Abstract
Microcrystal Electron Diffraction (MicroED) is the newest cryo-electron microscopy (cryo-EM) method, with over 70 protein, peptide, and several small organic molecule structures already determined. In MicroED, micro- or nanocrystalline samples in solution are deposited on electron microscopy grids and examined in a cryo-electron microscope, ideally under cryogenic conditions. Continuous rotation diffraction data are collected and then processed using conventional X-ray crystallography programs. The protocol outlined here details how to obtain and identify the nanocrystals, how to set up the microscope for screening and for MicroED data collection, and how to collect and process data to complete high-resolution structures. For well-behaving crystals with high-resolution diffraction in cryo-EM, the entire process can be achieved in less than an hour.
Keywords: MicroED, 3D Electron Crystallography, Cryo-EM, Electron Diffraction, Protein Structure, Grid Preparation, Microcrystal, Nanocrystal, Transmission Electron Microscopy
1. Introduction
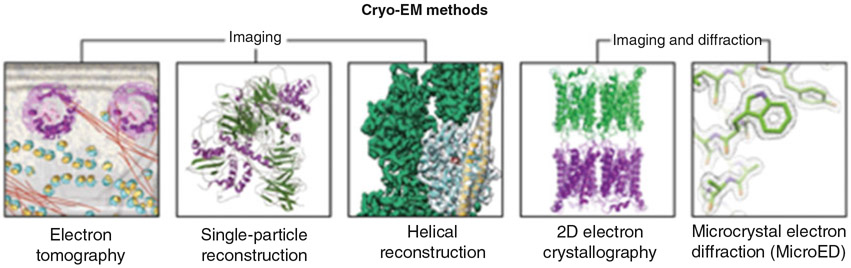
Modern cryo-EM encompasses at least five techniques: cryo-electron tomography [1], single-particle cryo-EM [2], helical reconstruction [3], electron crystallography of 2D crystals [4], and MicroED [5] (Fig. 1). In all these methods, frozen-hydrated samples [6] are examined in a transmission electron microscope (TEM); however, the data collection mode differs where the first three techniques use imaging of the biological sample exclusively, and the last two, use diffraction in addition to imaging.
Fig. 1.
Cryo-EM can be used to provide structural information from a wide range of samples. There are five major techniques: Cryo-electron tomography was developed to study whole cells and large organelles at resolutions of about 1 nm [30]. In single particle reconstruction isolated single particles are imagined and near atomic structures can be determined, [31] and similarly for helical assemblies helical image reconstruction can be used [32]. In 2D electron crystallography, the structures are obtained from the diffraction of highly ordered 2D crystals containing only one layer of the protein of interest [33] and in MicroED from the diffraction pattern of nano- or micro-sized three-dimensional crystals [8]
In MicroED, high-resolution electron diffraction data are collected from 3D micro- and nanocrystals, a billionth the size of those used for X-ray diffraction [5]. The use of such vanishingly small crystals has opened up for a new era in protein structural elucidation as previously unattainable “difficult to crystalize” samples are now accessible for investigation [5]. This includes several highly important targets such as various membrane proteins [7] and protein complexes [8], as well as small molecules [9] and natural products [10]. As compared with X-ray-free electron lasers (XFELS) [11] which also obtain data from microcrystals, MicroED is less expensive, requires substantially less sample, crystals can be substantially smaller, and the equipment needed is more accessible and easier to maintain and handle. Moreover, structures in MicroED can be completed with a single nanocrystal whereas in XFELs many thousands are necessary further complicating the procedure. A large part of the success of MicroED is owing to the use of continuous rotation during data collection [12], leading to well-defined rocking curve parameters and enabling data processing by well-established X-ray diffraction programs including MOSFLM [13, 14], XDS [15], the HKL suite [16], DIALS [17], CNS, [18, 19] PHENIX [20], Buster [21], SHELX [22], and the CCP4 [23] suite. Another important feature of MicroED is the use of an extremely low electron exposure (dose rate typically ~0.01 e−/Å2/s), minimizing radiation damage [24] during data collection and allowing for micro- and nanosized crystals to be examined. Since its initial demonstration in 2013 various proteins [7, 12, 25], protein complexes [8], protein–ligand complexes [26], peptides [27, 28], small molecules [9], natural products [10], and inorganic material [29] have been described by MicroED (Fig. 2). Future directions include developing imaging methods for phasing and high throughput structure determination [5].
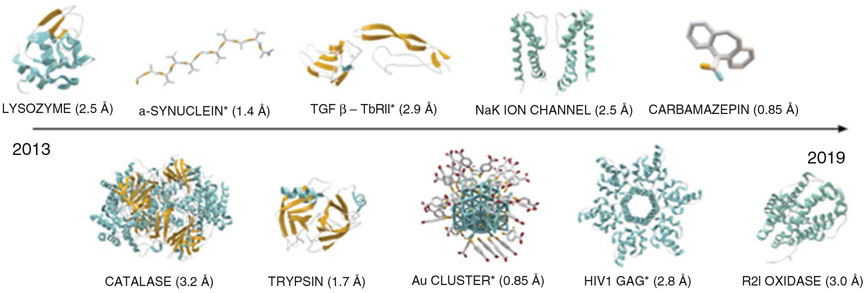
Fig. 2.
