Abstract
Medulloblastoma (MB) is a malignant pediatric brain tumor which shows upregulation of MYC and sonic hedgehog (SHH) signaling. SHH inhibitor face acquired resistance, which is a major cause of relapse. Further, direct MYC oncogene inhibition is a challenging therapeutic target, inhibition of MYC upstream insulin-like growth factor/phosphatidylinositol-4,5-bisphosphate 3-kinase (IGF/PI3K) is a promising alternative. While PI3K inhibition activates resistance mechanisms, simultaneous inhibition of bromodomain-containing protein 4 (BRD4) and PI3K can overcome resistance. We synthesized a new molecule 8-(2,3-dihydrobenzo[b][1,4] dioxin-6-yl)-2-morpholino-4H-chromen-4-one (MDP5) that targets both BRD4 and PI3K pathways. We used X-ray crystal structures and a molecular modeling approach to confirm the interactions between MDP5 with bromo domains (BDs) from both BRD2 and BRD4, and molecular modeling for PI3K binding. MDP5 was shown to inhibit target pathways and MB cell growth in vitro and in vivo. MDP5 showed higher potency in DAOY cells (IC50 5.5 μM) compared to SF2523 (IC50 12.6 μM), and its IC50 values in HD-MB03 cells were like SF2523. Treatment of MB cells with MDP5 significantly decreased colony formation, increased apoptosis, and halted cell cycle progression. Further, MDP5 was well tolerated in NSG mice bearing either xenograft or orthotopic MB tumors at the dose of 20 mg/kg, and significantly reduced tumor growth and prolonged animal survival.
Keywords: Medulloblastoma, BRD4 inhibitor, MDP5, Chemoresistance, Crystal structure
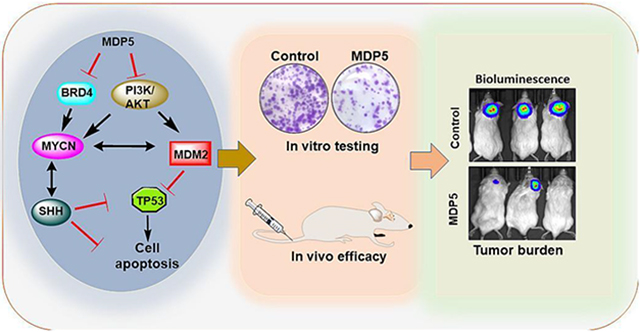
Graphical Abstract
1. Introduction
Medulloblastoma (MB) is the most common childhood brain tumor arising from the cerebellum. MB has four major dysregulated signaling pathways that form the primary basis of differentiation: sonic hedgehog (SHH), wingless (WNT), MYC (Group 3), and Group 4 [1]. These molecular subgroupings influence treatment decisions, and each type has a distinct prognosis. Overexpression of MYCN gene is associated with group 3 MB and significantly reduced survival outcomes [2]. MYC activation often derives from amplification at the MYC loci or genomic rearrangement of PVT1–MYC but can be caused by some unknown mechanisms [3]. MYC activity is upregulated in almost all MB subtypes and MYC targeting is speculated as a possible therapeutic strategy [4].
As a “super-transcription factor,” MYC regulates the transcription of over 15% of the human genome and promotes the transcription of many genes [5]. Targeting MYC directly is difficult due to the lack of a specific binding site in the encoded protein. However, inhibiting its upstream targeting proteins such as Bromo- and Extra-terminal domains (BET) provide a significant anticancer effect in preclinical models of MYC-amplified MB [4].
BET proteins share a tertiary structure, that includes four α-helices (αZ, αA, αB, and αC) and two loops (ZA and BC). Four α-helices form a left-handed α-helical bundle that, along with the two loops, creates a binding pocket that identifies acetylated lysine residues on N-terminal histone tails [6]. Among these BET proteins, BRD4 upregulation is well documented in various cancers. BRD4 interacts with CDK9 and cyclin T1, constituting the core complex that prevents RNA Pol II from pausing in the promoter-proximal region [7]. Several BRD4 inhibitors have entered clinical trials, in recent years, and demonstrated promising efficacy in inhibiting tumor growth. However, these inhibitors are non-selective toward all BET family members, which produces significant side effects [8].
Further, mutations in the catalytic subunit alpha of PI3K have been shown to accelerate the tumor growth in SHH-MB and metastasis of MB in mice [9]. PI3K activates AKT and mTOR to enhance mRNA translation, increases MYC protein half-life, and MYC transcriptional activity. Therefore, targeting any individual receptor may likely fail to provide therapeutic benefits, but directly targeting PI3K represents a potentially successful therapeutic strategy and is under investigation in different cancers [10]. Taken together, these findings suggest that simultaneous targeting BRD4 and PI3K pathways may be helpful in the treatment of human MB.
We have previously shown that BRD4 and PI3K/AKT dual inhibitor SF2523 reduced MB growth in mice [11]. Since SF2523 has a modest potency, delivery of sufficient dose to the brain may be challenging and higher dose could lead to systemic toxicity. Therefore, we synthesized and developed a new dual BRD4 and PI3K/AKTdual inhibitor MDP5, which represses MYC transcription resulting in cell cycle arrest and apoptotic cell death. Further in vivo studies confirmed that MDP5 significantly reduces tumor growth and enhances survival of mice, in an orthotopic MB tumor model.
2. Materials and methods
2.1. Reagents and cell culture
SF2523 (HY-101146) and LY294002 (HY-10108) were purchased from MedChem Express LLC. For in vitro studies, all the drugs were dissolved in dimethyl sulfoxide (DMSO) to make the desired stock concentrations. Cell culture media EMEM, DMEM, and RPMI were purchased from the ATCC, Hyclone Laboratories, and Gibco, respectively. FBS was purchased from BioTechne, antibiotic solution Anti-Anti was obtained from Sigma-Aldrich. DAOY, HD-MB03, and ONS-76 MB cells were cultured in EMEM, DMEM, and RPMI, respectively, with 10% FBS and 1% Anti-Anti. All cells were maintained in an incubator at 37 °C with relative humidity between 90 and 95% in the presence of 5% CO2. Primary antibodies were obtained from Abcam, Biomatik, Cell signaling technology (CST), and Santacruz Biotech. Horseradish peroxidase (HRP) conjugated and florescent labeled secondary antibodies were purchased from the Invitrogen (A16096) and Li-COR Biosciences (926-68070), respectively. All other reagents were obtained from Fisher Scientific.
2.2. Synthesis of 8-(2,3-dihydrobenzo[b][1,4] dioxin-6-yl)-2-morpholino-4H-chromen-4-one (MDP5)
The synthesis of MDP5 was carried out in the two steps. In step one, the chromenone intermediate was obtained by reacting phosphorous chloride to a mixture of 3-bromo-2-hydroxybenzoic acid (3.48 g, 16 mmol) and N-acetylmorpholine (4.12 g, 32 mmol) in chloroform (CHCl3; 10 mL).
In the second step, the 1,1′-Bis(diphenylphosphino)ferrocene palladium(II) dichloride was added to a mixture of the intermediate chromenone, 1,4-benzodioxane-6-boronic acid, and triethanolamine (TEA, 303 mg, 3 mmol) in dioxane (10 mL) and water (4 mL) under N2 to afford the desired product MDP5. The detailed synthesis method of MDP5 is given in supplementary methods.
2.3. Molecular docking
The Glide docking approach (Protein Preparation Wizard, Maestro, MacroModel, and Glide; Schrödinger, LLC: Portland, OR) was used to carry out docking study of LY294002, quercetin, SF2523, SF2535, and MDP1-MDP5. The first BRD4 bromodomain (BD1, PDB ID: 2U28) [12], the second BRD4 bromodomain (BD2, PDB ID: 5U2C), and the PI3Kα subtype protein human p110α/p85α (PDB ID: 2RD0) were retrieved from the Protein Data Bank (PDB) (http://www.rcsb.org) and were prepared and minimized using the MOE protein preparation module [13]. PI3K protein 2RD0 was prepared as outlined in our previous paper [14]. For clarity, H-atoms are not shown in the figures.
2.4. BRD2-BD2/BRD4-BD2 molecular cloning, expression, and purification
The genes encoding human BRD2-BD1, BRD2-BD2, and BRD4-BD2 were codon-optimized for expression in E. coli. The codon-optimized genes were PCR amplified, and the resulting PCR products were placed downstream of the ribosome binding site of a derivatized pET32 plasmid allowing for production of a protein possessing a cleavable polyhistidine tag at the N-terminus. This plasmid DNA was amplified and purified after transformation into E. coli. For Large-scale production of proteins, transformed E. coli were cultured, protein production induced by the addition of 0.5 mM IPTG, and cell lysate was used to afford purified proteins as described in the supplementary materials section. Proteins were purified to greater than 95 % purity as indicted by SDS-PAGE (Supplementary Figure S1) gel analysis using Image J (NIH). The presence or absence of nucleic acid contaminants was assessed by calculating the ratio of Abs 260 nm/Abs 280 nm. Tested samples gave values less than 0.7 suggest a lack of nucleic acid contamination.
2.5. Inhibitor binding using Protein Thermal Shift
Changes in BD thermal stability due to binding of SF2523 or MDP series molecules to the BDs employed a Protein Thermal Shift (PTS) assay. Each reaction used 20 μM concentration of recombinant BD protein in a 50 mM Tris pH 7.5 buffer containing 100 mM NaCl, 0.3 mM tris(2-carboxyethyl)phosphine) (TCEP), and 1 % DMSO. Protein Thermal Shift dye was used as the fluorophore (ThermoFisher).
2.6. Protein crystallization and X-ray diffraction experiments
The protein inhibitor complexes of BRD2-BD1/MDP5, BRD2-BD2/MDP5, BRD2-BD2/SF2523, and BRD4-BD2/SF2523 were prepared by mixing each protein with the compound separately. Each mixture consisted of 5 mg/mL of protein with respective compounds to maintain a 1:10 protein:inhibitor stoichiometric ratio. The LS-CAT beamline at the Advanced Photon Source of Argonne National Labs (IL), was used to perform the X-ray diffraction experiments. HKL2000 was used to index, integrate, and scale the collected diffraction data corresponding to each protein/inhibitor complex [15].
2.7. Cell viability and migration assays
For cell viability assay, DAOY and HD-MB03 cells were exposed to new analogs with a range of concentrations with method as described earlier [11]. Cell migration potential of MB cells in presence of MDP5 and SF2523 was assessed by using method described earlier with some minor modifications [16].
