Abstract
Purpose
Understanding the three-dimensional anatomy of percutaneous intervention in prostate cancer is essential to avoid complications. Recently, attempts have been made to use machine learning to automate the segmentation of functional structures such as the prostate gland, rectum, and bladder. However, a paucity of material is available to segment extracapsular structures that are known to cause needle deflection during percutaneous interventions. This research aims to explore the feasibility of the automatic segmentation of prostate and extracapsular structures to predict needle deflection.
Methods
Using pelvic magnetic resonance imagings (MRIs), 3D U-Net was trained and optimized for the prostate and extracapsular structures (bladder, rectum, pubic bone, pelvic diaphragm muscle, bulbospongiosus muscle, bull of the penis, ischiocavernosus muscle, crus of the penis, transverse perineal muscle, obturator internus muscle, and seminal vesicle). The segmentation accuracy was validated by putting intra-procedural MRIs into the 3D U-Net to segment the prostate and extracapsular structures in the image. Then, the segmented structures were used to predict deflected needle path in in-bore MRI-guided biopsy using a model-based approach.
Results
The 3D U-Net yielded Dice scores to parenchymal organs (0.61–0.83), such as prostate, bladder, rectum, bulb of the penis, crus of the penis, but lower in muscle structures (0.03–0.31), except and obturator internus muscle (0.71). The 3D U-Net showed higher Dice scores for functional structures (p <0.001) and complication-related structures (p <0.001). The segmentation of extracapsular anatomies helped to predict the deflected needle path in MRI-guided prostate interventions of the prostate with the accuracy of 0.9 to 4.9 mm.
Conclusion
Our segmentation method using 3D U-Net provided an accurate anatomical understanding of the prostate and extracapsular structures. In addition, our method was suitable for segmenting functional and complication-related structures. Finally, 3D images of the prostate and extracapsular structures could simulate the needle pathway to predict needle deflections.
Keywords: Prostate, Percutaneous intervention, Deep learning, 3D U-Net, Segmentation
Introduction
Transperineal percutaneous interventions, such as biopsies, brachytherapy, or ablative therapies, against prostate cancer assumes that the treatment devices are placed straight through template guides and in relation to the anatomy of the intended targets [1,2]. A deviation from the intended straight path leads to the inaccurate placement of devices, which causes ineffective diagnoses and treatments. For a prostate biopsy, failure to collect significant tissue that is detected during magnetic resonance imaging (MRI) or transrectal ultrasonography translates into a lower diagnostic yield and delays the appropriate treatment [3]. For brachytherapy involving the placement of radioactive seeds to optimally irradiate the tumor and protect the healthy tissue, deviation from the planned device paths means fewer treatment effects and more toxicity to the healthy tissues [4]. Lastly, in ablative therapies, where the resection probe is placed straight through the guiding template, deviation from the straight path can cause complications in healthy tissue [5]. Therefore, an attempt to predict needle deflection using the pre-procedural or intra-procedural MRI avoids complications due to the needle intervening in extra-prostate organs [6].
A transperineal biopsy is anecdotally known to be subject to needle deflection compared to transrectal biopsies since the needle must pass through skin, subcutaneous fat, and pelvic floor muscles [7]. Moreover, there has been documented evidence that devices deflect more in transperineal procedures than in transrectal approaches [8]. Blumenfeld et al. reported that needle deflection contributed to a targeting error of 3.2 mm to 8.7 mm depending on tissue type, and 0.6 mm to 1.1 mm in with an asymmetrically beveled tip [9]. Moreira et al. identified that needles deflect the most in extracapsular procedures using an MRI, especially the pelvic diaphragm, bulbospongiosus, and bulb of the penis. In fact, clinicians are trained to estimate the needle deflection near the target by mentally extrapolating the needle deflection at extracapsular structures [10]. This unspoken process refutes the engineering researchers’ assumption that needles mostly deflect in the prostate gland. There has been, however, no research conducted to objectively and computationally predict the needle deflection path and suggest to steer to counter the deflection helping physicians to aim lesion in percutaneous prostate intervention.
Therefore, the objective of this paper is to investigate the feasibility of multi-class segmentation of the prostate and extracapsular structure and use the segmentation results to computationally predict deflected needle paths by the model-based approach. A multi-class segmentation method for eight pelvic structures has been proposed for preoperative planning of radiotherapy. However, multi-organ segmentation has not been applied to segment extra-prostatic anatomies nor results of the multi-organ segmentation has never been applied to percutaneous needle intervention of the prostate. In this paper, we will validate the deep-learning-based method to segment extracapsular structures that are known to impact needle defection during percutaneous interventions. Second, we will present highlighted examples of MRI-guided cryotherapy where needle deflection is predicted using the detailed extracapsular segmentation labels produced in the first step.
Material and methods
Ethics statement
All procedures performed in the study were performed following the 1964 Declaration of Helsinki and were approved by the Brigham and Hospital’s institutional review board (IRB) and were anonymized following HIPPA guidelines. (License No.2000P001520) This study was performed retrospectively using consecutive patients’ preprocedural and intraprocedural MRI image data sets. Informed consent was obtained from all individual participants who were included in the study.
Imaging
Intra-procedural T2-weighted MRIs from in-bore MRI-guided interventions were used, and data from 73 patients were collected between November 2016 and May 2017 for training in deep-machine learning as part of automatic segmentation of the prostate and extracapsular structures (ASPE) [11]. In addition, MRIs from 20 subjects who underwent in-bore MRI-guided biopsy between December 2019 and December 2021 were separately collected to validate the ASPE. The biopsies were performed in a 3-T wide-bore MRI (Siemens Verio 3T; Siemens Healthcare, Erlangen, Germany) or a ceiling-mounted 3-T wide-bore MR imager (IMRIS/Siemens Verio; IMRIS, Minnetonka, Minn).
Manual segmentation
An MRI was used to segment the prostate and extracapsular structures. The extracapsular structures were defined as periprostatic structures containing the bladder, rectum, pubic bone, pelvic diaphragm muscle, bulbospongiosus muscle, bull of the penis, ischiocavernosus muscle, crus of the penis, transverse perineal muscle, obturator internus muscle, and seminal vesicle in this study. We prepared the dataset of the MRI segmentation as inputs for supervised learning using a convolutional neural network model.
The axial T2-weighted MRI was selected as it contains the prostate and all extracapsular structures in the pelvis and because most current research literature focuses on automatic segmentation using a T2-weighted MRI [12]. All annotations were performed on a slice-by-slice basis using a segmenting tool (3D Slicer version 4.11; Brigham and Women’s Hospital, Boston, MA, USA) by an experienced urologist for the training of machine learning and validation of the ASPE method [13].
Post-processing
Only subjects with labeled data for each structure in the pelvic region from the MRI image dataset were used for training and validation to evaluate and create a robust estimate of the model’s performance. The data were divided into training, validation, and testing sets using stratified sampling, and there was no patient overlap between the sets. Each image of the prostate and extracapsular structures was prepared in the nii.gz format, and the segmentation data were labeled before being fed into the network.
Multi-organ segmentation for extra-capsular structures
Our segmentation approach uses a 3D U-Net from the Medical Open Network for Artificial Intelligence (MONAI) framework to segment the prostate and extracapsular structures [14]. An overview of ASPE method is provided in Figure 1.
Fig. 1.
