Abstract
Global metagenomic surveys have revealed that bats host a diverse array of paramyxoviruses, including species from at least five major genera. An essential determinant of successful spillover is the entry of a virus into a new host. We evaluate the role of receptor usage in the zoonotic potential of bat-borne henipaviruses, morbilliviruses, pararubulaviruses, orthorubulaviruses, and jeilongviruses; successful spillover into humans depends upon compatibility of a respective viral attachment protein with its cognate receptor. We also emphasize the importance of post-entry restrictions in preventing spillover. Metagenomics and characterization of newly identified paramyxoviruses have greatly improved our understanding of spillover determinants, allowing for better forecasts of which bat-borne viruses may pose the greatest risk for cross-species transmission into humans.
Keywords: Bat paramyxovirus, viral entry, zoonosis, spillover, receptor usage
Graphical Abstract:
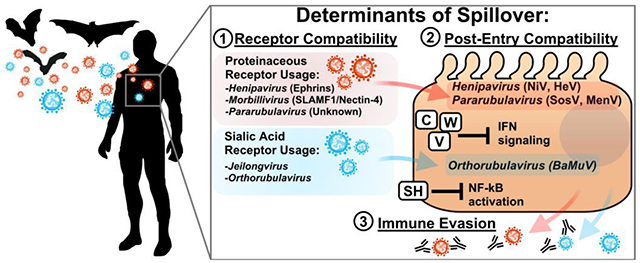
Successful spillover necessitates: (1) Compatibility of the virus receptor binding protein with its cognate receptor in a new host. Some paramyxoviruses, such as henipaviruses, morbilliviruses, and pararubulaviruses utilize proteinaceous receptors for entry; however, others, such as the orthorubulaviruses and jeilongviruses utilize sialic acids for entry. Efficient use of a target receptor is necessary for entry into the cell. (2) Post-entry compatibility between the viral life cycle and host factors, and ability of the virus to evade the innate immune system. Most paramyxoviruses encode proteins which antagonize the innate immune response; these include the C, V, and W genes, which can inhibit interferon signaling. Some species, such as members of Jeilongvirus and Orthorubulavirus, also encode SH proteins, which inhibit NF-kB signaling. Henipaviruses and pararubulaviruses have successfully spilled over from bats into humans, demonstrating post-entry compatibility. Experimental characterization of the bat mumps virus (BaMuV) has likewise demonstrated compatibility at the post-entry level in human cells. (3) Effective evasion of the immune response. Routine vaccination against measles virus and mumps virus generates broadly neutralizing antibodies which may prevent zoonosis of antigenically-similar species. Both bat-borne and morbilliviruses (MBaMV) and orthorubulaviruses (BaMuV) can be neutralized by sera from vaccinated individuals. A successful spillover requires fulfillment of all 3 of the above determinants.
Introduction
Across the globe, bats are the reservoir hosts of a myriad of RNA viruses, including paramyxoviruses (PMVs). Metagenomic surveys have revealed the great diversity of paramyxoviruses harbored by bats, including members of the genera henipavirus, pararubulavirus, orthorubulavirus, morbillivirus, and jeilongvirus[1–4]. With one of the highest rates of cross-species transmission among RNA viruses, paramyxoviruses are poised for zoonosis[5]. Concerningly, some deadliest viruses that have been identified to date include the bat-borne henipaviruses, which are highly adept at traversing the species barrier. Further, at least two bat-borne pararubulaviruses have caused severe disease in humans[6,7]. Mounting serological evidence has likewise indicated that unrecognized spillover events have occurred[8–10]. Receptor usage is a major determinant of the host range of zoonotic PMVs and their capacity for spillover[11]. However, post-entry determinants are likewise important for productive infection. Here, we summarize the receptor usage of diverse bat-borne paramyxoviruses, their potential to use human receptors, and the role of entry in zoonosis.
Paramyxoviruses have evolved separate proteins for the respective roles of (1) attachment and (2) entry into cells. The PMV receptor binding protein (RBP) forms a six-bladed β-propeller fold, and facilitates the binding of virions to the cell surface through interactions with a respective receptor[12,13]. Binding of a RBP to its cognate receptor results in triggering of the fusion (F) protein, which facilitates membrane fusion and concomitant entry of the viral genome into the cell. Triggering of F is dependent upon the successful interaction of a RBP with its respective receptor, and thus the fusion and subsequent entry of a given PMV necessitates the presence of a compatible receptor on the cell surface[14]. Attachment and entry are absolute requirements for a successful paramyxovirus spillover event, and concomitantly, receptor usage is a major determinant of the species tropism for a given virus[11].
Henipaviruses utilize highly-conserved ephrins for cell entry, maximizing opportunities for spillover.
By utilizing highly-conserved molecules as receptors, certain bat-borne paramyxoviruses, such as the henipaviruses, have effectively maximized the breadth of potential species which may serve as intermediate hosts. The prototypical henipaviruses, Nipah virus (NiV) and Hendra virus (HeV), are highly pathogenic in humans, causing severe encephalitic and respiratory disease with mortality rates upwards of 40%[15,16]. NiV, HeV, and the closely related, albeit non-pathogenic Cedar virus (CedV) circulate within bats in Southeast Asia and Australia[2,17–19]. Metagenomic surveys have further identified additional henipavirus species from bats, including Ghana virus (GhV), and more recently, Angavokely virus (AngV)[1,20]. While beyond the scope of this review, it is worth noting that metagenomic surveys have also recently identified several species of shrew- and rodent-borne paramyxoviruses which share a relatively close phylogenetic relationship with the bat-borne henipaviruses[21–24]. However, there is accumulating evidence that these viruses form a distinct clade, as they: (1) Do not utilize ephrins for entry[25,26]; (2) Possess unique genome structures[23]; and (3) circulate within non-chiropteran host reservoirs. Thus, such “henipa-like” viruses may require taxonomic reclassification.
The RBPs of NiV, HeV, GhV, and CedV all use the highly-conserved ephrin-B2 (EFNB2) as a receptor[27–30]. However, NiV and HeV also utilize ephrin-B3 (EFNB3)[31]. Further, CedV can make use of ephrin-B1 (EFNB1) in relevant primary cells such as HUVECs; in transfected cells, ephrin-A2 (EFNA2) and ephrin-A5 (EFNA5) have likewise been demonstrated as low-efficiency receptors, but the biological relevance of these remains unclear[30,32]. EFNB2 expression in neurons, endothelial cells, and smooth muscle in arterial vessels corresponds with the tissue tropism and pathology of NiV and HeV infections in humans. EFNB3 is expressed higher in the central nervous system (CNS) with expression profiles that are both distinct and overlapping with EFNB2. Expression of EFNB2 and EFNB3 permit NiV and HeV infection of the CNS, resulting in severe encephalitic disease[33]. EFNB3-blind henipaviruses, such as GhV, are likely to be less neuropathogenic. CedV, which utilizes EFNB1 and EFNB2, is non-pathogenic in multiple small-animal models, and it remains unclear how receptor usage corresponds to its tissue tropism and disease[19,34].