The array of MicroED structures published so far includes lysozyme [12], catalase [25], the amyloid core of α-synuclein [27], proteinase K [8], trypsin, [8], the complex of TGF-βm bound to its receptor TβRII [8], an inorganic gold cluster [29], the NaK ion channel [7], the HIV1 GAG protein bound to its ligand bevirimat [26], the R2lox enzyme [34], and several small organic molecules including carbamazepine [9]. Novel structures are marked with asterisk. There are today over 70 published MicroED structures
Herein, we describe step-by-step protocols with all the necessary details to prepare samples, collect data and solve structures using MicroED. The data is collected from vanishingly small crystals, with a thickness of ~500 nm or less. This makes the method highly attractive for anyone having difficulties in crystallizing their target, but MicroED can also be used for thicker crystals once they are broken into smaller crystalline domains, as described in Subheading 3.1. A general overview of the workflow is given in Fig. 3. The crystallization of microcrystals is carried out by the same methods as used for X-ray crystallography, such as vapor diffusion by hanging drop or sitting drop and liquid–liquid diffusion. If the crystals are too small to be verified by standard light microscopy, other methods such as negative staining EM can be used. The protocol below also explains how focused ion beam (FIB) milling in a scanning electron microscope (SEM) can be used to prepare crystalline lamella of desired thickness for MicroED. For grid preparation, the solution of microcrystals is deposited on cryo-EM grids followed by blotting and vitrification by plunge-freezing into liquid ethane. The grids are then screened, and data are collected and processed as described below. The structures can be solved from single crystals or by merging several data sets. For a crystal with high-quality diffraction, the entire process of data collection and processing can be achieved in less than an hour.
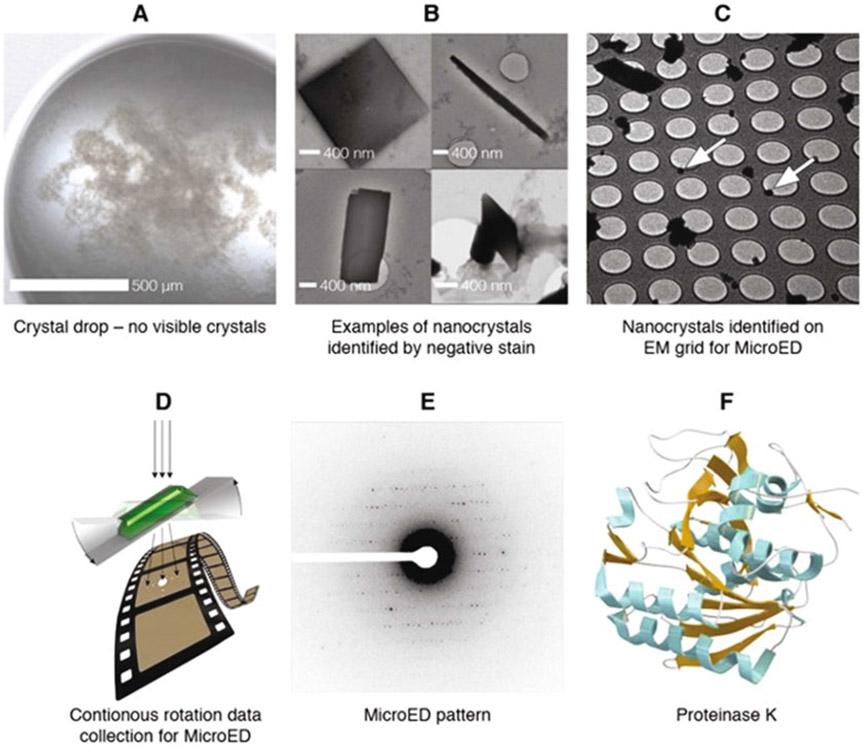
Fig. 3.
Overview of workflow. Crystal drops typically contain invisible crystals (a) that can be identified by, for example, negative stain (b). The nanocrystals are placed onto an electron microscopy grid (c, white arrows), and MicroED data are collected from a single crystal (d). The collected data (e) are processed using standard crystallographic software, yielding the structure, here exemplified by proteinase K (f) [8]
2. Materials
2.1. Preparing Crystals for MicroED
Protein sample.
Micropipette, capable of pipetting 1–10μL solution.
Pipette tips.
Ultrasonic water bath.
Microcentrifuge tubes.
Parafilm.
0.5 mm glass bead.
Vortex mixer.
Cryo-FIB DualBeam system, for example Aquilos from ThermoFisher Scientific.
Liquid nitrogen.
2.2. Identification of Microcrystals
Uranyl formate.
Buntzen burner.
MilliQ water.
Glass test tube.
Magnetic stirrer.
Aluminum-wrapped beaker.
5 M NaOH.
0.22μm syringe filter.
Aluminum-wrapped 8 mL falcon tube.
Anticapillary reverse (self-closing) tweezers.
Glow discharge cleaning system (e.g., PELCO easiGlow).
Parafilm.
Glass slide.
Whatman filter paper 1.
Cryo-grid storage boxes.
2.3. Grid Preparation
Glow discharge cleaning system (e.g., PELCO easiGlow).
Anticapillary reverse (self-closing) tweezers.
Micropipette, capable of pipetting 1.5–3μL solution.
300-Mesh copper holey carbon EM grids (Quantfoil, SPI supplies).
FEI Vitrobot Plunge-freezer System including Vitrobot coolant container.
Standard Vitrobot filter paper.
Locking Tweezers Assembly for Vitrobot.
Slide Warmer, for example, Premiere XH-2002.
Liquid nitrogen.
Ethane gas.
