Abstract
Colorectal cancer (CRC) is among the most common cancers globally and a major cause of cancer-related deaths. The American Cancer Society estimates that CRC will kill 1 in 60 Americans and CRC screening is recommended for all Americans ≥45 years of age. Current CRC screening methods are effective for preventing CRC and have been shown to reduce CRC-related mortality. However, none of the currently available tests is ideal, and many people are not compliant with CRC screening. Novel CRC screening tests based on advances in CRC molecular biology, genetics, and epigenetics combined with developments in sequencing technologies and computational analytic methods, have been developed to address the shortcomings of current CRC screening tests. These emerging tests include blood-based assays that use plasma-derived circulating tumor DNA and serum proteins to detect early CRC and advanced adenomas, assays that use stool DNA or mRNA, and methods for profiling the gut microbiome. Here we review current screening modalities, and we discuss the principles behind the most promising emerging CRC screening tests and the data supporting their potential to be used in clinical practice.
Keywords: colorectal cancer, screening, genetics, epigenetics, microbiome
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer death in the world in both men and women1. Considering this sobering fact, CRC screening is recommended in the US as well as most European countries, Canada, specific regions in North and South America, Asia, and Oceania2. Countries with widespread CRC screening have witnessed declines in CRC mortality1,3. In the US, the incidence of late-stage CRC has declined 2% per year from 2014–2018 in adults >50 years of age, likely secondary to screening4. However, although multiple CRC screening modalities are available, compliance with screening is below the National Colorectal Cancer Roundtable’s goal of 80%5. In addition, current screening modalities each fall short in different aspects with respect to access, cost, cultural acceptance, risk of complications, test performance, and patient and provider knowledge gaps. With advancing knowledge and understanding of the molecular pathology of precancerous adenomatous polyps, serrated sessile lesions (SSLs), and CRC, non-invasive screening modalities that use biomarkers of dysplasia and neoplasia are being developed and validated. Several may soon be approved for clinical use as CRC screening tests. These emerging tests have great potential to overcome existing barriers to CRC screening and improve compliance. The ideal test would be one that has a high sensitivity and specificity for detecting lesions that could progress to CRC as well as early-stage CRC, and is affordable, accessible, convenient, and low risk. CRC blood tests and second iteration stool DNA tests aspire to this ideal6. In this narrative review, we will briefly discuss gaps and barriers in current screening modalities and then examine promising classes of CRC screening tests that are under development and are being evaluated in clinical trials.
Opportunities for improvement with current screening modalities
There are a variety of CRC screening tests currently in clinical use, and none of them is an ideal modality. They include non-invasive tests (i.e., fecal immunochemical test (FIT), multi-target-stool DNA (MT-sDNA) test, plasma methylated SEPT9 test, CT colonography, and invasive tests (i.e., colonoscopy, sigmoidoscopy). Colonoscopy has superior sensitivity for CRC as well as for advanced adenomas (AA), but is invasive, inconvenient, associated with complications, and more expensive (and therefore less accessible) than alternatives such as FIT. Non-invasive tests are favorable with respect to cost, safety, and convenience, but the current tests have low sensitivity for AA (42% using MT-sDNA).
Despite there being a variety of test options, compliance with CRC screening recommendations is suboptimal. A recent survey of the MarketScan Commercial and Medicare Supplemental databases found 70% adherence with colon cancer screening, which is below the 80% screening rate goal set by the National Colorectal Cancer Roundtable5. Furthermore, utilization of screening tests is even lower in several groups including adults in the 45–49-year old age group, minority communities, the rural poor, and the uninsured. Barriers to compliance include limited time available for personal healthcare due to work and family obligations; psychosocial factors; socioeconomic factors including accessibility to care, insurance, and income to spend on discretionary services; limited healthcare access in rural communities; socioeconomic factors; language barriers and cultural factors7. The COVID-19 pandemic has underscored the importance of developing more non-invasive screening strategies, by revealing the impact of unanticipated barriers to accessing colonoscopy. An increase in non-invasive CRC screening occurred during the height of the pandemic as a result, which counterbalanced the drop in colonoscopies seen in 20208. In this review, we discuss emerging tests and their potential to enhance CRC screening compliance by overcoming some of these barriers.
Emerging insights into CRC pathogenesis form the basis for novel screening tests
Our understanding of the pathogenesis of CRC is central to the approaches being used in developing new screening tests. CRC develops with the transformation of normal epithelial cells into adenocarcinoma cells through a defined progression of both histological and genetic/epigenetic changes3. The progression of a polyp to cancer in classic tumorigenesis starts in aberrant crypts of the colonic villi that develop into either tubular or tubulovillous adenomas. These adenomas have the potential to progress to advanced adenomas and finally CRC. The adenoma-to-CRC progression is currently understood to occur as a result of the serial accumulation of genetic and epigenetic alterations in colonic epithelial cells in conjunction with alterations in the tissue microenvironment secondary to aging, inflammation, and the gut microbiome, which may all be interrelated3. The genetic and epigenetic alterations in colon adenomas and cancer are the basis of assays that use cfDNA as a tumor biomarker. Of note, circulating free DNA (cfDNA) is a broad term that describes DNA that is freely circulating in the bloodstream and is derived from cells but that is not associated with cells in the bloodstream. Circulating tumor DNA (ctDNA) is tumor-derived fragmented DNA in the bloodstream and is one type of cfDNA. ctDNA should not be confused with cell-free DNA (cfDNA), which is not necessarily of tumor origin. . Both colon adenomas and CRCs can be a source of ctDNA. ctDNA is derived from a number of sources, which include circulating tumor cells escaped from the primary tumor, from degraded tumor cell DNA release (secondary to tumor cell death) from cells in the primary tumor, and from secreted tumor cell exosomes 9,10. Analysis of ctDNA, commonly termed liquid biopsy, is a minimally invasive strategy that can be used for CRC screening and can be tailored to detect cancer related mutations, DNA methylation alterations, and chromatin alterations in ctDNA11,12.
In addition to tubular adenomas, SSLs have the potential to transform into CRC similar to tubular adenomas13,14. However, it is important to recognize that most SSLs have a CpG Island Methylator Phenotype (CIMP) and a unique mutation spectrum compared to tubular adenomas. SSLs that arise from the right colon commonly are CIMP positive and carry BRAF V600E mutation, while those that arise from the left colon tend to carry mutant KRAS and are CIMP low13,15. Approximately 20–30% of CRC are a result of the SSL pathway13. SSLs are more difficult to detect with the MT-sDNA test than adenomas and are not effectively detected by FIT.
Recent technologic advances and innovations in data analytics now permit detection of these genetic, epigenetic, and microenvironment alterations. These advances have improved our ability to detect pre-cancer and cancer specific molecular markers in blood, stool and urine samples, and this has set the stage for the emergence of an array of novel blood-, serum-, and stool-based CRC screening tests.
Current and next-generation stool-based screening assays
FIT and MT-sDNA have established the value of stool-based CRC screening. Stool based assays permit sampling the colonic milieu to capture key CRC-associated changes to the intestinal environment as well as shed tumor cells in the stool. A set of next-generation assays building on this principle are at different stages of development.
MT-sDNA test
The suboptimal sensitivity of FIT has motivated the development of tests that use more specific molecular biomarkers to detect colon polyps and CRCs such as gene mutations, abnormally methylated DNA loci, and micro RNAs3. Prior studies have shown that CRC and polyps shed neoplastic cells into the colonic lumen at a continuous rate, in contrast to the intermittent bleeding upon which FITs rely16. The MT-sDNA test, Cologuard (Exact Sciences), became clinically available in 2014 and includes assays for methylated DRG4 and BMP3, mutant KRAS and a FIT assay (Table 1). MT-sDNA was compared to FIT and colonoscopy in the “Deep-C” study in average risk patients and was found to have sensitivity of 92% for CRC at specificity of 87%, and sensitivity of 42.4% at specificity of 86.6% for advanced precancerous lesions (adenomas and SSLs), compared to 5.1% SSL sensitivity for FIT17. The Cologuard test is recommended to be performed every 3 years for CRC screening and is FDA-approved for use only in average-risk individuals. Limitations of the MT-sDNA test are the disagreeable aesthetics of stool-based tests to many people, high cost compared to FIT and lower sensitivity for advanced adenomas compared to colonoscopy.
