Abstract
LncRNA prostate androgen-regulated transcript 1 (PART1) is an important lncRNA in the carcinogenesis whose role has been firstly unraveled in prostate cancer. Expression of this lncRNA is activated by androgen in prostate cancer cells. In addition, this lncRNA has a role in the pathogenesis intervertebral disc degeneration, myocardial ischemia-reperfusion injury, osteoarthritis, osteoporosis and Parkinson’s disease. Diagnostic role of PART1 has been assessed in some types of cancers. Moreover, dysregulation of PART1 expression is regarded as a prognostic factor in a variety of cancers. The current review provides a concise but comprehensive summary of the role of PART1 in different cancers and non-malignant disorders.
Keywords: lncRNA, PART1, cancer, biomarker, diagnsotic marker
Introduction
Long non-coding RNAs (lncRNAs) have diverse roles in the carcinogenesis through modulation of gene expression. They can be localized in the nucleus or cytoplasm, thus regulating expression of genes through epigenetic, transcriptional and post-transcriptional mechanisms (Zhang et al., 2019a; Hussen et al., 2022). These effects are mediated through interactions with mRNAs, DNA molecules, proteins, and miRNAs (Zhang et al., 2019a; Ghafouri-Fard et al., 2022). The majority of identified lncRNAs are transcribed by RNA polymerase II; thus, they share several structural features with mRNAs, particularly in terms of having cap structure and poly A tail (Marchese et al., 2017). Yet, most lncRNAs lack coding capacity. The ENCODE project has annotated approximately 16,000 lncRNA genes in humans. These genes can produce more than 28,000 distinctive transcripts (Derrien et al., 2012).
LncRNAs have been shown to be involved in the carcinogenesis through modulation of expression of several tumor suppressor genes and oncogenes. Their altered expression in malignant cells have been associated with diverse abnormalities in the cell cycle regulation, cell proliferation, differentiation and apoptosis (Jiang et al., 2019). During the carcinogenesis process, lncRNAs regulate cell migration, invasion and stemness, thus they have prominent roles in the metastasis (Jiang et al., 2019).
LncRNA prostate androgen-regulated transcript 1 (PART1) is an important lncRNA in the carcinogenesis whose role has been firstly unraveled in prostate cancer. Expression of this lncRNA is activated by androgen in prostate cancer cells (Lin et al., 2000). Being encoded by a gene on gene to chromosome 5q12, PART1 has multiple alternatively transcripts none of them encoding a protein product (Figure 1). Expression assays have revealed biased expression of PART1 in brain, prostate, salivary gland, placenta and bladder (https://www.ncbi.nlm.nih.gov/gene/25859).
FIGURE 1.
The chromosomal location of the prostate androgen-regulated transcript 1 (PART1) was initially identified in PCa. The NCBI database reveals the existence of three transcripts: NR 024617.1 (2.5 kb), NR 028509.1 (5.9 kb), and NR 028508.1 (2.1 kb).
This lncRNA has dual roles in human tissues, being regarded as an oncogene in some tissues but tumor suppressor gene in others (Ran et al., 2022). The current review provides a concise but comprehensive summary of the role of PART1 in different cancers and non-malignant disorders.
Role of PART1 in cancers
Cell line studies
Functional studies in a variety of cancer-derived cell lines have assessed the consequences of up-regulation or silencing of PART1. Moreover, these studies have revealed a number of PART1 counterparts. In bladder cancer cells, enhanced expression of PART1 has promoted cell proliferation and invasiveness and suppressed cell apoptosis. On the other hand, PART1 silencing has suppressed cell proliferation and invasion and promoted apoptosis (Hu et al., 2019). In breast cancer cells, knockdown of PART1 has led to decreased proliferation, invasion and migration. Besides, miR-4516 has been found to be a direct counterpart of PART1. Suppression of miR-4516 has been found to rescue the effects of PART1 knockdown on breast cancer cells. Therefore, PART1 binding with miR-4516 promotes development of this type of cancer (Wang and Xu, 2020). Another study in breast cancer cells has shown that PART1 silencing improves the sensitivity of these cells to cisplatin, promotes cell apoptosis, and decreases expression proteins contributing in drug resistance (Lou et al., 2020). PART1 has also been found to be is enriched in triple negative breast cancer cells and in Aldefluorhigh cancer stem cells. PART1 silencing in these cell lines has reduced cell proliferation, migration, and mammosphere forming ability. This lncRNA has been able to affect expression of several genes, including myosin-Va, MYO5A, zinc fingers and homeoboxes protein 2 and ZHX2. In addition, expression of miR-190a-3p, miR-937-5p, miR-22-5p, miR-30b-3p, and miR-6870-5p has been shown to be affected by PART1. PART1 has a direct interaction with miR-937-5p (Cruickshank et al., 2021).
PART1 has also been among lncRNAs being targeted by the tumor suppressor protein ΔNp63α in cervical cancer cells (Liu et al., 2020).
In colorectal cancer cells, three independent studies have shown possible mechanisms for contribution of PART1 in the carcinogenesis. First, PART1 has been shown to regulate this process through targeting miR-150-5p/miR-520h/CTNNB1 axis and inducing activity of Wnt/β-catenin pathway (Zhou et al., 2020a). Moreover, PART1 can function as a molecular sponge for miR-143 in these cells (Hu et al., 2017). Finally, through sponging miR-150-5p, PART1 can increase expression of LRG1 in colorectal cancer cells (Lou et al., 2020).
In esophageal squamous cell carcinoma cells, PART1 has been shown to acts as a tumor suppressor lncRNA in a single study (Zhao et al., 2021). FOXP2 has been shown to bind to the promoter region of PART1 in these cells to regulate its expression. Up-regulation of PART1 could suppress cell proliferation and invasion, while its downregulation promotes cell proliferation and invasion in esophageal squamous cell carcinoma (Figure 2). From a mechanistical point of view, PART1 functions as a molecular sponge for miR-18a-5p, leading to over-expression of SOX6 and inactivation of the β-catenin/c-myc axis (Zhao et al., 2021). On the other hand, another study has shown that exosome-mediated transport of PART1 leads to induction of gefitinib resistance in esophageal squamous cell carcinoma cells through sponging miR-129 (Kang et al., 2018).
FIGURE 2.
An illustration shows the different signaling pathways of PART1 lncRNA with its expression and function in different types of cancer.
PART1 has been shown to restrain aggressive gastric cancer via decreasing expression of PDGFB through PLZF-mediated recruitment of EZH2 (Han et al., 2020). Similarly, PART1 has a tumor suppressor role in glioma through sponging miR-190a-3p and inactivating PTEN/AKT signals (Jin et al., 2020). Moreover, it can block carcinogenic process in glioma through modulation of miR-374b/SALL1 axis (Deng et al., 2022). Tables 1, 2 summarize the results of in vitro studies that reported up-regulation and down-regulation of PART1, respectively.
TABLE 1.
In vitro experiments to examine expression and function of PART1 in malignancies in which PART1 has been up-regulated (TCLs: tumor cell lines, NCL: normal cell line, ∆: knockdown or deletion, EMT: epithelial-mesenchymal transition, Brdu: Bromodeoxyuridine, DDP: cisplatin, ↑: increase, ↓: decrease).
