissue was frozen in liquid nitrogen and ground to powder before resuspending in nuclei isolation buffer to obtain a suspension of nuclei. The purified nuclei were digested with 100 units of HindIII and marked by incubating with biotin-14-dCTP. The ligated DNA was sheared into 300 to 600-bp fragments and then was blunt-end repaired and A-tailed, followed by purification through biotin–streptavidin-mediated pull-down. The Hi-C libraries were then quantified and sequenced using the Illumina Hiseq platform (Illumina, San Diego).
Nanopore Direct RNA Sequencing.
Total RNA from brain and spleen tissues was extracted by grinding tissue in TRIzol reagent (TIANGEN) on dry ice and processed following the protocol provided by the manufacturer. The direct RNA libraries were prepared using the Direct RNA Sequencing Kit SQK-RNA002 (Oxford Nanopore Technologies) according to the manufacturer’s protocol. Briefly, the poly-A RNAs were enriched using the NEBNext® Poly(A) mRNA Magnetic Isolation Module (CAT#E7490, NEB) with NEBNext Magnetic Oligo (dT)25 Beads. Next, 100 to 500 ng of the poly-A enriched RNAs were ligated to the reverse transcriptase adaptor using T4 DNA ligase, followed by reverse transcription. The reverse-transcribed RNAs were ligated to the sequencing adaptor and were purified using Agencourt RNAClean XP beads (Beckman Coulter). Finally, two libraries (for brain and spleen) were constructed and sequenced on two different R9.4.1 FlowCells using the PromethION sequencer (ONT, UK) at Grandomics Biosciences (Wuhan, China).
Genome Assembly.
We used Nextdenovo (v2.4.0) to assemble the Nanopore reads into primary contigs (ont_pri) with default parameters. The contigs were then polished by the reads with Nextpolish (v1.3.1) (53). To assemble the diploid contigs, Nanopore reads were partitioned into parental and maternal using the trio mode in canu (v2.1) (25). The Nextdenovo–Nextpolish pipeline was then applied to the maternal and paternal reads, respectively, to assemble the maternal (ont_mat) and paternal (ont_pat) contigs. For PacBio HiFi reads, hifiasm (0.16.0-r369) (54) with default parameter was used to assemble the primary contigs (hifi_pri). When the lists of paternal and maternal short-reads were supplied, hifiasm was used to produce paternal (hifi_pat) and maternal (hifi_mat) contigs. We aligned hifi_pri against ont_pri and replaced the homologous sequences in ont_pri with hifi_pri sequences; when a hifi_pri contig spans the ends of two ont_pri contigs, we manually joined the ont_pri contigs, producing the first version of primary contigs (pri.v1). Following this pipeline, we produced the replaced and gap-filled contigs for paternal (pat.final) and maternal (mat.final) genomes. Finally, we used pat.final and mat.final to fill gaps in the pri.v1 assembly, producing pri.v2 (GGswu1).
Hi-C Data Analysis.
To calculate interchromosomal interactions, Hi-C read pairs were mapped to the pri.v2 assembly using Bowtie2 (2.4.4) (55), with reads uniquely mapped and having mapping quality larger than 30 kept. We then binned and normalized the alignments using HiC-Pro (v2.10.0) (56) with default parameters at 10 Kb, 40 Kb, 100 Kb, 500 Kb, and 1 Mb resolutions under ICE-normalization (57). The interchromosomal contact frequency between each chromosome pair was determined by comparing the observed Hi-C contacts between two chromosomes to the expected contacts between them following (58), using a bin size of 40 Kb. The interactions between euchromatin were estimated after excluding all pericentromeric bins. The ratios of observed vs. expected interchromosomal contacts were log2 transformed.
To visualize the Hi-C heatmap and to do scaffolding, we used a juicer (1.7.6) (59) to process Hi-C read alignments. We then visualize the Hi-C heatmap with Juicebox (1.11.08) (60) where the order and orientation of contigs can be adjusted.
Methylation.
Nanopore reads were aligned against the reference by minimap2 (v2.24) using the “map-ont” setting. We used nanopolish (v0.13.2) (61) for detecting 5-methylcytosine bases in a CpG context. We used the script calculate_methylation_frequency from the nanopolish package to calculate methylation frequency at every called site and further summarize the methylation frequency at 500-bp windows.
ChIP-seq.