The receptor usage of AngV remains to be characterized, but in silico analysis demonstrates that it lacks several of the conserved ephrin-binding residues which are present in all other known bat-borne henipaviruses[20]. Thus, AngV may represent a new clade of henipavirus with differential receptor usage. Indeed, homology modeling of AngV-RBP (Fig 1A) and comparison with the sequences of NiV, HeV, CedV, and GhV-RBP (Fig 1B) demonstrate that AngV is missing key residues for EFNB2 recognition that are present in other henipaviruses[27,29,30,35]. Comparison of the respective interfaces between EFNB2 and the RBPs of NiV, CedV, and GhV demonstrates that the corresponding regions of AngV-RBP are highly dissimilar. Importantly, conserved residues which are required for interaction with EFNB2 in these binding pockets, indicated in magenta, are entirely absent in AngV-RBP (Fig 1A, 1B). Thus, AngV-RBP is not predicted to effectively recognize EFNB2 as an entry receptor. Until its entry receptor is characterized, the zoonotic potential of AngV remains unclear.
Figure 1. Comparison of bat-borne henipavirus RBP ephrin-binding residues.
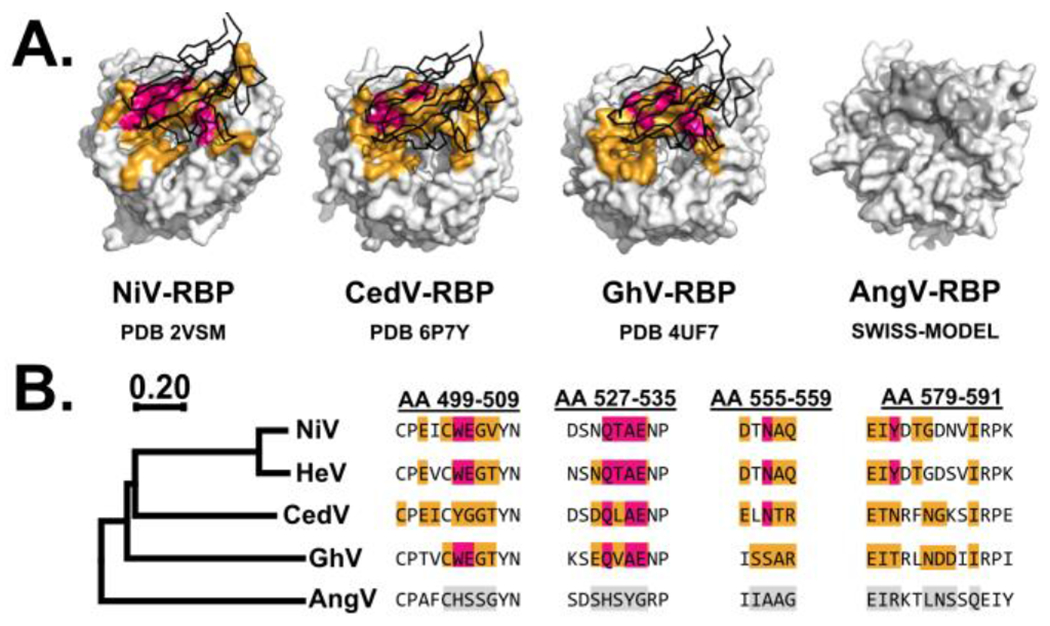
(A) Structures of NiV-RBP, CedV-RBP, and GhV-RBP bound to EFNB2 (PDB 2VSM, 6P7Y, and 4UF7 respectively). A homology model of AngV-RBP was generated via SWISS-MODEL by modeling AngV-RBP to 2VSM, 6P7Y, and 4UF7 and comparing resultant root mean square deviation (RMSD) values. Using 4UF7 as template yielded the lowest RMSD of 0.192, suggesting that GhV-RBP is the most compatible template for modeling AngV-RBP. (B) Sequence alignment of henipavirus RBP. Alignments were conducted using Clustal Omega with residue positions numbered in reference to the sequence of NiV-RBP. The phylogenetic tree was generated in MEGAX using the neighbor-joining method, with branch length representing amino acid substitutions by site. For (A) and (B), an orange coloration depicts occluded residues in the interface formed between respective RBP and EFNB2, which was predicted using PDBePISA. Residues highlighted in magenta are conserved residues required for binding of RBP to EFNB2. Gray coloration reflects residues in AngV-RBP which correspond by alignment/position with the binding pocket of the template model (4UF7). EFNB2 is shown in black.
Ephrins are highly conserved throughout mammals, allowing henipaviruses to infect a wide range of species; indeed, natural infections of NiV and HeV have been reported in bats, pigs, horses, dogs, and humans[15,33,36]. Likewise, experimental studies have demonstrated that ephrins from diverse species are capable of facilitating entry for NiV and HeV[37,38]. Due to the high level of receptor conservation, the ephrin-using henipaviruses will require little adaptation for entry into a human host. Although absent in humans, post-entry restrictions can protect against prototypical henipavirus infection in some hosts; for example, while mouse EFNB2 and EFNB3 can be utilized by both NiV and HeV for entry, infected mice do not develop productive infection nor manifest disease, whereas type I interferon receptor knockout mice succumb to infection[37–39]. The henipavirus V, W, and C proteins, which antagonize interferon signaling responses, have been demonstrated to be important virulence factors, as knockout of V or C can attenuate infection in susceptible animal models[40,41]. Similarly, CedV, which lacks an RNA editing site for transcribing innate immune antagonists, does not cause disease in ferrets, guinea pigs, nor hamsters[19,34]. Collectively, these findings emphasize an apparent requirement for effective control of the interferon signaling response post-entry to achieve productive henipavirus infection.
Bat-borne morbilliviruses require adaptation to use human SLAMF1.
Morbilliviruses characteristically recognize two receptors for entry: (1) Signaling lymphocytic activation molecule (SLAMF1/CD150) for entry into immune cells, and (2) nectin-4 for entry into polarized epithelial cells[42–46]. While the amino acid sequence of SLAMF1 varies among species, nectin-4 is relatively conserved. Species differences in SLAMF1 have driven a host-specific evolution of morbillivirus RBPs, which concomitantly possess inefficient usage of SLAMF1 outside of their respective reservoir hosts[47]. This limited recognition of ‘foreign’ SLAMF1 has been argued to restrict cross-species transmission of morbilliviruses, as immune cells in the upper respiratory tract are critical initial targets for productive infection[43,48]. Following intense, SLAMF1-dependent replication in lymphatic organs, nectin-4 is subsequently utilized for virus exit. Nectin-4 mediates the infection of lung epithelia from the basal-lateral surface, where virus is then shed into the airways[46]. Thus, despite its high conservation, nectin-4 usage alone is not suspected to be a driver of spillover for the morbilliviruses.
Canine distemper virus (CDV), however, is an exception to the species-restriction of morbilliviruses. CDV possesses an unusually broad host range for a morbillivirus, and is capable of infecting a diverse array of carnivores, and even some non-carnivores[47,49]. Further, CDV has spilled over into non-human primates, causing outbreaks with lethal disease[50,51]. Relatively few mutations in the RBP of CDV, as well as in the RBP of Peste des petits ruminants virus (PPRV), respectively, can confer these viruses with improved recognition of human SLAMF1[52–54]. As CDV can adapt to use human SLAMF1, there is concern that unvaccinated humans may serve as potential hosts[47]. In the event of animal morbillivirus spillover into humans, there is evidence that adaptive immunity in the population could hinder widespread transmission of zoonotic morbilliviruses; routine MeV immunization is known to induce cross-protective immunity against diverse morbilliviruses[53,55,56]. As such, continued measles vaccination efforts at a population level, even in the event of MeV eradication, could have a tremendous value in preventing zoonotic morbillivirus outbreaks[47].