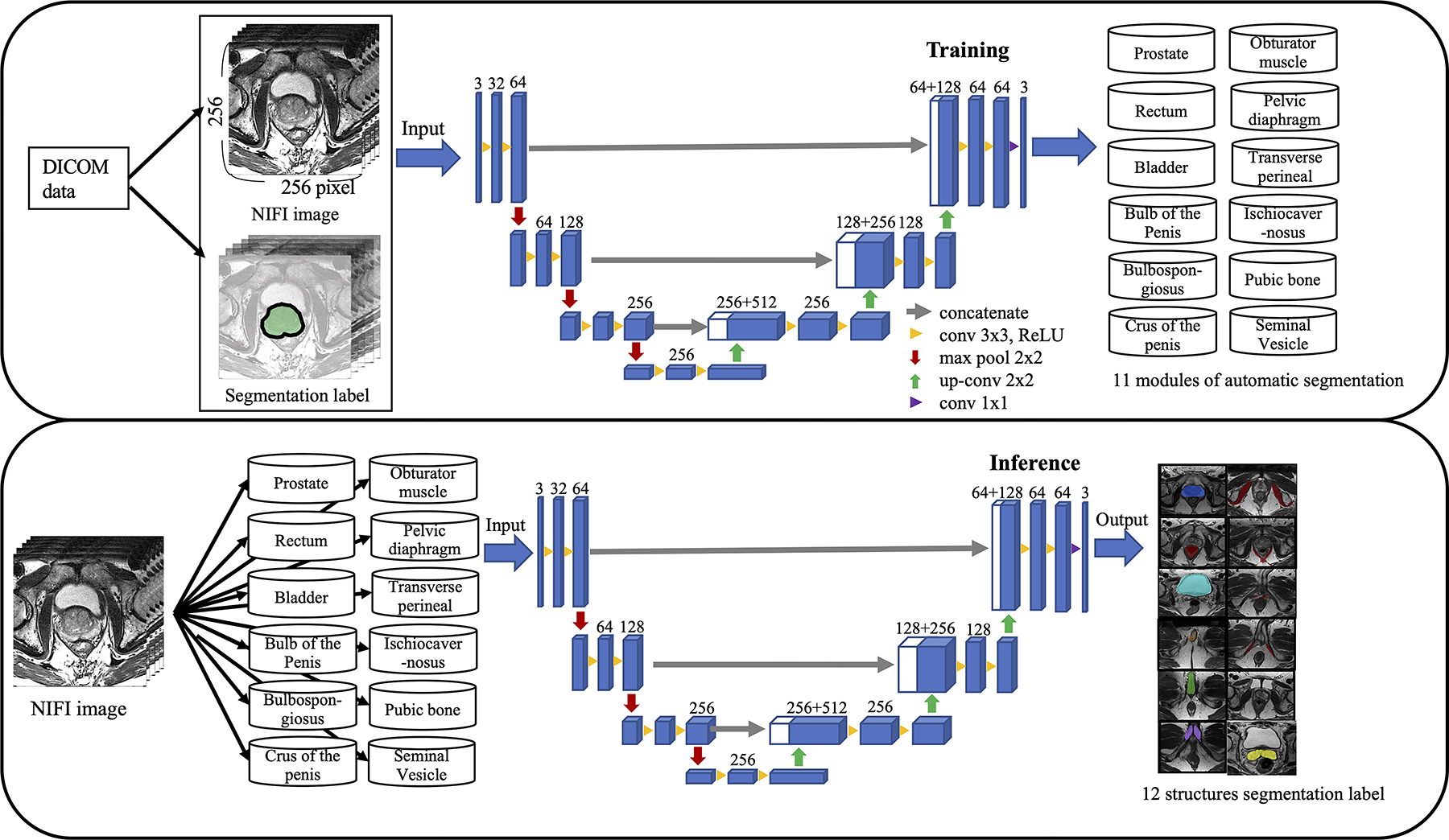
The overall framework of automatic segmentation for the prostate and extracapsular structures. Digital imaging and communications in medicine data were converted to the Neuroimaging Informatics Technology Initiative (NIfTI) images and manually segmented 12 structures. The training module was constructed by inputting the image data and label data of each of the 12 structures into the 3D U-Net as training data. Inference learned the label data by inputting the NIfTI image into the module of 12 structures
The 3D U-Net enabled the seamless segmentation of 3D volumes with high accuracy and performance, and the neural network was composed of a contractive and expanding path that aims to build a bottleneck in its center by combining convolution and pooling operations [15].
The training was conducted on a desktop computer with an NVIDIA GTX 1660 16GB graphics processing unit. The softmax function was used as the activation function in the output layer and was associated with the input image data of the MRIs. The loss function was measured by the cross-entropy error from the label data of manual segmentation and the label data of segmentation using the ASPE method, and the weights were optimized using the adaptive moment optimization (Adam) optimizer, as provided by PyTorch [16].
When fine-tuning the hyperparameters, the training and test results were compared to evaluate the best values of the loss function and the Dice score using 3D U-Net with the Adam optimizer and 500 epochs. The epochs were only added up to 1000 step-by-step to evaluate the best values if the loss function and Dice score were inadequate.
Verification
After the model was created in ASPE, additional 20 cases in which the prostate and extracapsular structures were included in MRI were input into the automatic segmentation module in the prostate and extracapsular structures after training for validation, and the result of the segmentation that was obtained from the label data of each structure is shown in Fig. 2.
Fig. 2.
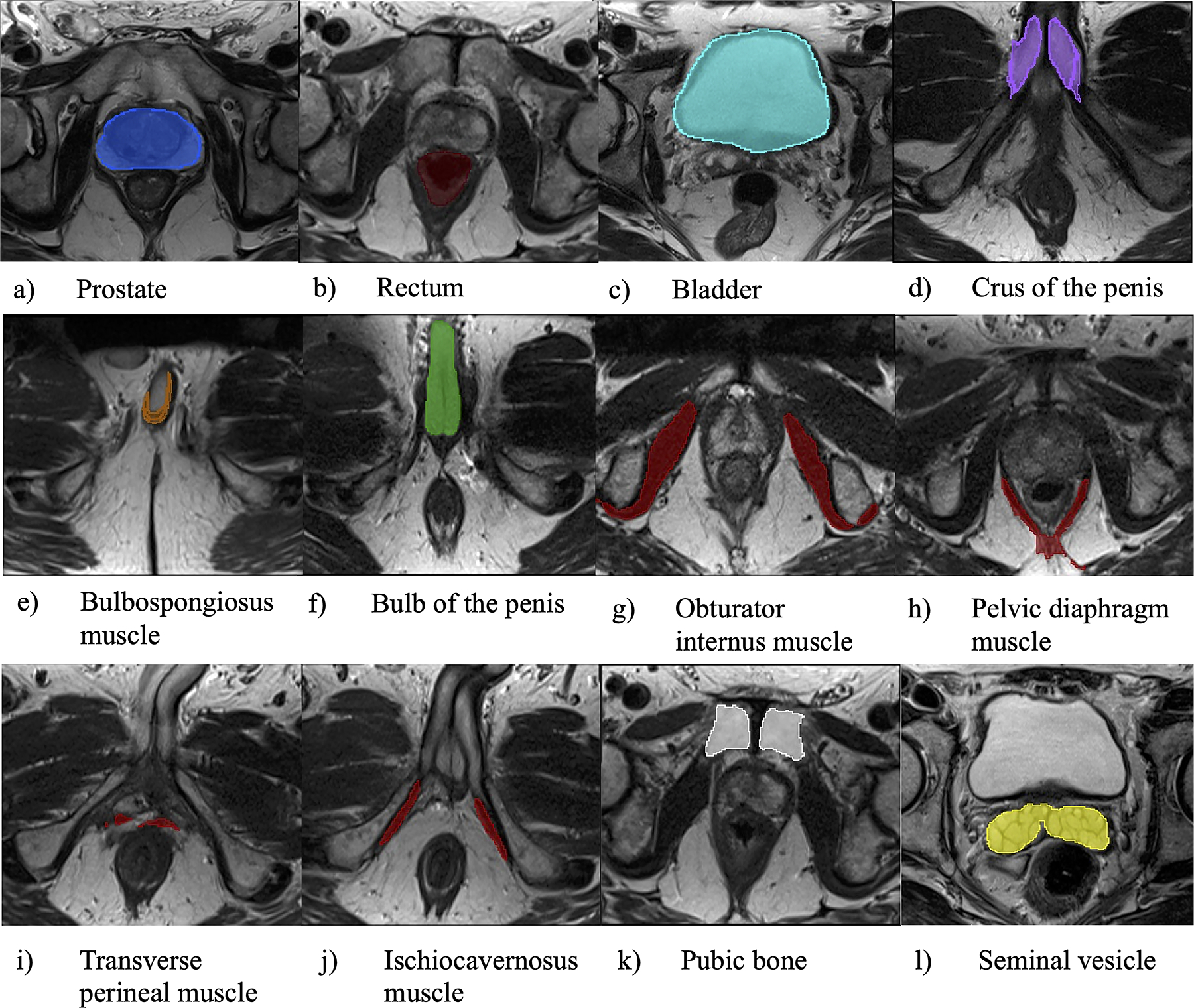
Segmentation results by automatic segmentation of prostate and extracapsular structures (ASPE) method. Label data obtained by the ASPE method for individual prostate and extracapsular structures are presented in figures a–l
Our verification study evaluated the following six metrics to assess the performance of the segmentation: Dice score [17], true positive rate (TP), true negative rate (TN), false positive rate (FP), false negative rate (FN), and 95 % Hausdorff distance (HD) [18]. These metrics were assessed by calculating the voxels between manual and automatic segmentation data.