Table 1:
Current blood and stool-based screening assays
| Current CRC genetic & epigenetic screening tests | Diagnostic Test | Study design | Sample size | Number of years recruited | Population | Age | Sensitivity for CRC | Specificity for CRC | Sensitivity for advanced adenomas |
|---|---|---|---|---|---|---|---|---|---|
| Blood-based screening assays | |||||||||
| Epi proColon, ColoVantage | mSEPT9 | Prospective Study | 7941 | 2 years | asymptomatic | ≥50 | 48.2% | 91.5% | 11.2% |
| Stool-based screening assays | |||||||||
| Cologuard | MT-sDNA | Prospective Study | 10,000 | 2 years | Average risk individuals undergoing screening colonoscopy | ≥50 | 92.3% | 86.6% | 42.4% |
MT-sDNA test 2.0
Currently the MT-sDNA test is the most sensitive non-invasive screening tool for CRC17.
Because there is opportunity to improve the sensitivity and specificity of current stool-based assays, the BLUE-C trial (NCT04144738) is a study that was initiated to determine if a MT-sDNA 2.0 assay being developed by Exact Sciences Co will improve the sensitivity and/or specificity of the currently approved MT-sDNA test. BLUE-C is a prospective cohort observational study whose primary objective is to assess the sensitivity and specificity for CRC of the MT-sDNA 2.0. Secondary outcomes include sensitivity for advanced adenomas, sensitivity for CRC and advanced adenomas compared to FIT, and specificity for no neoplastic findings.
With regards to possible approaches to improve the performance of the current MT-sDNA assay, it is worth noting that some stool-based protein biomarkers have shown higher discriminatory power than occult hemoglobin for CRC and advanced adenomas18. Based on a case-control study of novel stool-based protein biomarkers, 29 proteins were statistically significantly enriched in stool from patients with CRC compared to control stool samples. The sensitivities of these proteins were 80% for CRC and 45% for advanced adenomas with a 95% specificity, higher than hemoglobin alone18. These results are promising and suggest protein biomarkers may be able to improve current stool-based screening methods (Figure 2).
Figure 2.

Altered DNA and proteins in blood shed from CRC and AA can be detected in stool samples of individuals using the MT-sDNA test. The geneoscopy assay (RNA-FIT) uses 8 stool-derived eukaryotic RNA biomarkers along with a FIT test for CRC and AA detection.
Geneoscopy assay
Another stool-based assay under investigation is a multifactor assay (RNA-FIT test) that combines 8 stool-derived eukaryotic RNA biomarkers for CRC and advanced adenoma detection (Figure 2). This assay, termed Geneoscopy, in combination with a FIT test, has been assessed in a 1,305- patient, average-risk, prospective cohort study undergoing colonoscopy. Of note, this cohort was supplemented with a retrospective analysis of stool samples obtained from 22 patients diagnosed with AA or CRC before treatment or resection. The RNA-FIT test results were compared with colonoscopy findings. The assay was trained on a 5,939 subject dataset and then tested on 5,388 subjects. In the test set of subjects, the RNA-FIT test attained a 95% sensitivity for CRC (n= 522), 62% sensitivity for AA (n =552), 25% sensitivity for non-AA (n =5,139), 80% specificity for hyperplastic polyps (n= 574), and 85% specificity for no findings on a colonoscopy (n =5,101). This assay is a promising noninvasive option to screen for both CRC and precancerous adenomas and is currently being assessed in a large prospective cohort study (CRC-Prevent, NCT04739722).
Microbiome-based assays
Epidemiological and disease cohort studies implicate the gut microbiome as a major environmental determinant of CRC risk19–24. Preclinical studies have shown that ApcMin/+ mice, which are genetically engineered to develop multiple intestinal adenomas, have almost no tumor burden when reared in a microbe-free state25, highlighting the role of the gut microbiome in CRC pathogenesis.
Several feasibility studies have been recently published showing that gut microbes and/or metabolites may serve as biomarkers for early detection of adenomas and CRC23,26–29. Bork and colleagues have provided early evidence that metagenomic sequencing data may permit early detection of CRC through identification of a panel of CRC-associated bacterial species30. Further, they observed functional differences between CRC-associated and health-associated microbiomes, as predicted by metagenomic gene content, thus potentially offering insights into CRC pathogenesis secondary to the effects of the microbiome.
Based on their metagenomic model, they developed a classifier, which performed comparably to fecal occult blood testing (FOBT). Combining their classifier with FOBT led to significantly better predictions than either alone. Additionally, other studies have demonstrated the utility of microbiome-based screening using DNA in residual stool from FIT kits, thereby decreasing the need for collecting additional stool samples and reducing the overall cost of screening28,29. These findings support the development of combination tests that utilize microbiome profiling in conjunction with an established test (e.g. FOBT, FIT, or MT-sDNA) to improve overall test performance for CRC early detection (Figure 3).
Figure 3.
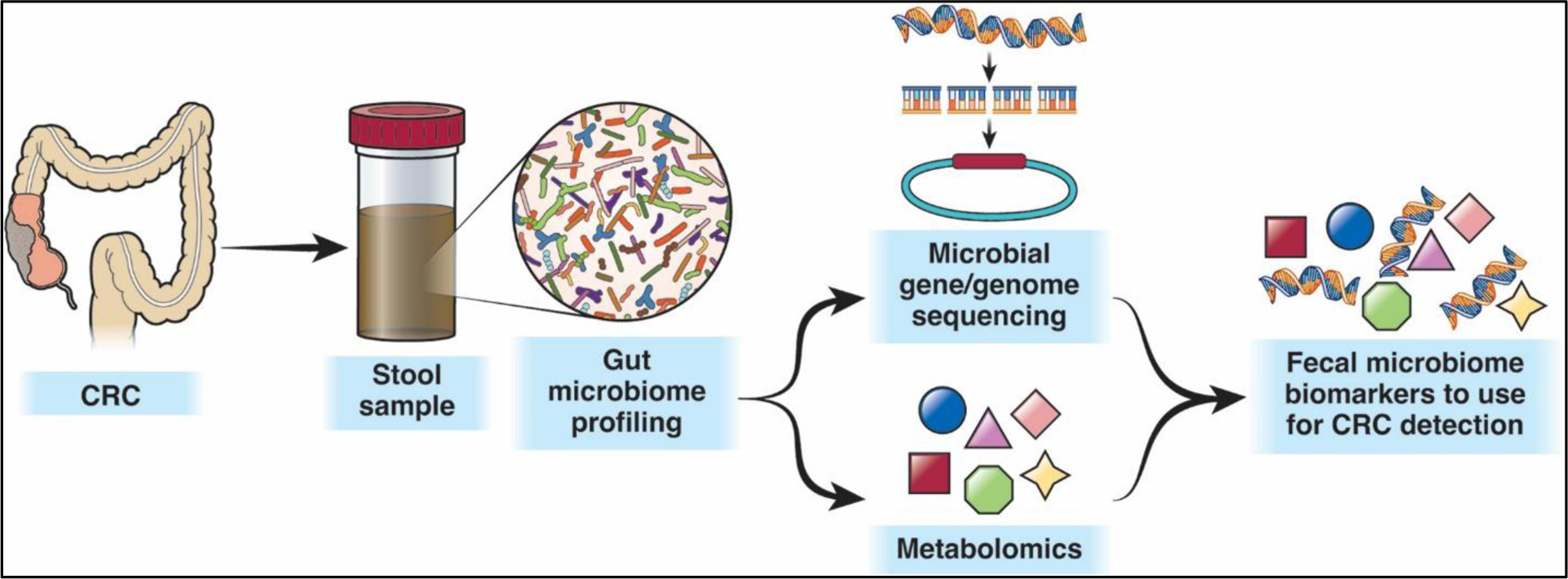
Microbiome-based assays for CRC detection take advantage of the differences in bacterial species, viruses, fungi, and microbiome-generated metabolites between stool from CRC/AA-associated and health-associated microbiomes to develop biomarkers that can be used for CRC or AA detection, either independently or along with established tests such as FOBT, FIT, or MT-sDNA.