| Tumor type | Cell line | Expression | Targets/Regulators and signaling pathways | Function | References |
|---|---|---|---|---|---|
| Bladder cancer | TCLs: 5637, T24 | — | — | ∆PART1: ↓proliferation, ↑apoptosis, ↓invasion | Hu et al. (2019) |
| Breast cancer | TCLs: MCF-7, SKBR3, BT-20, MDA-MB-231, ZR-75-1 | Up (TCLs vs. NCLs) | miR-4516 | ∆PART1: ↓proliferation, ↓migration, ↓invasion | Wang and Xu (2020) |
| NCL: MCF-10A | |||||
| TCLs: MCF-7, T47D, MDA-MB-435, BT-549 NCL: MCF-10A | Up (TCLs vs. NCLs) | — | ∆PART1: ↓proliferation (↓CDK2 and ↓cyclinE1, ↑P21), ↓migration and invasion (↓MMP3, ↓MMP10 and ↓MMP13), ↑cisplatin sensitivity | Lou et al. (2020) | |
| ∆PART1 (in cisplatin-treated cells) | |||||
| ↑apoptosis (↑Bax and cleaved caspase-3, ↓Bcl-2) | |||||
| ∆PART1 (in cisplatin-resistant cells) | |||||
| ↓Chemo-resistance: ↓MDP1, ↓MRP1, ↓GST-π, ↓ABCB1 (chemoresistance proteins) | |||||
| Triple-negative breast cancer (TNBC) | TCLs: such as HCC1806, HCC1395 | — | miRNAs-PART1 interactions→ gene expression alterations (genes like MYO5A, ZHX2, BICC1 and PPP2R3A) | ∆PART1: ↓proliferation, ↓migration, ↓mammosphere formation ability, ↓MYO5A, ZHX2 and BICC1 expression (oncogenes), ↑PPP2R3A expression (tumor suppressor) | Cruickshank et al. (2021) |
| Colorectal cancer (CRC) | TCLs: HCT-116, SW116, SW480, HT29 | Up (TCLs vs. NCLs) | miR-150-5p/miR-520h/CTNNB1, Wnt/β-catenin pathway | ∆PART1: ↓proliferation and cell viability, ↑apoptosis, ↓wound closure, ↓migration | Zhou et al. (2020a) |
| NCL: NCM460 | |||||
| TCLs: LoVo, HCT-116, SW620, SW480, HT29 | Up (TCLs vs. NCLs) | miR-143/DNMT3A | ∆PART1 (in SW620): ↓proliferation, ↓migration, ↓invasion | Hu et al. (2017) | |
| NCL: FHC | ↑PART1 (in LoVo) | ||||
| ↑cell growth, ↑migration, ↑invasion | |||||
| TCLs: HCT116, HT29, HEK-293T | — | miR-150-5p/LRG1 | ↑PART1 (in HCT116) | Lou et al. (2020) | |
| ↑proliferation (↑Brdu + cells), ↑migration, ↓apoptosis, ↑EMT (↑vimentin, ↓E-cadherin) | |||||
| ∆PART1 (in HT29) | |||||
| ↓proliferation (↓Brdu + cells), ↓migration, ↑apoptosis, ↓EMT (↓vimentin, ↑E-cadherin) | |||||
| Hepatocellular Carcinoma (HCC) | TCLs: SK-HEP-1, Huh-7, Huh-1, Hep3B | Up (TCLs vs. NCLs) | miR-149-5p/MAP2K1 | ∆PART1: ↓cell viability, ↓migration, ↓invasion | Zhou et al. (2020b) |
| NCL: THLE-2 | |||||
| TCLs: SMMC-7721, Huh-7 | Up (TCLs vs. NCLs) | miR-590-3p/HMGB2 | ∆PART1: ↓proliferation, ↓colony formation, ↓invasion | Pu et al. (2020) | |
| NCL: LO2 | |||||
| TCLs: HB611, Huh7, HCCLM3, Bel-7405 | Up (TCLs vs. NCLs) | miR-372-3p/TLR4 | ↑PART1 | Zhou et al. (2022) | |
| NCL: THLE-2, THP-1 | ↑cell viability, ↑migration, ↑invasion, ↑EMT (↓E-cadherin, ↑N-cadherin, ↑vimentin, ↑Twist, ↑Snail) | ||||
| ↑M2 macrophage polarization (↑M2 macrophage markers (Arg-1 and IL-10), ↓M1 macrophage markers (iNOS and TNF-α)) | |||||
| Liver cancer | TCLs: HepG2, HuH7, Hep3B | Up (TCLs vs. NCLs) | miR-3529-3p/FOXC2/AKT pathway (MMP-2 and MMP-9) | ∆PART1: ↓cell viability, ↓migration, ↓invasion | Weng et al. (2021) |
| NCL: LO2 | |||||
| Lung Squamous Cell Carcinoma (LSCC) | TCLs: H2170, H226, H520, SK-MES-1 | Up (TCLs vs. NCLs) | miR-185-5p/Six1 | ∆PART1 (in H2170) | Cao et al. (2021) |
| ↓colony formation ability, ↓cell viability, ↓migration, ↓invasion, ↓EMT (↑E-cadherin, ↓vimentin, ↓N-cadherin), ↑apoptosis | |||||
| NCL: BEAS-2B | ↑PART1 (in H520) | ||||
| ↑colony formation ability, ↑proliferation, ↑migration, ↑invasion, ↑EMT (↓E-cadherin, ↑vimentin, ↑N-cadherin), ↓apoptosis | |||||
| Non-small cell lung cancer (NSCLC) | TCLs: A549, H1650, H1975, SK-MES-1 | Up (TCLs vs. NCLs) | miR-635/JAK1 and JAK3 (JAK/STAT3 signaling pathway) | ↑PART1 (in H1975 & H1650) | Zhu et al. (2019) |
| ↑cell proliferation and viability (↓p21, ↓p27, ↑cyclin D2), ↑migration and invasion (↓E-cadherin, ↑MMP-2, ↑MMP-9) | |||||
| NCL: BEAS-2B, HEK-293T | ∆PART1 (in A549 & SK-MES-1) | ||||
| ↓proliferation and ↑G0/G1 cell cycle arrest (↑p21, ↑p27, ↓cyclin D2), ↓migration and invasion (↑E-cadherin, ↓MMP-2, ↓MMP-9) | |||||
| TCLs: SPC-A1, H1299, A549, H1650, H1975, PC-9 | Up (TCLs vs. NCLs) | miR-17-5p/TGFBETAR2 | ∆PART1: ↓proliferation, ↓migration, ↓invasion | Chen et al. (2021) | |
| NCL: 16HBE | |||||
| TCLs: A549, NCI-H2444, NCI-H647, NCI-H23 | Up (TCLs vs. NCLs) | — | ∆PART1: ↑erlotinib sensitivity (in TCLs with wild-type KRAS) | Chen et al. (2020) | |
| NCL: BEAS-2B | |||||
| Oral Squamous Cell Carcinoma (OSCC) | TCLs: Tca-8113, CAL27 | Up (TCLs vs. NCLs) | FUS/EZH2 | ∆PART1: ↓proliferation, ↓G2/M phase cells, ↑apoptosis, ↑G0/G1 phase cells | Yu et al. (2021) |
| NCL: NHOK | |||||
| Ovarian Cancer (OC) | TCLs: CaoV-3, SK-OV-3, HO-8910 | Up (TCLs vs. NCLs) | miR-503-5p/FOXK1 | ∆PART1: ↓viability, ↓migration, ↓invasion, ↑apoptosis | Li et al. (2022a) |
| NCL: IOSE80 | |||||
| TCLs: Caov3, OVCAR3, A2780, SKOV3 | Up (TCLs vs. NCLs) | miR-6884-5p/RACGAP1 and RRM2 | ∆PART1: ↓proliferation, ↓migration, ↓invasion | Li et al. (2022b) | |
| NCL: IOSE-386 | |||||
| TCLs: CAOV3, A2780 (DDP-resistant cell lines) | Up (DDP-resistant cell lines vs. control parental cell lines) | Transcriptional inducer of PART1: YY1 | ∆PART1 (in DDP-resistant cells) | Yang et al. (2021a) | |
| Targets of PART1: miR-512-3p/CHRAC1 | ↓proliferation, ↓migration, ↓invasion, ↑apoptosis, ↑chemosensitivity | ||||
| Pancreatic Cancer | TCLs: AsPC-1, Panc-1, SW 1990, BxPC-3 | Up (TCLs vs. NCLs) | miR-122 | ∆PART1: ↓proliferation, ↓invasion, ↑apoptosis (↓Bcl-2, ↑Bax) | Ghafouri-Fard et al. (2021) |
| NCL: HPDE6c7 | |||||
| TCL: PANC-1 | — | hsa-mir-21/SCRN1 | ∆PART1: ↓proliferation, ↓migration | Lu et al. (2022) | |
| Prostate cancer (PCa) | TCLs: LNCaP, PC3 | — | Target: TLR signaling pathway (TLR3, TNFSF10, CXCL13) | ∆PART1: ↓proliferation, ↑apoptosis | Sun et al. (2018) |
| Transcriptional modulators of PART1: androgens |
TABLE 2.
In vitro experiments to examine expression and function of PART1 in malignancies in which PART1 has been down-regulated (↑: increase, ↓: decrease).