Metagenomic surveys have detected morbillivirus RNA from bats, including two full-length sequences to date; these full-length species include Myotis bat morbillivirus (MBaMV) whose sequence was obtained from a Myotis riparius bat, and PBZ-1381, whose sequence was obtained from a Phyllostomus hastatus bat[1,4]. Employing a reverse genetics approach, we have successfully rescued MBaMV. Experimental characterization of MBaMV demonstrates that it preferentially uses Myotis SLAMF1 and possesses relatively poor usage of human SLAMF1[57]. To better understand incompatibilities between MBaMV-RBP and human SLAMF1, structural homology modeling was employed using the crystal structure of measles virus (MeV) RBP with marmoset SLAMF1 (PDB 3ALW) as a template on the SWISS-MODEL web server[58,59] (Fig 2A). All contact residues of human SLAMF1 with MeV-RBP are conserved between human and marmoset SLAMF1, with a single residue exception[59]. Analysis of the predicted interface between SLAMF1 with the respective attachment proteins of MeV, MBaMV, and PBZ-1381 was conducted using PDBepisa[60]. In agreement with our experimental findings, we observe that MBaMV RBP is predicted to be unable to form two of the three salt bridges that are present between MeV-RBP and marmoset SLAMF1 (Fig 2A and 2B). PBZ-1381, however, is predicted to form at least two of these salt-bridge interactions with SLAMF1 (Fig 2A and B). Concerningly, a single point mutation in PBZ-1381 RBP may be capable of restoring the absent salt-bridge (Fig 2B). Based on these models, PBZ-1381 RBP is predicted to be more compatible with human SLAMF1 than is MBaMV-RBP. While spillover of bat morbilliviruses will undoubtedly require adaptation of the RBP for improved recognition of human SLAMF1, species such as PBZ-1381, like CDV, may require relatively few mutations to gain usage of human SLAMF1.
Figure 2. Predicted compatibility of bat-borne morbillivirus RBPs with human SLAMF1.
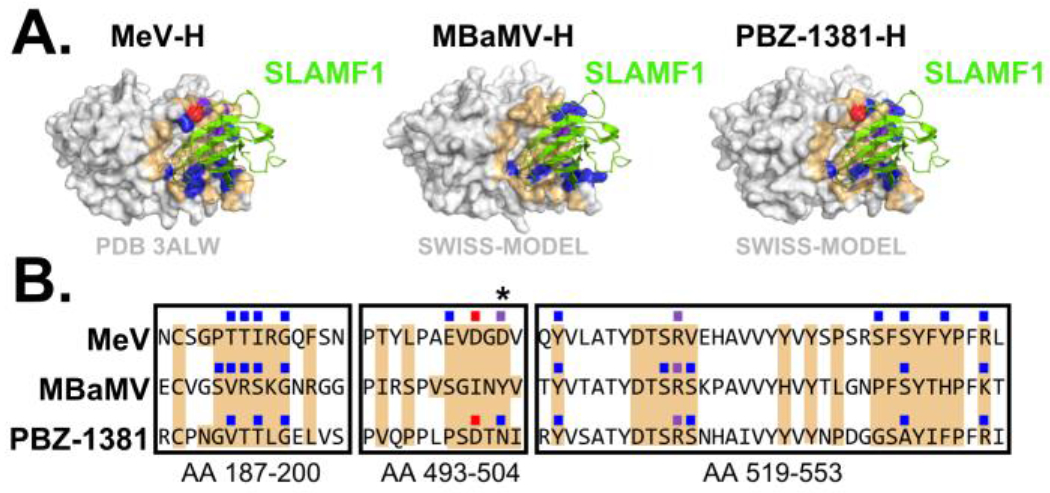
(A) MeV-RBP bound to SLAMF1 (PDB 3ALW) and homology modeling of the RBP from MBaMV and PBZ-1381. Models were made in SWISS-MODEL using 3ALW as template. RMSD values were 0.167 for MBaMV RBP and 0.143 for PBZ-1381 RBP. (B) Sequence alignments of the RBP from MeV, MBaMV, and PBZ-1381. Alignments were conducted using Clustal Omega with residue positions in reference to the PBZ-1381 RBP. Coloration depicts modeled interactions between each respective RBP and marmoset SLAMF1: Orange = occluded residue; Blue = hydrogen bond; Red = salt-bridge; Purple = hydrogen bond and salt bridge. Molecular interfaces were determined using PDBePISA. An asterisk (*) denotes a position in PBZ-1381 RBP in which the point mutation N503D could putatively confer usage of human/marmoset SLAMF1.
Pararubulaviruses use unidentified proteinaceous receptors that are expressed in the small intestines and secondary lymphoid tissue of mammals.
Pararubulaviruses have been identified exclusively in bats throughout Africa, Asia, and Australia[2,6,7,9,61,62]. To date, there are three documented cases of spillover of pararubulaviruses from bats into humans: In 1997, Menangle virus (MenV) spilled over from Pteropus bats into sows at a piggery in Australia, resulting in reproductive failure and severe malformations in stillborn piglets[6,63]. During this outbreak, two individuals with occupational exposure to pigs presented with fever, influenza-like symptoms, and a nonpruritic maculopapular rash[64]. Following the identification and isolation of MenV from infected piglets, serological studies of more than 250 laborers at the piggery revealed that only the two symptomatic individuals had seroconverted, implicating MenV as the etiological agent[64]. In 2012, a field biologist collecting biological specimens in South Sudan and Uganda fell ill with a fever, nasopharyngeal ulcerations, and rash. Sosuga virus (SosV), a novel pararubulavirus, was isolated from the blood of this patient[65]. Serological analysis of field specimens collected by the patient in the weeks immediately prior to symptom onset revealed that Rousettus aegypticus bats were the reservoir host of SosV[7]. Serological surveys of wildlife and human populations in close proximity to bat reservoirs of other pararubulaviruses have further suggested that there are undocumented instances of pararubulavirus spillover: Analysis of 169 human sera samples from Tioman Island, Malaysia, revealed that 3% of individuals were seropositive against Tioman virus (TioV)[8]. Similarly, sera from human donors from Ghana and Tanzania were found to neutralize Achimota virus 2 (AchiV-2)[9].
Although the pararubulaviruses are phylogenetically related to the sialic-acid using orthorubulaviruses, critical analysis of the RBP sequence revealed that these ‘rubula-like’ viruses lacked the characteristic ‘NRKSCS’ sialidase hexapeptide motif conserved among the sialic-acid using paramyxoviruses[66–68]. Further, experimental evidence has demonstrated that diverse pararubulaviruses, including MenV, Tioman virus (TioV), Teviot virus (TeV), and SosV, are not dependent upon sialic acids for cell entry[68,69]. These findings have resulted in the creation of the Pararubulavirus genus, distinguishing these viruses from the related orthorubulaviruses[70,71]. Of all pararubulaviruses identified to date, only the RBP of AchiV-2 encodes all seven of the conserved sialic acid active site residues; however, none of the species encode a complete sialidase hexapeptide motif. (Fig 3)[72,73]. Thus, proteinaceous receptors are suspected to be the targets of pararubulavirus RBP. The receptor(s) used by pararubulaviruses must possess relatively high sequence conservation across mammalian species as bats, pigs, and humans are all susceptible to natural pararubulavirus infection without adaptation of the RBP. Experimental infection of pigs with MenV, bats with SosV, and guinea pigs and ferrets with AchiV-1 and AchiV-2 have collectively revealed a tropism in which the small intestines and secondary lymphoid organs are major sites of viral replication[74–77]. Due to this shared tissue tropism, it is likely that receptor usage is conserved across the genus. The identification of such receptors will greatly enrich our understanding of the zoonotic potential of pararubulaviruses.