2.8. Effect of MDP5 on colony formation, cell cycle, and apoptosis
Colony formation ability, cell cycle analysis, and cell apoptosis assay were performed in MB cells in presence of drugs as described earlier [11]. For colony formation assay, after taking pictures, the colonies were dissolved in 10% acetic acid solution to measure the optical density (OD). Cell cycle distribution was determined in proliferating MB cells (0.2×106) in a 6 wells plate. Growth inhibition was induced by adding drug solutions at the concentration of 5.0 μM for 48 h. For apoptosis assay, the cells were treated/collected as above and stained in dark for 5 min with 5 μL Annexin V-FITC and 10μL propidium iodide (PI) in 500 μL of 1 × binding buffer.
2.9. RT-qPCR and Western blot analysis
DAOY and HD-MB03 cells were treated with SF2523, MDP1, and MDP5 at 5.0 μM concentration for 48 h. For RT-PCR, total mRNA was extracted using RNeasey kit and converted to cDNA using TaqMan Reverse Transcription Reagents (applied biosystems; N8080234). The PCR reaction was performed using reagents shown in supplementary Table S1. The gene expression levels were calculated using 2−ΔΔCq method with β-actin as the internal reference gene. Western blot analysis was performed using method described earlier [11]. For HRP detection, ECL™ start western blotting detection reagent was used (Supplementary Table S2a and b).
2.10. Tumor spheroid assay
Tumor spheroids were formed using MB cells following the method described before [16]. Briefly, cells were suspended in ultra-low attachment (ULA) 96-well plate for a week and when tumor spheroid size reached to 200 μm, then spheroids were treated with SF2523, MDP1, or MDP5 at a concentration of 5.0 μM. After 7 days of treatment, spheroids were stained and imaged with a confocal laser microscope (Zesis LSM 710).
2.11. Effect of drug treatment on tumor propagating cells and side population
The tumor propagating cell population in ONS-76 cells was identified based on CD15+ staining by fluorescence activated cell sorting (FACS) analysis. Briefly, ONS-76 cells were treated with SF2523, MDP1, and MDP5 with 5.0 μM concentrations for 48 h. Then, cells were trypsinized and 1×106 cells stained with a 5μL aliquot of APC-CD15 antibody (BioLegend; 301908) and analyzed with flow cytometry (BD, LSR Fortessa).
For the side population (SP) assay, ONS-76 cells were preincubated with SF2523, MDP1, and MDP5 for 0.5 h at RT. After washing thrice with staining buffer, 1×106 cells were stained with 5μg/mL of Hoechst 33342 (Thermo Scientific; 62249) for 1 h in an incubator with occasional shaking every 20 min. The PI solution 5μL was added to exclude dead cells and analyzed with flow cytometry (LSRII, BD Biosciences).
2.12. Antitumor efficacy of MDP5 in MB tumor-bearing mice
MB xenograft tumors were generated by injecting approximately 1 × 106 MB cells subcutaneously (S.C.) on the right flank (DAOY) and left flank (HD-MB03). The orthotopic MB tumors were generated by injecting 1 × 105 stereotaxically in the mouse cerebellum using our established protocol [11]. When the xenograft tumors volume reached a minimum size of 100 mm3 or the bioluminescence in orthotopic MB tumor reached ≥1×105 p/sec/cm2/sr, mice were randomly divided into the following three groups (n = 5): i) MB bearing mice with no treatment (control); ii) MB bearing mice treated intravenously (i.v.) with SF2523 cosolvent (mixture of propylene glycol 30%, Cremophor EL 20%, and water 50%) [17], and iii) MB bearing mice treated i.v. with MDP5 in cosolvent at the dose of 20 mg/kg every third day for two weeks. The mice were given seven injections in total, and their body weights were measured before every injection. In the case of the xenograft model, the tumor growth was monitored by calipers and by IVIS imaging in the case of the orthotopic model. For histological examination, the major organs of representative mice from each animal group were dissected and stained with H&E and different target proteins. Stained slides were imaged using an iScan HT Slide Scanner (Ventana Inc, AZ).
2.13. Statistical analysis
In vitro experiments were performed in triplicates. The student’s t-test was used to compare two different groups, and one-way ANOVA was performed for three or more groups. The data obtained were shown as the mean ± S.D., and p < 0.05 was considered statistically significant.
3. Results
3.1. Synthesis and Binding study of MDP series to BRD2 and BRD4-BD2
We have synthesized structural analogs of SF2523 termed as MDP series. The most potent molecule MDP5 was characterized with 1H NMR and mass spectrometry (Fig. 1A and b. The results from Protein Thermal Shift (PTS) assays illustrate that MDP1 through MDP4 do not significantly enhance the thermal stability of either BD2 (Supplementary Fig. S3a) as the change in BRD2-BD2 melting temperature (Tm) in the presence of these compounds resulted in Tm changes between −0.2 °C and 0.4 °C. The BRD4-BD2 experiments showed a Tm decrease of 1.3 °C with MDP1 and MDP3. A decrease in Tm of 0.6 °C was observed for MDP2 and MDP4 (Supplementary Fig. S3b). In contrast, both MDP5 and SF2523 increased the BRD2-BD2 Tm by 2.4 °C and 6.3 °C, respectively. Similarly, the Tm values for BRD4-BD2 with MDP5 and SF2523 increased by 1.7 and 3.4 °C, respectively (Supplementary Fig. S3c). These data suggest that MDP5 represents the best binder of MDP series and is comparable to SF2523 in terms of thermal stabilization of the BD2s. Additionally, compounds MDP1–4 destabilize BRD4-BD2 is intriguing while they have very little impact on BRD2-BD2 thermal stability. While the magnitude of the ΔTm is not indicative of strength of inhibitor binding, these results may suggest that BRD2-BD2 engages MDPs in a slightly different mode than does BRD4-BD2 or that a minor conformational change is required to promote inhibitor binding (Supplementary Fig. S3d).
Fig. 1.
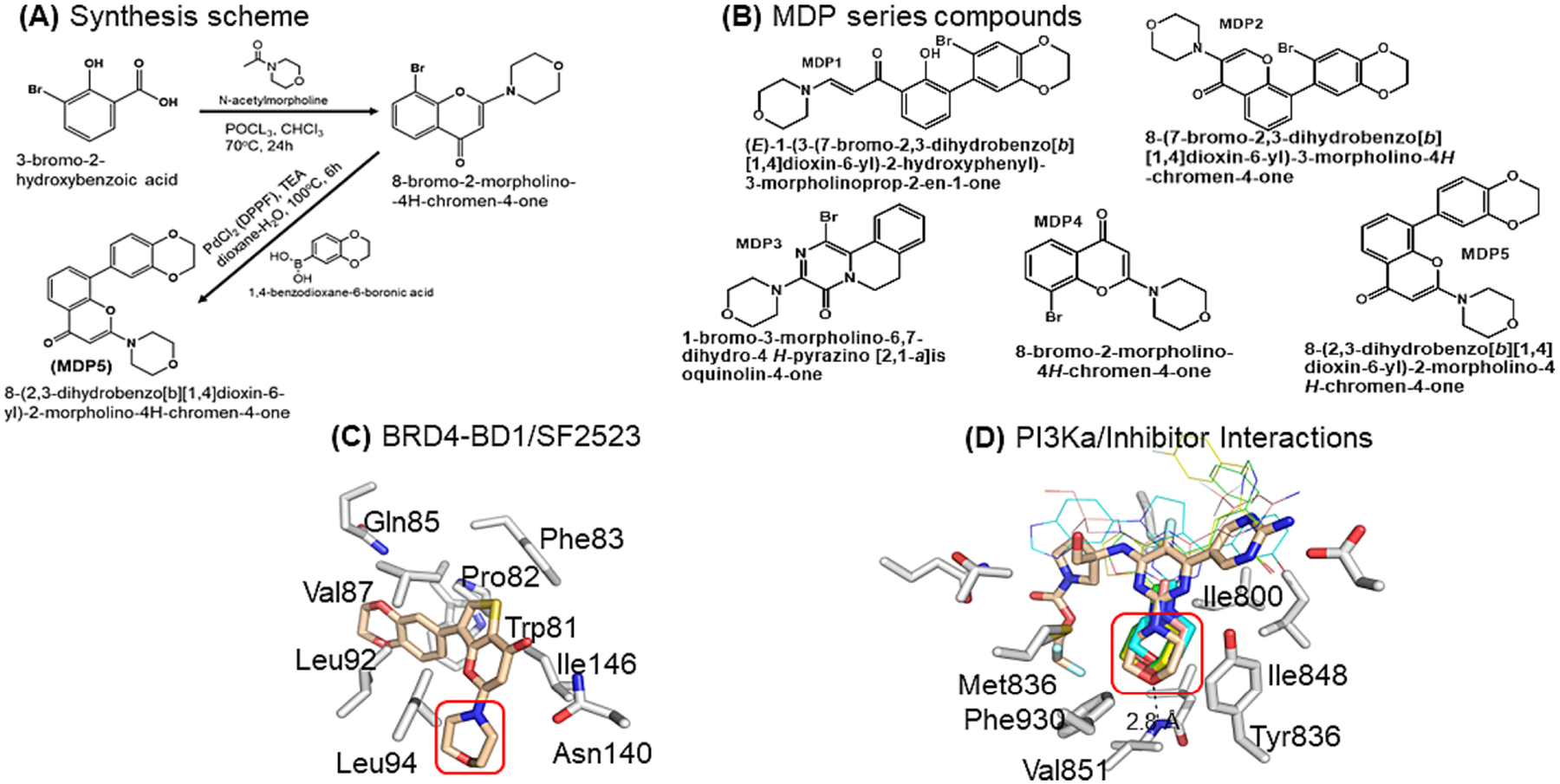
Inspirational structures for dual inhibitor of BRD-BDs and PI3Ka. A Synthesis scheme of MDP5. B Chemical structures of MDP series compounds. C X-ray crystal structure of BRD2-BD1/SF2523 complex (pdb accession: 5u28) highlighting the benzodioxane and thienopyranone interactions with the acetyllysine binding site and the lack of interaction between BD1 and the morpholino moiety of SF2523 (red box). D Xray crystal structures of PI3Ka/g (gray carbons, 7k6m) complexed with various morpholino-containing inhibitors (tan, cyan, pink, yellow, and green carbons representing pdb accession codes 7k6m, 3apf, 5jhb, 5xgh, 5xgj, respectively). The morpholino moiety binds in a hydrophobic pocket formed by the indicated PI3Ka residues and forms a pivotal hydrogen bonded interaction with the backbone amide of Val851 (red box). The narrow bonds indicate structurally variable regions of the 5 inhibitors.