| Tumor type | Cell line | Expression | Targets/Regulators and signaling pathways | Function | References |
|---|---|---|---|---|---|
| Cervical Squamous Cell Carcinoma (CSCC) | TCLs: SiHa, ME-180, C-33A, HeLa, HaCat, 293T | _ | Transcriptional regulator of PART1: ∆Np63α | ↑PART1 (in SiHa) | Liu et al. (2020) |
| ↓proliferation and colony formation, ↓S phase cells, ↑G1 phase cells, ↓migration, ↓invasion | |||||
| ∆PART1 (in ME-180) | |||||
| ↑proliferation and colony formation, ↑S phase cells, ↓G1 phase cells, ↑migration, ↑invasion | |||||
| Esophageal Squamous Cell Carcinoma (ESCC) | TCLs: Eca109, EC9706, TE1, KYSE70, KYSE450 | Down (TCLs vs. NCLs) | Regulator: FOXP2 | ↑PART1 | Zhao et al. (2021) |
| NCL: Het-1A | Target: miR-18a-5p/SOX6/β-catenin signaling pathway | ↓proliferation, ↓invasion | |||
| TCLs: TE1, TE6, TE8, TTn, KYSE-450 (gefitinib resistant cell lines) | Up (gefitinib resistant cell lines vs. parental cell lines) | Transcriptional inducer of PART1 in gefitinib resistant cells: STAT1 | ∆PART1 | Kang et al. (2018) | |
| Targets of PART1: miR-129, Bcl-2/Bax signaling pathway | ↑gefitinib chemotoxicity, ↑cell apoptosis | ||||
| Gastric cancer (GC) | TCLs: MGC-803, BGC-823, SGC-7901, NCI-N87, AGS, NUGC-3 | Down (TCLs vs. NCLs) | AR/PLZF/EZH2/PDGFB → PDGFRβ/PI3K/Akt signaling pathway | ↑PART1 (in MGC-803, BGC-823, SGC-7901) | Han et al. (2020) |
| ↓cell viability and colony formation, ↓migration, ↓invasion | |||||
| NCL: GES-1 | ∆PART1 (in AGS) | ||||
| ↑cell viability and colony formation, ↑migration, ↑invasion | |||||
| Glioma | TCLs: U87MG, LN-18, LN-428 | Down (TCLs vs. astrocytes) | miR-190a-3p, PTEN, PI3K/AKT signaling pathway | ↑PART1 | Jin et al. (2020) |
| ↓proliferation, ↑apoptosis (↓Bcl-2, ↑Bax) | |||||
| TCLs: A172, U373, LN229, U251 | Down (TCLs vs. NCLs) | miR-374b/SALL1 | ↑PART1 | Deng et al. (2022) | |
| NCL: NHA | ↓cell proliferation and viability, ↓migration, ↓EMT (↑E-cadherin, ↓N-cadherin, vimentin and Snail) | ||||
| Head and Neck Squamous Cell Carcinoma (HNSCC) | TCLs: CNE-2, C666-1, SCC-4 | Down (TCLs vs. NCLs) | — | — | Yang et al. (2021b) |
| NCL: HOK, NP69 | |||||
| Tongue Squamous Cell Carcinoma (TSCC) | TCLs: CAL-27, SCC9, SCC25 | Down (TCLs vs. NCLs) | miR-503-5p | ↑PART1 | Liu et al. (2020) |
| NCL: NHOK | ↓proliferation and viability, ↓migration, ↓invasion, ↓EMT (↓N-cadherin, ↓vimentin, ↑E-cadherin) |
Animal studies
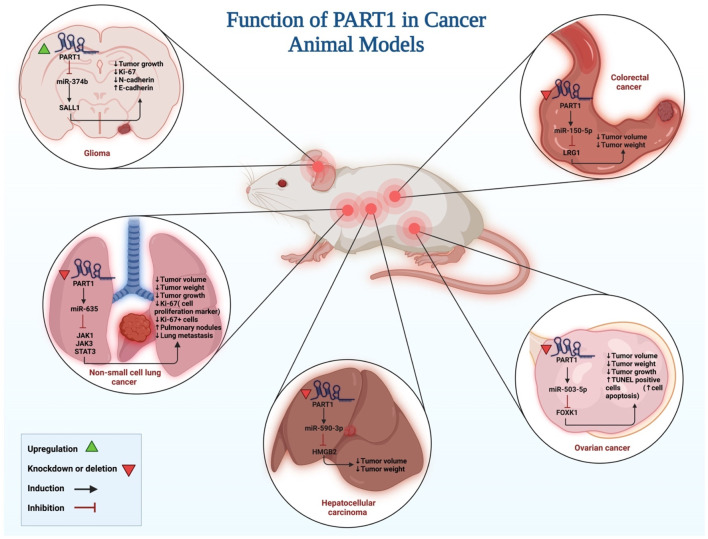
Different study groups have evaluated functional consequences of PART1 up-regulation or silencing on tumor formation in xenoraft models (Figure 3) (Table 3). Similar to in vitro studies, both tumor suppressor role and oncogenic role have been reported for PART1. Examples of the former type of function have been seen in animal models of cervical squamous cell carcinoma (Liu et al., 2020), gastric cancer (Han et al., 2020) and glioma (Deng et al., 2022) where up-regulation of PART1 has resulted in reduction of tumor growth. Animal models of colorectal carcinoma (Zhou et al., 2020a), hepatocellular carcinoma (Pu et al., 2020), lung cancer (Zhu et al., 2019), oral squamous cell carcinoma (Yu et al., 2021), ovarian cancer (Li et al., 2022b) and triple negative breast cancer (Cruickshank et al., 2021).
FIGURE 3.
An illustration depicts the roles of PART1 activation and silencing in tumor formation in xenograft models, as well as the signaling pathways involved.
TABLE 3.
Effects of PART1 in animal models for cancer (∆: knockdown or deletion, NR: not reported, CAM: chorioallantoic membrane, NOD-SCID mice: non-obese diabetic-severe combined immunodeficiency mice, TUNEL: terminal deoxynucleotidyl transferase dUTP nick end labeling, SPF: specific-pathogen-free, ↑: increase, ↓: decrease).
| Tumor type | Animal models (experimental and control group)/Number of studied animals | Target | Function | References |
|---|---|---|---|---|
| Cervical Squamous Cell Carcinoma (CSCC) | Female athymic nude mice/10 (5 for each group) | NR | ↑PART1 | Liu et al. (2020) |
| ↓tumor growth | ||||
| Colorectal cancer (CRC) | BALB/c nude mice/NR | ∆PART1 | ∆PART1 | Zhou et al. (2020a) |
| ↑miR-150-5p and miR-520 h | ↓tumor volume, ↓tumor weight, ↓tumor growth, ↓Ki-67, ↓β-catenin, ↓PCNA, ↓vimentin | |||
| BALB/c nude mice/NR | NR | ∆PART1 | Hu et al. (2017) | |
| ↓tumor growth, ↓tumor size, ↓tumor volume | ||||
| Male BALB/c nude mice/10 (5 for each group) | ∆PART1 | ∆PART1 | Lou et al. (2020) | |
| ↑miR-150-5p, ↓LRG1 | ↓tumor volume, ↓tumor weight | |||
| Esophageal Squamous Cell Carcinoma (ESCC) | Male BALB/c nude mice/NR | NR | ↑PART1 | Kang et al. (2018) |
| ↑gefitinib resistance, ↑Bcl-2, ↓Bax, ↓cleaved caspase-3, ↓cleaved PARP | ||||
| Gastric cancer | Chick embryo CAM/NR | NR | ↑PART1 | Han et al. (2020) |
| ↓tumor weight, ↓metastatic tumor colonies, ↓human Alu expression | ||||
| ∆PART1 | ||||
| ↑tumor weight, ↑lung metastasis | ||||
| NOD-SCID mice/6 (3 for each group) | NR | ↑PART1 | Han et al. (2020) | |
| ↓tumor growth, ↓lung metastasis, ↓human Alu expression | ||||
| ∆PART1 | ||||
| ↑tumor weight, ↑lung metastasis | ||||
| Glioma | SPF-grade nude mice/8 (4 for each group) | ↑PART1 | ↑PART1 | Deng et al. (2022) |
| ↑SALL1, ↓miR-374b | ↓tumor growth, ↓Ki-67, ↓N-cadherin, ↑E-cadherin | |||
| Hepatocellular Carcinoma (HCC) | BALB/c nude mice/NR | ∆PART1 | ∆PART1 | Pu et al. (2020) |
| ↓HMGB2, ↑miR-590-3p | ↓tumor volume, ↓tumor weight | |||
| Male nude mice/12 (6 for each group) | NR | ∆PART1: ↓tumorigenicity | Zhou et al. (2022) | |
| ↓tumor size, ↓tumor volume, ↓tumor mass, ↓Ki-67 positive cells, ↑apoptosis, ↑E-cadherin, ↓N-cadherin, ↓Twist, ↓Snail | ||||
| Non-small cell lung cancer (NSCLC) | BALB/c nude mice/NR | ∆PART1 | ∆PART1 | Zhu et al. (2019) |
| ↑miR-635, ↓JAK1, JAK3 and STAT3 | ↓tumor volume, ↓tumor weight, ↓tumor growth, ↓Ki-67 (cell proliferation marker), ↓Ki-67 + cells, ↑pulmonary nodules, ↓lung metastasis | |||
| Female athymic BALB/c nude mice/15 | NR | ∆PART1 | Chen et al. (2021) | |
| ↓tumor volume, ↓tumor weight, Ki-67 positive cells | ||||
| Oral Squamous Cell Carcinoma (OSCC) | BALB/c nude mice/16 (8 for each group) | ∆PART1 | ∆PART1 | Yu et al. (2021) |
| ↓EZH2 | ↓tumor volume, ↓PCNA, ↓cyclinD1, ↓Bcl-2, ↑Bax, ↑cleaved caspase-3 | |||
| Ovarian Cancer (OC) | BALB/c nude mice/10 (5 for each group) | ∆PART1 | ∆PART1 | Li et al. (2022a) |
| ↓FOXK1, ↑miR-503-5p | ↓tumor volume, ↓tumor weight, ↓tumor growth, ↑TUNEL positive cells (↑cell apoptosis) | |||
| BALB/c nude mice/NR | NR | ∆PART1 | Li et al. (2022b) | |
| ↓tumor volume, ↓tumor weight, ↓tumor growth | ||||
| Triple-negative breast cancer (TNBC) | NOD-SCID female mice/14 (7 for each group) | NR | ∆PART1 | Cruickshank et al. (2021) |
| ↓tumor volume, ↓tumor weight, ↓mammosphere formation ability |
Studies in clinical samples
Studies in clinical samples have shown up-regulation of PART1 in a variety of cancer tissues including bladder, breast and colorectal cancers (Tables 4, 5). However, there are a number of other cancerous tissues in which PART1 has been found to be down-regulated. For instance, expression of PART1 has been shown to be decreased in esophageal squamous cell carcinoma tissues parallel with down-regulation of SOX6. Notably, low expression of these two genes has been associated with TNM stage, lymph node metastasis and poor prognosis in these patients. Moreover, expression of FOXP2 has been reduced in these tissues in correlation with PART1 expression levels (Zhao et al., 2021). However, another study in this type of cancer has revealed up-regulation of PART1 in the sera samples of gefitinib non-responders versus responders (Kang et al., 2018). Moreover, PART1 is down-regulated in cervical squamous cell carcinoma tissues (Liu et al., 2020). In addition, dysregulation of PART1 has been associated with TNM stage, metastasis, tumor grade and diameter as well as histological type in a variety of cancers (Tables 4, 5).