Statistical analysis
First, the accuracy of segmentation prostate gland using ASPE was compared against Slice Tracker using 40 MRIs [19]. Here, the accuracy was defined as the percentage of correct answers to all segmentation predictions, and the precision was defined as the ratio of manual segmentation to the segmentation results of the proposed method.
Second, an analysis of variance (ANOVA) was performed to investigate if significant differences in the metrics exist between subgroups of the anatomies. Specifically, we divided the prostate and extracapsular structures into two subgroups, namely muscle tissues vs. parenchymal organs, and organs related to complications by percutaneous intervention vs. organs not related to complications were classified to compare the accuracy of automatic segmentation. The parenchymal organs were defined as the prostate, rectum, bladder, bulb of penis, crus of the penis, pubic bone and seminal vesicle. Organs related to complications were defined as the prostate, bladder, and rectum, and organs related to non-complications were defined as something other than the former.
All analyses were performed in combination with JMP Pro 13 software for Mac (version 13.0.0; SAS Institute Inc., Cary, NC, USA). A p value of < 0.05 indicated a statistical significance, and all p values were two-tailed [20].
Simulation to predict the needle deflection
A convolutional neural network using the Keras deep learning library and TensorFlow was implemented to predict the needle deflection from the segmented structures. The needle deflection model was trained using 287 needle deflection trajectories from 18 prostate cryoablation cases and tested in four cryoablation cases as well as the segmentation results produced from the proposed method. The assumption was made that needles are inserted as deep as the MRI-visible target. The distance between the predicted path and the deflected path was measured at the height of the tip for the predicted needle path using horizontal images of MRI, and the error was evaluated.
Result
Dice score, TP, TN, FP, and FN after training
2220 images from 73 patients were distributed for training, validation, and test sets, respectively, for machine learning in Table 1.
Table 1.
Training result of 3D U-Net using automatic segmentation of prostate and extracapsular structures method.
| Data split; training / validation / test, n | Loss coefficient | Dice score | |
|---|---|---|---|
|
| |||
| Prostate | 58 / 10 / 5 | 0.10 | 0.83 |
| Rectum | 58 / 10 / 5 | 0.15 | 0.76 |
| Bladder | 58 / 10 / 5 | 0.22 | 0.81 |
| Bulb of penis | 58 / 10 / 5 | 0.14 | 0.82 |
| Bulbospongiosus muscle | 48 / 5 / 3 | 0.17 | 0.44 |
| Crus of penis | 58 / 10 / 5 | 0.10 | 0.84 |
| Obturator internus muscle | 58 / 10 / 5 | 0.10 | 0.84 |
| Pelvic diaphragm muscle | 58 / 10 / 5 | 0.10 | 0.60 |
| Transverse perineal muscle | 58 / 10 / 5 | 0.2 | 0.15 |
| Ischiocavern muscle | 58 / 10 / 5 | 0.17 | 0.55 |
| Pubis bone | 58 / 10 / 5 | 0.10 | 0.73 |
| Seminal Vesicle | 58 / 10 / 5 | 0.2 | 0.43 |
The verification results showing the performance of segmenting prostatic and extracapsular structures are presented in Table 2 comprehensively, in and Fig. 3 to compare the Dice score of the anatomies.
Table 2.
Performance for automatic segmentation with the verification dataset
| Median (IQR) | Processing time for manual segmentation, sec | Processing time for our segmentation, sec | Dice score | TP rate, % | TN rate, % | FP rate, % | FN rate, % | 95% HD, mm |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Rectum | 184 (153–235) | 1 (1–1) | 0.78 (0.70–0.82) | 69.4 (56.2–75.5) | 99.5 (99.5–99.7) | 0.5 (0.3–1.8) | 30.6 (24.5–43.8) | 5.3 (4.2–10.4) |
| Bladder | 126 (86–194) | 1 (1–1) | 0.83 (0.83–0.90) | 85.4 (68.8–92.6) | 97.6 (92.1–98.7) | 2.4 (1.3–7.9) | 14.6 (7.4–31.2) | 5.8 (3.4–8.9) |
| Bulb of penis | 75 (63–90) | 2 (2–2) | 0.68 (0.56–0.74) | 56.0 (44.4–63.1) | 98.4 (95.0–99.6) | 1.6 (0.4–5.0) | 44.0 (36.9–55.6) | 4.6 (3.26.1) |
| Bulbospongiosus muscle | 62 (41–84) | 2 (2–2) | 0.28 (0.21–0.42) | 27.8 (17.8–49.2) | 99.1 (97.1–99.5) | 0.9 (0.5–0.9) | 72.2 (50.8–82.2) | 8.1 (4.8–12.5) |
| Crus of penis | 132 (104–200) | 2 (2–2) | 0.61 (0.58–0.74) | 53.0 (47.5–76.0) | 98.0 (95.4–99.3) | 2.0 (0.7–4.6) | 47.0 (24.0–52.5) | 4.8 (3.4–5.3) |
| Obturator internus muscle | 551 (392–645) | 1 (1–1) | 0.71 (0.64–0.76) | 61.2 (54.9–70.3) | 99.2 (99.0–99.4) | 0.8 (0.6–1.0) | 38.8 (29.7–45.1) | 7.9 (4.7–13.4) |
| Pelvic diaphragm muscle | 302 (248–413) | 2 (2–2) | 0.34 (0.30–0.42) | 28.6 (21.8–38.2) | 99.4 (98.7–99.8) | 0.6 (0.2–1.3) | 71.4 (61.8–78.2) | 8.9 (5.9–11.8) |
| Transverse perineal muscle | 56 (36–72) | 2 (2–2) | 0.03 (0.00–0.08) | 1.8 (0.0–5.9) | 99.8 (99.4–100.0) | 0.2 (0.0–0.6) | 98.2 (94.1–100.0) | 11.9 (8.3–17.6) |
| Ischiocavern muscle | 62 (53–82) | 2 (2–2) | 0.31 (0.16–0.46) | 25.2 (12.7–44.3) | 99.2 (98.0–99.7) | 0.8 (0.3–2.0) | 74.8 (55.7–87.3) | 6.7 (4.7–12.5) |
| Pubic bone | 331 (281–361) | 1 (1–1) | 0.65 (0.61–0.72) | 53.0 (46.0–61.3) | 99.6 (99.5–99.8) | 0.4 (0.2–0.5) | 47.0 (38.7–54.0) | 5.5 (4.6–7.5) |
| Seminal vesicle | 99 (93.5–117) | 2 (2–2) | 0.46 (0.32–0.46) | 45.3 (23.3–61.6) | 80.0 (63.0–84.0) | 28.0 (16.0–37.0) | 54.7 (38.4–76.6) | 7.4 (3.5–7.4) |
IQR; interquartile range, TP; true positive, TN; true negative, FP; false positive, FN; false negative, HD; Hausdorff distance, MSD; mean surface distance,
Fig. 3.