Cryo-grid storage boxes and gripper tool.
Whatman Filter paper 1.
2.4. Data Collection and Processing
Gold/graphitized calibration grids (Ted Pella, prod. no. 638).
Microscopes: Titan Krios or Talos Arctica transmission electron microscopes, both from ThermoScientific.
Cameras: Falcon 3EC Direct Electron Detector (ThermoFisher Scientific), K3 Direct Electron Detector (Gatan), Ceta 16 M camera (ThermoFisher Scientific), TVIPS TemCam-F416.
Image conversion tools: the conversion tools necessary for MRC or TVIPS file format to SMV are available on the Gonen lab webpage https://cryoem.ucla.edu/downloads
Software: EPU-D, CCP4, XDS, iMOSFLM, and Phenix.
3. Methods
3.1. Preparing Crystals for MicroED
Micro- and nanocrystals are prepared by the same crystallization methods used for larger crystals such as vapor diffusion by hanging drop or sitting drop and liquid–liquid diffusion. Crystals for MicroED should ideally be thinner than ~500 nm. If required, crystal growth can be restricted by using high salt concentration. In addition, larger crystals can easily and mechanically be broken into smaller crystal domains by pipetting, ultrasonication, or vortexing, as described below.
3.1.1. Fragment Crystals by Pipetting
Place the crystal drop under the light microscope.
Use a micropipette with a new tip and pipette up and down a couple of times directly into the drop while viewing the crystals in the microscope. Be careful not to create air bubbles.
Add well buffer/mother liquor if needed to dilute the sample.
3.1.2. Fragment Crystals by Sonication
Prepare an ultrasonic water bath by adding water to the container.
Transfer the crystal solution to a new microcentrifuge tube.
Add well buffer/mother liquor if needed to dilute the sample.
Close the microcentrifuge tube and wrap it with parafilm.
Set the sonicating bath to run continuously and dip the bottom end of the microcentrifuge tube into the water, make sure that the surface of the crystal solution is below the surface of the water bath (see Note 1).
3.1.3. Fragment Crystals by Vortexing
Transfer the crystal solution to a new 1.5μL microcentrifuge tube.
Add well buffer/mother liquor if needed to dilute the sample.
Add a glass bead of 0.5 mm diameter to the microcentrifuge tube. Close and wrap with parafilm.
Set the vortex mixer to auto mode at the highest speed setting. Use the flat rubberized head platform and place the bottom of the microcentrifuge tube in the center. Vortex for 2 s (see Note 2).
3.1.4. FIB Milling
Prepare the Aquilos by purging the lines with 10 L/min nitrogen gas flow, for at least 0.5 h.
Cool down the Aquilos and wait for the temperatures to stabilize, according to manufactures’ instructions.
Prepare grids according to Subheading 3.3.
Load grids into Aquilos under liquid nitrogen temperature.
Select a grid and move to deposition position to prepare for sputtering.
Select the time and current for sputtering. We usually use 7 mA, for 30 s, at 1 kV and pressure of 10 Pa.
When finished, move grid to mapping position to screen for crystals. We usually use 3.1 pA (5 kV) in SEM for screening (Fig. 4 a).
When a crystal has been identified, set the eucentric height by tilting to 18° and adjust with the stage Z bar.
Link Z height to FIB at 18 degrees and move to the FIB to mill crystal (Fig. 4b-c).
Place two rectangular milling patterns on the crystal and select a milling direction of top to bottom for the upper box, and bottom to top for the lower box. The milling is a stepwise process and the power and time is dependent on crystal size. In the first step, the boxes should be 3–5μm apart. A good starting point is 0.5 nA in the first round and then gradually reduce the current to 30 pA to get the final thickness of 200–300 nM (see Note 3).
When the lamella has been milled to the desired thickness, move to the next crystal until all sites have been milled. Keep the grids under liquid nitrogen temperature when transferring to the cryo-electron microscope (Fig. 4 e).
Fig. 4.
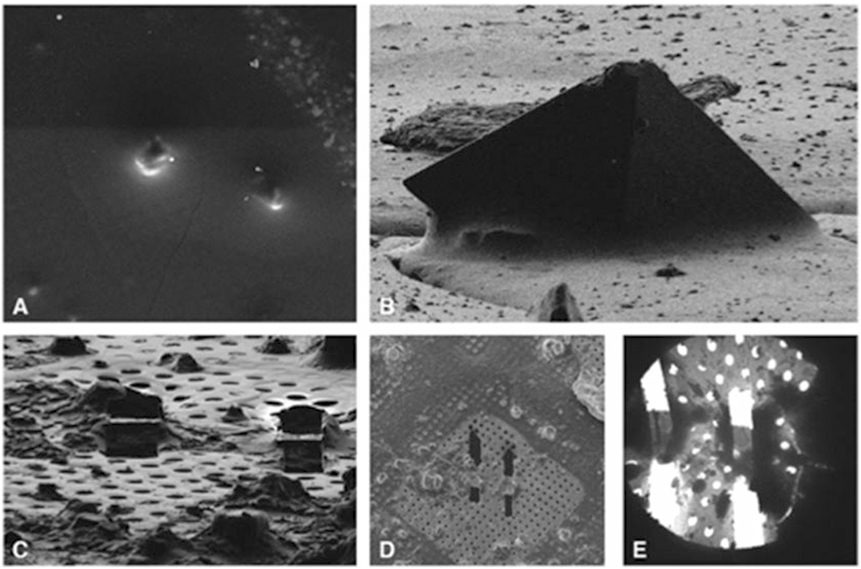
Images of crystals in SEM (a) and FIB (b) which are stepwise milled to a final thickness of about 200–300 nm, generating lamella here shown in FIB (c), SEM (d), and in the cryo-TEM (e)
3.2. Identification of Microcrystals
Crystals that due to their small dimensions are invisible by optical microscopy can be identified by ultraviolet (UV) light, fluorescence, or Second Order Nonlinear Imaging of Chiral Crystals (SONICC), using for example a Rock Imager, or by negative stain. Outlined below is our protocol for identification of microcrystals using negative stain, and in Subheading 3.5 it is described how the cryo-electron microscope can be used to screen for micro- and nanocrystals.