TABLE 4.
Function of PART1 up-regulation in the development of malignancy on the basis of studies in clinical samples (ANTs: adjacent normal tissues, TCGA: the cancer genome atlas, METABRIC: molecular taxonomy of breast cancer international consortium, GEPIA: gene expression profiling interactive analysis, GEO: gene expression omnibus, GTEx: genotype–tissue expression, ENCORI: encyclopedia of RNA interaction, GBM: high-grade glioma, LGG: low-grade glioma, ER: early recurrence, BCLC: Barcelona clinic liver cancer, OS: overall survival, DFS: disease-free survival, FIGO: international federation of gynecology and obstetrics, TNM: tumor-node-metastasis, T stage: tumor stage, T classification: tumor classification).
| Tumor type | Samples | Expression (tumor vs. normal control) | Kaplan-meier analysis | Univariate cox regression analysis | Multivariate cox regression analysis | Association of dysregulation of PART1 with clinicopathologic characteristics | References |
|---|---|---|---|---|---|---|---|
| Bladder Cancer | 30 pairs of tumor tissues and ANTs + GEO database | Up-regulated | — | — | — | — | Hu et al. (2019) |
| Breast Cancer | 31 pairs of tumor tissues and ANTs | Up-regulated | High PART1 expression correlated with poorer OS | — | — | Metastasis, tumor stage | Wang and Xu (2020) |
| 30 pairs of tumor tissues and ANTs | Up-regulated | — | — | — | — | Lou et al. (2020) | |
| Triple-negative breast cancer (TNBC) | Datasets from METABRIC, Cell 2015 and TCGA PanCancer | Up-regulated (basal-like and TNBC vs. other subtype tumors) | High PART1 expression correlated with poorer survival (in basal-like BC) | — | — | — | Cruickshank et al. (2021) |
| Luminal Breast Cancer | 10 pairs of tumor tissues and ANTs + TCGA data | Up-regulated | High PART1 expression correlated with poorer OS | — | — | Ki-67, tumor grade, tumor diameter | Jiang et al. (2020) |
| Clear cell Renal Cell Carcinoma (ccRCC) | 254 tumor samples and 71 normal controls (from TCGA database) | _ | Low PART1 expression correlated with longer OS | — | — | Tumor metastasis | Liu et al. (2019) |
| Colorectal Cancer (CRC) | 38 pairs of tumor tissues and ANTs | Up-regulated | — | — | — | — | Zhou et al. (2020a) |
| 50 pairs of tumor tissues and ANTs | Up-regulated | High PART1 expression correlated with poorer OS | — | — | Tumor invasion, TNM stage | Hu et al. (2017) | |
| 56 pairs of tumor tissues and ANTs | Up-regulated | — | — | — | Lymph node metastasis, invasion depth, TNM stage | Lou et al. (2020) | |
| 10 patient blood samples and 10 normal blood samples | Up-regulated | — | — | — | — | Lou et al. (2020) | |
| Esophageal Squamous Cell Carcinoma (ESCC) | 79 serum samples from patients receiving gefitinib therapy (42 responding patients and 37 non-responding patients) | Up-regulated (non-responding vs. responding samples) | — | — | — | — | Kang et al. (2018) |
| Hepatocellular Carcinoma (HCC) | 48 pairs of tumor tissues and ANTs | Up-regulated | — | — | — | — | Zhou et al. (2020b) |
| 374 tumor and 50 normal samples (from ENCORI website) | Up-regulated | — | — | — | — | Pu et al. (2020) | |
| 255 HCC patients: 133 ER and 92 non-ER patients (from TCGA) | Up-regulated (ER vs. non-ER) | — | — | — | — | Lv et al. (2018) | |
| 51pairs of tumor tissues and ANTs + TCGA data | Up-regulated | High PART1 expression correlated with poorer OS and DFS | — | — | — | Zhou et al. (2022) | |
| Liver Cancer | 30 patient blood samples and 30 normal blood samples | Up-regulated | — | — | — | Tumor size, TNM stage, BCLC stage | Weng et al. (2021) |
| Lung Squamous Cell Carcinoma (LSCC) | 51 pairs of tumor tissues and ANTs | Up-regulated | High PART1 expression correlated with poorer OS | — | ↑PART1, ↓miR-185-5P, ↑Six1, differentiation, lymph node metastasis (independent risk factors for OS) | Tumor size, histological stage, lymph node metastasis, differentiation | Cao et al. (2021) |
| Non-small cell lung cancer (NSCLC) | 60 pairs of tumor tissues and ANTs | Up-regulated | High PART1 expression correlated with poorer OS | Histology and EGFR mutation (shorter OS) | PART1 expression, histology (independent prognostic factors for OS) | Histologic type (↑PART1 in squamous NSCLC tumors) | Zhu et al. (2019) |
| 208 pairs of tumor tissues and ANTs | Up-regulated | High PART1 expression correlated with poorer OS and DFS | High PART1 expression, high T stage, lymph node metastasis, poor differentiation (poor OS and DFS) | PART1 expression (independent prognostic factor for OS and DFS) | Histologic type (↑PART1 in squamous tumors) | Li et al. (2017) | |
| 30 pairs of tumor tissues and ANTs | Up-regulated | — | — | — | — | Chen et al. (2021) | |
| Oral Squamous Cell Carcinoma (OSCC) | 36 pairs of tumor tissues and ANTs | Up-regulated | — | — | — | Tumor size, node metastasis, clinical stage | Yu et al. (2021) |
| Ovarian Cancer (OC) | 50 pairs of tumor tissues and ANTs | Up-regulated | — | — | — | Lymph node metastasis, FIGO stage | Li et al. (2022a) |
| 426 tumor samples and 88 normal samples (from GEPIA) | Up-regulated | — | — | — | — | Li et al. (2022b) | |
| TCGA datasets | Up-regulated | — | — | — | — | Yang et al. (2021a) | |
| Pancreatic Cancer | 45pairs of tumor tissues and ANTs | Up-regulated | High PART1 expression correlated with poorer 5-year OS | — | — | Tumor size, T classification, clinical stage, vascular invasion | Ghafouri-Fard et al. (2021) |
| Pancreatic Neuroendocrine Tumors (PanNETs) | 17 tumor tissues and 8 ANTs | Up-regulated | — | — | — | — | Xiao et al. (2019) |
| Prostate Cancer (PCa) | 30 pairs of tumor tissues and ANTs | Up-regulated | — | — | — | Tumor stage, Gleason score | Sun et al. (2018) |
| 27 pairs of tumor tissues and ANTs | Up-regulated (in 18 patients), Down-regulated (in 7 patients), Similar expression (in 2 patients) | — | — | — | — | Sidiropoulos et al. (2001) |
TABLE 5.