Further studies have highlighted the potential of using fecal metagenomic biomarkers for early CRC diagnosis. A study conducted by Yu et al in 2017 was the first to show the presence of certain fecal microbiome gene markers associated with CRC across multiple ethnicities through the use of aggregated information from 4 different countries26. They studied cohorts from China, Denmark, Austria, and France, and although the populations differed in their microbial community structures, the microbiome CRC signatures were found universally26. A recent cross-sectional retrospective case-control study in China looked at 4 different microbial markers, Fusobacterium nucleatum (Fn), Lachnoclostridium sp. (m3), Bacteroides clarus (Bc), and Clostridium hathewayi (Ch) as potential screening markers for the diagnosis of CRC and AA. The fecal samples were collected from 210 patients with CRC, 115 patients with advanced adenomas, 86 patients with non-advanced adenomas, and 265 non-neoplastic control subjects. Their results showed that Fn performed the best as an individual marker for CRC and that Lachnoclostridium sp. (m3) alone was best at diagnosing adenomas27. The sensitivity and specificity for CRC were 84.9% and 83.3% respectively, and the sensitivity and specificity for adenomas were 38.6% and 98.6%27. This study helps set the stage for future investigation of microbiome-based assays for CRC screening, as it shows their potential to be used in combination with FIT or as individual tests for the sensitive and specific detection of CRC and possibly adenomas. Limitations of the study include that it is retrospective, has a small sample size, and is not generalizable for general screening populations as participants were already known to have CRC or advanced adenomas.
Another recent study focused on identifying adenoma-specific microbial markers that can be used for early CRC screening. This was a case-control study of over 1000 fecal samples that used 16S rRNA sequencing data to identify models that were able to distinguish between the microbiome composition in people with colorectal adenomas versus healthy control subjects. The models predicted colorectal adenoma with a sensitivity of 82% and specificity of 62%, and CRC with 66% sensitivity and 90% specificity31. As with the prior study by Liang et al, this assay class has the potential to be used as standalone screening test or in combination with FIT to enhance sensitivity and possibly specificity.
Informative CRC biomarkers need not include only microbial species but may also include downstream effects of those microbes. For example, a carcinogenic strain of E. coli has been found to induce mutational signatures shared by many patients with CRC, suggesting generalizable early genotoxicity32. Alterations in microbiome-generated serum and fecal metabolites have been reported and proposed as the basis for non-invasive screening tests33–32.
A recent meta-analysis assessed the diagnostic capacity for sets of bacterial species and metabolic processes that are significantly altered in individuals with CRC23. Bacteria such as Fusobacterium nucleatum, Porphyromonas, Parvimonas, Peptostreptococcus, Gemella, Prevotella, and Solobacterium, which have been previously associated with CRC, were part of the investigators’ proposed core set of CRC-associated bacteria. They found evidence of altered amino acid, carbohydrate, bile acid, and mucin metabolism by the gut microbiome, which could be incorporated into CRC screening tests based on panels of microbiome and microbial metabolite biomarkers.
In aggregate, these studies suggest that metagenomic sequencing can establish generalizable microbiome CRC signatures that can be used for the detection of CRC. However, based on a review of patents filed globally and registered clinical trials at clinicaltrials.gov, there appear to be no current microbiome-based screening tests that are close to being clinically available despite the rich published literature demonstrating the potential of microbiome based CRC screening tests to be used clinically. Of interest and consistent with this potential, there are ongoing microbiome-based clinical studies that assess the effect of microbiome modulation on improving the efficacy of immunotherapy for the treatment of cancer (e.g. NCT04208958, a trial of VE800, a biologic agent manufactured by Vedanta, in patients with advanced cancer treated with nivolumab). Successful development of microbiome-based interventions could potentially be repurposed for treating or preventing dysplasia or early neoplasia as well.
In conclusion, studies to date demonstrate the potential for the gut microbiome to be used to develop novel stool-based tests that could complement the repertoire of current non-invasive CRC screening tests, either independently or in combination with FIT. However, there are many hurdles and potential limitations that remain to be addressed to ultimately develop a gut microbiome biomarker stool assay that is robust enough to be used for CRC screening and to guide clinical care. These issues include: 1) determining whether the gut microbiome changes precede or result from the formation of pre-cancerous colon polyps or CRC; 2) if microbiome changes precede dysplasia or neoplasia, determining if CRC-associated gut bacteria are causal in colon polyp and/or CRC formation; 3) in light of the tremendous global diversity and variation in microbiome structure and function, identifying generalizable and population-specific features of the gut microbiome that are drivers and/or biomarkers of CRC and can be leveraged in screening tests; 4) developing unified, documented, and reproducible protocols for studying the human gut microbiome from fecal samples, which will lead to comparable results across studies and can be externally validated in independent cohorts; and 5) determining the combinations of metabolites, bacterial genetic markers, and human mutation signatures that are most useful and cost-effective in screening. Once these issues are resolved not only can inexpensive and non-invasive CRC screening tests be developed, but there is the potential to also develop novel prevention therapies based on diet, the use of antibiotics, or probiotics, and prescribed lifestyle changes.
Conclusions
Stool based tests are already widely used in the US for noninterventional CRC screening. They have modest sensitivity for advanced adenomas and SSLs. The emerging tests using CRC related molecular alterations have reasonable potential to improve the sensitivity and specificity of this class of tests. Ongoing clinical trials will likely yield results in the next 1–2 years and may change the types of stool based tests used as tier 1 CRC screening tests. The stool based assays based on the gut microbiome are a new class and are showing potential to improve the accuracy of stool-based assays, particularly when used in combination with FIT. However, gut microbiome based assays are likely several years from clinical use.
Current and emerging blood-based CRC screening tests
As discussed earlier, several barriers to CRC screening have motivated the development of non-invasive modalities that are easily accessible to the general population. In this section, we will discuss the currently available blood-based screening test and some of the most promising emerging tests, which include blood-based biomarker assays and are often referred to as “liquid biopsies” (Table 1 and 2).