3.2. Verification of the docking method
Inhibitor docking scores against the three models are listed in Supplementary Table S3, and the differences (ΔΔG) between predicted (BD1_d, BD2_d) and observed binding affinity (BD1_o, BD2_o) were also given in Supplementary Table S2. Results show that the mean errors between predicted and observed binding affinity, as measured by ΔΔG_BD1 and ΔΔG_BD2, were −1.73 and −0.46 for BD1 and BD2, respectively. Thus, the Glide docking results reasonably predict the affinity of molecules to target protein in this case with the negative mean errors suggesting that the Glide docking scores slightly underestimated the observed binding affinity in these model proteins (Supplementary Table S4). The IC50 of SF2523 against PI3Kα was reported to be 16 nM, using the above conversion factor, −10.63 kcal/mol. The mean error between the predicted docking score and the observed ΔG is −2.27 kcal/mol, suggesting a reasonably good prediction in PI3Kα protein (RD0 model).
Docking analysis of synthesized compounds against model proteins BD1, BD2, and PI3K.
To gain detailed insight into the potential binding interactions of our analogs MDP1–5 (Fig. 1B) with PI3Kα and BRD4 (BD1 and BD2 proteins), we carried out molecular docking studies using the Glide docking approach (Fig. 1C and D). Our docking analysis predicts that quercetin and MDP1 may have the best activities against PI3Kα, followed by MDP3 and MDP5 [18]. MDP4 may have similar activity as LY294002, a known PI3Kα inhibitor [19]. The docking studies further suggest that LY294002 and quercetin may be dual inhibitors of PI3Kα and BD1 and BD2. MDP1 and MDP5 are predicted by docking studies to be dual inhibitors of PI3K and BD1 and BD2, whereas MDP3 may be a weak inhibitor of BD2 but exhibit greater selectivity toward BD1. Therefore, MDP2 and MDP3 may be more selective toward BD1. Overall, MDP1 shows the best binding toward PI3Kα, MDP5 shows the best binding toward BD1 and BD2 while MDP2 and MDP3 show more selectivity toward BD1 over BD2.
3.3. MDP5 and SF2523 have different binding modes in BRD2-BD2/MDP5, BRD2-BD2/SF2523, and BRD4-BD2/SF2523 complexes
The protein-inhibitor interactions were characterized by determining the X-ray crystal structures of ligand free BRD4-BD2, and complexes representing BRD2-BD2/MDP5, BRD2-BD2/SF2523, BRD2-BD1-MDP5, and BRD4-BD2/SF2523, which were resolved to resolutions of 1.22 Å, 1.20 Å, 1.27 Å, 2.50 Å, and 2.08 Å, respectively (Supplementary Table S5). In all cases of protein/ligand complexes, difference in density for the respective ligand was observed in the acetyl lysine binding site.
In all inhibitor bound structures, the carbonyl oxygen of the 4H-pyran-4-one ring forms a hydrogen-bonded interaction with ND2 of Asn429/433 (BRD2-BD2:429 and BRD4-BD2: 433) and a water-mediated hydrogen-bonded interaction with Tyr386/390 (BRD2-BD2:386 and BRD4-BD2: 390) (Fig. 2A and C). Those two interactions were consistent with the previously published acetyl lysine and inhibitor-bound BRD2-BD2/BRD4-BD2 crystal structures [20]. Furthermore, the NE2 nitrogen atom of His433/437 (BRD2-BD2 residue 433 and BRD4-BD2 residue 437) forms a water-mediated hydrogen-bonded interaction with the oxygen atom of the morpholine moiety. This interaction is observed in all three structures described here although the bonding lengths vary slightly. Specifically, the distances are 3.1 Å in BRD2-BD2/MDP5, 2.5 Å in BRD2-BD2/SF2523, and 3.5 Å in BRD4-BD2/SF2523 complexes, respectively.
Fig. 2.
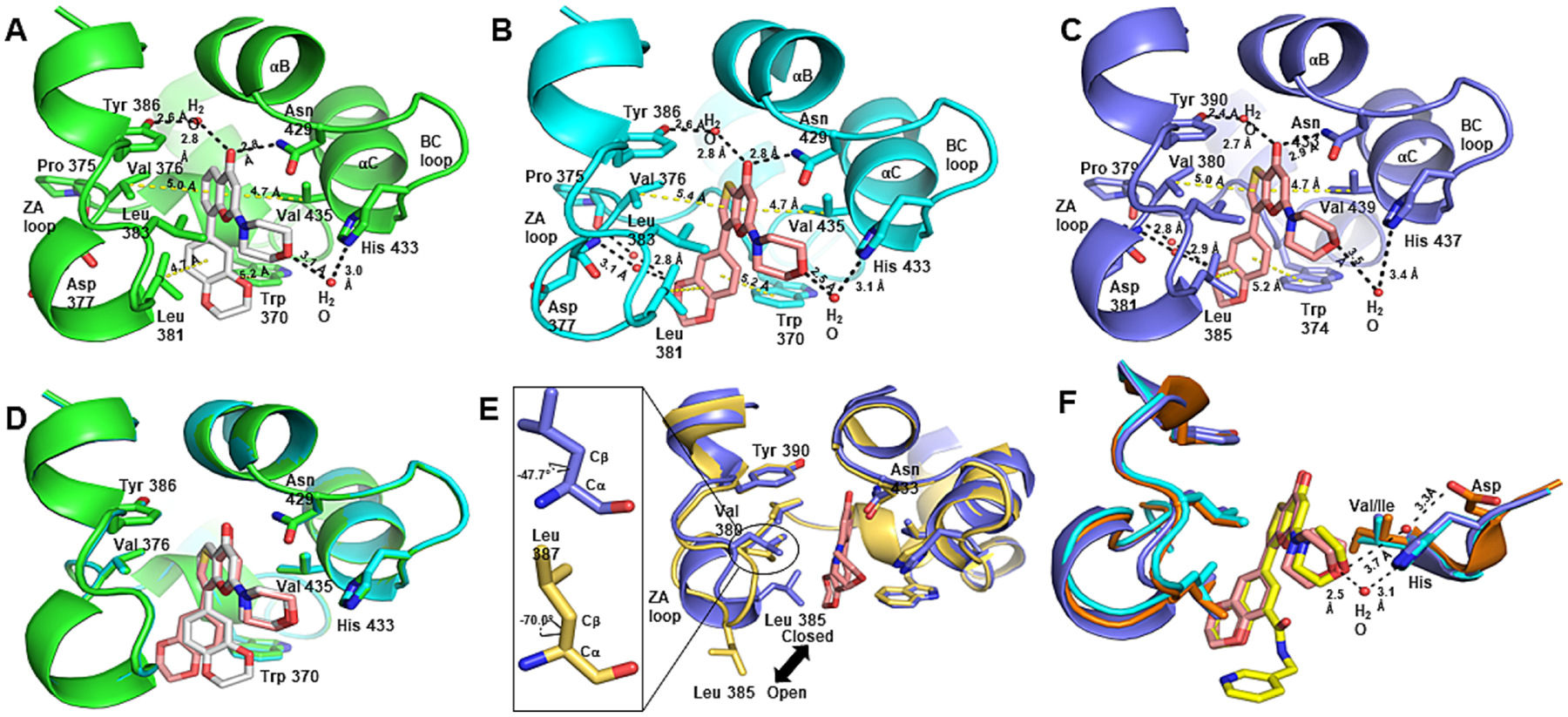
BRD2-BD2/BRD4-BD2 binding interactions with MDP5 and SF2523. A Hydrogen bonded interactions (black dashed bonds) and van der Waals interactions (yellow dashed bonds) formed in the acetyllysine binding pocket of BRD2-BD2 (green carbon atoms) with MDP5 (gray carbon atoms). B Hydrogen bonded interactions (black dashed bonds) and van der Waals interactions (yellow dashed bonds) formed in the acetyllysine binding pocket of BRD2-BD2 (cyan carbon atoms) with SF2523 (pink carbon atoms). C Hydrogen bonded interactions (black dashed bonds) and van der Waals interactions (yellow dashed bonds) formed in the acetyllysine binding pocket of BRD4-BD2 (slate carbon atoms) with SF2523. D Superimposed acetyllysine binding pockets of BRD2-BD2/MPD5 and BRD2-BD2/SF2523 illustrating the difference in benzodioxane binding modes. E Superimposed acetyllysine binding pockets of ligandfree BRD4-BD2 (gold carbon atoms) and BRD4-BD2/SF2523 highlighting the ligandinduced conformational change in BRD4-BD2. F Superimposed acetyllysine binding pockets of the BRD4-BD1 (orange carbon atoms)/SRX3212 (yellow carbon atoms) complex, the BRD2-BD2/SF2523 complex, and the BRD4-BD2/SF2523 complex structures suggesting inclusion of an additional ring off the benzodioxane moiety of MDP5.
Although the polar interactions are important for inhibitor recognition and specificity of binding, acetyllysinelysine binding pocket is predominantly hydrophobic, and the interactions of the inhibitors reflect that fact. The BRD2-BD2/MDP5 and BRD2-BD2/SF2523 complex structures illustrate that the side chains of Val376, Leu383, and Val435 form van der Waals interactions with the chromone moiety of MPD5 and thienopyranone moiety of SF2523 (Fig. 2A and B). Similar interactions are formed by Val380, Leu387, and Val439 side chains with the thienopyranone moiety of SF2523 in the BRD4-BD2/SF2523 complex (Fig. 2C). When considering the benzodioxane moiety of MDP5 and SF2523, the benzene ring forms π-π interactions with the side chain of Trp370/374 (BRD2-BD2:370 and BRD4-BD2: 374) and van der Waals interactions with the side chains of Leu381/Leu385 (BRD2-BD2:381 and BRD4-BD2 385) (Fig. 2A and C). It is clear from the superimposed structures of BRD2-BD2/MDP5 and BRD4-BD2/SF2523 complexes that each compound possesses a different binding mode for the respective benzodioxane moieties (Fig. 2D).
When comparing the chromone moiety of MDP5 and thienopyranone of SF2523, the 4H-pyran-4-one ring is common, but the ring fused to the pyranone differs in that SF2523 contains a five-membered thiophene ring instead of the six-membered benzene ring in MDP5. As a result of the geometric difference between the five- and six-membered rings, the bond vector connecting to the benzodioxane moiety of SF2523 versus that of MDP5 differs. Due to this slight orientational change of the bond and a 186.4° rotation of the dihedral angle, the benzodioxane moiety of SF2523 is positioned closer to the ZA loop of the acetyl lysine binding pocket. The different binding mode significantly alters the interactions formed between the dioxane moiety and the protein. Specifically, the SF2523 complex structures show the benzodioxane moiety forming two water-mediated hydrogen-bonded interactions with the carbonyl oxygen of Pro375/379 (BRD2-BD2: 375 and BRD4-BD2: 379) and the backbone nitrogen atom of Asp377/381(BRD2-BD2: 377 and BRD4-BD2: 381) (Fig. 2B and C). In contrast, the benzodioxane moiety of MDP5 in the BRD2-BD2/MDP5 complex structure does not form any interactions with those same protein backbone atoms (Fig. 2A). Indeed, the benzodioxane moiety of MDP5 interacts directly with the side chain of Trp370.