3.2.1. Identification of Microcrystals Using Negative Stain EM
Weigh 50 mg uranyl formate (see Note 4) into a beaker. Use a Bunsen burner to heat up MilliQ water in a glass test tube (see Note 5) and dissolve the uranyl formate in 5 mL of the water. Stir the mixture under an aluminum-wrapped beaker for 5 min.
Add 5μl 5 M NaOH into the mixture and stir for another 5 min.
Filter the uranyl formate solution through a 0.22μm filter and transfer to aluminum-wrapped 8 mL falcon tube (see Note 6).
Use tweezers to transfer carbon-coated grids onto glass slides wrapped in parafilm. Place the glass slide with grids in a glow discharge chamber and glow discharge for 30 s (see Subheading 3.3.1).
Prepare three drops of MiliQ water and 2 drops of uranyl solution for each grid sample on parafilm.
Use tweezers to pick up a grid and add 2μL sample to the carbon-coated side. Wait 20 s and then blot excess solution onto filter paper.
Wash the grid by gently touching the carbon-coated side of the grid on the surface of the first MilliQ water drop, without disrupting the surface tension (see Note 7). Blot on filter paper and repeat for the other two drops.
Wash the grid in the same way as in the first uranyl drop and blot on filter paper.
Stain the grid by gently rotating the grid in the last uranyl drop for 20 s, again without disrupting the surface tension.
Dry the grid by aspirating off the side of the grid for a few seconds using a vacuum. Store the grids in grid boxes.
3.3. Grid Preparation for MicroED
3.3.1. Glow Discharge
Place grids with the carbon side up in a glow discharge cleaning system and run it with negative polarity to make grids more accessible for the crystal drop. We typically use 1 × 10−1 mbar vacuum and 15 mA power and glow discharge with negative polarity for 30 s.
3.3.2. Automated Vitrification
The procedure described below is a typical procedure using the Vitrobot from ThermoFisher Scientific; however, other vitrification systems can be used as well.
Assemble the Vitrobot coolant container by placing the cryo-gridbox and the inner ethane container into the box holder and put the aluminum bridge on top (Fig. 5) and add liquid nitrogen to the outer reservoir to cool it down (see Note 8).
Attach filter paper to the blotting pads of the Vitrobot and set the temperature to and humidity (see Note 9). Put all tools that will be used on a 38 °C slide warmer.
Slowly fill the inner brass reservoir of the Vitrobot coolant container with ethane gas, the low temperature will cause it to condense (see Note 10).
When the reservoir is filled, wait for the ethane to begin freezing and then remove the aluminum bridge and place the container into the Vitrobot (see Note 11).
Use the locking tweezers to pick up a glow discharged grid and place in the Vitrobot with the carbon side to the left. Insert grid into the chamber of the Vitrobot.
Carefully pipette 1.5–3μL microcrystal solution onto the carbon side of the grid. Select blotting time and force and blot the sample to remove excess solution (see Note 12).
Plunge freeze into the liquid ethane and carefully remove the tweezers and quickly transfer the grid to a cryo-gridbox in the liquid nitrogen outer reservoir. Make sure to keep grid under liquid ethane or nitrogen all the time.
The grids can either be transferred to the microscope for examination or they can be stored in liquid nitrogen.
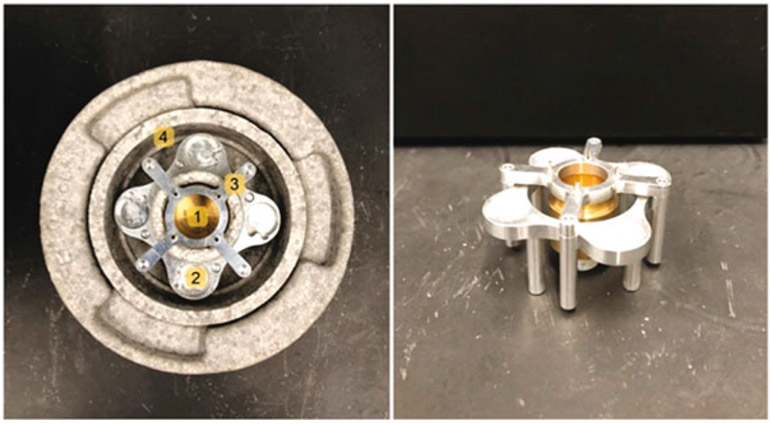
Fig. 5.
Assembled Vitrobot coolant container, with inner brass ethane container (1) cryo-gridbox holder (2), aluminum bridge (3), and outer reservoir for liquid nitrogen (4)
3.3.3. Manual Blotting and Freezing
Prepare the coolant container as described in Subheading 3.3.2, preferably in a fume hood.