ChIP assays were performed by Shanghai Jiayin Biotechnology Co., Ltd, according to the standard cross-linking ChIP protocol with modifications. Briefly, cells were harvested and cross-linker with 1% formaldehyde for 10 min at room temperature. After sonication, immunoprecipitation was performed with anti-histone H3K9me3 (Abcam, ab8898, 5 μg). The immunoprecipitated complex was washed, and DNA was extracted and purified by Universal DNA Purification Kit (#DP214). The ChIP-Seq library was prepared using a ChIP-seq DNA sample preparation kit (NEBNext® UltraTMII DNA) according to the manufacturer’s instructions. Extracted DNA was ligated to specific adaptors followed by deep sequencing in the Illumina Novaseq 6000 (Annoroad Gene Technology company). ChIP-seq for CENP-A (DRR018430) (62), H3K27me3 (SRR12697592) (63), and H3K36me3 (SRR15150478) (64) data was downloaded from NCBI SRA. We aligned the ChIP-seq reads with the BWA-MEM algorithm with options “-k 50 -c 1000000”. Alignment duplications were marked with sambamba (0.6.3) (65) and were filtered with samtools (view -q 30 -F 2308). We counted the reads with BEDTools genomecov (2.29.2) (66). ChIP/input ratios were calculated in 10-kb windows. To calculate relative H3K9me3 levels for repetitive sequences, we divided the ChIp/input ratios of repeats over those of unique sequences.
Genome Annotation.
We used Trinity (2.8.4) (67) to assemble transcripts with RNA-seq data from 10 different tissues (SI Appendix, Table S9). Protein sequences of chicken and humans were downloaded from the RefSeq database. We used maker (2.31.10) (68) to predict gene models with evidence from both assembled transcripts and protein homology. We further used the HISAT2 (2.1.0) (69)-StringTie (2.1.1) (70) pipeline to assemble the transcripts through a genome-guided method. We performed the Augustus (3.4.0) (71) gene model training through the BUSCO (4.0.5) (72) pipeline and predicted the gene models using the trained profile. For Nanopore full-length transcriptome data, we corrected the reads with TranscriptClean (v2.0.2) (73), mapped the clean reads with minimap2 (2.21-r1071, -x splice) (74), and predicted the gene models with StringTie. For PacBio full-length transcriptome sequencing data downloaded from SRA (75), we followed the IsoSeq3 pipeline to obtain clustered and nonredundant transcripts and their alignments against the genome. The abovementioned predicted gene model or aligned transcripts were integrated by EVM (1.1.1) (76) to predict gene models. We then used the PASApipeline (2.4.1) (77) to polish the gene models with the transcripts obtained from Trinity assembly and IsoSeq3. Repeats were masked with RepeatMasker (4.1.2) using an avian repeat library (78). To determine the repeat units of tandem repeats, we divided the repetitive sequences into 30-kb windows and used TideHunter (1.4.2) (79) to predict tandem repeats. StainedGlass (v0.1) (80) was used to visualize tandem repeats.
Gene Expression.
The RNA-seq datasets we used for analyses are shown in SI Appendix, Table S9. We used HiSAT2 to map raw RNA-seq reads with the options “-k 4 --max-intronlen 40000 --min-intronlen 30”. To quantify the expression level, we counted the mapped reads using featureCounts (1.6.2) (81) with options “-M -C” and calculated the TPM (transcripts per million) values. For each gene, a mean expression level across tissues was calculated. The expression breadth was measured by calculating the tau values (82). To estimate expression levels for dot-chromosomes amplicons, we calculated the mean read counts across all copies which were then normalized by the total mRNA lengths.
Vertebrate Karyotype Evolution.
We used OrthoFinder (2.5.2) (83) to group homologous genes from chicken, zebra finch (Taeniopygia guttata) (7), human, spotted gar (Lepisosteus oculatus) (84), white-spotted bamboo shark (Chiloscyllium plagiosum) (85), and amphioxus (Branchiostoma belcheri, Bb) (15). The Bb-chicken orthologous pairs were extracted from the pairwise relationships. For each Bb chromosome, we calculated the relative abundance of homologous genes for each chicken chromosome to determine the homologous chromosomes. In each orthologous group, we extracted genes from homologous chromosomes to identify ohnologs. We followed the analyses in Huang et al. (15) to reconstruct the evolutionary history of chordate chromosomes.
FISH Experiments.