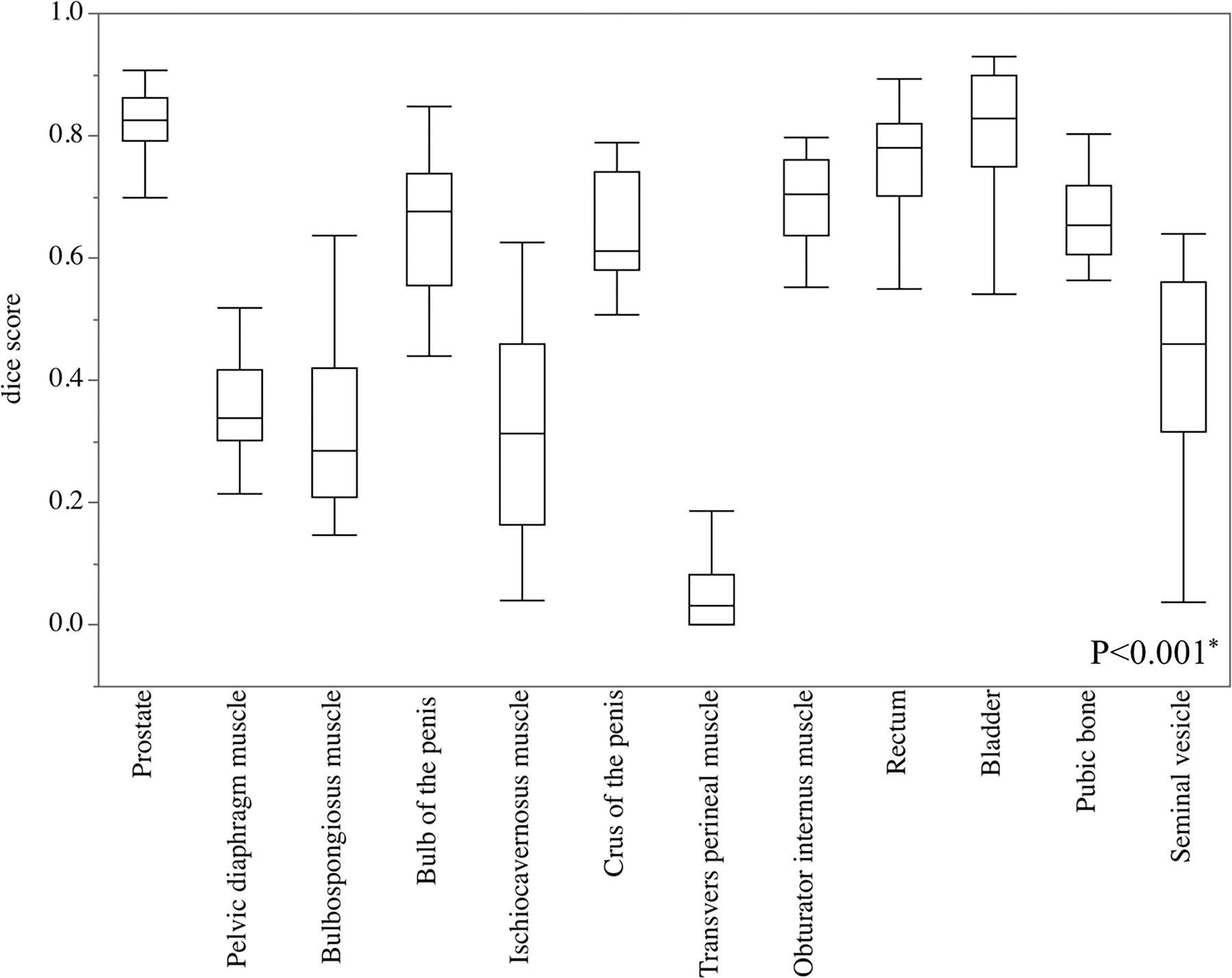
Analysis of variance in the Dice score of prostate and extracapsular structures. The Dice scores in the prostate, bulb of the penis, crus of the penis, obturator internus muscle, rectum, bladder, and pubic bone were significantly higher than in other extracapsular structures
A representative result of the segmentation in the verification study is shown in Fig. 4. Similarly, the 3D reconstruction of the segmented data using the anatomies with Dice score of 0.8 or higher (prostate, rectum, bladder, pubic bone, obturator internus muscle, bulb of the penis, and crus of the penis) is presented in Fig. 5.
Fig. 4.
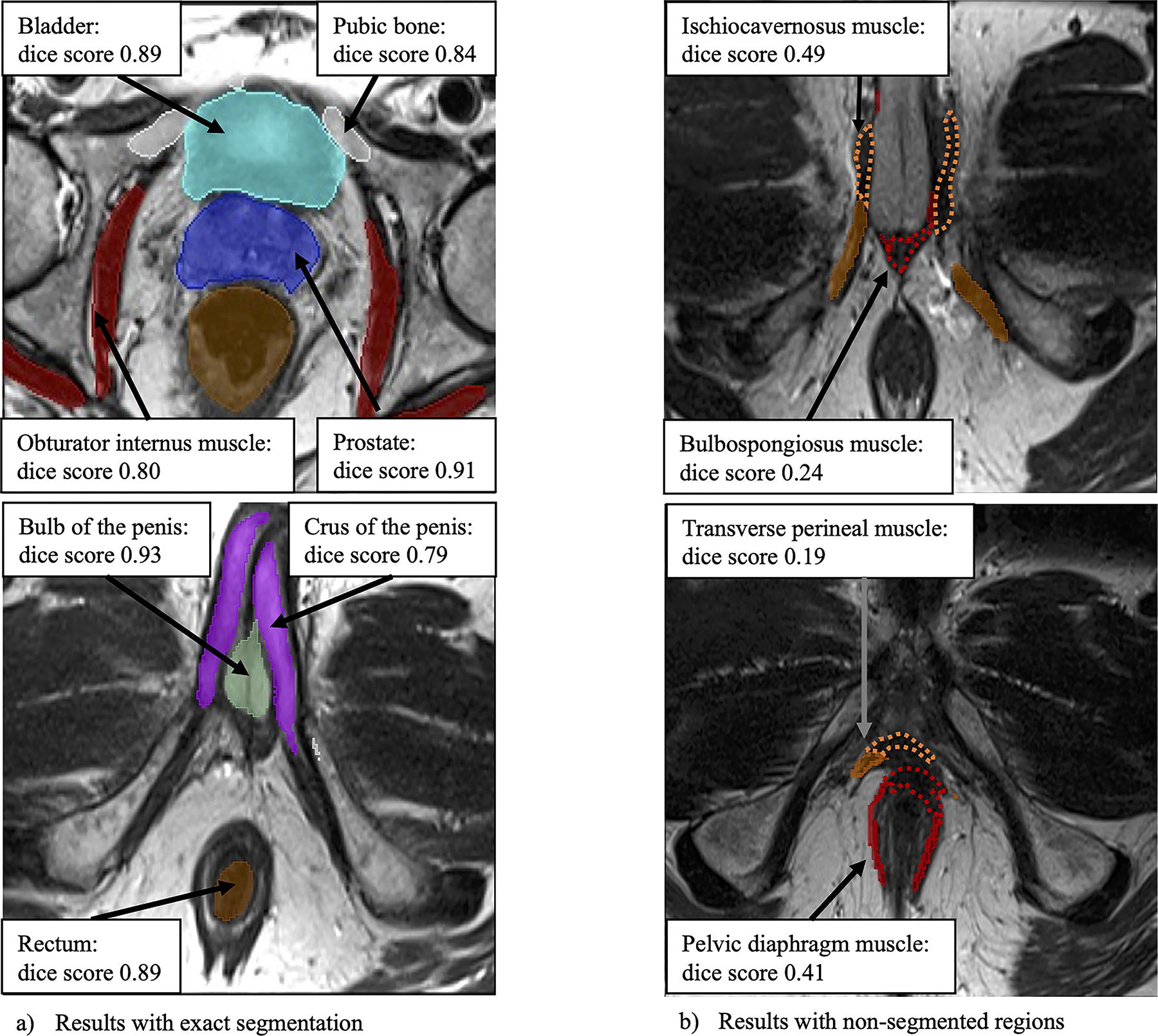
Clinical case using automatic segmentation of the prostate and extracapsular structures method (case number 16, 61-year-old man, prostate specific antigen values 4.8 ng/dL). Figure in the top and bottom left, marked as a contained correct segmentation of the prostate, bladder, pubic bone, obturator internus muscle, rectum, bulb of the penis, and crus of the penis. Figure in the top and bottom right marked as b contained unsegmented areas of ischiocavernosus muscle, bulbospongiosus muscle, transverse perineal muscle, and pelvic diaphragm muscle (wavy line)
Fig. 5.