Function of PART1 down-regulation in the development of malignancy on the basis of studies in clinical samples.
| Tumor type | Samples | Expression (tumor vs. normal control) | Kaplan-meier analysis | Univariate cox regression analysis | Multivariate cox regression analysis | Association of dysregulation of PART1 with clinicopathologic characteristics | References |
|---|---|---|---|---|---|---|---|
| Cervical Squamous Cell Carcinoma (CSCC) | 15 samples: 5 cervical cancer and 10 uterine myoma | Down-regulated (tumor vs. normal tissues) | — | — | — | — | Liu et al. (2020) |
| Esophageal Squamous Cell Carcinoma (ESCC) | 75 pairs of tumor tissues and ANTs + TCGA database and GEO dataset | Down-regulated | Low PART1 expression correlated with shorter survival | — | — | TNM stage, lymph node metastasis | Zhao et al. (2021) |
| Gastric Cancer | 15 pairs of tumor tissues and ANTs | Down-regulated | — | — | — | — | Gu et al. (2019) |
| 136 tumor tissues and 94 ANTs | Down-regulated | Low PART1 expression correlated with shorter OS | — | — | Distant tumor metastasis, liver metastasis | Han et al. (2020) | |
| Glioma | 50 tumor tissues and 6 normal brain tissues | Down-regulated | — | — | — | — | Jin et al. (2020) |
| GEPIA and TCGA dataset | Down-regulated (GBM vs. normal, LGG vs. normal, and GBM vs. LGG) | — | — | — | — | Jin et al. (2020) | |
| 665 tumor samples (from TCGA) and 188 normal control samples (from GTEx) | Down-regulated | — | — | — | — | Yang et al. (2021c) | |
| Head and Neck Squamous Cell Carcinoma (HNSCC) | 10 patient blood samples and 10 normal blood samples + GEPIA database | Down-regulated | — | — | — | — | Yang et al. (2021b) |
| Tongue Squamous Cell Carcinoma (TSCC) | 40 pairs of tumor tissues and ANTs | Down-regulated | Low PART1 expression correlated with poorer OS | — | — | Tumor classification, clinical stage, lymph node metastasis | Liu et al. (2020) |
| 147 tumor samples and 15 normal samples (from TCGA database) | _ | High PART1 expression correlated with longer OS | — | — | — | Song et al. (2019) | |
| 122 tumor samples and 15 normal samples (from TCGA database) | Down-regulated | — | — | — | — | Zhang et al. (2019b) |
Diagnostic value of PART1
Diagnostic value of PART1 has been evaluated in the context of esophageal squamous cell carcinoma (Kang et al., 2018) and lung squamous cell carcinoma (Cao et al., 2021) (Table 6). In the former type of cancer, PART1 levels could differentiate between gefitinib responders and non-responders with AUC value of 0.839 (Kang et al., 2018). In the latter type of cancer, this lncRNA could separate cancerous and non-cancerous tissues with AUC value of 0.7857 (Cao et al., 2021).
TABLE 6.
Value of PART1 in cancer diagnosis (ANTs: adjacent normal tissues).
| Tumor type | Samples | Distinguish between | Area under the curve (AUC) | Sensitivity (%) | Specificity (%) | References |
|---|---|---|---|---|---|---|
| Esophageal Squamous Cell Carcinoma (ESCC) | 79 serum samples from patients receiving gefitinib therapy | 37 non-responding patients vs. 42 responding patients | 0.839 | 78.6 | 86.5 | Kang et al. (2018) |
| Lung Squamous Cell Carcinoma (LSCC) | 51 pairs of tumor tissues and ANTs | LSCC tissues vs. ANTs | 0.7857 | 66.67 | 86.27 | Cao et al. (2021) |
Role of PART1 in non-malignant disorders
Cell line studies
PART1 is among lncRNAs that are dysregulated in SARS-CoV-2 infected cells as revealed by an in silico analysis of GSE147507 dataset. Expression of PART1 has been found to reduced in at least two independent SARS-CoV-2-infected cell lines. Dysregulated lncRNAs have been shown to interact with a variety of genes/proteins and miRNAs which have been linked with signaling pathways regulating viral infection, inflammatory responses and immune function (Laha et al., 2021). PART1 is alos involved in the pathogenesis of intervertebral disc degeneration via regulation of the miR-93/MMP2 axis (Gao et al., 2020) as well as miR-190a-3p expression (Zhang et al., 2021a). Tables 7, 8 summarize the role of PART1 in other non-malignant disorders based on cell line studies that reported up-regulation and down-regulation of PART1, respectively.
TABLE 7.
Cell line studies on PART1 function in non-malignant illnesses in which PART1 has been up-regulated (∆: knockdown or deletion, NP cells: nucleus pulposus cells, ECM: extracellular matrix, LPS: lipopolysaccharide, MPP+: methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine, H/R: hypoxia/reoxygenation, ROS: reactive oxygen species, MMP13: matrix metallopeptidase13, MMP: mitochondrial membrane potential, ↑: increase, ↓: decrease).
| Disorder | Cell line | Expression | Targets/Regulators and signaling pathways | Function | References |
|---|---|---|---|---|---|
| Intervertebral Disc Degeneration (IDD) | NP cells (derived from IDD patients) | — | miR-93-5p/MMP2 | ∆PART1 | Gao et al. (2020) |
| ↑proliferation (↑Ki-67), ↑colony formation ability, ↓apoptosis (↓cleaved caspase-3), ↑ECM synthesis (↑aggrecan and collagen II), ↓ECM degradation (↓ADAMTS4 and MMP13) | |||||
| In vitro IDD models: LPS-stimulated NP cells | High (LPS-induced NP cells vs. normal NP cells) | miR-190a-3p | ∆PART1 | Zhang et al. (2021a) | |
| Controls: NP cells | ↑cell viability, ↓apoptosis, ↓inflammatory response (↓TNF-α, ↓IL-1β, ↓IL-6), ↓ECM degradation (↑aggrecan, ↑collagen II) | ||||
| Osteoarthritis (OA) | C20/A4 (the immortalized human chondrocytes cell lines) | — | miR-590-3p, TGFBR2/Smad3 signaling pathway | ∆PART1 | Lu et al. (2019) |
| ↓cell viability, ↑apoptosis (↑cleaved caspase-3 and caspase-9, ↑Bax) | |||||
| ↑PART1 | |||||
| ↓effects of IL-1β | |||||
| ↑cell viability and ↓apoptosis rate | |||||
| OA chondrocytes and normal chondrocytes | High (OA cells vs. normal cells) | miR-373-3p/SOX4 | ∆PART1 | Zhu and Jiang (2019) | |
| ↓Cell proliferation and viability, ↓ECM degradation (↓MMP13, ↑collagen II, ↑aggrecan), ↑apoptosis (↓Bcl-2, ↑Bax, ↑cleaved caspase-3) | |||||
| Osteoporosis (OP) | hBMSCs (human bone marrow-derived mesenchymal stem cells) | High (osteogenesis-induced BMSCs vs. controls) | Targets: miR-185-5p/RUNX3 | ∆PART1 (in hBMSCs) | Zhang et al. (2021b) |
| Transcriptional activator of PART1: RUNX3 | ↓osteogenic differentiation (↓osteogenesis markers such as OCN, OSX and COL1A, ↓ALP activity, ↓matrix mineralization), ↑apoptosis |
TABLE 8.
Non-malignant illnesses in which PART1 has been down-regulated (↑: increase, ↓: decrease).
| Disorder | Cell line | Expression | Targets/Regulators and signaling pathways | Function | References |
|---|---|---|---|---|---|
| COVID-19 (coronavirus disease 19) | A549, Calu3 | Down-regulated (SARS-CoV-2 infected cells vs. control cells) | — | — | Laha et al. (2021) |
| Myocardial Ischemia-Reperfusion Injury (MI/RI) | In vitro H/R model: H/R NMVCs (neonatal mice ventricle cells) | Down-regulated (H/R cells vs. controls) | miR-503-5p/BIRC5 | ↑PART1 | Guo et al. (2021) |
| Controls: NMVCs | ↑cell viability, ↓apoptosis (↓ H/R injury) | ||||
| ↑mitochondrial function (↓ROS, ↑ATP level, ↑GSH level, ↑MMP level) | |||||
| Parkinson’s disease (PD) | In vitro PD models: MPP(+)-treated SH-SY5Y cells | Down-regulated (PD model cells vs. controls) | microRNA-106b-5p/MCL1 | ↑PART1: ↓effects of MPP + treatment | Shen et al. (2021) |
| Control group: SH-SY5Y cells | ↑cell viability, ↓apoptosis (↓cleaved caspase-3), ↓inflammatory response (↓TNF-α, IL-1β and IL-6), ↓oxidative stress (↓LDH and ROS, ↑SOD) |
Animal studies
Two different studies in animal models have shown the importance of PART1 in myocardial ischemia-reperfusion injury (Guo et al., 2021) and Parkinson’s disease (Shen et al., 2021) (Table 9). In animal models of myocardial ischemia-reperfusion injury, up-regulation of PART1 has resulted in the alleviation of tissue injury, enhancement of cardiac function and reduction of infarction size (Guo et al., 2021).
TABLE 9.