Furthermore, the acetyl lysine binding pockets of the BRD4-BD2 and BRD4-BD2/SF2523 structures were superimposed to identify any protein structural changes resulting from binding of SF2523 (Fig. 2E). This comparison highlights the restructuring of the ZA loop in the BRD4-BD2/SF2523 complex structure. As shown in Figure 2E, the side chain of Leu385 has changed from an open conformation in the ligand-free BRD4-BD2 structure to a closed conformation in the BRD4-BD2/SF2523 structure. This structural difference was observed in both molecules in the asymmetric unit of BRD4-BD2/SF2523 complex structure is unlikely to represent a crystallographic artifact. Also, when comparing molecule B of BRD4-BD2/SF2523 and the ligand-free BRD4-BD2 structures, the ZA loop region around Leu 385 of the BRD4-BD2/SF2523 complex structure has rearranged to form a single-turn α-helix. The superimposed structures further indicate a dihedral rotation in the Leu 387 of BRD4-BD2/SF2523 complex structure. The χ1 dihedral angle (between Cα and Cβ atoms) differs by 22.3° for residue Leu 387 in the BRD4-BD2 and BRD4-BD2/SF2523 structures. All the observed changes in this region strongly suggest BRD4-BD2 ligand recognition differs from other BDs in BRD2 and BRD4, which do not exhibit a similar conformational change upon ligand binding. This difference may play a vital role in developing potent inhibitors that bind selectively to the acetyllysine binding site of BRD4-BD2.
For further understanding regarding inhibitor selectivity, MDP5 and SF2523 complex structures were compared to that of SRX3212, which is a derivative of SF2523. In a study by Vann et al., SRX3212 exhibited a 65-fold and 48-fold better binding affinity than SF2523 for BRD4-BD1 and BRD4-BD2, respectively [21]. The BRD4-BD2/SF2523 and BRD2-BD2/SF2523 complex structures were superimposed with BRD4-BD1/SRX3212 (PDB ID:6X7C) to gain insight regarding the difference in potency of these two compounds (Fig. 2F). SRX3212 contains an additional pyrid-3-ylmethylaminocarbonyl group attached to the benzodioxane moiety but this extension does not appear to form any additional interactions with BD1. Indeed, the only additional interaction is an intramolecular hydrogen-bond between the nitrogen of SRX3212 amide linker with an oxygen atom of benzodioxane. This may lower rotational entropy of SRX3212 that strengthens SRX3212 binding within the acetyllysine binding pocket. Furthermore, the superimposed BRD4-BD2/SF2523, BRD2-BD2/SF2523, and BRD4-BD1/SRX3212 structures indicate the water-mediated hydrogen bonded interaction of oxygen atoms in benzodioxane are consistent in all three structures suggesting that protein desolvation upon ligand binding is unlikely to account for the affinity difference. One final difference in the BRD4-BD1/SRX3212 complex structure is the presence of a water-mediated hydrogen bonded interaction between the oxygen atom of the morpholine moiety and the side chain of Asp141 with hydrogen bond lengths of 3.3 Å and 3.7 Å. While th SF2523 bound BRD2-BD2 and BRD4-BD2 structures also exhibit a water-mediated interaction between the oxygen of the morpholine moiety and the NE2 of the His side chain (Fig. 2F), these hydrogen bonds are only 2.5 and 3.1 Å.
The BRD2-BD2/MDP5 complex oxygen atom of morpholine moiety forms a water-mediated hydrogen bonded interaction with the side chain His433. Due to the hydrogen bonding distance between the water molecule to Asp160, and the Asp side chain’s orientation illustrate that the above-mentioned water-mediated hydrogen bonded interaction is relatively weak in the BRD2-BD1/MDP5 complex. The superimposed structures further suggest that sequence variation, Val435 in BRD2-BD2 versus Ile162 in BRD2-BD1, could impact inhibitor affinity. The additional methyl moiety increases the surface area of Ile162 and facilitates the formation of additional van der Waals interactions with the entire chromone moiety of MDP5 in the BRD2-BD1/MDP5 structure.
3.4. MDP5 inhibits MB cell growth and migration
MDP series compounds MDP1 and MDP5 significantly inhibited the proliferation in both cell lines with the IC50 values in DAOY and HD-MB03 cells for MDP1 were 5.51 μM and 5.56 μM, for MDP5, IC50 values were 5.57 μM and 5.14 μM, respectively (Fig. 3A and B). In comparison, the IC50 for SF2523 was 12.6 μM in DAOY and 5.14 μM in HD-MB03 cells.
Fig. 3.
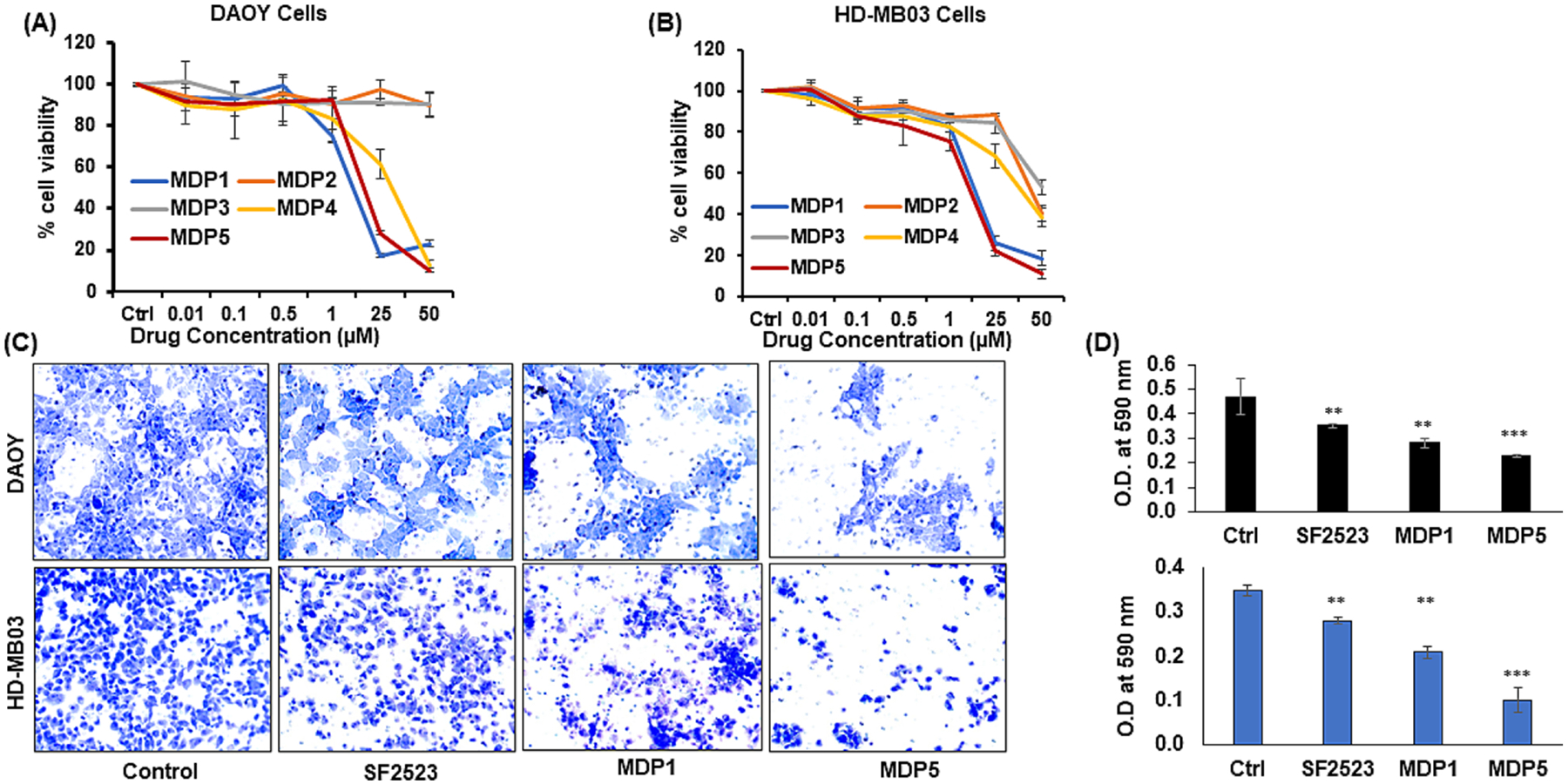
BRD4 and PI3K dual inhibitor SF2523 and its novel analogs MDP1 and MDP5 inhibit MB cell proliferation and migration in culture. A&B Cell viability assay in DAOY and HD-MB03 cells after incubation with different concentrations of MDP series compounds. C In the Transwell migration assay, cells were treated with SF2523, MDP1, and MDP5 at 5.0 μM concentration for 24 h. Cells were trypsinized and placed in apical chamber of insert in 24 well plate to 24 h. Next, migrated cells were fixed in formalin and stained with 0.5% crystal violet for 30 min. Images were taken using a microscope at 20x. D Next, stained cells were dissolved in 10% acetic acid solution and measured optical density (OD) at 590 nm. Scale bar, 100 μm; ** P < 0.01 and *** P < 0.001 vs. Control.
After 24 h the untreated DAOY and HD-MB03 cells migrated towards the center and closed the scratched width approximately 80% and 70%, respectively, as compared to initial. However, lesser migration was observed in SF2523, MDP1, and MDP5 treated cells at 24 h (Supplementary Fig. S4). Further, SF2523, MDP1, and MDP5 decreased the extent of migration in DAOY and HD-MB03 cells across the transwells insert membrane compared to control cells. (Fig. 3C). However, only MDP5 was able to decrease migration significantly, than control in both the cell lines (Fig. 3D).
3.5. MDP5 inhibits cell cycle progression, colony formation, and apoptosis in MB cells
We observed that upon the treatment of MB cells with MDP5 and SF2523 was arrested in the G1 phase after treatment compared to control and MDP1 (Fig. 4A and B). In DAOY cells, 52.8% of untreated cells was found in the G1 phase, whereas their population was 73.1%, 40.1%, and 73.2% in SF2523, MDP1 and MDP5 treated cells, respectively. In the case of HD-MB03 cells, 44.7% of untreated cells were in the G1 phase, while SF2523, MDP1, and MDP5 treated cells were observed in the G1 phase at 58.9%, 26.2%, and 52.2%, respectively. In clonogenic assays, both the cell lines and treatment groups showed fewer colonies than the untreated control cells (Fig. 4C). However, MDP5 was most effective in inhibition of colony formation potential compared to SF2523 and MDP1 groups.
Fig. 4.