Figure 3. Conservation of the sialic acid active site and sialidase hexapeptide motif across diverse bat-borne paramyxovirus RBPs.
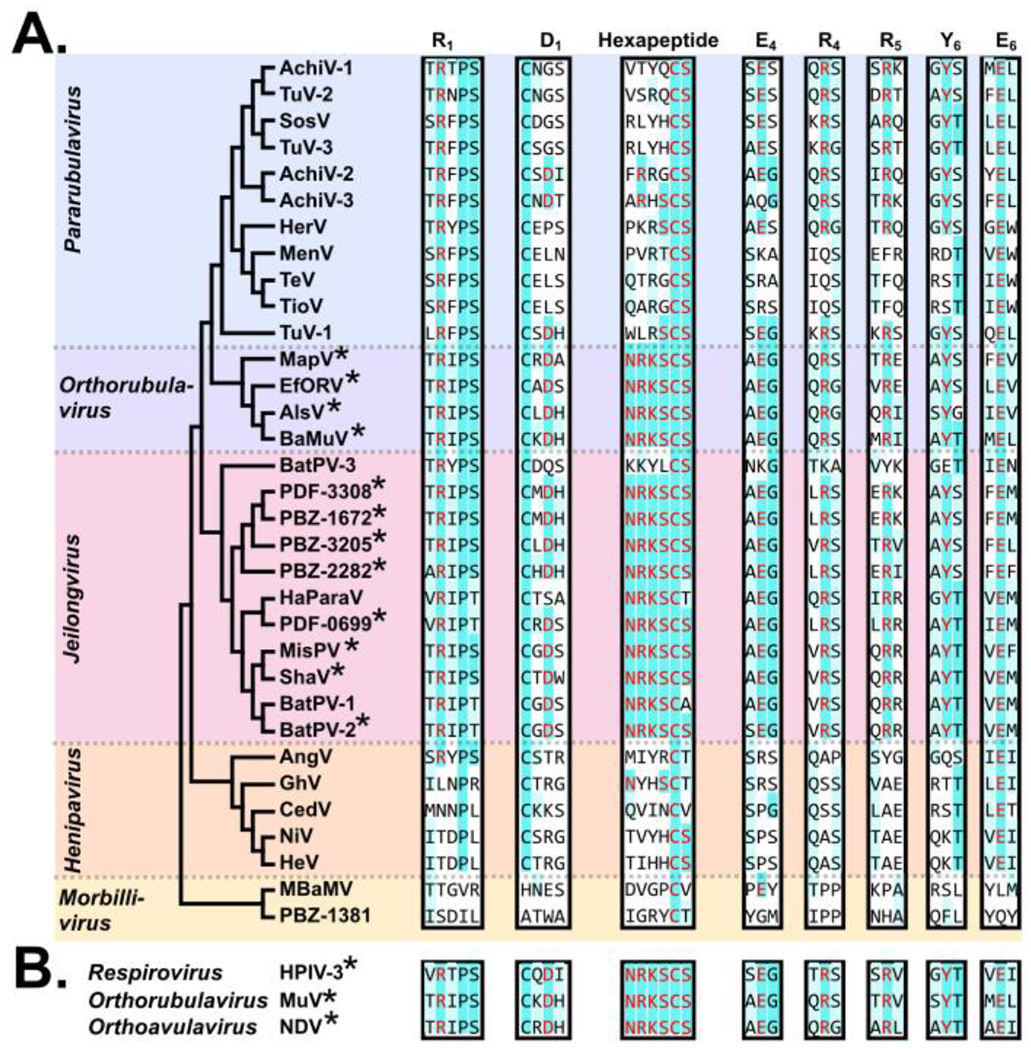
Sequence alignment of RBP from diverse bat-borne paramyxoviruses was conducted in MegaX using Clustal Omega. The phylogenetic tree demonstrates topology only, and was generated in MegaX using the neighbor-joining method. The seven conserved sialic acid active site residues and hexapeptide motif are labeled by residue and blade location. An asterisk (*) denotes that a respective species encodes all seven conserved residues and a full hexapeptide motif. Coloration represents comparison of a respective species to known sialic acid using viruses (HPIV-3, MuV, and NDV), with red = an exact match to a given active site or hexapeptide motif residue, cyan = residues conserved across all three sialic-acid-using viruses at a given position, and light-blue = a biochemically-similar amino acid relative to one or more of the sialic-acid using viruses at a given position.
Bat-borne Orthorubulaviruses and Jeilongviruses use sialic acids as entry receptors.
Some paramyxoviruses recognize sialic acids as entry receptors, as opposed to proteinaceous receptors. The RBP of all known sialic-acid-using paramyxoviruses encode a highly conserved sialic-acid binding site, as well as the neuraminidase hexapeptide motif which is critical for virus release[72,78–80]. These residues are not conserved across protein-using paramyxoviruses, and their presence is an indicator of sialic-acid usage (Fig 3). Paramyxoviruses of the genus Orthorubulavirus encode all seven key residues of the sialic acid binding site as well as a full NRKSCS hexapeptide motif, and have been demonstrated to use sialic acids for cell entry[81]. Orthorubulaviruses have been identified in bats, including a species with high sequence similarity to the human pathogen mumps virus (MuV)[1,82]. The bat mumps virus (BaMuV) has been rescued using reverse genetics, and was found to efficiently infect human cells[83]. Similar to the MBaMV, human sera was found to neutralize BaMuV, suggesting that routine MuV immunization in humans can afford cross-protective immunity against some zoonotic orthorubulavirus species[83–85].
Jeilongviruses were initially identified in rodents; however, numerous related species have since been identified in bats[3,4,86–88]. Differences in host range, genome structure, and phylogenetic relatedness have resulted in proposals for a re-classification of bat-borne jeilongviruses into a new genus, Shaanvirus[4,87]. Characterization of bat-borne jeilongviruses have revealed that the RBP of most members possess both hemmagglutinin and neuraminidase activities, implicating the use of sialic acids as receptors[3,86,89]. In agreement with experimental findings, sequence alignment of bat-borne jeilongvirus attachment proteins reveals conservation of the motifs required for both sialic acid recognition and sialidase activity across diverse species; however, bat paramyxovirus 3 (BatPV-3) appears to be an exception, as its RBP does not encode a functional hexapeptide motif nor key residues for sialic acid binding (Fig 3). At least four other jeilongviruses have been identified which lack both the sialidase hexapeptide and sialic acid binding motifs[3]. Thus, a subset of bat-borne jeilongviruses might employ a sialic-acid-independent means of attachment, requiring further characterization to better elucidate their respective requirements for entry.
The zoonotic potential of bat-borne orthorubulaviruses and jeilongviruses remains uncertain, as there are no documented spillovers of these species into humans. However, there is phylogenetic evidence of jeilongvirus host-switching between bats and rodents[88]. As sialic acid structures are common amongst all mammals, the species restriction of the sialic-acid using paramyxoviruses will be heavily dependent upon the post-entry compatibility of newfound hosts with paramyxovirus replication.
Successful zoonosis requires post-entry compatibility with the biology of a new host.