Animal studies on the involvement of PART1 in non-malignant disorders (MPTP: methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine hydrochloride, I/R: Ischemia-Reperfusion, EF: ejection fraction, FS: fraction shortening, ↑: increase, ↓: decrease).
| Disorder | Animal model (experimental and control group)/Number of studied animals | Expression | Result | References |
|---|---|---|---|---|
| Myocardial Ischemia-Reperfusion Injury (MI/RI) | Male C57BL/6 mice (in vivo I/R model)/40 | Down-regulated (I/R models vs. controls) | ↑PART1 | Guo et al. (2021) |
| I/R injury alleviation | ||||
| ↑left ventricular EF and FS, ↓infract size, ↓Bax, ↓cytochrome-c, ↑Bcl-2 | ||||
| Parkinson’s disease (PD) | C57BL/6 mice (in vivo PD model through receiving MPTP)/10 for each group | Down-regulated (MPTP group vs. controls) | PART1 alleviates MPP(+)-associated neuronal damage through modulation of miR-106b-5p/MCL1 axis | Shen et al. (2021) |
Studies in clinical samples
Experiments in clinical samples have shown down-regulation of PART1 in biological samples obtained from patients with Alzheimer’s disease (Huaying et al., 2020), Parkinson’s disease (Chi et al., 2019) and preeclampsia (Peñailillo et al., 2022). On the other hand, PART1 has been found to be up-regulated in nucleus pulposus samples of patients with intervertebral disc degeneration (Gao et al., 2020). Table 10 shows the results of studies on humans samples to ascertain how PART1 is expressed in non-cancerous disorders.
TABLE 10.
Studies on humans samples to ascertain how PART1 is expressed in non-cancerous disorders (NP: nucleus pulposus).
| Disorder | Samples | Expression (disease group vs. normal controls) | References |
|---|---|---|---|
| Alzheimer’s disease (AD) | AD and normal serum samples | Down-regulated | Huaying et al. (2020) |
| Intervertebral Disc Degeneration (IDD) | 30 NP tissues from IDD patients and 30 control NP tissues | Up-regulated | Gao et al. (2020) |
| Osteoarthritis (OA) | 30 OA cartilage tissues and 30 normal cartilage tissues | Down-regulated | Lu et al. (2019) |
| 35 cartilage tissues from OA patients and 15 cartilage tissues from patients without OA) | Up-regulated | Zhu and Jiang (2019) | |
| Parkinson’s disease (PD) | 50 PD blood samples and 22 controls | Down-regulated | Chi et al. (2019) |
| Preeclampsia | 7 preeclampsia placentas and 7 control placenta samples | Down-regulated | Peñailillo et al. (2022) |
Discussion
PART1 is an lncRNA with diverse functions in the carcinogenesis (Lin et al., 2000). It can affect maintenance of cancer stem cells (Cruickshank et al., 2021) and epithelial to mesenchymal transition (Lou et al., 2020) in a variety of tissues. Moreover, it has a role in modulation of response of cancer cells to cisplatin, erlotinib and gefitinib. Mechanistically, PART1 can act as molecular sponge for a variety of miRNAs such as miR-4516, miR-150-5p, miR-143, miR-18a-5p, miR-129, miR-190a-3p, miR-374b, miR-149-5p, miR-590-3p, miR-372-3p, miR-3529-3p, miR-185-5p, miR-17-5p, miR-503-5p, miR-6884-5p, miR-512-3p, miR-122 and miR-503-5p. It can regulate activity of some cancer-related signaling pathways such as Wnt/β-catenin, PI3K/AKT, PTEN and JAK/STAT3 (Zhu et al., 2019).
Transcription of PART1 can be regulated by a number of transcription factors such as androgens, ∆Np63α, FOXP2, STAT1 and YY1. However, the importance of methylation marks in its promotor on PART1 expression has not been elucidated.
An important feature of PART1 participation in the carcinogenesis is its diverse roles and possibly its tissue-dependent functions in this process. Future studies should identify the mechanism of such tissue-dependent functions and determinants its oncogenic versus tumor suppressor roles.
Since dysregulation of PART1 in tumor tissues has been associated with aggressive behavior of cancer cells, PART1 can be regarded as a prognostic factor in different types of cancers. However, data regarding the application of PART1 as a diagnostic tool in cancer is not sufficient. Since abnormal expression of PART1 has been reported in a variety of cancers, it is possible that expression levels of PART1 can differentiate cancerous tissues from normal counterparts with appropriate diagnostic power.
Taken together, PART1 participates in the pathogenesis of cancer and a variety of non-cancerous conditions including neurodegenerative disorders. Diagnostic value of PART1 has been assessed in few types of cancers, including esophageal (Kang et al., 2018) and lung (Cao et al., 2021) cancers revealing promising results. Moreover, modulation of expression of PART1 in cancer cell lines or animal models of cancers have been associated with therapeutic benefits. However, this filed lacks sufficient data from clinical models. Future functional studies can provide important information about the underlying mechanisms and consequences of PART1 dysregulation in these disorders. The results of such studies can help in design of novel therapeutic modalities based on this lncRNA, particularly in cancers.
Acknowledgments
The authors would like to thank the Clinical Research Development Unit (CRDU) of Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran for their support, cooperation and assistance throughout the period of study.
Author contributions
SG-F wrote the draft and revised it. MT and AB designed and supervised the study. BH, GS, AH, and SA collected the data and designed the figures and tables. All the authors read the submitted version and approved it.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Cao Y., Zhang R., Luo X., Yang Y. (2021). LncRNA PART1 promotes lung squamous cell carcinoma progression via miR-185-5p/Six1 axis. Hum. Exp. Toxicol. 40 (6), 960–976. PubMed PMID: 33300377. Epub 2020/12/11. eng. 10.1177/0960327120979032 [DOI] [PubMed] [Google Scholar]
- Chen S. C., Diao Y. Z., Zhao Z. H., Li X. L. (2020). Inhibition of lncRNA PART1 chemosensitizes wild type but not KRAS mutant NSCLC cells. Cancer Manag. Res. 12, 4453–4460. PubMed PMID: 32606939. Pubmed Central PMCID: PMC7293907. Epub 2020/07/02. eng. 10.2147/CMAR.S245257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhou X., Huang C., Li L., Qin Y., Tian Z., et al. (2021). LncRNA PART1 promotes cell proliferation and progression in non-small-cell lung cancer cells via sponging miR-17-5p. J. Cell. Biochem. 122 (3-4), 315–325. PubMed PMID: 33368623. Epub 2020/12/29. eng. 10.1002/jcb.29714 [DOI] [PubMed] [Google Scholar]
- Chi L. M., Wang L. P., Jiao D. (2019). Identification of differentially expressed genes and long noncoding RNAs associated with Parkinson's disease. Park. Dis. 2019, 6078251. PubMed PMID: 30867898. Pubmed Central PMCID: PMC6379850. Epub 2019/03/15. eng. 10.1155/2019/6078251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank B. M., Wasson M. D., Brown J. M., Fernando W., Venkatesh J., Walker O. L., et al. (2021). LncRNA PART1 promotes proliferation and migration, is associated with cancer stem cells, and alters the miRNA landscape in triple-negative breast cancer. Cancers (Basel) 13 (11), 2644. PubMed PMID: 34072264. Pubmed Central PMCID: PMC8198907. Epub 2021/06/03. eng. 10.3390/cancers13112644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y. W., Shu Y. G., Sun S. L. (2022). LncRNA PART1 inhibits glioma proliferation and migration via miR-374b/SALL1 axis. Neurochem. Int. 157, 105347. PubMed PMID: 35490895. Epub 2022/05/02. eng. 10.1016/j.neuint.2022.105347 [DOI] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22 (9), 1775–1789. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Hao L., Zhao Z. (2020). Long non-coding RNA PART1 promotes intervertebral disc degeneration through regulating the miR-93/MMP2 pathway in nucleus pulposus cells. Int. J. Mol. Med. 46 (1), 289–299. PubMed PMID: 32319551. Pubmed Central PMCID: PMC7255469. Epub 2020/04/23. eng. 10.3892/ijmm.2020.4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafouri-Fard S., Hussen B. M., Gharebaghi A., Eghtedarian R., Taheri M. (2021). LncRNA signature in colorectal cancer. Pathology-Research Pract. 222, 153432. 10.1016/j.prp.2021.153432 [DOI] [PubMed] [Google Scholar]
