Abstract
Two xylanase-encoding genes, named xyn11A and xyn10B, were isolated from a genomic library of Cellulomonas pachnodae by expression in Escherichia coli. The deduced polypeptide, Xyn11A, consists of 335 amino acids with a calculated molecular mass of 34,383 Da. Different domains could be identified in the Xyn11A protein on the basis of homology searches. Xyn11A contains a catalytic domain belonging to family 11 glycosyl hydrolases and a C-terminal xylan binding domain, which are separated from the catalytic domain by a typical linker sequence. Binding studies with native Xyn11A and a truncated derivative of Xyn11A, lacking the putative binding domain, confirmed the function of the two domains. The second xylanase, designated Xyn10B, consists of 1,183 amino acids with a calculated molecular mass of 124,136 Da. Xyn10B also appears to be a modular protein, but typical linker sequences that separate the different domains were not identified. It comprises a N-terminal signal peptide followed by a stretch of amino acids that shows homology to thermostabilizing domains. Downstream of the latter domain, a catalytic domain specific for family 10 glycosyl hydrolases was identified. A truncated derivative of Xyn10B bound tightly to Avicel, which was in accordance with the identified cellulose binding domain at the C terminus of Xyn10B on the basis of homology. C. pachnodae, a (hemi)cellulolytic bacterium that was isolated from the hindgut of herbivorous Pachnoda marginata larvae, secretes at least two xylanases in the culture fluid. Although both Xyn11A and Xyn10B had the highest homology to xylanases from Cellulomonas fimi, distinct differences in the molecular organizations of the xylanases from the two Cellulomonas species were identified.
Xylan, a major component of the plant cell wall, consists of a backbone of β-1,4-linked d-xylosyl residues with substitutions of arabinosyl, acetyl, and glucuronosyl residues (39, 40). The hydrolysis of the xylan backbone involves the action of endo-β-1,4-d-xylanases (EC 3.2.1.8) and β-d-xylosidases (EC 3.2.1.37). Recently, a (hemi)cellulolytic bacterium belonging to the genus Cellulomonas was isolated from the hindgut of Pachnoda marginata larvae (5). The larvae of these herbivorous insects digest their lignocellulose-rich diet (e.g., beech litter) with the aid of intestinal microorganisms (1, 4). The bacterium Cellulomonas pachnodae secretes both xylanase and endoglucanase activities into the culture medium (5). Thus far, xylanase-encoding genes from several Cellulomonas species have been cloned and sequenced (2, 7, 8), and detailed molecular and biochemical studies were carried out with the xylanases from Cellulomonas fimi (3, 9, 24, 30).
Microbial endo-β-1,4-xylanases may consist of multiple discrete domains joined by linker sequences (16, 17, 42). In addition to one or more catalytic domains, they may contain domains of mainly three types: polysaccharide binding domains, thermostabilizing domains, and domains homologous to the NodB protein from nitrogen-fixing bacteria. Catalytic domains, cellulose binding domains (CBDs), and xylan binding domains (XBDs) of glycosyl hydrolases are grouped into families on the bases of amino acid similarity and hydrophobic cluster analysis (18, 26). Since the hindgut of P. marginata larvae might be a new source of bacterial (hemi)cellulolytic enzymes, a gene library of C. pachnodae was constructed in Escherichia coli to investigate in more detail the xylanases produced by this bacterium. Here, the isolation of two xylanase-encoding genes and the comparison of the deduced amino acid sequences with other xylanases are described. A few biochemical characteristics of the recombinant xylanases were further characterized.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
C. pachnodae (DSM 12657) was isolated from the hindgut of P. marginata larvae as described earlier (5). For DNA extraction, C. pachnodae was cultivated under aerobic conditions at 30°C in basal medium (6). For zymogram analysis, C. pachnodae was cultivated in basal medium containing 5 g of CMC (carboxymethyl cellulose, sodium salt, low viscosity; Sigma)/liter, NaOH-treated beech litter, or xylan (from oat spelts; Sigma) as described previously (5).
A BamHI-digested and CIAP-treated ZAP Express vector (Stratagene) was used for the construction of a genomic library of C. pachnodae by using E. coli XL1-Blue MRF′ (Stratagene) as a host. The plasmids pGEM-T Easy (Promega), pTZ 18R (27), and pUC19 (43) were used for subcloning. E. coli cells were cultivated in Luria-Bertani (LB) medium (33) at 30°C or 37°C, supplemented with 50 μg of ampicillin or kanamycin/ml if appropriate. Solid media contained 1.5% (wt/vol) agar (Difco).
Molecular techniques.
All molecular techniques were performed essentially as outlined by Sambrook et al. (33). Genomic DNA of C. pachnodae was extracted as described by Johnson (21). DNA fragments were purified from agarose gels with the QIAEXII Gel Extraction Kit (Qiagen). Plasmid DNA was purified with the QIAprep Spin Miniprep Kit (Qiagen). Direct purification of PCR products was carried out with the WIZARD PCR Preps DNA purification system (Promega). All procedures were carried out as described by the manufacturers. Plasmid DNA was introduced into E. coli cells by electroporation using a Bio-Rad Gene Pulser. Southern blot analyses using digoxygenin (DIG)-labeled probes were carried out as described in the DIG High Prime User’s Guide (32). Probes were labeled using the DIG-labeling Probe Synthesis Kit or the DIG High Prime Kit (Roche Molecular Biochemicals).
Primers and PCR conditions.
The following primers were used for the amplification of the different DNA fragments that are described in more detail in the following sections. The position on the derived xyn11A and xyn10B nucleotide sequences are given between brackets. Primers derived from xyn11A were as follows (Fig. 1): pAC3, 5′-ACA-GCA-CCG-GGA-GCA-GCG-GC-3′ (positions 143 to 162); pAC4, 5′-GCC-GAT-GGT-GAT-GTT-CGA-CG-3′ (positions 671 to 690, antisense); and XCatD, 5′-TTT-TCT-GCA-GTC-AGG-GCG-GCG-TCG-TCG-TCC-CGC-CG-3′ (positions 717 to 738, antisense). Primers derived from xyn10B were as follows: pAC13, 5′-GGC-GGG-CAT-GGT-GAA-CGT-GCC-3′ (positions 996 to 1016, antisense), and pAC14, 5′-GTA-CAA-CTC-GGG-CAA-CGT-CTC-3′ (positions 1073 to 1093). The standard primers used were T3 primer, T7 primer, and SP6 (Gibco).