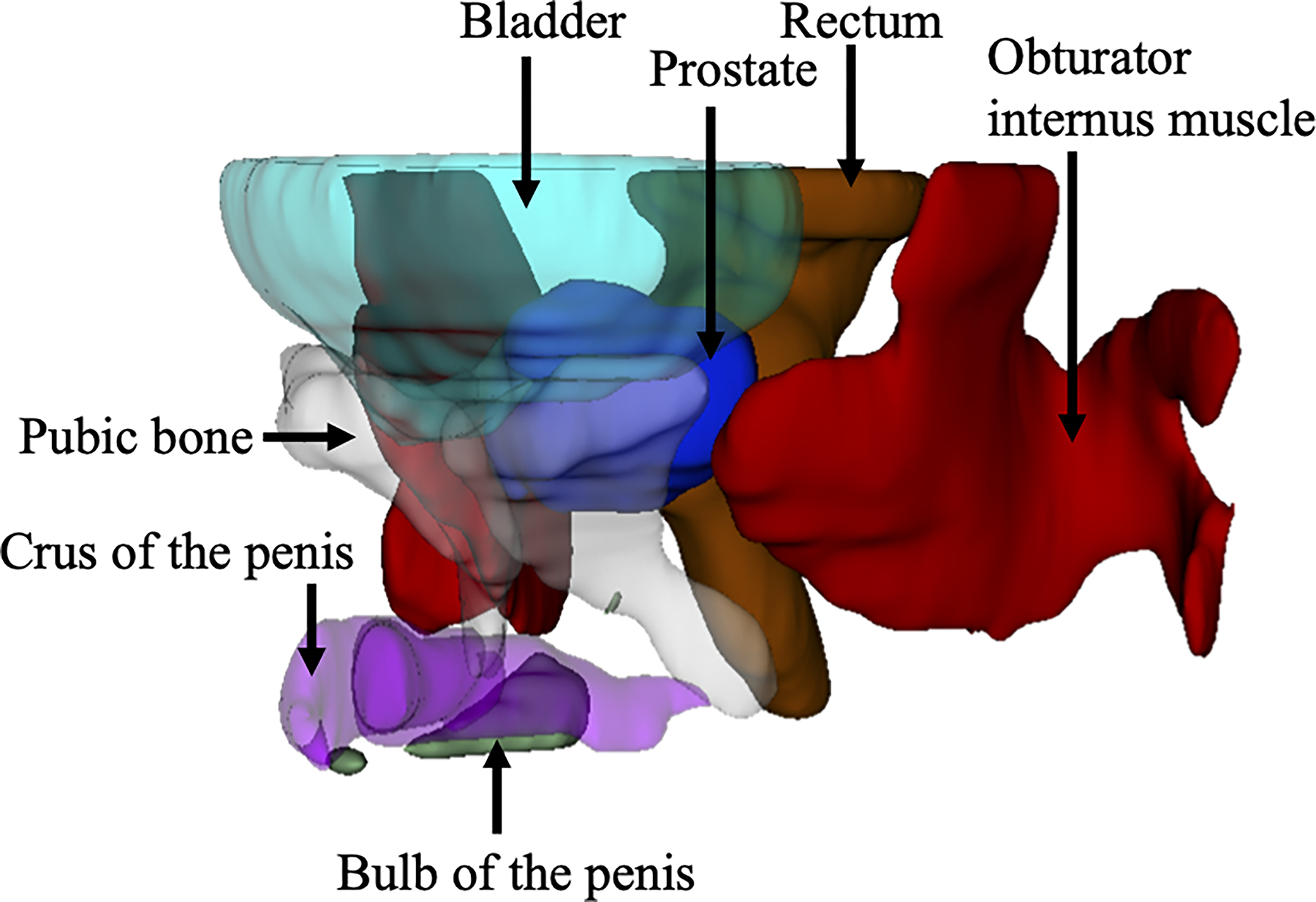
A 3D image with 0.80 or more Dice score (Case number 20, 79-year-old man, prostate specific antigen values 5.4 ng/dL). Label data obtained by automatic segmentation of the prostate and extracapsular structures were reconstructed three-dimensionally
Statistical analysis
The ASPE method significantly reduced the processing time for segmentation (Slicer Tracker vs. ASPE; 84 sec vs. 5 sec, p < 0.001), the TP (71.1 % vs. 78.9 %, p =0.002) was significantly higher, and the FN (27.3 % vs. 21.1 %, p = 0.001) and 95 % HD (5.1 mm vs. 4.9 mm, p = 0.023) was lower than the Slicer Tracker. Hence, the accuracy (0.81 vs. 0.86, p = 0.001), the precision (0.89 vs. 0.94, p = 0.029) and the Dice score (0.79 vs. 0.82, p < 0.001) were improved significantly in the ASPE method group (Table 3).
Table 3.
Comparison the accuracy of the prostate between Slicer Tracker and MONAI segmentation.
| Slice Tracker [19] (n=40) Median (IQR) | ASPE (n=40) Median (IQR) | p | |
|---|---|---|---|
|
| |||
| Processing time, sec | 840 (79–90) | 5 (5–5) | <0.001* |
| Dice score | 0.79 (0.44–0.86) | 0.82 (0.77–0.86) | <0.001* |
| TP rate, % | 71.1 (29.2–87.1) | 78.9 (71.5–83.9) | 0.002* |
| TN rate, % | 90.1 (85.5–98.5) | 94.2 (90.3–97.8) | 0.064 |
| FP rate, % | 7.5 (0.73–14.4) | 5.8 (90.3–97.8) | 0.11 |
| FN rate, % | 27.3 (11.8–69.3) | 21.1 (16.1–28.5) | 0.001* |
| Accuracy | 0.81 (0.63–0.87) | 0.86 (0.82–0.89) | <0.001* |
| Precision | 0.89 (0.85–0.95) | 0.94 (0.89–0.97) | 0.29 |
| 95% HD, mm | 5.1 (3.8–13) | 4.9 (3.8–6.0) | 0.023* |
IQR; interquartile range, ASPE; automatic segmentation of prostate and extraprostatic structure, TP; true positive, TN; true negative, FP; false positive, FN; false negative, HD; Hausdorff distance,
The Dice score of parenchymal organs was higher than muscle tissues in Table 4. When analyzing the organs related to procedural complications as in Table 5, the Dice score was higher in organs related to complications than ones not related to complications.
Table 4.
Comparison the accuracy between muscle tissue and parenchymal organ.
| Median (IQR) | Muscle tissue n=80 | Parenchymal organ n=156 | p |
|---|---|---|---|
|
| |||
| Dice score | 0.32 (0.11–0.56) | 0.68 (0.53–0.79) | <0.001* |
| TP rate, % | 22.1 (6.4–55.1) | 58.8 (45.4–77.3) | <0.001* |
| TN rate, % | 99.4 (98.9) | 98.3 (94.9–99.6) | <0.001* |
| FP rate, % | 0.6 (0.2–1.1) | 1.7 (0.5–5.1) | <0.001* |
| FN rate, % | 72.9 (44.9–93.6) | 41.2 (22.7–54.6) | <0.001* |
| 95% HD, mm | 9.0 (5.8–13.4) | 5.2 (4.0–8.0) | <0.001* |
IQR; interquartile range, TP; true positive, TN; true negative, FP; false positive, FN; false negative, HD; Hausdorff distance
Table 5.