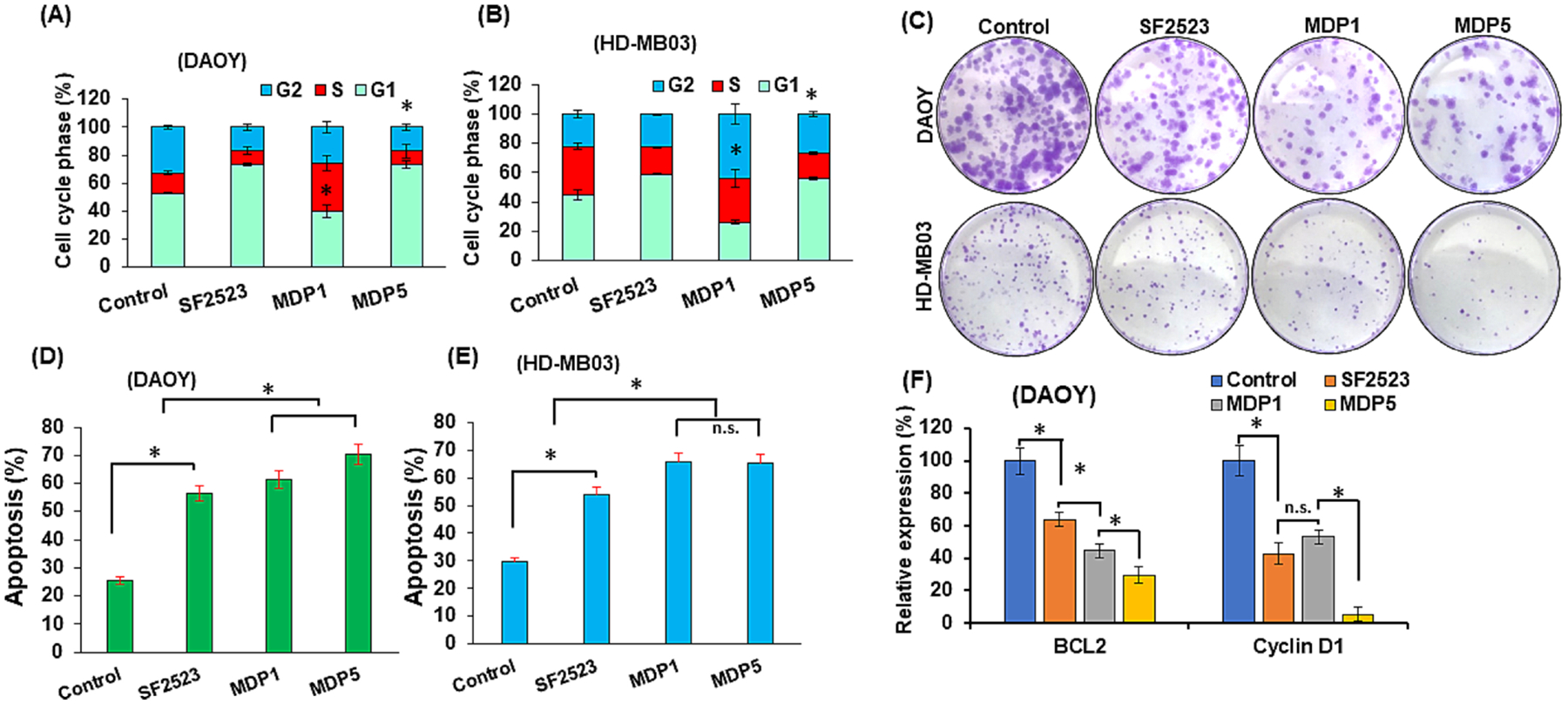
BRD4 and PI3K dual inhibitors MDP1 and MDP5 promote MB cell arrest in G1 phase and apoptosis but inhibits colony formation more efficiently than SF2523. DAOY and HD-MB03 cells were treated with SF2523, MDP1, and MDP5 at the dose of 5.0 μM for 48 h. A and B Histogram shows the quantitative analysis of cell cycle assay (n = 5, mean ± S.D., p < 0.05). Cells treated with SF2523 and MDP5 show cell cycle arrest in G1 phase as compared to control and MDP1. C Treatment with MDP5 showed lower number of colonies in both the cell lines. D and E Apoptotic assay (n = 5, mean ± S.D., *p < 0.05). F Relative mRNA expression levels (%) of BCL2 and Cyclin D1 genes after 48 h incubation with SF2523, MDP1 and MDP5.
Further, the apoptotic assay results revealed that, in DAOY cells, percentages of apoptotic cells after 48 h of treatment with SF2523 and MDP1 were 56.50% and 61.31%, respectively, but it increased to 70.35% after MDP5 treatment compared to only 25.61% in untreated cells (Fig. 4D). Similarly, in HD-MB03 cells percentages of apoptotic cells were 54% and 65.66% after 48h of treatment with SF2523 and MDP1, respectively, but increased to 65.15% in MDP5 treated cells versus 29.76% in untreated cells (Fig. 4E).
3.6. MDP5 treatment decreases target gene expression at mRNA and protein levels
We further determined expression of cell death related gene BCL2 in MB cells. and observed that BCL2 mRNA expression was significantly reduced after MDP5 treatments (Fig. 4F). Further, cyclin D1 gene expression was also decreased significantly compared to the control, which highlights that MDP5 was the most potent among the treatment groups. Further, we confirmed these results by Western blot analysis, and observed that cyclin D1, p-AKT (Ser473) and p-PI3K protein levels were reduced while p-MYCN (Ser54) level was explicitly induced in MDP5 treated cells (Fig. 5A and B). The phosphorylation at Ser-54 in MYC protein is known to control its degradation, while phosphorylation at Ser473 in AKT protein is known to control its activation [22].
Fig. 5.
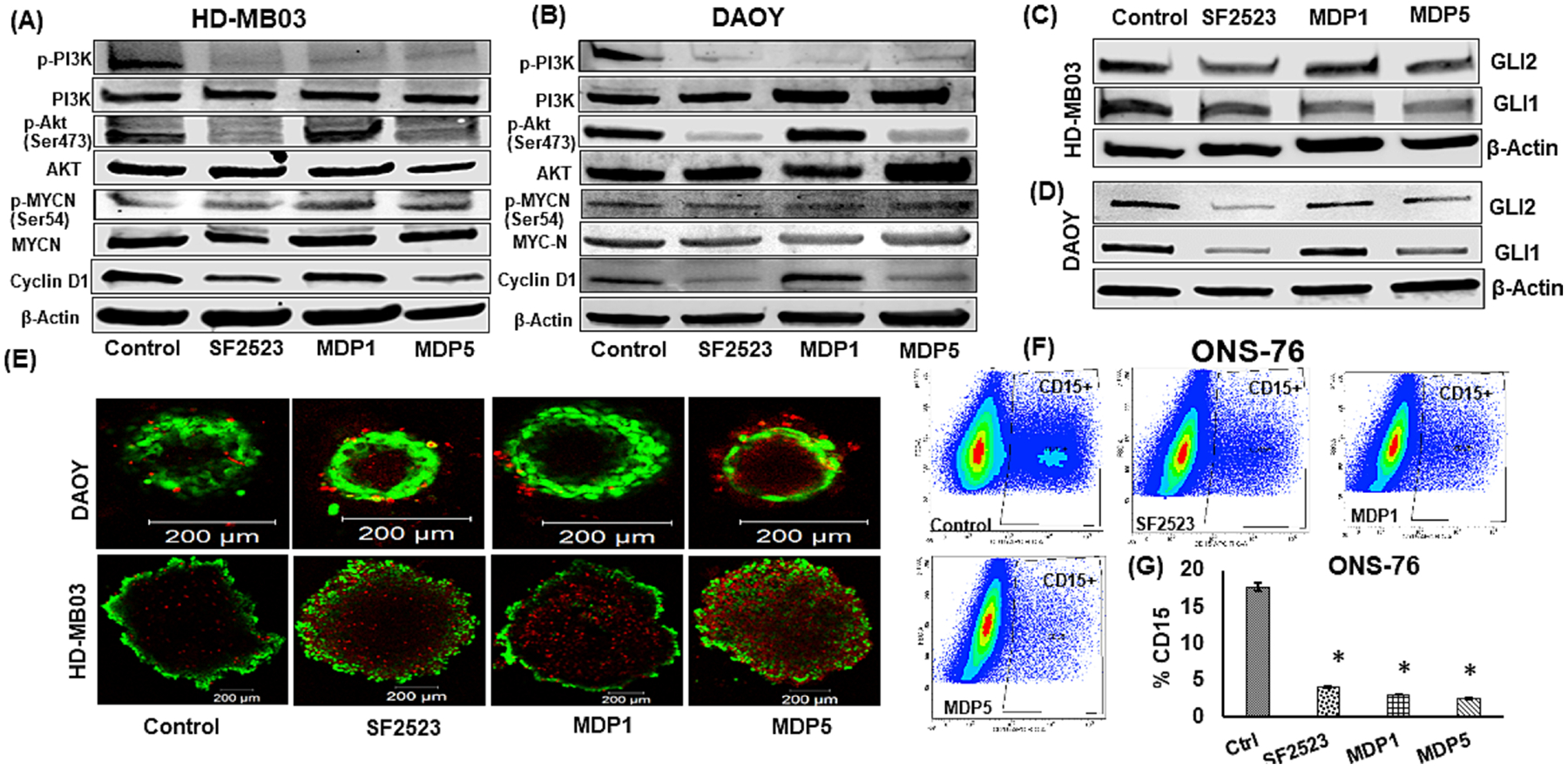
BRD4 and PI3K dual inhibitor MDP5 treatment decreases target gene expression at protein levels. A and B Western blot analysis of target proteins after HDMB03 and DAOY cells were incubated with 5.0 μM of these drugs for 48 h. C and D Effects of MDP5 treatment on target protein including GLI1 and GLI2. E Representative confocal images of DAOY and HD-MB03 spheroids after MDP5 treatment with 5.0 μM). MDP5 decreased the tumor spheroid formation in both MB cells. Spheroids were stained for live (Calcein AM; in green) and dead (ethidium homodimer-1; in red) cells after 7 days of treatment. (Scale bars = 200 μm). F Effect of MDP5 treatment on CD15+ population in MB cells ONS-76. SHH group (ONS-76 cells) were treated with SF2523, MDP1, and MDP5 for 48 h treated with 5.0 μM. Next, cells were incubated with APC-CD15 for 30 min on ice in dark. Before FACS analysis, samples were washed thrice with staining buffer. G Quantitation of CD15+ cells (n = 3. p < 0.05, control vs. treatment groups).
We next tested if MDP1 and MDP5 could decrease GLI1 and GLI2 protein expression as previously observed following SF2523 treatment [11]. We observed that MDP1 and MDP5 showed a decrease in both (Fig. 5C and D). MDP5 displayed the largest reduction in GLI1 protein expression among treated HD-MB03 cells. In contrast, SF2523 exceeded all other treatment options in terms of lowering GLI2 protein accumulation.