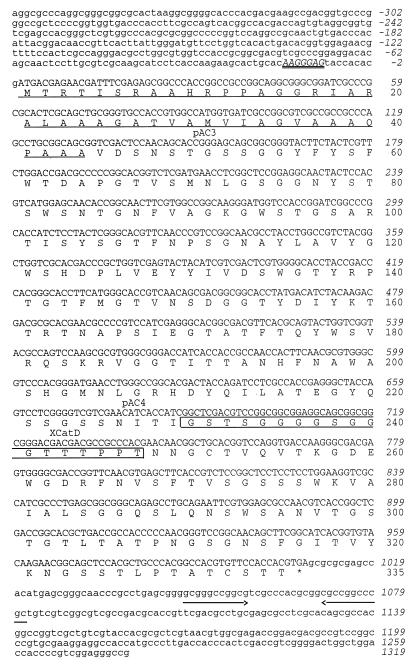
FIG. 1.
Nucleotide sequence of xyn11A and its flanking regions and deduced amino acid sequence. The putative Shine-Dalgarno-type ribosome binding site is indicated in capital italics and is double underlined. The positions of the primers pAC3, pAC4, and XCatD are indicated. The amino acids underlined at the N terminus are the deduced signal peptide. The linker sequence between the family 11 catalytic domain and the XBD is boxed. The translational stop codon is indicated by an asterisk (∗). A palindrome is indicated by arrows.
Unless otherwise noted, the mixtures for the different PCRs contained 1× Super Taq reaction buffer (from the supplier), 0.2 mM deoxynucleoside triphosphates, 500 ng of each primer, 10 to 100 ng of plasmid DNA, and 1 U of Super Taq polymerase (Pharmacia) in a total volume of 50 μl. For the amplification of the putative catalytic domain of xyn11A, Pwo polymerase (Roche Molecular Biochemicals) was used, and 2 mM MgSO4 was added to the reaction mixture. If not mentioned otherwise, the PCRs started with a 5-min denaturation at 94°C, followed by 25 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C. A final extension of 10 min at 72°C was performed. The amplified PCR products were analyzed by agarose gel electrophoresis.
Construction and screening of a C. pachnodae gene library.
Genomic DNA of C. pachnodae was partially digested with Bsp143I (MBI, Fermentas, isoschizomer of Sau3A) and fragments were separated by gel electrophoresis. Fragments in the range of 2 to 14 kb were purified and ligated into the ZAP Express vector in accordance with the supplier’s protocol (Stratagene). Phage DNA was packaged with Packagene Lambda DNA Packaging System (Promega) and was used to infect E. coli XL1-Blue MRF′. The total number of PFU in the primary library was 4 × 104, and 80% of the isolated plasmids contained an insert with an average size of 2.8 kb. It was calculated that the library harbored 20 times the size of the genome (±4.5 Mb; reference 15). Subsequently, the library was spread on LB plates containing IPTG (isopropyl-β-d-thiogalactopyranoside) and kanamycin and incubated overnight at 37°C. For the identification of xylanase-producing E. coli transformants, plates were covered with a soft agar overlay containing 7 g of agar/liter and 10 g of AZCL (azurine cross-linked)-xylan blue (Megazyme)/liter. Plates were reincubated at 37°C and xylanase activity was indicated by the formation of a blue zone around the colonies. Pure cultures of xylanase-positive transformants were obtained after repeated streaking of the colonies to fresh LB plates with appropriate antibiotics.
DNA sequencing and sequence analysis.
The nucleotide sequences of both strands were determined by AmpliTaq FS DNA polymerase fluorescent dye terminator reactions as recommended by the supplier (Perkin-Elmer). Sequencing products were detected by using an Applied Biosystems 373 stretch automated sequencer (Applied Biosystems Inc., Foster City, Calif.). Nucleotide and deduced amino acid sequences were analyzed with the PCGENE program from Intelligenetics. Related sequences were obtained from database searches (Swissprot, PIR, EMBL, and GenBank) using the programs BLASTP 2.0 and FASTA. The sequences were aligned with the program PILEUP (10).
Inverse PCR.
SmaI-digested genomic DNA of C. pachnodae was separated on an agarose gel. Fragments of 0.8 to 1.5 kb were purified from the gel and self-ligated at room temperature for 4 h by using T4 ligase (Gibco). The ligated SmaI fragments (10 to 200 ng) and the primers pAC13 and (in the opposite direction) pAC14, corresponding to the 5′ end of the truncated xylanase gene in pXyl19 (Fig. 2B), were used in the PCR. The PCR conditions were as described above, except that 35 cycles were carried out and an annealing temperature of 55°C was used. In addition, 2.5% of dimethyl sulfoxide was added to the reaction mixture, and prior to the PCR the reaction mixture was heated for 3.5 min at 98°C.
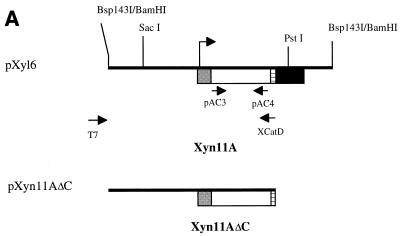
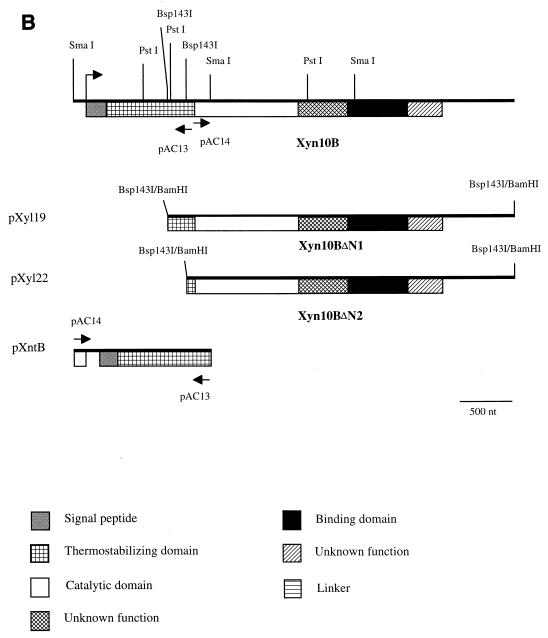
FIG. 2.