Comparison the accuracy between organs related to non-complications and complication.
| Median (IQR) | Non-complication n=156 | Complications n=80 | p |
|---|---|---|---|
|
| |||
| Dice score | 0.45 (0.24–0.63) | 0.78 (0.68–0.84) | <0.001* |
| TP rate, % | 40.3 (18.7–58.8) | 73.8 (53.9–83.3) | <0.001* |
| TN rate, % | 99.1 (97.4–99.6) | 98.9 (96.7–99.6) | 0.11 |
| FP rate, % | 0.9 (0.4–2.6) | 1.1 (0.4–3.3) | 0.11 |
| FN rate, % | 59.7 (41.2–81.3) | 26.2 (16.7–46.1) | <0.001* |
| 95% HD, mm | 7.2 (4.6–11.0) | 4.9 (4.0–7.8) | 0.18 |
IQR; interquartile range, TP; true positive, TN; true negative, FP; false positive, FN; false negative, HD; Hausdorff distance
Simulation to predict the needle deflection
The model was retrospectively tested in four prostate cryoablation cases. In four cases, the model was observed to generate predicted needle deflection paths that was typically toward the direction of deflection observed from the inserted needle in the cases (Fig. 6). The needle in the predicted path and the deflected path penetrated the pelvic diaphragm muscle or the bulb of the penis, the error between the predicted path and the deflected path was 0.9 mm to 4.9 mm at the height of the tip for the predicted path (Table 6).
Fig. 6.
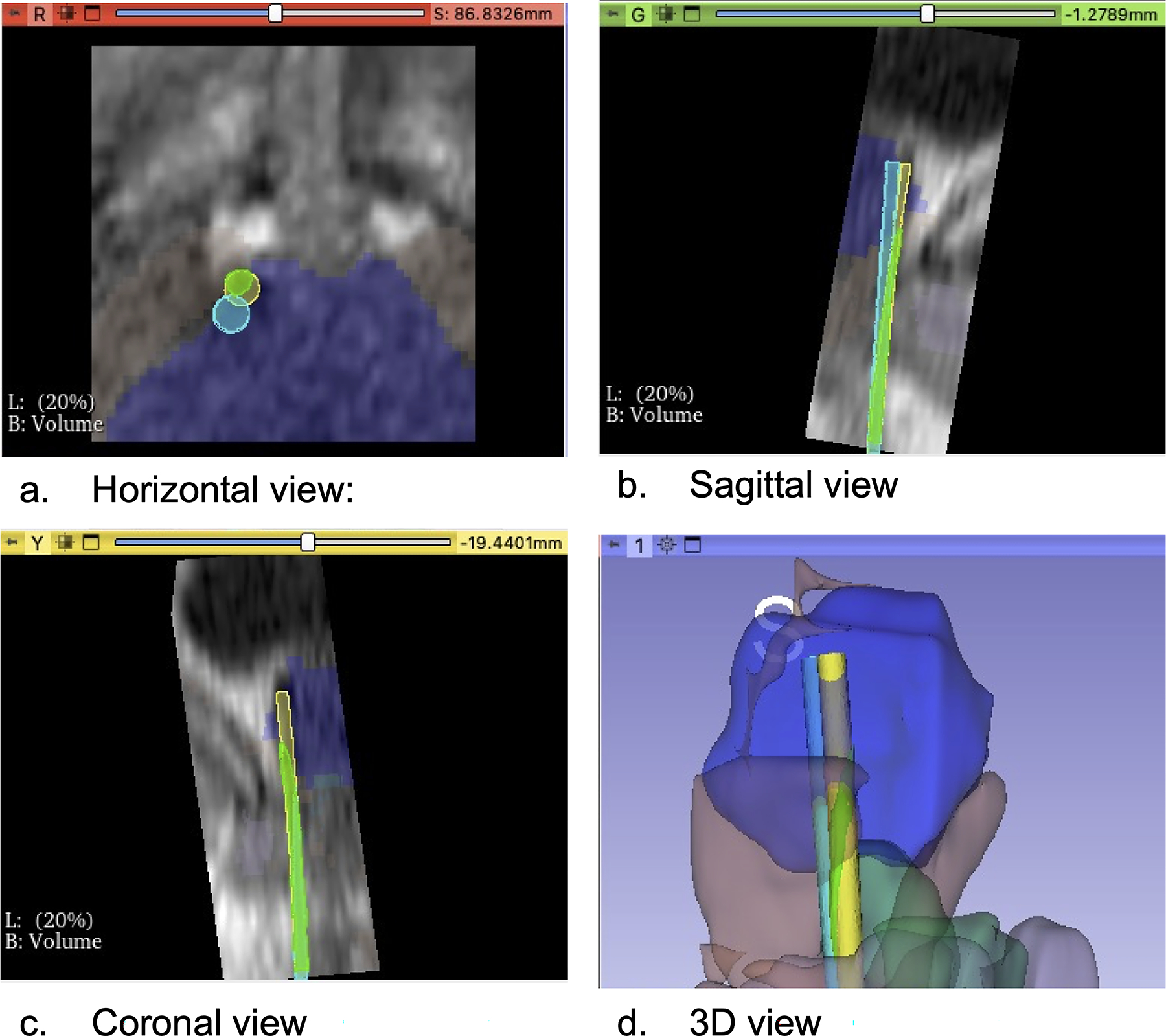
A segmentation label was automatically created using the MRI of case no.4, and needle deflection was predicted. a Horizontal view: The light blue circle represented the preoperatively planned path, the light green circle represented the needle path including prediction of needle deflection, and the yellow circle represented the deflected path through which the actual needle passed. b Sagittal view: The deflected path deviated from the planned path. c Coronal view: The predicted path overlapped with the deflected path, and we were able to predict the needle deflection. d 3D view: We visualized three paths in a 3D image: the blue structure was the prostate, the brown structure was the pelvic diaphragm muscle, the green structure was the bulb of the penis
Table 6.
Predict the needle deflection.
| Case No. | Type of penetrating tissue | Distance between the predicted path and the deflected path, [mm] |
|---|---|---|
| 1 | Pelvic diaphragm muscle | 1.7 |
| 2 | Bulb of the penis | 4.9 |
| 3 | Pelvic diaphragm muscle | 3.8 |
| 4 | Pelvic diaphragm muscle | 0.9 |
Discussion
This study is the first to report that the proposed automatic segmentation accurately identified and segmented the prostate and extracapsular structures from an MRI. In addition, the ASPE method we presented proved to be suitable for the automatic segmentation of the prostate, parenchymal organs and complication-related structures. Finally, we presented highlighted images where the needle deflection was predicted using automatic segmented labels of extracapsular structures.
A transperineal prostate needle biopsy is the standard test for patients with elevated prostate specific antigen levels among percutaneous interventions. As randomized trials have confirmed the superiority of MRI-guided prostate biopsies to detect significant cancers, MRIs could be used as a triage test to lead patients with significant prostate cancer to the proper treatment strategy [21,22]. Likewise, excellent oncological outcomes of brachytherapy and cryotherapy have been reported for significant prostate cancer, and it is one of the standard treatments for low- and intermediate-risk prostate cancers [23,24]. Hence, the accurate insertion of the needle into the targeted lesion of the prostate gland is essential for diagnosing and treating significant prostate cancer, and a 3D understanding of anatomy using pre-procedural MRI while predicting needle deflection leads to accurate needle placement in the target lesion [25]. Moreover, the 3D construction of pelvic structures in every detail may accelerate the spatial recognition of the physicians’ prostate and its surrounding structures, minimizing complications and improving the patient’s quality of life after the procedures [26]. Complications that are associated with prostate biopsy, brachytherapy, and ablation therapy have been reported as 3.1%, 5%, and 19%, respectively, and the invasion of the tip of the needle into extracapsular structures by needle deflection causes serious complications, such as a rectal injury, severe infection, bladder injury, bleeding, and associated urinary retention [27–29]. Despite the benefits of 3D images of the prostate and extracapsular structures, preprocedural has taken a great deal of time to segment these structures in MRI. Furthermore, few attempts exist to automate multi-segmentation for prostate and extracapsular structures in percutaneous interventions. Moreover, clinical challenges using multi-segmentation have not been researched comprehensively to predict needle deflection.
Regarding the multi-class segmentation method, Kuisma et al. reported a multi-organ segmentation method using an MRI and reported Dice scores of the prostate (0.84), bladder (0.92), and rectum (0.86) [30]. Furthermore, Nuo et al. reported that a multi-task edge-recalibrated network attains superior performance of the quantitative Dice score for seminal vesicle (76.4 %) [31]. This paper, however, handles more detailed extra-prostatic tissue than these prior-art.