Table 2:
Summary of ongoing studies of stool and blood based screening
| Emerging blood based screening tests | Diagnostic test | Study design | Sample size | Target duration | Population | Age | Primary Outcome | Secondary Outcome |
|---|---|---|---|---|---|---|---|---|
| CancerSEEK - MCED | ctDNA alterations+ protein expression patterns | Prospective Observational Study | 6399 | 3 yrs | 1000 volunteers with cancer; 2000 volunteers with no known cancer
|
≥50 | Develop and validate the classification algorithm used by the CancerSEEK cancer screening test by collecting clinically annotated peripheral blood specimens from subjects with cancer and no known cancer. | |
| Galleri - MCED | ctDNA | Prospective Observational Study | 35000 | 5 yrs | Any adult | ≥18 | Cancer detection rate; Performance of Galleri: sensitivity, specificity, positive predictive value; Population cancer detection rate; Cancer signal detected; Distribution of cancer stage in those who received a cancer diagnosis following a Galleri cancer signal detected test result | Time from test administration to cancer diagnosis; Healthcare resource utilization; Adherence to cancer screening guidelines; To describe the repeated use of Galleri test in patients |
| ECLIPSE (ctDNA LUNAR 2) | ctDNA | Prospective Observational Study | 20000 | 5 yrs | Asymptomatic, average risk individuals; screening colonoscopy population | 45–84 | Sensitivity for CRC detection, specificity of advanced neoplasia detection | Positive and negative predictive values for CRC detection; sensitivity and specificity for advanced adenoma detection |
| PREEMPT CRC | ctDNA | Prospective Observational Study | 25000 | 2 yrs | Asymptomatic, average risk individuals; screening colonoscopy population | 45–85 | Sensitivity for CRC of freenome test and specificity | |
| Emerging stool based screening tests | ||||||||
| BLUE-C | MT-sDNA 2.0 | Prospective Observational Study | 24000 | 3 years | Asymptomatic, average risk individuals; screening colonoscopy population | ≥40 | Sensitivity for CRC with the mt-sDNA 2.0 test; Specificity with the mt-sDNA 2.0 test | Sensitivity for advanced precancerous lesions; Sensitivity for CRC compared to a commercially available FIT; Sensitivity for advanced precancerous lesions compared to commercially available FIT; Specificity for no colorectal neoplastic findings |
| Colonosight Test | MT-sRNA (FIT-RNA) | Prospective Clinical Trial | 10000 | Asymptomatic, average risk; screening colonoscopy population | 45–84 | Sensitivity for CRC, advanced adenomas, high grade dysplasia and other adenomas; Specificity of negative test |
Plasma methylated SEPT9 DNA assay
This assay detects cfDNA using a PCR based assay for plasma-derived methylated SEPT9 cfDNA. It is marketed under the trade names Epi proColon and ColoVantage and was FDA approved in 2016 for individuals refusing other CRC screening tests. This blood-based screening method addresses limitations of other CRC screening tests that currently impede adherence and participation in screening, including convenience, access, and patient acceptability. Although this test is FDA approved it is not widely used in screening because of its low sensitivity for colon adenomas and CRC. In a large prospective study, SEPT9 had 48% sensitivity for the detection of CRC and was unable to detect most pre-cancerous polyps (sensitivity 11.2%)36. Despite its convenience, its poor performance compared to FIT and colonoscopy and cost have made this a test of last resort for CRC screening. Furthermore, it is unclear how often this test should be performed for screening, and it is not currently recommended by United States Multi-Society Task Force of Colorectal Cancer (US MSTF)37.
ctDNA and protein-based assays
Because of the lack of sufficiently accurate blood-based screening tests, many proof of principle studies using a myriad of different circulating tumor cell DNA (ctDNA), protein-, and/or glycoprotein-based blood tests have been done and have shown the potential of this class of assays to be used for CRC screening. None have had sufficient sensitivity to be a first-line CRC screening test or have been validated in a screening setting38–40. In fact, several years ago, in a study of precancerous adenomas in human cohorts and in a preclinical model, it was estimated that the genetic heterogeneity and low levels of ctDNA associated with adenomas made it unlikely that ctDNA would be detectable in liquid biopsies based on the technologies available then12. The low sensitivity and specificity38 of these prior tests has been addressed by recent progress in the technologies related to biomarker detection, which has led to several promising blood-based CRC tests under development.
The new technologies that have advanced the field are primarily based on the detection of tumor cell derived nucleic acids (i.e. genomic DNA, mRNA, miRNA) in circulation. It has long been known that tumor cell DNA is present in the plasma and serum of patients with cancer, which is derived from circulating tumor cells, degraded tumor cell DNA release, and tumor cell exosomes9,10. Importantly, because the ctDNA is massively diluted in a background of normal cell DNA, there is a low signal-to-noise ratio, which is a proverbial needle in a haystack that has prevented the development of sensitive early detection assays for cancer until recently. Because of the technologies developed in the last 10 years, signal-to-noise ratio, which is called the mutant allele fraction (MAF), that is as small as 1:10,000 or lower can now be reliably detected. The sensitivity of these technologies is critical for the use of ctDNA in cancer screening, as the amount of DNA shed by tumors is directly related to their size and early tumors are typically <3 cm in size41,42.
One of the central technologies that has spurred the development of ctDNA-based cancer screening studies is next generation sequencing (NGS), which has the capacity to profile billions of DNA fragments in circulation and can be used to detect ctDNA fragments of varying sizes as well as methylated ctDNA (Figure 1). Methods that have increased the fidelity of NGS sequencing and reading the fragments, as well as the dramatically decreased costs that have accompanied advances in the technology, have created an opportunity for the development of blood-based cancer screening tests for use in clinical care. In parallel with NGS, advances in PCR based technologies, such as droplet digital PCR (ddPCR) have broadened the range of methods used in emerging cancer screening tests.
Figure 1.
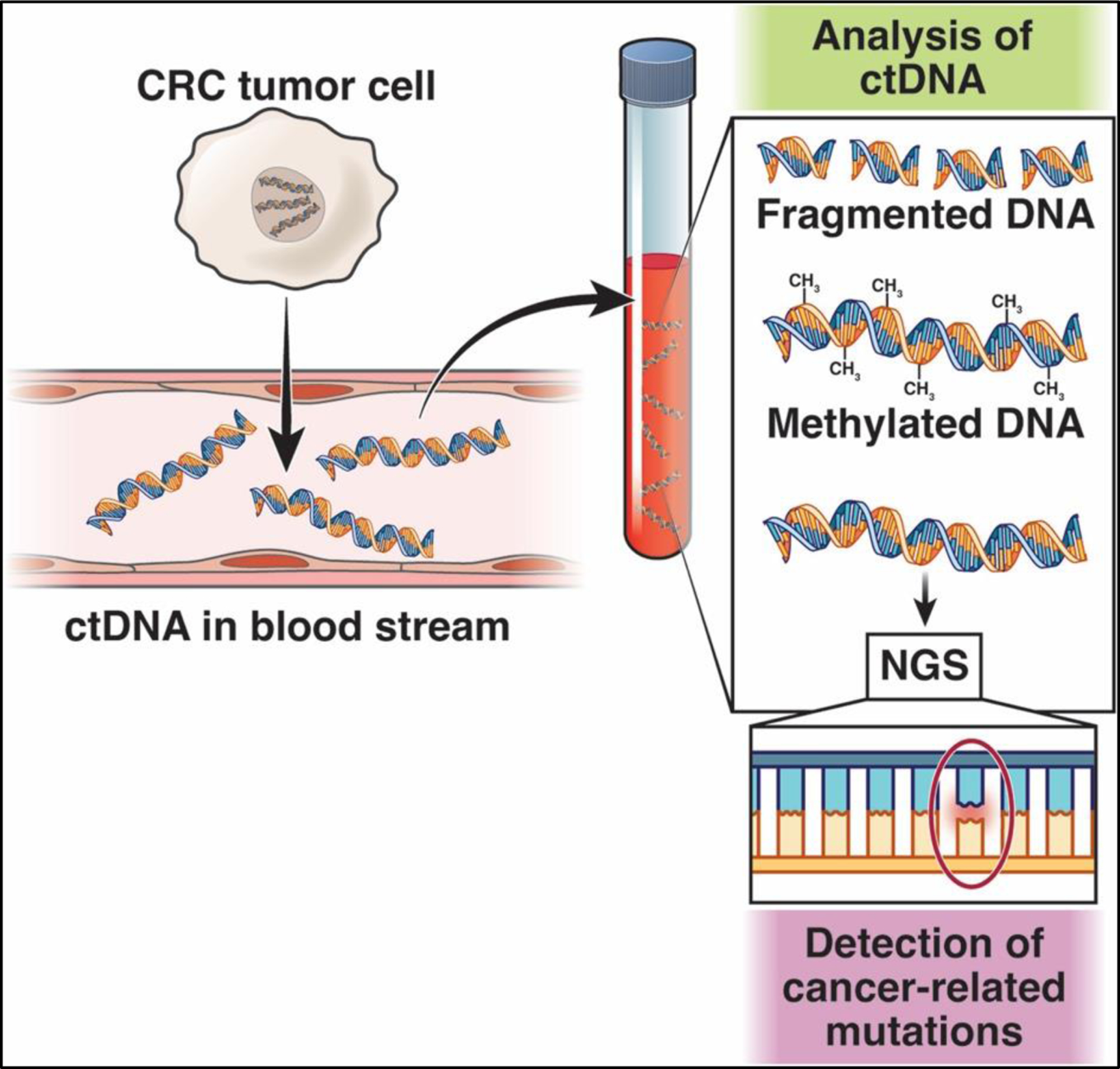
ctDNA-based assays are based on the detection of tumor cell-derived nucleic acids in the circulation. Next-generation sequencing is used to detect ctDNA fragments (with cancer-related mutations) and aberrantly methylated ctDNA.