While many zoonotic viruses are well-equipped to ‘sample’ a diverse range of potential hosts, successful entry into a host cell is not always sufficient for productive replication and spread. Not all cells are equally amenable to viral replication, as specific host factors can either promote or inhibit the viral life cycle. Following successful entry, the innate immune response and restriction factors may hinder viral replication and prevent secondary transmission[90]. To overcome these hurtles, paramyxoviruses often encode accessory proteins that have evolved to effectively antagonize the innate immune system of their natural hosts, and are critical for overcoming cellular antiviral responses[91]. These accessory proteins, such as the p-editing products V and W, or the small hydrophobic (SH) proteins, must be effective in a newfound host for a productive infection to occur.
The henipavirus accessory proteins are capable of efficiently antagonizing the human toll-like receptor and interferon pathways, and contribute towards pathogenicity during spillover[19,41,92]. Despite receptor compatibility, intraperitoneal infection of wild-type laboratory mouse strains with NiV does not cause encephalitis; however, NiV infection of IFNAR knock-out mice results in neurological disease, demonstrating an inability of NiV to effectively antagonize the mouse innate immune system[38,39,93]. Similarly, CedV, which cannot make V nor W, fails to cause disease in numerous small animal models[19,34]. In addition to the henipaviruses, pararubulaviruses have been documented to cause disease in humans[64,65]. While the V and W proteins of pararubulaviruses have not been extensively characterized, the pathogenesis during human infection suggests that there is effective antagonization of human innate immune signaling.
Because sialic acid molecules are conserved across all species, one would anticipate that the sialic-acid using paramyxoviruses would be adept at spillover. Despite this expectation, to date there have been no major outbreaks of sialic-acid using paramyxoviruses from animals to humans, suggesting post-entry incompatibilities with newfound hosts. For example, while human and bovine parainfluenza virus 3 (HPIV-3 and BPIV-3, respectively) are antigenically and genetically similar, BPIV-3 replication is attenuated in humans while HPIV-3 results in disease[94,95]. Similarly, experimental infection of MuV in mice fails to overcome rodent innate immunity, despite causing disease in human hosts[96,97]. It is worth noting, however, that characterization of the V protein of BaMuV has demonstrated that it can effectively block human interferon signaling, suggesting that BaMuV could be pathogenic in humans[83]. Collectively, these phenomena demonstrate the need for post-entry compatibility of paramyxovirus accessory proteins with their targets in newfound hosts.
Conclusions
Many bat-borne paramyxoviruses have evolved to exploit broadly-conserved molecules as receptors. Because ephrins are highly conserved across species, the ephrin-using henipaviruses can easily traverse the species barrier to explore compatibility with new hosts. Although the receptor usage of the pararubulaviruses remains uncharacterized, their broad species range and conserved tissue tropism suggests that members of this genus likewise recognize well-conserved proteinaceous mammalian receptor(s). Thus, the spillover of bat-borne henipaviruses and pararubulaviruses into humans is unlikely to be restricted at the level of entry. By contrast, the bat-borne morbilliviruses have evolved to recognize the less-conserved SLAMF1 as a primary receptor; consequently, morbilliviruses are more restricted to their reservoir hosts. Bat morbilliviruses will likely require adaptation of the RBP for efficient use of human SLAMF1; however, some species, such as PBZ-1381, may require relatively few mutations and could be more readily poised for spillover into humans. While entry is an absolute requirement for spillover, it does not guarantee productive replication. The host environment must be amenable to viral infection, with host factors that are compatible with the viral life cycle. Paramyxoviruses must be able to evade host antiviral responses. Incompatibilities between viral accessory proteins and their targets will impair productive replication and limit the potential species tropism. Such post-entry restriction appears to be protective against the sialic-acid using paramyxoviruses; despite utilizing a broadly conserved receptor, the bat-borne orthorubulaviruses and jeilongviruses have not been observed to cause disease in humans. However, BaMuV, which has been demonstrated to effectively evade human innate immunity in vitro, may still pose a risk for spillover[83]. The continued identification and characterization of bat-borne paramyxoviruses, both at entry and post-entry stages, will be key for effective pandemic prevention and preparedness.
Acknowledgements.
This work is supported in part by National Institutes of Health (USA) R01AI123449 and the Bill and Melinda Gates Foundation Pandemic Antiviral Discovery (PAD) initiative for henipavirus to B.L. G.D.H is supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. 1842169. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. We apologize to any authors whose work could not be included in this article due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Annotated Bibliography.
- 1.**.Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Cottontail VM, Rasche A, Yordanov S, et al. Bats host major mammalian paramyxoviruses. Nat Commun 2012, 3:796. [DOI] [PMC free article] [PubMed] [Google Scholar]; An extensive metagenomic survey identified 66 novel bat- and rodent-borne paramyxoviruses, including a bat mumps virus and a Ghanaian bat-borne henipavirus.
- 2.Barr J, Smith C, Smith I, de Jong C, Todd S, Melville D, Broos A, Crameri S, Haining J, Marsh G, et al. Isolation of multiple novel paramyxoviruses from pteropid bat urine. J Gen Virol 2015, 96:24–29. [DOI] [PubMed] [Google Scholar]
- 3.*.Larsen BB, Gryseels S, Otto HW, Worobey M: Evolution and diversity of bat and rodent Paramyxoviruses from North America. J Virol 2022, 96:e0109821. [DOI] [PMC free article] [PubMed] [Google Scholar]; Metagenomic survey that identified previously undescribed jeilongviruses and members of the proposed genus shaanvirus in North American bats and rodents.
- 4.*.Wells HL, Loh E, Nava A, Solorio MR, Lee MH, Lee J, Sukor JRA, Navarrete-Macias I, Liang E, Firth C, et al. Classification of new morbillivirus and jeilongvirus sequences from bats sampled in Brazil and Malaysia. Arch Virol 2022, doi: 10.1007/s00705-022-05500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identified novel bat-borne paramyxoviruses, including jeilongviruses and two fully sequenced bat-borne morbilliviruses.