(A) Schematic representation of the Bsp143I fragment containing Xyn11A, which was ligated into the BamHI site of the multiple cloning site of the pBK-CMV phagemid vector. The translational start codon is indicated by an arrow. (B) Schematic representation of Xyn10B and its derivatives Xyn10BΔN1 and Xyn10BΔN2 encoded by Bsp143I fragments in pXyl19 and pXyl22, respectively. The translational start codon is indicated by an arrow. The N-terminal fragment of xyn10B was obtained by cloning of an inverse PCR product, yielding pXntB.
Enzyme assays.
Cells from 100-ml E. coli cultures, harboring pXyl6 or pXyl19, grown overnight were harvested at 15,000 × g for 5 min, resuspended in 5 ml of 100 mM phosphate buffer (pH 6.8), and sonicated on ice for 10 min with 50% pulse duration (Branson B12; tip diameter, 3 mm; output, 40 W). The suspension was centrifuged (15,000 × g) and the supernatant (cell-free extract [CFE]) was used for enzyme assays. The recombinant enzymes were tested for the ability to hydrolyze CMC and xylan. CMC-ase and xylanase activities were assayed at 40°C for 30 min in 100 mM phosphate-citrate buffer (pH 6.0), as described by Teunissen et al. (38). Reducing sugars were determined by the dinitrosalicylic acid method (29) using glucose or xylose as a standard. Protein concentrations were determined with the Bio-Rad protein assay (Bio-Rad Laboratories, Richmond, Calif.), using bovine serum albumin as a standard.
Binding capacity, apparent Km, and pH and temperature profiles.
The substrates xylan (from oat spelts), Avicel PH 105 (diameter, 0.019 mm; Serva), and homogenized beech litter were washed five times in 100 mM Tris-HCl (pH 7.5) and suspended in the same buffer (5% [wt/vol]) prior to the binding assay. Beech litter (collected in a forest near Nijmegen) was homogenized in a Waring blender until pieces passed through a 4-mm sieve. The capacity of the recombinant xylanases to bind to the substrates was assayed as follows. CFE was incubated in 1.5-ml Eppendorf tubes on ice for 1 h with an equal volume of substrate and mixed every 5 min. The substrates were removed by centrifugation (15,000 × g), and the supernatant was assayed for residual enzyme activity. The remaining substrates were washed four times with 100 mM Tris-HCl (pH 7.5). After the last washing step, bound protein was eluted from the substrates with 0.5 to 1.0 ml of demineralized water. For the determination of the apparent Km, pH, and temperature optima, recombinant Xyn11A was partially enriched. The enzyme was incubated with xylan and subsequently eluted with demineralized water as described above. This procedure resulted in a threefold increase in specific activity. The apparent Km value of Xyn11A against insoluble xylan was determined from Lineweaver-Burk plots; the concentration of insoluble xylan (from oat spelts) used was 0.1 to 50 mg/ml.
SDS-PAGE.
The supernatant of the C. pachnodae cultures and CFE of E. coli harboring pXyl6 or pXyl19 were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). SDS-PAGE was performed in vertical 10% (wt/vol) polyacrylamide gels using a Mini Protean II system (Bio-Rad) in the presence of SDS (0.1% [wt/vol]), as described by Laemmli (23). The substrate, Remazol brilliant blue xylan (0.2% [wt/vol]; Sigma), was added before polymerization. Enzyme samples were pretreated with sample buffer (62.5 mM Tris-HCl [pH 6.8], 2.3% [wt/vol] SDS, 10% [wt/vol] glycerol, 5% [vol/vol] β-mercaptoethanol, and 0.01% [wt/vol] bromophenol blue) for 18 h at 20°C for zymogram analysis. The HMW-SDS calibration kit (Pharmacia) and LMW Dalton Mark VII-L (MicalCo) were used as molecular weight standards. Electrophoresis was conducted at room temperature at a constant current (40 mA) until the tracking dye reached the bottom of the gel. After electrophoresis, the lanes containing the molecular weight standards were stained separately for protein with Coomassie brilliant blue G-250 (Serva). The remaining part of the gel was subjected to zymogram analysis for the detection of xylanase activity. After three washes with 100 mM phosphate-citrate solution (pH 6.0), the gel was incubated for 16 h at 37°C in the same buffer. Xylanase activity is visualized as light bands against a dark background. The molecular masses of the proteins were estimated by using the Bio-Rad GelDoc 1000 Multi Scan program.
Nucleotide sequence accession numbers.
The nucleotide sequences for xyn11A and xyn10B have been deposited in the EMBL, GenBank, and DDBJ databases under the accession no. AF120156 and AF120157, respectively.
RESULTS
Cloning of C. pachnodae xylanase-encoding genes.
About 11,000 E. coli transformants from the amplified genomic library of C. pachnodae were spread on AZCL-xylan blue plates. This constituted about five times the size of the genome. Among the transformants, 22 colonies showed xylanase activity. Plasmid DNA was isolated from the colonies with xylanase activity. Restriction enzyme analysis showed that the plasmids, named pXyl1 to pXyl22, contained nine different inserts ranging in size from 2.1 to 5.6 kb (results not shown). A PstI site was found in all inserts. The plasmid harboring the smallest insert, pXyl6, was chosen for detailed analyses.
Nucleotide sequence of xyn11A.
The complete nucleotide sequence of the pXyl6 insert was determined (Fig. 1). Translation of the nucleotide sequence revealed a single open reading frame (ORF) of 1,008 bp, encoding a polypeptide of 335 amino acid residues, with a calculated molecular mass of 34,383 Da and a pI of 9.9. The proposed translational ATG start codon is preceded by a putative ribosome binding site (AAGGGAG). Downstream of the translational stop codon an inverted repeat was found which could function as a transcription termination signal. Searches in the databases revealed that the deduced amino acid sequence showed extensive homology to xylanases belonging to family 11 glycosyl hydrolases (cellulase family G). The highest homology (62% identity) was with XynD of C. fimi (30). The xylanase from C. pachnodae was designated Xyn11A (gene xyn11A), in accordance with the suggestions of Henrissat et al. (19).
Domain structure of Xyn11A.