An important issue regarding mutation based cancer screening tests that arises due to the ability to do massive-scale sequencing is the recognition that DNA mutations in cancer genes is common in normal tissues, particularly in the elderly and in cancer survivors43–45, which can increase the false positive rate of the assays. Leukocytes in the peripheral blood acquire mutations as part of the normal aging process, DNA from leukocytes (buffy coat) from the patient’s blood samples is tested in parallel with the cell free plasma DNA from the patient to exclude mutations originating from benign clonal expansions, such as clonal hematopoiesis of indeterminate potential (CHIP)46.
It is also important to note that although the technology and mutation knowledge advances have improved the specificity of NGS and advanced PCR technologies for detecting cancer relevant mutations, the sensitivity of the methods has been typically too low to be of use for detecting early-stage cancers. Key limitations of DNA mutation-based assays are the relatively small number of genes that are commonly mutated in any individual cancer and the cancer specificity of the gene mutations47. Furthermore, the marked intertumoral heterogeneity among the specific cancers further impairs the sensitivity of mutation-based screening studies. For example, CRCs vary from having <1 mutation per 106 base-pairs to >100 mutations per 106 base-pairs, which makes a subset of CRCs essentially undetectable, and there are only 32 genes that are recurrently mutated in CRCs, which further impedes the sensitivity of the mutation-based assays48.
The solution to the intrinsic limitation of DNA mutation-based screening assays came from the recognition that epigenomic alterations, such as aberrant DNA methylation and chromatin structure alterations, are much more frequent than mutations, by orders of magnitude, in most cancers and occur very early in the tumorigenesis process, often at tumor initiation26,47,49. The incorporation of epigenetic markers into the emerging screening testshas yielded blood-based assays that can detect early-stage cancers50–52. The emerging plasma DNA based assays use artificial intelligence (AI) informed cancer specific methylation signatures derived from NGS based high-depth, targeted bisulfite sequencing of panels of >100,000 methylation sites. They demonstrate substantial improvements in sensitivity compared to mutation-based assays without loss of specificity53–55. An observational study of a cfDNA methylome based assay in a real-world setting has been initiated based on the performance of this class of assays to determine the feasibility of a methylome multi-cancer early detection assay (NCT05205967).
In addition to DNA methylation, other epigenetic alterations have been incorporated into ctDNA based assays to further improve sensitivity and specificity. Differential fragmentation patterns of ctDNA (called fragmentomics) in patients with cancer compared with those in individuals without cancer have been assessed and included in many ctDNA based assays. These fragments are detected using whole genome NGS data and have been used to develop cancer specific fragmentation signatures that have been shown to be present in early-stage cancers and to be specific for the site of origin of the cancer56. (The interested reader is directed to recent reviews on this subject.56,57)
In summary, the development of modern NGS and advanced PCR technologies, in combination with the creation of machine learning and AI informed cancer biomarker panels that include DNA mutations, DNA methylation and DNA fragmentomics, has led to a generation of ctDNA detection assays that have potential to be used for the sensitive and specific detection of early-stage cancers and potentially even pre-cancers. The major outstanding questions that remain to be addressed are the performance of the assays in a screening population setting, the clinical utility of the assays, and their cost effectiveness.
Another class of blood-based CRC screening tests that has seen promising advances is protein biomarker panel assays. The incorporation of glycoproteins into protein biomarker panels has led to improvements in the performance of these assays. Although many serum protein biomarker panels have been studied in case-control studies using symptomatic CRC patients and shown AUCs ranging from 0.62–0.996, very few have been assessed in screening population-based validation studies58. The few that have been assessed show marked decreases in assay sensitivity for CRC in the validation studies, with one assay having only a 17% sensitivity at 90% specificity40,59. A panel that included a signature with CYFRA21–1, ferritin, Osteopontin, anti-p53, seprase, and CEA decreased from 70% to 42% in sensitivity at 95% specificity58 when validated in a true screening setting. The highest performing assay panel tested in a screening population to date has a sensitivity of 76.9% for CRC60, which notably does not match that of other currently used CRC screening modalities61. At this time, there are no protein-based CRC screening tests that are poised to be adopted into clinical care soon.
ctDNA blood-based assays for CRC early detection
The use of ctDNA based blood tests for CRC detection was first advanced in monitoring for minimal residual disease (MRD) in patients after colorectal cancer resection. (Examples of commercially available MRD assays include Guardant360 (Guardant Health), Signatera (Natera), FoundationOne Liquid CDx (Foundation Medicine), and many others.) The emergence of these technologies has led to assays that have enough potential to be used in CRC screening that there are three ongoing large screening population based randomized clinical trials comparing blood-based assays to colonoscopy and FIT test.
There are currently two emerging ctDNA blood-based tests that are expected to have data from large prospective screening population-based studies in the next year. The first is the ctDNA LUNAR-2 test (Guardant Health) which is currently being studied in the ECLIPSE (Evaluation of the ctDNA LUNAR-2 Test in an Average Patient Screening Episode; NCT04136002) trial which is a 24-month prospective, observational, multicenter study (130 centers in the US).
Of note, the LUNAR-2 test performance has been assessed in a cohort study of 699 Korean individuals with newly diagnosed CRC, stage I-III with an age range of 20–89 (median 63) prior to surgical resection, compared to controls (n=279; age range 20–91, median 57) who were confirmed negative for CRC by screening colonoscopy. The sensitivity and specificity of the test were found to be 96% and 94%, respectively. Of note, the sensitivity for asymptomatic stage I/II CRC was 90%62. Following this study, subsequent research in the US, Canada and EU was carried out with 309 subjects, including those with normal colons (N=166), advanced adenomas (N=51) and CRC (MN=51). The subjects were balanced by age (a mean of 64 years old) and gender. The assay demonstrated 91% sensitivity (detection rate) for CRC (95% CI 84% – 95%), including 90% for Stage I, 97% for Stage II, and 86% for Stage III CRC. The assay also demonstrated 20% sensitivity for advanced adenomas (95% CI; 11% – 32%) and 92% specificity in the normal cases (data from Guardant Health website). Based on these results, a blood based CRC screening assay, marketed under the name Shield™, was made available for clinical use in May 2022.
A major limitation of these case control studies is that they are not large observational trials in a screening population. The ECLIPSE trial addresses these limitations by being a prospective screening population based study that assesses the performance of the LUNAR-2 test in average risk individuals compared to colonoscopy, with the primary outcome being sensitivity for CRC detection and specificity for advanced neoplasia detection63. Approximately 13,000 average risk adults between the ages of 45–84 recruited from sites throughout the U.S. were included in the study64. Participants do not have a history of cancer, inflammatory bowel disease or genetic predisposition to CRC and had not received recent CRC screening. The blood sample was obtained prior to undergoing colonoscopy preparation via mobile phlebotomy. The results of the blood draw and the colonoscopy will be retroactively compared from the prospectively collected samples64.