Chromosome-specific oligo probes were developed using a previously published pipeline (86). The single-copy oligos (45 nt or 58 nt) were screened using the Chorus2 (1.1) software (87), followed by filtering for repeat sequence by applying ChorusNGSfilter.py and ChorusNGSselect.py script (−q 0.1; -p 0.9; −d 25, 45 or 58). The oligo library was synthesized by CustomArray (Genscript, Nanjing, China). A 23 nt forward primer (T7 RNA polymerase promoter sequence) and a 20 nt reverse primer were flanked by the synthetic oligo sequences (Dataset S2). PCR amplification of the library was performed to generate double-stranded DNA templates, and then the amplified products were transcribed into RNA. Labeling of the library was performed via reverse transcription of the RNA using 5′ digoxigenin-labeled primers. Unincorporated primers and the RNA template were degraded to obtain a labeled, single-stranded oligo probe.
Chromosome suspension was prepared according to a previously described method (52). The chromosome suspension was added dropwise onto glass slides, and the slide was air-dried and then kept at −20 °C until use. Oligo probes and chromosome slides were simultaneously denatured for 1 min on 70 °C hotplates prior to hybridization in a humidified chamber at 37 °C for 48 h. Slides were washed for 3 min in 2 × SSC, 10 min in 2 × SSC, and 3 min in 1 × PBS, respectively. Hybridization signals were detected with rhodamine-conjugated anti-digoxigenin (Roche Diagnostics, Basel, Switzerland) for digoxigenin-labeled probes. Then, the slides were dried and counterstained with DAPI. An Olympus BX53 epifluorescence microscope was used to observe metaphase plates with fluorescent signals that were photographed with a cooled CCD camera and visualized using cellSens Dimension 1.9 software (Olympus Corporation).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Acknowledgments
We thank Guojie Zhang, Timothy Sackton, Qi Zhou, and Weihao Shang for helpful comments on the manuscript and Panyi Li for her assistance in sample collections. This study is supported by the start-up grant from Southwest Univerity and FWF Erwin Schrödinger fellowship (J4477-B) to Luohao Xu. S.L. is supported by the 2115 Talent Development Program of China Agricultural University. G.C. is supported by the earmarked fund for CARS (CARS-41).
Author contributions
G.C. and Luohao Xu designed research; Z.H., Z.X., Y.H., J.L., C.Y., and Luohao Xu performed research; Z.H., Z.X., H.B., Y.H., W.C., L. Xue, X.Z., Li Xu, M.C., H.F., Y.C., T.F., S.L., G.C., and Luohao Xu contributed new reagents/analytic tools; Z.H., Z.X., H.B., N.K., X.D., J.L., H.L., C.Y., W.C., Q.G., Z.Y., T.F., S.L., G.C., and Luohao Xu analyzed data; and S.L. and Luohao Xu wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. D.W.B. is a guest editor invited by the Editorial Board.
Contributor Information
Guobin Chang, Email: gbchang@yzu.edu.cn.
Luohao Xu, Email: luohaox@gmail.com.
Data, Materials, and Software Availability
The assemblies and raw sequencing data are available under the NCBI accession PRJNA693184. All accessions are listed in the SI Appendix, Table S9. The custom codes used in the study were reposited at Github (https://github.com/lurebgi/chicken-T2T) (88). All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Zhang G., et al. , Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M., et al. , 863 genomes reveal the origin and domestication of chicken. Cell Res. 30, 693–701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bravo G. A., Jonathan Schmitt C., Edwards S. V., What have we learned from the first 500 Avian genomes? Annu. Rev. Ecol. Evol. Syst. 52, 611–639 (2021). [Google Scholar]
- 4.International Chicken Genome Sequencing Consortium, Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, 695–716 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Schmid M., et al. , Third report on chicken genes and chromosomes 2015. Cytogenet. Genome Res. 145, 78–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren W. C., et al. , A new chicken genome assembly provides insight into Avian genome structure. G3 7, 109–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhie A., et al. , Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin D. K., Robertson L. B. W., Tempest H. G., Skinner B. M., The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet. Genome Res. 117, 64–77 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Romanov M. N., et al. , Reconstruction of gross avian genome structure, organization and evolution suggests that the chicken lineage most closely resembles the dinosaur avian ancestor. BMC Genom. 15, 1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatani Y., Takeda H., Kohara Y., Morishita S., Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 17, 1254–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burt D. W., Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 96, 97–112 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Uno Y., et al. , Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS One 7, e53027 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Connor R. E., et al. , Patterns of microchromosome organization remain highly conserved throughout avian evolution. Chromosoma 128, 21–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waters P. D., et al. , Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc. Natl. Acad. Sci. U.S.A. 118, e2112494118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Z., et al. , Three amphioxus reference genomes reveal gene and chromosome evolution of chordates. bioRxiv [Preprint] (2022). 10.1101/2022.01.04.475009 (Accessed on 5 January 2022). [DOI] [PMC free article] [PubMed]