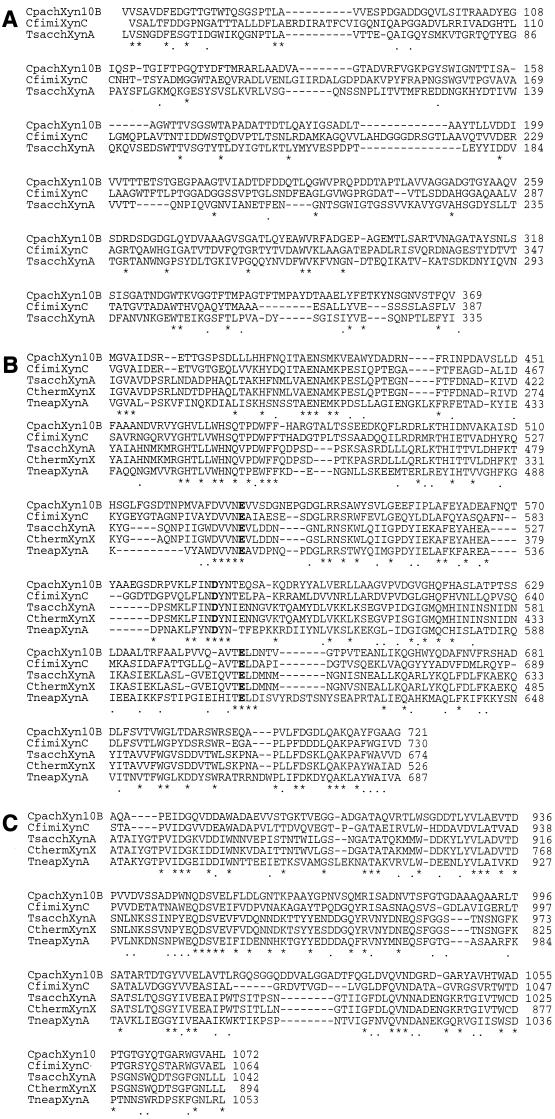
The N terminus of Xyn11A comprises a typical prokaryotic signal peptide of 44 amino acids (Fig. 1) (41). Furthermore, two distinct domains connected by a linker sequence rich in proline, glycine, and threonine residues were identified (Fig. 1). The amino acids 45 to 233 exhibited significant homology to the catalytic domains of family 11 glycosyl hydrolases (Fig. 3A). The highest score (77% identity) was found with the catalytic domain of a xylanase from Streptomyces thermoviolaceus (unpublished results). The amino acid residues 248 to 335 had highest identities with the XBDs and CBDs (63 and 65%, respectively) of XynD from C. fimi (30). An alignment of this region with family II binding domains is shown in Fig. 3B.
FIG. 3.
Alignment of the amino acid sequences of family 11 catalytic domains (A) and XBDs (B) of Xyn11A of C. pachnodae (this study), XynD of C. fimi (accession no. P54865), a xylanase of S. thermoviolaceus (accession no. D85897), XynB of Streptomyces lividans (accession no. P26515), and a xylanase of Thermomonospora fusca (accession no. U01242). Numbering of the amino acids starts at the N termini of the proteins. Conserved and identical amino acids are indicated by asterisks (∗) and points (.), respectively. Highly conserved amino acid residues are shown in boldface letters. Gaps are indicated by dashes.
The function of the putative catalytic domain identified in Xyn11A was investigated in more detail. The 5′ region of xyn11A was amplified by PCR from pXyl6, using the primers XCatD and T7 and with pXyl6 as a template (Fig. 2A). The expected fragment of about 1,600 bp was synthesized and subsequently incubated with the endonucleases HindIII and PstI. The fragment was isolated and cloned in the corresponding sites of pBK-CMV, yielding pXyn11AΔC. E. coli transformants, harboring pXyn11AΔC, showed xylanase activity. This indicated that the first 248 amino acids in the derived polypeptide of Xyn11A harbor the catalytic domain.
Analysis of xylanase-producing transformants.
A PCR was set up to determine whether the eight remaining plasmids with different insert sizes comprised the same gene as pXyl6. The PCR was performed using the xyn11A-specific primers pAC3 and pAC4 and each plasmid as a template. Analysis of the PCR mixtures by gel electrophoresis showed that from six plasmids a fragment of 550 bp was synthesized, which was identical to the size of the fragment from pXyl6. The nucleotide sequences of the 550-bp fragments and nucleotide sequence data from using T3 and T7 primers showed that the six plasmids harbored the same xyn11A gene. Furthermore, these plasmids hybridized with a DIG-labeled probe synthesized on a 1,800-bp SacI-HindIII fragment from pXyl6 (Fig. 2A, HindIII site of the pBK-CMV multiple cloning site) in a dot blot analysis.
Two plasmids, pXyl19 and pXyl22, did not hybridize with this probe, and a larger fragment (600 bp) was obtained from the PCR. The fragments were cloned into pGEM-T Easy and the nucleotide sequence and deduced amino acid sequence were determined. These data revealed that pXyl19 and pXyl22 harbored similar xylanase genes which were different from xyn11A.
Nucleotide sequence of xyn10B.
The insert of pXyl19 was subcloned by using PstI and SacI and the complete nucleotide sequence was determined. The nucleotide sequence of the pXyl19 insert was 3,321 bp long and a single ORF could be deduced. However, a translational start codon preceded by transcriptional elements could not be identified. Apparently, the fragment in pXyl19 encodes an active but truncated xylanase. To clone the 5′ region of the xylanase-encoding gene, an inverse PCR approach was used. A DIG-labeled probe with T3 primer and pAC13 was developed to clone the 5′ end of the xylanase gene. Southern blot analysis of C. pachnodae genomic DNA digested with different restriction enzymes (PvuI, SmaI, KpnI, SstI, and SalI) was carried out. A SmaI fragment of about 1,200 kb hybridized with the DIG-labeled probe, corresponding to the N-terminal region of the xylanase gene. Sequence analysis revealed the presence of two SmaI sites in the pXyl19 insert (Fig. 2B). Because of the position of the SmaI site at position 556 it was concluded that the 1,200-bp SmaI fragment obtained after digestion of genomic DNA comprised the 5′ end of the xylanase gene. The fragment was isolated from the gel and ligated to obtain a circular SmaI fragment. The 5′ region was amplified using the primers pAC13 and pAC14 and the circular fragment as a template. A PCR product of approximately 1,100 bp was obtained and cloned into pGEM-T Easy, yielding pXntB (Fig. 2B), and sequenced. The nucleotide sequence of the insert in pXntB was aligned with the nucleotide sequence from pXyl19, which revealed an ORF of 3,552 bp, encoding an amino acid sequence of 1,183 amino acid residues, with a calculated molecular mass of 124,136 Da and a pI of 4.06. Upstream (6 bp) of the ATG translational start codon the sequence GAAGGAG could function as the ribosome binding site. The deduced amino acid sequence showed homology to xylanases belonging to glycosyl hydrolase family 10 (cellulase family F). Therefore, this xylanase from C. pachnodae was designated Xyn10B (gene xyn10B; reference 19). The truncated derivatives of Xyn10B from pXyl19 and pXyl22 were designated Xyn10BΔN1 and Xyn10BΔN2.