- 5.Kitchen A, Shackelton LA, Holmes EC: Family level phylogenies reveal modes of macroevolution in RNA viruses. Proc Natl Acad Sci U S A 2011, 108:238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philbey AW, Kirkland PD, Ross AD, Davis RJ, Gleeson AB, Love RJ, Daniels PW, Gould AR, Hyatt AD: An apparently new virus (family Paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg Infect Dis 1998, 4:269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amman BR, Albariño CG, Bird BH, Nyakarahuka L, Sealy TK, Balinandi S, Schuh AJ, Campbell SM, Ströher U, Jones MEB, et al. A Recently Discovered Pathogenic Paramyxovirus, Sosuga Virus, is Present in Rousettus aegyptiacus Fruit Bats at Multiple Locations in Uganda. J Wildl Dis 2015, 51:774–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yaiw KC, Crameri G, Wang L, Chong HT, Chua KB, Tan CT, Goh KJ, Shamala D, Wong KT: Serological evidence of possible human infection with Tioman virus, a newly described paramyxovirus of bat origin. J Infect Dis 2007, 196:884–886. [DOI] [PubMed] [Google Scholar]
- 9.Baker KS, Todd S, Marsh GA, Crameri G, Barr J, Kamins AO, Peel AJ, Yu M, Hayman DTS, Nadjm B, et al. Novel, potentially zoonotic paramyxoviruses from the African straw-colored fruit bat Eidolon helvum. J Virol 2013, 87:1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pernet O, Schneider BS, Beaty SM, LeBreton M, Yun TE, Park A, Zachariah TT, Bowden TA, Hitchens P, Ramirez CM, et al.: Evidence for henipavirus spillover into human populations in Africa. Nat Commun 2014, 5:5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeltina A, Bowden TA, Lee B: Emerging Paramyxoviruses: Receptor Tropism and Zoonotic Potential. PLoS Pathog 2016, 12:e1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowden TA, Crispin M, Jones EY, Stuart DI: Shared paramyxoviral glycoprotein architecture is adapted for diverse attachment strategies. Biochem Soc Trans 2010, 38:1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plemper RK, & Lamb RA: Paramyxoviridae: The Viruses and Their Replication. In Fields Virology: Emerging Viruses. Edited by Howley PM, Knipe DM. Lippincott Williams & Wilkins; 2020. [Google Scholar]
- 14.Navaratnarajah CK, Generous AR, Yousaf I, Cattaneo R: Receptor-mediated cell entry of paramyxoviruses: Mechanisms, and consequences for tropism and pathogenesis. J Biol Chem 2020, 295:2771–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eaton BT, Broder CC, Middleton D, Wang L-F: Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol 2006, 4:23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh GA, Wang L-F: Hendra and Nipah viruses: why are they so deadly? Curr Opin Virol 2012, 2:242–247. [DOI] [PubMed] [Google Scholar]
- 17.Young PL, Halpin K, Selleck PW, Field H, Gravel JL, Kelly MA, Mackenzie JS: Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus. Emerg Infect Dis 1996, 2:239–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yob JM, Field H, Rashdi AM, Morrissy C, van der Heide B, Rota P, bin Adzhar A, White J, Daniels P, Jamaluddin A, et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg Infect Dis 2001, 7:439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh GA, De Jong C, Barr JA, Tachedjian M, Smith C: Cedar virus: a novel Henipavirus isolated from Australian bats. 2012, PLoS Pathog 2012, 8:e1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.*.Madera S, Kistler A, Ranaivoson HC, Ahyong V, Andrianiaina A, Andry S, Raharinosy V, Randriambolamanantsoa TH, Ravelomanantsoa NAF, Tato CM, et al. Discovery and Genomic Characterization of a Novel Henipavirus, Angavokely Virus, from Fruit Bats in Madagascar. J Virol 2022, 96:e0092122. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identifiied a bat-borne henipavirus from Madagascar which appears to utilize an ephrin-independent mechanism of entry.
- 21.Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J, Sun L, Zhu Y, Zhou H, Jin Q: Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg Infect Dis 2014, 20:1064–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S-H, Kim K, Kim J, No JS, Park K, Budhathoki S, Lee SH, Lee J, Cho SH, Cho S, et al. Discovery and Genetic Characterization of Novel Paramyxoviruses Related to the Genus in Species in the Republic of Korea. Viruses 2021, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.*.Vanmechelen B, Meurs S, Horemans M, Loosen A, Joly Maes T, Laenen L, Vergote V, Koundouno FR, Magassouba N, Konde MK, et al. The characterization of multiple novel paramyxoviruses highlights the diverse nature of the subfamily. Virus Evol 2022, 8:veac061. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identified diverse rodent- and shrew-borne paramyxoviruses, including novel Mojiang-like henipaviruses, from animals in Belgium and Guinea.
- 24.Zhang X-A, Li H, Jiang F-C, Zhu F, Zhang Y-F, Chen J-J, Tan C-W, Anderson DE, Fan H, Dong L-Y, et al. A Zoonotic Henipavirus in Febrile Patients in China. N Engl J Med 2022, 387:470–472. [DOI] [PubMed] [Google Scholar]
- 25.Rissanen I, Ahmed AA, Azarm K, Beaty S, Hong P, Nambulli S, Duprex WP, Lee B, Bowden TA: Idiosyncratic Mòjiāng virus attachment glycoprotein directs a host-cell entry pathway distinct from genetically related henipaviruses. Nat Commun 2017, 8:16060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheliout Da Silva S, Yan L, Dang HV, Xu K, Epstein JH, Veesler D, Broder CC: Functional analysis of the fusion and attachment glycoproteins of mojiang henipavirus. Viruses 2021, 13:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negrete OA, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian R, Tajyar S, Lee B: EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 2005, 436:401–405. [DOI] [PubMed] [Google Scholar]
- 28.MI Bonaparte, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, Choudhry V, Dimitrov DS, Wang L-F, Eaton BT, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A 2005, 102:10652–10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee B, Pernet O, Ahmed AA, Zeltina A, Beaty SM, Bowden TA: Molecular recognition of human ephrinB2 cell surface receptor by an emergent African henipavirus. Proc Natl Acad Sci U S A 2015, 112:E2156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laing ED, Navaratnarajah CK, Cheliout Da Silva S, Petzing SR, Xu Y, Sterling SL, Marsh GA, Wang L-F, Amaya M, Nikolov DB, et al. Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus. Proc Natl Acad Sci U S A 2019, 116:20707–20715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negrete OA, Wolf MC, Aguilar HC, Enterlein S, Wang W, Mühlberger E, Su SV, Bertolotti-Ciarlet A, Flick R, Lee B: Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog 2006, 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pryce R, Azarm K, Rissanen I, Harlos K, Bowden TA, Lee B: A key region of molecular specificity orchestrates unique ephrin-B1 utilization by Cedar virus. Life Sci Alliance 2020, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pernet O, Wang YE, Lee B: Henipavirus receptor usage and tropism. Curr Top Microbiol Immunol 2012, 359:59–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.*.Schountz T, Campbell C, Wagner K, Rovnak J, Martellaro C, DeBuysscher BL, Feldmann H, Prescott J: Differential Innate Immune Responses Elicited by Nipah Virus and Cedar Virus Correlate with Disparate In Vivo Pathogenesis in Hamsters. Viruses 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Compared Cedar virus and Nipah virus, demonstrating that the inability of Cedar virus to antagonize the type I interferon response is in part responsible for its lack of pathogenicity.