- 16.Nakatani Y., et al. , Reconstruction of proto-vertebrate, proto-cyclostome and proto-gnathostome genomes provides new insights into early vertebrate evolution. Nat. Commun. 12, 4489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McQueen H. A., Siriaco G., Bird A. P., Chicken microchromosomes are hyperacetylated, early replicating, and gene rich. Genome Res. 8, 621–630 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M., et al. , De novo assembly of 20 chicken genomes reveals the undetectable phenomenon for thousands of core genes on micro-chromosomes and sub-telomeric regions. Mol. Biol. Evol. 39, msac066 (2022), 10.1093/molbev/msac066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naish M., et al. , The genetic and epigenetic landscape of the centromeres. Science 374, eabi7489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue L., et al. , Telomere-to-telomere assembly of a fish Y chromosome reveals the origin of a young sex chromosome pair. Genome Biol. 22, 203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain M., et al. , Linear assembly of a human centromere on the Y chromosome. Nature Biotechnology 36, 321–323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vollger M. R., et al. , Segmental duplications and their variation in a complete human genome. Science 376, eabj6965 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nurk S., et al. , The complete sequence of a human genome. Science 376, 44–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao Y., Zhang G., Publisher correction: A complete, telomere-to-telomere human genome sequence presents new opportunities for evolutionary genomics. Nat. Methods 19, 899 (2022), 10.1038/s41592-022-01551-x. [DOI] [PubMed] [Google Scholar]
- 25.Koren S., et al. , De novo assembly of haplotype-resolved genomes with trio binning. Nat. Biotechnol. 36, 1174–1182 10.1038/nbt.4277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masabanda J. S., et al. , Molecular cytogenetic definition of the chicken genome: The first complete avian karyotype. Genetics 166, 1367–1373 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solinhac R., et al. , Integrative mapping analysis of chicken microchromosome 16 organization. BMC Genom. 11, 616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riddle N. C., Elgin S. C. R., The dot chromosome: Where genes flourish amidst repeats. Genetics 210, 757–772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pichugin A. M., et al. , Estimation of the minimal size of chicken Gallus gallus domesticus microchromosomes via pulsed-field electrophoresis. Russ. J. Genet. 37, 535–538 (2001). [Google Scholar]
- 30.Perry B. W., Schield D. R., Adams R. H., Castoe T. A., Microchromosomes exhibit distinct features of vertebrate chromosome structure and function with underappreciated ramifications for genome evolution. Mol. Biol. Evol. 38, 904–910 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J., et al. , A new emu genome illuminates the evolution of genome configuration and nuclear architecture of avian chromosomes. Genome Res. 31, 497–511 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Habermann F. A., et al. , Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res. 9, 569–584 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Sacerdot C., Louis A., Bon C., Berthelot C., Crollius H. R., Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 19, 166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simakov O., et al. , Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 4, 820–830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang W., et al. , Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 20, 1219–1228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piégu B., Arensburger P., Guillou F., Bigot Y., But where did the centromeres go in the chicken genome models? Chromosome Res. 26, 297–306 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Wicker T., et al. , The repetitive landscape of the chicken genome. Genome Res. 15, 126–136 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krasikova A., et al. , On the positions of centromeres in chicken lampbrush chromosomes. Chromosome Res. 14, 777–789 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Matzke M. A., et al. , A 41–42 bp tandemly repeated sequence isolated from nuclear envelopes of chicken erythrocytes is located predominantly on microchromosomes. Chromosoma 99, 131–137 (1990). [DOI] [PubMed] [Google Scholar]
- 40.Nanda I., et al. , Distribution of telomeric (TTAGGG)(n) sequences in avian chromosomes. Chromosoma 111, 215–227 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Maslova A., Zlotina A., Kosyakova N., Sidorova M., Krasikova A., Three-dimensional architecture of tandem repeats in chicken interphase nucleus. Chromosome Res. 23, 625–639 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Hori T., et al. , Constitutive centromere-associated network controls centromere drift in vertebrate cells. J. Cell Biol. 216, 101–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukagawa T., Earnshaw W. C., The centromere: Chromatin foundation for the kinetochore machinery. Dev. Cell 30, 496–508 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altemose N., et al. , Complete genomic and epigenetic maps of human centromeres. Science 376, eabl4178 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Q., et al. , Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 346, 1246338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu L., Zhou Q., The female-specific W chromosomes of birds have conserved gene contents but are not feminized. Genes 11, 1126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellott D. W., et al. , Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466, 612–616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarvis E. D., et al. , Semi-automated assembly of high-quality diploid human reference genomes. Nature 611, 519–531 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Browne P. D., et al. , GC bias affects genomic and metagenomic reconstructions, underrepresenting GC-poor organisms. Gigascience 9, giaa008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo H., et al. , Retrieving the near-complete genome of a threatened bird from wild frozen samples. bioRxiv [Preprint] (2022). 10.1101/2022.12.22.521577 (Accessed 22 December 2022). [DOI]