Domain structure of Xyn10B.
The deduced N-terminal sequence of 58 amino acids contains a sequence similar to the signal peptides found in proteins secreted by prokaryotes. The best scores for the putative cleavage site (41) were identified at position 53, between the Thr and Ser residues, or at position 58, between the Ala and Glu residues. Comparison of the amino acid sequence of Xyn10B to those in the protein databases revealed that the mature Xyn10B comprises five different domains (Fig. 2B). The region between amino acid residues 59 and 388 showed some homology to thermostabilizing domains in xylanases of thermophilic bacteria (Fig. 4A), like XynA of Thermoanaerobacterium saccharolyticum (25% identity; reference 25) and the thermostabilizing domain identified in XynC of C. fimi (32% identity; reference 9). The apparent catalytic domain extended from the amino acids 389 to 720, which showed extensive homology to the catalytic domains of family 10 of the glycosyl hydrolases (Fig. 4B). The highest sequence identity (50%) was found with the catalytic domain in XynC of C. fimi (9). Comparison of the amino acid sequence from 720 to 880 to other peptides in the databases did not reveal a possible function for this region. The amino acids 881 to 1071 showed homology to the CBDs of XylC of C. fimi (47% identity; reference 9), XynA of Thermotoga neapolitana (34% identity; reference 44), and XynX of Clostridium thermocellum (36% identity; unpublished results and Fig. 4C). The C-terminal 113 amino acids from Xyn10B showed homology to C termini from chitinases from, e.g., Aeromonas caviae (36) and cellulases from an alkaliphilic Bacillus strain (14). These amino acid sequences were considered to be involved in the binding of the protein to hydrophobic substrates (14, 36). Typical linker sequences, as found in Xyn11A, were not present in Xyn10B. The highest score for homology of the complete deduced amino acid sequence of xyn10B was found with XynC of C. fimi (39% identity; reference 9).
FIG. 4.
Alignment of the amino acid sequences of thermostabilizing domains (A), family 10 catalytic domains (B), and CBDs (C) of Xyn10B of C. pachnodae (this study), XynC of C. fimi (accession no. Z50866), XynA of T. saccharolyticum (accession no. P36917), XynX of Clostridium thermocellum (accession no. P38535), and a xylanase of Thermotoga neapolitana (accession no. Q60042). Numbering starts at the N termini of the proteins. Conserved and identical amino acids are indicated by asterisks (∗) and points (.), respectively. Highly conserved glutamic acid residues are shown in boldface letters. Gaps are indicated by dashes.
Binding capacity of Xyn11A, Xyn11AΔC, and Xyn10BΔN1.
The binding capacity of the cloned xylanases was analyzed by incubating the different recombinant enzymes with several substrates (Table 1). Fifty percent of Xyn11A bound to xylan, whereas no binding was identified to Avicel or homogenized beech litter. The truncated derivative of Xyn11A, Xyn11AΔC, did not bind to any of the substrates. Apparently, the region between the amino acid residues 248 and 335 in Xyn11A harbors an XBD. Binding studies of Xyn11A in Tris-HCl or phosphate-citrate buffer at the optimum pH for activity (pH 6.0) gave similar results (data not shown). In Xyn10B, a binding domain was identified at the C terminus on the basis of homology. The binding capacity was assayed with Xyn10BΔN1 from E. coli harboring pXyl19. Approximately 100 and 20% of the truncated Xyn10B was bound after adsorption to Avicel and homogenized beech litter, respectively, but no adsorption to xylan was found. Apparently, Xyn10BΔN1 contains a CBD. It was possible to elute the bound Xyn10BΔN1 from beech litter with demineralized water (10% recovery), but not from Avicel.
TABLE 1.
Binding of Xyn11A, Xyn11AΔC, and Xyn10BΔN1 to insoluble substrates
| Substrate | Residual activity after binding (total units)a
|
||
|---|---|---|---|
| Xyn11A | Xyn11AΔC | Xyn10BΔN1 | |
| Beech litter | 0.50b | 0.59 | 0.11 |
| Xylan | 0.27 | 0.54 | 0.58 |
| Avicel | 0.51 | 0.55 | 0.005 |
| Controlc | 0.52 | 0.53 | 0.56 |
The following amounts (total units) were added to the solid substrates: Xyn11A, 0.51 ± 0.01 U; Xyn11AΔC, 0.55 ± 0.03 U; and Xyn10BΔN1, 0.57 ± 0.02 U.
Values are the means of results from duplicate experiments and are expressed in total units of residual xylanase activity present in the supernatant after adsorption to the substrates on ice for 1 h and centrifugation. Individual enzyme activities did not differ by more than 2%.
Controls were without addition of substrate.
Activity of the recombinant enzymes.
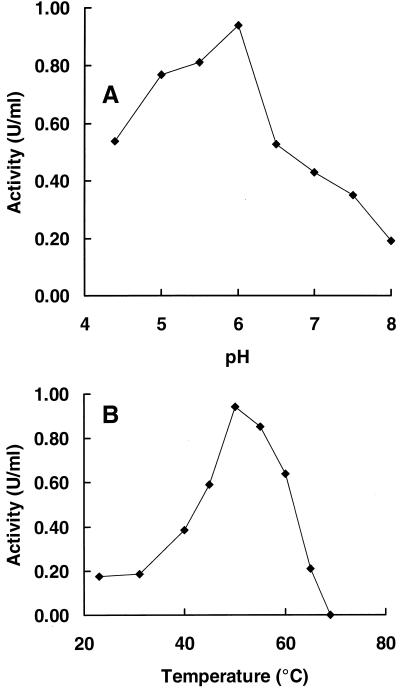
Recombinant Xyn11A and Xyn10BΔN1 showed activity towards xylan, but no activity towards CMC was found. The recombinant Xyn11A, produced by E. coli harboring pXyl6, was partially purified as indicated in Materials and Methods. The enzyme preparation obtained was used to determine the apparent Km and the effect of pH and temperature on xylanase activity. The recombinant enzyme had a pH optimum of 6.0 and the xylanase activity was highest at a temperature between 50 and 55°C (Fig. 5). The apparent Km value of Xyn11A against oat spelt xylan was 6.2 ± 1.4 mg/ml (n = 3). The maximum reaction rate of this enzyme preparation was 6.1 ± 1.6 U/mg.