- Ghafouri-Fard S., Sohrabi B., Hussen B. M., Mehravaran E., Jamali E., Arsang-Jang S., et al. (2022). Down-regulation of MEG3, PANDA and CASC2 as p53-related lncRNAs in breast cancer. Breast Dis. 41 (1), 137–143. PubMed PMID: 35034894. eng. 10.3233/BD-210069 [DOI] [PubMed] [Google Scholar]
- Gu W., Ren J. H., Zheng X., Hu X. Y., Hu M. J. (2019). Comprehensive analysis of expression profiles of long non-coding RNAs with associated ceRNA network involved in gastric cancer progression. Mol. Med. Rep. 20 (3), 2209–2218. PubMed PMID: 31322220. Pubmed Central PMCID: PMC6691204. Epub 2019/07/20. eng. 10.3892/mmr.2019.10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Zhao M., Jia G., Ma R., Li M. (2021). LncRNA PART1 alleviated myocardial ischemia/reperfusion injury via suppressing miR-503-5p/BIRC5 mediated mitochondrial apoptosis. Int. J. Cardiol. 338, 176–184. PubMed PMID: 34082009. Epub 2021/06/04. eng. 10.1016/j.ijcard.2021.05.044 [DOI] [PubMed] [Google Scholar]
- Han H., Wang S., Meng J., Lyu G., Ding G., Hu Y., et al. (2020). Long noncoding RNA PART1 restrains aggressive gastric cancer through the epigenetic silencing of PDGFB via the PLZF-mediated recruitment of EZH2. Oncogene 39 (42), 6513–6528. PubMed PMID: 32901105. Epub 2020/09/10. eng. 10.1038/s41388-020-01442-5 [DOI] [PubMed] [Google Scholar]
- Hu Y., Ma Z., He Y., Liu W., Su Y., Tang Z. (2017). PART-1 functions as a competitive endogenous RNA for promoting tumor progression by sponging miR-143 in colorectal cancer. Biochem. Biophys. Res. Commun. 490 (2), 317–323. PubMed PMID: 28619512. Epub 2017/06/18. eng. 10.1016/j.bbrc.2017.06.042 [DOI] [PubMed] [Google Scholar]
- Hu X., Feng H., Huang H., Gu W., Fang Q., Xie Y., et al. (2019). Downregulated long noncoding RNA PART1 inhibits proliferation and promotes apoptosis in bladder cancer. Technol. Cancer Res. Treat. 18, 1533033819846638. PubMed PMID: 31311442. Pubmed Central PMCID: PMC6636221. Epub 2019/07/18. eng. 10.1177/1533033819846638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huaying C., Xing J., Luya J., Linhui N., Di S., Xianjun D. (2020). A signature of five long non-coding RNAs for predicting the prognosis of Alzheimer's disease based on competing endogenous RNA networks. Front. Aging Neurosci. 12, 598606. PubMed PMID: 33584243. Pubmed Central PMCID: PMC7876075. Epub 2021/02/16. eng. 10.3389/fnagi.2020.598606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussen B. M., Kheder R. K., Abdullah S. T., Hidayat H. J., Rahman H. S., Salihi A., et al. (20222022). Functional interplay between long non-coding RNAs and Breast CSCs. Cancer Cell. Int. 22 (1), 233. 10.1186/s12935-022-02653-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M. C., Ni J. J., Cui W. Y., Wang B. Y., Zhuo W. (2019). Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 9 (7), 1354–1366. PubMed PMID: 31392074. Pubmed Central PMCID: PMC6682721. Epub 2019/08/09. eng. [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Cheng P., Luo B., Huang J. (2020). Construction and analysis of a long non-coding RNA-associated competing endogenous RNA network identified potential prognostic biomarkers in luminal breast cancer. Onco Targets Ther. 13, 4271–4282. PubMed PMID: 32547061. Pubmed Central PMCID: PMC7244246. Epub 2020/06/18. eng. 10.2147/OTT.S240973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Piao L., Sun G., Lv C., Jing Y., Jin R. (2020). Long non-coding RNA PART1 exerts tumor suppressive functions in glioma via sponging miR-190a-3p and inactivation of PTEN/AKT pathway. Onco Targets Ther. 13, 1073–1086. PubMed PMID: 32099409. Pubmed Central PMCID: PMC7007780. Epub 2020/02/27. eng. 10.2147/OTT.S232848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Ren M., Li Y., Fu Y., Deng M., Li C. (2018). Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J. Exp. Clin. Cancer Res. 37 (1), 171. PubMed PMID: 30049286. Pubmed Central PMCID: PMC6063009. Epub 2018/07/28. eng. 10.1186/s13046-018-0845-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Laha S., Saha C., Dutta S., Basu M., Chatterjee R., Ghosh S., et al. (2021). In silico analysis of altered expression of long non-coding RNA in SARS-CoV-2 infected cells and their possible regulation by STAT1, STAT3 and interferon regulatory factors. Heliyon 7 (3), e06395. PubMed PMID: 33688586. Pubmed Central PMCID: PMC7914022. Epub 2021/03/11. eng. 10.1016/j.heliyon.2021.e06395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhang W., Zhang S., Wang C., Lin Y. (2017). PART1 expression is associated with poor prognosis and tumor recurrence in stage I-III non-small cell lung cancer. J. Cancer 8 (10), 1795–1800. PubMed PMID: 28819376. Pubmed Central PMCID: PMC5556642. Epub 2017/08/19. eng. 10.7150/jca.18848 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li B., Lou G., Zhang J., Cao N., Yu X. (2022). Repression of lncRNA PART1 attenuates ovarian cancer cell viability, migration and invasion through the miR-503-5p/FOXK1 axis. BMC Cancer 22 (1), 124. PubMed PMID: 35100978. Pubmed Central PMCID: PMC8802513. Epub 2022/02/02. eng. 10.1186/s12885-021-09005-x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li H., Lei Y., Li S., Li F., Lei J. (2022). LncRNA PART1 stimulates the development of ovarian cancer by up-regulating RACGAP1 and RRM2. Reprod. Sci. 29 (8), 2224–2235. PubMed PMID: 35553409. Epub 2022/05/14. eng. 10.1007/s43032-022-00905-2 [DOI] [PubMed] [Google Scholar]
- Lin B., White J. T., Ferguson C., Bumgarner R., Friedman C., Trask B., et al. (2000). PART-1: A novel human prostate-specific, androgen-regulated gene that maps to chromosome 5q12. Cancer Res. 60 (4), 858–863. PubMed PMID: 10706094. Epub 2000/03/08. eng. [PubMed] [Google Scholar]
- Liu B., Ma T., Li Q., Wang S., Sun W., Li W., et al. (2019). Identification of a lncRNA-associated competing endogenous RNA-regulated network in clear cell renal cell carcinoma. Mol. Med. Rep. 20 (1), 485–494. PubMed PMID: 31180525. Pubmed Central PMCID: PMC6580006. Epub 2019/06/11. eng. 10.3892/mmr.2019.10290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhu C., Xu Z., Wang J., Qian L., Zhou Q., et al. (2020). lncRNA PART1 and MIR17HG as ΔNp63α direct targets regulate tumor progression of cervical squamous cell carcinoma. Cancer Sci. 111 (11), 4129–4141. PubMed PMID: 32920922. Pubmed Central PMCID: PMC7648017. Epub 2020/09/14. eng. 10.1111/cas.14649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou T., Ke K., Zhang L., Miao C., Liu Y. (2020). LncRNA PART1 facilitates the malignant progression of colorectal cancer via miR-150-5p/LRG1 axis. J. Cell. Biochem. 121 (10), 4271–4281. PubMed PMID: 31898365. Epub 2020/01/04. eng. 10.1002/jcb.29635 [DOI] [PubMed] [Google Scholar]
- Lu C., Li Z., Hu S., Cai Y., Peng K. (2019). LncRNA PART-1 targets TGFBR2/Smad3 to regulate cell viability and apoptosis of chondrocytes via acting as miR-590-3p sponge in osteoarthritis. J. Cell. Mol. Med. 23 (12), 8196–8205. PubMed PMID: 31571401. Pubmed Central PMCID: PMC6850963. Epub 2019/10/02. eng. 10.1111/jcmm.14690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. Y., Hua J., Liu J., Wei M. Y., Liang C., Meng Q. C., et al. (2022). Construction of a paclitaxel-related competitive endogenous RNA network and identification of a potential regulatory axis in pancreatic cancer. Transl. Oncol. 20, 101419. PubMed PMID: 35413498. Pubmed Central PMCID: PMC9018166. Epub 2022/04/13. eng. 10.1016/j.tranon.2022.101419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y., Wei W., Huang Z., Chen Z., Fang Y., Pan L., et al. (2018). Long non-coding RNA expression profile can predict early recurrence in hepatocellular carcinoma after curative resection. Hepatol. Res. 48 (13), 1140–1148. PubMed PMID: 29924905. Epub 2018/06/21. eng. 10.1111/hepr.13220 [DOI] [PubMed] [Google Scholar]