This study relates to prior arts that argue the clinical impact of needle deflection caused by anatomical structures within the pelvis in percutaneous intervention. The prior prediction study for needle deflection focused on biomechanical modeling within the prostate and physically modeling the needle-tissue interactions within the prostatic tissues. Du et al. reported that needle-tissue interaction was modeled by a series of nonlinear springs and the needle deflection was predicted with the Rayleigh-Ritz method [32]. Patil et al. reported that by developing a new needle steering system that could automatically reach the target in a 3D environment, the needle could be delivered to the target with an average error of 3 mm or less [33]. Stone et al. reported a straight sampling method in the majority of the prostate gland by combining a 3-point trocar tip on the needle with a 20 vet tip on the cutting cannula [34]. The current study is the first attempt to use extra-prostatic segmentation as a material for a data-driven approach, not a biomechanical approach to study needle-tissue interaction, to simulate needle deflection. We anticipate the data-driven approach is potentially more clinically feasible and realistic than the biomechanical modeling approach or approaches focusing on intra-prostatic tissue and needle-tissue interactions [35,36]. Our decision to focus on a deflection in extra-prostatic tissues grew out of our real clinical experience with MRI-guided biopsies where the retrospective analysis indicated that it is not intra-prostateic tissue, but extra-prostatic tissue that has the most impact on needle deflection [10].
This study has several limitations. First, it was a non-randomized observational study with a retrospective design. Therefore, there was a risk of inherent selection bias in the training and validation datasets. Second, our study was conducted using a single MRI machine at a single institution. Therefore, it has not been verified using images taken by other types of MRI equipment. In this study, the needle tip was displaced differently from the expected position in the prostate puncture by needle deflection. In actual, the needle penetrated the pelvic diaphragm muscle or bulb of the penis, and needle deflection occurred. The striated muscles related to urinary function and sexual function, such as transverse perineal muscle, bulbospongiosus muscle, pelvic diaphragm muscle, and ischiocavernosus muscle, have individual differences in mass and development, and there were several variations. The abundant variations in muscle tissue may be the factors that reduce the accuracy of segmentation using machine learning. Nevertheless, the pelvic muscles are tissues that constitute the urethra in front of the prostate and are likely to be penetrated by a needle that has inserted transperineally. Since these muscles can be factors that cause needle deflection, improving the accuracy of segmentation of these muscle tissues using the ASPE method may help identify the tissue that causes needle deflection. Furthermore, the label data of muscle tissue with improved Dice scores may contribute to the elucidation of tissue-needle interactions in percutaneous prostate needle interventions and to predict the needle deflection. Therefore, our results should be interpreted with caution.
Despite these limitations, the ASPE method automatically reconstructed the prostate, rectum, bladder, bulb of the penis, crus of the penis, obturator internus muscle, and pubic bone, which were important for the needle to enter the target lesion. Our approach was suitable for segmenting the prostate, parenchymal structures, and complication-related structures and could help predict interference with these important structures by the needle.
Conclusions
In conclusion, the ASPE method builds machine-learned modules from MRIs, and these modules automatically segment the prostate and extracapsular structures. The validation results showed that the segmentation method was suitable for segmenting functional structures and structures involved in complications.
Funding
The research reported in this study was supported by NIH/NIBIB of the National Institutes of Health under award numbers P41EB028741, R01EB030539, R01CA235134, and R01CA232174.
Footnotes
Declarations
Informed consent Informed consent was obtained from all individual participants who were included in the study. The participants consented to the submission of the case report to the journal.
References
- 1.van der Poel HG, van den Bergh RC, Briers E, Cornford P, Govorov A, Henry AM, Lam TB, Mason MD, Rouviere O, De Santis M, Willemse PPM, van Popple H, Mottet N (2018) Focal therapy in primary localised prostate cancer: the European Association of Urology position in 2018. Eur Urol 74(1):84–91. 10.1016/j.eururo.2018.01.001 [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Xu S, Squires A, Seifabadi R, Turkbey IB, Pinto PA, Choyke P, Wood B, Tse ZTH (2017) MRI-guided robotically assisted focal laser ablation of the prostate using canine cadavers. IEEE Trans Biomed Eng 65(7):1434–1442. 10.1109/TBME.2017.2756907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin PR, Cool DW, Fenster A, Ward AD (2018) A comparison of prostate tumor targeting strategies using magnetic resonance imaging-targeted, transrectal ultrasound-guided fusion biopsy. Med Phys 45(3):1018–1028. 10.1002/mp.12769 [DOI] [PubMed] [Google Scholar]
- 4.Lehmann T, Rossa C, Usmani N, Sloboda RS, Tavakoli M (2017) Intraoperative tissue young’s modulus identification during needle insertion using a laterally actuated needle. IEEE Trans Instrum Meas 67(2):371–381. 10.1109/TIM.2017.2774182 [DOI] [Google Scholar]
- 5.Bass R, Fleshner N, Finelli A, Barkin J, Zhang L, Klotz L (2019) Oncologic and functional outcomes of partial gland ablation with high intensity focused ultrasound for localized prostate cancer. J Urol 201(1):113–119. 10.1016/j.juro.2018.07.040 [DOI] [PubMed] [Google Scholar]
- 6.Jarosek SL, Virnig BA, Chu H, Elliott SP (2015) Propensity-weighted long-term risk of urinary adverse events after prostate cancer surgery, radiation, or both. Eur Urol 67(2):273–280. 10.1016/j.eururo.2014.08.061 [DOI] [PubMed] [Google Scholar]
- 7.Li AD, Plott J, Chen L, Montgomery JS, Shih A (2020) Needle deflection and tissue sampling length in needle biopsy. J Mech Behav Biomed Mater 104:103632. 10.1016/j.jmbbm.2020.103632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peter A, Mohantha D (2013) Prostate biopsy: will transperineal replace transrectal? BJU Int 112:533–534. 10.1111/bju.12299 [DOI] [PubMed] [Google Scholar]
- 9.Blumenfeld P, Hata N, DiMaio S, Zou K, Haker S, Fichtinger G, Tempany CM (2007) Transperineal prostate biopsy under magnetic resonance image guidance: a needle placement accuracy study. J Magn Reson Imaging Off J Int Soc Magn Reson Med 26(3):688–694. 10.1002/jmri.21067 [DOI] [PubMed] [Google Scholar]
- 10.Moreira P, Patel N, Wartenberg M, Li G, Tuncali K, Heffter T, Burdette EC, Iordachita I, Fischer GS, Hata N (2018) Evaluation of robot-assisted MRI-guided prostate biopsy: needle path analysis during clinical trials. Phys Med Biol 63(20):20NT02. 10.1088/1361-6560/aae214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penzkofer T, Tuncali K, Fedorov A, Song SE, Tokuda J, Fennessy FM, Vangel MG, Kibel AS, Mulkern RV, Wells WM, Hata N, Tempany CMC (2015) Transperineal in-bore 3-T MR imaging-guided prostate biopsy: a prospective clinical observational study. Radiology 274(1):170–180. 10.1148/radiol.14140221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Lei Y, Tian S, Wang T, Liu Y, Patel P, Jani AB, Mao H, Curran WJ, Liu T, Yang X (2019) Deeply supervised 3D fully convolutional networks with group dilated convolution for automatic MRI prostate segmentation. Med Phys 46(4):1707–1718. 10.1002/mp.13416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Alyward S, Miller JV, Pieper S, Kikinis R (2012) 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30(9):1323–1341. 10.1016/j.mri.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaliyugarasan SK, Lundervold A, Lundervold AS (2021) Pulmonary nodule classification in lung cancer from 3D thoracic CT scans using fastai and MONAI. Int J Interact Multimed Artif Intell 6(7):83–89. 10.9781/ijimai.2021.05.002 [DOI] [Google Scholar]