FIG. 5.
Influence of the pH (A) and temperature (B) on the activity of partially purified recombinant Xyn11A. For the pH profile, the enzyme activity was measured at 50°C in 100 mM phosphate-citrate buffer adjusted to the correct pH. For the temperature profile, enzyme activity was measured in 100 mM phosphate-citrate buffer (pH 6.0) at different temperatures. Values are the means of results of duplicate experiments; separate values do not differ more than 10%.
Comparison of C. pachnodae xylanases with recombinant Xyn11A and Xyn10B.
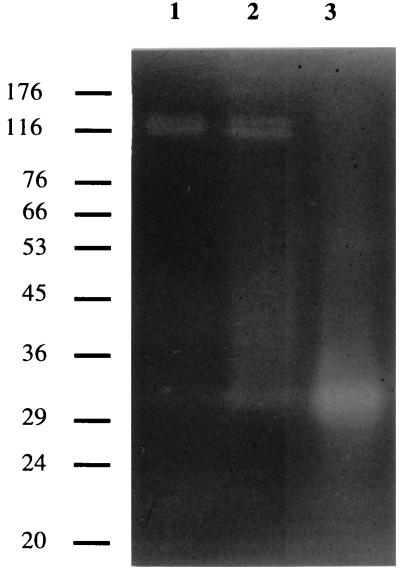
Zymogram analysis was carried out to compare the different xylanases present in C. pachnodae culture fluid with the recombinant Xyn11A. Furthermore, a comparison with the molecular masses of the deduced polypeptides xyn11A and xyn10B was made. In the supernatant of C. pachnodae cultures grown with NaOH-treated beech litter or xylan as a substrate, two xylanases with high molecular masses (111 and 120 kDa) and a xylanase of approximately 32 kDa were present (Fig. 6). In the supernatant of the xylan cultures, the 32-kDa activity band was faint and difficult to identify. The activity band of the recombinant Xyn11A corresponded to the 32-kDa activity band in C. pachnodae cultures. These activity bands are in agreement with the calculated molecular mass of the deduced amino acid sequence of Xyn11A (34 kDa). The molecular weight of the deduced amino acid sequence of Xyn10B (124 kDa) corresponds to the size of the largest activity band found in C. pachnodae cultures.
FIG. 6.
Zymogram of xylanases found in C. pachnodae culture fluids and of recombinant Xyn11A. Lanes: 1, supernatant xylan culture; 2, supernatant beech litter culture; 3, CFE of E. coli harboring pXyl6. Samples contained 0.5 to 1 mU of xylanase activity. Position of molecular mass markers are indicated by horizontal lines and are expressed in kilodaltons.
DISCUSSION
In this study, two new xylanase-encoding genes from the (hemi)cellulolytic bacterium C. pachnodae were isolated and characterized. The genes of C. pachnodae, designated xyn11A and xyn10B, have G+C contents of 70 and 72%, respectively, which correspond to the high G+C levels found in other Cellulomonas species (37). Both Xyn11A and Xyn10B of C. pachnodae appeared to be modular xylanases which did not show endoglucanase activity with CMC as a substrate. The presence of an N-terminal catalytic domain and a C-terminal XBD in Xyn11A was confirmed by the truncated derivative, Xyn11AΔC. The polypeptide Xyn11AΔC, which did not contain the C-terminal 90 amino acids, still showed xylanase activity but had lost the capacity to bind to xylan. There are a few reports on XBDs in xylanases. They were also identified in XynA of T. fusca (20), XynB of S. lividans (35), and XynD of C. fimi (3, 30). According to the classification of Tomme et al. (40), these domains belong to family II of substrate binding domains. Family II CBDs contain four strictly conserved tryptophan and two conserved cysteine residues (28, 40). Amino acid sequence alignment of family II XBDs showed also two conserved cysteine residues, corresponding to Cys-251 and Cys-332 in the deduced amino acid sequence of Xyn11A (Fig. 3B). In contrast, only three highly conserved tryptophan residues, corresponding to Trp-261, Trp-277, and Trp-293 of Xyn11A, could be identified. Likely, the substrate specificity of family II binding domains is influenced by the presence of a fourth tryptophan residue.
In contrast to Xyn11A, linker sequences separating the different domains were not identified in Xyn10B, nor were they found in the endoglucanase Cel6A from C. pachnodae (6). Apparently, such linker sequences are not essential for the function of distinct domains in glycosyl hydrolases. One of the domains identified in Xyn10B was a CBD. On the basis of similarity and length (190 amino acids), this CBD can be classified as a family IX binding domain (40). The Xyn10B CBD bound tightly to Avicel, and it was not possible to elute Xyn10BΔN1 from Avicel (Table 1). Apparently, Xyn10B harbors a high-affinity CBD (12, 26, 30). Although most high-affinity CBDs were found in cellulases, they were also present in xylanases from, e.g., Pseudomonas fluorescens subsp. cellulosa (12), Cellvibrio mixtus (31) and C. fimi (30). The different specificities of CBDs and/or XBDs of cellulases and xylanases produced by many microorganisms would provide these microorganisms with multiple ways to attack complex plant fibers.