Carefully pipette 1.5–3μL microcrystal solution to a glow discharged grid and blot by placing it carbon side up on a filter paper.
When the excess liquid has been blotted, quickly freeze in the liquid ethane and transfer to cryo-gridbox as described in Subheading 3.3.2.
The grids can either be transferred to the microscope for examination or they can be stored in liquid nitrogen.
3.4. Setting up the Microscope (ThermoFisher Cryo-TEMs Such as Glacios, Talos Arctica, and Titan Krios)
3.4.1. Calibrating the Electron Dose of the Microscope
Find a large space such as an area of broken carbon film or unload the grid.
Go to exposure/bright field mode and keep the same illumination settings (C2 condenser and intensity) as will be used in diffraction mode. The beam must be parallel and illuminate the entire sensor.
Use a direct electron detector such as the Falcon 3EC and record a 10 s exposure.
Note the dose rate and convert to e−/Å2/s.
3.4.2. Calibrating Detector Distance
Load a TEM grid with a standard sample for calibration, for example, gold/graphitized grids (Ted Pella, prod. no. 638).
Align the microscope as described in Subheading 3.4.3.
Measure the diffraction pattern and calculate the distance according to the formula Lλ ≈ Rd, where L is the calibrated detector distance, λ is the relativistic wavelength of electrons (0.0251 Å at 200 kV acceleration voltage), R is the radius of the diffraction ring (see Note 13), and d is the known lattice spacing of the standard sample.
3.4.3. Setting up the Microscope for Low-Dose Electron Diffraction (Talos Arctica)
Load the frozen grids into the microscope under liquid nitrogen temperature.
Set the microscope to diffraction mode and activate the low-dose mode (see Note 14). The low-dose mode consists of search, focus, and exposure modes.
In search mode, use the magnification control to set the camera distance. Set the spot size and the C2 condenser. These settings will be different for every microscope (see Note 15).
With the microscope in search mode and the screen inserted, find a feature on the grid such as a thick ice or black blob and center on it.
Switch to exposure mode. Use the magnification control to set the detector distance; for protein samples, we typically use 2–3 m and for small molecules 0.85–1.1 m (see Note 16).
Center the beam using the diffraction shift controls and make it as small as possible using the intensity control.
Switch back to search mode and center to the burn mark using the diffraction shift controls.
Repeat steps 6 and 7 until the beam stays centered after changing between the two modes.
In search mode, decrease the intensity so that the image of the grid is slightly larger than the outermost of the red circles.
Using the focus control, reduce focus until an oval shape is visible within the red circles.
Using the beam shift, adjust the beam to the center of the oval. Decrease the focus further until a three-pointed star is visible and align the star using the stigmator controls.
Make the beam as small as possible using the focus and then reset defocus. Set the defocus to 1.2 e−6.
Set the intensity to its original value.
Repeat step 6–7 to check that the beam did not move.
3.5. Grid Screening
For low magnification screening of the grids, an Atlas can be collected using EPU-D (Subheading 3.5.1). The grids can also be screened using the search mode (Subheading 3.5.2), within the low-dose settings (Fig. 6).
Fig. 6.
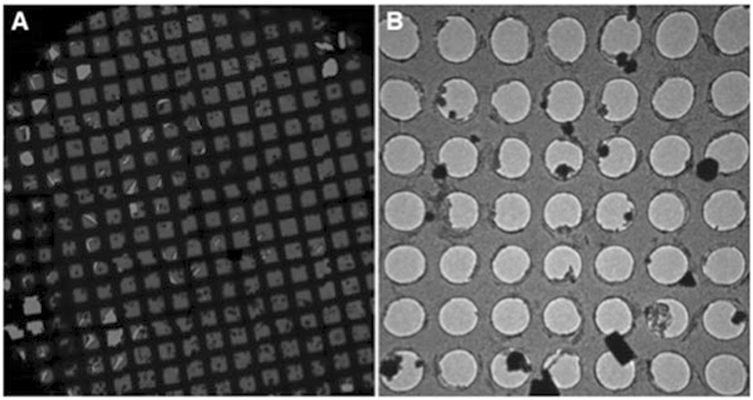
To identify crystals on the grids, an overview or Atlas (a) can be collected or the grids can be screened using the search mode (b)
3.5.1. Atlas
Align the beam in imaging mode. Set the spot size, magnification, and C2 condenser. Again, these will be different for different microscopes (see Note 17).
In search mode, decrease the intensity so that it is slightly larger than the middle of the red circles.
Center using the beam shift.
Increase the intensity so that it fits the outermost of the red circles.
Center to the ring using the C2 lens aperture controls.
Repeat steps 2–5 until and check that the beam did not move.
Lift the screen and start EPU Atlas.
Under the preparation tab set presets to Atlas, binning to 1,readout to full, exposure time to 3, noise reduction to yes, frames summed to 1 and intensity to 100. Also set the magnification and spot size to the values used for imaging mode (see Note 17).
Use the preview to quickly check that the settings are fine.
Go to atlas acquisition and acquire to record the full atlas.
Use “move stage to” and “save position” to save the positions of the potential microcrystals.
3.5.2. Grid Screen in Search Mode
In search mode, move around to look for areas of thin ice and well-separated microcrystals.
To check for diffraction, center on the potential crystal and insert the selected area aperture.
Blank the beam and go to exposure mode.
Insert the beam stop and lift the screen.
Under low dose, set the integration time to 3, sampling to 2, and read out area to full.