- Marchese F. P., Raimondi I., Huarte M. (2017). The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 18 (1), 206–213. 10.1186/s13059-017-1348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñailillo R., Monteiro L. J., Acuña-Gallardo S., García F., Velásquez V., Correa P., et al. (2022). Identification of LOC101927355 as a novel biomarker for preeclampsia. Biomedicines 10 (6), 1253. PubMed PMID: 35740273. Pubmed Central PMCID: PMC9219905. Epub 2022/06/25. eng. 10.3390/biomedicines10061253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Tan C., Shao Z., Wu X., Zhang Y., Xu Z., et al. (2020). Long noncoding RNA PART1 promotes hepatocellular carcinoma progression via targeting miR-590-3p/HMGB2 Axis. Onco Targets Ther. 13, 9203–9211. PubMed PMID: 32982307. Pubmed Central PMCID: PMC7502387. Epub 2020/09/29. eng. 10.2147/OTT.S259962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran R., Gong C. Y., Wang Z. Q., Zhou W. M., Zhang S. B., Shi Y. Q., et al. (2022). Long non-coding RNA PART1: Dual role in cancer. Hum. Cell. 35 (5), 1364–1374. PubMed PMID: 35864416. Epub 2022/07/22. eng. 10.1007/s13577-022-00752-y [DOI] [PubMed] [Google Scholar]
- Shen Y., Cui X., Xu N., Hu Y., Zhang Z. (2021). lncRNA PART1 mitigates MPP(+)-induced neuronal injury in SH-SY5Y cells via micRNA-106b-5p/MCL1 axis. Am. J. Transl. Res. 13 (8), 8897–8908. PubMed PMID: 34540003. Pubmed Central PMCID: PMC8430160. Epub 2021/09/21. eng. [PMC free article] [PubMed] [Google Scholar]
- Sidiropoulos M., Chang A., Jung K., Diamandis E. P. (2001). Expression and regulation of prostate androgen regulated transcript-1 (PART-1) and identification of differential expression in prostatic cancer. Br. J. Cancer 85 (3), 393–397. PubMed PMID: 11487271. Pubmed Central PMCID: PMC2364080. Epub 2001/08/07. eng. 10.1054/bjoc.2001.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Pan Y., Liu J. (2019). Functional analysis of lncRNAs based on competitive endogenous RNA in tongue squamous cell carcinoma. PeerJ 7, e6991. PubMed PMID: 31179185. Pubmed Central PMCID: PMC6544013. Epub 2019/06/11. eng. 10.7717/peerj.6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Geng D., Li S., Chen Z., Zhao W. (2018). LncRNA PART1 modulates toll-like receptor pathways to influence cell proliferation and apoptosis in prostate cancer cells. Biol. Chem. 399 (4), 387–395. PubMed PMID: 29261512. Epub 2017/12/21. eng. 10.1515/hsz-2017-0255 [DOI] [PubMed] [Google Scholar]
- Wang Z., Xu R. (2020). lncRNA PART1 promotes breast cancer cell progression by directly targeting miR-4516. Cancer Manag. Res. 12, 7753–7760. PubMed PMID: 32922076. Pubmed Central PMCID: PMC7457826. Epub 2020/09/15. eng. 10.2147/CMAR.S249296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Z., Peng J., Wu W., Zhang C., Zhao J., Gao H. (2021). Downregulation of PART1 inhibits proliferation and differentiation of Hep3B cells by targeting hsa-miR-3529-3p/FOXC2 Axis. J. Oncol. 2021, 7792223. PubMed PMID: 34484336. Pubmed Central PMCID: PMC8410447. Epub 2021/09/07. eng. 10.1155/2021/7792223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Yang Y., Wang Y., Li X., Wang W. (2019). Five novel genes related to the pathogenesis and progression of pancreatic neuroendocrine tumors by bioinformatics analysis with RT-qPCR verification. Front. Neurosci. 13, 937. PubMed PMID: 31607839. Pubmed Central PMCID: PMC6771308. Epub 2019/10/15. eng. 10.3389/fnins.2019.00937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Zhang X., Zhu L., Yang Y., Yin X. (2021). YY1-Induced lncRNA PART1 enhanced resistance of ovarian cancer cells to cisplatin by regulating miR-512-3p/CHRAC1 Axis. DNA Cell. Biol. 40 (6), 821–832. PubMed PMID: 34030482. Epub 2021/05/26. eng. 10.1089/dna.2021.0059 [DOI] [PubMed] [Google Scholar]
- Yang L., Lu P., Yang X., Li K., Chen X., Qu S. (2021). Excavating novel diagnostic and prognostic long non-coding RNAs (lncRNAs) for head and neck squamous cell carcinoma: An integrated bioinformatics analysis of competing endogenous RNAs (ceRNAs) and gene co-expression networks. Bioengineered 12 (2), 12821–12838. PubMed PMID: 34898376. Pubmed Central PMCID: PMC8810019. Epub 2021/12/14. eng. 10.1080/21655979.2021.2003925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Gong W., Zhang T., Gao H. (2021). Molecular features of glioma determined and validated using combined TCGA and GTEx data analyses. Front. Oncol. 11, 729137. PubMed PMID: 34660294. Pubmed Central PMCID: PMC8516354. Epub 2021/10/19. eng. 10.3389/fonc.2021.729137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Du Y., Wang S., Zheng X. (2021). LncRNA PART1 promotes cell proliferation and inhibits apoptosis of oral squamous cell carcinoma by blocking EZH2 degradation. J. Biochem. 169 (6), 721–730. PubMed PMID: 33725092. Epub 2021/03/17. eng. 10.1093/jb/mvab026 [DOI] [PubMed] [Google Scholar]
- Zhang X., Wang W., Zhu W., Dong J., Cheng Y., Yin Z., et al. (2019). Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 20 (22), 5573. PubMed PMID: 31717266. Pubmed Central PMCID: PMC6888083. Epub 2019/11/14. eng. 10.3390/ijms20225573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Cao R., Li Q., Yao M., Chen Y., Zhou H. (2019). Comprehensive analysis of lncRNA-associated competing endogenous RNA network in tongue squamous cell carcinoma. PeerJ 7, e6397. PubMed PMID: 30755833. Pubmed Central PMCID: PMC6368841. Epub 2019/02/14. eng. 10.7717/peerj.6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Huo Y., Zhou Z., Zhang P., Hu J. (2021). Role of lncRNA PART1 in intervertebral disc degeneration and associated underlying mechanism. Exp. Ther. Med. 21 (2), 131. PubMed PMID: 33376513. Pubmed Central PMCID: PMC7751492. Epub 2020/12/31. eng. 10.3892/etm.2020.9563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Xu N., Yu C., Miao K., Wang Q. (2021). LncRNA PART1/miR-185-5p/RUNX3 feedback loop modulates osteogenic differentiation of bone marrow mesenchymal stem cells. Autoimmunity 54 (7), 422–429. PubMed PMID: 34431433. Epub 2021/08/26. eng. 10.1080/08916934.2021.1966771 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhang Q., Liu H., Wang N., Zhang X., Yang S. (2021). lncRNA PART1, manipulated by transcriptional factor FOXP2, suppresses proliferation and invasion in ESCC by regulating the miR-18a-5p/SOX6 signaling axis. Oncol. Rep. 45 (3), 1118–1132. PubMed PMID: 33432363. Pubmed Central PMCID: PMC7859983. Epub 2021/01/13. eng. 10.3892/or.2021.7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Wu L., Ma N., Tang F., Zong Z., Chen S. (2020). LncRNA PART1 regulates colorectal cancer via targeting miR-150-5p/miR-520h/CTNNB1 and activating Wnt/β-catenin pathway. Int. J. Biochem. Cell. Biol. 118, 105637. PubMed PMID: 31669140. Epub 2019/11/02. eng. 10.1016/j.biocel.2019.105637 [DOI] [PubMed] [Google Scholar]
- Zhou C., Wang P., Tu M., Huang Y., Xiong F., Wu Y. (2020). Long non-coding RNA PART1 promotes proliferation, migration and invasion of hepatocellular carcinoma cells via miR-149-5p/MAP2K1 Axis. Cancer Manag. Res. 12, 3771–3782. PubMed PMID: 32547213. Pubmed Central PMCID: PMC7248804. Epub 2020/06/18. eng. 10.2147/CMAR.S246311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Che J., Xu L., Yang W., Zhou W., Zhou C. (2022). Tumor-derived extracellular vesicles containing long noncoding RNA PART1 exert oncogenic effect in hepatocellular carcinoma by polarizing macrophages into M2. Dig. Liver Dis. 54 (4), 543–553. PubMed PMID: 34497040. Epub 2021/09/10. eng. 10.1016/j.dld.2021.07.005 [DOI] [PubMed] [Google Scholar]
- Zhu D., Yu Y., Wang W., Wu K., Liu D., Yang Y., et al. (2019). Long noncoding RNA PART1 promotes progression of non-small cell lung cancer cells via JAK-STAT signaling pathway. Cancer Med. 8 (13), 6064–6081. PubMed PMID: 31436388. Pubmed Central PMCID: PMC6792487. Epub 2019/08/23. eng. 10.1002/cam4.2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. J., Jiang D. M. (2019). LncRNA PART1 modulates chondrocyte proliferation, apoptosis, and extracellular matrix degradation in osteoarthritis via regulating miR-373-3p/SOX4 axis. Eur. Rev. Med. Pharmacol. Sci. 23 (19), 8175–8185. PubMed PMID: 31646607. Epub 2019/10/28. eng. 10.26355/eurrev_201910_19124 [DOI] [PubMed] [Google Scholar]