Extensive homology of Xyn11A with family 11 catalytic domains was found. Amino acid sequence alignment showed that the amino acids Glu-128 and Glu-217 of Xyn11A correspond to the highly conserved Glu residues important for catalytic activity in family 11 glycosyl hydrolases (Fig. 3A; reference 22). Also, alignment of Xyn10B with other family 10 catalytic domains showed a few regions of highly conserved amino acid sequences (Fig. 4B). The amino acid residues important for the catalytic activity of family 10 glycosyl hydrolases correspond with Glu-530, Asp-585, and Glu-648 in the deduced amino acid sequence of Xyn10B (25). E. coli cells harboring the plasmid pXyl19 or pXyl22, which was devoid of the N-terminal 325 or 351 amino acids of Xyn10B, respectively, showed xylanase activity. This was in agreement with the location of the putative catalytic domain identified in Xyn10B on the basis of homology. The N terminus of Xyn10B comprised a domain which was homologous to thermostabilizing domains of xylanases from thermophilic bacteria (Fig. 4A; references 25 and 44). Also in XynC of C. fimi, which showed an optimum temperature for xylanase activity at 60°C, a thermostabilizing domain was identified upstream of the catalytic domain (9). There seems to be no obvious reason why a thermostabilizing domain should be present in enzymes of mesophilic bacteria like C. pachnodae and C. fimi. However, it has been shown that thermostabilizing domains also result in a general increased stability of these enzymes against proteolytic attack and extremes of pH (13). In both XynD and XynC of C. fimi, a region homologous to the NodB protein from nitrogen-fixing rhizobia was identified (9, 11, 31). In XynD, this region appeared to be essential for the removal of acetyl groups from acetylated xylan (24). Although the highest overall sequence identities of Xyn11A and Xyn10B of C. pachnodae were found with XynD and XynC, respectively, of C. fimi (9, 30), such NodB domains were not identified in Xyn11A or Xyn10B.
Zymogram analysis showed three bands with xylanase activity in the supernatant of C. pachnodae cultures (Fig. 6). Our screening of the C. pachnodae genomic library revealed the presence of two different xylanases. The calculated molecular masses of these proteins corresponded very well to the 120- and 32-kDa activity bands. It is not clear whether in C. pachnodae the xylanase visible at 111 kDa is encoded by a third gene. In C. fimi cultures, protease activity was observed, which resulted in smaller but still active proteins (34). Previously, this was also observed in endoglucanase zymograms of C. pachnodae culture fluids (6). Therefore, the 111-kDa xylanase may be a product of proteolytic cleavage of the 120-kDa protein and not of a third gene. Alternatively, this xylanase may not be found in the library due to low enzyme activity or low expression level of the encoding gene in E. coli. In addition, the zymograms showed that the growth substrate influenced the amount of the different xylanases in C. pachnodae cultures (Fig. 6). On a complex substrate like NaOH-treated beech litter, the intensities of the activity bands of Xyn11A and Xyn10B were comparable, whereas Xyn11A was hardly detected in the culture fluid with xylan as a substrate. However, further studies are needed to elucidate the mechanism that regulates the production of the different xylanases in C. pachnodae.
ACKNOWLEDGMENTS
We thank J. J. Bos and A. J. A. van Kampen for assistance in nucleotide sequence determination.
The work was supported by IOP Senter, a division of Environmental Biotechnology, project no. IMB93004.
REFERENCES
- 1.Bayon C, Mathelin J. Carbohydrate fermentation and by-product adsorption studied with labelled cellulose in Oryctes nasicornis larvae (Coleoptera: Scrarabaeidae) J Insect Physiol. 1980;26:833–840. [Google Scholar]
- 2.Bhalerao J, Patki A H, Bhave M, Khurana I, Deobagkar D N. Molecular cloning and expression of a xylanase gene from Cellulomonas sp. into Escherichia coli. Appl Microbiol Biotechnol. 1990;34:71–76. [Google Scholar]
- 3.Black G W, Hazlewood G P, Millward-Sadler S J, Laurie J I, Gilbert H J. A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem J. 1995;307:191–195. doi: 10.1042/bj3070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cazemier A E, Op den Camp H J M, Hackstein J H P, Vogels G D. Fibre digestion in arthropods. Comp Biochem Physiol. 1997;118A:101–109. [Google Scholar]
- 5.Cazemier, A. E., H. J. M. Op den Camp, J. C. Verdoes, J. H. P. Hackstein, and G. D. Vogels. Cellulomonas pachnodae sp. nov., a member of the (hemi)cellulolytic hindgut flora of larvae of the scarab beetle Pachnoda marginata. Submitted for publication. [DOI] [PubMed]
- 6.Cazemier, A. E., J. C. Verdoes, H. J. M. Op den Camp, J. H. P. Hackstein, and A. J. J. van Ooyen. A β-1,4-endoglucanase encoding gene from Cellulomonas pachnodae. Appl. Microbiol. Biotechnol, in press. [DOI] [PubMed]
- 7.Chaudhary P, Deobagkar D N. Characterization of cloned endoglucanase from Cellulomonas sp. NCIM 2353 expressed in Escherichia coli. Curr Microbiol. 1997;34:273–279. doi: 10.1007/s002849900181. [DOI] [PubMed] [Google Scholar]
- 8.Clarke J H, Laurie J I, Gilbert H J, Hazlewood G P. Multiple xylanases of Cellulomonas fimi are encoded by distinct genes. FEMS Microbiol Lett. 1991;67:305–310. doi: 10.1016/0378-1097(91)90493-t. [DOI] [PubMed] [Google Scholar]
- 9.Clarke J H, Davidson K, Gilbert H J, Fontes C M G A, Hazlewood G P. A modular xylanase from mesophilic Cellulomonas fimi contains the same cellulose-binding and thermostabilizing domains as xylanases from thermophilic bacteria. FEMS Microbiol Rev. 1996;139:27–35. doi: 10.1111/j.1574-6968.1996.tb08175.x. [DOI] [PubMed] [Google Scholar]
- 10.Devereux J, Haeberli P, Smithies D. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egelhoff T T, Fischer R F, Jacobs T W, Mulligan J T, Long S R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA. 1985;4:241–248. doi: 10.1089/dna.1985.4.241. [DOI] [PubMed] [Google Scholar]