Unblank the beam and press acquire.
To continue to search for crystals, go back to search mode, remove the selected area and the beam stop, and insert the screen.
3.6. Data Collection
As the stage in MicroED data collection is continuously rotated, the frame rate of the detector has to be fast enough. Therefore, charge-coupled device (CCD) detectors are not generally recommended and complementary metal-oxide-semiconductor (CMOS)-based detector (e.g., TVIPS F416) or direct electron detectors (e.g., Falcon 3EC) should be used. To set the rotation, we use the Delphi scripting interface, but this can also be done in EPU or TEMspy.
In search mode, center on the crystal for data collection and set the eucentric height by changing the α-tilt between the desired angles for recording data and adjusting with the Z-height.
Insert the selected area aperture and check the α-tilt again.
Set the α-tilt value of the stage to the starting value for data collection.
Blank the beam and go to exposure mode.
Insert the beam stop.
Set the rotation to continuous and the rotation speed to 0.3° / s in Delphi.
Set the final tilt value that will be used.
Lift the screen and unblank the beam.
Press acquire and start the rotation to record data.
3.7. Data Processing
MicroED data are most commonly processed with the CCP4 programs, XDS, SHELX, and Phenix although other programs, such as HKL-2000/3000 and DIALS, can be used as well. For the processing of MicroED data, MRC or TVIPS file format has to be converted to SMV. The conversion tools necessary are available on the Gonen lab webpage https://cryoem.ucla.edu/downloads.
3.7.1. Indexing and Integration with iMosflm
Start iMosflm and select processing options from the settings menu. Go to the indexing tab and uncheck automatically index after spot finding.
Select add images from the session menu. Load the dataset by double-clicking one of the images.
Go to settings and experiment settings in the main iMosflm window. If the rotation rate during data collection was negative, check the reverse direction of spindle rotation box in the experiment tab.
Go to the detector tab and set the gain and ADC offset according to the detector.
Go to settings and processing options. In the integration section of the advanced integration tab, change null pixel threshold to −1.
In the image display window, adjust the red, green, and blue masks (see Note 18).
Go to the indexing task. Choose a set of images spanning a wedge of approximately 10°–20° by adding them in the image window (see Note 19).
Initially, use the automatic estimation for mosaicity and press index. Then change it to make sure, there are no full reflections in the integration.
Go to the integration task. Change the filename and check the fix tilt and twist boxes in the fix column.
Process the data by pressing process. Adjust the mosaicity and optionally the mosaic block size in the images task if needed.
Perform a QuickScale.
3.7.2. Indexing and Integration with XDS
Start xdsgui and select the desired directory using the choose or create new folder button.
Go to the frame tab and click load. Go to the project folder, select an image, and click open.
Click generate XDS.INP to generate the initial input file for XDS.
Go to the XDS.INP tab to view and adjust the input file (see Note 20).
Adjust values for the beam center assigning the corresponding x and y values in ORGX=x and ORGY=y.
Set REFINE(CORRECT)=CELL BEAM ORIENTATION AXIS.
Add OFFSET=adc offset used for image conversion.
Set ROTATION_AXIS according to the rotation direction. For example, for forward direction the default value of ROTATION_AXIS=1 0 0 is used.
Save the modified input file and click run XDS.
3.7.3. Merging and Phasing Preparation
Start the CCP4 interface.
Go to directories & project dir. and fill in the project. Browse for the data to be used in the “uses directory” field.
Download the coordinates and structure factors for the molecular replacement search model, i.e., PDB-formatted coordinates and CIF-formatted structure factors. Move the downloaded files to the project directory.
Convert the CIF structure factors to MTZ file format. Go to the reflection data utilities task in the CCP4 interface and choose convert to/modify/extend MTZ. Choose import reflection file in mmCIF format and create MTZ file. Set the in path to that of the downloaded CIF file. Change the out path and choose run now from the run drop-down menu.
3.7.4. Merging iMosflm Processed Data with AIMLESS
Go to the data reduction and analysis task in the CCP4 interface and select symmetry, scale, merge (aimless).
Set the path of HKLIN #1 to the output MTZ file from the integration step described above using the browse button.
Set the project, crystal, and dataset names.
Check to ensure unique data and add FreeR column for 0.05 fraction of the data box.
Check copy FreeR from another MTZ box.
Set the MTZ with FreeR to the MTZ file created previously from the CIF file (see Subheading 3.7.3) using the browse button.
Click run now from the run drop-down menu.
3.7.5. Merging XDS Processed Data with XSCALE
Go to the XSCALE tab in XDS. Add an INPUT_FILE line with the path to XDS_ASCII.HKL for each additional dataset to be included.
If the data will be phased by molecular replacement, convert the merged intensities to MTZ file format. Go to the XDSCONV tab and set OUTPUT_FILE to, e.g., temp.hkl CCP_I + F. Click run XDSCONV to produce temp.mtz in the project directory.
To add the free flags from the molecular replacement search model, open CCP4 and go to reflection data utilities and then merge MTZ files (cad). Use the browse buttons to give the merged MTZ as the first file. Click add input MTZ file to add the MTZ from the search model as the second file. Use all columns for the merged data and selected columns (R-free-flags) for the search model’s data.
3.7.6. Molecular Replacement Using MOLREP
Start the CCP4 interface.
Go to the molecular replacement task and select run Molrep - auto MR.