- 15.Blanc-Durand P, Van Der Gucht A, Schaefer N, Itti E, Prior JO (2018) Automatic lesion detection and segmentation of 18F-FET PET in gliomas: a full 3D U-Net convolutional neural network study. PLoS ONE 13(4):e0195798. 10.1371/journal.pone.0195798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khaire UM, Dhanalakshmi R (2020) High-dimensional microarray dataset classification using an improved adam optimizer (iAdam). J Ambient Intell Humaniz Comput 11(11):5187–5204. 10.1007/s12652-020-01832-3 [DOI] [Google Scholar]
- 17.Gibson E, Giganti F, Hu Y, Bonmati E, Bandula S, Gurusamy K, Davidson B, Pereira SP, Clarkson MJ, Barratt DC (2018) Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Trans Med Imaging 37(8):1822–1834. 10.1109/TMI.2018.2806309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali MH, Al Mohammed BAD, Ismail A, Zolkipli MF (2018) A new intrusion detection system based on fast learning network and particle swarm optimization. IEEE Access 6:20255–20261. 10.1109/ACCESS.2018.2820092 [DOI] [Google Scholar]
- 19.Mehrtash A, Ghafoorian M, Pernelle G, Ziaei A, Heslinga FG, Tuncali K, Fedorov A, Kikinis R, Tempany CM, Wells WM, Abolmaesumi P, Kapur T (2018) Automatic needle segmentation and localization in MRI with 3-D convolutional neural networks: application to MRI-targeted prostate biopsy. IEEE Trans Med Imaging 38(4):1026–1036. 10.1109/TMI.2018.2876796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assel M, Sjoberg D, Elders A, Wang X, Huo D, Botchway A, Delfino K, Fan Y, Zhao Z, Koyama T, Hollenbeck B, Qin R, Zahnd W, Zabor EC, Kattan MW, Vickers AJ (2019) Guidelines for reporting of statistics for clinical research in urology. J Urol 201(3):595–604. 10.1097/JU.0000000000000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, Vaarala MH, Briganti A, Budäus L, Hellawell G, Hindley RG, Roobol MJ, Eggener S, Ghei M, Villers A, Bladou F, Villeirs GM, Virdi J, Boxler S, Robert G, Singh PB, Venderink W, Hadaschik BA, Ruffion A, Hu JC, Margolis D, Crouzet S, Klotz L, Taneja SS, Pinto P, Gill I et al. (2018) MRI-targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med 378(19):1767–1777. 10.1056/NEJMoa1801993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Leest M, Cornel E, Israël B, Hendriks R, Padhani AR, Hoogenboom M, Zamecnik P, Bakker D, Setiasti AY, Veltman J, van den Hout HV, van der Lelij H, van Oort I, Klaver S, Debruyne F, Sedelaar M, Hannink G, Rovers M, Hulsbergen-van de Kaa C, Barentsz OJ (2019) Head-to-head comparison of transrectal ultrasound-guided prostate biopsy versus multiparametric prostate resonance imaging with subsequent magnetic resonance-guided biopsy in biopsy-naive men with elevated prostate-specific antigen: a large prospective multicenter clinical study. Eur Urol 75(4):570–578. 10.1016/j.eururo.2018.11.023 [DOI] [PubMed] [Google Scholar]
- 23.Chin J, Rumble RB, Kollmeier M, Heath E, Efstathiou J, Dorff T, Berman B, Feifer A, Jacques A, Loblaw DA (2017) Brachytherapy for patients with prostate cancer: American Society of Clinical Oncology/Cancer Care Ontario joint guideline update. J Clin Oncol 35(15):1737–1743. 10.1200/JOP.2016.020610 [DOI] [PubMed] [Google Scholar]
- 24.Shah TT, Peters M, Eldred-Evans D, Miah S, Yap T, Faure-Walker NA, Hosking-Jervis F, Thomas B, Dudderidge T, Hindley RG, McCracken S, Greene D, Nigam R, Valerio M, Minhas S, Winkler M, Arya M, Ahmed HU (2019) Early-medium-term outcomes of primary focal cryotherapy to treat nonmetastatic clinically significant prostate cancer from a prospective multicentre registry. Eur Urol 76(1):98–105. 10.1016/j.eururo.2018.12.030 [DOI] [PubMed] [Google Scholar]
- 25.Huang MD, Weng HH, Hsu SL, Hsu LS, Lin WM, Chen CW, Tsai YH (2019) Accuracy and complications of CT-guided pulmonary core biopsy in small nodules: a single-center experience. Cancer Imaging 19(1):1–10. 10.1186/s40644-019-0240-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernhard JC, Isotani S, Matsugasumi T, Duddalwar V, Hung AJ, Suer E, Baco E, Satkunasivam R, Djaladat H, Metcalfe C et al. (2016) Personalized 3D printed model of kidney and tumor anatomy: a useful tool for patient education. World J Urol 34(3):337–345. 10.1007/s00345-015-1632-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagenlehner FM, van Oostrum E, Tenke P, Tandogdu Z, Cek M, Grabe M, Wullt B, Pickard R, Naber KG, Pilatz A, Weidner W, Bjerklund-Johansen TE, Investigators G (2013) Infective complications after prostate biopsy: outcome of the Global Prevalence Study of Infections in Urology (GPIU) 2010 and 2011, a prospective multinational multicentre prostate biopsy study. Eur Urol 63(3):521–527. 10.1016/j.eururo.2012.06.003 [DOI] [PubMed] [Google Scholar]
- 28.Chargari C, Deutsch E, Blanchard P, Gouy S, Martelli H, Guérin F, Dumas I, Bossi A, Morice P, Viswanathan AN, Haie-Meder C (2019) Brachytherapy: an overview for clinicians. CA A Cancer J Clin 69(5):386–401. 10.3322/caac.21578 [DOI] [PubMed] [Google Scholar]
- 29.Durand M, Barret E, Galiano M, Rozet F, Sanchez-Salas R, Ahallal Y, Macek P, Gaya JM, Cerruti J, Devilliers H, Loeffler J, Amiel J, Vallancien G, Cathelineau X (2014) Focal cryoablation: a treatment option for unilateral low-risk prostate cancer. BJU Int 113(1):56–64. 10.1111/bju.12370 [DOI] [PubMed] [Google Scholar]
- 30.Kuisma A, Ranta I, Keyriläinen J, Suilamo S, Wright P, Pesola M, Warner L, Löyttyniemi E, Minn H (2020) Validation of automated magnetic resonance image segmentation for radiation therapy planning in prostate cancer. Phys Imaging Radiat Oncol 13:14–20. 10.1016/j.phro.2020.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong N, Gou S, Chen S, Yao Y, Yang S, Cao M, Kishan A, Sheng K (2021) Multi-task edge-recalibrated network for male pelvic multi-organ segmentation on CT images. Phys Med Biol 66(3):035001. [DOI] [PubMed] [Google Scholar]
- 32.Du H, Zhang Y, Jiang J, Zhao Y (2015) Needle deflection during insertion into soft tissue based on virtual spring model. Int J Multimed Ubiquitous Eng 10(1):209–218. 10.14257/IJMUE.2015.10.1.20 [DOI] [Google Scholar]
- 33.Patil S, Burgner J, Webster RJ, Alterovitz R (2014) Needle steering in 3-D via rapid replanning. IEEE Trans Rob 30(4):853–864. 10.1109/TRO.2014.2307633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stone NN, Mouraviev V, Schechter D, Goetz J, Lucia MS, Smith EE, Crawford ED (2017) Deflection analysis of different needle designs for prostate biopsy and focal therapy. Technol Cancer Res Treat 16(5):654–661. 10.1177/1533034616671007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carriere J, Khadem M, Rossa C, Usmani N, Sloboda R, Tavakoli M (2017) Surgeon-in-the-loop 3-d needle steering through ultrasound-guided feedback control. IEEE Robot Autom Lett 3(1):469–476. 10.1109/LRA.2017.2768122 [DOI] [Google Scholar]
- 36.Seifabadi R, Aalamifar F, Iordachita I, Fichtinger G (2016) Toward teleoperated needle steering under continuous MRI guidance for prostate percutaneous interventions. Int J Med Robot Comput Assist Surg 12(3):355–369. 10.1002/rcs.1692 [DOI] [PubMed] [Google Scholar]