- 51.Organ C. L., Shedlock A. M., Meade A., Pagel M., Edwards S. V., Origin of avian genome size and structure in non-avian dinosaurs. Nature 446, 180–184 (2007). [DOI] [PubMed] [Google Scholar]
- 52.O’Connor R. E., et al. , Reconstruction of the diapsid ancestral genome permits chromosome evolution tracing in avian and non-avian dinosaurs. Nat. Commun. 9, 1883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J., Fan J., Sun Z., Liu S., NextPolish: A fast and efficient genome polishing tool for long-read assembly. Bioinformatics 36, 2253–2255 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Cheng H., Concepcion G. T., Feng X., Zhang H., Li H., Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Servant N., et al. , HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imakaev M., Fudenberg G., McCord R. P., Naumova N., Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nature 9, 999–1003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., et al. , Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148, 908–921 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Durand N. C., et al. , Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durand N. C., et al. , Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loman N. J., Quick J., Simpson J. T., A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 12, 733–735 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Hori T., et al. , Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev. Cell 29, 740–749 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kern C., et al. , Functional annotations of three domestic animal genomes provide vital resources for comparative and agricultural research. Nat. Commun. 12, 1821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tarsalainen A., et al. , Ig enhancers increase RNA polymerase II stalling at somatic hypermutation target sequences. J. Immunol. 208, 143–154 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tarasov A., Vilella A. J., Cuppen E., Nijman I. J., Prins P., Sambamba: Fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grabherr M. G., et al. , Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holt C., Yandell M., MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pertea M., et al. , StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stanke M., et al. , AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Wyman D., Mortazavi A., TranscriptClean: Variant-aware correction of indels, mismatches and splice junctions in long-read transcripts. Bioinformatics 35, 340–342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ren J., Sun C., Clinton M., Yang N., Dynamic transcriptional landscape of the early chick embryo. Front. Cell Dev. Biol. 7, 196 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haas B. J., et al. , Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haas B. J., et al. , Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peona V., et al. , Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird-of-paradise. Mol. Ecol. Resour. 21, 263–286 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao Y., Liu B., Wang Y., Xing Y., TideHunter: Efficient and sensitive tandem repeat detection from noisy long-reads using seed-and-chain. Bioinformatics 35, i200–i207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vollger M. R., Kerpedjiev P., Phillippy A. M., Eichler E. E., StainedGlass: Interactive visualization of massive tandem repeat structures with identity heatmaps. Bioinformatics 38, 2049–2051 (2022), 10.1093/bioinformatics/btac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Yanai I., et al. , Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21, 650–659 (2005). [DOI] [PubMed] [Google Scholar]
- 83.Emms D. M., Kelly S., OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genom. Biol. 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Braasch I., et al. , The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 48, 427–437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y., et al. , The white-spotted bamboo shark genome reveals chromosome rearrangements and fast-evolving immune genes of cartilaginous fish. iScience 23, 101754 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Albert P. S., et al. , Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc. Natl. Acad. Sci. U.S.A. 116, 1679–1685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang T., Liu G., Zhao H., Braz G. T., Jiang J., Chorus2: Design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization. Plant Biotechnol. J. 19, 1967–1978 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang, et al. Chicken T2T custom scripts, Github, https://github.com/lurebgi/chicken-T2T, deposited on Jul 23, 2022
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Data Availability Statement
The assemblies and raw sequencing data are available under the NCBI accession PRJNA693184. All accessions are listed in the SI Appendix, Table S9. The custom codes used in the study were reposited at Github (https://github.com/lurebgi/chicken-T2T) (88). All study data are included in the article and/or SI Appendix.