- 35.Negrete OA, Chu D, Aguilar HC, Lee B: Single Amino Acid Changes in the Nipah and Hendra Virus Attachment Glycoproteins Distinguish EphrinB2 from EphrinB3 Usage. Journal of Virology 2007, 81:10804–10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mills JN, Alim ANM, Bunning ML, Lee OB, Wagoner KD, Amman BR, Stockton PC, Ksiazek TG: Nipah virus infection in dogs, Malaysia, 1999. Emerg Infect Dis 2009, 15:950–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bossart KN, Tachedjian M, McEachern JA, Crameri G, Zhu Z, Dimitrov DS, Broder CC, Wang L-F: Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology 2008, 372:357–371. [DOI] [PubMed] [Google Scholar]
- 38.Wong KT, Grosjean I, Brisson C, Blanquier B, Fevre-Montange M, Bernard A, Loth P, Georges-Courbot M-C, Chevallier M, Akaoka H, et al. A golden hamster model for human acute Nipah virus infection. Am J Pathol 2003, 163:2127–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhondt KP, Mathieu C, Chalons M, Reynaud JM, Vallve A, Raoul H, Horvat B: Type I interferon signaling protects mice from lethal henipavirus infection. J Infect Dis 2013, 207:142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoneda M, Guillaume V, Sato H, Fujita K, Georges-Courbot M-C, Ikeda F, Omi M, Muto-Terao Y, Wild TF, Kai C: The nonstructural proteins of Nipah virus play a key role in pathogenicity in experimentally infected animals. PLoS One 2010, 5:e12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satterfield BA, Cross RW, Fenton KA, Agans KN, Basler CF, Geisbert TW, Mire CE: The immunomodulating V and W proteins of Nipah virus determine disease course. Nature Communications 2015, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatsuo H, Ono N, Tanaka K, Yanagi Y: SLAM (CDw150) is a cellular receptor for measles virus. Nature 2000, 406:893–897. [DOI] [PubMed] [Google Scholar]
- 43.Tatsuo H, Ono N, Yanagi Y: Morbilliviruses use signaling lymphocyte activation molecules (CD150) as cellular receptors. J Virol 2001, 75:5842–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noyce RS, Bondre DG, Ha MN, Lin L-T, Sisson G, Tsao M-S, Richardson CD: Tumor Cell Marker PVRL4 (Nectin 4) Is an Epithelial Cell Receptor for Measles Virus. PLoS Pathogens 2011, 7:e1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noyce RS, Richardson CD: Nectin 4 is the epithelial cell receptor for measles virus. Trends in Microbiology 2012, 20:429–439. [DOI] [PubMed] [Google Scholar]
- 46.Mühlebach MD, Mateo M, Sinn PL, Prüfer S, Uhlig KM, Leonard VHJ, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 2011, 480:530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.**.Takeda M, Seki F, Yamamoto Y, Nao N, Tokiwa H: Animal morbilliviruses and their cross-species transmission potential. Curr Opin Virol 2020, 41:38–45. [DOI] [PubMed] [Google Scholar]; Reviews the potential for animal morbilliviruses to spill over from natural hosts into new species, with an analysis of receptor compatibilities among several morbilliviruses and their natural host species.
- 48.Ohishi K, Maruyama T, Seki F, Takeda M: Marine Morbilliviruses: Diversity and Interaction with Signaling Lymphocyte Activation Molecules. Viruses 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beineke A, Baumgärtner W, Wohlsein P: Cross-species transmission of canine distemper virus-an update. One Health 2015, 1:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu W, Zheng Y, Zhang S, Fan Q, Liu H, Zhang F, Wang W, Liao G, Hu R: Canine distemper outbreak in rhesus monkeys, China. Emerg Infect Dis 2011, 17:1541–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Z, Li A, Ye H, Shi Y, Hu Z, Zeng L: Natural infection with canine distemper virus in hand-feeding Rhesus monkeys in China. Vet Microbiol 2010, 141:374–378. [DOI] [PubMed] [Google Scholar]
- 52.Sakai K, Yoshikawa T, Seki F, Fukushi S, Tahara M, Nagata N, Ami Y, Mizutani T, Kurane I, Yamaguchi R, et al. Canine distemper virus associated with a lethal outbreak in monkeys can readily adapt to use human receptors. J Virol 2013, 87:7170–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdullah N, Kelly JT, Graham SC, Birch J, Gonçalves-Carneiro D, Mitchell T, Thompson RN, Lythgoe KA, Logan N, Hosie MJ, et al. Structure-Guided Identification of a Nonhuman Morbillivirus with Zoonotic Potential. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bieringer M, Han JW, Kendl S, Khosravi M, Plattet P, Schneider-Schaulies J: Experimental adaptation of wild-type canine distemper virus (CDV) to the human entry receptor CD150. PLoS One 2013, 8:e57488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Vries RD, Ludlow M, Verburgh RJ, van Amerongen G, Yüksel S, Nguyen DT, McQuaid S, Osterhaus ADME, Duprex WP, de Swart RL: Measles vaccination of nonhuman primates provides partial protection against infection with canine distemper virus. J Virol 2014, 88:4423–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beauverger P, Buckland R, Wild TF: Measles virus antigens induce both type-specific and canine distemper virus cross-reactive cytotoxic T lymphocytes in mice: localization of a common Ld-restricted nucleoprotein epitope. J Gen Virol 1993, 74 (Pt 11):2357–2363. [DOI] [PubMed] [Google Scholar]
- 57.Ikegame S, Carmichael JC, Wells H, Furler RL, Acklin JA, Chiu H-P, Oguntuyo KY, Cox RM, Patel AR, Kowdle S, et al. Zoonotic potential of a novel bat morbillivirus. bioRxiv 2021, doi: 10.1101/2021.09.17.460143. [DOI] [Google Scholar]
- 58.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 2018, 46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y: Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol 2011, 18:135–141. [DOI] [PubMed] [Google Scholar]
- 60.Krissinel E, Henrick K: Inference of Macromolecular Assemblies from Crystalline State. Journal of Molecular Biology 2007, 372:774–797. [DOI] [PubMed] [Google Scholar]
- 61.Baker KS, Tachedjian M, Barr J, Marsh GA, Todd S, Crameri G, Crameri S, Smith I, Holmes CEG, Suu-Ire R, et al. Achimota Pararubulavirus 3: A New Bat-Derived Paramyxovirus of the Genus. Viruses 2020, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chua KB, Wang L-F, Lam SK, Crameri G, Yu M, Wise T, Boyle D, Hyatt AD, Eaton BT: Tioman Virus, a Novel Paramyxovirus Isolated from Fruit Bats in Malaysia. Virology 2001, 283:215–229. [DOI] [PubMed] [Google Scholar]
- 63.Barr JA, Smith C, Marsh GA, Field H, Wang L-F: Evidence of bat origin for Menangle virus, a zoonotic paramyxovirus first isolated from diseased pigs. J Gen Virol 2012, 93:2590–2594. [DOI] [PubMed] [Google Scholar]
- 64.Chant K: Probable Human Infection with a Newly Described Virus in the Family Paramyxoviridae. Emerging Infectious Diseases 1998, 4:273–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Albariño CG, Foltzer M, Towner JS, Rowe LA, Campbell S, Jaramillo CM, Bird BH, Reeder DM, Vodzak ME, Rota P, et al. Novel paramyxovirus associated with severe acute febrile disease, South Sudan and Uganda, 2012. Emerg Infect Dis 2014, 20:211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bowden TR, Westenberg M, Wang LF, Eaton BT, Boyle DB: Molecular characterization of Menangle virus, a novel paramyxovirus which infects pigs, fruit bats, and humans. Virology 2001, 283:358–373. [DOI] [PubMed] [Google Scholar]
- 67.Chua KB, Wang L-F, Lam SK, Eaton BT: Full length genome sequence of Tioman virus, a novel paramyxovirus in the genus Rubulavirus isolated from fruit bats in Malaysia. Archives of Virology 2002, 147:1323–1348. [DOI] [PubMed] [Google Scholar]
- 68.**.Stelfox AJ, Bowden TA: A structure-based rationale for sialic acid independent host-cell entry of Sosuga virus. Proc Natl Acad Sci U S A 2019, 116:21514–21520. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provides structural and experimental evidence to demonstrate that the Sosuga virus RBP does not utilize sialic acid for host-cell entry.