Another emerging ctDNA based CRC screening assay has been developed by Freenome and is being assessed in the PREEMPT CRC trial (Prevention of Colorectal Cancer Through Multiomics Blood Testing; NCT04369053). The assay uses a machine learning determined pattern of genomic alterations, epigenomic alterations and protein expression alterations to identify CRC DNA55,65–67. The trial is a prospective multi-center observation study across the US looking at the Freenome blood-based assay for early detection of CRC in average risk individuals undergoing routine screening with colonoscopy. More than 35,000 average-risk individuals between the ages of 45–84 years old undergoing routine screening colonoscopy participated in this trial. The primary outcome of the study is the sensitivity and specificity for CRC of the Freenome test with results from the trial expected soon. Results from the ECLIPSE trial and PREEMPT trial will provide useful data regarding the place of the blood-based assays in the current menu of CRC screening test options.
In addition to these two tests, there is a substantial number of other tests (>5 at the time that of writing, (e.g.FirstSight test68) that are under investigation or being offered as CRC screening laboratory developed tests (LDTs) based on small cohort or case control studies. None of these tests have been evaluated in observational cohort studies in a CRC screening population, and most have no peer reviewed studies to support their claims. It is also notable that Exact Sciences is developing a blood based screening test and plans to evaluate this in subjects participating in the BLUE-C CRC observational screening study (NCT04144738), which will include >25,000 average risk individuals who are >40 years of age, eligible for CRC screening, and scheduled for a screening colonoscopy.
Conclusions
ctDNA based CRC screening tests are a promising and exciting class of tests. The emerging tests are showing potential to substantially improve the accuracy of the current CRC blood based screening test (methyl SEPT9) for CRC. The results to date suggest that ongoing large trials will demonstrate similar accuracy to FIT for CRC but lower sensitivity for advanced adenomas. If this potential is realized, then these assays will have role in CRC screening due to their convenience and appeal to the public., and could increase compliance rates for CRC screening. The position among other CRC screening modalities (as tier 1, 2 or 3 options), will depend on their relative sensitivity and specificity for CRC and adenomas. It is expected that such information will be available in the next 1–3 years.
Multi-cancer early detection assays (MCED)
An exciting recent development in cancer early detection assays is a new class of biomarkers called multi-cancer early detection (MCED) tests. MCEDs are being developed because of their potential to significantly improve compliance with cancer screening recommendations as they are blood-based assays and simplify screening by combining many cancer screening tests in one convenient test. Like the emerging blood based CRC screening tests described above, MCED use ctDNA +/− protein biomarker patterns to reliably identify cancer specific signatures in plasma derived DNA and serum or plasma proteins. The cancer-specific signatures are derived using individually or combined patterns of genomic alterations, epigenomic alterations (e.g. DNA methylation, DNA chromatin structure as represented by DNA fragment size), and plasma/serum protein expression levels that are trained using artificial intelligence (AI) methods. An example of this class of biomarker assays is a test developed by scientists at GRAIL, Inc that uses DNA methylation signatures and that has been shown capable of detecting 25 different cancer signatures, including CRC. A second representative MCED test was developed by scientists at Thrive/Exact Sciences and uses genomic alterations, epigenomic alterations and protein expression patterns. This test, named CancerSEEK demonstrated the ability to detect 8 different cancer types using plasma, = with sensitivities ranging from 69–98% for liver, stomach, pancreas, esophagus and ovarian cancer and a >99% specificity for all these cancers. The sensitivity varied substantially for stage I cancers from 100% (liver cancer) to 20% (esophageal cancer)53. In addition to these two MCED tests, a variety of other MCED tests are being developed, but there is no literature beyond feasibility studies. Limitations of the MCED tests are the false positive rates, which ranged from 8% to 17%, if the inability to correctly determine the site of origin of the cancer signal is considered a false positive, and the lack of evaluation in screening populations that have matched control subjects, which is needed to accurately determine the true specificity of the test when used for cancer screening. Another major issue that needs to be defined is the care pathway for individuals with positive MCED assay results. A positive result will need to be confirmed using additional diagnostic studies that have well characterized levels of accuracy. The confirmatory diagnostic pathway is not established at this time for many of the cancer types detected by the MCEDs and will likely be complex and associated with additional costs and risks. Thus, perhaps the most significant unresolved issue is the lack of demonstration of clinical utility and cost effectiveness of MCED tests, based ideally on controlled clinical trial data. At this time, it is not clear how to optimally design such studies for MCED tests, and this has led the NCI to develop advisory boards on this topic. These current barriers to the appropriate use of the MCED tests, especially when the MCED test assesses for currently unscreened cancer types, has led to efforts to selectively use specific components for cancers for which screening is currently recommended. So, while MCED tests have exciting potential to dramatically alter how cancer screening is conducted and to improve the compliance with current cancer screening efforts, there are still many questions that need answering before this can be realized in clinical practice.
Conclusions
CRC is a common but preventable cancer. CRC screening is predominantly done using colonoscopy or stool-based tests such as FIT and MT-DNA, and has been shown to prevent CRC related deaths. However, the currently available tests have a variety of drawbacks making none of them ideal. Furthermore, compliance with CRC screening in the US is well below the target of 80%. Emerging CRC screening tests have the potential to address the drawbacks of current modalities and to improve compliance. These tests are the result of advancements in the technology used to detect cell free nucleic acids (e.g. DNA, mRNA, etc.), analysis methods, and in our understanding of the molecular pathology of CRC. These tests are in different stages of development with plasma cfDNA based assays being the class of assay that is most mature. The sensitivity and specificity of cfDNA based assays from large clinical trials done in CRC screening populations are expected in the next year and likely will result in the addition of these tests to the menu of CRC screening options currently available. Screening assays that are based on the gut microbiome also appear to be promising but are likely a few years from being tested in large prospective observational studies in a CRC screening population. It is clear that there will be additional CRC screening tests available in the near future, although their place in the menu of screening options remains to be defined (Table 3).
Table 3:
Colorectal Cancer Screening Test Type Strengths and Weaknesses
| Screening Test | Sensitivity/Specificity for Cancer | Sensitivity/Specificity for Advanced Adenomas | Convenience | Safety | Cost |
|---|---|---|---|---|---|
| FIT | +++ | + | +++ | ++++ | + |
| Colonoscopy | ++++ | ++++ | + | ++ | ++++ |
| Stool DNA | +++ | ++ | +++ | ++++ | ++ |
| Stool RNA | +++ | ++ | +++ | ++++ | ? |
| ct DNA | +++ | +/? | ++++ | ++++ | ++/? |
| Serum proteome | ++/? | ? | ++++ | ++++ | ++/? |
| Microbiome | +++/? | ? | ++++ | ++++ | ? |
?=insufficient data at this time to determine reliably
Funding:
This work was supported by the NIH (U54 CA274374 to ND and WG).
Abbreviations used:
- CRC
colorectal cancer
- FIT
fecal immunochemical test
- SSLs
serrated sessile lesions
- MT-sDNA
multi-target-stool DNA
- MT-sRNA
multi-target-stool RNA
- CIMP
CpG Island Methylator Phenotype
- cfDNA
circulating free DNA
- ctDNA
circulating tumor cell DNA
- ddPCR
droplet digital PCR
- CHIP
clonal hematopoiesis of indeterminate potential
- MCED
multi-cancer early detection
- AA
advanced adenomas
- FOBT
fecal occult blood testing
- NGS
next generation sequencing
- AI
artificial intelligence
- MRD
minimal residual disease
- LDTs
laboratory development tests
- MAF
mutant allele fraction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors declare no conflicts of interest.