Set data to the MTZ file with the merged intensities from Subheading 3.7.4 or Subheading 3.7.5. Set model to the coordinate file downloaded in Subheading 3.7.3. Set solution to sample_name.pdb.
If the space group was not defined during any of the previous steps, click on search options and set SG to use Laue class instead of the default as is. The correct space group should then be found during molecular replacement. Then rerun MOLREP setting SG to use the correct space group.
Click run now from the run drop-down menu.
When MOLREP has finished, inspect the log file by double-clicking the job in the central panel of the main CCP4i window. Check the contrast (see Note 21).
3.7.7. Molecular Replacement Using Phaser
Start the Phenix graphical user interface.
Go to new project button and set the project ID and select the project directory.
Go to molecular replacement in the right part of the frame, and select Phaser-MR (simple one-component interface).
Add the downloaded search model file (see Subheading 3.7.3) and the MTZ data file to search (see Subheadings 3.7.4 and 3.7.5) in the input files tab.
Go to the search options tab and click on other settings. Set scattering form factor type to use electron scattering.
If the space group was not defined during any of the previous steps, try alternative space group drop down menu and select “all possible”. The correct space group should be identified during molecular replacement.
Click the run icon and select Run Now.
When Phaser is finished, check the translation function Z-score (TFZ) in the run status tab (see Note 22).
3.7.8. Refinement Using REFMAC
Start the CCP4 interface.
Go to the refinement task and choose run refmac5. Assign paths to the MTZ and PDB input files using the browse buttons, and set where the MTZ out and PDB out files are to be written.
Select Run&View Com File from the run drop-down menu. A window will open where the command line arguments and the input script can be edited. To enable electron-scattering factors, open a new line before any keyword and enter source EC MB. Click Continue without display.
3.7.9. Refinement with phenix.refine
Start the Phenix graphical user interface.
Choose refinement in the right part of the frame, and select phenix.refine.
Go to the input data tab. Add the molecular replacement solution (see Subheadings 3.7.6 and 3.7.7) and the data to refine against (see Subheadings 3.7.4 and 3.7.5).
Go to the refinement settings tab and set the scattering table to electron in the other options section.
Click Run. When refinement is done, inspect the R-factors. After the first round of refinement, they may be high.
4. Notes
The intensity and time used have to be optimized for each sample but a good starting point is 1–10 s, at the lowest intensity setting.
Ensure that bead and tube temperatures are stabilized at sample temperature before starting. The speed and time used might have to be optimized depending on sample.
The lamella will usually be thinner than what is indicated in the settings; therefore, we usually set the lamella thickness to ~350–300 nm to obtain a final thickness of ~200 nm.
Uranyl formate is light sensitive, keep dark.
Keep tube pointed away from you, might splash.
The uranyl solution should be a light yellow color. It can be kept for 4–5 days. After this, it will begin to precipitate and should be discarded.
Do not dip the entire grid into the drop which will disturb the carbon coating.
During grid preparation, make sure the level of liquid nitrogen does not fall too low and fill the reservoir with additional liquid nitrogen whenever necessary.
The temperature and humidity for blotting has to be optimized for each sample, 17 °C and 50% humidity are good starting points.
Be careful, liquid ethane is explosive. Make sure to include the aluminum bridge, it will transfer heat from the ethane to the liquid nitrogen.
The ethane needs to be almost frozen, not just a liquid. Before plunging the grid, check that the top of the ethane is not completely frozen.
Good starting points are blotting force of 1 and blotting time of 5 s. However, it is recommended to try a range of blotting times, for example 2–12 s, to obtain the optimal ice thickness. Alternatively, the grids can be blotted manually from the copper side using a filter paper strip. The filter paper strip is folded into a tear drop shape and held with the tweezers by its thinner end. The grid is then blotted gently with the thicker end.
The radius of the diffraction ring can be obtained by measuring the radius in pixels and then multiplying it by the pixel size of the detector.
The microscope is operated in low-dose mode, meaning a low number of electrons per Å2 per second will minimize exposure of the sample to damaging radiation.
The settings are different for every microscope and depend on what the target dose is. We usually set the camera distance to 6.9 m, spot size to 9 and C2 to 100μm in search mode.
The detector should be close enough to capture all high-resolution data, yet at a distance long enough to keep the reflections well separated.
Use the biggest aperture and the lowest magnification possible and the largest spot size. We usually set the spot size to 11, magnification to LM155X and the C2 condenser to 100μm for collecting an atlas.
The red mask should be centered on the halo around the circular part of the beam stop shadow. The red rectangle corresponds to the area used for estimation of the image background and should not cover any areas that deviate significantly from the overall image, such as the beam stop shadow. The green mask should be adjusted so it covers the shadow of the beam stop. The blue mask indicates the resolution limits. For initial assessment of a high-resolution dataset, it may be set to the edge of the image, whereas for final integration, it may be dragged off the image entirely.
The number of spots in the wedge should be in the hundreds. If the wedge is too small, autoindexing might fail. If the wedge is too large, experimental errors may accumulate and prevent successful indexing.
The input file is a plain text file with keywords and values separated by an equals (=) sign. Any text following an exclamation mark (!) is a comment that will be ignored.
According to the MOLREP manual page [http://www.ccp4.ac.uk/html/molrep.html#score], a contrast greater than 3 means that MOLREP has found a solution.
According to the PhaserWiki [http://www.phaser.cimr.cam.ac.uk/index.php/Molecular_Replacement], a value greater than 8 means Phaser has found a solution.