- 69.Johnson RI, Tachedjian M, Clayton BA, Layton R, Bergfeld J, Wang L-F, Marsh GA: Characterization of Teviot virus, an Australian bat-borne paramyxovirus. J Gen Virol 2019, 100:403–413. [DOI] [PubMed] [Google Scholar]
- 70.Rima B, Balkema-Buschmann A, Dundon WG, Duprex P, Easton A, Fouchier R, Kurath G, Lamb R, Lee B, Rota P, et al. ICTV Virus Taxonomy Profile: Paramyxoviridae. J Gen Virol 2019, 100:1593–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rima B, Collins P, Easton A, Fouchier R, Kurath G, Lamb RA, Lee B, Maisner A, Rota P, Wang L-F: Problems of classification in the family Paramyxoviridae. Arch Virol 2018, 163:1395–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langedijk JP, Daus FJ, van Oirschot JT: Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J Virol 1997, 71:6155–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jorgensen ED, Collins PL, Lomedico PT: Cloning and nucleotide sequence of Newcastle disease virus hemagglutinin-neuraminidase mRNA: identification of a putative sialic acid binding site. Virology 1987, 156:12–24. [DOI] [PubMed] [Google Scholar]
- 74.Bowden TR, Bingham J, Harper JA, Boyle DB: Menangle virus, a pteropid bat paramyxovirus infectious for pigs and humans, exhibits tropism for secondary lymphoid organs and intestinal epithelium in weaned pigs. J Gen Virol 2012, 93:1007–1016. [DOI] [PubMed] [Google Scholar]
- 75.*.Kirejczyk SGM, Amman BR, Schuh AJ, Sealy TK, Albariño CG, Zhang J, Brown CC, Towner JS: Histopathologic and Immunohistochemical Evaluation of Induced Lesions, Tissue Tropism and Host Responses following Experimental Infection of Egyptian Rousette Bats () with the Zoonotic Paramyxovirus, Sosuga Virus. Viruses 2022, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]; Histopathologic examination of tissue from experimentally infected Rousettus aegyptiacus bats reveals a tropism for the small intestines and salivary glands for Sosuga virus in its natural reservoir host.
- 76.Amman BR, Schuh AJ, Sealy TK, Spengler JR, Welch SR, Kirejczyk SGM, Albariño CG, Nichol ST, Towner JS: Experimental infection of Egyptian rousette bats (Rousettus aegyptiacus) with Sosuga virus demonstrates potential transmission routes for a bat-borne human pathogenic paramyxovirus. PLOS Neglected Tropical Diseases 2020, 14:e0008092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barr J, Todd S, Crameri G, Foord A, Marsh G, Frazer L, Payne J, Harper J, Baker KS, Cunningham AA, et al. Animal infection studies of two recently discovered African bat paramyxoviruses, Achimota 1 and Achimota 2. Sci Rep 2018, 8:12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crennell S, Takimoto T, Portner A, Taylor G: Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol 2000, 7:1068–1074. [DOI] [PubMed] [Google Scholar]
- 79.Iorio RM, Field GM, Sauvron JM, Mirza AM, Deng R, Mahon PJ, Langedijk JP: Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J Virol 2001, 75:1918–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Connaris H, Takimoto T, Russell R, Crennell S, Moustafa I, Portner A, Taylor G: Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J Virol 2002, 76:1816–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kubota M, Takeuchi K, Watanabe S, Ohno S, Matsuoka R, Kohda D, Nakakita S-I, Hiramatsu H, Suzuki Y, Nakayama T, et al. Trisaccharide containing α2,3-linked sialic acid is a receptor for mumps virus. Proc Natl Acad Sci U S A 2016, 113:11579–11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hause BM, Nelson E, Christopher-Hennings J: Eptesicus fuscus Orthorubulavirus, a Close Relative of Human Parainfluenza Virus 4, Discovered in a Bat in South Dakota. Microbiol Spectr 2021, 9:e0093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.**.Krüger N, Sauder C, Hüttl S, Papies J, Voigt K, Herrler G, Hardes K, Steinmetzer T, Örvell C, Drexler JF, et al. Entry, Replication, Immune Evasion, and Neurotoxicity of Synthetically Engineered Bat-Borne Mumps Virus. Cell Rep 2018, 25:312–320.e7. [DOI] [PubMed] [Google Scholar]; Demonstrated that a recombinant bat mumps virus (BaMuV) can effectively replicate in human cells and is able to antagonize human innate immune responses.
- 84.Beaty SM, Nachbagauer R, Hirsh A, Vigant F, Duehr J, Azarm KD, Stelfox AJ, Bowden TA, Duprex WP, Krammer F, et al. Cross-Reactive and Cross-Neutralizing Activity of Human Mumps Antibodies Against a Novel Mumps Virus From Bats. J Infect Dis 2017, 215:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katoh H, Kubota T, Ihara T, Maeda K, Takeda M, Kidokoro M: Cross-Neutralization between Human and African Bat Mumps Viruses. Emerg Infect Dis 2016, 22:703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Noh JY, Jeong DG, Yoon S-W, Kim JH, Choi YG, Kang S-Y, Kim HK: Isolation and characterization of novel bat paramyxovirus B16-40 potentially belonging to the proposed genus Shaanvirus. Scientific Reports 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu Z, Yang L, Ren X, He G, Zhang J, Yang J, Qian Z, Dong J, Sun L, Zhu Y, et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J 2016, 10:609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu W, Huang Y, Yu X, Chen H, Li D, Zhou L, Huang Q, Liu L, Yang J, Lu S: Discovery and evolutionary analysis of a novel bat-borne Paramyxovirus. Viruses 2022, 14:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jang SS, Noh JY, Kim MC, Lim HA, Song MS, Kim HK: α2,3-Linked Sialic Acids Are the Potential Attachment Receptor for Shaan Virus Infection in MARC-145 Cells. Microbiol Spectr 2022, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Farrukee R, Ait-Goughoulte M, Saunders PM, Londrigan SL, Reading PC: Host Cell Restriction Factors of Paramyxoviruses and Pneumoviruses. Viruses 2020, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang C, Wang T, Duan L, Chen H, Hu R, Wang X, Jia Y, Chu Z, Liu H, Wang X, et al. Evasion of Host Antiviral Innate Immunity by Paramyxovirus Accessory Proteins. Front Microbiol 2022, 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shaw ML: Henipaviruses employ a multifaceted approach to evade the antiviral interferon response. Viruses 2009, 1:1190–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dups J, Middleton D, Long F, Arkinstall R, Marsh GA, Wang L-F: Subclinical infection without encephalitis in mice following intranasal exposure to Nipah virus-Malaysia and Nipah virus-Bangladesh. Virol J 2014, 11:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karron RA, Wright PF, Hall SL, Makhene M, Thompson J, Burns BA, Tollefson S, Steinhoff MC, Wilson MH, Harris DO: A live attenuated bovine parainfluenza virus type 3 vaccine is safe, infectious, immunogenic, and phenotypically stable in infants and children. J Infect Dis 1995, 171:1107–1114. [DOI] [PubMed] [Google Scholar]
- 95.Thibault PA, Watkinson RE, Moreira-Soto A, Drexler JF, Lee B: Zoonotic Potential of Emerging Paramyxoviruses: Knowns and Unknowns. Adv Virus Res 2017, 98:1–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsurudome M, Yamada A, Hishiyama M, Ito Y: Replication of mumps virus in mouse: transient replication in lung and potential of systemic infection. Archives of Virology 1987, 97:167–179. [DOI] [PubMed] [Google Scholar]
- 97.Cusi MG, Correale P, Valassina M, Sabatino M, Valensin PE, Donati M, Glück R: Comparative study of the immune response in mice immunized with four live attenuated strains of mumps virus by intranasal or intramuscular route. Archives of Virology 2001, 146:1241–1248. [DOI] [PubMed] [Google Scholar]