References
- 1.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14(2):89–103. doi: 10.5114/pg.2018.8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navarro M, Nicolas A, Ferrandez A, Lanas A. Colorectal cancer population screening programs worldwide in 2016: An update. World J Gastroenterol. May 28 2017;23(20):3632–3642. doi: 10.3748/wjg.v23.i20.3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickinson BT, Kisiel J, Ahlquist DA, Grady WM. Molecular markers for colorectal cancer screening. Gut. Sep 2015;64(9):1485–94. doi: 10.1136/gutjnl-2014-308075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. January 2022;72(1):7–33. doi: 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 5.Fisher DA, Princic N, Miller-Wilson LA, Wilson K, Fendrick AM, Limburg P. Utilization of a Colorectal Cancer Screening Test Among Individuals With Average Risk. JAMA Netw Open. September 01 2021;4(9):e2122269. doi: 10.1001/jamanetworkopen.2021.22269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melson JE, Imperiale TF, Itzkowitz SH, et al. AGA White Paper: Roadmap for the Future of Colorectal Cancer Screening in the United States. Clin Gastroenterol Hepatol. November 2020;18(12):2667–2678.e2. doi: 10.1016/j.cgh.2020.06.053 [DOI] [PubMed] [Google Scholar]
- 7.Sly JR, Edwards T, Shelton RC, Jandorf L. Identifying barriers to colonoscopy screening for nonadherent African American participants in a patient navigation intervention. Health Educ Behav. Aug 2013;40(4):449–57. doi: 10.1177/1090198112459514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fedewa SA, Star J, Bandi P, et al. Changes in Cancer Screening in the US During the COVID-19 Pandemic. JAMA Netw Open. June 01 2022;5(6):e2215490. doi: 10.1001/jamanetworkopen.2022.15490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. Feb 7 2020;367(6478)doi: 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alix-Panabieres C, Schwarzenbach H, Pantel K. Circulating tumor cells and circulating tumor DNA. Annu Rev Med. 2012;63:199–215. doi: 10.1146/annurev-med-062310-094219 [DOI] [PubMed] [Google Scholar]
- 11.Zhao X, Dai F, Mei L, et al. The Potential Use of Dynamics Changes of ctDNA and cfDNA in the Perioperative Period to Predict the Recurrence Risk in Early NSCLC. Front Oncol. 2021;11:671963. doi: 10.3389/fonc.2021.671963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myint NNM, Verma AM, Fernandez-Garcia D, et al. Circulating tumor DNA in patients with colorectal adenomas: assessment of detectability and genetic heterogeneity. Cell Death Dis. Aug 30 2018;9(9):894. doi: 10.1038/s41419-018-0934-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology. Feb 2013;62(3):367–86. doi: 10.1111/his.12055 [DOI] [PubMed] [Google Scholar]
- 14.Goldstein NS. Serrated pathway and APC (conventional)-type colorectal polyps: molecular-morphologic correlations, genetic pathways, and implications for classification. Am J Clin Pathol. Jan 2006;125(1):146–53. [PubMed] [Google Scholar]
- 15.Noffsinger AE. Serrated polyps and colorectal cancer: new pathway to malignancy. Annu Rev Pathol. 2009;4:343–64. doi: 10.1146/annurev.pathol.4.110807.092317 [DOI] [PubMed] [Google Scholar]
- 16.Ahlquist DA, Harrington JJ, Burgart LJ, Roche PC. Morphometric analysis of the “mucocellular layer” overlying colorectal cancer and normal mucosa: relevance to exfoliation and stool screening. Hum Pathol. Jan 2000;31(1):51–7. [DOI] [PubMed] [Google Scholar]
- 17.Imperiale TF, Ransohoff DF, Itzkowitz SH. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. July 10 2014;371(2):187–8. doi: 10.1056/NEJMc1405215 [DOI] [PubMed] [Google Scholar]
- 18.Bosch LJW, de Wit M, Pham TV, et al. Novel Stool-Based Protein Biomarkers for Improved Colorectal Cancer Screening: A Case-Control Study. Ann Intern Med. Dec 19 2017;167(12):855–866. doi: 10.7326/M17-1068 [DOI] [PubMed] [Google Scholar]
- 19.Burkitt DP. Epidemiology of cancer of the colon and rectum. Cancer. Jul 1971;28(1):3–13. doi: [DOI] [PubMed] [Google Scholar]
- 20.Flemer B, Gaci N, Borrel G, et al. Fecal microbiota variation across the lifespan of the healthy laboratory rat. Gut Microbes. September 03 2017;8(5):428–439. doi: 10.1080/19490976.2017.1334033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burns MB, Montassier E, Abrahante J, et al. Colorectal cancer mutational profiles correlate with defined microbial communities in the tumor microenvironment. PLoS Genet. June 2018;14(6):e1007376. doi: 10.1371/journal.pgen.1007376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong TNY, Wang X, Nakatsu G, et al. Association Between Bacteremia From Specific Microbes and Subsequent Diagnosis of Colorectal Cancer. Gastroenterology. August 2018;155(2):383–390.e8. doi: 10.1053/j.gastro.2018.04.028 [DOI] [PubMed] [Google Scholar]
- 23.Wirbel J, Pyl PT, Kartal E, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. April 2019;25(4):679–689. doi: 10.1038/s41591-019-0406-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yachida S, Mizutani S, Shiroma H, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. Jun 2019;25(6):968–976. doi: 10.1038/s41591-019-0458-7 [DOI] [PubMed] [Google Scholar]
- 25.Tomkovich S, Yang Y, Winglee K, et al. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer Res. May 15 2017;77(10):2620–2632. doi: 10.1158/0008-5472.CAN-16-3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. January 2017;66(1):70–78. doi: 10.1136/gutjnl-2015-309800 [DOI] [PubMed] [Google Scholar]
- 27.Liang JQ, Wong SH, Szeto CH, et al. Fecal microbial DNA markers serve for screening colorectal neoplasm in asymptomatic subjects. J Gastroenterol Hepatol. Apr 2021;36(4):1035–1043. doi: 10.1111/jgh.15171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baxter NT, Koumpouras CC, Rogers MA, Ruffin MT, Schloss PD. DNA from fecal immunochemical test can replace stool for detection of colonic lesions using a microbiota-based model. Microbiome. November 14 2016;4(1):59. doi: 10.1186/s40168-016-0205-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krigul KL, Aasmets O, Lüll K, Org T, Org E. Using fecal immunochemical tubes for the analysis of the gut microbiome has the potential to improve colorectal cancer screening. Sci Rep. October 01 2021;11(1):19603. doi: 10.1038/s41598-021-99046-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. Nov 28 2014;10:766. doi: 10.15252/msb.20145645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Jiao N, Zhu R, et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat Commun. May 24 2021;12(1):3063. doi: 10.1038/s41467-021-23265-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, et al. Mutational signature in colorectal cancer caused by genotoxic pks. Nature. April 2020;580(7802):269–273. doi: 10.1038/s41586-020-2080-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J, Djukovic D, Deng L, et al. Colorectal cancer detection using targeted serum metabolic profiling. J Proteome Res. Sep 05 2014;13(9):4120–30. doi: 10.1021/pr500494u [DOI] [PubMed] [Google Scholar]
- 34.Coker OO, Liu C, Wu WKK, et al. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. February 21 2022;10(1):35. doi: 10.1186/s40168-021-01208-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J, Djukovic D, Deng L, et al. Targeted serum metabolite profiling and sequential metabolite ratio analysis for colorectal cancer progression monitoring. Anal Bioanal Chem. Oct 2015;407(26):7857–63. doi: 10.1007/s00216-015-8984-8 [DOI] [PubMed] [Google Scholar]