To determine the cytotoxicity effect of these compounds in three-dimensional (3D) tumor spheroids that mimic MB tumor microenvironment, live and dead cell assays were carried out. As shown in Figure 5E, all the treatments reduced the size of spheroids after 7 days of treatment compared to DMSO-treated control group. Dead cells along the margin of tumor spheroids were easily separated during staining, resulting in a tooth-shaped morphology. Cell viability of the tumor spheroids was significantly decreased following treatment with SF2523 and MDP5. The dead cells distributed across the inner core and the superficial regions of spheroids when exposed to either SF2523 or MDP5 at 5.0 μM concentration. A higher number of dead cells in the core of all the treated spheroids is possibly due to the diffusion of the drugs and high levels of cellular stress inside the 3D structure [23].
We calculated the proportion of CD15+ cells in various MB cells. SHH group derived DAOY cells demonstrated 3.87 ± 0.2%, ONS-76 cells shown 23.05 ±3%, Group 3 cells HDMB-03 demonstrated 1.73 ±0.4%, and D283 cells demonstrated 12.97 ±3% CD15+ cells among them. Based on these results, we selected ONS-76 cells for further experiment. Upon treatment with MDP5, the percentage of CD15+ cell population in ONS-76 cells was declined compared to control cells (Fig. 5F). Even though the percentage of CD15+ cells dropped significantly in all the treatment groups when compared to the control group, but no discernible difference was seen between the treated groups (Fig. 5G). We investigated whether treatment with SF2523 and MDP5 for 30 min decreases the proportion of SP in ONS-76 cell cultures (Supplementary Fig. 5a). Treatment with SF2523, MDP1, and MDP5 the percentage of SP proportion significantly decreased to 2.57 %, 0.7%, and 0.9%, respectively, as compared to 4.57% for the control cells (Supplementary Fig. S5b).
3.7. MDP5 treatment decreases xenograft tumor burden in mice
We next evaluated the effect of MDP5 treatment on the inhibition of tumor growth in mice. First, we utilized a subcutaneous (S.C) tumor xenograft model with DAOY and HD-MB03 cells (Fig. 6). At a dose of 20 mg/kg, animals showed no sign of toxicity, as evident by little change in body weight during the treatment (Fig. 6A). Moreover, histologic examination of major organs including heart, lung, liver, and kidneys, did not demonstrate toxicity after completion of the treatment (Supplementary Fig. S6). In both DAOY and HD-MB03 cells generated xenograft tumors, MDP5 treatment dramatically reduced tumor growth relative to the control group (Fig. 6B and G). Further, H&E staining and immunohistochemical (IHC) analysis of Ki67 showed that MDP5 treatment suppressed cell proliferation and decreased tumor burden in DAOY and HD-MB03 cells generated tumors (Supplementary Fig. S7). Together, these data suggest that MDP5 shows potent anti-tumor activity in vivo in the xenograft model and decreased MB tumor burden and cellular proliferation without significant toxicity.
Fig. 6.
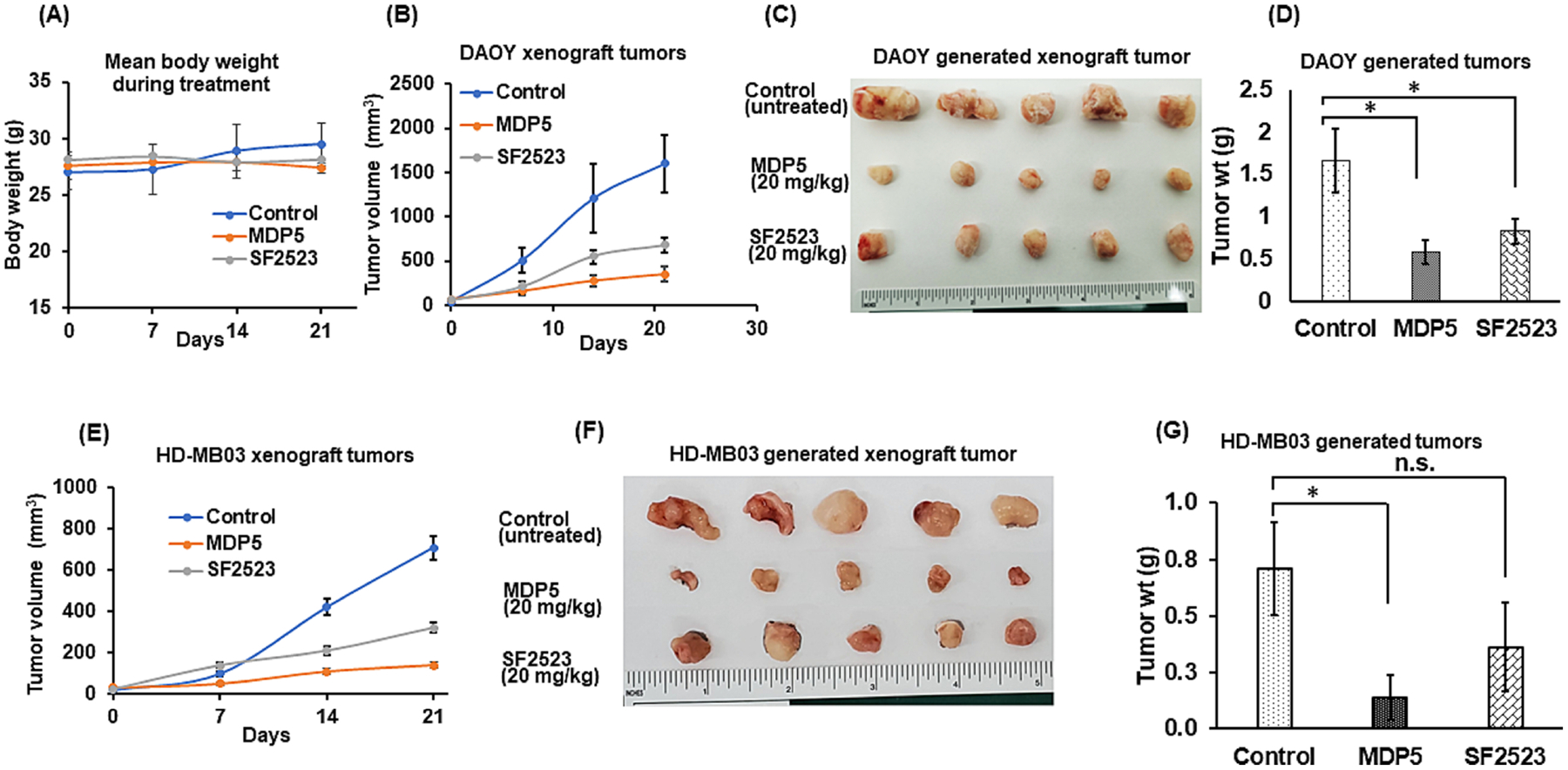
BRD4 and PI3K dual inhibitor MDP5 inhibits tumor growth more effectively than SF2523 when administered systemically into MB xenograft mice. A Animal body weight during the treatment period. B Tumor burden curve of DAOY cell generated tumors. C Representative pictures of harvested tumors. D Tumor weights (mean ± S.D., n = 5, *p < 0.05). E Tumor burden curve of HD-MB03 cell generated tumors. F Representative pictures of harvested tumors. G Tumor weights (mean ± S.D., n = 5, *p < 0.05).
Next, we tested whether MDP5 treatment suppresses MB growth in an intracranial orthotopic xenograft model. IVIS imaging on day 1 (the start of injection) and on day 28 (after the last injection) of these mice from each group show that treatment with MDP5 had a significant effect on the suppression of tumor growth in vivo (Fig. 7A). The bioluminescence (BLI) signal measurement shows a rapid increase in control animals, while a low BLI signal was detected in MDP5 treated mice (Fig. 7B).
Fig. 7.
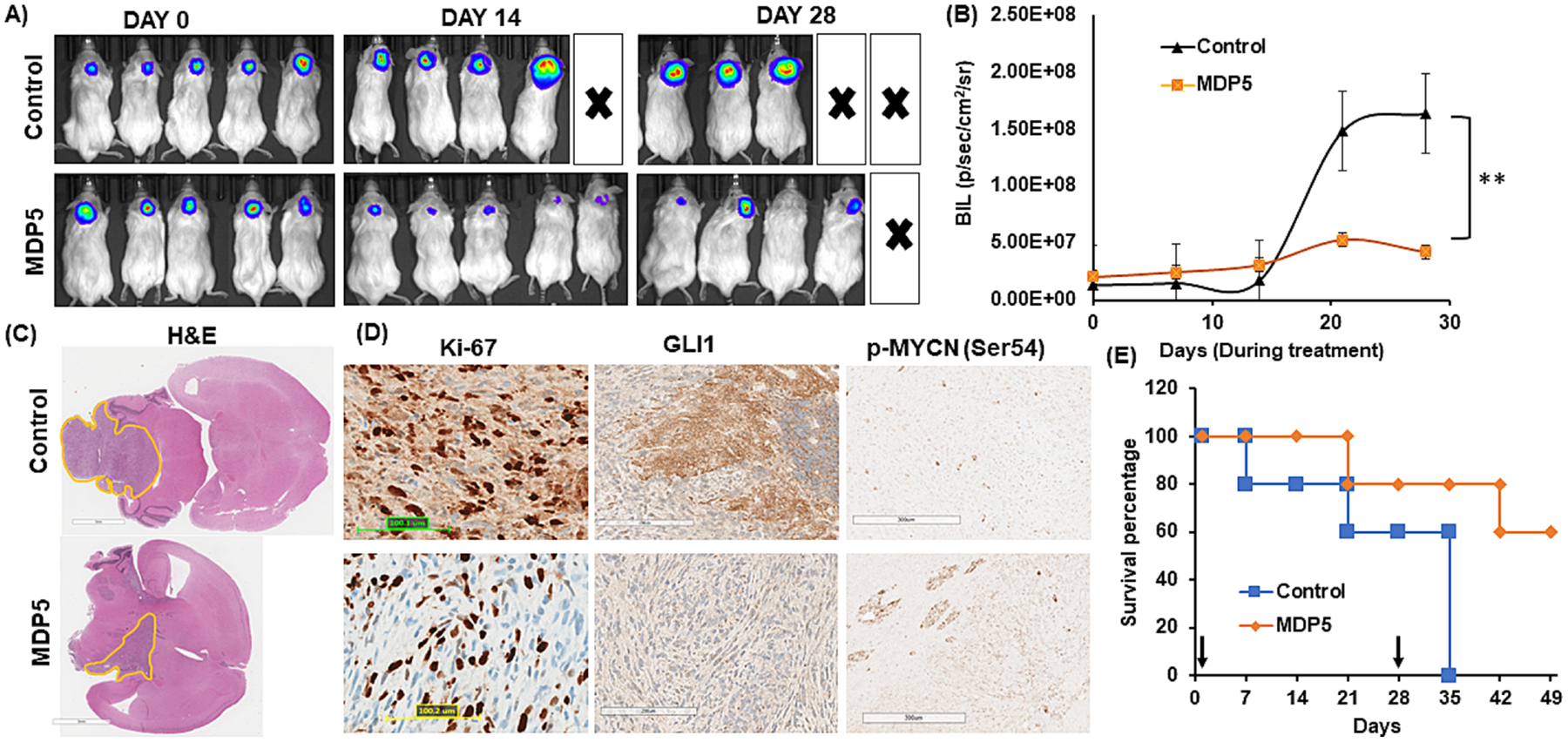
Antitumor efficacy of MDP5 after systemic administration into DAOY cell generated orthotopic MB bearing NSG mice. A Bioluminescence imaging of orthotopic MB bearing mice during the treatment. B Quantitation of BLI in MDP5 treated and control group (n = 3, **P < 0.01). C H&E staining of brain harvested from mice bearing orthotopic MB after treatment (Scale bar 3 mm). D IHC staining of tumor tissues for Ki-67, GLI1, and p-MYCN (Ser54) (Scale bar 100–300 μM). E Overall survival curve showing MDP5 treated mice survived significantly longer than the control group (n = 3).