Acknowledgments
We thank all members of the Gonen laboratory, current and past and all collaborators who worked with us on MicroED applications. This work was supported by the National Institutes of Health P41GM136508. The Gonen laboratory is supported by the Howard Hughes Medical Institute.
References
- 1.Koning RI, Koster AJ, Sharp TH (2018) Advances in cryo-electron tomography for biology and medicine. Ann Anat 217:82–96 [DOI] [PubMed] [Google Scholar]
- 2.Glaeser RM (2019) How good can single-particle Cryo-EM become? What remains before it approaches its physical limits? Annu Rev Biophys 48:45–46 [DOI] [PubMed] [Google Scholar]
- 3.Fromm SA, Sachse C (2016) Chapter twelve - Cryo-EM structure determination using segmented helical image reconstruction. In: Crowther RA (ed) The resolution revolution: recent advances in cryoEM, vol 579. Academic Press, pp 307–328 [DOI] [PubMed] [Google Scholar]
- 4.Righetto RD, Biyani N, Kowal J et al. (2019) Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM. Nat Commun 10:1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nannenga BL, Gonen T (2109) The cryo-EM method microcrystal electron diffraction (MicroED). Nat Methods 16:369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iancu CV, Tivol WF, Schooler JB et al. (2006) Electron cryotomography sample preparation using the Vitrobot. Nat Protoc 1:2813–2819 [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Gonen T (2018) MicroED structure of the NaK ion channel reveals a Na+ partition process into the selectivity filter. Commun. Biol 1:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De La Cruz MJ, Hattne J, Shi D et al. (2107) Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat Methods 14:399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones CG, Martynowycz MW, Hattne J et al. (2018) The CryoEM method MicroED as a powerful tool for small molecule structure determination. ACS Cent Sci 4:1587–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ting CP, Funk MA, Halaby SL et al. (2019) Use of a scaffold peptide in the biosynthesis of amino acid-derived natural products. Science 365:280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman HN (2019) X-ray free-electron lasers for the structure and dynamics of macromolecules. Annu Rev Biochem 88:35–58 [DOI] [PubMed] [Google Scholar]
- 12.Nannenga BL, Shi D, Leslie AG, Gonen T (2014) High-resolution structure determination by continuous-rotation data collection in MicroED. Nat Methods 11:927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Battye TG, Kontogiannis L, Johnson O et al. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leslie AG, Powell HR (2007) Processing diffraction data with mosflm BT. In: Read RJ, Sussman JL (eds) Evolving methods for macromolecular crystallography. Springer; Netherlands, pp 41–51 [Google Scholar]
- 15.Kabsch W (2010) XDS. Acta Crystallogr Sect D 66:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor WB (1997) Processing of X-ray diffraction data collected in oscillation mode. In: Macromolecular crystallography part a, vol 276. Academic Press, pp 307–326 [DOI] [PubMed] [Google Scholar]
- 17.Waterman DG, Winter G, Parkhurst JM et al. (2013) The DIALS framework for integration software. CCP4 Newsl PROTEIN Crystallogr 49:16–19 [Google Scholar]
- 18.Brünger AT et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D 54:905–921 [DOI] [PubMed] [Google Scholar]
- 19.Brunger AT, Adams PD, Clore GM et al. (2007) Version 1.2 of the crystallography and NMR system. Nat Protoc 2:2728–2733 [DOI] [PubMed] [Google Scholar]
- 20.Adams PD, Afonine PV, Bunkóczi G et al. (2010) PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr Sect D 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanc E, Roversi P, Vonrhein C et al. (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr Sect D 60:2210–2221 [DOI] [PubMed] [Google Scholar]
- 22.Sheldrick GM (2010) Experimental phasing with SHELXC: combining chain tracing with density modification. Acta Crystallogr Sect D 66:479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winn MD, Ballard CC, Cowtan KD et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nannenga BL, Gonen T (2014) Protein structure determination by MicroED. Curr Opin Struct Biol 27:24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nanneng BL, Shi D, Hattne J et al. (2014) Structure of catalase determined by MicroED. elife 3:e03600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purdy MD, Shi D, Chrustowicz J et al. (2018) MicroED structures of HIV-1 gag CTD-SP1 reveal binding interactions with the maturation inhibitor bevirimat. Proc Natl Acad Sci U S A 115:13258–13263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez JA, Ivanova MI, Sawaya MR et al. (2015) Structure of the toxic core of α-synuclein from invisible crystals. Nature 525:486–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawaya MR, Rodriguez J, Cascio D et al. (2016) Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED. Proc Natl Acad Sci U S A 113:11232–11236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vergara S, Lukes DA, Martynowycz MW et al. (2017) MicroED structure of au 146 (p-MBA) 57 at subatomic resolution reveals a twinned FCC cluster. J Phys Chem Lett 8:5523–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahamid J, Pfeffer S, Schaffer M et al. (2016) Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351:969–972 [DOI] [PubMed] [Google Scholar]
- 31.Kasinath V, Faini M, Poepsel S et al. (2018) Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359:940–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von der Ecken J, Müller M, Lehman W et al. (2015) Structure of the F-actin–tropomyosin complex. Nature 519:114–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonen T, Cheng Y, Sliz P et al. (2005) Lipid–protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438:633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, Lebrette H, Clabbers MT et al. (2019) Solving a new R2lox protein structure by microcrystal electron diffraction. Sci Adv 5:eaax4621. [DOI] [PMC free article] [PubMed] [Google Scholar]