- 12.Ferreira L, Durrant A, Hall J, Hazlewood G, Gilbert H. Spatial separation of protein domains is not necessary for catalytic activity or substrate binding in xylanase. Biochem J. 1990;269:261–264. doi: 10.1042/bj2690261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontes C M G A, Hazlewood G P, Morag E, Hall J, Hirst B H, Gilbert H J. Evidence for a general role for non-catalytic thermostabilizing domains in xylanases from thermophilic bacteria. Biochem J. 1995;307:151–158. doi: 10.1042/bj3070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukumori F, Sashihar N, Kudo T, Horikoshi K. Nucleotide sequences of two cellulase genes from alkalophilic Bacillus sp. strain N-4 and their strong homology. J Bacteriol. 1986;168:479–485. doi: 10.1128/jb.168.2.479-485.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fülop L, Trân S L P, Prágai Z, Felföldi F, Ponyi T. Cloning and expression of a β-1,4-endoglucanase gene from Cellulomonas sp. CelB7 in Escherichia coli; purification and characterization of the recombinant enzyme. FEMS Microbiol Lett. 1996;145:355–360. doi: 10.1111/j.1574-6968.1996.tb08600.x. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert H J, Hazelwood G P. Bacterial cellulases and xylanases. J Gen Microbiol. 1993;139:187–194. [Google Scholar]
- 17.Gilkes N R, Henrissat B, Kilburn D G, Miller R C, Jr, Warren R A J. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarity. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henrissat B, Teeri T T, Warren R A J. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 1998;425:352–354. doi: 10.1016/s0014-5793(98)00265-8. [DOI] [PubMed] [Google Scholar]
- 20.Irwin D, Jung E D, Wilson D B. Characterization and sequence of a Thermomonospora fusca xylanase. Appl Environ Microbiol. 1994;60:763–770. doi: 10.1128/aem.60.3.763-770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson J L. Similarity analysis of DNAs. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods in general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 655–682. [Google Scholar]
- 22.Ko E P, Akatsuka H, Moriyama H, Shinmyo A, Hata Y, Katsube Y, Urabe I, Okada H. Site-directed mutagenesis at aspartate and glutamate residues of xylanase from Bacillus pumilus. Biochem J. 1992;288:117–121. doi: 10.1042/bj2880117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Laurie J I, Clarke J H, Ciruela A, Faulds G B, Williamson G, Gilbert H J, Rixon J E, Millward-Sadler S J, Hazlewood G P. The NodB domain of a multidomain xylanase from Cellulomonas fimi deacetylates acetylxylan. FEMS Microbiol Lett. 1997;148:261–264. [Google Scholar]
- 25.Lee Y E, Lowe E, Henrissat B, Zeikus J G. Characterization of the active site and thermostability regions of endoxylanase from Thermoanaerobacterium saccharolyticum B6A-RI. J Bacteriol. 1993;175:5890–5898. doi: 10.1128/jb.175.18.5890-5898.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linder M, Teeri T T. The roles and function of cellulose-binding domains. J Biotechnol. 1997;57:15–28. doi: 10.1016/s0168-1656(97)00088-6. [DOI] [PubMed] [Google Scholar]
- 27.Mead D A, Szczesna-Skorupa E, Kemper B. Single-stranded DNA ‘blue’ T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986;1:67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- 28.Meinke A, Gilkes N R, Kilburn D G, Miller R C, Jr, Warren R A J. Cellulose-binding polypeptides from Cellulomonas fimi: endoglucanase D (CenD), a family A β-1,4-glucanase. J Bacteriol. 1993;175:1910–1918. doi: 10.1128/jb.175.7.1910-1918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller G L. Use of dinitrosalicylic as reagent for the determination of reducing sugars. Anal Chem. 1959;31:426–428. [Google Scholar]
- 30.Millward-Sadler S J, Poole D M, Henrissat B, Hazlewood G P, Clarke J H, Gilbert H J. Evidence for a general role for high-affinity non-catalytic cellulose binding domains in microbial plant cell wall hydrolases. Mol Microbiol. 1994;11:375–382. doi: 10.1111/j.1365-2958.1994.tb00317.x. [DOI] [PubMed] [Google Scholar]
- 31.Millward-Sadler S J, Davidson K, Hazlewood G P, Black G W, Gilbert H J, Clarke J H. Novel cellulose-binding domains, NodB homologues and conserved modular architecture in xylanases from the aerobic soil bacteria Pseudomonas fluorescens subsp. cellulosa and Cellvibrio mixtus. Biochem J. 1995;312:39–48. doi: 10.1042/bj3120039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roche Molecular Biochemicals. DIG High Prime user’s guide. Almere, The Netherlands: Roche Molecular Biochemicals; 1995. [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sandercock L E, Meinke A, Gilkes N R, Kilburn D G, Warren R A J. Degradation of cellulases in cultures of Cellulomonas fimi. FEMS Microbiol Lett. 1996;143:7–12. [Google Scholar]
- 35.Shareck F, Roy C, Yaguchi M, Morosoli R, Kluepfel D. Sequences of three genes specifying xylanases in Streptomyces lividans. Gene. 1991;107:75–82. doi: 10.1016/0378-1119(91)90299-q. [DOI] [PubMed] [Google Scholar]
- 36.Sitrit Y, Vorgias C E, Chet I, Oppenheim A B. Cloning and primary structure of the chiA gene from Aeromonas caviae. J Bacteriol. 1995;177:4187–4189. doi: 10.1128/jb.177.14.4187-4189.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stackebrand E, Prauser H. The family Cellulomonadaceae. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. Vol. 2. New York, N.Y: Springer-Verlag Inc.; 1992. pp. 1323–1345. [Google Scholar]
- 38.Teunissen M J, Smits A M, Op den Camp H J M, Vogels G D. Fermentation of cellulose and production of cellulolytic and xylanolytic enzymes by anaerobic fungi from ruminant and non-ruminant herbivores. Arch Microbiol. 1991;137:1401–1408. doi: 10.1007/BF00263000. [DOI] [PubMed] [Google Scholar]
- 39.Thomson J A. Molecular biology of xylan degradation. FEMS Microbiol Rev. 1993;104:65–82. doi: 10.1111/j.1574-6968.1993.tb05864.x. [DOI] [PubMed] [Google Scholar]
- 40.Tomme P, Warren R A J, Gilkes N R. Cellulose hydrolysis by bacteria and fungi. Adv Microbiol Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 41.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong K K Y, Tan L U L, Saddler J N. Multiplicity of β-1,4-xylanases in microorganisms: functions and applications. Microbiol Rev. 1988;52:305–317. doi: 10.1128/mr.52.3.305-317.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanish-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 44.Zverlov V, Piotukh K, Dakhova O, Velikodvorskaya G, Borriss R. The multidomain xylanase A of hyperthermophilic bacterium Thermotoga neapolitana is extremely thermoresistant. Appl Microbiol Biotechnol. 1996;45:245–247. doi: 10.1007/s002530050678. [DOI] [PubMed] [Google Scholar]