These mice were kept without any further treatment for survival study. When the animal started showing distress and tilted head due to the high tumor burden, the animal was sacrificed, and its brain was harvested. The H&E results corroborated the data obtained by IVIS and showed that mice treated with MDP5 had an overall smaller tumor volume when compared to untreated mice (Fig. 7C). We determined the relative expression of proliferation marker Ki67, and MDP5 target genes p-MYCN (Ser54) and GLI1 by IHC staining (Fig. 7D). As shown in Figure 7E, in MDP5 treatment group, four animals out of five survived up to 55 days in an orthotopic xenograft model, while all the control animals were dead by day 35.
4. DISCUSSION
BRD4 is a super enhancer that mediate expression of several genes involved in inflammation and tumors including the Hh signaling pathway components and MYC [8]. Since the first small molecular BRD4 inhibitor JQ1 was discovered, several categories of inhibitors with different chemical scaffolds (I-BET762, OTX015, and CPI-0610) have been identified. However, most of these drugs are nonspecific inhibitors for members of the bromodomain and extra-terminal (BET) family and induce numerous adverse effects [8].
A common occurrence in MB is an upregulation of the MYC and SHH signaling pathways. Although these are classified as distinct groups, MYCN and MYCL1 are highly expressed in the SHH subgroup [4]. Therefore, BRD4 inhibitors are quite useful from a therapeutic standpoint in the treatment of MB. However, treating MB with BRD4 inhibition is difficult due to the development of chemoresistance by PI3K, which also plays important roles in MB cell growth and tumorigenesis. PI3K mediated chemoresistance can be overcome by modulating MYC using PI3K/BRD4 dual inhibitors.
Toward this end, we synthesized novel BRD4/PI3K dual inhibitors MDP1–5, that, are structural analogs of SF2523. Among these molecules our study predicted that MDP1 would exhibit the strongest binding to PI3K, MDP5 would exhibit the strongest binding to BD1 and BD2, while MDP2 and MDP3 have more selectivity for BD1 over BD2 (Supplementary Table S3). Inhibiting PI3K solely in MB has previously been demonstrated to be unsuccessful, especially in resistant cells, but selectively targeting the BD2 domain of BET family proteins has been shown to exhibit robust antiproliferative activity in different cancers [24], with one contrary report where BD1 inhibitors showed better efficacy [25]. Taken together, this suggests that MDP1 alone would be an unfavorable strategy and MDP2 or MPD3 might have some efficacy as a monotherapy. However, in our cell viability studies, we found MDP2 and MDP3 less effective and chose to focus on comparing MDP5 and SF2523.
The X-ray crystal structures of bromodomains complexed with MDP5 and SF2523indicated a small but intriguing change in the binding modes of MDP5 and SF2523 in the acetyllysine binding pocket. The benzodioxane moiety of SF2523 in the BRD2-BD2/SF2523 structure has moved towards the ZA loop and forms additional water-mediated hydrogen-bonded interactions with the carbonyl oxygen of Pro375 and backbone nitrogen of the Asp377 in contrast to that observed in the BRD2-BD2/MDP5 complex. Those additional hydrogen-bonded interactions further contribute to the stabilization of SF2523 within the BRD2-BD2 acetyllysine binding pocket (Supplementary Fig. S8).
The observed conformational change of Leu385 (open to closed-form), the ZA loop rearrangement, and the dihedral rotation of Leu387 upon binding of SF2523 to BRD4-BD2 may account for the PTS results. In contrast to the induced fit for ligand binding to BRD4-BD2, structural data for BRD2-BD2 suggest that the acetyllysine binding site does not change structure upon binding of ligands. This is made clear by superimposing the ligand-free BRD2-BD2 structure with the numerous BRD2-BD2/ligand complexes described here. Therefore, the modest thermal stabilization observed in the BRD2-BD experiments suggest that the MDP compounds are binding to BRD2-BDs in a manner consistent with other known ligands but form only a minimum of interactions. In contrast, when the same MDP compounds bind to BRD4-BD, they induce structural dynamics in the ZA loop but fail to form alternative stabilizing interactions, which results in the 0.6 to 1.3 °C thermal destabilization of those complexes. This further suggests that future design of MDP derivatives should account for the structural differences between the ligand-free forms of BRD2-BD2 and BRD4-BD2 and seek to bind BRD4-BD2 but prevent the conformational change. This would result in a more potent inhibitor of BRD4-BD2 but significantly weakly binding to BRD2-BD2 due to steric hindrance with the ZA loop (Supplementary Fig. S9).
Our structural results indicate that MDP5 retains much of the essential structural properties of parent compound SF2523 to support BD binding. It is clear from a comparison of the BRD4-BD1 complexes with MDP5 that it competes with acetyllysine binding and maintains the conserved hydrogen bond with Asn140 found in acetyllysine-BD complexes but does not interfere with the distinctive water shell of BD1. The morpholino ring of MDP5 is parallel to the α-helices of BD1 and buried deep in the pocket to support binding. The benzodioxane group of MDP5 faces the ZA loop in BD1, which places it at right angles to the α-helices and interacts with the hydrophobic cage formed by the side chains of Trp81, Pro82, and Leu92. The chromone component of MDP5 is stabilized by the side chains of Val87 and Ile146, while the amide nitrogen of the Asn140 side chain and the hydroxyl group of Tyr97 in BD1, interacts with the carbonyl oxygen in the chromone group by hydrogen bonding. Further, additional hydrogen bonds between Trp 81, Gln85, and Asp88 in the ZA loop of BD1 hold the dioxane ring of MDP5 in position.
The determined structures also offer rationale for the relative lack of desired efficacy for MDP1–4. MDP2 differs from MDP5 only by the inclusion of the 2-bromo group on the benzo dioxane. The binding mode of MDP5 limits the positioning of that bromine atom to only 1 of 2 locations and both are detrimental to BD binding due to either steric hindrance or electronic repulsion by positioning near a cluster of 3 backbone carbonyl groups. The MDP1 compound possesses the same bromo-group as MDP2 but has additional rotatable bonds due the elimination of the chromone moiety. Therefore, MDP1 either pays a larger entropic penalty of binding or interacts with the BDs in a completely different binding mode than MDP5. MDP4 replaces the benzodioxane moiety, and all of the important BD interactions, with a bromine. The installed bromine of MDP4 is unlikely to form any supporting interactions with the BDs. The change in carbonyl positioning of MDP3 forces a detrimental interaction with the Asn140 side chain in BRD2-BD1 or the Asn 429/433 side chain carbonyl in the BD2s that likely weakens binding.
While MDP5 (5.5 μM) showed higher potency in DAOY cells compared to SF2523 (12.6 μM) but showed similar potency on HD-MB03 (MYC amplified) cells. In, cell migration, apoptosis, cell cycle, and wound healing assays MDP5 showed higher potency compared to SF2523 in both DAOY and HD-MB03 cell lines. MDP5 decreased the target downstream proteins like p-AKT, MYCN, and Cyclin D1. MDP5 increased the degradation of MYCN protein as indicated by upregulation of p-MYCN (Ser54). These results agree with a previous report where SF2523 blocks PI3K activation and down-regulates MYCN and Cyclin D1 in vivo [12]. It is well established that the SHH-GLI signaling pathway is activated by multiple snon-canonical activation method in cancers. Consequently, SMO inhibition is less effective against tumors with non-canonical SHH-GLI signaling, which jeopardizes the therapeutic effectiveness of these antagonists. The PI3K/AKT/mTOR pathway is known to upregulate GLI1/GLI2 transcription and enhance protein levels [26]. Accordingly, MDP5 showed downregulation of these proteins in our results (Fig. 5 C&D), indicative of a promising anti-cancer therapy.
CSCs, often called as tumor initiating cells, are capable of self-renewal which contributes to tumor growth and resistance. Human MB and Patched mutant mouse model cells that express the progenitor markers Math1 and CD15 are known to have the ability to proliferate and a reduced propensity to undergo apoptosis and differentiate. Our results confirmed that BRD4 and PI3K inhibition decreased CSCs and SP cells, demonstrating it could be effective in eradicating chemoresistant cell populations.
For in vivo efficacy and safety studies, DAOY and HD-MB03 cells generated xenograft bearing NSG mice were treated with 20 mg/kg dose of SF2523, MDP1, and MDP5. It was observed that MDP5 was able to significantly reduce the tumor burden compared to other treatments and vehicle treated control group. Further, mice treated with MDP5 at the tested dose showed no significant differences in body weight or histological slides of important organs when compared to mice treated with a vehicle.
Although MDP5 biodistribution remains to be experimentally evaluated, the physiochemical properties of MDP5 are consistent with compounds shown to cross the BBB based in earlier reports. For example, the mass is lower than 450 a.m.u., the compound is lipophilic with a CLogP of 2.99, has less than 3 hydrogen bond donors, and a topological polar surface area (tPSA) of 57.23. Each of these physicochemical parameters are known to contribute to BBB penetrability [26, 27]. Indeed, the tPSA, is a descriptor to define the sum of surfaces of polar atoms in a molecule, which is one of the most useful parameters for predicting molecular transport properties [28]. Further, the 1,4-dioxane ring fused onto the aromatic ring reduces the tPSA with respect to other analogs. Since these properties are indirect indicators of BBB penetration ability of a molecules, we tested whether MDP5 treatment suppresses MB growth in an intracranial orthotopic MB model and observed that MDP5 significantly suppressed tumor growth in vivo. These mice were allowed to survive after the final treatment and their brains were harvested, after the mice exhibited moribund behavior. H&E staining and IHC analysis of Ki-67 reveal a much larger tumor at the cerebellum region of the vehicle-treated mouse compared to MDP5 and SF2523 treated mice. Although PI3K inhibitors showed early promise as a MB target, therapeutic resistance is a frequent problem, and that resistance is linked to MYC up-regulation by hyperactivity of BRD4 transcriptional control.