- 36.Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. Feb 2014;63(2):317–25. doi: 10.1136/gutjnl-2012-304149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rex DK, Boland CR, Dominitz JA, et al. Colorectal Cancer Screening: Recommendations for Physicians and Patients From the U.S. Multi-Society Task Force on Colorectal Cancer. Gastroenterology. July 2017;153(1):307–323. doi: 10.1053/j.gastro.2017.05.013 [DOI] [PubMed] [Google Scholar]
- 38.Almeida-Lousada H, Mestre A, Ramalhete S, et al. Screening for Colorectal Cancer Leading into a New Decade: The “Roaring ‘20s” for Epigenetic Biomarkers? Curr Oncol. November 20 2021;28(6):4874–4893. doi: 10.3390/curroncol28060411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rho YS, Gilabert M, Polom K, et al. Comparing Clinical Characteristics and Outcomes of Young-onset and Late-onset Colorectal Cancer: An International Collaborative Study. Clin Colorectal Cancer. December 2017;16(4):334–342. doi: 10.1016/j.clcc.2017.03.008 [DOI] [PubMed] [Google Scholar]
- 40.Kleif J, Jørgensen LN, Hendel JW, et al. Early detection of colorectal neoplasia: application of a blood-based serological protein test on subjects undergoing population-based screening. Br J Cancer. June 2022;126(10):1387–1393. doi: 10.1038/s41416-022-01712-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parkinson CA, Gale D, Piskorz AM, et al. Exploratory Analysis of TP53 Mutations in Circulating Tumour DNA as Biomarkers of Treatment Response for Patients with Relapsed High-Grade Serous Ovarian Carcinoma: A Retrospective Study. PLoS Med. Dec 2016;13(12):e1002198. doi: 10.1371/journal.pmed.1002198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart CM, Tsui DWY. Circulating cell-free DNA for non-invasive cancer management. Cancer Genet. December 2018;228–229:169–179. doi: 10.1016/j.cancergen.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li N, Lu B, Luo C, et al. Incidence, mortality, survival, risk factor and screening of colorectal cancer: A comparison among China, Europe, and northern America. Cancer Lett. December 01 2021;522:255–268. doi: 10.1016/j.canlet.2021.09.034 [DOI] [PubMed] [Google Scholar]
- 44.Matas J, Kohrn B, Fredrickson J, et al. Colorectal Cancer Is Associated with the Presence of Cancer Driver Mutations in Normal Colon. Cancer Res. April 15 2022;82(8):1492–1502. doi: 10.1158/0008-5472.CAN-21-3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee-Six H, Olafsson S, Ellis P, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. Oct 2019;574(7779):532–537. doi: 10.1038/s41586-019-1672-7 [DOI] [PubMed] [Google Scholar]
- 46.Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol. September 2018;15(9):577–586. doi: 10.1038/s41571-018-0058-3 [DOI] [PubMed] [Google Scholar]
- 47.Fitzgerald RC, Antoniou AC, Fruk L, Rosenfeld N. The future of early cancer detection. Nat Med. April 2022;28(4):666–677. doi: 10.1038/s41591-022-01746-x [DOI] [PubMed] [Google Scholar]
- 48.Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. Jul 18 2012;487(7407):330–7. doi: 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo Y, Wang L, Wang J. Developing proteomics-based biomarkers for colorectal neoplasms for clinical practice: opportunities and challenges. Proteomics Clin Appl. Jan 2013;7(1–2):30–41. doi: 10.1002/prca.201200071 [DOI] [PubMed] [Google Scholar]
- 50.Shen Y, Tong M, Liang Q, et al. Epigenomics alternations and dynamic transcriptional changes in responses to 5-fluorouracil stimulation reveal mechanisms of acquired drug resistance of colorectal cancer cells. Pharmacogenomics J. January 2018;18(1):23–28. doi: 10.1038/tpj.2016.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen SY, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. November 2018;563(7732):579–583. doi: 10.1038/s41586-018-0703-0 [DOI] [PubMed] [Google Scholar]
- 52.Luo H, Zhao Q, Wei W, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med. January 01 2020;12(524)doi: 10.1126/scitranslmed.aax7533 [DOI] [PubMed] [Google Scholar]
- 53.Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. Feb 23 2018;359(6378):926–930. doi: 10.1126/science.aar3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein EA, Richards D, Cohn A, et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann Oncol. September 2021;32(9):1167–1177. doi: 10.1016/j.annonc.2021.05.806 [DOI] [PubMed] [Google Scholar]
- 55.Wan N, Weinberg D, Liu TY, et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer. Aug 23 2019;19(1):832. doi: 10.1186/s12885-019-6003-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lo YMD, Han DSC, Jiang P, Chiu RWK. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science. Apr 9 2021;372(6538)doi: 10.1126/science.aaw3616 [DOI] [PubMed] [Google Scholar]
- 57.Gai W, Sun K. Epigenetic Biomarkers in Cell-Free DNA and Applications in Liquid Biopsy. Genes (Basel). Jan 9 2019;10(1)doi: 10.3390/genes10010032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhardwaj M, Erben V, Schrotz-King P, Brenner H. Cell Line Secretome and Tumor Tissue Proteome Markers for Early Detection of Colorectal Cancer: A Systematic Review. Cancers (Basel). Nov 16 2017;9(11)doi: 10.3390/cancers9110156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilhelmsen M, Christensen IJ, Rasmussen L, et al. Detection of colorectal neoplasia: Combination of eight blood-based, cancer-associated protein biomarkers. Int J Cancer. March 15 2017;140(6):1436–1446. doi: 10.1002/ijc.30558 [DOI] [PubMed] [Google Scholar]
- 60.Wen L, Li J, Guo H, et al. Genome-scale detection of hypermethylated CpG islands in circulating cell-free DNA of hepatocellular carcinoma patients. Cell research. 2015;25(11):1250–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harlid S, Harbs J, Myte R, et al. A two-tiered targeted proteomics approach to identify pre-diagnostic biomarkers of colorectal cancer risk. Sci Rep. March 04 2021;11(1):5151. doi: 10.1038/s41598-021-83968-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee J, Kim HC, Kim ST, et al. Multimodal circulating tumor DNA (ctDNA) colorectal neoplasia detection assay for asymptomatic and early-stage colorectal cancer (CRC). Journal of Clinical Oncology. 2021;39(15_suppl):3536–3536. doi: 10.1200/JCO.2021.39.15_suppl.3536 [DOI] [Google Scholar]
- 63.Kim HC, Kim ST, He Y, et al. Multimodal circulating tumor DNA blood-based colorectal cancer screening test demonstrates clinically meaningful sensitivity across multiple clinical parameters. presented at: American College of Gastroenterology; 2021; Las Vegas, NV. [Google Scholar]
- 64.Raymond VM, Higashi L, Marino E, Lang K. Evaluation of the ctDNA LUNAR-2 Test in an Average Patient Screening Episode (ECLIPSE). Journal of Clinical Oncology. 2021;39(3) [Google Scholar]
- 65.Lin J, Ariazi E, Dzamba M, et al. Evaluation of a sensitive blood test for the detection of colorectal advanced adenomas in a prospective cohort using a multiomics approach. J Clin Oncol. 2021;39:43. [Google Scholar]
- 66.Liu Y, Liu T-Y, Weinberg DE, et al. Spatial co-fragmentation pattern of cell-free DNA recapitulates in vivo chromatin organization and identifies tissues-of-origin. BioRxiv. 2019:564773. [Google Scholar]
- 67.Putcha G, Liu T-Y, Ariazi E, et al. Blood-based detection of early-stage colorectal cancer using multiomics and machine learning. 2020:23–25. [Google Scholar]
- 68.Friedland S, Watson D, Pan J, et al. Development and Clinical Validation of a Blood Test for Early Detection of Colorectal Adenomas and Cancer for Screening and Postpolypectomy Surveillance. Gastro Hep Advances. 2022;1(2):223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]