5. CONCLUSION
In conclusion, we report a highly potent dual functional PI3K and BRD4 inhibitor molecule, MDP5, that promotes MYC degradation by simultaneous inhibition of AKT function and inhibition of BRD4 acetyllysine binding to suppress MYC transcription. Our study found that among all the MDP series compounds, MDP5 has the highest potency in controlling MB cell growth in vitro as well as in vivo, and our structural analysis sheds light on its mode of action and future scaffold optimization for improving anticancer efficacy.
Supplementary Material
Appendix A. Supplementary material and data.
Highlights.
Inhibition of SHH and group 3 MB with newly synthesized MDP5; a dual inhibitor of BDR4 and PI3K is a viable strategy to overcome resistance and effective MB treatment.
In vitro results showed that MDP5 has the ability to target the dysregulated genes at RNA and protein levels. MDP5 was effective in targeting tumor propagating cells and tumor spheroid.
MDP5 reduced tumor growth and prolonged the life span of MB tumor bearing orthotopic mouse model.
Acknowledgments
The X-ray diffraction datasets were obtained using resources of the Advanced Photon Source, a U.S. department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne. National Laboratory under Contract No.DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economics Development Corporation and Michigan Technology Tri-Corridor (Grant 085P1000817).
Funding
The NIH (1R01NS128336 and 1R01NS116037), Pediatric Cancer Research Group of the University of Nebraska Medical Center and Children’s Hospital (LB805), Nebraska DHHS LB506, and Nebraska Research Initiative (NRI) Program to Ram Mahato are duly acknowledged for providing financial support for this work.
Abbreviations
- BDs
Bromo domains
- BET
bromodomain and extra-terminal
- CSCs
cancer stem cells
- DMSO
dimethyl sulfoxide
- IHC analysis
immunohistochemical analysis
- MB
Medulloblastoma
- PI3K
phosphatidylinositol-4,5-bisphosphate 3-kinase
- MDP5
8-(2,3-dihydrobenzo[b] [1,4] dioxin-6-yl)-2-morpholino-4H-chromen-4-one
- SHH
sonic hedgehog
- SP
side population
- TEA
triethanolamine
- TCEP
tris(2-carboxyethyl)phosphine)
- 3D
three-dimensional
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethics statement
Animals used in this study has been approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Nebraska Medical Center (UNMC). Animals were housed in Durham Research Building (DRC) II for subcutaneous animal model and orthotopic generated animals were transferred to Buffet cancer center for IVIS imaging at UNMC.
Declaration of competing interest
All the authors declared that they do not have any competing interests.
Data availability statement
The paper and/or the Supplementary Materials contain all the information required to assess the conclusions made in the paper. You may ask the authors for more information about this paper.
REFERENCES
- [1].Kumar V, Kumar V, McGuire T, Coulter DW, Sharp JG, Mahato RI, Challenges and Recent Advances in Medulloblastoma Therapy, Trends Pharmacol Sci, 38 (2017) 1061–1084. [DOI] [PubMed] [Google Scholar]
- [2].Borgenvik A, Cancer M, Hutter S, Swartling FJ, Targeting MYCN in Molecularly Defined Malignant Brain Tumors, Front Oncol, 10 (2020) 626751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Olivero CE, Martinez-Terroba E, Zimmer J, Liao C, Tesfaye E, Hooshdaran N, Schofield JA, Bendor J, Fang D, Simon MD, Zamudio JR, Dimitrova N, p53 Activates the Long Noncoding RNA Pvt1b to Inhibit Myc and Suppress Tumorigenesis, Mol Cell, 77 (2020) 761–774 e768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Roussel MF, Robinson GW, Role of MYC in Medulloblastoma, Cold Spring Harb Perspect Med, 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen H, Liu H, Qing G, Targeting oncogenic Myc as a strategy for cancer treatment, Signal Transduct Target Ther, 3 (2018) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, Dey A, Ozato K, Singer DS, BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin, Nat Struct Mol Biol, 23 (2016) 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Taniguchi Y, The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins, Int J Mol Sci, 17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhou Z, Li X, Liu Z, Huang L, Yao Y, Li L, Chen J, Zhang R, Zhou J, Wang L, Zhang QQ, A Bromodomain-Containing Protein 4 (BRD4) Inhibitor Suppresses Angiogenesis by Regulating AP-1 Expression, Front Pharmacol, 11 (2020) 1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, Virtanen C, Bradner JE, Bader GD, Mills GB, Pe’er D, Moffat J, Neel BG, Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance, Cell, 164 (2016) 293–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X, Targeting PI3K in cancer: mechanisms and advances in clinical trials, Mol Cancer, 18 (2019) 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kumar V, Wang Q, Sethi B, Lin F, Kumar V, Coulter DW, Dong Y, Mahato RI, Polymeric nanomedicine for overcoming resistance mechanisms in hedgehog and Myc-amplified medulloblastoma, Biomaterials, 278 (2021) 121138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Andrews FH, Singh AR, Joshi S, Smith CA, Morales GA, Garlich JR, Durden DL, Kutateladze TG, Dual-activity PI3K-BRD4 inhibitor for the orthogonal inhibition of MYC to block tumor growth and metastasis, Proc Natl Acad Sci U S A, 114 (2017) E1072–E1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM, The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations, Science, 318 (2007) 1744–1748. [DOI] [PubMed] [Google Scholar]
- [14].Sabbah DA, Vennerstrom JL, Zhong H, Docking studies on isoform-specific inhibition of phosphoinositide-3-kinases, J Chem Inf Model, 50 (2010) 1887–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Otwinowski Z, Minor W, [20] Processing of X-ray diffraction data collected in oscillation mode, in: Methods in Enzymology, Academic Press, 1997, pp. 307–326. [DOI] [PubMed] [Google Scholar]
- [16].Bhattarai RS, Kumar V, Romanova S, Bariwal J, Chen H, Deng S, Bhatt VR, Bronich T, Li W, Mahato RI, Nanoformulation design and therapeutic potential of a novel tubulin inhibitor in pancreatic cancer, J Control Release, 329 (2021) 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kumar V, Mundra V, Peng Y, Wang Y, Tan C, Mahato RI, Pharmacokinetics and biodistribution of polymeric micelles containing miRNA and small-molecule drug in orthotopic pancreatic tumor-bearing mice, Theranostics, 8 (2018) 4033–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pisonero-Vaquero S, Garcia-Mediavilla MV, Jorquera F, Majano PL, Benet M, Jover R, Gonzalez-Gallego J, Sanchez-Campos S, Modulation of PI3K-LXRalpha-dependent lipogenesis mediated by oxidative/nitrosative stress contributes to inhibition of HCV replication by quercetin, Lab Invest, 94 (2014) 262–274. [DOI] [PubMed] [Google Scholar]
- [19].Wang Y, Kuramitsu Y, Baron B, Kitagawa T, Tokuda K, Akada J, Maehara SI, Maehara Y, Nakamura K, PI3K inhibitor LY294002, as opposed to wortmannin, enhances AKT phosphorylation in gemcitabine-resistant pancreatic cancer cells, Int J Oncol, 50 (2017) 606–612. [DOI] [PubMed] [Google Scholar]
- [20].Romero FA, Taylor AM, Crawford TD, Tsui V, Côté A, Magnuson S, Disrupting Acetyl-Lysine Recognition: Progress in the Development of Bromodomain Inhibitors, Journal of Medicinal Chemistry, 59 (2016) 1271–1298. [DOI] [PubMed] [Google Scholar]
- [21].Vann KR, Pal D, Morales GA, Burgoyne AM, Durden DL, Kutateladze TG, Design of thienopyranone-based BET inhibitors that bind multiple synthetic lethality targets, Scientific reports, 10 (2020) 12027–12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kapeli K, Hurlin PJ, Differential regulation of N-Myc and c-Myc synthesis, degradation, and transcriptional activity by the Ras/mitogen-activated protein kinase pathway, J Biol Chem, 286 (2011) 38498–38508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Galateanu B, Hudita A, Negrei C, Ion RM, Costache M, Stan M, Nikitovic D, Hayes AW, Spandidos DA, Tsatsakis AM, Ginghina O, Impact of multicellular tumor spheroids as an in vivo-like tumor model on anticancer drug response, Int J Oncol, 48 (2016) 2295–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, Hsiao K, Yuan J, Green J, Ospina B, Yu Q, Ostrom L, Fordjour P, Anderson DL, Monahan JE, Kelleher JF, Peukert S, Pan S, Wu X, Maira SM, Garcia-Echeverria C, Briggs KJ, Watkins DN, Yao YM, Lengauer C, Warmuth M, Sellers WR, Dorsch M, Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma, Sci Transl Med, 2 (2010) 51ra70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gilan O, Rioja I, Knezevic K, Bell MJ, Yeung MM, Harker NR, Lam EYN, Chung CW, Bamborough P, Petretich M, Urh M, Atkinson SJ, Bassil AK, Roberts EJ, Vassiliadis D, Burr ML, Preston AGS, Wellaway C, Werner T, Gray JR, Michon AM, Gobbetti T, Kumar V, Soden PE, Haynes A, Vappiani J, Tough DF, Taylor S, Dawson SJ, Bantscheff M, Lindon M, Drewes G, Demont EH, Daniels DL, Grandi P, Prinjha RK, Dawson MA, Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation, Science, 368 (2020) 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kenny PW, Hydrogen-Bond Donors in Drug Design, J Med Chem, 65 (2022) 14261–14275. [DOI] [PubMed] [Google Scholar]
- [27].Sethi B, Kumar V, Mahato K, Coulter DW, Mahato RI, Recent advances in drug delivery and targeting to the brain, J Control Release, 350 (2022) 668–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xiong B, Wang Y, Chen Y, Xing S, Liao Q, Chen Y, Li Q, Li W, Sun H, Strategies for Structural Modification of Small Molecules to Improve Blood-Brain Barrier Penetration: A Recent Perspective, J Med Chem, 64 (2021) 13152–13173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix A. Supplementary material and data.
Data Availability Statement
The paper and/or the Supplementary Materials contain all the information required to assess the conclusions made in the paper. You may ask the authors for more information about this paper.


